Introduction
Lung cancer has been reported to be among the most
deadly cancers worldwide (1).
Non-small-cell lung cancer (NSCLC) accounts for most primary lung
cancer (2). Instead, the
small-cell lung cancer (SCLC) is a distinct form of lung cancer
with unique clinical and histological characteristics, representing
~15% of all new cases of lung cancer (2). The reported prognosis and five-year
survival rates are terribly poor (1). In China, it has been shown that
approximately over half a million people died from lung cancer each
year (3).
The microRNAs (miRNAs) denote myriads of noncoding
RNAs with short length which suppress gene expression by base
paring to the 3′-untranslated region (3′-UTR) of targets (4,5).
The miRNAs can either exist individually or as a cluster in the
whole genome. Meanwhile, they can also reside in introns or exons
of functional genes (6). To date,
more than one thousand miRNAs have been defined. They can regulate
multiple pathways associated with cancer by binding numerous target
mRNAs (7). Although the detailed
clues for the exact molecular mechanisms underlying lung cancer
cell proliferation or metastasis are still elusive, recent
researches have implied that miRNAs may play pivotal roles in lung
cancer progression (6,8,9).
For instance, miR-451 was shown to be lowered in NSCLC tissues
while raised levels of miR-451 are capable of suppressing NSCLC
proliferation by inhibiting ras-related protein 14 (RAB14) and
triggering apoptosis (10).
miR-31 was found to be an oncogenic miRNA through promoting NSCLC
growth and repressing tumor suppressor genes such as large tumor
suppressor 2 (LATS2) and PP2A regulatory subunit B α isoform
(PPP2R2A) (11). Instead, miR-25
was reported to regulate SCLC progression by targeting CDK2
(12). The miR-34b/c has also
been suggested to play an oncogenic role in SCLC (13). However, whether a single microRNA
can play a role in both NSCLC and SCLC is largely elusive.
The interferon regulator factor 2 (IRF2) is a member
of the interferon regulatory family and participates in various
pathways related to tumorigenesis via modulating target gene
expression (14). IRF2 was also
reported to be the functional antagonist for IRF1 and regulate
those genes with interferon regulatory element binding sites either
as positive or negative factors (15). IRF2 has recently become an active
area of research owing to its role in tumor progression (16,17). For example, IRF2 can inhibit p21
and promote proliferation of cells as shown in leukemogenesis
(18). In some type of cancer
cells, aberrant expression of IRF2 was correlated with tumor
growth, invasion and stage (19).
Meanwhile, reducing IRF2 can decrease the expression of cyclin D1
while elevating IRF2 expression can enhance the malignancy of ESCC
cells (19,20). Some studies have implicated the
role of IRF2 in tumor progression such as in pancreatic cancer and
breast cancer although the exact molecular mechanisms remain to be
clarified (21).
In this study, we first showed that downregulating
miR-450 in both NSCLC and SCLC cells and human tumors. We then
explored the functional roles of miR-450 in shaping malignant
phenotypes of NSCLC such as proliferation and invasion.
Furthermore, we investigated the targeting effect of miR-450 on
IRF2 and direct regulatory roles of IRF2 in miR-450 induced NSCLC
and SCLC growth inhibition. Our present study may extend current
understanding on the epigenetic regulation in human lung
cancer.
Materials and methods
Lung cancer cells and human samples
There were 7 lung cancer cell lines used in this
study, H1703, SPC-A1, H510A, H1299, H920, H522 and H2291. The
SPC-A1 cell line was purchased from the Shanghai Institute of Cell
Biology (Shanghai, China). The other cell lines were all
commercially available from the American Type Culture Collection
(ATCC, Shanghai, China). All the cell lines were NSCLC, apart from
H510A, which was SCLC. A control cell line, normal human bronchial
epithelial cell line (NuLi-1) was obtained from ATCC. All the cells
were cultured in RPMI-1640 medium supplemented with 10% fetal
bovine serum and 150 U/ml penicillin plus 150 µg/ml
streptomycin (all from Sigma, Shanghai, China) in a culture chamber
with 5% CO2 at 37°C. Human lung cancer samples were
surgically retrieved from patients registered at Shanghai General
Hospital between October 2014 and April 2015. The tumor (T) tissues
and corresponding adjacent non-tumor (ANT) tissues were all paired
samples. All patients provided written informed consent for the use
of their samples. All surgical and experimental procedures related
with human subjects were approved by the Human Research and Ethics
Committee of Shanghai General Hospital, Shanghai Jiaotong
University School of Medicine.
Reverse transcription-quantitative RT-PCR
(RT-qPCR)
To evaluate the expression of miR-450 and IRF2 in
lung cancer, RT-qPCR was carried out. In brief, total RNA was
extracted from both the NSCLC and SCLC cell lines, and human
samples using TRIzol reagent (Sigma), and reverse transcribed into
cDNA using a SYBR Premix Ex Taq™ kit (Takara Bio, Inc., Shiga,
Japan) following the manufacturer's instructions. To detect the
gene for miR-450, a TaqMan miRNA qRT-PCR kit (Applied Biosystems,
Foster City, CA, USA) was applied. For quantifying IRF2 mRNA, a
SYBR-Green PCR Master mix kit (Applied Biosystems) was used. In
both cases, GAPDH was used as an internal control. All reactions
were performed using an ABI PRISM® 7000 Sequence
Detection system (Applied Biosystems) following the manufacturer's
instructions. Relative expression levels were shown which denote
the fold change relative to the levels under control
conditions.
MicroRNA-450 transfection
We used a lentiviral transfection system to
ectopically upregulate miR-450 in the SCLC and NSCLC cell lines
(H510A and H2291 cells, respectively). The lentivirus covering
human miR-450 mimics (Lenti-miR-450), or a negative control miRNA
(Lenti-C) were purchased from Sigma. The sequences were as follows:
miR-450 sense, 5′-TTTTTGCGATGTGTTCCTAATG-3′ and antisense,
5′-GATATGCCACGGGTTAGATT-3′; Lenti-C sense, 5′-CTCGCTTCTCGAGCACA-3′
and antisense, 5′-AACGCTTCACGAAGGTTCGT-3′. The transduction of
lentivirus into the H510A and H2291 were performed using
Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer's instructions. After 24 h,
the cultured medium was removed and RT-qPCR was used to confirm the
efficiency of transfection.
