Introduction
Chronic lymphocytic leukemia (CLL) is characterized
by the proliferation and accumulation of malignant CD5+
CD19+ and CD23+ mature, monoclonal B
lymphocytes in the peripheral blood, bone marrow, lymph nodes, and
other secondary lymphoid organs (1,2).
The biological behaviors and the clinical features of the disease
are significantly heterogeneous (3,4).
Several factors, including immunoglobulin heavy chain variable
region (IGHV) gene mutation, cytogenetic abnormalities,
zeta-chain-associated protein kinase 70 (ZAP70) and CD38 expression
at diagnosis, predict prognosis and help guide therapeutic
decisions (1). Recurrent gene
mutations such as NOTCH1, MYD88, BIRC3 and
SF3B1 confer drug resistance and adverse prognosis in CLL
(2,5,6).
Notch homolog 1, translocation-associated
(Drosophila), also known as NOTCH1, encodes a
single-pass class I trans-membrane protein which exists as a
non-covalently linked heterodimer (7). Functioning as a ligand-activation
transcription factor, it is composed of an extracellular domain
mediating ligand binding and an intracellular domain mediating
signaling. Ligands, Delta-like and Jagged, following binding to the
NOTCH receptor, induce proteolytic cleavage of the receptor, and
result in the release and translocation of the intracellular domain
to the nucleus, thus leading to transcriptional activation of
multiple target genes including nuclear factor
κ-light-chain-enhancer of activated B cells (NF-κB) (8–10).
NOTCH1 mutations in exon 34, selectively
disrupt the carboxy-terminal
proline-glutamate-serine-threonine-rich (PEST) domain of the
protein, resulting in a truncated protein which is more stable,
impaired NOTCH1 degradation, and constitutive transcriptional
activation of the NOTCH1 downstream signaling including canonical
and noncanonical NF-κB pathways (5,11–18). Mutations in NOTCH1 are
reported to be associated with a particularly poor outcome and may
play a pivotal role in the pathogenesis and treatment resistance of
CLL, and contribute to poor patient prognosis (19). Based on current research,
activating mutations of NOTCH1 occur at a low frequency in
CLL patients at diagnosis (8.3–12.6%), but at a significantly
higher frequency in patients with the more clinically aggressive
IGHV unmutated subtype of CLL (20.4%), Richter syndrome
(31.0%) and chemo-refractory CLL (20.8%) (5,13–15,20–23). Among all NOTCH1 mutations,
~80% are located in exon 34 which are selected to disrupt the PEST
domain.
High-resolution melting (HRM) assay is a technique
for fast genotyping and high-throughput mutation of germ-line and
somatic mutation analysis, which has been established since 2003
(24–26). The HRM melting profile provides a
specific sequence-related pattern that differentiates wild-type
sequences from homozygote or heterozygote variants. This method is
based on real-time PCR amplification in the presence of a
saturating intercalating fluorescent dye (27–29); mutations residing in the melting
domain are visualized as alteration of the melting curve derived
after PCR amplification by increasing the temperature and measuring
the decrease of fluorescence emitted from within the double helix
while the DNA strands separate (25,30,31).
In the present study, we developed an accurate and
sensitive HRM assay for detecting somatic NOTCH1 mutations
in a total of 133 CLL patients. These gene mutations were further
confirmed by direct sequencing, the gold standard. The results of
HRM analysis achieved 100% concordance with those from direct
sequencing. The HRM method we developed proved to be an effective,
rapid and sensitive approach for NOTCH1 screening with
higher sensitivity and shorter turn-around time (TAT). It may be
routinely used for the high-throughput screening of NOTCH1
in clinical CLL patients at diagnosis or at any clinical course of
CLL, which is significant for decision-making regarding therapeutic
strategies.
Materials and methods
Clinical specimens
A total of 133 CLL bone marrow samples were obtained
after informed consent from patients fulfilling diagnostic and
immune-phenotypic criteria for CLL at the Hematologic Department,
The First Affiliated Hospital of Soochow University. Only patients
at initial diagnosis without prior therapy were included in the
study. Positive cut-off values were 30 and 20% for CD38 and ZAP70
expression. The mutation status of the IGHV gene and
cytogenetic alterations for all the patients included in this study
were also analyzed. Germline IGHV was defined as ≥98%
homology. Research was performed upon approval of the Ethics
Committee of The First Affiliated Hospital of Soochow
University.
Genomic DNA (gDNA) extraction
To obtain somatic DNA from CLL cells, the cells were
isolated from bone marrow of CLL patients by density gradient
centrifugation over lymphocyte cell separation media (Cedarlane
Laboratories, Shanghai, China). After isolation, the cells were
stained with anti-CD19, anti-CD5 and anti-CD20 antibodies (all from
BD Biosciences, Shanghai, China), which were further analyzed by
flow cytometry. Only CLL samples containing ≥95%
CD5+CD19+ cells were included in the study.
gDNA was extracted using gDNA isolation kits (Omega BioTek
Guangzhou, Ltd., Guangzhou, China) according to the manufacturer's
instructions. DNA was quantified using a NanoDrop ND-1000
fluorospectrometer (Thermo Fisher Scientific, Shanghai, China) and
the A260/280 value was ensured between 1.8 and 2.0.
