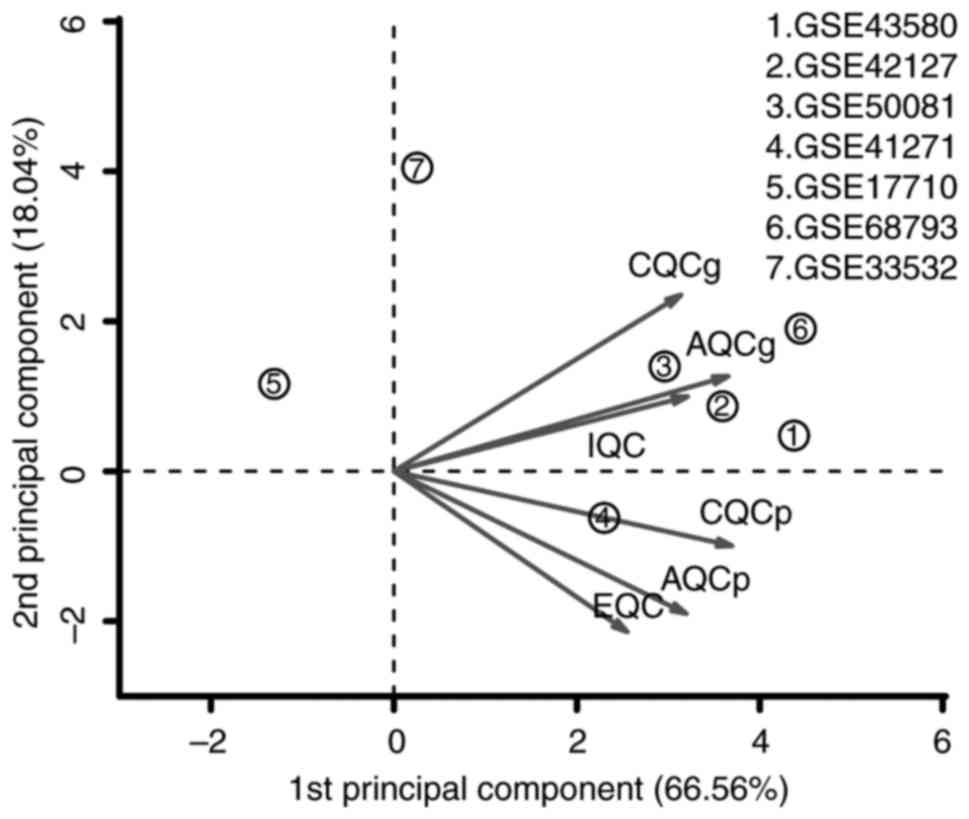
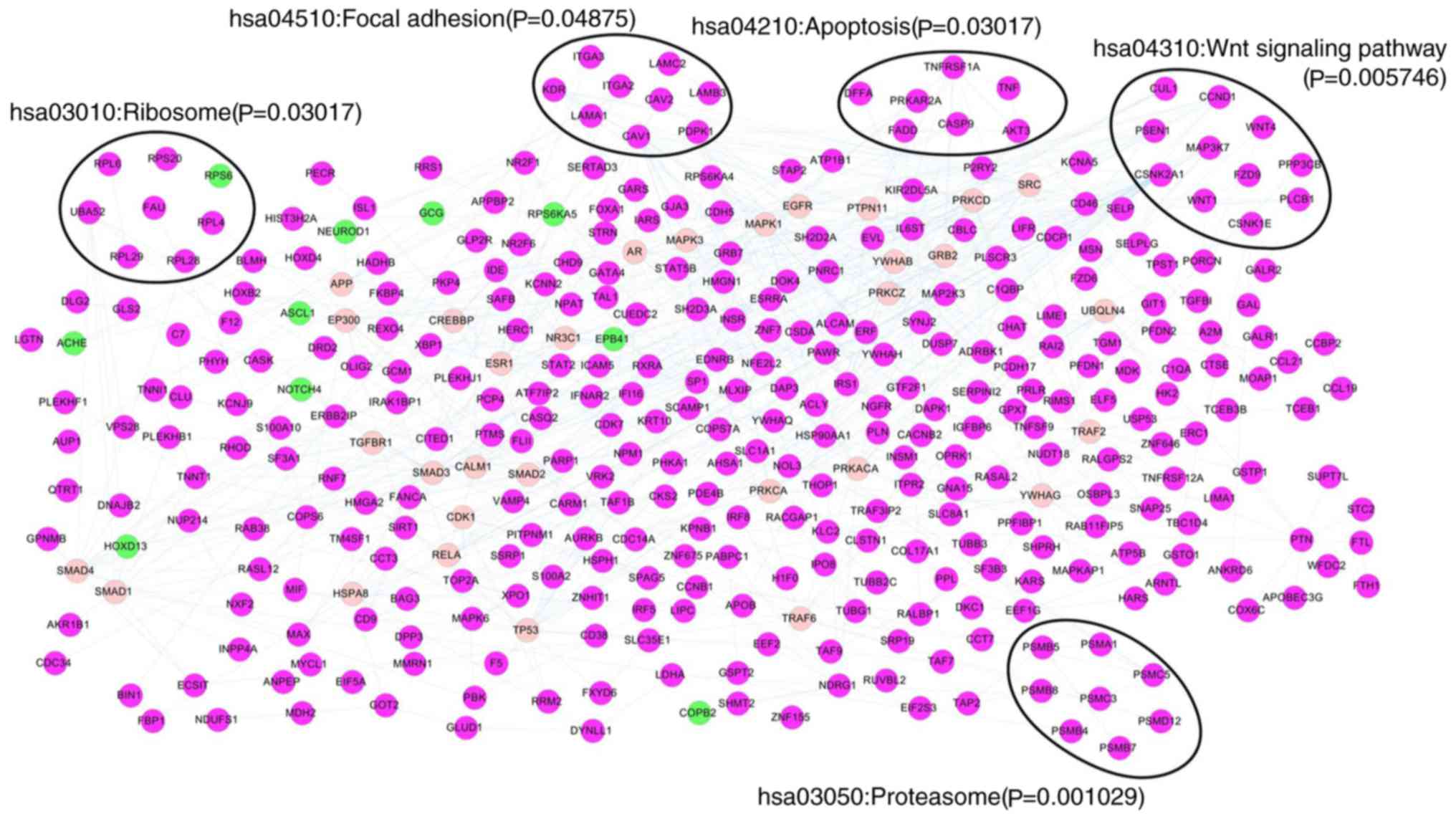
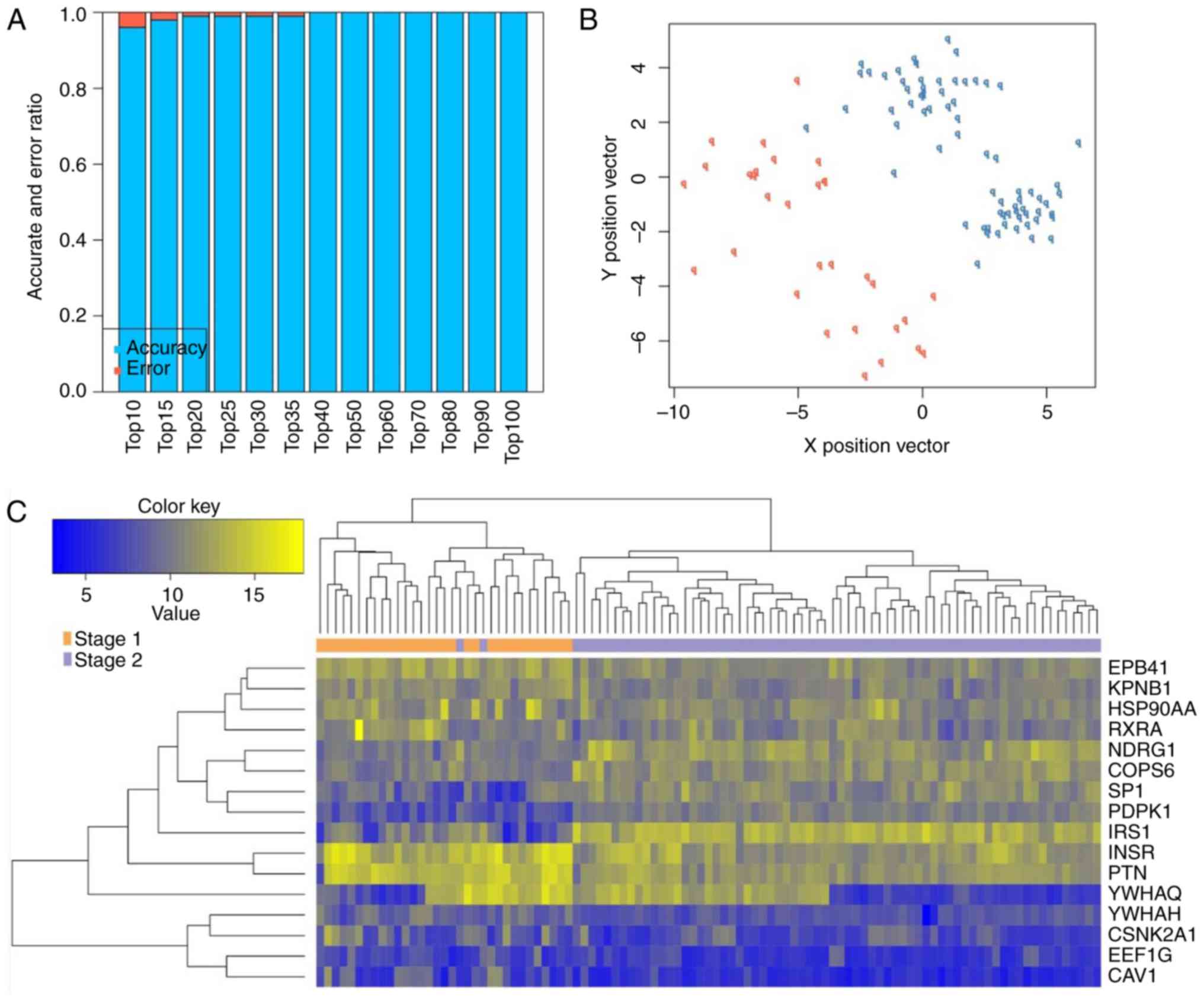
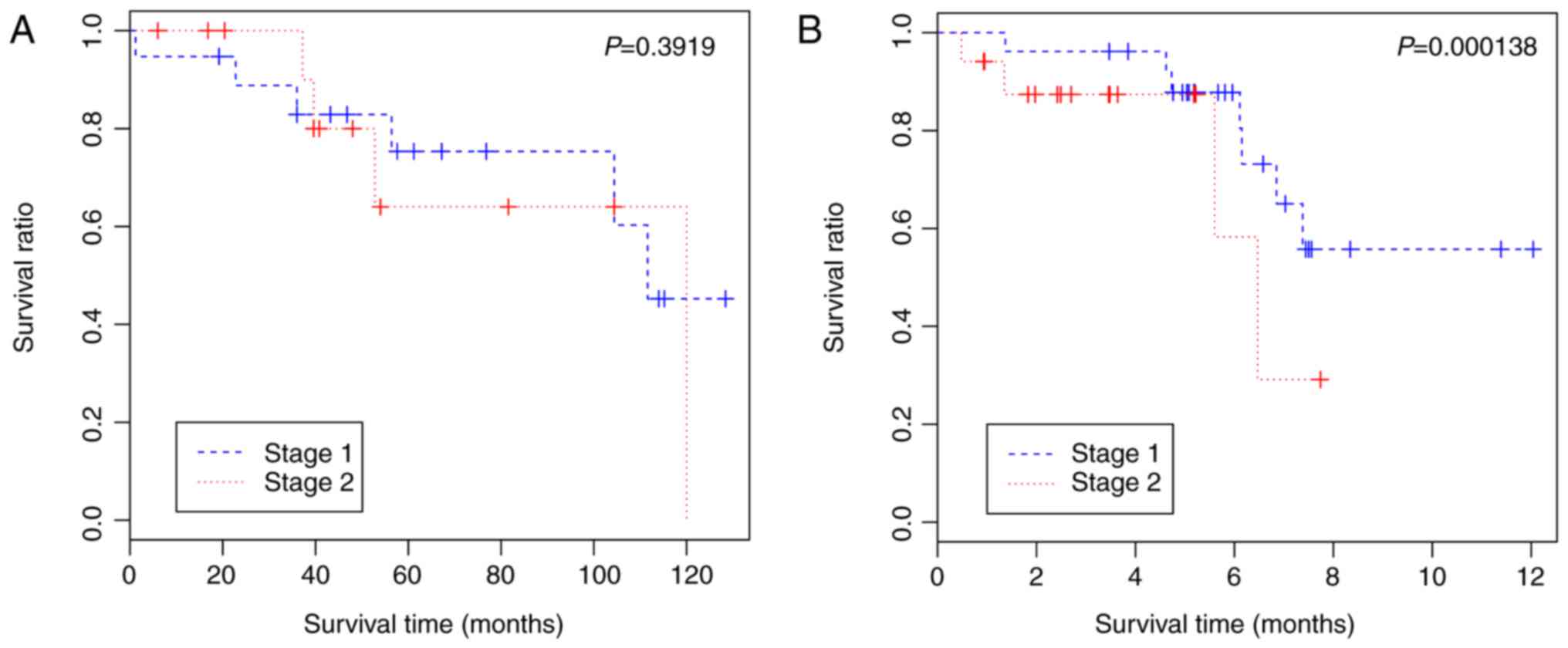
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 66:7–30. 2017. View Article : Google Scholar
|
|
2
|
McPhail S, Johnson S, Greenberg D, Peake M
and Rous B: Stage at diagnosis and early mortality from cancer in
England. Br J Cancer. 112(Suppl 1): S108–S115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Walters S, Maringe C, Coleman MP, Peake
MD, Butler J, Young N, Bergström S, Hanna L, Jakobsen E, Kölbeck K,
et al: Lung cancer survival and stage at diagnosis in Australia,
Canada, Denmark, Norway, Sweden and the UK: A population-based
study, 2004–2007. Thorax. 68:551–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klarod K, Hongsprabhas P, Khampitak T,
Wirasorn K, Kiertiburanakul S, Tangrassameeprasert R, Daduang J,