Cell proliferation assay
A
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma) was utilized to examine cell proliferation. In brief,
24 h following transfection, the H510A and H2291 cells were all
suspended and plated in 96-well plates (104 cells/well)
for 5 days. Subsequently, 20 µl MTT solution (5 mg/ml) were
added to the culture every 24 h and maintained for 4 h. Crystalline
formazan was dissolved in 150 µl sodium dodecyl sulfate
sodium salt (SDS, 15%) solution for 24 h. The optical density (OD)
at 490 nm was evaluated using a Spectramax M5 microplate reader
(Molecular Devices, Sunnyvale, CA, USA) following the instructions
provided by the manufacturer.
Cell invasion assays
Chemotaxis 96-well Transwell assay (Qiangen, Inc.,
Valencia, CA, USA) was utilized to evaluate cell invasion. The
upper chamber was first coated with Matrigel (Invitrogen, Shanghai,
China) overnight. The lentivirus-transfected H510A or H2291 cells
were suspended and plated into the upper chamber (104
cells/well) in RPMI-1640 medium (Sigma) without serum. The lower
chambers were filled with RPMI-1640 medium plus additional 2% fetal
bovine serum (FBS). After 24 h of incubation, all upper chambers
were replenished and the cells migrating into lower chambers were
fixed with 5% PFA and immunostained with crystal violet. The
results of Transwell assay were subsequently quantified using a
Leica inverted microscope fluorescent microscope (DM-IRB; Leica
Microsystems GmbH, Wetzlar, Germany). The invasive capability was
determined by counting the total number of invaded cells under each
experimental condition and normalized to the number for the control
condition.
In vivo implantation of tumors
The H510A cells were transduced with the
lentiviruses for 8 h and then cultured for an additional 24 h. The
cells were then re-suspended and 106 cells were
implanted subcutaneously into the rear flanks of nude mice. The
nude mice were obtained from Model Animal Research Center (Nanjing,
China). In total, 12 mice (age, 4–6 weeks; average weight, 16.1 g)
were used in this study (i.e., 6 in the Lenti-C and 6 in the
Lenti-450 group). The animal experiments and procedures were
approved and reviewed by the Ethics Committee of Shanghai General
Hospital, Shanghai Jiaotong University School of Medicine. The
in vivo volume of tumors was determined weekly (length ×
width × height). After 6 weeks, all mice were sacrificed by an
overdose of sodium amobarbital for Ki-67 immunostaining
(Sigma).
Dual-luciferase reporter assay
To identify potential targets of miR-450, we
utilized several miRNAtargets prediction tools, such as TargetScan
(www.targetscan.org) PicTar
(pictar.mdc-berlin.de) and miRDB (www.mirdb.org). The IRF2 gene was amplified from a
human lung cDNA library and verified. The 3′-UTR of IRF2 with
predicted binding sites to hsa-miR-450 was cloned into the
Xbal immediately downstream of Renilla luciferase
reporter plasmid phRL-TK (Promega, Madison, WI, USA) leading to the
wild-type IRF2 luciferase reporter plasmids (IRF2 3′ UTR WT). The
binding site of hsa-miR-450 on the IRF2 3′-UTR was also mutated by
a Quik-Change™ Site-Directed Mutagenesis kit (Stratagene, La Jolla,
CA, USA). The mutated IRF2 3′-UTR was then incorporated into
phRL-TK to create the mutated IRF2 luciferase reporter plasmid
(IRF2 3′ UTR MUT). In 293T cells (obtained from the Shanghai
Institute of Cell Biology), transfection with Lenti-miR-450 with
IRF2 3′ UTR (WT), IRF2 3′ UTR (MUT) or an empty Renilla
luciferase reporter plasmid (control) was performed for 36 h. The
relative luciferase units (RLU) were then measured by a
dual-luciferase reporter assay (Promega) following the
manufacturer's instructions.
Western blot analysis
The H510A and H2291 cells were harvested with cell
lysis buffer containing 15% glycerol and 2% (Sigma). The protein
extracts (100 µg each) were dissolved by 10% SDS-PAGE and
transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA,
USA). The membranes were then incubated with antibodies against
human IRF2 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA)
at 4°C overnight, and HRP-conjugated secondary antibodies (1:103)
at 20°C for 1.5 h. β-actin was used as an internal control. The
blots were visualized with an enhanced chemiluminescence film
system (Amersham Pharmacia Biotech, Shanghai, China).
IRF2 overexpression assay
Whole sequences of IRF2 were cloned into a
recombinant plasmid eukaryotic expression plasmid pcDNA3.1 (Sigma)
to create the IRF2 overexpression plasmid, pcDNA3.1/IRF2, following
the manufacturer's instructions. The transfection of pcDNA3.1/IRF2
and an empty pcDNA3.1 plasmid pcDNA3.1/(+) into H510A and H2291
cells were performed with Lipofectamine 2000 (Invitrogen).
Twenty-four hours after transfection, the cells were re-suspended
and plated in 96-well plates.
Statistical analysis
All experiments were performed at least in
triplicate. The results are presented as the mean ± standard error.