Cell culture
The acute lymphoblastic leukemia cell line Molt4 and
the acute T cell leukemia cell line Jurkat, purchased from the
Shanghai Institute for Biological Sciences (Shanghai, China), were
cultured in RPMI-1640 medium (HyClone, Logan, UT, USA) supplemented
with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin and
100 µg/ml streptomycin, 2 mM L-glutamine and 1 mM sodium
pyruvate at 37°C in a humidified incubator at 5%
CO2.
HRM assay
The primers of NOTCH1 used in this study are
listed in Table I. The PEST
domain of NOTCH1 was amplified in two fragments (Ex34a and
Ex34b) with product sizes of 131 and 116 bp. The HRM assay was
performed using Fast EvaGreen® qPCR Master Mix (Biotium,
Hayward, CA, USA) on a LightCycler 480 instrument (Roche
Diagnostics, Beijing, China). The reaction mixture in a 20
µl final volume contained 1X Fast EvaGreen qPCR Master Mix,
200 nM forward primer and 200 nM reverse primer, 50 ng of genomic
DNA and PCR grade water. The cycling and melting conditions were as
follows: 95°C for 2 min (95°C for 5 sec, 60°C for 35 sec, 72°C for
25 sec) ×50 cycles; HRM: 95°C for 1 min, 40°C for 1 min, 65°C for 1
sec, with a continuous increase in temperature from 65°C to 95°C at
the rate of 0.02°C/sec with 25 signal acquisitions per degree; and
cooling: 40°C for 30 sec. The melting profiles of the amplicons
were analyzed using LightCycler 480 Gene-Scanning software to
detect wild-type and mutations. All samples were tested in
triplicate.
 | Table IPrimers for direct sequencing or HRM
assay of the PEST domain of the NOTCH1 gene. |
Table I
Primers for direct sequencing or HRM
assay of the PEST domain of the NOTCH1 gene.
| Method | Primer name | Sequences | Amplicon size |
|---|
| Sequencing | NOTCH1
Ex34_F |
5′-CTGGCGGTGCACACTATTCTG-3′ | 327 bp |
| NOTCH1
Ex34_R |
5′-GCGCGCCGTTTACTTGAAG-3′ |
| HRM | NOTCH1
Ex34a_F |
5′-ACAGCTACTCCTCGCCTGTG-3′ | 131 bp |
| NOTCH1
Ex34a_R |
5′-GTCGGAGACGTTGGAATGCG-3′ |
| NOTCH1
Ex34b_F |
5′-GTGCACACTATTCTGCCCCAG-3′ | 116 bp |
| NOTCH1
Ex34b_R |
5′-GAGTAGCTGTGCTGCGAGG-3′ |
Direct sequencing
NOTCH1 screening (PEST domain; RefSeq
NM_017617.4) in CLL was carried out by PCR amplification and direct
sequencing. The primers used are listed in Table I. The PCR reaction was amplified
using Platinum Taq DNA polymerase (Invitrogen, Beijing,
China) and conducted under the following conditions: 94°C for 5
min, (94°C for 30 sec, 60°C for 30 sec, 72°C for 45 sec) ×40
cycles; 72°C for 10 min. The PCR products were checked on 2%
agarose gels. PCR products were purified and followed by
bi-directional sequencing using an ABI 3730 DNA Analyzer (Applied
Biosystems, Inc., Beijing, China). Sequencing chromatograms were
analyzed using DNA Baser 3.0. Nucleotide changes detected by
sequencing were all checked in Sanger's COSMIC database, and
diagnosed as mutations accordingly.
Sensitivity determination
Cancer cell lines, Molt4 and Jurkat, were used.
Molt4 cells harbor deletion of a CT dinucleotide in the PEST domain
of the NOTCH1 gene (heterozygous for c.7541_7542delCT).
Jurkat cells are wild-type of the NOTCH1 gene. Serial
dilutions of the NOTCH1 mutant Molt4 cells with Jurkat cells
were used to determine the sensitivity of the direct sequencing and
HRM.
Statistical analysis
Data analysis was performed using GraphPad Prism 5
program (GraphPad Software, Inc., La Jolla, CA, USA). The
Chi-square test was used to analyze biological features between
NOTCH1-mutated and unmutated CLL groups. Differences were
considered to be statistically significant when the p-value was
<0.05.