Yongvanit P and Boonsiri P: Serum antioxidant levels and
nutritional status in early and advanced stage lung cancer
patients. Nutrition. 27:1156–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lim W, Zhang WH, Miller CR, Watters JW,
Gao F, Viswanathan A, Govindan R and McLeod HL: PTEN and
phosphorylated AKT expression and prognosis in early-and late-stage
non-small cell lung cancer. Oncol Rep. 17:853–857. 2007.PubMed/NCBI
|
|
6
|
Lemon Lu Y, Liu W, Yi PY, Morrison Y, Yang
C, Sun P, Szoke Z, Gerald J, Watson WLM, et al: A gene expression
signature predicts survival of patients with stage I non-small cell
lung cancer. PLoS Med. 3:e4672006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Der SD, Sykes J, Pintilie M, Zhu CQ,
Strumpf D, Liu N, Jurisica I, Shepherd FA and Tsao MS: Validation
of a histology-independent prognostic gene signature for
early-stage, non-small-cell lung cancer including stage IA
patients. J Thorac Oncol. 9:59–64. 2014. View Article : Google Scholar
|
|
8
|
Huang S, Reitze NJ, Ewing AL, McCreary S,
Uihlein AH, Brower SL, Wang D, Wang T, Gabrin MJ, Keating KE, et
al: Analytical performance of a 15-gene prognostic assay for
early-stage non-small-cell lung carcinoma using RNA-stabilized
tissue. J Mol Diagn. 17:438–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skrzypski M, Jassem E, Taron M, Sanchez
JJ, Mendez P, Rzyman W, Gulida G, Raz D, Jablons D, Provencio M, et
al: Three-gene expression signature predicts survival in
early-stage squamous cell carcinoma of the lung. Clin Cancer Res.
14:4794–4799. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krzystanek M, Moldvay J, Szüts D, Szallasi
Z and Eklund AC: A robust prognostic gene expression signature for
early stage lung adenocarcinoma. Biomarker Res. 4:12016. View Article : Google Scholar
|
|
11
|
Hwang JA, Song JS, Yu DY, Kim HR, Park HJ,
Park YS, Kim WS and Choi CM: Peroxiredoxin 4 as an independent
prognostic marker for survival in patients with early-stage lung
squamous cell carcinoma. Int J Clin Exp Pathol. 8:6627–6635.
2015.PubMed/NCBI
|
|
12
|
Zhu CQ, Strumpf D, Li CY, Li Q, Liu N, Der
S, Shepherd FA, Tsao MS and Jurisica I: Prognostic gene expression
signature for squamous cell carcinoma of lung. Clin Cancer Res.
16:5038–5047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim TH, Choi SJ, Lee YH, Song GG and Ji
JD: Gene expression profile predicting the response to anti-TNF
treatment in patients with rheumatoid arthritis; analysis of GEO
datasets. Joint Bone Spine. 81:325–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolstad BM, Irizarry RA, Åstrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
16
|
Yang ZH, Zheng R, Gao Y and Zhang Q:
Identification of suitable genes contributes to lung adenocarcinoma
clustering by multiple meta-analysis methods. Clin Respir J.