Survival was evaluated with Kaplan-Meier curves and compared using
a log-rank test. Statistical differences were measured using a
Student's t-test using SPSS 16.0 software (SPSS Inc., Chicago, IL,
USA) and a value of P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-450 expression is decreased in lung
cancer cells and correlates with survival
In the present study, we first used RT-qPCR to
demonstrate the downregulation of miR-450 in established lung
cancer cell lines in vitro, as well as in human samples
in vivo. The results revealed that miR-450 was
down-regulated in the 7 lung cancer cell lines examined, H510A,
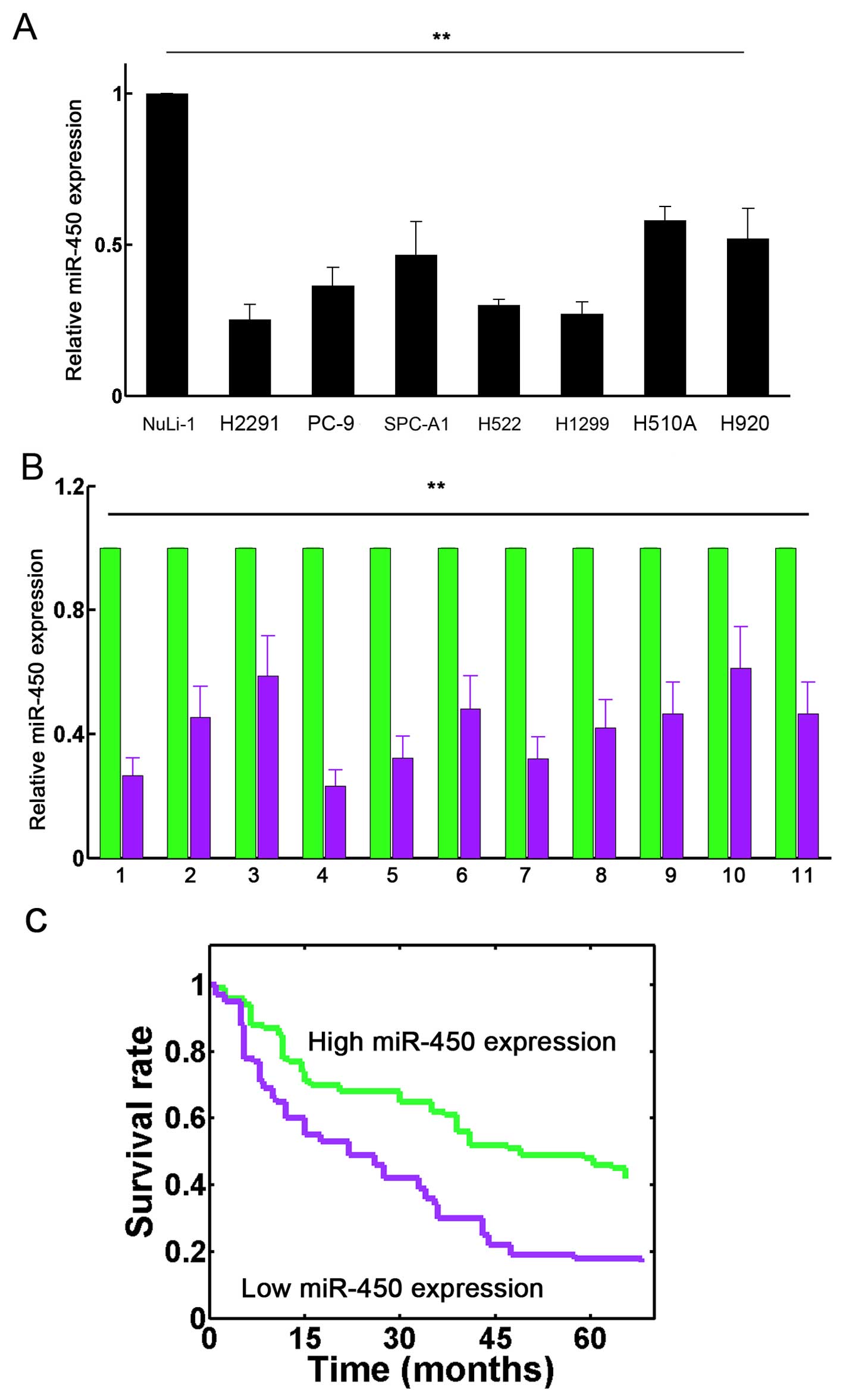
H1703, SPC-A1, H522, H1299, SK-MES-1, H920 and H2291 compared with
the normal human bronchial epithelial cell line (NuLi-1) (Fig. 1A; P<0.01). We also demonstrated
that miR-450 expression was significantly decreased in the tumor
(T) tissues than in the corresponding non-tumor (ANT) tissues in
the 11 patients with lung cancer (Fig. 1B; P<0.01). In addition, we also
found that the intrinsic expression of miR-450 exhibited a
significant correlation with the survival rates of the patients
with lung cancer (P=0.004; Fig.
1C). These results suggest that miR-450 is frequently
downregulated in lung cancer and that its downregulation correlates
with poor survival.
Upregulation of miR-450 inhibits lung
cancer cell invasion and proliferation
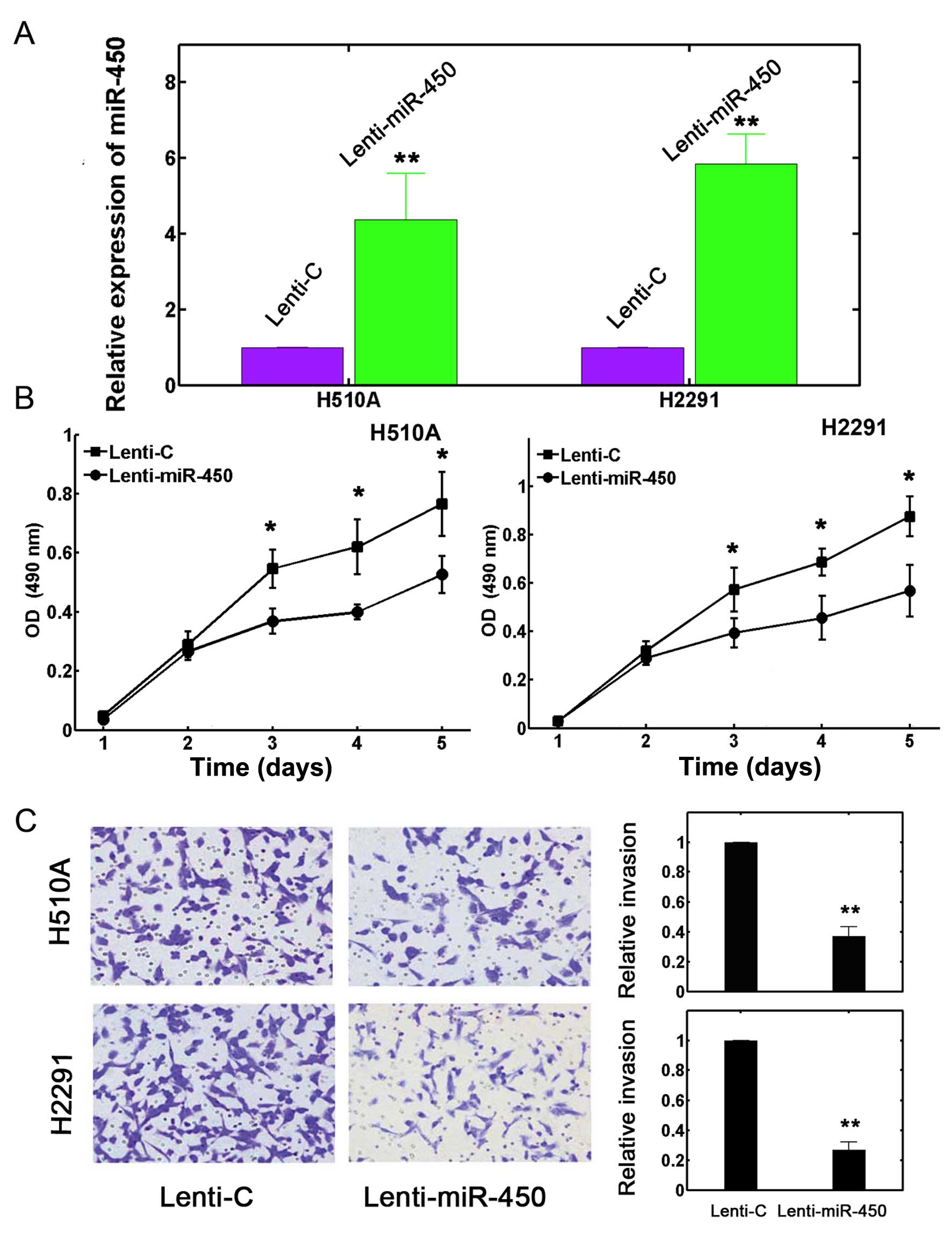
We transfected the H510A and H2291 cells with
lentiviruses in order to increase the expression of miR-450. After