Results
Detection of NOTCH1 mutations by HRM
analysis
For the HRM assay, we chose the optimal temperature
and the primer concentration to generate specific products with
efficient amplification and melting with an acceptable profile. The
normalized and shifted melting curves provide the basic
representation of the different genotypes, while difference plots
show the difference between fluorescence of a patient's sample and
a wild-type template at each temperature transition. The normalized
and shifted melting curves and difference plots of 6.02% (8/133) of
the patients were significantly different from those of the
wild-type control. There were two types of shifted melting curves
and difference plots in the 8 CLL samples with NOTCH1
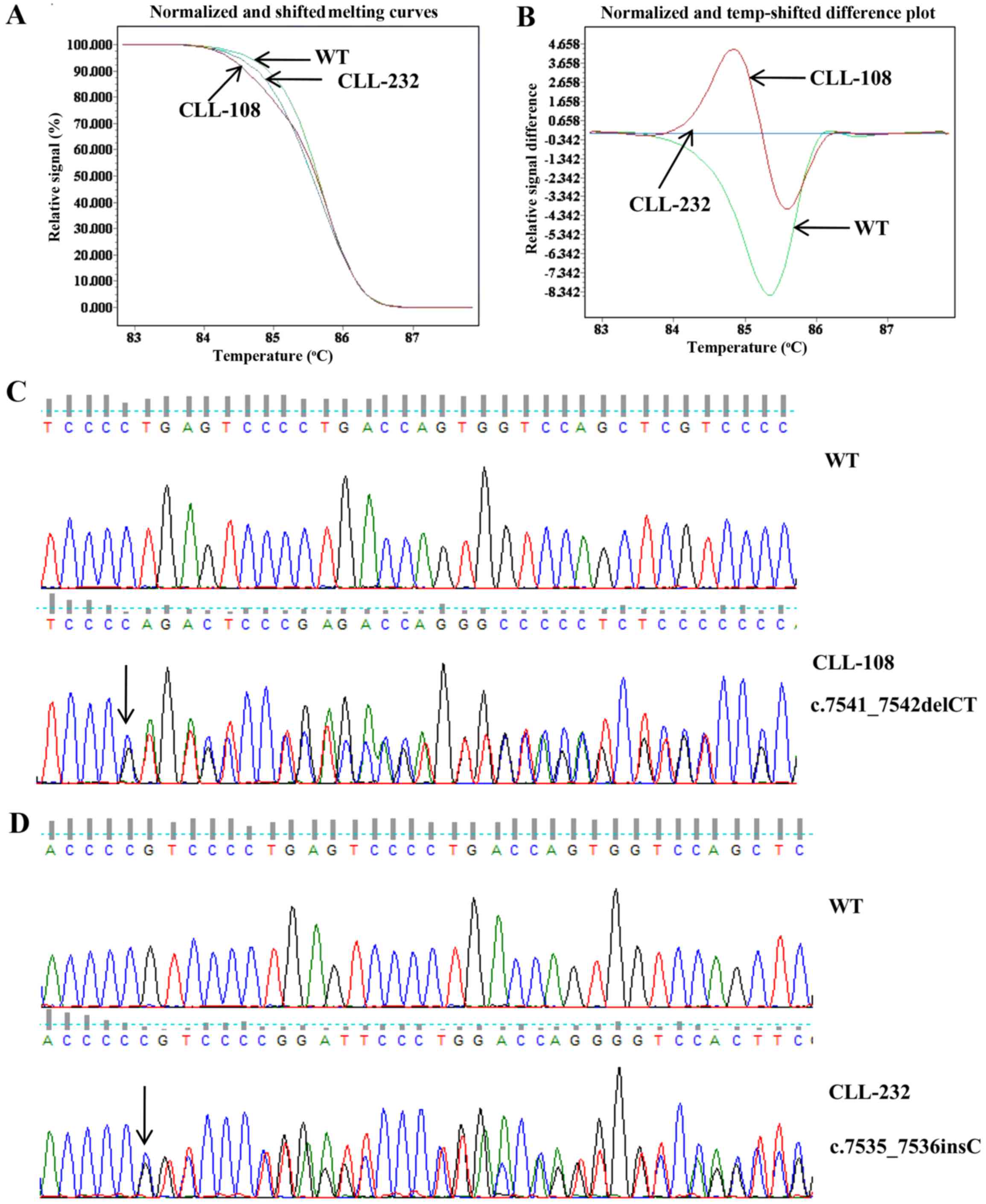
mutations, represented by CLL-108 and CLL-232 (Fig. 1A and B). The melting curves of 7
out of 8 NOTCH1-mutated patients were exactly the same. The
7 patients were confirmed to be c.7541_7542delCT (p.
P2514fs*4) mutated by direct sequencing represented by
CLL-108 (Fig. 1C). The melting
curve of 1 out of 8 NOTCH1-mutated patients was different.
This patient was confirmed to be c.7535_7536insC mutated
(p.S2513fs*3) represented by CLL-232 (Fig. 1D).
Screening for NOTCH1 mutations in the
PEST domain by direct sequencing
We also analyzed the NOTCH1 mutation status
of 133 samples with CLL by direct sequencing and then compared the
findings to that of the HRM assay. Among the 133 patients, 8
(6.02%) patients, who scored positively in the HRM assay, also
turned out to carry somatic mutations of the NOTCH1 gene,
indicated by direct sequencing (Table II). All the mutations were
heterozygous. Of them, 7 (87.5%) cases had the 2-bp frame-shift
deletion, c.7541_7542delCT (p.P2514fs*4) and 1 case had
a frame-shift insertion, c.7535_7536insC (p.S2513fs*3).
