Introduction
Preeclampsia (PE) is a pregnancy-specific
complication associated with maternal and fetal morbidity and
mortality; however, the exact etiology of this disease is not yet
clearly understood (1). Until
recently, PE has generally been described to be initiated by
reduced placental perfusion, followed by impaired placentation,
which in turn results in the release of molecules and factors
leading to systemic endothelial activation and maternal clinical
manifestations (1,2).
Placental extracellular vesicles, including
microvesicles, exosomes and syncytiotrophoblast-derived
microparticles, can interact with endothelial cells and contribute
to the development of PE (3,4).
Exosomes are nanovesicles (30–100 nm) that contain a wide range of
functional proteins, mRNAs and microRNAs (miRNAs/miRs), and can be
viewed as mediators of intercellular communication without the need
for cell-cell contact (5,6). There has been growing interest in
exosomal miRNAs, since exosomal miRNA-mediated signaling has been
reported to serve a significant role in various cell functions,
including immune cell proliferation and endothelial cell migration
(7,8).
miRNAs are known to suppress gene expression
post-transcription by binding the 3′-untranslated region (3′-UTR)
of target mRNA. Numerous miRNAs have been demonstrated to affect
placentation (9,10), and miRNAs expressed in the
placenta can be transferred into the maternal circulation via
exosomes during human pregnancy (11). miR-155 is a well-known miRNA that
possesses inflammatory and anti-angiogenic functions (12,13). Our previous study, and other
reported research, demonstrated that miR-155 is upregulated in the
plasma and placentas from patients with PE, thus suggesting that
miR-155 is associated with the pathogenesis of PE (14,15).
Reduced nitric oxide (NO) bioavailability is
involved in the pathophysiological alterations of PE, and decreased
endothelial nitric oxide synthase (eNOS) expression has been
detected in the placentas and sera of patients with PE (16,17). Our previous study indicated that
miR-155 inhibits endothelium-dependent vasorelaxation by
suppressing eNOS expression in human umbilical vein endothelial
cells (HUVECs) (18). Therefore,
it may be hypothesized that upregulation of placenta-associated
serum exosomal miR-155 suppresses the expression of eNOS in
endothelial cells during PE development.
The present study initially isolated and identified
placenta-associated exosomes from the maternal sera of patients
with PE and normal pregnant women. Subsequently, the effects of
placenta-associated serum exosomes from patients with PE on NO
production and eNOS expression in HUVECs were determined. In
addition, the expression levels of miR-155 in these exosomes were
detected. Finally, the expression of eNOS in endothelial cells
treated with miR-155-overexpressed exosomes from BeWo cells was
investigated.
Materials and methods
Human serum samples
Eligible participants were enrolled from the Nanjing
Drum Tower Hospital (Nanjing, China) from May 1, 2015 to April 30,
2016. A total of 20 samples were obtained, 10 from patients with PE
and 10 from gestational age-matched normal pregnant women (Table I). The diagnostic criteria of PE
are described by Cunningham et al (19). Pregnant patients with PE alongside
chronic hypertension, cardiovascular diseases, gestational diabetes
mellitus or chronic nephritis were excluded from the present study.
Sera samples (2 ml) were obtained from each pregnant woman prior to
treatment and were maintained at −80°C. The present study was
approved by the Scientific Research Ethics Committee of the Drum
Tower Hospital (2009041), and informed consent was obtained from
all of the participants.
 | Table IClinical characteristics of patients
with PE and normal pregnant women. |
Table I
Clinical characteristics of patients
with PE and normal pregnant women.
|
Characteristica | Normal (n=10) | PE (n=10) | P-value |
|---|
| Maternal age
(years) | 27.56±3.21 | 29.11±5.01 | n.s. |
| Gestational age
(weeks) | 39.14±0.66 | 38.30±2.15 | n.s. |
| Systolic BP
(mmHg)b | 120.11±8.78 | 149.56±17.17 | <0.05 |
| Diastolic BP
(mmHg)b | 80.22±8.50 | 97.67±6.95 | <0.05 |
| Proteinuriab | – | + – +++ | <0.05 |
| AST (U/l)b | 10.28±1.87 | 12.42±3.51 | n.s. |
| ALT (U/l)b | 18.75±2.96 | 24.13±7.60 | n.s. |
| Cr
(µmol/l)b | 49.5±7.05 | 42.6±1.34 | n.s. |
| Platelets
(×109/l)b | 194.57±56.73 | 187.8±32.99 | n.s. |
| Fetal weight
(g) |
3,252.22±377.97 |
2,822.78±400.67 | <0.05 |
| Placenta weight
(g) | 630.00±76.03 | 548.33±56.01 | <0.05 |
Cell lines
BeWo cells (GDC072) were purchased from the China
Center for Type Culture Collection (CCTCC, Wuhan, China). Cells
were maintained at 37°C in a humidified incubator containing 5%
CO2 and were cultured in RPMI-1640 medium (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS; HyClone; GE Healthcare
Life Sciences), 100 U/ml penicillin and 100 µg/ml
streptomycin. COS-7 cells (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) were cultured as described previously (18).