24 h, the transfection efficiency was verified by RT-qPCR. The
results revealed that the expression level of miR-450 was
significantly increased by transfection with lentivirus containing
miR-450 mimics (Lenti-miR-450) in both the H510A and H2291 cells
compared with the control (Lenti-C) (Fig. 2A; P<0.05).
The H510A and H2291 cells transfected with
lentivirus were re-suspended and plated in a 96-well plate for 120
h. Cell proliferation assay was carried out every 24 h to determine
the effects of miR-450 upregulation on lung cancer cell growth. The
results revealed that the upregulation of miR-450 significantly
attenuated lung cancer cell proliferation in both the H510A and
H2291 cells (Fig. 2B; P<0.05).
We also used a Transwell assay to examine the effects of miR-450
upregulation on lung cancer cell invasion. The results revealed
that transfection with miR-450 mimics markedly reduced cell
invasion (Fig. 2C, left panel).
Quantification also confirmed that the upregulation of miR-450
reduced the invasive capabilities of the H510A and H2291 cells by
>50% (Fig. 2C, right panel;
P<0.05). Taken together, these data suggest that the
upregulation of miR-450 inhibits the proliferation and invasion of
lung cancer cells.
Upregulation of miR-450 inhibits the
growth of implanted lung tumors in vivo
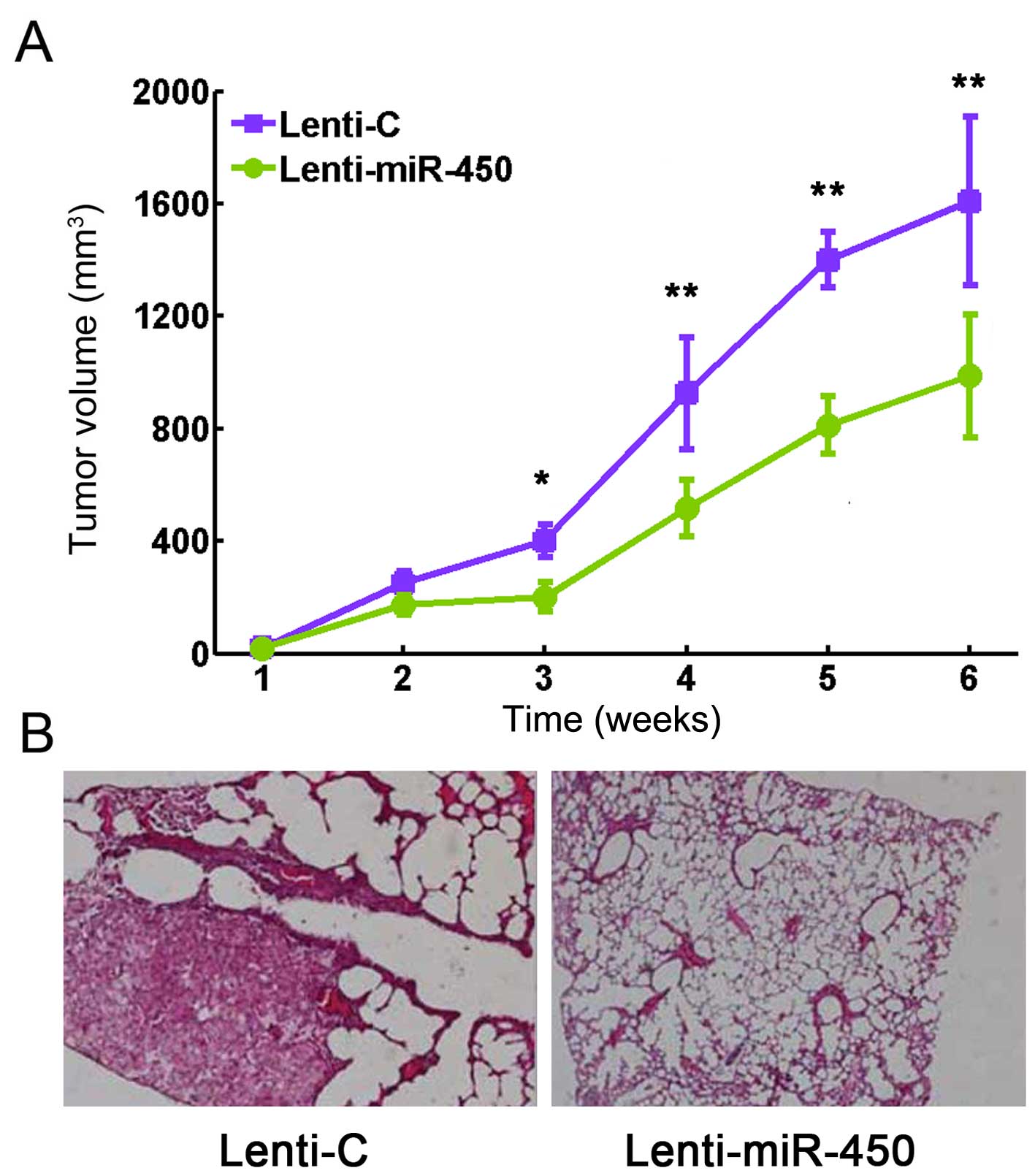
As we found that the upregulation of miR-450
inhibits lung cancer cell, we wished to determine whether miR-450
has a similar effect on lung tumor growth in vivo. Thus, we
transfected the H510A cells with Lenti-miR-450 or Lenti-C for 24 h.
