Congenital heart disease (CHD) involves the
anatomical structure abnormality caused by the formation of
obstacles or the abnormal development of the heart and great
vessels during the period of embryonic development, or a group of
congenital malformations with actual or potential influence on
heart function arising from the open tunnels which should have
self-closed after childbirth. CHD mostly occurs during 2–8 weeks
after impregnation, and it is the most common cardiovascular
malformation affecting children; it severely affects the health of
infants and young children (1–4).
At present, CHD is regarded as a multigene disease influenced by
the environment and heredity; however, the pathogenesis of the
disease and the underlying molecular mechanisms and interations
between genes remain unclear (5–8).
Heart development is a very complex process, involving the
expression of numerous genes at specific time points in the process
of embryonic development, and it is regulated by many transcription
factors (9,10). The realization of this process is
not only determined by gene sequences, but is also largely
generated by the transformation of epigenetics. In addition, an
increasing number of studies have found that children with CHD have
an extremely low occurrence rate of gene mutation, which can only
explain a small number of CHD cases, as there is no pathogenic gene
transformation for the majority of CHD cases (11–13). Some recent studies have found that
'epigenetics' may very likely participate or play an important role
in the occurrence of CHD (14–16).
Epigenetics suggests that DNA sequence undergoes no
transformation, but the gene expression occurs by heritable
transformation, which is the other heritable material
transformations in the cells apart from the heritage information
and with stable heredity in the process of cell development and
proliferation (17–19). Epigenetics is mainly the
reversible and heritable transformation of gene function with the
DNA sequences of the nucleus unchanged, and these transformations
include DNA modifications (such as methylation), various kinds of
histone modifications (such as methylation, acetylation,
ubiquitination and phosphorylation), chromatin remodeling,
non-encoding RNA regulation (20–26). In the field of 'epigenetics', by
transforming the chromatin structure with the help of related
enzymes and interacting with other regulator protein, histone
modifications regulate gene expression, and influence occurrence
and development of diseases, which is also known as 'the second
heritage code', mainly including methylation, acetylation,
phosphorylation, ubiquitin, adenosine, small ubiquitin related
modification, ADP ribosylation and proline isomerization (21,27,28). Histone methylation is one of the
most common histone modifications, and mainly includes arginine
methylation and lysine methylation (29–31). Comparatively speaking, histone
methylation modification transforms loosen or agglutination state
of chromatin mainly through influencing the affinity of histone and
DNA, regulates gene expression by influencing the affinity of other
transcription factors and structural gene promoters. Thus, it can
be seen that histone methylation modification has the gene
expression regulatory function similar to the DNA genetic code, and
plays an important role in the process of growth and development
(32–34).
In recent years, studies have shown that histone
lysine methylation is not only closely related to tumor occurrence
and development, developmental defects, senile dementia, cardiac
hypertrophy and other clinical diseases, but also participates in
the occurrence of CHD, influencing the development of heart
structure and CHD candidate gene expression (35–45). For this reason, this review aims
to summarize the new progress of CHD epigenetic mechanism research
from the aspect of histone lysine methylation modification, in
order to provide a new scientific basis for the prevention and
treatment of CHD.
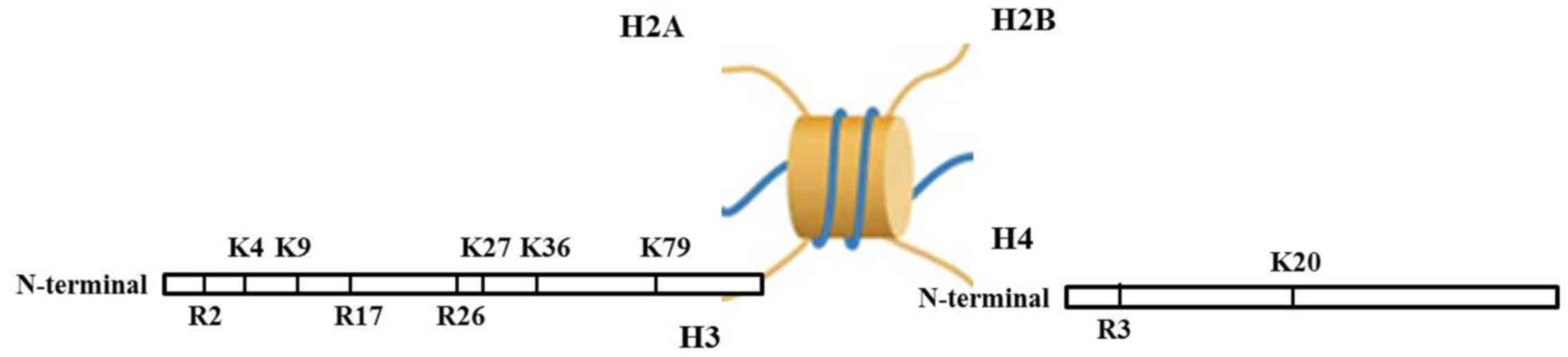
Histone methylation involves the methylation
occurring at histone H3 and the N-terminal of H4 arginine (Arg) or
lysine (Lys) residues, catalyzed by histone methyltransferase. The
function of histone methylation is mainly reflected in
heterochromatin forming, genomic imprinting, X chromosome
inactivation and transcriptional control (46–52). Apart from histone
methyltransferase, the histone demethylase is also found (53,54). At first, it was considered that
the histone methylation effect was stable and irreversible;
however, the existence of methyltransferase renders the process of
histone methylation more dynamical.
Histone methylation can occur on of Lys and Arg
histone residues. Lysine residues can be single, double and triple
methylated, while arginine residues can be single and double
methylated (55–57). This varying degree of methylation
largely increases the complexity of histone modifications and
regulator gene expressions (29,58). Methylation action sites are in the
N atoms at the side chains of Lys and Arg. Lys locus 4, 9, 27, 36
and 79 of histone H3 (H3K4, H3K9, H3K27, H3K36 and H3K79), Lys
locus 20 of histone H4 (H4K20), Arg locus 2, 17, 26 of histone H3
(H3R2, H3R17 and H3R26) and Arg locus 3 of histone H4 (H4R3) are
the common loci of methylation (Fig.
1). Studies have shown that histone Arg methylation is a
comparatively dynamical mark, and is related to gene activation;
however, Arg methylation lacking H3 and H4 is related to gene
silencing (59–61). On the contrary, Lys methylation
seems to be a stable mark of gene expression regulation. For
example, H3K4 methylation is related to gene activation, while H3K9
and H3K27 methylation are related to gene silencing (38,62–66). In addition, H4K20 methylation is
related to gene silencing, H3K36 and H3K79 are related to gene
activation (67–72).
Arginine methylation occurs at the equal locus of
histone H3R2/R17/R26 and H4R3, and plays a promoting effect on gene
expression (60). When the
methylation process occurs, histone arginine methyltransferase or
protein arginine methyltransferase (PRMT), as the collaborative
activity factor, is recruited into the promoter region of the
target gene and thus activates gene expression (73–75). The PRMT of catalyzing histone
arginine methyltransferase includes two categories: the first type
of PRMT being catalyzed to form single methylation arginine and
unsymmetrical double methylation arginine, and the second type of
PRMT being catalyzed to form single methylation arginine and
symmetrical double methylation arginine (76,77). The family of PRMT includes PRMTl,
PRMT3, RMTI/HMT, PRMT4/CAMRl and PRMT5. K4, K9, K27, K36, K79 of
histone H3 and K20 of H4 all can be methylated. The methylation
level is regulated and executed by a type of methyltransferase with
SET structural domain with highly conserved nucleus and the pre-SET
and post-SET structural domain with abundant cysteine sequence
(31,78,79). The denomination of SET structural
domain is constituted with 3 initial letters of 3 genes from
expressing SET structural domain found at the earliest, which are
Su(var)3–9, E(z) and Trx (80).
Furthermore, histone demethylase can catalyze histone lysine to
demethylation, and then affect the level of methylation. At
present, the demethylase of histone mainly has two types: lysine
specific demethylase (LSD1) and Jmjc domain-containing histone
demethylase (JHDM) (81,82).
Epigenetic modification, including methylation and
acetylization plays an important role in the regulation of gene
expression. Studies have indicated that histone methylation can
reduce or increase its affinity for charged DNA, loosen or tighten
the chromatin structure to affect the accessibility and
interactions between transcriptional factors and DNA templates,
ultimately promote or inhibit gene expression (83,84). Recent evidence supports a
prominent role for histone methylation in normal and aberrant heart
development (85–87). A recent study demonstrated that
changes in histone methylation levels in histone H3 that binds with
critical promoter parts of the ssTnI gene can cause the
corresponding changes in ssTnI gene expression, which indicated
that histone methylation was involved in the regulation of
myofibril gene expression in the heart during development (88). Additionally, Hand2 and Irx4
transcription factors have been shown to be reduced in
SMYD1-deficient mice, suggesting that SMYD1-mediated histone
methylation is necessary for the expression of these essential
cardiac transcription factors (89). Hence, these findings illustrate
the pervasive roles of histone methylation in the process of heart
development.