Another 93.98% (125/133) of the CLL samples, showing consistency in
melting curves and temp-shifted plots from those generated by the
wild-type template, were implicated to be NOTCH1 wild-type
in the direct sequencing. Above all, the results of the
NOTCH1 mutations detected by HRM analysis were 100%
consistent with the findings from the direct sequencing.
 | Table IINOTCH1 mutations of the CLL
patients. |
Table II
NOTCH1 mutations of the CLL
patients.
| Mutation | CLL patients | Direct
sequencing | HRM analysis |
|---|
| CLL-48 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-108 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-109 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-185 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-213 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-232 |
c.7535_7536insC |
p.S2513fs*3 | Mutated |
| CLL-250 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
| CLL-295 |
c.7541_7542delCT |
p.P2514fs*4 | Mutated |
NOTCH1 mutations are associated with
adverse biological features in CLL patients
The main biological features of the CLL cohort
according to NOTCH1 mutations are listed in Table III. NOTCH1-mutated CLL
patients presented with higher frequencies of germline IGHV
unmutated (6/8, 75%) status and trisomy 12 (5/8, 62.5%) than these
frequencies noted in the NOTCH1-unmutated patients. It is
noteworthy that there were significant correlations between
NOTCH1 mutations and IGHV status (p=0.0053) or
trisomy 12 (p=0.0053). There were no significant correlations
between expression level of CD38 (p=0.4612) or ZAP70 (p=0.5066)
with NOTCH1 mutations.
 | Table IIICharacteristics of the CLL patients
according to NOTCH1 mutations. |
Table III
Characteristics of the CLL patients
according to NOTCH1 mutations.
|
Characteristics | All
(n=133)
| NOTCH1
wild-type
(n=125)
| NOTCH1
mutated
(n=8)
| P-value |
|---|
| N (%) | N (%) | N (%) |
|---|
| IGHV
unmutated | 41 (30.8) | 35 (28) | 6 (75) | 0.0053 |
| Trisomy 12 | 30 (22.6) | 25 (20) | 5 (62.5) | 0.0053 |
| CD38+
expression | 21 (15.8) | 19 (15.2) | 2 (25) | 0.4612 |
| ZAP70+
expression | 22 (16.5) | 20 (15) | 2 (25) | 0.5066 |
Sensitivity evaluation by HRM and direct
sequencing
Cancer cell lines with known NOTCH1 genotype
were used for the validation and sensitivity testing for the HRM
assay and direct sequencing. The acute lymphoblastic leukemia cell
line Molt4 harboring a heterozygous 7541_7542delCT
(p.P2514fs*4) in NOTCH1 was used as a positive
control and the acute T-cell leukemia cell line Jurkat which has a
wild-type NOTCH1 genotype was used as a negative control.
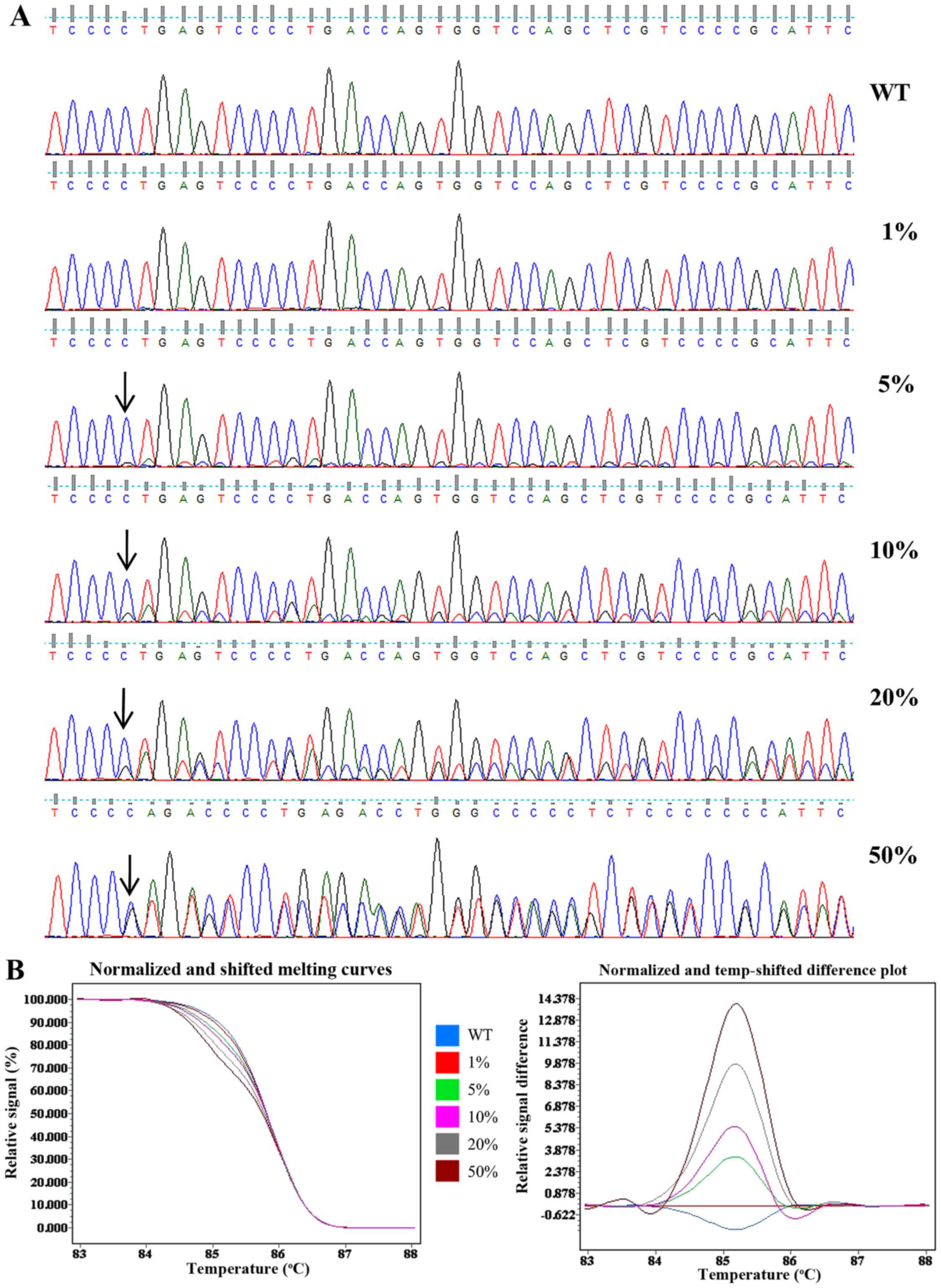
The gDNA of the Molt4 cells was serially diluted into Jurkat gDNA
at ratios of 100, 40, 20, 10 and 2% to yield mutant allele
frequencies of 50, 20, 10, 5 and 1%. The relative sensitivities of
direct sequencing and HRM were evaluated using the diluted gDNA.
The mutation was detectable at a low peak by direct sequencing when
the mutant frequency was >10%. However, when the mutation
frequency was at 5%, it was only distinguishable from the
background. When the mutant frequency was <5%, the mutation was
not detectable (Fig. 2A).
Fig. 2B shows a normalized and
shifted melting curves plot and normalized and temp-shifted
difference plot of the HRM data. The melting curve from 1% mutant
template sufficiently differed from the wild-type template, and
this distinct melting profile was consistently observed across all
other templates measured (5, 10, 20 and 50%). Thus, the sensitivity
of direct sequencing and HRM was found to be 10 and 1%,
respectively.
Discussion
The presence of NOTCH1 mutations disrupting
the carboxy-terminal PEST domain appears to confer adverse
prognosis in CLL (11,12,14,15). Evaluating the NOTCH1
mutation status is useful in clinical practice for patients with
CLL and may facilitate therapeutic decision-making (16,32,33). Therefore, selection of method for
detecting somatic mutation with high sensitivity and specificity
are of great importance. In this study, we successfully developed a
powerful, highly sensitive, cost-effective and easy to perform
approach for NOTCH1 mutation screening in CLL. Most
important, this is the first study to report NOTCH1 mutation
detection in bone marrow samples of Chinese CLL patients using the
HRM assay.
In the 133 newly diagnosed CLL patients, the melting
curves of 6.02% (8/133) of the CLL patients were markedly different
compared with the other 125 patients. The melting curves of 7 out
of 8 NOTCH1-mutated patients were exactly the same. The 7
patients were confirmed to be c.7541_7542delCT mutated. The melting
curve of 1 out of 8 NOTCH1-mutated patients was different.
This patient was confirmed to be c.7535_7536insC mutated. All of
the 133 CLL samples lacked single nucleotide polymorphisms (SNPs)
according to the direct sequencing. SNPs are the result of genomic
variation or changes of a single nucleotide in the genomic DNA. The
HRM method has been successfully used in SNP genotyping and
mutation detection (34,35). Indeed, there are certain SNPs in
the region of our PCR amplicons according to the dbSNP database.
Unexpected SNPs may generated unique amplicon melting patterns
(36,37) (shifted melting curves and
difference plot patterns) and be differentiated from either the
c.7541_7542delCT or c.7535_7536insC mutation. Altogether, the HRM
analysis used by us generated specific melting profiles that
allowed the discrimination between wild-type and mutated samples
(Fig. 1). Our HRM assay was
confirmed to be reliable since all mutations detected by the HRM
assay were also confirmed by direct sequencing.
In the present study, direct sequencing confirmed
that 6.02% of the 133 Chinese CLL cases at diagnosis harbored
NOTCH1 mutations in the PEST domain, and there were two
genotypic heterozygous variations, c.7541_7542delCT
(p.P2514fs*4) and c.7535_7536insC
(p.S2513fs*3). The former mutation type comprised up to
87.5% (7/8) of the mutations. This frequency is consistent with a
report in the Chinese population by Xia et al (38), but lower than that found in a
series of studies of European CLL patients (13,14,39). Therefore, there are discrepancies
between Asian and European populations. CLL patients with mutated
NOTCH1 showed a higher frequency of unmutated IGHV
and trisomy 12, with statistically significant differences. This is
in concordance with several previous studies (40–42).
The high sensitivity of the method was also
confirmed in our experiments. Using cell lines with wild-type and
heterozygous c.7541_7542delCT NOTCH1 mutation, we found that
the sensitivity of HRM in our experimental setting was 1%, while
direct sequencing analysis had a sensitivity of 10%. However, the
frequency of the NOTCH1 mutation detected by HRM was the
same as that of direct sequencing, indicating that the mutational
burden in our CLL cohort was >10%. Actually, the samples we
analyzed were CLL monoclonal cells. Only CLL samples containing
≥95% of CD5+CD19+ cells were included in the
study.
Direct sequencing, known as the 'gold standard', has
been used to detect somatic mutations for many years. It is able to
detect any mutation in the DNA sequence being analyzed. But its
limited sensitivity, high cost and long TAT have prompted the
development of alternative methods for routine clinical testing
which have greater diagnostic practicality for somatic mutation
detection.
HRM is more sensitive, faster, less expensive and
time saving than direct sequencing. In this study, the entire
procedure needs ~50 ng gDNA and requires 0.5 days for the TAT, one
sixth of the time compared to direct sequencing (Table IV). Another major advantage for
HRM over direct sequencing is that it is performed in a 'closed
tube' system. This eliminates the risk of post-PCR product
contamination during scanning while also reducing processing time,
since our PCR and HRM assay were performed within one instrument in
the present study.
 | Table IVComparison of direct sequencing and
HRM assay. |
Table IV
Comparison of direct sequencing and
HRM assay.
| Methods | Facts |
Sensitivity
(%) | Mutation
analysis
methods TAT, da |
|---|
| HRM | Detects both known
and unknown mutations; inexpensive; requires sequencing
validation | 1 | 0.5 |
| Direct
sequencing | Detects every
nucleotide change; expensive; criterion standard | 10 | 3 |
However, it has also been reported that this method
can produce false-positive results due to bad DNA quality,
particularly when the starting material is formalin-fixed,
paraffin-embedded (FFPE) tissue (43). In the present study, the gDNA we
detected was extracted from the bone marrow of the CLL patients,
therefore the DNA quality was controllable. The concordance rate
between HRM and sequencing was 100%. There was only a slight
false-positive chance for our analysis.
To the best of our knowledge, HRM analysis has
already been applied for the diagnosis of tumors and genetic
diseases. HRM analysis has been confirmed to a reliable, accurate,
and rapid screening method for APC mutations in oral squamous cell
carcinoma (44). BRCA1 gene
screening using HRM analysis has been demonstrated to be useful for
the diagnosis of lung adenocarcinoma (45) and Moroccan breast cancer (46); Studies using HRM analysis showed
heterozygous mutations in COL1A1 and COL1A2 genes are associated
with osteogenesis imperfecta (47).
In conclusion, our HRM assay opens a new avenue for
the detection of NOTCH1 gene mutations in CLL. It is a valid
and promising tool for high-throughput NOTCH1 screening,
enabling real-time evaluation of CLL progression, which is
significant for decision-making regarding treatment.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81400154), the Natural
Science Foundation of Jiangsu Province (no. BK20151211), and the
Ninth Science and Technology Development Program of Suzhou (no.
SYS201337).
References
|
1
|
Zenz T, Mertens D, Küppers R, Döhner H and
Stilgenbauer S: From pathogenesis to treatment of chronic
lymphocytic leukaemia. Nat Rev Cancer. 10:37–50. 2010.
|
|
2
|
Gaidano G, Foà R and Dalla-Favera R:
Molecular pathogenesis of chronic lymphocytic leukemia. J Clin
Invest. 122:3432–3438. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiorazzi N, Rai KR and Ferrarini M:
Chronic lymphocytic leukemia. N Engl J Med. 352:804–815. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tam CS and Keating MJ: Chemoimmunotherapy
of chronic lymphocytic leukemia. Nat Rev Clin Oncol. 7:521–532.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Puente XS, Pinyol M, Quesada V, Conde L,
Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz
M, et al: Whole-genome sequencing identifies recurrent mutations in
chronic lymphocytic leukaemia. Nature. 475:101–105. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rossi D, Fangazio M, Rasi S, Vaisitti T,
Monti S, Cresta S, Chiaretti S, Del Giudice I, Fabbri G, Bruscaggin
A, et al: Disruption of BIRC3 associates with fludarabine
chemore-fractoriness in TP53 wild-type chronic lymphocytic
leukemia. Blood. 119:2854–2862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blaumueller CM, Qi H, Zagouras P and
Artavanis-Tsakonas S: Intracellular cleavage of Notch leads to a
heterodimeric receptor on the plasma membrane. Cell. 90:281–291.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan JS, Kousis PC, Suliman S, Visan I and
Guidos CJ: Functions of notch signaling in the immune system:
Consensus and controversies. Annu Rev Immunol. 28:343–365. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lobry C, Oh P and Aifantis I: Oncogenic
and tumor suppressor functions of Notch in cancer: It's NOTCH what
you think. J Exp Med. 208:1931–1935. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lobry C, Oh P, Mansour MR, Look AT and
Aifantis I: Notch signaling: Switching an oncogene to a tumor
suppressor. Blood. 123:2451–2459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rosati E, Sabatini R, Rampino G, Tabilio
A, Di Ianni M, Fettucciari K, Bartoli A, Coaccioli S, Screpanti I
and Marconi P: Constitutively activated Notch signaling is involved
in survival and apoptosis resistance of B-CLL cells. Blood.