The cells were then re-suspended (the final cell number is
106) and implanted subcutaneously into the rear flanks
of nude mice. The sizes of the lung tumors, incuding length, width
and height were measured each week and the total tumor volumes were
determined. The results revealed that the growth capacity of the
implanted lung tumors was significantly attenuated by the
upregulation of miR-450 (Fig. 3A;
P<0.05). Six weeks after implantation, the lung tumors were
extracted and Ki-67 immunostaining was then performed. The results
revealed that Ki-67 staining was substantially decreased with the
upregulation of miR-450 (Fig.
3B).
miR-450 directly targets IRF2 in
NSCLC
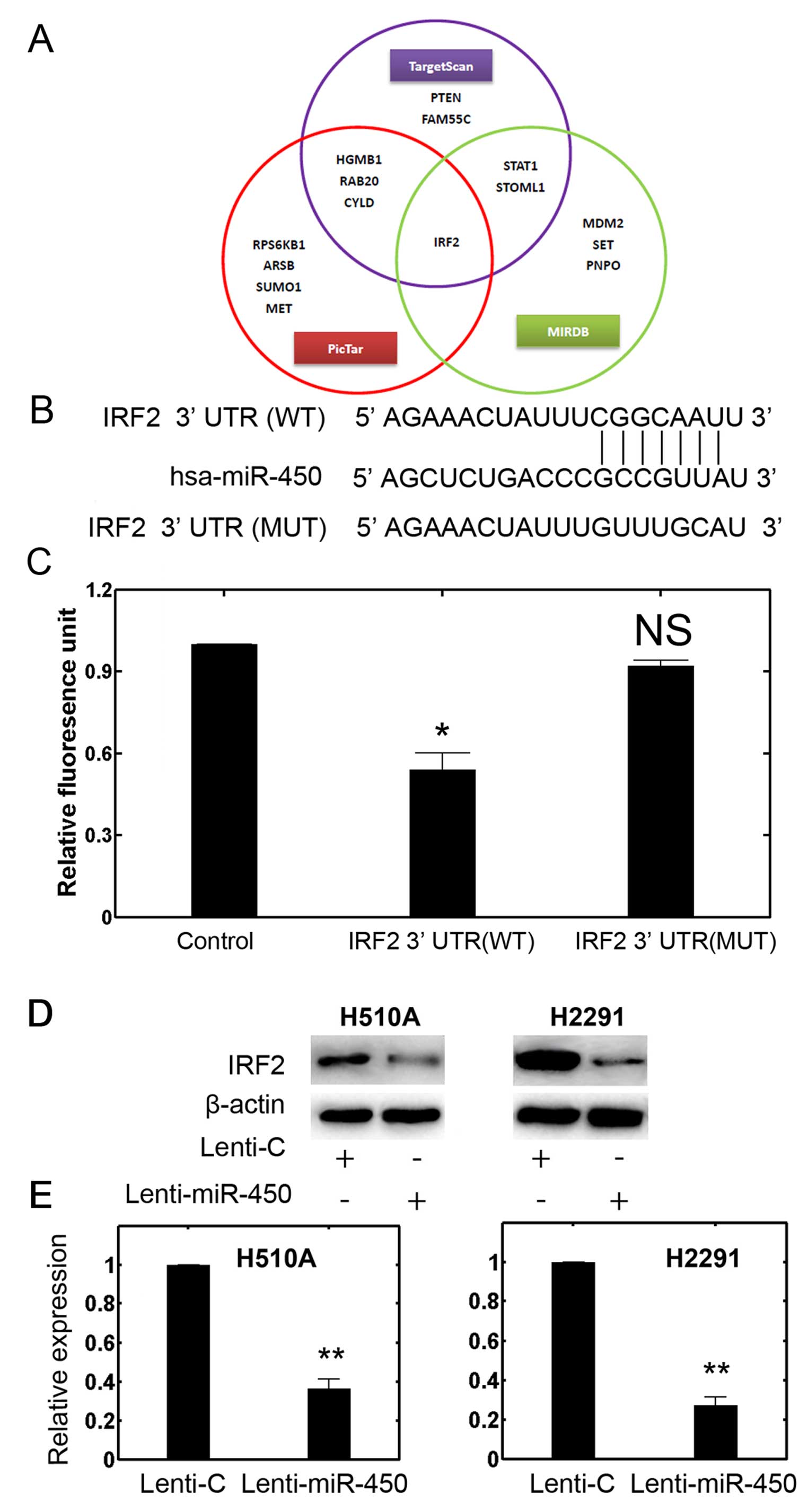
To identify potential targets of miR-450, we
utilized several miRNAtargets prediction tools, such as TargetScan
(www.targetscan.org) PicTar
(pictar.mdc-berlin.de) and miRDB (www.mirdb.org)
(Fig. 4A). The cross-verification
of several databases indicated that that the oncogene, IRF2, may be
a promising candidate (Fig. 4A and
B). We then performed dual-luciferase reporter assays using the
293T cells. The results revealed that miR-450 can target luciferase
plasmids with an intact or WT IRF2 3′-UTR and significantly reduced
the RLU, whereas miR-450 had little effect on the luciferase
plasmid containing a mutation (MUT) in the IRF2 3′ UTR (Fig. 4C; P<0.05). We then paid more
attention to two established NSCLC cell lines, H510A and H2291
cells, using western blot analysis and RT-qPCR to evaluate the
profiles of IRF2 in the cells transfected with Lenti-miR-450. The
results revealed that in both the H510A and H2291 cells, the
protein level of IRF2 was markedly downregulated by the
upregulation of miR-450 (Fig.