10:631–646. 2016. View Article : Google Scholar
|
|
17
|
Kang DD, Sibille E, Kaminski N and Tseng
GC: MetaQC: Objective quality control and inclusion/exclusion
criteria for genomic meta-analysis. Nucleic Acids Res. 40:e152012.
View Article : Google Scholar :
|
|
18
|
Chang LC, Lin HM, Sibille E and Tseng GC:
Meta-analysis methods for combining multiple expression profiles:
Comparisons, statistical characterization and an application
guideline. BMC Bioinformatics. 14:3682013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar
|
|
20
|
Peri S, Navarro JD, Kristiansen TZ,
Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN,
Deshpande N, Suresh S, et al: Human protein reference database as a
discovery resource for proteomics. Nucleic Acids Res. 32:D497–D501.
2004. View Article : Google Scholar :
|
|
21
|
Meyer D and Wien FT: Support vector
machines. The interface to libsvm in package. e10712015.
|
|
22
|
Therneau T: A package for survival
analysis in S. R package version 2.37-4. URL http://CRANR-project.org/package=survivalBox980032.
pp. 23298–20032. 2013
|
|
23
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Soc Series B. 57:289–300.
1995.
|
|
24
|
Sunaga N, Miyajima K, Suzuki M, Sato M,
White MA, Ramirez RD, Shay JW, Gazdar AF and Minna JD: Different
roles for caveolin-1 in the development of non-small cell lung
cancer versus small cell lung cancer. Cancer Res. 64:4277–4285.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen D, Shen C, Du H, Zhou Y and Che G:
Duplex value of caveolin-1 in non-small cell lung cancer: A meta
analysis. Familial Cancer. 13:449–457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ho CC, Kuo SH, Huang PH, Huang HY, Yang CH
and Yang PC: Caveolin-1 expression is significantly associated with
drug resistance and poor prognosis in advanced non-small cell lung
cancer patients treated with gemcitabine-based chemotherapy. Lung
Cancer. 59:105–110. 2008. View Article : Google Scholar
|
|
27
|
Matassa D, Amoroso MR, Agliarulo I,
Maddalena F, Sisinni L, Paladino S, Romano S, Romano MF, Sagar V,
Loreni F, et al: Translational control in the stress adaptive
response of cancer cells: A novel role for the heat shock protein
TRAP1. Cell Death Dis. 4:e8512013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng Y, McFarland BC, Drygin D, Yu H,
Bellis SL, Kim H, Bredel M and Benveniste EN: Targeting protein
kinase CK2 suppresses prosurvival signaling pathways and growth of
glioblastoma. Clin Cancer Res. 19:6484–6494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O-charoenrat P, Rusch V, Talbot SG,
Sarkaria I, Viale A, Socci N, Ngai I, Rao P and Singh B: Casein
kinase II alpha subunit and C1-inhibitor are independent predictors
of outcome in patients with squamous cell carcinoma of the lung.
Clinical Cancer Res. 10:5792–5803. 2004. View Article : Google Scholar
|
|
30
|
Jäger R, List B, Knabbe C, Souttou B,
Raulais D, Zeiler T, Wellstein A, Aigner A, Neubauer A and Zugmaier
G: Serum levels of the angiogenic factor pleiotrophin in relation
to disease stage In lung cancer patients. Br J Cancer. 86:858–863.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X and
Jin GH: Lung cancer cell migration is regulated via repressing
growth factor PTN/RPTP β/ζ signaling by menin. Oncogene.
29:5416–5426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao S, Feng Z, Xu B, Wu Y, Yin P, Yang Y,
Hua X and Jin GH: Suppression of lung adenocarcinoma through menin
and poly-comb gene-mediated repression of growth factor
pleiotrophin. Oncogene. 28:4095–4104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Du ZY, Shi MH, Ji CH and Yu Y: Serum
pleiotrophin could be an early indicator for diagnosis and
prognosis of non-small cell lung cancer. Asian Pac J Cancer Prev.