113:856–865. 2009. View Article : Google Scholar
|
|
12
|
Arruga F, Gizdic B, Serra S, Vaisitti T,
Ciardullo C, Coscia M, Laurenti L, D'Arena G, Jaksic O, Inghirami
G, et al: Functional impact of NOTCH1 mutations in chronic
lymphocytic leukemia. Leukemia. 28:1060–1070. 2014. View Article : Google Scholar
|
|
13
|
Fabbri G, Rasi S, Rossi D, Trifonov V,
Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al:
Analysis of the chronic lymphocytic leukemia coding genome: Role of
NOTCH1 mutational activation. J Exp Med. 208:1389–1401. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rossi D, Rasi S, Fabbri G, Spina V,
Fangazio M, Forconi F, Marasca R, Laurenti L, Bruscaggin A, Cerri
M, et al: Mutations of NOTCH1 are an independent predictor of
survival in chronic lymphocytic leukemia. Blood. 119:521–529. 2012.
View Article : Google Scholar :
|
|
15
|
Weissmann S, Roller A, Jeromin S,
Hernández M, Abáigar M, Hernández-Rivas JM, Grossmann V, Haferlach
C, Kern W, Haferlach T, et al: Prognostic impact and landscape of
NOTCH1 mutations in chronic lymphocytic leukemia (CLL): A study on
852 patients. Leukemia. 27:2393–2396. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
López-Guerra M, Xargay-Torrent S, Rosich
L, Montraveta A, Roldán J, Matas-Céspedes A, Villamor N, Aymerich
M, López-Otín C, Pérez-Galán P, et al: The γ-secretase inhibitor
PF-03084014 combined with fludarabine antagonizes migration,
invasion and angiogenesis in NOTCH1-mutated CLL cells. Leukemia.
29:96–106. 2015. View Article : Google Scholar
|
|
17
|
Osipo C, Golde TE, Osborne BA and Miele
LA: Off the beaten pathway: The complex cross talk between Notch
and NF-kappaB. Lab Invest. 11–17. 2008. View Article : Google Scholar
|
|
18
|
Di Ianni M, Baldoni S, Rosati E, Ciurnelli
R, Cavalli L, Martelli MF, Marconi P, Screpanti I and Falzetti F: A
new genetic lesion in B-CLL: A NOTCH1 PEST domain mutation. Br J
Haematol. 146:689–691. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sportoletti P, Baldoni S, Cavalli L, Del
Papa B, Bonifacio E, Ciurnelli R, Bell AS, Di Tommaso A, Rosati E,
Crescenzi B, et al: NOTCH1 PEST domain mutation is an adverse
prognostic factor in B-CLL. Br J Haematol. 151:404–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sportoletti P, Baldoni S, Del Papa B,
Aureli P, Dorillo E, Ruggeri L, Plebani S, Amico V, Di Tommaso A,
Rosati E, et al: A revised NOTCH1 mutation frequency still impacts
survival while the allele burden predicts early progression in
chronic lymphocytic leukemia. Leukemia. 28:436–439. 2014.
View Article : Google Scholar
|
|
21
|
Stilgenbauer S, Schnaiter A, Paschka P,
Zenz T, Rossi M, Döhner K, Bühler A, Böttcher S, Ritgen M, Kneba M,
et al: Gene mutations and treatment outcome in chronic lymphocytic
leukemia: Results from the CLL8 trial. Blood. 123:3247–3254. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oscier DG, Rose-Zerilli MJ, Winkelmann N,
Gonzalez de Castro D, Gomez B, Forster J, Parker H, Parker A,
Gardiner A, Collins A, et al: The clinical significance of NOTCH1
and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 121:468–475.
2013. View Article : Google Scholar
|
|
23
|
Puente XS, Beà S, Valdés-Mas R, Villamor
N, Gutiérrez-Abril J, Martín-Subero JI, Munar M, Rubio-Pérez C,
Jares P, Aymerich M, et al: Non-coding recurrent mutations in
chronic lymphocytic leukaemia. Nature. 526:519–524. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gundry CN, Vandersteen JG, Reed GH, Pryor
RJ, Chen J and Wittwer CT: Amplicon melting analysis with labeled
primers: A closed-tube method for differentiating homozygotes and
hetero-zygotes. Clin Chem. 49:396–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reed GH, Kent JO and Wittwer CT:
High-resolution DNA melting analysis for simple and efficient
molecular diagnostics. Pharmacogenomics. 8:597–608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taylor CF: Mutation scanning using
high-resolution melting. Biochem Soc Trans. 37:433–437. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liew M, Pryor R, Palais R, Meadows C,
Erali M, Lyon E and Wittwer C: Genotyping of single-nucleotide
polymorphisms by high-resolution melting of small amplicons. Clin
Chem. 50:1156–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reed GH and Wittwer CT: Sensitivity and
specificity of single-nucleotide polymorphism scanning by
high-resolution melting analysis. Clin Chem. 50:1748–1754. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Graham R, Liew M, Meadows C, Lyon E and
Wittwer CT: Distinguishing different DNA heterozygotes by
high-resolution melting. Clin Chem. 51:1295–1298. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Herrmann MG, Durtschi JD, Bromley LK,
Wittwer CT and Voelkerding KV: Amplicon DNA melting analysis for
mutation scanning and genotyping: Cross-platform comparison of
instruments and dyes. Clin Chem. 52:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Montgomery J, Wittwer CT, Palais R and
Zhou L: Simultaneous mutation scanning and genotyping by
high-resolution DNA melting analysis. Nat Protoc. 2:59–66. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wei P, Walls M, Qiu M, Ding R, Denlinger
RH, Wong A, Tsaparikos K, Jani JP, Hosea N, Sands M, et al:
Evaluation of selective gamma-secretase inhibitor PF-03084014 for
its antitumor efficacy and gastrointestinal safety to guide optimal
clinical trial design. Mol Cancer Ther. 9:1618–1628. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de
Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al:
Therapeutic antibody targeting of individual Notch receptors.
Nature. 464:1052–1057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmidt U, Hulkkonen J and Naue J:
Detection of a G>C single nucleotide polymorphism within a
repetitive DNA sequence by high-resolution DNA melting. Int J Legal
Med. Mar 14–2016.Epub ahead of print. View Article : Google Scholar
|
|
35
|
Hong Y, Pandey MK, Liu Y, Chen X, Liu H,
Varshney RK, Liang X and Huang S: Identification and evaluation of
single-nucleotide polymorphisms in allotetraploid peanut (Arachis
hypogaea L.) based on amplicon sequencing combined with high
resolution melting (HRM) analysis. Front Plant Sci. 6:10682015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
de Juan I, Esteban E, Palanca S, Barragán
E and Bolufer P: High-resolution melting analysis for rapid
screening of BRCA1 and BRCA2 Spanish mutations. Breast Cancer Res
Treat. 115:405–414. 2009. View Article : Google Scholar
|
|
37
|
Er TK and Chang JG: High-resolution
melting: Applications in genetic disorders. Clin Chim Acta.
414:197–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia Y, Fan L, Wang L, Gale RP, Wang M,
Tian T, Wu W, Yu L, Chen YY, Xu W, et al: Frequencies of SF3B1,
NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in
Chinese with chronic lymphocytic leukemia: Disparities with
Europeans. Oncotarget. 6:5426–5434. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shedden K, Li Y, Ouillette P and Malek SN:
Characteristics of chronic lymphocytic leukemia with somatically
acquired mutations in NOTCH1 exon 34. Leukemia. 26:1108–1110. 2012.
View Article : Google Scholar
|
|
40
|
Balatti V, Bottoni A, Palamarchuk A, Alder
H, Rassenti LZ, Kipps TJ, Pekarsky Y and Croce CM: NOTCH1 mutations
in CLL associated with trisomy 12. Blood. 119:329–331. 2012.
View Article : Google Scholar :
|
|
41
|
Del Giudice I, Rossi D, Chiaretti S,
Marinelli M, Tavolaro S, Gabrielli S, Laurenti L, Marasca R, Rasi
S, Fangazio M, et al: NOTCH1 mutations in +12 chronic lymphocytic
leukemia (CLL) confer an unfavorable prognosis, induce a
distinctive transcriptional profiling and refine the intermediate
prognosis of +12 CLL. Haematologica. 97:437–441. 2012. View Article : Google Scholar :
|
|
42
|
Villamor N, Conde L, Martínez-Trillos A,
Cazorla M, Navarro A, Beà S, López C, Colomer D, Pinyol M, Aymerich
M, et al: NOTCH1 mutations identify a genetic subgroup of chronic
lymphocytic leukemia patients with high risk of transformation and
poor outcome. Leukemia. 27:1100–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Do H, Krypuy M, Mitchell PL, Fox SB and
Dobrovic A: High resolution melting analysis for rapid and
sensitive EGFR and KRAS mutation detection in formalin fixed
paraffin embedded biopsies. BMC Cancer. 8:1422008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Marino M, Monzani ML, Brigante G, Cioni K,
Madeo B, Santi D, Maiorana A, Bettelli S, Moriondo V, Pignatti E,
et al: High-resolution melting is a sensitive, cost-effective,
time-saving technique for BRAF V600E detection in thyroid FNAB
washing liquid: A prospective cohort study. Eur Thyroid J. 4:73–81.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wright GM, Do H, Weiss J, Alam NZ, Rathi
V, Walkiewicz M, John T, Russell PA and Dobrovic A: Mapping of
actionable mutations to histological subtype domains in lung
adenocarcinoma: Implications for precision medicine. Oncotarget.
5:2107–2115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
El Khachibi M, Diakite B, Hamzi K, Badou
A, Senhaji MA, Bakhchane A, Jouhadi H, Barakat A, Benider A and
Nadifi S: Screening of exon 11 of BRCA1 gene using the high
resolution melting approach for diagnosis in Moroccan breast cancer
patients. BMC Cancer. 15:812015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang J, Ren X, Bai X, Zhang T, Wang Y, Li
K and Li G: Identification of gene mutation in patients with
osteogenesis imperfecta using high resolution melting analysis. Sci
Rep. 5:134682015. View Article : Google Scholar
|