4D). The results of RT-qPCR confirmed this finding and showed
that the expression of IRF2 in the H510A and H2291 cells was also
downregulated by the upregulation of miR-450 (Fig. 4E; P<0.05). Taken together, our
data from dual-luciferase reporter assay, western blot analysis and
RT-qPCR suggest that IRF2 may be the direct downstream target of
miR-450 in both NSCLC and SCLC cells.
Upregulation of IRF2 restores the
malignant phenotypes of lung cancer cells following transfection
with miR-450 mimics
We then conjectured that IRF2 may directly affect
the miR-450-mediated inhibition of lung cancer cell proliferation
and invasion. To verify this hypothesis, we constructed a mammalian
plasmid to ectopically overexpress IRF2 (pcDNA3.1/IRF-2) in the
H510A and H2291 cells. The efficiency of transfection was confirmed
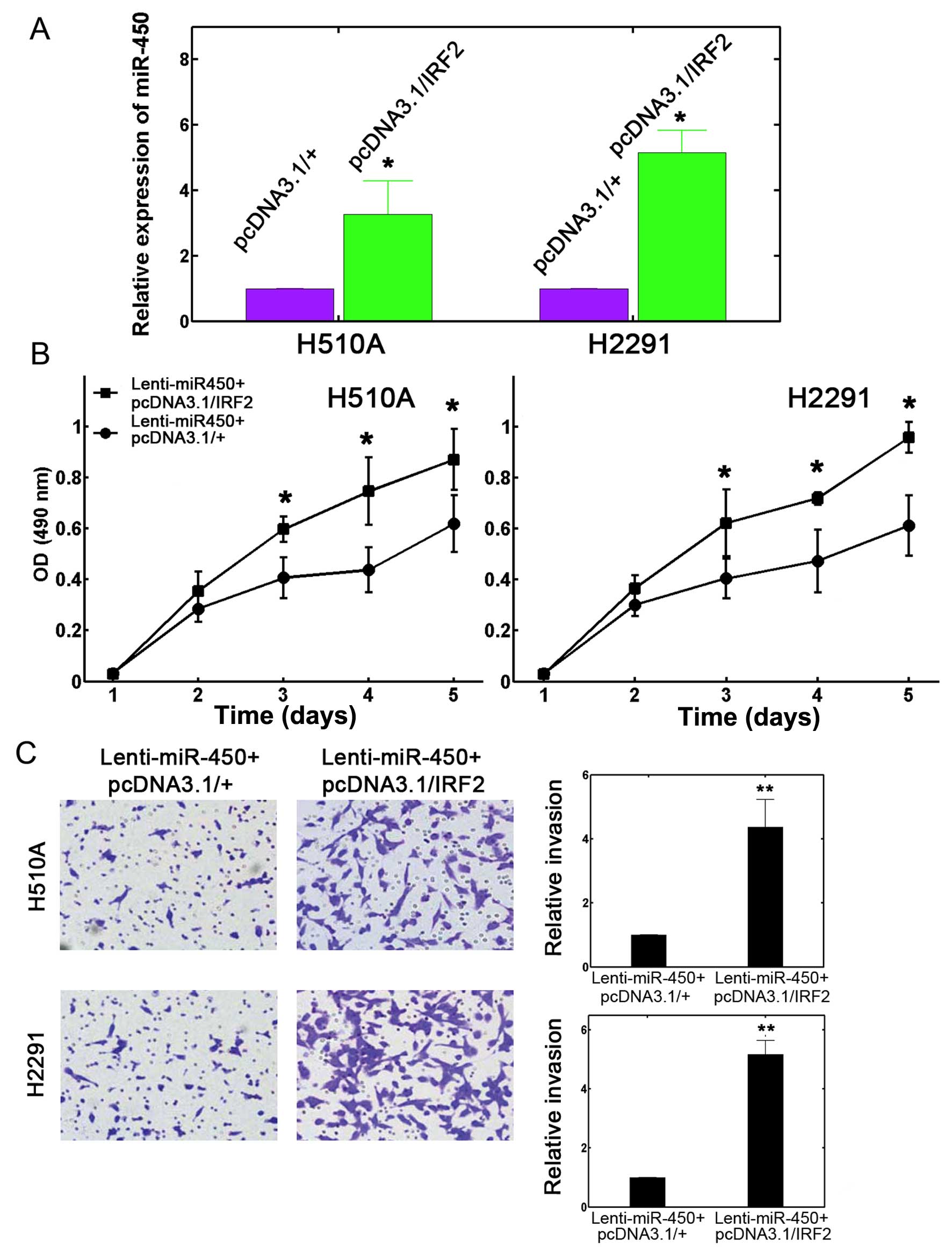
by RT-qPCR (Fig. 5A; P<0.05).
We then performed a dual transfection assay. First, the H510A and
H2291 cells were transfected with Lenti-miR-450. After 24 h, the
cells were transfected with either pcDNA3.1/IRF2 or the empty
control plasmid pcDNA3.1/(+) for an additional 24 h. A 5-day
proliferation assay was then performed. The results revealed that
IRF2 overexpression effectively restored lung cancer cell
proliferation, which was initially attenuated by the upregulation
of miR-450 in both the H510A and H2291 cells (Fig. 5B; P<0.05). Transwell assay also
revealed that IRF2 overexpression significantly increased the
number of invading cells into the lower chambers for both the H510A
and H2291 cells (Fig. 5C, left
panel). The invasive capability which was originally inhibited by
miR-450 upregulation, ws elevated by >200% with the
overexpression of IRF2 (Fig. 5C,
right panel; P<0.05). Therefore, all these results suggest that
IRF2 directly mediates the regulatory effects of miR-450 on the
proliferation and invasion of both NSCLC and SCLC cells.
Discussion
The miRNAs are small and non-coding RNAs, which
serve as key regulators of gene expression by directly targeting
mRNAs for translational repression. Deregulated miRNAs have been
reported to be associated with the malignant progression of
numerous types of cancer (22).
These miRNAs may play regulatory and pivotal roles in the
pathogenesis of tumors (23,24). As previously reported, lung cancer
patients usually suffer from tumor cell invasion and metastasis
before diagnosis, rendering many present treatments such as
surgery, radiotherapy and chemotherapy ineffective (25). Therefore, exploring the detailed
mechanisms of lung cancer is critically important for development
of novel therapeutics to improve overall survival rates.
Only a few studies to date have implied a role for
miR-450. It has been demonstrated that miR-450a-3p regulates
embryonic development by targeting Bub1 protein in mouse embryonic
fibroblasts (MEFs) (26). Another
study also implicated miR-450 in ERα signaling pathways (27). Furthermore, miR-450 can also be
upregulated in TGF-9-treated PH5CH8 hepatocytes (28). However, a functional role of
miR-450 in lung cancer has rarely been reported. In the present
study, we first evaluated the pattern of miR-450 expression in
several established lung cancer cell lines. We found that miR-450
was ubiquitously downregulated in all the tested lung cancer cell
lines compared to normal bronchial cells. We also confirmed that
miR-450 expression was significantly reduced in tumor tissues from
lung cancer patients in comparison with normal adjacent tissues
(Fig. 1). Moreover, transection
with miR-450 mimics can stably increased miR-450 expression in the
H510A or H2291 lung cancer cell lines (Fig. 2). Furthermore, the results of MTT
and Transwell assays demonstrated that miR-450 suppresses the
progression of lung cancer by inhibiting proliferation and invasion
(Figs. 2 and 3). Therefore, from our preliminary
experiments, it can be argued that miR-450 is a tumor suppressor,
at least in NSCLC and SCLC cells.
The role of miRNAs in lung cancer development has
been an active area of research. However, the majority of studies
have mainly focused on either SCLC or NSCLC. In a previous study,
Du et al argued that miR-337-3p can sensitize NSCLC cells to
taxanes by targeting STAT3 and RAP1A, whereas the inhibition of
miR-139-5p decreases SCLC cell viability through an unknown target
(29). Furthermore, a recent
miRNA profiling study aimed to distinguish NSCLC from SCLC by
identifying unique miRNA expression patterns (30). However, few reports have
investigated whether a single miRNA species can play a concordant
role in both NSCLC and SCLC. In the current study, we found that
miR-450 can mediate the tumor suppression of both SCLC and NSCLC
cells by targeting IRF2. The overexpression of miR-450 inhibits the
proliferation of both H510A and H2291 cells. Therefore, our study
may instead unravel a novel facet of miRNA regulation in cancer
development.
We further demonstrated that the transcription
factor, IRF2, serves as a direct molecular target of miR-450
(Fig. 4). Upon miR-450
upregulation, not only lung cancer cell proliferation or invasion
was inhibited, but IRF2 was also downregulated. That prompted us to
further investigate whether IRF2 is directly involved in the
regulation of miR-450 in lung cancer. Our IRF2 overexpression
experiment partially confirmed this hypothesis, showing that the
miR-450-induced inhibition on lung cancer cell proliferation and
invasion was substantially restored by IRF2 overexpression. IRF1
and IRF2 have been shown to be upregulated by type I and type II
interferons (31). In return,
IFN-α and -β can also be regulated by IRF family members (31). IRF1 has been reported to inhibit
viral infection, particularly HCV (32). However, IRF2 was identified as a
transcriptional repressor and can compete with the role of IRF1
(33). More importantly, IRF2 can
also function as an oncogenic regulator. The deregulation of IRF2
has been reported in numerous tumor types. For example, Cui et
al demonstrated that IRF2 was significantly upregulated in
primary pancreatic cancer cells and correlated with a poor survival
(34). Connett et al
further argued that the level of IRF2 was maintained in tumor
tissues and was associated with malignancy (35). In addition, it was also shown that
IRF2 is involved in the negative feedback in the IFN-γ pathway and
has been implicated in tumor progression (36). In a more recent study, IRF2 was
directly implicated in gastric cancer by modulating p53 expression
(16). Therefore, miR-450 may
target a universal oncogenic factor and exert its tumor suppressive
role. Whether miR-450 is involved in other signaling pathways and
plays undetermined roles remains to be clarified in future
studies.
In conclusion, in this study, we established for the
first time, to the best of our knowledge, a functional associatoin
between miR-450 and human lung cancer. The upregulation of miR-450
expression exerts tumor suppressive effects on lung cancer. IRF2
was identified as the downstream target of miR-450 in lung cancer
as it can counteract the effects of miR-450-induced tumor
inhibition. The miR-450 and IRF2 signaling pathway should be
further investigated in depth to uncover the epigenetic regulation
of miRNAs in human lung cancer. This may help establish more
elaborate targets in tumor intervention.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xue C, Hu Z, Jiang W, Zhao Y, Xu F, Huang
Y, Zhao H, Wu J, Zhang Y, Zhao L, et al: National survey of the
medical treatment status for non-small cell lung cancer (NSCLC) in
China. Lung Cancer. 77:371–375. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009. View Article : Google Scholar
|
|
6
|
Feng B, Zhang K, Wang R and Chen L:
Non-small-cell lung cancer and miRNAs: novel biomarkers and
promising tools for treatment. Clin Sci (Lond). 128:619–634. 2015.
View Article : Google Scholar
|
|
7
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalia M: Biomarkers for personalized
oncology: Recent advances and future challenges. Metabolism.