CHD is the most common type of birth defect,
manifesting as obstacles in the process of embryonic heart or blood
vessel development, which may result in the morphology, structure,
function and metabolic abnormalities of heart and blood vessels
(2,3,5).
According to the statistics, CHD has become the first reason for
birth defects and the main cause of perinatal death and death in
children (2). The causes of CHD
are not yet completely clear; however, most scholars consider that
many types of CHD are caused by a single gene mutation and
chromosome aberration, and most types of CHD belong to complex
genetic diseases, which are caused by the interaction between
genetic factors and environmental factors (90,91). Studies have shown that histone
lysine methylation modification as part of the epigenetic
regulation, is involved in the development of heart and blood
vessels, which is also one of the causes of CHD (92,93). The level of histone lysine
methylation is determined by the balance of histone methylation and
demethylation, which is a process by which methyl groups are
transferred onto or removed from the amino acids of histone
proteins. Histone methyltransferase and histone demethylase
catalyze histone methylation and demethylation, respectively. In
most cases, under the action of the methylation and demethylation,
the histone tails relax or surround, which can loosen or inhibit
DNA of transcription factors so as to turn the genes in DNA 'off'
and 'on', resulting in the normal or aberrant expression of related
genes and leading to abnormal heart development. To understand the
research progress of histone lysine methylation and CHD, we
summarize the known histone lysine modifying enzymes which
regulates CHD in Table I.
During heart development, several cardiac progenitor
pools give rise to diverse cell lineages, such as cardiomyocytes,
vascular smooth muscle cells, fibroblasts that form the connective
tissues and endothelial cells of the endocardium. The heart
expresses many epigenetic factors, including both histone modifying
proteins and chromatin remodelers. Among the epigenetic factors,
TrxG proteins are special family of chromatin factors that regulate
developmental gene expression in the heart (94). TrxG proteins function in
multi-subunit complexes, three TrxG complexes, the MLL complex, the
BRM/BAF complex and a supercomplex, and have been purified in
mammalian cells (95). TrxG
proteins are evolutionarily conserved H3K4 methyltransferases that
maintain the transactivation states of lineage-specific genes
during embryonic development. Multiple TrxG genes are normally
expressed in the mouse heart. Due to the essential function of TrxG
genes, constitutive knockouts of key TrxG genes often result in
lethality during early embryogenesis before cardiac phenotypes can
be analyzed. The differentiation of mouse embryonic stem cells
(ESCs) toward mesodermal and endodermal lineages is severely
altered and, in particular, the cardiac lineage differentiation of
ESCs is completely abolished in the absence of MLL2, a TrxG member.
Moreover, the expression of core cardiac transcription factors and
the levels of H3K4 trimethylation of these cardiac-specific
promoters are significantly decreased by the loss of MLL2 (96). Taken together, these results
reveal a critical role for MLL2 in the proliferation and cardiac
lineage differentiation of mouse ESCs, and provide critical insight
not only into the novel role of the TrxG protein in cardiac
development, but also into their clinical significance in related
CHD.
SET and myeloid, nervy and DEAF-1 (MYND)
domain-containing proteins (SMYDs), including SMYD1–5, have two
functional protein domains, SET (mediates histone lysine
methylation activity) and MYND (mediates the protein-protein
interaction and binds to DNA motifs) domains (80). SET-MYND-domain 1 (SMYD1/BOP)
encodes an evolutionary conserved histone methyltransferase
containing a split SET domain interrupted by a MYND domain, which
includes two members SMYD1a and SMYD1b, can catalyze H3K4
methylation (97). The expression
of SMYD1 is restricted to skeletal and cardiac muscles in humans,
fish, chickens and mice. There is evidence to indicate that SMYD1
plays important roles in cardiac differentiation, development and
function (98). The global
knockdown of SMYD1a and SMYD1b in zebrafish has been shown to
result in the disruption of myofibril formation and an absence of
beating of the heart. Molecular and cellular experiments showed
that myofibers in embryos in which SMYD1 was knocked down appeared
as immature myofibers with centrally located nuclei and
disorganized myofibrils, indicating that SMYD1 played a critical
role in myofibers maturation and contraction (97). Conventional null SMYD1 mice die
in utero around embryonic day 10.5 (E10.5) due to heart
defects, including disrupted maturation of ventricular
cardiomyocytes and malformation of the right ventricle (99). However, Diehl et al
recently reported that SET and MYND domain containing 2 (SMYD2), is
capable of H3K4 methylation when bound to Hsp90a and acts on
non-histone targets by inhibiting the functional activity of p53
via methylation of p53, lysine 370, which was differentially
expressed during cardiac development with highest expression in the
neonatal heart (100). To
elucidate the functional role of SMYD2 in the heart, they generated
knockout mice harboring a cardiomyocyte-specific deletion of SMYD2
and performed histological, functional and molecular experiments.
Unexpectedly, cardiac deletion of SMYD2 was dispensable for proper
morphological and functional development of the murine heart
(100). H3K4 methyltransferase
SMYD3 is highly expressed within developing zebrafish heart and
knockdown of it led to severe defects such as pericardial edema and
abnormal expression of three heart-chamber markers in cardiac
morphogenesis (101). These
results indicate that SMYD3 plays an important role in heart
development and its proper functioning is essential for normal
heart morphogenesis during development.
TBX transcription factors share a highly conserved
DNA-binding domain and play critical roles in embryonic development
(102). Six members of TBX
family (TBX1, TBX18 and TBX20 of the TBX1 subfamily, and TBX2, TBX3
and TBX5 of the TBX2 subfamily) are required for the cardiac
morphogenesis in mammals (103).
TBX1 interacts with H3K4 methyltransferase to enhance its H3K4
monomethylation status through T-box, regulates expression of
related genes by epigenetic patterns (104). TBX1 mutation can lead to
DiGeorge syndrome (DGS), which is the most common microdeletion
syndrome, and is characterized by congenital cardiac, craniofacial
and immune system abnormalities (105). Additionally, Pan et al
reported that a novel heterozygous TBX1 mutation, p.Q277X, was
identified in an index patient with double outlet right ventricle
and ventricular septal defect (106). TBX2 gene is expressed in the
myocardium of the atrioventricular canal, outflow tract and inflow
tract and plays a critical role in heart chamber formation
(107). The genomic deletion and
duplication of TBX2 gene have been found to be associated with
ventricular septal defects (108). The evolutionary conserved TBX3
gene encodes T-box transcription factors and locus forms a CTCF
independent autonomous regulatory domain with multiple
combinatorial regulatory elements, which plays crucial roles in the
development and homeostasis of the cardiac conduction system in
humans and mice (109). Previous
studies have found that TBX5 is expressed in the proepicardial
organ or septum transversum, which is required for the normal
development of proepicardium/proepicardial organ cells, as well as
proper epicardial formation and maturation (110). Additionally, TBX5 deficiency
delays epicardiac cell attachment to the myocardium and impairs
production of epicardial-derived cells and their migration into the
myocardium, and results in abnormal coronary vasculogenesis and
murine ischemic cardiomyopathy (111). Clinical studies have shown that
Holt-Oram syndrome is caused by mutations in TBX5, which is a human
inherited disorder and manifests as left pericardium agenesis and
anomalous coronary arteries along with ventricular septal defects
(112–114). These findings all demonstrate
that TBX5 is essential for epicardial development in hearts and
establishment of the coronary vasculature. Similar to TBX5, TBX18
is also highly expressed in proepicardial cells and proepicardium,
TBX18-deficient proepicardium produces an epicardium and coronary
vasculature with structural and functional defects, and that
remodeling of the disorganized subepicardial plexus in
TBX18-deficient hearts produced a mature coronary artery network
with fewer distributing conduit vessels and smaller lumen profiles,
which indicates that TBX18 plays critical role in coronary
development (115). However,
TBX20 is necessary in heart development by regulating cardiomyocyte
proliferation and regional specification and formation of cardiac
chambers and valves; TBX20 mutations in mice can result in the
failure of heart looping, developmental arrest, and the lack of
chamber differentiation, and loss of TBX20 in mice leads to
cardiomyopathy with associated arrhythmias and death (116,117). More seriously, mutations in
human TBX20 result in cardiac malformations including septal
defects, double outlet right ventricle and cardiomyopathy (118,119). These findings provide novel
insight into the molecular mechanism underlying CHD and suggest
potential implications for the development of novel preventive and
therapeutic strategies for CHD.
DPF3 is a member of the highly conserved d4 protein
family, which is characterized by a double PHD finger in the
C-terminal and has two splice variants DPF3a and DPF3b in human and
mice (120). In the process of
embryonic development, DPF3 is expressed both in heart and somites
of mouse, chicken and zebrafish, which is important epigenetic
regulation factor for heart and muscle development by associated
with the BAF chromatin remodeling complex and binds methylated
lysine residues of H3K4 (121).
Previous studies have found that DPF3 mutation or deletion leads to
incomplete cardiac looping, attenuated ventricular contractility
and disassembled muscular fibers caused by the transcriptional
deregulation of structural and regulatory proteins in the heart,
which all demonstrate that DPF3 is responsible for cardiac
development imbalance, ventricular septal defect and other cardiac
disorders (121).
PTIP is an essential cofactor for H3K4me by KMT2C/D,
which is encoded by the Paxip1 gene and is essential for embryonic
development in mice and flies (122–124). As a critical component of the
KMT2C/D complex, the loss of PTIP leads to reduced levels of
H3K4me3 in whole embryos, ESCs and Drosophila larvae
(125,126). Stein et al demonstrated
that temporal and tissue-specific deletion of PTIP reduces H3K4
methylation level and alters the transcriptional program in
nondividing cardiomyocytes. It is suggested that a role for KMT2
complexes not just in establishing active chromatin domains but
also in the maintenance of the differentiated state over time.
Furthermore, the loss of PTIP-mediated H3K4me results in
significant changes in the physiology of the cardiomyocytes,
suggesting that PTIP deletion is the direct cause of premature
ventricular beats, a harbinger of lethal ventricular arrhythmias in
nondividing cardiomyocytes (42,127).
SETD7 also termed as SET7/9, is another type of
histone lysine methyltransferase and only has SET domain for
methyltransferase activity, but not MYND domain, which is initially
discovered as a specific methyltransferase for nonmethylated H3K4.
Tao et al found that the knockdown of SETD7 showed the
defects in skeletal muscle formation and myofibril structures in a
zebrafish developmental model (128). To examine the function of SETD7
in heart development, Kim et al firstly demonstrated that
SETD7 was highly expressed in developing zebrafish heart and
knockdown of it led to severe defects in cardiac morphogenesis such
as developmental heart edema. Furthermore, the double knockdown of
SMYD3 and SETD7 caused synergistic defects in heart development.
Similar to the knockdown effect, the overexpression of SETD7 also
caused the heart morphogenesis defects in zebrafish (85). These results indicate that the
histone modifying enzyme, SETD7, plays an important role during
heart development and its proper functioning is essential for
normal heart morphogenesis during development.
LSD1 (also known as AOF2/KDM1A), is a member of a
group of enzymes with lysine specific demethylase activity. LSD1
performs enzymatic activity toward di- and monomethyl H3K4 and H3K9
respectively; the specificity for H3K9 arises when LSD1 binds to
the androgen receptor, resulting in a shift of its activity from
H3K4 (129). LSD1 interacts with
proteins mostly through the tower domain, an extended helical
structure. Furthmore, there is evidence to indicate that
LSD1-interacting proteins can regulate the activity and specificity
of LSD1 in developmental processes (130,131). Nicholson et al found that
mice homozygous for a hypomorphic LSD1 allele exhibit a failure to
survive after birth perinatally due to heart defects, with the
majority of animals suffering from ventricular septal defects
(132). Therefore, the
above-mentioned studies thereby illuminate a novel role for LSD1 in
the development of the mammalian heart.
It was found that BCOR inhibited gene transcription
by interacting with BCL-6, and BCOR mutation resulted in abnormal
activation of AP-2a, which was a key factor that mediated the
differentiation of bone marrow mesenchymal stem cells (MSCs)
(133). Fan et al also
pointed out that BCOR recruited a histone demethylase JHDM1B to the
target gene promoter, resulting in the demethylation of H3K4me3 and
H3K36me2 and transcription repression of genes; however, BCL-6
mutation may impair the recruitment of JHDM1B to chromatin,
resulted in increased methylation levels of H3K4 and H3K36
(133). Abnormal histone
methylation due to BCOR mutation may affect BCL-6 binding to the
AP-2a promoter, causing aberrant activation of gene and resulting
in the in occurrence of oculo-facio-cardio-dental (OFCD) syndrome,
which is a rare genetic disorder characterized by teeth with
extremely long roots, and craniofacial, eye and congenital cardiac
abnormalities include septal defect and mitral valve defect
abnormalities (133–135). On the whole, it was identified
that BCOR mutation affected heart development and AP-2α played a
role in congenial heart defects associated with OFCD patients, and
indicated that BCOR may be a novel target for diagnostic and
treatment strategies of OFCD syndrome.
G9a and GLP are known as major H3K9 mono-and
di-methyltransferases and contribute to transcriptional silencing,
which play critical biological roles in various cells and tissues.
For example, G9a and GLP are indispensable for mouse early
development; G9a or GLP knockout mice exhibit embryonic lethality
around E9.5 due to severe growth defects (38,136,137). In order to clarify the roles of
G9a and GLP in cardiac development, Inagawa et al analyzed
the phenotypes of cardiomyocyte specific GLP knockout and G9a
knockdown mice, it was shown that the H3K9me2 level decreased
markedly in the nuclei of the cardiomyocytes of these mice, and the
mice exhibited neonatal lethality and severe cardiac defects
characterized by atrioventricular septal defects (138). These data indicated that G9a and
GLP were required for H3K9me2 in cardiomyocytes and performed an
essential role in normal morphogenesis of the atrioventricular
septum through regulation of the size of the atrioventricular
cushion.
EHMT1 is located on chromosome 9 and encodes
Eu-HMTase1, which is a type of methyltransferase found in the
region of the chromatin region. EHMT1 could specifically modify
H3K9 methylation and thus inhibit the activity of related genes.
Some studies have found EHMT1 mutation or deletion is the main
cause of chromosome 9q subtelomere deletion syndrome (9qSTDS),
approximately half of affected individuals have congenital heart
defects primarily characterized by atrial septal defect or
ventricular septal defect (139,140).
The PR/SET domain zinc-finger transcriptional
repressor Blimp-1/PRDM is initially cloned as a negative regulator
of key transcription factors expression and encoded by PRDM1, which
contains PR/SET domain in N-terminal and C2H2 zinc finger structure
in C-terminal, and thus plays essential roles in primordial germ
cell specification, placental, heart, and forelimb development,
plasma cell differentiation, and T-cell homeostasis through
regulating DNA binding, nuclear input and recruitment of histone
modifying enzymes (141).
Blimp-1/PRDM causes H3K4 methylation by recruiting of histone
modifying enzymes, which inhibits relevant genes expression and
thus regulates the development of embryos. In the development of
embryos, Blimp-1/PRDM deficiency caused serious cardiac defects
including ventricular septal defect and persistent arterial trunk
(141,142).
Jarid2, also termed as Jumonji (jmj), the founding
member of the Jumonji family, all of which contain the JmjC domain
that generally confers histone demethylase activities, which can
catalyze H3K9 methylation and function as a transcriptional
repressor and to interact with other nuclear factors. Lee et
al found that the Jarid2 homozygous mouse embryos show heart
malformations, including ventricular septal defect, noncompaction
of the ventricular wall, double-outlet right ventricle, and dilated
atria; furthermore, expression of Jarid2 in the interventricular
septum, ventricular wall and outflow tract, which is correlated
well with the locations of defects observed in the hearts of mutant
mice. These results indicate that Jarid2 plays an important role in
embryonic heart development (143). At the molecular level, Kim et
al demonstrated that Jarid2 can inhibit the proliferation of
cardiomyocytes, and inhibit the expression of atrial natriuretic
peptide (ANP) by repressing the interaction with transcription
factor Nkx2.5 and GATA4 (144).
Other studies have also found that mutations or deletions of Jarid2
could increase H3K9 and H3K36 methylation level, resulting in the
abnormal expression of development-related genes and thus inducing
atrial septal defect or ventricular septal defect and other cardiac
defects (145,146).
Polycomb group proteins are key regulators of gene
expression during development and differentiation, silencing genes
via regulation of the chromatin structure, which act in complexes
that have specific catalytic functions important for
transcriptional repression. In mammals, 2 major Polycomb group
complexes exist: polycomb repressive complex 1 (PRC1) and PRC2.
Whereas PRC1 ubiquitinates histone H2A on Lys119,1 PRC2 catalyzes
dimethylation and trimethylation of H3K27, generating H3K27me2/3
(147). Weston et al
pointed out that Rae28 protein, the core component of PRC1, which
made PRC1 bind to H3K27me3 and then formed chromatin tight
structure to prevent the occurrence of transcription. Rae28
mutation or deletion mice tend to perform bone dysplasia and heart
development defects (148).
However, Ezh2, the major histone methyltransferase of PRC2,
trimethylates H3K27 and is essential for embryonic development
(149). Delgado-Olguín et
al have shown that Ezh2 stabilizes cardiac gene expression and
prevents cardiac pathology, but Ezh2 deletion in cardiac
progenitors causes postnatal myocardial pathology and destabilizes
cardiac gene expression, which suggests that Ezh2 is essential for
stable postnatal heart gene expression and homeostasis (150). Furthermore, Chen et al
demonstrated that a variety of cardiovascular structural
malformations were observed in the Ezh2 mutant mice, including
double outlet right ventricle, persistent truncus arteriosus,
membranous and muscular ventricular septal defects, atrial septal
defects, atrioventricular canal defects and enlarged aortic valves,
which defined an indispensible role of Ezh2 in normal
cardiovascular development (151).
Histone demethylase UTX, also known as KDM6A, that
specifically targets the repressive H3K27me3 modification plays an
important role in the activation of 'bivalent' genes in response to
specific developmental cues. Welstead et al showed that
UTX-deficient embryos had reduced somite counts, neural tube
closure defects and heart malformation that presented between E9.5
and E13.5 (152). Other studies
have also found that UTX-deficient ESCs failed to develop
heart-like rhythmic contractions under a cardiac differentiation
condition; UTX deficient mice exhibited severe defects in heart
development and embryonic lethality; these data establish that UTX
is required for heart development acts as a critical switch to
activate the cardiac developmental program (153,154).
Jmjd3 (KDM6B), another H3K27 demethylase, functions
redundantly with UTX. Jmjd3 is induced and participates in Hox gene
expression during development, neuronal differentiation and
inflammation, and recent data suggest that Jmjd3 inhibits
reprogramming by inducing cellular senescence (155). Jmjd3 deficient mice showed
embryonic lethality before E6.5, suggesting a crucial role of Jmjd3
in early embryonic development (156,157). The ablation of Jmjd3 in mouse
ESCs impaired mesoderm and subsequent endothelial and cardiac
differentiation. These results clarify that Jmjd3 is necessary for
mesoderm differentiation and cardiovascular lineage commitment
(158).
NSD1 is a structure containing the SET domain
proteins, with specific H3K36 and H4K20 methyltransferase activity,
which is associated with Sotos syndrome by haploinsufficiency
(159,160). A very frequent feature among
Sotos syndrome patients with intragenic mutations was the presence
of congenital heart defects or heart conduction defects, including
isolated atrial septal defect, atrial septal defect in association
with other structural abnormalities (patent ductus arteriosus;
aortic valve dysplasia; ventricular septal defect and aortic
coarctation) (161). This
evidence indicates that NSD1 may be a cause of a higher prevalence
of congenital heart defect in Sotos syndrome patients, which may be
a novel target of diagnosis and treatment strategy for Sotos
syndrome.
Jmjd5 is a histone demethylase that specifically
removes methyl moieties from dimethylated H3K36 and exerts a
pro-proliferative effect on a large of cells. Strong Jmjd5
expression was observed only in the yolk sac at E8.5, Jmjd5 was
robustly expressed in E10.5 embryos at several sites, including the
heart and eye, which indicated that Jmjd5 may play an important
role in heart and eye development. Jmjd5 deficiency mice embryos
showed delayed development already at E8.5, embryonic lethal around
E10 and were actively resorbed at E10.5 (164,165). Collectively, these data indicate
that Jmjd5 is essential during embryonal development including
heart development.
H3K79 methylation is related to gene activation and
DNA damage repair. Histone methylation occurs at H3K79 is catalyzed
by yeast disruptor of telomeric silencing (DOT1) and its mammalian
homolog, DOT1L. DOT1 is a kind of evolutionary highly conservative
histone methyltransferase, which does not contain the SET domain
structure, can be specific to different methylation levels in the
H3K79. Compared with other histone lysine methylation, in yeast,
DOT1 activity is positively regulated during transcription
elongation through Rad6-Bre1 mono-ubiquitination of H2B (166). Recently, loss-of-function
experiments revealed a critical role of DOT1L during mouse
embryogenesis, as germline Dot1L knockout caused lethality at E10.5
with growth impairment, yolk sac angiogenesis defects, and cardiac
dilation (167). In addition,
cardiac-specific knockout of DOT1L resulted in increased mortality
rate with chamber dilation, increased cardiomyocyte cell death,
systolic dysfunction and conduction abnormalities (168). These phenotypes mimic those
exhibited in patients with dilated cardiomyopathy. Interestingly,
Nguyen et al demonstrated that DOT1L is downregulated in
idiopathic DCM patient samples compared with normal controls
(168). Therefore, the above
studies not only establish a critical role for DOT1L-mediated H3K79
methylation in cardiomyocyte function, but also open new avenues
for the diagnosis and treatment of CHD.
H4K20 can be catalyzed to different forms of
monomethylation, dimethylation and trimethylation, PR-SET7 can only
single methylate H4K20, but double and triple methylation of H4K20
are catalyzed by two other methyltransferases SUV4-20h1 and
SUV4-20h2 (169). It is shown
that H4K20 methylation is related to transcription silence,
H4K20me3 plays a vital role in the regulation of DNA damage but not
directly regulates the expression of genes (170). In addition, Tatton-Brown and
Rahman have found that NSD1 is a protein containing the SET domain
structure, which with specific H4K20 and H3K36 methyltransferase
activity (171). NSD1 mutation
or deficiency is the main cause of Sotos syndrome which with a high
incidence of CHD characterized by ventricular septal defect, atrial
septal defects and patent ductus arteriosus (159,160).
Histone modification is the important content of
epigenetics, which is not only showed as directly regulating gene
expressions, but also influencing gene activity through DNA
modification because of its intimate touching with DNA. However,
single histone modification usually cannot come into effect
individually, and it determines together the gene expression of
genome through collaborative effect of multiple histone
modifications; but, yet so far, about the mechanism of histone
modification, especially the specific mechanism of regulation of
histone modification is not quite clear. Therefore, although the
histone lysine methylation modification is related to CHD, further
intensive research is still needed to illuminate relationship at
the level of molecule. At the same time, besides the genetic
factors of CHD pathogenesis, there still exist outer environmental
factors, so that the complexity of diagnosing and treatment of
diseases has been increased. However, it is clear that the
comprehensive and meticulous investigation of histone lysine
methylation modifications may provide new insight and understaning
into the exploration of CHD pathogenesis and targeted
prevention.
We apologize to the colleagues whose studies could
not be cited in this review due to space limitations. This study
was supported by grants from the Fundamental Research Funds for the
Central Universities (no. 2042016kf0074), the National Natural
Science Foundation of China (no. 81170307) and Specialized Research
Fund for the Doctoral Program of Higher Education (no.
20120141110013).
|
1
|
Campos CM, Zanardo EA, Dutra RL,
Kulikowski LD and Kim CA: Investigation of copy number variation in
children with conotruncal heart defects. Arq Bras Cardiol.
104:24–31. 2015.
|
|
2
|
Barros TL, Dias MdeJ and Nina RV:
Congenital cardiac disease in childhood x socioeconomic conditions:
A relationship to be considered in public health? Rev Bras Cir
Cardiovasc. 29:448–454. 2014.PubMed/NCBI
|
|
3
|
Steffensen TS and Spicer DE: Congenital
coronary artery anomalies for the pathologist. Fetal Pediatr
Pathol. 33:268–288. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanchez-Castro M, Pichon O, Briand A,
Poulain D, Gournay V, David A and Le Caignec C: Disruption of the
SEMA3D gene in a patient with congenital heart defects. Hum Mutat.
36:30–33. 2015. View Article : Google Scholar
|
|
5
|
Vecoli C, Pulignani S, Foffa I and
Andreassi MG: Congenital heart disease: The crossroads of genetics,
epigenetics and environment. Curr Genomics. 15:390–399. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gittenberger-de Groot AC, Calkoen EE,
Poelmann RE, Bartelings MM and Jongbloed MR: Morphogenesis and
molecular considerations on congenital cardiac septal defects. Ann
Med. 46:640–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Li P, Chen S, Xi L, Guo Y, Guo A
and Sun K: Influence of genes and the environment in familial
congenital heart defects. Mol Med Rep. 9:695–700. 2014. View Article : Google Scholar
|
|
8
|
Lalani SR and Belmont JW: Genetic basis of
congenital cardiovascular malformations. Eur J Med Genet.
57:402–413. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kathiriya IS, Nora EP and Bruneau BG:
Investigating the transcriptional control of cardiovascular
development. Circ Res. 116:700–714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meganathan K, Sotiriadou I, Natarajan K,
Hescheler J and Sachinidis A: Signaling molecules, transcription
growth factors and other regulators revealed from in-vivo and
in-vitro models for the regulation of cardiac development. Int J
Cardiol. 183:117–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ,
Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, et al: Global
genetic analysis in mice unveils central role for cilia in
congenital heart disease. Nature. 521:520–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andersen TA, Troelsen KL and Larsen LA: Of
mice and men: Molecular genetics of congenital heart disease. Cell
Mol Life Sci. 71:1327–1352. 2014. View Article : Google Scholar :
|
|
13
|
Yuan S, Zaidi S and Brueckner M:
Congenital heart disease: Emerging themes linking genetics and
development. Curr Opin Genet Dev. 23:352–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Serra-Juhé C, Cuscó I, Homs A, Flores R,
Torán N and Pérez-Jurado LA: DNA methylation abnormalities in
congenital heart disease. Epigenetics. 10:167–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang QJ and Liu ZP: Histone methylations
in heart development, congenital and adult heart diseases.
Epigenomics. 7:321–330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang CP and Bruneau BG: Epigenetics and
cardiovascular development. Annu Rev Physiol. 74:41–68. 2012.
View Article : Google Scholar
|
|
17
|
D'Urso A and Brickner JH: Mechanisms of
epigenetic memory. Trends Genet. 30:230–236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Becker PB and Workman JL: Nucleosome
remodeling and epigenetics. Cold Spring Harb Perspect Biol.
5:a0179052013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnakumar R and Blelloch RH:
Epigenetics of cellular reprogramming. Curr Opin Genet Dev.
23:548–555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singmann P, Shem-Tov D, Wahl S, Grallert
H, Fiorito G, Shin SY, Schramm K, Wolf P, Kunze S, Baran Y, et al:
Characterization of whole-genome autosomal differences of DNA
methylation between men and women. Epigenetics Chromatin. 8:432015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ziegler-Birling C, Daujat S, Schneider R
and Torres-Padilla ME: Dynamics of histone H3 acetylation in the
nucleosome core during mouse pre-implantation development.
Epigenetics. 11:553–562. 2016. View Article : Google Scholar :
|
|
22
|
Dudakovic A, Camilleri ET, Xu F, Riester
SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler
R, Deyle DR, et al: Epigenetic control of skeletal development by
the histone methyltransferase Ezh2. J Biol Chem. 290:27604–27617.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang F, Ramakrishnan S, Pokhrel S,
Pflueger C, Parnell TJ, Kasten MM, Currie SL, Bhachech N, Horikoshi
M, Graves BJ, et al: Interaction of the Jhd2 H3K4 demethylase with
chromatin is controlled by histone H2A surfaces and restricted by
H2B ubiquitination. J Biol Chem. 290:28760–28777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Casas-Mollano JA, Xu J, Riethoven
JJ, Zhang C and Cerutti H: Osmotic stress induces phosphorylation
of histone H3 at threonine 3 in pericentromeric regions of
Arabidopsis thaliana. Proc Natl Acad Sci USA. 112:8487–8492. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao Y, Des Marais TL and Costa M:
Chromatin memory in the development of human cancers. Gene Technol.
3:1142014.
|
|
26
|
Heo JB and Sung S: Encoding memory of
winter by noncoding RNAs. Epigenetics. 6:544–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Torres IO and Fujimori DG: Functional
coupling between writers, erasers and readers of histone and DNA
methylation. Curr Opin Struct Biol. 35:68–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jaworska J, Ziemka-Nalecz M and Zalewska
T: Histone deacetylases 1 and 2 are required for brain development.
Int J Dev Biol. 59:171–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carr SM, Poppy Roworth A, Chan C and La
Thangue NB: Post-translational control of transcription factors:
Methylation ranks highly. FEBS J. 282:4450–4465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gupta N, Madapura MP, Bhat UA and Rao MR:
Mapping of post-translational modifications of transition proteins,
TP1 and TP2, and identification of protein arginine
methyltransferase 4 and lysine methyltransferase 7 as
methyltransferase for TP2. J Biol Chem. 290:12101–12122. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Binda O: On your histone mark, SET,
methylate! Epigenetics. 8:457–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lesne A, Foray N, Cathala G, Forné T, Wong
H and Victor JM: Chromatin fiber allostery and the epigenetic code.
J Phys Condens Matter. 27:0641142015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zuchegna C, Aceto F, Bertoni A, Romano A,
Perillo B, Laccetti P, Gottesman ME, Avvedimento EV and Porcellini
A: Mechanism of retinoic acid-induced transcription: Histone code,
DNA oxidation and formation of chromatin loops. Nucleic Acids Res.
42:11040–11055. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gayatri S and Bedford MT: Readers of
histone methylarginine marks. Biochim Biophys Acta. 1839:702–710.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Casciello F, Windloch K, Gannon F and Lee
JS: Functional role of G9a histone methyltransferase in cancer.
Front Immunol. 6:4872015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ortega-Molina A, Boss IW, Canela A, Pan H,
Jiang Y, Zhao C, Jiang M, Hu D, Agirre X, Niesvizky I, et al: The
histone lysine methyltransferase KMT2D sustains a gene expression
program that represses B cell lymphoma development. Nat Med.
21:1199–1208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cho MH, Park JH, Choi HJ, Park MK, Won HY,
Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al: DOT1L cooperates
with the c-Myc-p300 complex to epigenetically derepress CDH1
transcription factors in breast cancer progression. Nat Commun.
6:78212015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu N, Zhang Z, Wu H, Jiang Y, Meng L,
Xiong J, Zhao Z, Zhou X, Li J, Li H, et al: Recognition of H3K9
methylation by GLP is required for efficient establishment of H3K9
methylation, rapid target gene repression, and mouse viability.
Genes Dev. 29:379–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nicetto D, Hahn M, Jung J, Schneider TD,
Straub T, David R, Schotta G and Rupp RA: Suv4-20h histone
methyltransferases promote neuroectodermal differentiation by
silencing the pluripotency-associated Oct-25 gene. PLoS Genet.
9:e10031882013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Maes T, Mascaró C, Ortega A, Lunardi S,
Ciceri F, Somervaille TC and Buesa C: KDM1 histone lysine
demethylases as targets for treatments of oncological and
neurodegenerative disease. Epigenomics. 7:609–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Belakavadi M, Dell J, Grover GJ and
Fondell JD: Thyroid hormone suppression of β-amyloid precursor
protein gene expression in the brain involves multiple epigenetic
regulatory events. Mol Cell Endocrinol. 339:72–80. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Stein AB, Goonewardena SN, Jones TA,
Prusick PJ, Bazzi AA, Belyavskaya JM, McCoskey MM and Dandar RA:
The PTIP-associated histone methyltransferase complex prevents
stress-induced maladaptive cardiac remodeling. PLoS One.
10:e01278392015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mathiyalagan P, Keating ST, Du XJ and
El-Osta A: Chromatin modifications remodel cardiac gene expression.
Cardiovasc Res. 103:7–16. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hohl M, Wagner M, Reil JC, Müller SA,
Tauchnitz M, Zimmer AM, Lehmann LH, Thiel G, Böhm M, Backs J, et
al: HDAC4 controls histone methylation in response to elevated
cardiac load. J Clin Invest. 123:1359–1370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bingham AJ, Ooi L, Kozera L, White E and
Wood IC: The repressor element 1-silencing transcription factor
regulates heart-specific gene expression using multiple
chromatin-modifying complexes. Mol Cell Biol. 27:4082–4092. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cabrera JR, Olcese U and Horabin JI: A
balancing act: Heterochromatin protein 1a and the polycomb group
coordinate their levels to silence chromatin in Drosophila.
Epigenetics Chromatin. 8:172015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sautel CF, Cannella D, Bastien O, Kieffer
S, Aldebert D, Garin J, Tardieux I, Belrhali H and Hakimi MA:
SET8-mediated methylations of histone H4 lysine 20 mark silent
heterochromatic domains in apicomplexan genomes. Mol Cell Biol.
27:5711–5724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo X, Wang L, Li J, Ding Z, Xiao J, Yin
X, He S, Shi P, Dong L, Li G, et al: Structural insight into
autoinhibition and histone H3-induced activation of DNMT3A. Nature.
517:640–644. 2015. View Article : Google Scholar
|
|
49
|
Minkovsky A, Sahakyan A, Rankin-Gee E,
Bonora G, Patel S and Plath K: The Mbd1-Atf7ip-Setdb1 pathway
contributes to the maintenance of X chromosome inactivation.
Epigenetics Chromatin. 7:122014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Simon MD, Pinter SF, Fang R, Sarma K,
Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE
and Lee JT: High-resolution Xist binding maps reveal two-step
spreading during X-chromosome inactivation. Nature. 504:465–469.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J, Telese F, Tan Y, Li W, Jin C, He
X, Basnet H, Ma Q, Merkurjev D, Zhu X, et al: LSD1n is an H4K20
demethylase regulating memory formation via transcriptional
elongation control. Nat Neurosci. 18:1256–1264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sen P, Dang W, Donahue G, Dai J, Dorsey J,
Cao X, Liu W, Cao K, Perry R, Lee JY, et al: H3K36 methylation
promotes longevity by enhancing transcriptional fidelity. Genes
Dev. 29:1362–1376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kang SC, Kim SK, Chai JC, Kim SH, Won KJ,
Lee YS, Jung KH and Chai YG: Transcriptomic Profiling and H3K27me3
Distribution reveal both demethylase-dependent and independent
regulation of developmental gene transcription in cell
differentiation. PLoS One. 10:e01352762015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Copur Ö and Müller J: The histone H3-K27
demethylase Utx regulates HOX gene expression in Drosophila in a
temporally restricted manner. Development. 140:3478–3485. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
You L, Nie J, Sun WJ, Zheng ZQ and Yang
XJ: Lysine acetylation: Enzymes, bromodomains and links to
different diseases. Essays Biochem. 52:1–12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Migliori V, Phalke S, Bezzi M and Guccione
E: Arginine/lysinemethyl/methyl switches: Biochemical role of
histone arginine methylation in transcriptional regulation.
Epigenomics. 2:119–137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Davie JK and Dent SY: Transcriptional
control: An activating role for arginine methylation. Curr Biol.
12:R59–R61. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tessarz P and Kouzarides T: Histone core
modifications regulating nucleosome structure and dynamics. Nat Rev
Mol Cell Biol. 15:703–708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Strahl BD, Briggs SD, Brame CJ, Caldwell
JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et
al: Methylation of histone H4 at arginine 3 occurs in vivo and is
mediated by the nuclear receptor coactivator PRMT1. Curr Biol.
11:996–1000. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang H, Huang ZQ, Xia L, Feng Q,
Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst
P, et al: Methylation of histone H4 at arginine 3 facilitating
transcriptional activation by nuclear hormone receptor. Science.
293:853–857. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Schurter BT, Koh SS, Chen D, Bunick GJ,
Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR and
Aswad DW: Methylation of histone H3 by coactivator-associated
arginine methyltransferase 1. Biochemistry. 40:5747–5756. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Aoshima K, Inoue E, Sawa H and Okada Y:
Paternal H3K4 methylation is required for minor zygotic gene
activation and early mouse embryonic development. EMBO Rep.
16:803–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shinsky SA, Monteith KE, Viggiano S and
Cosgrove MS: Biochemical reconstitution and phylogenetic comparison
of human SET1 family core complexes involved in histone
methylation. J Biol Chem. 290:6361–6375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen CW, Koche RP, Sinha AU, Deshpande AJ,
Zhu N, Eng R, Doench JG, Xu H, Chu SH, Qi J, et al: DOT1L inhibits
SIRT1-mediated epigenetic silencing to maintain leukemic gene
expression in MLL-rearranged leukemia. Nat Med. 21:335–343. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiao L and Liu X: Structural basis of
histone H3K27 trimethylation by an active polycomb repressive
complex 2. Science. 350:aac43832015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Foda BM and Singh U: Dimethylated H3K27 is
a repressive epigenetic histone mark in the protist Entamoeba
histolytica and is significantly enriched in genes silenced via the
RNAi pathway. J Biol Chem. 290:21114–21130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Vieira FQ, Costa-Pinheiro P, Almeida-Rios
D, Graça I, Monteiro-Reis S, Simões-Sousa S, Carneiro I, Sousa EJ,
Godinho MI, Baltazar F, et al: SMYD3 contributes to a more
aggressive phenotype of prostate cancer and targets Cyclin D2
through H4K20me3. Oncotarget. 6:13644–13657. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bierhoff H, Dammert MA, Brocks D,
Dambacher S, Schotta G and Grummt I: Quiescence-induced LncRNAs
trigger H4K20 trimethylation and transcriptional silencing. Mol
Cell. 54:675–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang X, Tanaka K, Yan J, Li J, Peng D,
Jiang Y, Yang Z, Barton MC, Wen H and Shi X: Regulation of estrogen
receptor α by histone methyltransferase SMYD2-mediated protein
methylation. Proc Natl Acad Sci USA. 110:17284–17289. 2013.
View Article : Google Scholar
|
|
70
|
Hossain MA, Chung C, Pradhan SK and
Johnson TL: The yeast cap binding complex modulates transcription
factor recruitment and establishes proper histone H3K36
trimethylation during active transcription. Mol Cell Biol.
33:785–799. 2013. View Article : Google Scholar :
|
|
71
|
Ontoso D, Acosta I, van Leeuwen F, Freire
R and San-Segundo PA: Dot1-dependent histone H3K79 methylation
promotes activation of the Mek1 meiotic checkpoint effector kinase
by regulating the Hop1 adaptor. PLoS Genet. 9:e10032622013.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kim SK, Jung I, Lee H, Kang K, Kim M,
Jeong K, Kwon CS, Han YM, Kim YS, Kim D, et al: Human histone H3K79
methyltransferase DOT1L protein [corrected] binds actively
transcribing RNA polymerase II to regulate gene expression. J Biol
Chem. 287:39698–39709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao XX, Zhang YB, Ni PL, Wu ZL, Yan YC
and Li YP: Protein arginine methyltransferase 6 (Prmt6) is
essential for early zebrafish development through the direct
suppression of Gadd45alphaa stress sensor gene. J Biol Chem.
291:402–412. 2016. View Article : Google Scholar
|
|
74
|
Feng Y, Hadjikyriacou A and Clarke SG:
Substrate specificity of human protein arginine methyltransferase 7
(PRMT7): The importance of acidic residues in the double E loop. J
Biol Chem. 289:32604–32616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B,
Wu W and Li H: Molecular basis underlying histone H3
lysine-arginine methylation pattern readout by Spin/Ssty repeats of
Spindlin1. Genes Dev. 28:622–636. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Nguyen HC, Wang M, Salsburg A and Knuckley
B: Development of a plate-based screening assay to investigate the
dubstrate dpecificity of the PRMT gamily of rnzymes. ACS Comb Sci.
17:500–505. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tini M, Naeem H and Torchia J: Biochemical
analysis of arginine methylation in transcription. Methods Mol
Biol. 523:235–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Del Rizzo PA and Trievel RC: Substrate and
product specificities of SET domain methyltransferases.
Epigenetics. 6:1059–1067. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Couture JF and Trievel RC:
Histone-modifying enzymes: Encrypting an enigmatic epigenetic code.
Curr Opin Struct Biol. 16:753–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Spellmon N, Holcomb J, Trescott L,
Sirinupong N and Yang Z: Structure and function of SET and MYND
domain-containing proteins. Int J Mol Sci. 16:1406–1428. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Anand R and Marmorstein R: Structure and
mechanism of lysine-specific demethylase enzymes. J Biol Chem.
282:35425–35429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Tsukada Y and Zhang Y: Purification of
histone demethylases from HeLa cells. Methods. 40:318–326. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Liu L, Jin G and Zhou X: Modeling the
relationship of epigenetic modifications to transcription factor
binding. Nucleic Acids Res. 43:3873–3885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Bai H, Li Y, Gao H, Dong Y, Han P and Yu
H: Histone methyltransferase SMYD3 regulates the expression of
transcriptional factors during bovine oocyte maturation and early
embryonic development. Cytotechnology. 68:849–859. 2016. View Article : Google Scholar :
|
|
85
|
Kim JD, Kim E, Koun S, Ham HJ, Rhee M, Kim
MJ and Huh TL: Proper activity of histone H3 lysine 4 (H3K4)
methyltransferase is required for morphogenesis during zebrafish
cardiogenesis. Mol Cells. 38:580–586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Martinez SR, Gay MS and Zhang L:
Epigenetic mechanisms in heart development and disease. Drug Discov
Today. 20:799–811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Dorn GW II and Matkovich SJ:
Epitranscriptional regulation of cardiovascular development and
disease. J Physiol. 593:1799–1808. 2015. View Article : Google Scholar :
|
|
88
|
Zhao W, Liu L, Pan B, Xu Y, Zhu J, Nan C,
Huang X and Tian J: Epigenetic regulation of cardiac myofibril gene
expression during heart development. Cardiovasc Toxicol.
15:203–209. 2015. View Article : Google Scholar
|
|
89
|
Park CY, Pierce SA, von Drehle M, Ivey KN,
Morgan JA, Blau HM and Srivastava D: skNAC, a Smyd1-interacting
transcription factor, is involved in cardiac development and
skeletal muscle growth and regeneration. Proc Natl Acad Sci USA.
107:20750–20755. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Drake KM, Comhair SA, Erzurum SC, Tuder RM
and Aldred MA: Endothelial chromosome 13 deletion in congenital
heart disease-associated pulmonary arterial hypertension
dysregulates SMAD9 signaling. Am J Respir Crit Care Med.
191:850–854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Geng J, Picker J, Zheng Z, Zhang X, Wang
J, Hisama F, Brown DW, Mullen MP, Harris D, Stoler J, et al:
Chromosome microarray testing for patients with congenital heart
defects reveals novel disease causing loci and high diagnostic
yield. BMC Genomics. 15:11272014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zaidi S, Choi M, Wakimoto H, Ma L, Jiang
J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown
KK, et al: De novo mutations in histone-modifying genes in
congenital heart disease. Nature. 498:220–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ozanne SE and Constância M: Mechanisms of
disease: The developmental origins of disease and the role of the
epigenotype. Nat Clin Pract Endocrinol Metab. 3:539–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang QT: Epigenetic regulation of cardiac
development and function by polycomb group and trithorax group
proteins. Dev Dyn. 241:1021–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Geisler SJ and Paro R: Trithorax and
Polycomb group-dependent regulation: A tale of opposing activities.
Development. 142:2876–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wan X, Liu L, Ding X, Zhou P, Yuan X, Zhou
Z, Hu P, Zhou H, Li Q, Zhang S, et al: Mll2 controls cardiac
lineage differentiation of mouse embryonic stem cells by promoting
H3K4me3 deposition at cardiac-specific genes. Stem Cell Rev.