16:1421–1425. 2014. View Article : Google Scholar
|
|
34
|
Brown KC, Perry HE, Lau JK, Jones DV,
Pulliam JF, Thornhill BA, Crabtree CM, Luo H, Chen YC and Dasgupta
P: Nicotine induces the up-regulation of the α7-nicotinic receptor
(α7-nAChR) in human squamous cell lung cancer cells via the
Sp1/GATA protein pathway. J Biol Chem. 288:33049–33059. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li M, Ling B, Xiao T, Tan J, An N, Han N,
Guo S, Cheng S and Zhang K: Sp1 transcriptionally regulates BRK1
expression in non-small cell lung cancer cells. Gene. 542:134–140.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cha N, Lv M, Zhao YJ, Yang D, Wang EH and
Wu GP: Diagnostic utility of VEGF mRNA and SP1 mRNA expression in
bronchial cells of patients with lung cancer. Respirology.
19:544–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao S, Fang L, Phan LM, Qdaisat A, Yeung
SC and Lee MH: COP9 signalosome subunit 6 (CSN6) regulates
E6AP/UBE3A in cervical cancer. Oncotarget. 6:28026–28041. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Azuma K, Kawahara A, Hattori S, Taira T,
Tsurutani J, Watari K, Shibata T, Murakami Y, Takamori S, Ono M, et
al: NDRG1/Cap43/Drg-1 may predict tumor angiogenesis and poor
outcome in patients with lung cancer. J Thorac Oncol. 7:779–789.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang D, Tian X and Jiang Y: NDRG1/Cap43
overexpression in tumor tissues and serum from lung cancer
patients. J Cancer Res Clin Oncol. 138:1813–1820. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Robb VA, Li W, Gascard P, Perry A,
Mohandas N and Gutmann DH: Identification of a third Protein 4.1
tumor suppressor, Protein 4.1 R, in meningioma pathogenesis.
Neurobiol Dis. 13:191–202. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou L, Luan H, Liu Q, Jiang T, Liang H,
Dong X and Shang H: Activation of PI3K/Akt and ERK signaling
pathways antagonized sinomenine-induced lung cancer cell apoptosis.
Mol Med Rep. 5:1256–1260. 2012.PubMed/NCBI
|
|
42
|
Zhang C, Lan T, Hou J, Li J, Fang R, Yang
Z, Zhang M, Liu J and Liu B: NOX4 promotes non-small cell lung
cancer cell proliferation and metastasis through positive feedback
regulation of PI3K/Akt signaling. Oncotarget. 5:4392–4405. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gallegos Ruiz MI, Floor K, Roepman P,
Rodriguez JA, Meijer GA, Mooi WJ, Jassem E, Niklinski J, Muley T,
van Zandwijk N, et al: Integration of gene dosage and gene
expression in non-small cell lung cancer, identification of HSP90
as potential target. PLoS One. 3:e00017222008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han L, Zhang G, Zhang N, Li H, Liu Y, Fu A
and Zheng Y: Prognostic potential of microRNA-138 and its target
mRNA PDK1 in sera for patients with non-small cell lung cancer. Med
Oncol. 31:1292014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Taniguchi CM, Emanuelli B and Kahn CR:
Critical nodes in signalling pathways: Insights into insulin
action. Nat Rev Mol Cell Biol. 7:85–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Heidegger I, Kern J, Ofer P, Klocker H and
Massoner P: Oncogenic functions of IGF1R and INSR in prostate
cancer include enhanced tumor growth, cell migration and
angiogenesis. Oncotarget. 5:2723–2735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim JS, Kim ES, Liu D, Lee JJ, Solis L,
Behrens C, Lippman SM, Hong WK, Wistuba II and Lee HY: Prognostic
impact of insulin receptor expression on survival of patients with
nonsmall cell lung cancer. Cancer. 118:2454–2465. 2012. View Article : Google Scholar
|
|
48
|
Wang GH, Jiang FQ, Duan YH, Zeng ZP, Chen
F, Dai Y, Chen JB, Liu JX, Liu J, Zhou H, et al: Targeting
truncated retinoid X receptor-α by CF31 Induces TNF-α-dependent
apoptosis. Cancer Res. 73:307–318. 2013. View Article : Google Scholar
|
|
49
|
Radhakrishnan VM and Martinez JD:
14-3-3gamma induces oncogenic transformation by stimulating MAP
kinase and PI3K signaling. PLoS One. 5:e114332010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang
H, Shen J, Zhao RY, Caraway NP, Katz RL, et al: Up-regulation of
14-3-3zeta in lung cancer and its implication as prognostic and
therapeutic target. Cancer Res. 67:7901–7906. 2007. View Article : Google Scholar : PubMed/NCBI
|