64(Suppl 1): S16–S21. 2015. View Article : Google Scholar
|
|
9
|
Zhang Y, Yang Q and Wang S: MicroRNAs: A
new key in lung cancer. Cancer Chemother Pharmacol. 74:1105–1111.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Sempere LF, Ouyang H, Memoli VA,
Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al:
MicroRNA-31 functions as an oncogenic microRNA in mouse and human
lung cancer cells by repressing specific tumor suppressors. J Clin
Invest. 120:1298–1309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Z, Liu J, Wang C, Wang Y, Jiang Y and
Guo M: MicroRNA-25 regulates small cell lung cancer cell
development and cell cycle through cyclin E2. Int J Clin Exp
Pathol. 7:7726–7734. 2014.
|
|
13
|
Tanaka N, Toyooka S, Soh J, Kubo T,
Yamamoto H, Maki Y, Muraoka T, Shien K, Furukawa M, Ueno T, et al:
Frequent methylation and oncogenic role of microRNA-34b/c in
small-cell lung cancer. Lung Cancer. 76:32–38. 2012. View Article : Google Scholar
|
|
14
|
Harada H, Taniguchi T and Tanaka N: The
role of interferon regulatory factors in the interferon system and
cell growth control. Biochimie. 80:641–650. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xi H, Eason DD, Ghosh D, Dovhey S, Wright
KL and Blanck G: Co-occupancy of the interfearon regulatory element
of the class II transactivator (CIITA) type IV promoter by
interferon regulatory factors 1 and 2. Oncogene. 18:5889–5903.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen YJ, Wu H, Zhu JM, Li XD, Luo SW, Dong
L, Liu TT and Shen XZ: MicroRNA-18a modulates P53 expression by
targeting IRF2 in gastric cancer patients. J Gastroenterol Hepatol.
31:155–63. 2016. View Article : Google Scholar
|
|
17
|
Sakai T, Mashima H, Yamada Y, Goto T, Sato
W, Dohmen T, Kamada K, Yoshioka M, Uchinami H, Yamamoto Y and
Ohnishi H: The roles of interferon regulatory factors 1 and 2 in
the progression of human pancreatic cancer. Pancreas. 43:909–916.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choo A, Palladinetti P, Holmes T, Basu S,
Shen S, Lock RB, O'Brien TA, Symonds G and Dolnikov A: siRNA
targeting the IRF2 transcription factor inhibits leukaemic cell
growth. Int J Oncol. 33:175–183. 2008.PubMed/NCBI
|
|
19
|
Nicolini A, Carpi A and Rossi G: Cytokines
in breast cancer. Cytokine Growth Factor Rev. 17:325–337. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Liu DP, Chen PP, Koeffler HP, Tong
XJ and Xie D: Involvement of IFN regulatory factor (IRF)-1 and
IRF-2 in the formation and progression of human esophageal cancers.
Cancer Res. 67:2535–2543. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xi H and Blanck G: Interferon regulatory
factor-2 point mutations in human pancreatic tumors. Int J Cancer.
87:803–808. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dykxhoorn DM: MicroRNAs and metastasis:
Little RNAs go a long way. Cancer Res. 70:6401–6406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar
|
|
25
|
Zhao Y, Wei Q, Hu L, Chen F, Hu Z, Heist
RS, Su L, Amos CI, Shen H and Christiani DC: Polymorphisms in
MicroRNAs are associated with survival in non-small cell lung
cancer. Cancer Epidemiol Biomarkers Prev. 23:2503–2511. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo M, Weng Y, Tang J, Hu M, Liu Q, Jiang
F, Yang D, Liu C, Zhan X, Song P, et al: MicroRNA-450a-3p represses
cell proliferation and regulates embryo development by regulating
Bub1 expression in mouse. PLoS One. 7:e479142012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Castellano L, Giamas G, Jacob J, Coombes
RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman
J, et al: The estrogen receptor-alpha-induced microRNA signature
regulates itself and its transcriptional response. Proc Natl Acad
Sci USA. 106:15732–15737. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brockhausen J, Tay SS, Grzelak CA,
Bertolino P, Bowen DG, d'Avigdor WM, Teoh N, Pok S, Shackel N,
Gamble JR, et al: miR-181a mediates TGF-β-induced hepatocyte EMT
and is dysregulated in cirrhosis and hepatocellular cancer. Liver
Int. 35:240–253. 2015. View Article : Google Scholar
|
|
29
|
Du L and Pertsemlidis A: microRNA
regulation of cell viability and drug sensitivity in lung cancer.
Expert Opin Biol Ther. 12:1221–1239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du L, Schageman JJ, Irnov, Girard L,
Hammond SM, Minna JD, Gazdar AF and Pertsemlidis A: MicroRNA
expression distinguishes SCLC from NSCLC lung tumor cells and
suggests a possible pathological relationship between SCLCs and
NSCLCs. J Exp Clin Cancer Res. 29:752010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Masumi A, Ito M, Mochida K, Hamaguchi I,
Mizukami T, Momose H, Kuramitsu M, Tsuruhara M, Takizawa K, Kato A
and Yamaguchi K: Enhanced RIG-I expression is mediated by
interferon regulatory factor-2 in peripheral blood B cells from
hepatitis C virus-infected patients. Biochem Biophys Res Commun.
391:1623–1628. 2010. View Article : Google Scholar
|
|
32
|
Ciccaglione AR, Stellacci E, Marcantonio
C, Muto V, Equestre M, Marsili G, Rapicetta M and Battistini A:
Repression of interferon regulatory factor 1 by hepatitis C virus
core protein results in inhibition of antiviral and
immunomodulatory genes. J Virol. 81:202–214. 2007. View Article : Google Scholar :
|
|
33
|
Ikushima H, Negishi H and Taniguchi T: The
IRF family transcription factors at the interface of innate and
adaptive immune responses. Cold Spring Harb Symp Quant Biol.
78:105–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui L, Deng Y, Rong Y, Lou W, Mao Z, Feng
Y, Xie D and Jin D: IRF-2 is over-expressed in pancreatic cancer
and promotes the growth of pancreatic cancer cells. Tumour Biol.
33:247–255. 2012. View Article : Google Scholar
|
|
35
|
Connett JM, Badri L, Giordano TJ, Connett
WC and Doherty GM: Interferon regulatory factor 1 (IRF-1) and IRF-2
expression in breast cancer tissue microarrays. J Interferon
Cytokine Res. 25:587–594. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Liu D, Chen P, Koeffler HP, Tong X
and Xie D: Negative feedback regulation of IFN-gamma pathway by IFN
regulatory factor 2 in esophageal cancers. Cancer Res.
68:1136–1143. 2008. View Article : Google Scholar : PubMed/NCBI
|