HUVEC isolation
Primary HUVECs were isolated from the vascular wall
of umbilical veins obtained from normal pregnant women in the third
trimester (not from the normal group). HUVECs were extracted by
treatment of umbilical veins with 0.2% collagenase I (Worthington
Biochemical Corporation, Lakewood, NJ, USA) for 10 min at 37°C
(20); the cells were then seeded
into 0.15% gelatin-coated plates (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and were cultured for no more than six
generations in M199 medium (HyClone; GE Healthcare Life Sciences)
containing 20% FBS and antibiotics (100 U/ml penicillin and 100
µg/ml streptomycin) at 37°C in a humidified incubator
containing 5% CO2.
Adenovirus and plasmid construction
pcDNA3-eNOS-3′-UTR plasmid and Ad-miR-155 adenovirus
were constructed as previously described (18). Ad-LacZ served as the control for
Ad-miR-155 infection. The COS-7 cells were plated and cultured
overnight in the 6-well plates and at 70–80% confluence, the cells
were transfected with pcDNA-eNOS-3′-UTR (2.0 µg) using
Lipofectamine 2000 transfection reagent (Life Technologies, Grand
Island, NY, USA). The relative intensity of eNOS protein expression
was assessed by ImageJ 2X software (National Institutes of Health,
Bethesda, MD, USA).
Exosome isolated from human peripheral
sera
Exosomes were obtained by a series of centrifugation
steps, combined with polyethylene glycol (PEG) precipitation.
Briefly, serum (200 µl) was centrifuged at 3,000 × g for 20
min at 4°C, after which the supernatant was passed through a
0.22-µm filter and was incubated with 8% PEG 6000 overnight
(21,22). Following further centrifugation at
10,000 × g for 1 h at 4°C, the 8% PEG 6000-serum mixture
precipitate was resuspended in 0.25 M sucrose. Subsequently, the
suspension was layered onto a linear sucrose gradient,
ultracentrifuged at 100,000 × g for 5 h at 4°C and divided into six
fractions: F1 (supernatant), 1.03; F2, 1.06; F3, 1.09; F4, 1.11;
F5, 1.14; and F6, 1.18 g/ml. These fractions were incubated with 8%
PEG 6000 overnight and were then centrifuged at 10,000 × g for 1 h
at 4°C. Finally, the exosomes were suspended in 200 µl PBS.
An equal volume of sample from each fraction was used for western
blotting. Sonicated exosomes were the control group since the
sonicate would damage the exosomes.
Exosome isolation from culture
medium
BeWo cells (1.5×106) were plated into a
100-mm plate, infected with Ad-miR-155 or Ad-LacZ (100 multiplicity
of infection) for 8 h at 37°C and were then incubated in medium
containing 2% exosome-free FBS for a further 48 h at 37°C. The
culture supernatants were sequentially centrifuged at 300 × g for
10 min at 4°C, 2,000 × g for 10 min at 4°C and 10,000 × g for 30
min at 4°C. Subsequently, the cleared supernatants were
ultracentrifuged at 100,000 × g for 70 min at 4°C and the pellets
were collected. Finally, the pellet suspension was ultracentrifuged
for a second time at 100,000 × g for 70 min at 4°C and the pellets
were resuspended in 200 µl PBS (23).
Primary HUVECs (1.0×105) in 12-well
plates were incubated with serum exosomes or with exosomes obtained
from BeWo cell supernatants (5.0 µg protein/ml) in
exosome-free medium for the indicated durations.
Transmission electron microscopy
(TEM)
Exosome-TEM-easy kit (cat. no. P130; Seajet
Scientific, Inc., Beijing, China) was used to detect exosomes using
Formvar/carbon-coated TEM grids (24). Freshly isolated exosome suspension
(5–10 µl) was dropped onto clean parafilm. Subsequently, the
grids were floated on the drop with the coated side facing the
suspension; the grid membranes were allowed to absorb exosomes for
10 min. The grids were then transferred to the wash buffer for 30
sec, and were washed twice. The grids were then transferred to EM
solution for 10 min, and were washed twice. Finally, the grids were
transferred to filter paper, coated side up, and were air dried
overnight. The exosomes were visualized under TEM (JEM-1010; JEOL,
Ltd., Tokyo, Japan) and 10 images were analyzed from each
sample.
Dynamic light scattering (DLS)
DLS measurements were performed using a Zetasizer
3000-HA (Malvern Instruments Ltd., Malvern, UK). Exosome samples
were diluted in PBS and 0.05% Tween-20 to a total volume of 1.0 ml.
The samples were diluted with PBS (1:10, 1:100 and 1:1,000). The
samples with various dilutions in a total volume of 1.0 ml were
performed with standard settings (refractive index, 1.330;
viscosity, 0.89; temperature, 25°C) in the software.
Exosome labeling
Exosomes from BeWo cells or serum exosomes (5.0
µg protein/ml used to treat HUVECs in 12-well plates) were
resuspended in PBS (1 ml) and mixed with the green fluorescent dye
PKH67 (Sigma-Aldrich; Merck KGaA) in 1 ml Diluent C,
4×10−6 M (25). After
a 3 min incubation, 1% bovine serum albumin (2 ml) was added to
terminate the labeling reaction, and the exosomes were harvested
and washed twice with PBS by centrifugation (100,000 × g for 1 h).
The exosomes were resuspended in medium (1 ml) supplemented with
10% ultrasonicated FBS and were then added to HUVECs in 12-well
plates for 6 h incubation. The cells were washed three times with
PBS, fixed with 4% paraformaldehyde for 30 min at room temperature,
and were then stained with DAPI (1:5,000; Invitrogen; Thermo Fisher
Scientific, Inc.) for 5 min. After washing with PBS, the cells were
visualized under a fluorescence microscope (Leica DMR; Leica
Microsystems GmbH, Wetzlar, Germany).
Western blotting
Western blotting was performed according to standard
procedures (26). Equal amounts
of protein preparations were separated by SDS-PAGE, transferred to
polyvinylidene fluoride membranes, immunoblotted with primary
antibodies and visualized using a chemiluminescence detection kit
(EMD Millipore, Bedford, MA, USA). The primary antibodies were as
follows: Anti-eNOS (c-20; 1:1,000; sc-654) and anti-cluster of
differentiation (CD)81 (1.3.3.22; 1:1,000; sc-7637) (both from
Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-placental
alkaline phosphatase (PLAP; 1:1,000; AB21938b; Sangon Biotech Co.,
Ltd., Shanghai, China), anti-CD63 (K156; 1:1,000; BS3474) and
anti-α-tubulin (G436; 1:5,000; BS1699) (both from Bioworld
Technology Inc., St. Louis Park, MN, USA). For the examination of
CD63, CD81 and PLAP levels in the serum exosomes from patients with
PE, an equal volume of sample from each fraction was respectively
used for western blotting, referring to the study by Putz et
al (24). The expression
levels of CD81, CD63 and PLAP were detected to be the highest level
in the F3 fraction, indicating that placenta-associated serum
exosomes were mostly accumulated in that fraction. For the
examination of the levels of CD63, CD81 and PLAP in exosomes
obtained from cell medium, an equal total protein of exosomes from
the Ad-LacZ-infected cell medium and Ad-miR-155-infected cell
medium were loaded for western blotting. The protein concentrations
of the isolated exosomes from 2 groups were detected
(Ad-LacZ-exosome, 0.35 µg/µl; Ad-miR-155-exosome,
0.33 µg/µl). And the total protein (10 µg)
from each group was loaded for western blotting to detect the
expression levels of CD81, CD63 and PLAP.
Silver staining of polyacrylamide
gels
The main steps of silver staining following PAGE (at
room temperature) included: Fixing in acetic acid and ethanol
mixture (v/v, 4/1) for 30 min, permeabilization with 8 mM
Na2S2O3 for 30 min and staining
with 7.5 mM AgNO3 in 37% formaldehyde solution for 30
min. The bands would then be invisible when the gels were incubated
in 0.25 M Na2CO3 for 2–15 min. Finally, 50 mM
glycine solution was added to terminate the staining process for 10
min.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) and was reverse
transcribed into cDNA with primers in a 20 µl reaction using
a PrimerScript RT regent kit (Takara, Otsu, Japan). Total RNA was
extracted from exosomes using an Ambion mirVana PARIS kit (Ambion;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. miR-39 (1.6×108 copy/µl, 2 µl;
Qiagen, