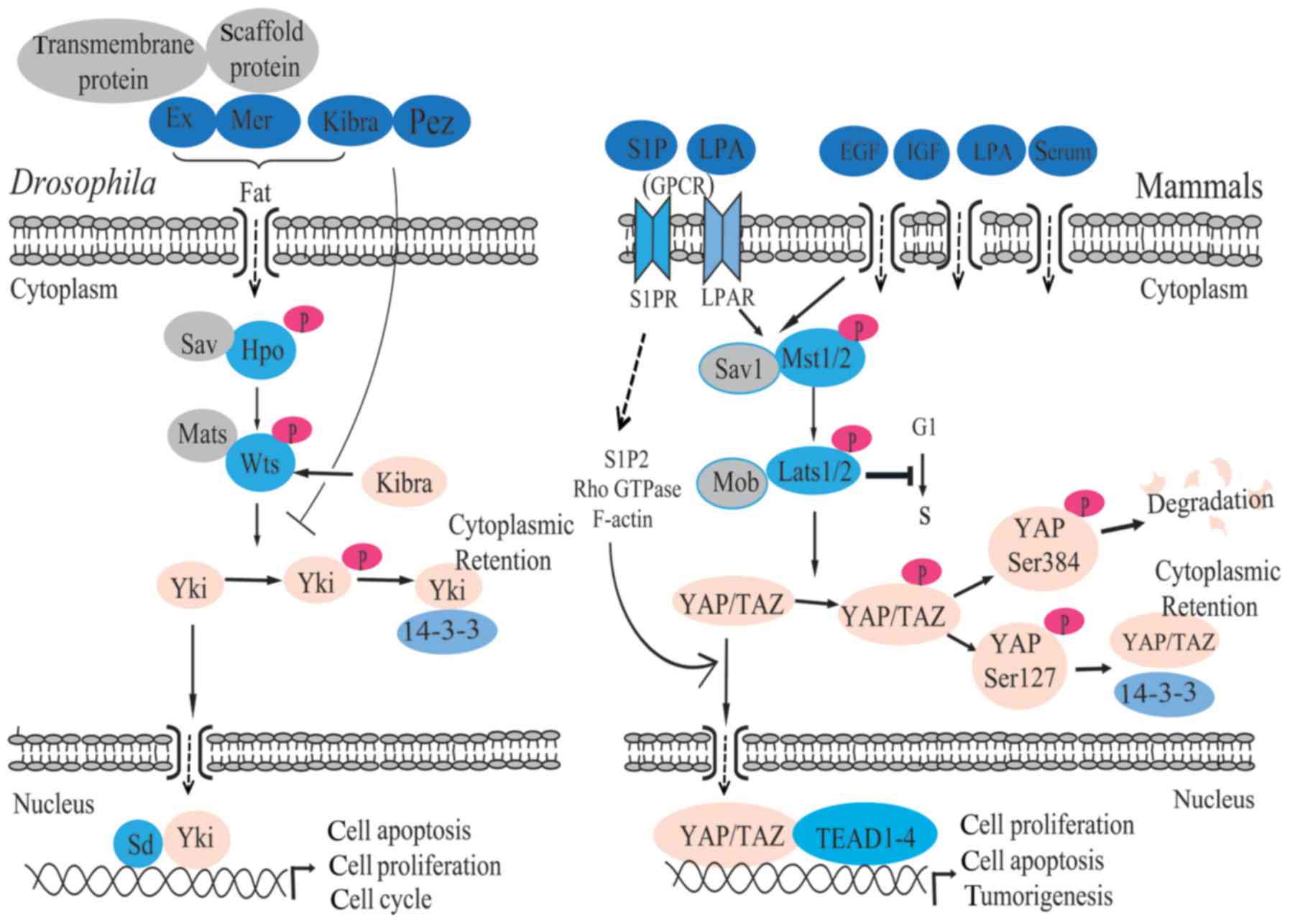
Fat is a kind of protein encoded by the fat gene.
Pez is an evolutionarily conserved FERM domain protein containing a
protein tyrosine phosphatase (PTP) domain. Fat (25,26), Pez (27) and junction-related proteins,
including Expanded (Ex) (28),
Merlin (Mer) (28,29), Kibra (30), neurofibromin 2 (NF2) (28,31) and Crumbs (Crb) (32), have been identified as upstream
regulators of the Hippo pathway. Although the regulatory mechanism
of the Hippo pathway remains unclear, an increasing number of
studies have provided evidence that the Hippo pathway is tightly
correlated with the external environment and regulated by the
extracellular factors (15,33,34). The Fat complex that is composed of
Ex, Mer and Kibra can activate the Hippo pathway through
phosphorylating Hpo and increase the stability of Ex protein
(35,36). Ex and Mer, membrane-associated
proteins, contain the four-point-one, ezrin, radixin, moesin (FERM)
domain and can interact with a variety of cell scaffold proteins
and transmembrane proteins through the N-terminal region of Ex and
the C-terminal domain of Mer, which is characteristic of the
protein 4.1 family. It has been reported that their interactions
are involved in regulating cell apoptosis, cell proliferation and
the cell cycle (37). Kibra
protein, which contains WW and C2 domains, activates Wts via
interaction with Ex and Mer (38). In addition, NF2, as a homologous
protein of Mer and a tumor suppressor gene, is a negative regulator
of cell growth. The mutation of NF2 causes central nervous system
tumors, such as neurofibromatosis (39). With conditional knockout of the
NF2 gene in the mouse liver, the risk of cancer is increased
greatly (40). Additionally, Crb,
a transmembrane protein, can also regulate the Hippo pathway
(41). Crb has a large
extracellular domain and a short intracellular domain. For the Crb
cytoplasmic tail, it has two recognized motifs. One links to the
spectra and actin cytoskeleton (PDZ-binding motif) and the other is
the juxtamembrane FERM-domain binding motif (JM), which interacts
with polarity regulatory factors, including Stardust (Sdt),
PALS1-associated tight junction protein (Patj) and partitioning
defective 6 (Par6). By interacting with the JM of Crb, Ex
expression is inhibited, which ultimately results in an increase in
Yki activity and increased organ growth (32,41). Pez, a newly found binding partner
of Kibra (27,38), is a type of FERM domain protein
containing the protein tyrosine phosphatase domain. Pez, as a
component of upstream Hippo signaling, serves an important role in
cell metabolism, proliferation and differentiation, particularly
with regard to restricting the activity of Yki in the adult midgut
epithelium (27). Meanwhile,
transcriptional levels of the target gene of Yki have been elevated
in Pez mutant guts (42).
The core transduction pathway of Hippo is a kinase
cascade. As a tumor suppressor, Mst1/2 was the first factor to be
identified (43). Mst1/2, a
member of the nuclear Dbf2-related protein family, encodes kinase
and can phosphorylate Sav1, Lats1/2 and Mob (44,45). Stimulus signals of the Hippo
pathway activate Mst1/2, then Mst1/2 phosphorylates Lats1/2 through
its interaction with Sav1, which increases the activity of Mst1/2
(45). Sav1 serves a pivotal role
in connecting Msts1/2 to Lats1/2 (17), and it can increase or decrease the
activity of Lats1/2 after being phosphorylated by Mst1/2 (46). Lats1/2 can negatively regulate the
cell cycle, the ectopic expression of Lats2 can inhibit the
transformation from the cellular G1 period to the S period
(47), and the overexpression of
Lats1/2 will lead to the stagnation of the cellular G2/M period and
apoptosis (48,49). Subsequent to being phosphorylated
by Mst1/2, Mob1 combines with Lats1/2, which leads to the
activation of Lats1/2 (45).
Afterward, Lats1/2 phosphorylates YAP/TAZ (20) (Fig.
1). An increasing number of studies have illustrated that the
highly conservative peculiarity of Hippo pathway is involved in the
growth of tissues and organs, and tumorigenesis (10,13,16,28,31,50,51). Deficiency of Mst1/2 will lead to
the enlargement of the liver in mice, finally inducing a tumor
(52). However, when upstream
kinase is not activated, YAP/TAZ translocates into the nucleus to
serve a role in gene expression (Fig.
1).
Found in 1994, YAP binds to the SH3 region of the
Yes protein in the Src protein kinase family through the sequence
‘PVKQPPPLAP’ and is a protein with a molecular weight of about 65
kDa (53,54). YAP is located on chromosome 11q22
in humans (55,56). The YAP protein contains one or two
WW structure domains, a 14-3-3 protein binding site, a SH3 binding
motif, a PDZ binding motif of the C-terminal, an insertion in the
N-terminal TEAD-binding region, a proline-rich domain of the
amino-terminal and a coiled-coil structure domain of the
transcriptional regulation structure (57). YAP can interact with a variety of
proteins, includimg angiomotin family proteins, TEADs and Sd, and
it can participate in multiple signaling transduction pathways in
cells through the structure domains and amino acid sequences
(57). YAP is a type of
multifunctional cell connection protein and transcription
activating factor, with a variety of structural domains and
specific sequences that determine the diversity of its role. The
transcriptional activity of YAP depends on its intracellular
localization (24). When YAP is
located in the nucleus, it can activate gene transcription through
interaction with transcription activation factors, and then the
phosphorylated YAP transposes into the cytoplasm and accumulates
there (58). The cytoplasmic
localization of phosphorylated YAP can prevent or terminate its
activation activity in the nucleus (51,59). Due to the interaction with
phosphorylated Ser127 and 14-3-3 proteins, accumulating YAP cannot
transpose into the nucleus, causing YAP to lose its role as a
transcription activation factor (58,60).
First identified in 2000, TAZ can also combine with
14-3-3 proteins and is a type of protein composed of 400 amino
acids containing a WW domain and a PDZ domain (61). Through sequence comparison, it has
been revealed that TAZ is the homologous protein of YAP, as well as
exhibiting the function of transcriptional activation (20). The coding gene of TAZ is located
on chromosome 3q24, and its molecular weight is 45 kDa (61). The combination of 14-3-3 proteins
and TAZ requires phosphorylation of the 89th TAZ serine, as the TAZ
Ser89 area is one of the best combination sequences of 14-3-3
proteins and is known as RSXpSXP. The other is RXY/FXpSXP, and both
can be recognized by the varyiong types of 14-3-3 proteins
(62). YAP possesses the same
feature of the combination sequences (61,62). TAZ has the same transcriptional
activation function as YAP. YAP, TAZ and Yorkie all lack DNA
binding domains. Therefore, they can only regulate transcription
via activation (little inhibition) of the activity of transcription
factors. The upstream regulatory mechanisms produce the 14-3-3
protein binding sites via phosphorylation of TAZ Ser89 with a
kinase, and then control TAZ cytoplasmic retention. Thus, the
transcriptional regulation of TAZ is inhibited (61,63).
YAP/TAZ are the major downstream effectors of the
Hippo pathway. Due to the lack of DNA-binding domains, YAP/TAZ act
by connecting with other transcription factors, including TEAD/TEF
family transcription factors (50). When YAP Ser127 is phosphorylated
by Lats1/2, it generates a 14-3-3 protein binding site (58,63). Lats2 promotes the cytoplasmic
accumulation of YAP by promoting YAP Ser127 phosphorylation and the
combination of YAP and 14-3-3 proteins (60,64). YAP Ser384, which is phosphorylated
by Lats1/2, promotes casein kinase 1 (CK1) to phosphorylate YAP
Ser384, and leads to the degradation of YAP (65,66) (Fig.
1). Furthermore, the activity of YAP/TAZ can be regulated by
serum-derived sphingosine-1-phosphate (S1P), LPA, GPCR signaling
and the actin-myosin cytoskeleton (6). S1P and LPA can induce YAP
dephosphorylation, nuclear localization, activation through the
S1P2 receptor, Rho GTPase activation and F-actin
polymerization (6).
The Hippo pathway serves a crucial role in
regulating organ size, cell proliferation and apoptosis, and it can
maintain a stable internal environment. The inactivation of Mst1/2,
Sav1, Lats1/2 and Mob1, and the overexpression of YAP/TAZ can cause
an increase in the organ size, cell proliferation (14) and even tumorigenesis, including
liver cancer (67), gastric
cancer (68), breast cancer
(69), high-grade dysplasia and
adenocarcinoma of the esophagus, gastric adenocarcinoma and
metastatic gastric disease (70).
Furthermore, the Hippo pathway regulates the inhibition of cell
contact (71). Studies have shown
that normal cells, which highly express YAP, grow continuously due
to mutual contact; hyperexpression of YAP can decrease contact
inhibition and promote cell proliferation (14,60,67). Cell contact inhibits cell growth,
which is a physiological characteristic of cells. Previous studies
have found that soluble growth factors, including epidermal growth
factor (EGF), insulin-like growth factor, serum and LPA, can
inhibit YAP/TAZ and reduce the effect of cell contact on the
majority of occasions (72–74). Two newly identified downstream
factors, phosphoinositide 3-kinase (PI3K) and
phosphoinositide-dependent protein kinase 1, for the Hippo pathway
were found to inhibit the pathway and induce YAP nuclear retention
via interaction with soluble growth factors, and finally, by
activating growth genes (72)
(Fig. 1). Cell contact is
correlated with cell density. Cell density can control the
localization and phosphorylation of YAP (60). YAP locates in the nucleus at low
cell density in NIH-3T3 cells and the human breast epithelial
MCF10A cell line. The decrease in cell density will increase the
cell area. While at high cell densities, YAP locates in the
cytoplasm and exhibits slower electrophoretic migration. The
electrophoretic migration is density-dependent due to the
phosphorylation. Phosphorylated YAP migrates faster (60,71).
The Wnt/β-catenin signaling pathway is an extremely
conservative pathway in biological evolution (75,76). Wnt can secrete glycoprotein
through binding with receptors on the cell membranes (77) and can activate intracellular
signaling pathways, which regulate the expression of target genes.
Thus, cell proliferation, differentiation and apoptosis can be
regulated, and various diseases will be caused, even tumorigenesis
(75,78,79). Wnt pathways contain the canonical
(Wnt/β-catenin), non-canonical (Wnt/JNK pathway) and
Ca2+ pathways (80,81). At present, the mechanism of the
canonical pathway is the most exhaustively researched. It has been
found that β-catenin is the principal regulation point of the
canonical Wnt pathway, and it is also the major component of cell
adhesion (75). Interaction
between β-catenin and E-cadherin can connect the intracytoplasmic
cadherins to the actin cytoskeleton, thus allowing cell contact,
adhesion and motility (75).
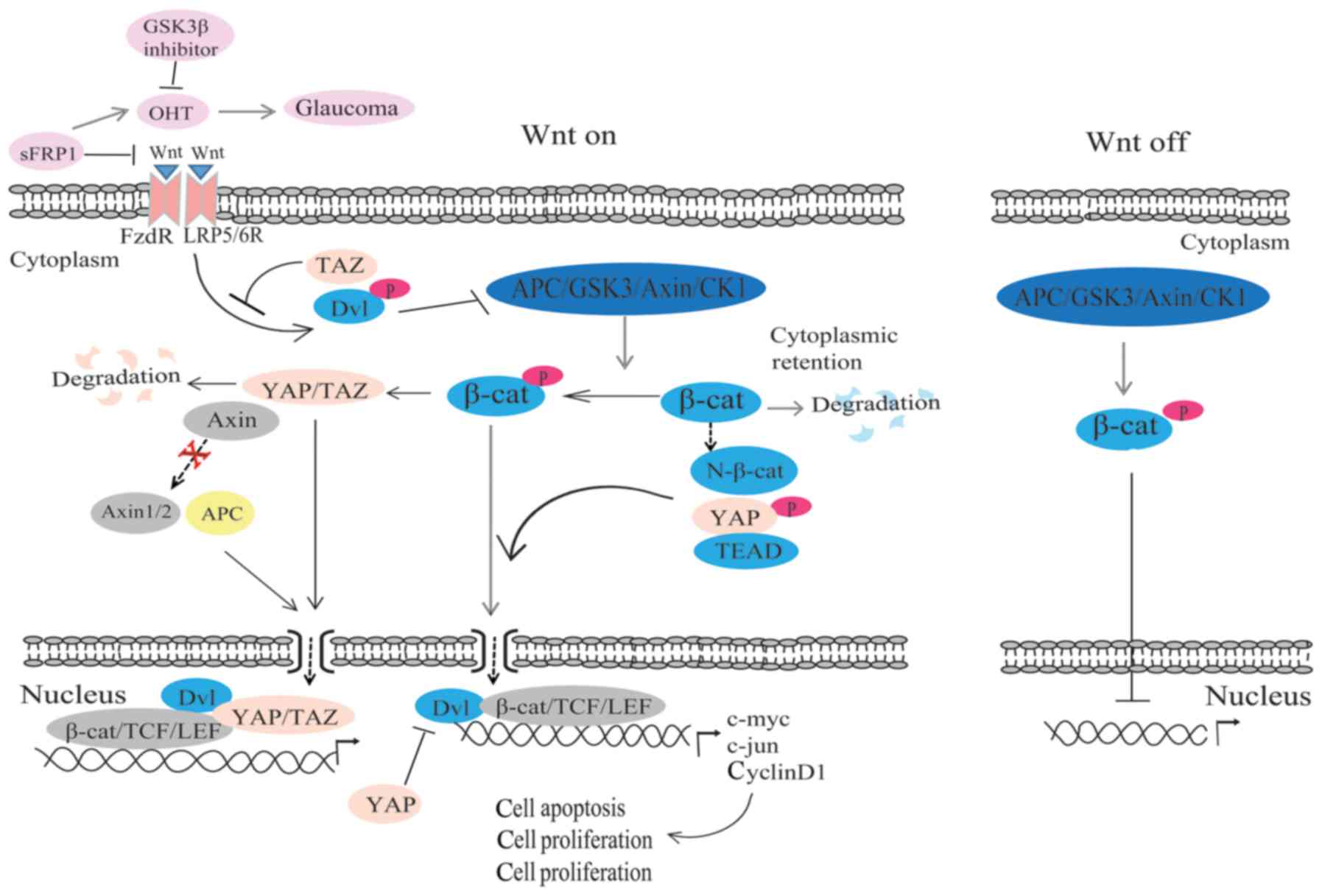
The Wnt/β-catenin and Hippo pathways regulate the
expression of genes, cell proliferation, cell differentiation and
cell apoptosis, so they interact mutually to a certain extent. On
one hand, studies have reported that phosphorylated β-catenin can
promote YAP/TAZ degradation by serving as the presenting factor
(90,91). On the other hand, through
controlling β-catenin stability, Wnt mediates YAP/TAZ stabilization
and promotes YAP/TAZ to translocate into the nucleus in a manner
independent of the Hippo pathway (90) (Fig.
2). Furthermore, Axin is not only associated with the β-catenin
protein destruction complex, but is also strongly associated with
endogenous YAP/TAZ (92,93). When Axin1/2 or APC is knocked out,
YAP/TAZ will accumulate in the nucleus, which indicates that Wnt
regulates YAP/TAZ through promoting their nuclear accumulation
(80,83).
YAP is the target gene of Wnt/β-catenin. TEAD binds
with the domain of phosphorylated YAP, and then directly connects
to the N-terminal of β-catenin and promotes β-catenin nuclear
translocation. It has been demonstrated that the Hippo pathway
antagonizes signal transduction of the Wnt pathway (94). Meanwhile, TAZ is a downstream
regulator of Wnt signaling (90).
Based on the study by Varelas et al (8), TAZ can combine with Dvl protein and
prevent the phosphorylation of Dvl by Wnt, resulting in the
inhibition of the Wnt pathway (Fig.
2). The Hippo pathway may inhibit the expression of the target
Wnt gene through altering the activity of β-catenin. Studies have
revealed that TAZ degradation requires β-catenin phosphorylation,
and the combination of phosphorylated YAP/TAZ and β-catenin, which
will cause the accumulation of β-catenin in the cytoplasm; thus,
the activity of β-catenin transcription is inhibited (90,95). The localization of β-catenin is
enhanced in the cytoplasm and nucleus in TAZ mutant kidneys in mice
(8). Together, these results
suggest that the Hippo pathway can negatively regulate the activity
of β-catenin transcription.
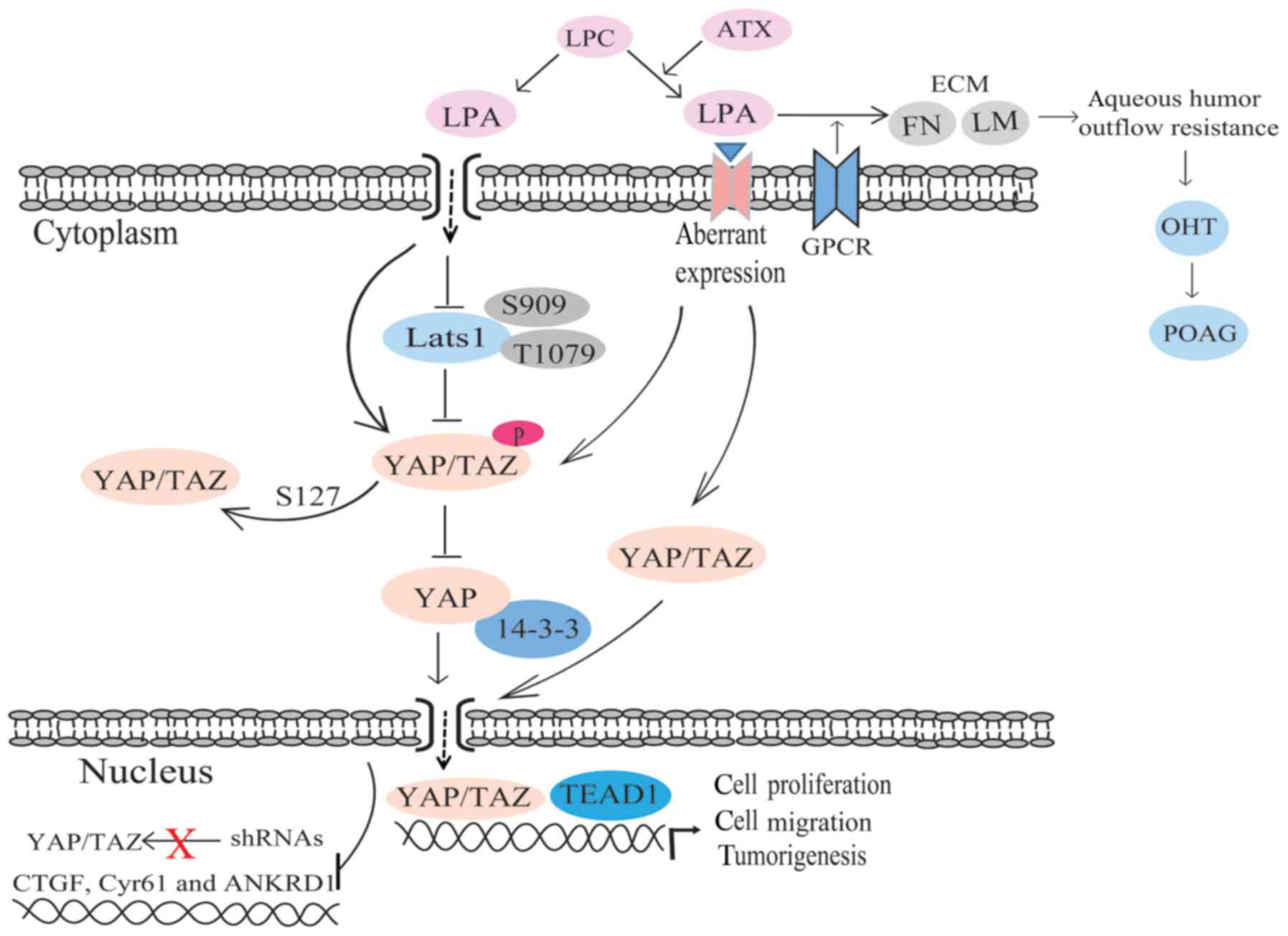
Studies have indicated that the Wnt/β-catenin
pathway is the chief Wnt pathway in human trabecular meshwork (HTM)
cells and that it serves a role in regulating IOP (5). Secreted frizzled-related protein 1
(sFRP1), the Wnt pathway inhibitor, is found to be elevated in the
glaucomatous trabecular mesh-work, and exogenous sFRP1 brings about
high IOP (96,97). In sFRP1-perfused human eyes, the
level of β-catenin was shown to be significantly decreased.
According to a recent study, sFRP1 is associated with cell
stiffness (97). HTM cells have
different responses to the stimulus of different concentrations of
sFRP1 (97). It has been
illustrated that sFRP1 is elevated in normal HTM cells grown on
substrates that simulate the stiffness of the glaucomatous HTM. The
increase in stiffness of the HTM enhances the aqueous humor outflow
resistance and is conducive to elevated IOP (97). Additionally, one study revealed
that a small molecule GSK3β inhibitor can increase the activity of
the Wnt/β-catenin pathway and remit ocular hypertension, which is
caused by sFRP1 in mouse eyes (96) (Fig.
2). It has been reported that there may be two effects of Wnt
in glaucoma (5). On one hand, the
nuclear translocation of β-catenin influences gene expression
(5). The glaucoma gene myocilin
(MYOC) has been reported to be a regulator of Wnt/β-catenin
signaling (98). However, whether
MYOC mutation affects Wnt in the HTM remains unclear. On the other
hand, the aqueous humor outflow resistance is affected by the
change in adhesion junctions and cell contact, and then IOP is
regulated (5).
The Wnt/β-catenin pathway is believed to be a novel
interventional route for the treatment of glaucoma (99). The Wnt/β-catenin pathway is
influenced by YAP/TAZ, and YAP/TAZ exert an adverse effect on Wnt
(5). Therefore, we speculate that
there is a close association among the Wnt/β-catenin pathway, the
Hippo pathway and glaucoma. YAP/TAZ may be novel target genes for
the treatment of glaucoma.
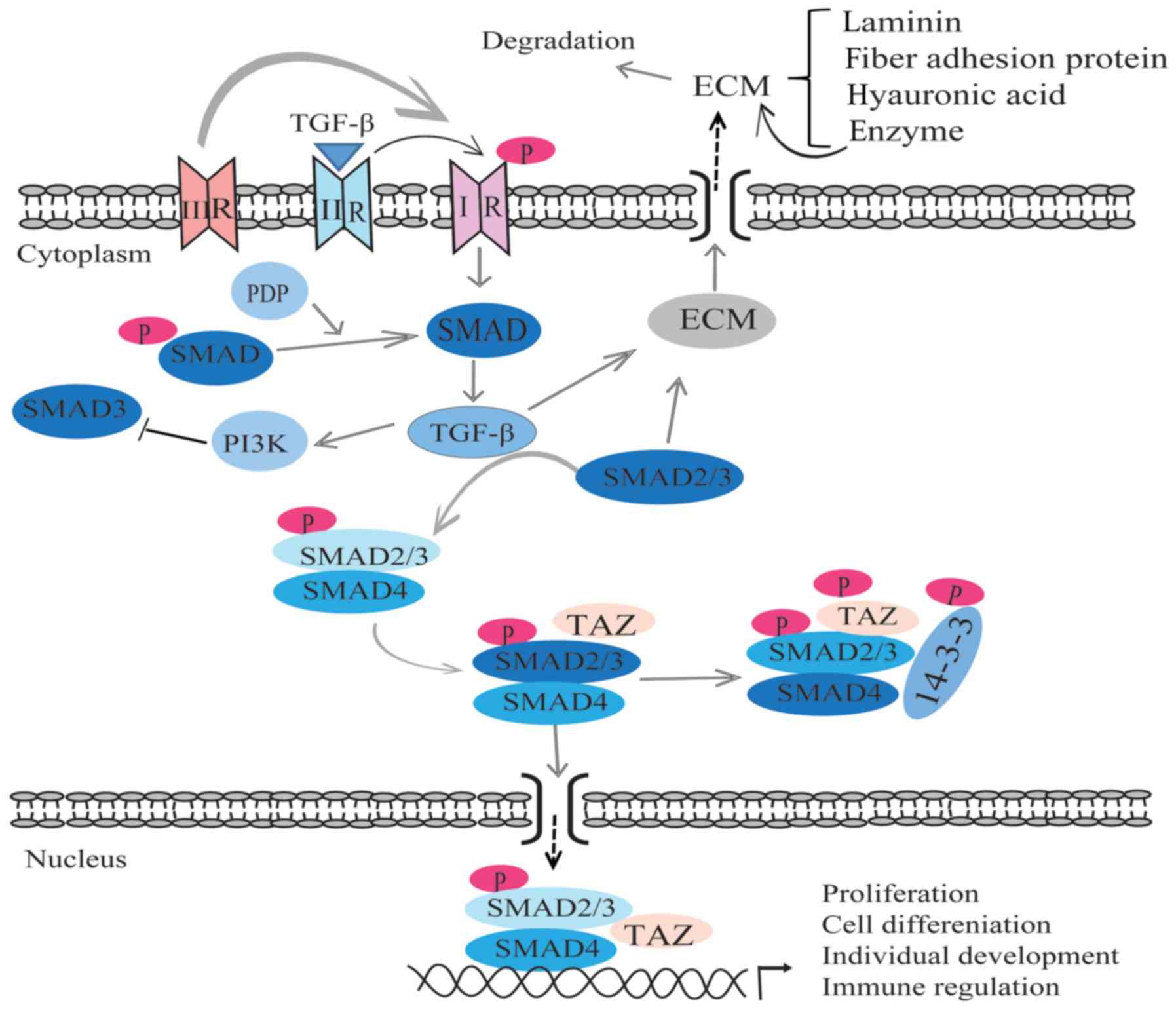
TGF-β, which was first identified by Delarco and
Todar in 1978, is a type of multifunctional cytokine (110–112). TGF-β has been found in almost
all human cells, where it has a significant role in apoptosis,
differentiation, the cell cycle, immunoregulation and the synthesis
of the ECM via binding with its receptors (113). TGF-β has five subtypes, and
three of these, TGF-β1, TGF-β2 and TGF-β3, are present in mammals,
with homology of 70–80% (114).
The signaling transduction of TGF-β acts by combining its ligand
with receptors that are the membrane-associated proteins on the
cell membrane. Receptor types I, II and III participate in
TGF-β-mediated signaling transduction. However, receptor type III
only has a supporting role during the incorporation of ligand and
receptors I/II in the transduction, as it has no serine/threonine
activity. TGF-β combines with the type II receptor, then the latter
phosphorylates the type I receptor, which leads to the activation
of this receptor (115). Later,
activated type I receptor activates target protein, such as SMAD
protein, resulting in the activation of the signaling transduction
of TGF-β (115). The Smad family
of proteins are key proteins in the process of TGF-β signal
transduction, and has numerous subtypes (116) (Fig. 4). The PI3K signaling pathway can
be regulated by TGF-β. Inhibiting PI3K can reduce the activity of
Smad3 (receptor activation SMAD protein) (117). Pyruvate dehydrogenase
phosphatase can specifically dephosphorylate SMAD in
Drosophila, indicating that SMAD is the core protein that
mediates the signal transduction of TGF-β (118). TGF-β has numerous biological
effects and can be expressed in various types of cells, with an
important role in regulating cell proliferation, cell
differentiation, synthesis, deposition and degradation of the ECM
(119). In addition, TGF-β
mediates the individual development of growing, inflammatory
process and the immune regulation (119).
Increasing aqueous outflow resistance is associated
with the abnormal contraction and movement of the trabecular
meshwork, as well the abnormal synthesis and secretion of the ECM.
These abnormal activities cause the deposition of the ECM, and
finally lead to a high IOP, and the damage of the structure and
function of the eyes (4). A
previous study proved that the human corneal endothelial layer,
ciliary body, iris and trabecular meshwork can express and
transcribe the RNA of TGF-β1 and TGF-β2 (120). Evidence has shown that the
concentration of TGF-β in the aqueous humor of patients with POAG
is markedly higher than that in patients without POAG (121). In HTM cells, the constitutive
expression and release of biologically active TGF-β2 can be
mediated by ϱ-GTPase signaling (122). ϱ-associated kinase inhibitors
directly affect the TM and SC by controlling the synthesis of actin
stress fibers, focal adhesion and cellular contraction (123,124). Trabecular meshwork cells
synthesize and secrete the components of the ECM, including
laminin, fiber adhesion proteins, hyaluronic acid and enzymes that
can degrade the ECM (7). Research
has revealed that Smad2/3, which can stimulate the cell to
synthesize ECM, is the key protein in the process of ECM
reconstruction (115). The
increasing secretion of the ECM and the reduction of Smad2/3 will
lead to the accumulation of ECM and stop the aqueous outflow from
increasing. Studies have indicated that TGF-β can promote the
synthesis of the ECM via regulation of trabecular meshwork cells,
and then affect the aqueous human outflow resistance (7,120,125). Experiments in pig eyes have
found that TGF-β2 can promote the synthesis of collagen protein and
fiber connection protein. The synthesis of collagen protein and
fiber connection protein inhibits the synthesis of hyaluronic acid
and regulates the molecular structure of fiber connection protein.
The proteolytic enzyme reduces the degradation of fiber link
protein and then leads to the deposition of fiber connection
protein, ultimately causing glaucoma (126) (Fig. 4). TGF-β can also promote the
synthesis of metalloproteinase tissue inhibitors to inhibit matrix
metalloproteinases and the degradation of the ECM, which causes the
deposition of ECM in the trabecular meshwork (7,125). Overall, TGF-β promotes the
deposition of the ECM by promoting the synthesis of the ECM,
regulating trabecular cells and altering the structure of the ECM.
Enhancing the levels of inhibitors of protease can reduce the
degradation of the ECM.
It has been found that TGF-β can inhibit the
proliferation of human trabecular cells stimulated by EGF (127). The reduction in the number of
trabecular meshwork cells indicates that TGF-β can inhibit
trabecular cell proliferation and then participate in the
pathogenesis of glaucoma. TGF-β1 can enhance the expression of
α-actin mRNA in human and monkey trabecular meshwork cells. TGF-β1
can promote the transfomation of the trabecular meshwork into
fibroblasts, resulting in an increase in the contraction and motor
function of the trabecular meshwork (128). Trabecular cell microfilament
actin levels increased by TGF-β1 can promote the ability of cell
shrinkage and movement, which is conducive to the outflow of the
aqueous humor. If the abilities of shrinkage and movement are
excessively enhanced, the trabecular meshwork shrinks and loses
elasticity. Atrophy and inelasticity of the trabecular meshwork act
against the outflow of the aqueous humor and may cause the IOP to
increase, leading to glaucoma.
Activated TGF-β receptors phosphorylate SMAD2/3 on
the C-terminal SSXS motif, and then the phosphorylated SMAD2/3
binds with SMAD4 to form a complex. TAZ can bind with the SMAD
complex to promote the nuclear accumulation of SMAD (129). Lats can regulate the activity of
the Hippo pathway and SMAD2/3 localization via YAP/TAZ. Studies of
the regulation of TGF-β/SMAD signaling in cells indicate that the
Hippo kinase cascade may regulate SMAD nuclear localization by
YAP/TAZ phosphorylation (9)
(Fig. 4).
It has been demonstrated that YAP/TAZ exist in all
levels of the trabecular meshwork (134), including juxtacanalicular
tissue, which is considered to be the fundamental part of aqueous
humor outflow resistance. YAP was confirmed to be reduced in the
trabecular meshwork incubated in a stiff substrate, while TAZ level
was increased (134). According
to this difference, YAP principally serves an important role in
normal trabecular tissue, while TAZ markedly affects glaucoma
trabecular tissue (134). This
result is different from that found in the study by Dupont et
al (133), which observed
that YAP and TAZ were both increased in mammalian epithelial cells
that were cultured on a stiff substrate (133). This result may caused by
different cell types exhibiting different reactions to YAP/TAZ
(135). YAP acts as a power
transduction factor, and its positive reaction in trabecular cells
shows that they may be involved in the development of glaucoma.
There is a mutual regulation among the newly discovered target
paths of glaucoma treatment and YAP/TAZ, although there is no
experimental report to verify this.
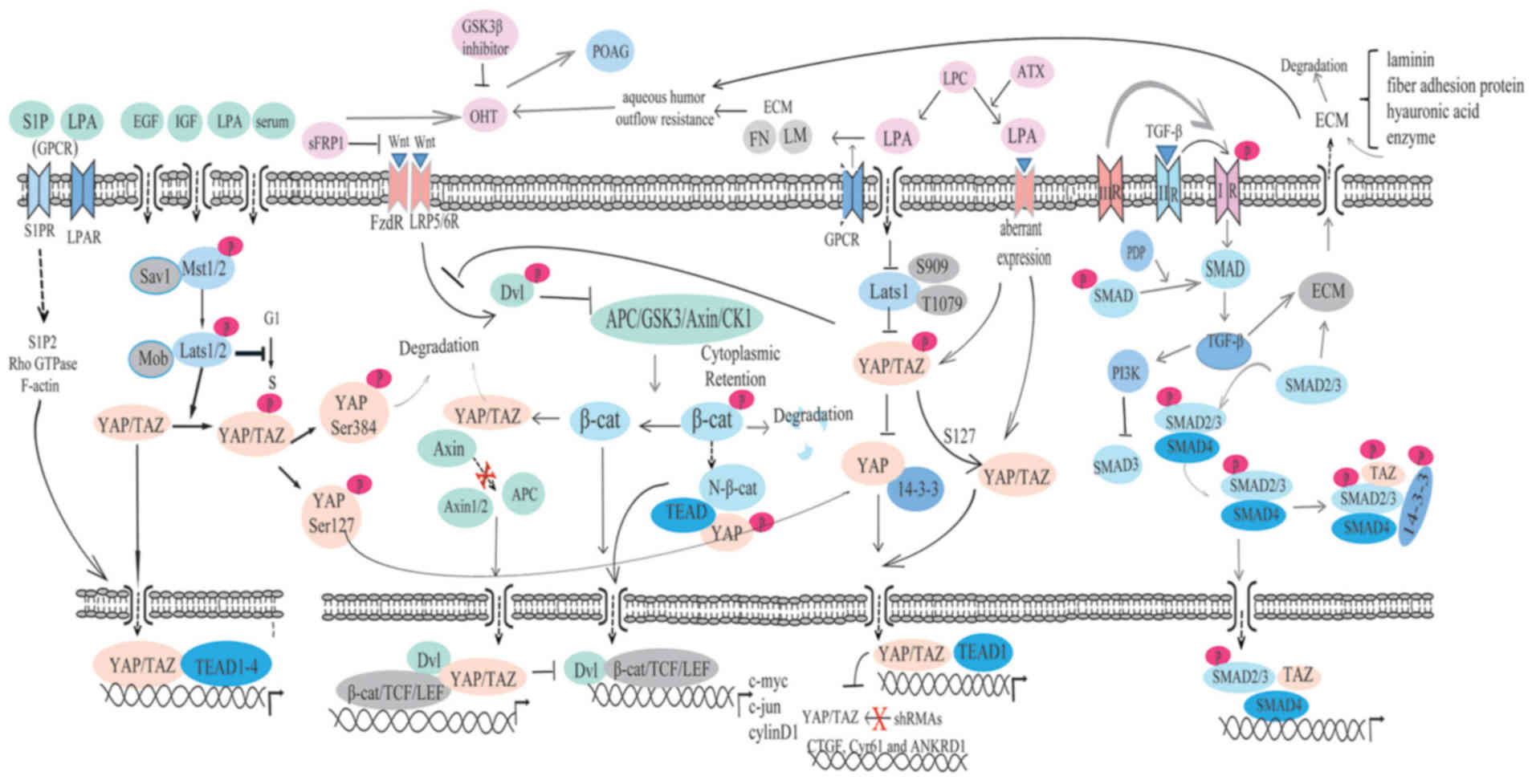
The present review attempted to highlight the
important role of YAP/TAZ and the Wnt/β-catenin, LPA and TGF-β/Smad
pathways, which are all involved in the regulation of the
pathogenesis of glaucoma (Fig.
5). These signaling pathways have become novel target pathways
in the treatment of glaucoma and they also regulate the Hippo
pathway. We hypothesize that the Hippo pathway may serve an
important role in the pathogenesis of glaucoma, and YAP/TAZ may
become novel target genes in the treatment of glaucoma. However,
certain problems require solving, including determining the
specific pathogenesis of glaucoma, the manner in which upstream
factors regulate the Hippo signaling pathway and investigating the
definitive regulating mechanism of YAP/TAZ in glaucoma. Further
study is also required into whether there any other relevant
regulators mediating the Wnt/β-catenin, LPA and TGF-β/Smad
pathways, and whether MYOC mutations affect Wnt signaling in the
trabecular meshwork.
This review was supported by the National Science
Foundation of China (grant no. 81300768), the Sichuan Science
Foundation (grant no. 2015JY0183), the Sichuan Health and Family
Planning Commission (grant no. 16ZD025) and the National Key
Speciality Construction Project of Clinical Pharmacy (grant no.
30305030698).
|
1
|
Quigley HA: Open-angle glaucoma. N Engl J
Med. 328:1097–1106. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson M: ‘What controls aqueous humour
outflow resistance?’. Exp Eye Res. 82:545–557. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Johnstone MA and Grant WG:
Pressure-dependent changes in structures of the aqueous outflow
system of human and monkey eyes. Am J Ophthalmol. 75:365–383. 1973.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Knepper PA, Goossens W, Hvizd M and
Palmberg PF: Glycosaminoglycans of the human trabecular meshwork in
primary open-angle glaucoma. Invest Ophthalmol Vis Sci.
37:1360–1367. 1996.
|
|
5
|
Mao W, Millar JC, Wang WH, Silverman SM,
Liu Y, Wordinger RJ, Rubin JS, Pang IH and Clark AF: Existence of
the canonical Wnt signaling pathway in the human trabecular
meshwork. Invest Ophthalmol Vis Sci. 53:7043–7051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miller E, Yang J, DeRan M, Wu C, Su AI,
Bonamy GM, Liu J, Peters EC and Wu X: Identification of
serum-derived sphin-gosine-1-phosphate as a small molecule
regulator of YAP. Chem Biol. 19:955–962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fleenor DL, Shepard AR, Hellberg PE,
Jacobson N, Pang IH and Clark AF: TGFbeta2-induced changes in human
trabecular meshwork: implications for intraocular pressure. Invest
Ophthalmol Vis Sci. 47:226–234. 2006. View Article : Google Scholar
|
|
8
|
Varelas X, Miller BW, Sopko R, Song S,
Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill
H, et al: The Hippo pathway regulates Wnt/beta-catenin signaling.
Dev Cell. 18:579–591. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Varelas X, Samavarchi-Tehrani P, Narimatsu
M, Weiss A, Cockburn K, Larsen BG, Rossant J and Wrana JL: The
Crumbs complex couples cell density sensing to Hippo-dependent
control of the TGF-β-SMAD pathway. Dev Cell. 19:831–844. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kango-Singh M and Singh A: Regulation of
organ size: insights from the Drosophila Hippo signaling pathway.
Dev Dyn. 238:1627–1637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saucedo LJ and Edgar BA: Filling out the
Hippo pathway. Nat Rev Mol Cell Biol. 8:613–621. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Buttitta LA and Edgar BA: How size is
controlled: from Hippos to Yorkies. Nat Cell Biol. 9:1225–1227.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan D: Hippo signaling in organ size
control. Genes Dev. 21:886–897. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao B, Lei QY and Guan KL: The Hippo-YAP
pathway: new connections between regulation of organ size and
cancer. Curr Opin Cell Biol. 20:638–646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu FX and Guan KL: The Hippo pathway:
regulators and regulations. Genes Dev. 27:355–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Justice RW, Zilian O, Woods DF, Noll M and
Bryant PJ: The Drosophila tumor suppressor gene warts encodes a
homolog of human myotonic dystrophy kinase and is required for the
control of cell shape and proliferation. Genes Dev. 9:534–546.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tapon N, Harvey KF, Bell DW, Wahrer DC,
Schiripo TA, Haber D and Hariharan IK: Salvador promotes both cell
cycle exit and apoptosis in Drosophila and is mutated in human
cancer cell lines. Cell. 110:467–478. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Udan RS, Kango-Singh M, Nolo R, Tao C and
Halder G: Hippo promotes proliferation arrest and apoptosis in the
Salvador/Warts pathway. Nat Cell Biol. 5:914–920. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lai ZC, Wei X, Shimizu T, Ramos E,
Rohrbaugh M, Nikolaidis N, Ho LL and Li Y: Control of cell
proliferation and apoptosis by mob as tumor suppressor, mats. Cell.
120:675–685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang J, Wu S, Barrera J, Matthews K and
Pan D: The Hippo signaling pathway coordinately regulates cell
proliferation and apoptosis by inactivating Yorkie, the Drosophila
homolog of YAP. Cell. 122:421–434. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goulev Y, Fauny JD, Gonzalez-Marti B,
Flagiello D, Silber J and Zider A: SCALLOPED interacts with YORKIE,
the nuclear effector of the hippo tumor-suppressor pathway in
Drosophila. Curr Biol. 18:435–441. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu
J, Lin JD, Wang CY, Chinnaiyan AM, et al: TEAD mediates
YAP-dependent gene induction and growth control. Genes Dev.
22:1962–1971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hilman D and Gat U: The evolutionary
history of YAP and the hippo/YAP pathway. Mol Biol Evol.
28:2403–2417. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao B, Li L and Guan KL: Hippo signaling
at a glance. J Cell Sci. 123:4001–4006. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rauskolb C, Pan G, Reddy BV, Oh H and
Irvine KD: Zyxin links fat signaling to the hippo pathway. PLoS
Biol. 9:e10006242011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bryant PJ, Huettner B, Held LI Jr, Ryerse
J and Szidonya J: Mutations at the fat locus interfere with cell
proliferation control and epithelial morphogenesis in Drosophila.
Dev Biol. 129:541–554. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Poernbacher I, Baumgartner R, Marada SK,
Edwards K and Stocker H: Drosophila Pez acts in Hippo signaling to
restrict intestinal stem cell proliferation. Curr Biol. 22:389–396.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hamaratoglu F, Willecke M, Kango-Singh M,
Nolo R, Hyun E, Tao C, Jafar-Nejad H and Halder G: The
tumour-suppressor genes NF2/Merlin and Expanded act through Hippo
signalling to regulate cell proliferation and apoptosis. Nat Cell
Biol. 8:27–36. 2006. View Article : Google Scholar
|
|
29
|
Zhao B, Tumaneng K and Guan KL: The Hippo
pathway in organ size control, tissue regeneration and stem cell
self-renewal. Nat Cell Biol. 13:877–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu J, Zheng Y, Dong J, Klusza S, Deng WM
and Pan D: Kibra functions as a tumor suppressor protein that
regulates Hippo signaling in conjunction with Merlin and Expanded.
Dev Cell. 18:288–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harvey KF, Zhang X and Thomas DM: The
Hippo pathway and human cancer. Nat Rev Cancer. 13:246–257. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robinson BS, Huang J, Hong Y and Moberg
KH: Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila
via the FERM-domain protein Expanded. Curr Biol. 20:582–590. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng Z, Moroishi T and Guan KL: Mechanisms
of Hippo pathway regulation. Genes Dev. 30:1–7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun S and Irvine KD: Cellular organization
and cytoskeletal regulation of the Hippo signaling network. Trends
Cell Biol. 26:694–704. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tyler DM and Baker NE: Expanded and fat
regulate growth and differentiation in the Drosophila eye through
multiple signaling pathways. Dev Biol. 305:187–201. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Willecke M, Hamaratoglu F, Kango-Singh M,
Udan R, Chen CL, Tao C, Zhang X and Halder G: The fat cadherin acts
through the Hippo tumor-suppressor pathway to regulate tissue size.
Curr Biol. 16:2090–2100. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCartney BM, Kulikauskas RM, LaJeunesse
DR and Fehon RG: The neurofibromatosis-2 homologue, Merlin, and the
tumor suppressor expanded function together in Drosophila to
regulate cell proliferation and differentiation. Development.
127:1315–1324. 2000.PubMed/NCBI
|
|
38
|
Baumgartner R, Poernbacher I, Buser N,
Hafen E and Stocker H: The WW domain protein Kibra acts upstream of
Hippo in Drosophila. Dev Cell. 18:309–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tikoo A, Varga M, Ramesh V, Gusella J and
Maruta H: An anti-Ras function of neurofibromatosis type 2 gene
product (NF2/Merlin). J Biol Chem. 269:23387–23390. 1994.PubMed/NCBI
|
|
40
|
Yi C and Kissil JL: Merlin in organ size
control and tumorigenesis: Hippo versus EGFR? Genes Dev.
24:1673–1679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen CL, Gajewski KM, Hamaratoglu F,
Bossuyt W, Sansores-Garcia L, Tao C and Halder G: The apical-basal
cell polarity determinant Crumbs regulates Hippo signaling in
Drosophila. Proc Natl Acad Sci USA. 107:15810–15815. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Edgar BA: From cell structure to
transcription: Hippo forges a new path. Cell. 124:267–273. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Avruch J, Zhou D, Fitamant J and Bardeesy
N: Mst1/2 signalling to Yap: gatekeeper for liver size and tumour
development. Br J Cancer. 104:24–32. 2011. View Article : Google Scholar :
|
|
44
|
Wu S, Huang J, Dong J and Pan D: Hippo
encodes a Ste-20 family protein kinase that restricts cell
proliferation and promotes apoptosis in conjunction with salvador
and warts. Cell. 114:445–456. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chan EH, Nousiainen M, Chalamalasetty RB,
Schäfer A, Nigg EA and Silljé HH: The Ste20-like kinase Mst2
activates the human large tumor suppressor kinase Lats1. Oncogene.
24:2076–2086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Callus BA, Verhagen AM and Vaux DL:
Association of mammalian sterile twenty kinases, Mst1 and Mst2,
with hSalvador via C-terminal coiled-coil domains, leads to its
stabilization and phosphorylation. FEBS J. 273:4264–4276. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Pei J, Xia H, Ke H, Wang H and Tao
W: Lats2, a putative tumor suppressor, inhibits G1/S transition.
Oncogene. 22:4398–4405. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xia H, Qi H, Li Y, Pei J, Barton J,
Blackstad M, Xu T and Tao W: LATS1 tumor suppressor regulates G2/M
transition and apoptosis. Oncogene. 21:1233–1241. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang X, Li DM, Chen W and Xu T: Human
homologue of Drosophila lats, LATS1, negatively regulate growth by
inducing G(2)/M arrest or apoptosis. Oncogene. 20:6516–6523. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pan D: The Hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao B, Li L, Lei Q and Guan KL: The
Hippo-YAP pathway in organ size control and tumorigenesis: an
updated version. Genes De. 24:862–874. 2010. View Article : Google Scholar
|
|
52
|
Zhang X, George J, Deb S, Degoutin JL,
Takano EA, Fox SB, Bowtell DD and Harvey KF; AOC S Study group: The
Hippo pathway transcriptional co-activator, YAP, is an ovarian
cancer oncogene. Oncogene. 30:2810–2822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sudol M: Yes-associated protein (YAP65) is
a proline-rich phosphoprotein that binds to the SH3 domain of the
Yes proto-oncogene product. Oncogene. 9:2145–2152. 1994.PubMed/NCBI
|
|
54
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in Drosophila and
mammals. Cell. 130:1120–1133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zender L, Spector MS, Xue W, Flemming P,
Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et
al: Identification and validation of oncogenes in liver cancer
using an integrative oncogenomic approach. Cell. 125:1253–1267.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Overholtzer M, Zhang J, Smolen GA, Muir B,
Li W, Sgroi DC, Deng CX, Brugge JS and Haber DA: Transforming
properties of YAP, a candidate oncogene on the chromosome 11q22
amplicon. Proc Natl Acad Sci USA. 103:12405–12410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen L, Chan SW, Zhang X, Walsh M, Lim CJ,
Hong W and Song H: Structural basis of YAP recognition by TEAD4 in
the Hippo pathway. Genes Dev. 24:290–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Oh H and Irvine KD: In vivo regulation of
Yorkie phosphorylation and localization. Development.
135:1081–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Oh H and Irvine KD: Yorkie: the final
destination of Hippo signaling. Trends Cell Biol. 20:410–417. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim
J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP
oncoprotein by the Hippo pathway is involved in cell contact
inhibition and tissue growth control. Genes Dev. 21:2747–2761.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kanai F, Marignani PA, Sarbassova D, Yagi
R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley
LC, et al: TAZ: a novel transcriptional co-activator regulated by
interactions with 14-3-3 and PDZ domain proteins. EMBO J.
19:6778–6791. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yaffe MB, Rittinger K, Volinia S, Caron
PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ and Cantley LC: The
structural basis for 14-3-3:phosphopeptide binding specificity.
Cell. 91:961–971. 1997. View Article : Google Scholar
|
|
63
|
Lei QY, Zhang H, Zhao B, Zha ZY, Bai F,
Pei XH, Zhao S, Xiong Y and Guan KL: TAZ promotes cell
proliferation and epithelial-mesenchymal transition and is
inhibited by the Hippo pathway. Mol Cell Biol. 28:2426–2436. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Basu S, Totty NF, Irwin MS, Sudol M and
Downward J: Akt phosphorylates the Yes-associated protein, YAP, to
induce interaction with 14-3-3 and attenuation of p73-mediated
apoptosis. Mol Cell. 11:11–23. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhao B, Li L, Tumaneng K, Wang CY and Guan
KL: A coordinated phosphorylation by Lats and CK1 regulates YAP
stability through SCF(beta-TRCP). Genes Dev. 24:72–85. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu CY, Zha ZY, Zhou X, Zhang H, Huang W,
Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al: The Hippo tumor
pathway promotes TAZ degradation by phosphorylating a phosphodegron
and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem.
285:37159–37169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Camargo FD, Gokhale S, Johnnidis JB, Fu D,
Bell GW, Jaenisch R and Brummelkamp TR: YAP1 increases organ size
and expands undifferentiated progenitor cells. Curr Biol.
17:2054–2060. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Da CL, Xin Y, Zhao J and Luo XD:
Significance and relationship between Yes-associated protein and
survivin expression in gastric carcinoma and precancerous lesions.
World J Gastroenterol. 15:4055–4061. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang X, Su L and Ou Q: Yes-associated
protein promotes tumour development in luminal epithelial derived
breast cancer. Eur J Cancer. 48:1227–1234. 2012. View Article : Google Scholar
|
|
70
|
Lam-Himlin DM, Daniels JA, Gayyed MF, Dong
J, Maitra A, Pan D, Montgomery EA and Anders RA: The Hippo pathway
in human upper gastrointestinal dysplasia and carcinoma: a novel
oncogenic pathway. Int J Gastrointest Cancer. 37:103–109. 2006.
|
|
71
|
Wada K, Itoga K, Okano T, Yonemura S and
Sasaki H: Hippo pathway regulation by cell morphology and stress
fibers. Development. 138:3907–3914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Straßburger K, Tiebe M, Pinna F, Breuhahn
K and Teleman AA: Insulin/IGF signaling drives cell proliferation
in part via Yorkie/YAP. Dev Biol. 367:187–196. 2012. View Article : Google Scholar
|
|
73
|
Yu FX, Zhao B, Panupinthu N, Jewell JL,
Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al:
Regulation of the Hippo-YAP pathway by G-protein-coupled receptor
signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fan R, Kim NG and Gumbiner BM: Regulation
of Hippo pathway by mitogenic growth factors via phosphoinositide
3-kinase and phosphoinositide-dependent kinase-1. Proc Natl Acad
Sci USA. 110:2569–2574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kikuchi A, Yamamoto H and Sato A:
Selective activation mechanisms of Wnt signaling pathways. Trends
Cell Biol. 19:119–129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
He X, Semenov M, Tamai K and Zeng X: LDL
receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling:
arrows point the way. Development. 131:1663–1677. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hoffmeyer K, Raggioli A, Rudloff S, Anton
R, Hierholzer A, Del Valle I, Hein K, Vogt R and Kemler R:
Wnt/β-catenin signaling regulates telomerase in stem cells and
cancer cells. Science. 336:1549–1554. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ouyang H, Zhuo Y and Zhang K: WNT
signaling in stem cell differentiation and tumor formation. J Clin
Invest. 123:1422–1424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xing Y, Clements WK, Kimelman D and Xu W:
Crystal structure of a beta-catenin/axin complex suggests a
mechanism for the beta-catenin destruction complex. Genes Dev.
17:2753–2764. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Habas R and Dawid IB: Dishevelled and Wnt
signaling: is the nucleus the final frontier? J Biol. 4:22005.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Tolwinski NS, Wehrli M, Rives A, Erdeniz
N, DiNardo S and Wieschaus E: Wg/Wnt signal can be transmitted
through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta
activity. Dev Cell. 4:407–418. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa
N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al:
AXIN1 mutations in hepatocellular carcinomas, and growth
suppression in cancer cells by virus-mediated transfer of AXIN1.
Nat Genet. 24:245–250. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
84
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
85
|
Cong F, Schweizer L and Varmus H: Wnt
signals across the plasma membrane to activate the beta-catenin
pathway by forming oligomers containing its receptors, Frizzled and
LRP. Development. 131:5103–5115. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cliffe A, Hamada F and Bienz M: A role of
Dishevelled in relocating Axin to the plasma membrane during
wingless signaling. Curr Biol. 13:960–966. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Saito-Diaz K, Chen TW, Wang X, Thorne CA,
Wallace HA, Page-McCaw A and Lee E: The way Wnt works: components
and mechanism. Growth Factors. 31:1–31. 2013. View Article : Google Scholar :
|
|
88
|
Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP and Li
L: Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading
to stabilization of beta-catenin-TCF interaction. J Cell Biol.
180:1087–1100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Itoh K, Brott BK, Bae GU, Ratcliffe MJ and
Sokol SY: Nuclear localization is required for Dishevelled function
in Wnt/beta-catenin signaling. J Biol. 4:32005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Azzolin L, Zanconato F, Bresolin S,
Forcato M, Basso G, Bicciato S, Cordenonsi M and Piccolo S: Role of
TAZ as mediator of Wnt signaling. Cell. 151:1443–1456. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Heallen T, Zhang M, Wang J,
Bonilla-Claudio M, Klysik E, Johnson RL and Martin JF: Hippo
pathway inhibits Wnt signaling to restrain cardiomyocyte
proliferation and heart size. Science. 332:458–461. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Willert K, Shibamoto S and Nusse R:
Wnt-induced dephosphorylation of axin releases beta-catenin from
the axin complex. Genes Dev. 13:1768–1773. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li VS, Ng SS, Boersema PJ, Low TY,
Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi
T, et al: Wnt signaling through inhibition of β-catenin degradation
in an intact Axin1 complex. Cell. 149:1245–1256. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Konsavage WM Jr, Kyler SL, Rennoll SA, Jin
G and Yochum GS: Wnt/β-catenin signaling regulates Yes-associated
protein (YAP) gene expression in colorectal carcinoma cells. J Biol
Chem. 287:11730–11739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Azzolin L, Panciera T, Soligo S, Enzo E,
Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V,
et al: YAP/TAZ incorporation in the β-catenin destruction complex
orchestrates the Wnt response. Cell. 158:157–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang WH, McNatt LG, Pang IH, Millar JC,
Hellberg PE, Hellberg MH, Steely HT, Rubin JS, Fingert JH,
Sheffield VC, et al: Increased expression of the WNT antagonist
sFRP-1 in glaucoma elevates intraocular pressure. J Clin Invest.
118:1056–1064. 2008.PubMed/NCBI
|
|
97
|
Morgan JT, Raghunathan VK, Chang YR,
Murphy CJ and Russell P: Wnt inhibition induces persistent
increases in intrinsic stiffness of human trabecular meshwork
cells. Exp Eye Res. 132:174–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kwon HS, Lee HS, Ji Y, Rubin JS and
Tomarev SI: Myocilin is a modulator of Wnt signaling. Mol Cell
Biol. 29:2139–2154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tovar-Vidales T, Roque R, Clark AF and
Wordinger RJ: Tissue transglutaminase expression and activity in
normal and glaucomatous human trabecular meshwork cells and
tissues. Invest Ophthalmol Vis Sci. 49:622–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Choi JW, Herr DR, Noguchi K, Yung YC, Lee
CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, et al: LPA
receptors: subtypes and biological actions. Annu Rev Pharmacol
Toxicol. 50:157–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
van Corven EJ, Groenink A, Jalink K,
Eichholtz T and Moolenaar WH: Lysophosphatidate-induced cell
proliferation: identification and dissection of signaling pathways
mediated by G proteins. Cell. 59:45–54. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Shida D, Kitayama J, Yamaguchi H, Okaji Y,
Tsuno NH, Watanabe T, Takuwa Y and Nagawa H: Lysophosphatidic acid
(LPA) enhances the metastatic potential of human colon carcinoma
DLD1 cells through LPA1. Cancer Res. 63:1706–1711. 2003.PubMed/NCBI
|
|
103
|
Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W,
Zhang F, Yu S, Stephens LC, Cui X, Murrow G, et al: Expression of
autotaxin and lysophosphatidic acid receptors increases mammary
tumorigenesis, invasion, and metastases. Cancer Cell. 15:539–550.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Rohen JW: Why is intraocular pressure
elevated in chronic simple glaucoma? Anatomical considerations.
Ophthalmology. 90:758–765. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
No authors listed. The Advanced Glaucoma
Intervention Study (AGIS): 7. The relationship between control of
intraocular pressure and visual field deterioration. The AGIS
Investigators. Am J Ophthalmol. 130:429–440. 2000. View Article : Google Scholar
|
|
106
|
Gasiorowski JZ and Russell P: Biological
properties of trabecular meshwork cells. Exp Eye Res. 88:671–675.
2009. View Article : Google Scholar
|
|
107
|
Iyer P, Lalane R III, Morris C, Challa P,
Vann R and Rao PV: Autotaxin-lysophosphatidic acid axis is a novel
molecular target for lowering intraocular pressure. PLoS One.
7:e426272012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Li AF, Tane N and Roy S: Fibronectin
overexpression inhibits trabecular meshwork cell monolayer
permeability. Mol Vis. 10:750–757. 2004.PubMed/NCBI
|
|
109
|
Willier S, Butt E and Grunewald TG:
Lysophosphatidic acid (LPA) signalling in cell migration and cancer
invasion: a focussed review and analysis of LPA receptor gene
expression on the basis of more than 1700 cancer microarrays. Biol
Cell. 105:317–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
De Larco JE and Todaro GJ: Growth factors
from murine sarcoma virus-transformed cells. Proc Natl Acad Sci
USA. 75:4001–4005. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Todaro GJ and De Larco JE: Growth factors
produced by sarcoma virus-transformed cells. Cancer Res.
38:4147–4154. 1978.PubMed/NCBI
|
|
112
|
Roberts AB, Lamb LC, Newton DL, Sporn MB,
De Larco JE and Todaro GJ: Transforming growth factors: isolation
of polypeptides from virally and chemically transformed cells by
acid/ethanol extraction. Proc Natl Acad Sci USA. 77:3494–3498.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Pena RA, Jerdan JA and Glaser BM: Effects
of TGF-beta and TGF-beta neutralizing antibodies on
fibroblast-induced collagen gel contraction: implications for
proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci.
35:2804–2808. 1994.PubMed/NCBI
|
|
114
|
Border WA, Noble NA, Yamamoto T, Harper
JR, Yamaguchi Yu, Pierschbacher MD and Ruoslahti E: Natural
inhibitor of transforming growth factor-beta protects against
scarring in experimental kidney disease. Nature. 360:361–364. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zode GS, Sethi A, Brun-Zinkernagel AM,
Chang IF, Clark AF and Wordinger RJ: Transforming growth factor-β2
increases extracellular matrix proteins in optic nerve head cells
via activation of the Smad signaling pathway. Mol Vis.
17:1745–1758. 2011.
|
|
116
|
Itoh S, Itoh F, Goumans MJ and Ten Dijke
P: Signaling of transforming growth factor-beta family members
through Smad proteins. Eur J Biochem. 267:6954–6967. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Dupont J, McNeilly J, Vaiman A, Canepa S,
Combarnous Y and Taragnat C: Activin signaling pathways in ovine
pituitary and LbetaT2 gonadotrope cells. Biol Reprod. 68:1877–1887.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Chen HB, Shen J, Ip YT and Xu L:
Identification of phosphatases for Smad in the BMP/DPP pathway.
Genes Dev. 20:648–653. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Eisenstein R and Grant-Bertacchini D:
Growth inhibitory activities in avascular tissues are recognized by
anti-transforming growth factor beta antibodies. Curr Eye Res.
10:157–162. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Tripathi RC, Li J, Chan WF and Tripathi
BJ: Aqueous humor in glaucomatous eyes contains an increased level
of TGF-beta 2. Exp Eye Res. 59:723–727. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Inatani M, Tanihara H, Katsuta H, Honjo M,
Kido N and Honda Y: Transforming growth factor-beta 2 levels in
aqueous humor of glaucomatous eyes. Graefes Arch Clin Exp
Ophthalmol. 239:109–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Pervan CL, Lautz JD, Blitzer AL, Langert
KA and Stubbs EB Jr: Rho GTPase signaling promotes constitutive
expression and release of TGF-β2 by human trabecular meshwork
cells. Exp Eye Res. 146:95–102. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Rao PV, Deng PF, Kumar J and Epstein DL:
Modulation of aqueous humor outflow facility by the Rho
kinase-specific inhibitor Y-27632. Invest Ophthalmol Vis Sci.
42:1029–1037. 2001.PubMed/NCBI
|
|
124
|
Inoue T and Tanihara H: Rho-associated
kinase inhibitors: a novel glaucoma therapy. Prog Retin Eye Res.
37:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Takai Y, Tanito M and Ohira A: Multiplex
cytokine analysis of aqueous humor in eyes with primary open-angle
glaucoma, exfoliation glaucoma, and cataract. Invest Ophthalmol Vis
Sci. 53:241–247. 2012. View Article : Google Scholar
|
|
126
|
Li J, Tripathi BJ and Tripathi RC:
Modulation of pre-mRNA splicing and protein production of
fibronectin by TGF-beta2 in porcine trabecular cells. Invest
Ophthalmol Vis Sci. 41:3437–3443. 2000.PubMed/NCBI
|
|
127
|
Wordinger RJ, Clark AF, Agarwal R, Lambert
W, McNatt L, Wilson SE, Qu Z and Fung BK: Cultured human trabecular
meshwork cells express functional growth factor receptors. Invest
Ophthalmol Vis Sci. 39:1575–1589. 1998.PubMed/NCBI
|
|
128
|
Tamm ER, Siegner A, Baur A and
Lütjen-Drecoll E: Transforming growth factor-beta 1 induces
alpha-smooth muscle-actin expression in cultured human and monkey
trabecular meshwork. Exp Eye Res. 62:389–397. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Varelas X, Sakuma R, Samavarchi-Tehrani P,
Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW and Wrana JL:
TAZ controls Smad nucleocytoplasmic shuttling and regulates human
embryonic stem-cell self-renewal. Nat Cell Biol. 10:837–848. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Quigley HA and Broman AT: The number of
people with glaucoma worldwide in 2010-2020. Br J Ophthalmol.
90:262–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Tamm ER: The trabecular meshwork outflow
pathways: structural and functional aspects. Exp Eye Res.
88:648–655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Last JA, Pan T, Ding Y, Reilly CM, Keller
K, Acott TS, Fautsch MP, Murphy CJ and Russell P: Elastic modulus
determination of normal and glaucomatous human trabecular meshwork.
Invest Ophthalmol Vis Sci. 52:2147–2152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Dupont S, Morsut L, Aragona M, Enzo E,
Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M,
Bicciato S, et al: Role of YAP/TAZ in mechanotransduction. Nature.
474:179–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Raghunathan VK, Morgan JT, Dreier B,
Reilly CM, Thomasy SM, Wood JA, Ly I, Tuyen BC, Hughbanks M, Murphy
CJ, et al: Role of substratum stiffness in modulating genes
associated with extracellular matrix and mechanotransducers YAP and
TAZ. Invest Ophthalmol Vis Sci. 54:378–386. 2013. View Article : Google Scholar :
|
|
135
|
Comes N, Buie LK and Borras T: Evidence
for a role of angiopoietin-like 7 (ANGPTL7) in extracellular matrix
formation of the human trabecular meshwork: implications for
glaucoma. Genes Cells. 16:243–259. 2011. View Article : Google Scholar : PubMed/NCBI
|