10:643–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Tan X, Rotllant J, Li H, De Deyne P and Du
SJ: SmyD1, a histone methyltransferase, is required for myofibril
organization and muscle contraction in zebrafish embryos. Proc Natl
Acad Sci USA. 103:2713–2718. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Du SJ, Tan X and Zhang J: SMYD proteins:
Key regulators in skeletal and cardiac muscle development and
function. Anat Rec (Hoboken). 297:1650–1662. 2014. View Article : Google Scholar
|
|
99
|
Rasmussen TL, Ma Y, Park CY, Harriss J,
Pierce SA, Dekker JD, Valenzuela N, Srivastava D, Schwartz RJ,
Stewart MD, et al: Smyd1 facilitates heart development by
antagonizing oxidative and ER stress responses. PLoS One.
10:e01217652015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Diehl F, Brown MA, van Amerongen MJ,
Novoyatleva T, Wietelmann A, Harriss J, Ferrazzi F, Böttger T,
Harvey RP, Tucker PW, et al: Cardiac deletion of Smyd2 is
dispensable for mouse heart development. PLoS One. 5:e97482010.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fujii T, Tsunesumi S, Yamaguchi K,
Watanabe S and Furukawa Y: Smyd3 is required for the development of
cardiac and skeletal muscle in zebrafish. PLoS One. 6:e234912011.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Papaioannou VE: The T-box gene family:
Emerging roles in development, stem cells and cancer. Development.
141:3819–3833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Greulich F, Rudat C and Kispert A:
Mechanisms of T-box gene function in the developing heart.
Cardiovasc Res. 91:212–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chen L, Fulcoli FG, Ferrentino R,
Martucciello S, Illingworth EA and Baldini A: Transcriptional
control in cardiac progenitors: Tbx1 interacts with the BAF
chromatin remodeling complex and regulates Wnt5a. PLoS Genet.
8:e10025712012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Caprio C and Baldini A: p53 Suppression
partially rescues the mutant phenotype in mouse models of DiGeorge
syndrome. Proc Natl Acad Sci USA. 111:13385–13390. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Pan Y, Wang ZG, Liu XY, Zhao H, Zhou N,
Zheng GF, Qiu XB, Li RG, Yuan F, Shi HY, et al: A novel TBX1
loss-of-function mutation associated with congenital heart disease.
Pediatr Cardiol. 36:1400–1410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Sedletcaia A and Evans T: Heart chamber
size in zebrafish is regulated redundantly by duplicated tbx2
genes. Dev Dyn. 240:1548–1557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pang S, Liu Y, Zhao Z, Huang W, Chen D and
Yan B: Novel and functional sequence variants within the TBX2 gene
promoter in ventricular septal defects. Biochimie. 95:1807–1809.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
van Weerd JH, Badi I, van den Boogaard M,
Stefanovic S, van de Werken HJ, Gomez-Velazquez M, Badia-Careaga C,
Manzanares M, de Laat W, Barnett P, et al: A large permissive
regulatory domain exclusively controls Tbx3 expression in the
cardiac conduction system. Circ Res. 115:432–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hatcher CJ, Diman NY, Kim MS, Pennisi D,
Song Y, Goldstein MM, Mikawa T and Basson CT: A role for Tbx5 in
proepicardial cell migration during cardiogenesis. Physiol
Genomics. 18:129–140. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Diman NY, Brooks G, Kruithof BP, Elemento
O, Seidman JG, Seidman CE, Basson CT and Hatcher CJ: Tbx5 is
required for avian and Mammalian epicardial formation and coronary
vasculogenesis. Circ Res. 115:834–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhou L, Liu J, Olson P, Zhang K, Wynne J
and Xie L: Tbx5 and Osr1 interact to regulate posterior second
heart field cell cycle progression for cardiac septation. J Mol
Cell Cardiol. 85:1–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Al-Qattan MM and Abou Al-Shaar H:
Molecular basis of the clinical features of Holt-Oram syndrome
resulting from missense and extended protein mutations of the TBX5
gene as well as TBX5 intragenic duplications. Gene. 560:129–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Kimura M, Kikuchi A, Ichinoi N and Kure S:
Novel TBX5 duplication in a Japanese family with Holt-Oram
syndrome. Pediatr Cardiol. 36:244–247. 2015. View Article : Google Scholar
|
|
115
|
Wu SP, Dong XR, Regan JN, Su C and Majesky
MW: Tbx18 regulates development of the epicardium and coronary
vessels. Dev Biol. 383:307–320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Cai X, Zhang W, Hu J, Zhang L, Sultana N,
Wu B, Cai W, Zhou B and Cai CL: Tbx20 acts upstream of Wnt
signaling to regulate endocardial cushion formation and valve
remodeling during mouse cardiogenesis. Development. 140:3176–3187.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Shen T, Aneas I, Sakabe N, Dirschinger RJ,
Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al: Tbx20
regulates a genetic program essential to adult mouse cardiomyocyte
function. J Clin Invest. 121:4640–4654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang W, Chen H, Wang Y, Yong W, Zhu W,
Liu Y, Wagner GR, Payne RM, Field LJ, Xin H, et al: Tbx20
transcription factor is a downstream mediator for bone
morphogenetic protein-10 in regulating cardiac ventricular wall
development and function. J Biol Chem. 286:36820–36829. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Kirk EP, Sunde M, Costa MW, Rankin SA,
Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, et al:
Mutations in cardiac T-box factor gene TBX20 are associated with
diverse cardiac pathologies, including defects of septation and
valvulogenesis and cardiomyopathy. Am J Hum Genet. 81:280–291.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ishizaka A, Mizutani T, Kobayashi K, Tando
T, Sakurai K, Fujiwara T and Iba H: Double plant homeodomain (PHD)
finger proteins DPF3a and -3b are required as transcriptional
co-activators in SWI/SNF complex-dependent activation of NF-κB
RelA/p50 heterodimer. J Biol Chem. 287:11924–11933. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lange M, Kaynak B, Forster UB, Tönjes M,
Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, et
al: Regulation of muscle development by DPF3, a novel histone
acetylation and methylation reader of the BAF chromatin remodeling
complex. Genes Dev. 22:2370–2384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liu Y, Huang Y, Fan J and Zhu GZ: PITX2
associates with PTIP-containing histone H3 lysine 4
methyltransferase complex. Biochem Biophys Res Commun. 444:634–637.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kim D, Patel SR, Xiao H and Dressler GR:
The role of PTIP in maintaining embryonic stem cell pluripotency.
Stem Cells. 27:1516–1523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Fang M, Ren H, Liu J, Cadigan KM, Patel SR
and Dressler GR: Drosophila ptip is essential for
anterior/posterior patterning in development and interacts with the
PcG and trxG pathways. Development. 136:1929–1938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Callen E, Faryabi RB, Luckey M, Hao B,
Daniel JA, Yang W, Sun HW, Dressler G, Peng W, Chi H, et al: The
DNA damage- and transcription-associated protein paxip1 controls
thymocyte development and emigration. Immunity. 37:971–985. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Daniel JA, Santos MA, Wang Z, Zang C,
Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, et
al: PTIP promotes chromatin changes critical for immunoglobulin
class switch recombination. Science. 329:917–923. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Stein AB, Jones TA, Herron TJ, Patel SR,
Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, et
al: Loss of H3K4 methylation destabilizes gene expression patterns
and physiological functions in adult murine cardiomyocytes. J Clin
Invest. 121:2641–2650. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Tao Y, Neppl RL, Huang ZP, Chen J, Tang
RH, Cao R, Zhang Y, Jin SW and Wang DZ: The histone
methyltransferase Set7/9 promotes myoblast differentiation and
myofibril assembly. J Cell Biol. 194:551–565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Nicholson TB and Chen T: LSD1 demethylates
histone and non-histone proteins. Epigenetics. 4:129–132. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
He Y, Tang D, Cai C, Chai R and Li H: LSD1
is required for hair cell regeneration in zebrafish. Mol Neurobiol.
53:2421–2434. 2016. View Article : Google Scholar
|
|
131
|
van Riel B, Pakozdi T, Brouwer R, Monteiro
R, Tuladhar K, Franke V, Bryne JC, Jorna R, Rijkers EJ, van Ijcken
W, et al: A novel complex, RUNX1-MYEF2, represses hematopoietic
genes in erythroid cells. Mol Cell Biol. 32:3814–3822. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Nicholson TB, Singh AK, Su H, Hevi S, Wang
J, Bajko J, Li M, Valdez R, Goetschkes M, Capodieci P, et al: A
hypomorphic lsd1 allele results in heart development defects in
mice. PLoS One. 8:e609132013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan
G, Shi S and Wang CY: BCOR regulates mesenchymal stem cell function
by epigenetic mechanisms. Nat Cell Biol. 11:1002–1009. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Hilton E, Johnston J, Whalen S, Okamoto N,
Hatsukawa Y, Nishio J, Kohara H, Hirano Y, Mizuno S, Torii C, et
al: BCOR analysis in patients with OFCD and Lenz microphthalmia
syndromes, mental retardation with ocular anomalies, and cardiac
laterality defects. Eur J Hum Genet. 17:1325–1335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Di Stefano C, Lombardo B, Fabbricatore C,
Munno C, Caliendo I, Gallo F and Pastore L:
Oculo-facio-cardio-dental (OFCD) syndrome: The first Italian case
of BCOR and co-occurring OTC gene deletion. Gene. 559:203–206.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Huang XJ, Ma X, Wang X, Zhou X, Li J, Sun
SC and Liu H: Involvement of G9A-like protein (GLP) in the
development of mouse preimplantation embryos in vitro. Reprod
Fertil Dev. May 18–2015.Epub ahead of print. View Article : Google Scholar
|
|
137
|
Tachibana M, Ueda J, Fukuda M, Takeda N,
Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T and Shinkai Y:
Histone methyltransferases G9a and GLP form heteromeric complexes
and are both crucial for methylation of euchromatin at H3-K9. Genes
Dev. 19:815–826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Inagawa M, Nakajima K, Makino T, Ogawa S,
Kojima M, Ito S, Ikenishi A, Hayashi T, Schwartz RJ, Nakamura K, et
al: Histone H3 lysine 9 methyltransferases, G9a and GLP are
essential for cardiac morphogenesis. Mech Dev. 130:519–531. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Kleefstra T, van Zelst-Stams WA, Nillesen
WM, Cormier-Daire V, Houge G, Foulds N, van Dooren M, Willemsen MH,
Pfundt R, Turner A, et al: Further clinical and molecular
delineation of the 9q subtelomeric deletion syndrome supports a
major contribution of EHMT1 haploinsufficiency to the core
phenotype. J Med Genet. 46:598–606. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Stewart DR and Kleefstra T: The chromosome
9q subtelomere deletion syndrome. Am J Med Genet C Semin Med Genet.
145C:383–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Bikoff EK, Morgan MA and Robertson EJ: An
expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev.
19:379–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Morgan MA, Mould AW, Li L, Robertson EJ
and Bikoff EK: Alternative splicing regulates Prdm1/Blimp-1 DNA
binding activities and corepressor interactions. Mol Cell Biol.
32:3403–3413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Lee Y, Song AJ, Baker R, Micales B, Conway
SJ and Lyons GE: Jumonji, a nuclear protein that is necessary for
normal heart development. Circ Res. 86:932–938. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Kim TG, Chen J, Sadoshima J and Lee Y:
Jumonji represses atrial natriuretic factor gene expression by
inhibiting transcriptional activities of cardiac transcription
factors. Mol Cell Biol. 24:10151–10160. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Mysliwiec MR, Carlson CD, Tietjen J, Hung
H, Ansari AZ and Lee Y: Jarid2 (Jumonji, AT rich interactive domain
2) regulates NOTCH1 expression via histone modification in the
developing heart. J Biol Chem. 287:1235–1241. 2012. View Article : Google Scholar :
|
|
146
|
Mysliwiec MR, Bresnick EH and Lee Y:
Endothelial Jarid2/Jumonji is required for normal cardiac
development and proper Notch1 expression. J Biol Chem.
286:17193–17204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Dobreva G and Braun T: When silence is
broken: Polycomb group proteins in heart development. Circ Res.
110:372–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Weston AD, Ozolins TR and Brown NA:
Thoracic skeletal defects and cardiac malformations: A common
epigenetic link? Birth Defects Res C Embryo Today. 78:354–370.
2006. View Article : Google Scholar
|
|
149
|
Sanulli S, Justin N, Teissandier A,
Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST,
Offer J, et al: Jarid2 methylation via the PRC2 complex regulates
H3K27me3 deposition during cell differentiation. Mol Cell.
57:769–783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Delgado-Olguín P, Huang Y, Li X,
Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A and Bruneau
BG: Epigenetic repression of cardiac progenitor gene expression by
Ezh2 is required for postnatal cardiac homeostasis. Nat Genet.
44:343–347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Chen L, Ma Y, Kim EY, Yu W, Schwartz RJ,
Qian L and Wang J: Conditional ablation of Ezh2 in murine hearts
reveals its essential roles in endocardial cushion formation,
cardiomyocyte proliferation and survival. PLoS One. 7:e310052012.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Welstead GG, Creyghton MP, Bilodeau S,
Cheng AW, Markoulaki S, Young RA and Jaenisch R: X-linked H3K27me3
demethylase Utx is required for embryonic development in a
sex-specific manner. Proc Natl Acad Sci USA. 109:13004–13009. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Morales Torres C, Laugesen A and Helin K:
Utx is required for proper induction of ectoderm and mesoderm
during differentiation of embryonic stem cells. PLoS One.
8:e600202013. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Lee S, Lee JW and Lee SK: UTX, a histone
H3-lysine 27 demethylase, acts as a critical switch to activate the
cardiac developmental program. Dev Cell. 22:25–37. 2012. View Article : Google Scholar
|
|
155
|
Agger K, Cloos PA, Christensen J, Pasini
D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE and Helin
K: UTX and JMJD3 are histone H3K27 demethylases involved in HOX
gene regulation and development. Nature. 449:731–734. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Li Q, Wang HY, Chepelev I, Zhu Q, Wei G,
Zhao K and Wang RF: Stage-dependent and locus-specific role of
histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of
lung development. PLoS Genet. 10:e10045242014. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Burgold T, Voituron N, Caganova M,
Tripathi PP, Menuet C, Tusi BK, Spreafico F, Bévengut M, Gestreau
C, Buontempo S, et al: The H3K27 demethylase JMJD3 is required for
maintenance of the embryonic respiratory neuronal network, neonatal
breathing, and survival. Cell Rep. 2:1244–1258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Ohtani K, Zhao C, Dobreva G, Manavski Y,
Kluge B, Braun T, Rieger MA, Zeiher AM and Dimmeler S: Jmjd3
controls mesodermal and cardiovascular differentiation of embryonic
stem cells. Circ Res. 113:856–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Chen CP, Lin CJ, Chern SR, Liu YP, Kuo YL,
Chen YN, Wu PS, Town DD, Chen LF, Yang CW, et al: Prenatal
diagnosis and molecular cytogenetic characterization of a 1.07-Mb
microdeletion at 5q35.2-q35.3 associated with NSD1
haploinsufficiency and Sotos syndrome. Taiwan J Obstet Gynecol.
53:583–587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Park SH, Lee JE, Sohn YB and Ko JM: First
identified Korean family with Sotos syndrome caused by a novel
intragenic mutation in NSD1. Ann Clin Lab Sci. 44:228–231.
2014.PubMed/NCBI
|
|
161
|
Sheth K, Moss J, Hyland S, Stinton C, Cole
T and Oliver C: The behavioral characteristics of Sotos syndrome.
Am J Med Genet A. 167A:2945–2956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Vallaster M, Vallaster CD and Wu SM:
Epigenetic mechanisms in cardiac development and disease. Acta
Biochim Biophys Sin (Shanghai). 44:92–102. 2012. View Article : Google Scholar
|
|
163
|
Nimura K, Ura K, Shiratori H, Ikawa M,
Okabe M, Schwartz RJ and Kaneda Y: A histone H3 lysine 36
trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome.
Nature. 460:287–291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Oh S and Janknecht R: Histone demethylase
JMJD5 is essential for embryonic development. Biochem Biophys Res
Commun. 420:61–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Kawakami E, Tokunaga A, Ozawa M, Sakamoto
R and Yoshida N: The histone demethylase Fbxl11/Kdm2a plays an
essential role in embryonic development by repressing cell-cycle
regulators. Mech Dev. 135:31–42. 2015. View Article : Google Scholar
|
|
166
|
Mohan M, Herz HM, Takahashi YH, Lin C, Lai
KC, Zhang Y, Washburn MP, Florens L and Shilatifard A: Linking
H3K79 trimethylation to Wnt signaling through a novel
Dot1-containing complex (DotCom). Genes Dev. 24:574–589. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Jones B, Su H, Bhat A, Lei H, Bajko J,
Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al: The histone
H3K79 methyltransferase Dot1L is essential for mammalian
development and heterochromatin structure. PLoS Genet.
4:e10001902008. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li
J, Chen T, Wang DZ, Xiao X and Zhang Y: DOT1L regulates dystrophin
expression and is critical for cardiac function. Genes Dev.
25:263–274. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Beck DB, Burton A, Oda H, Ziegler-Birling
C, Torres-Padilla ME and Reinberg D: The role of PR-Set7 in
replication licensing depends on Suv4-20h. Genes Dev. 26:2580–2589.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Lennartsson A and Ekwall K: Histone
modification patterns and epigenetic codes. Biochim Biophys Acta.
1790:863–868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Tatton-Brown K and Rahman N: Clinical
features of NSD1-positive Sotos syndrome. Clin Dysmorphol.
13:199–204. 2004. View Article : Google Scholar : PubMed/NCBI
|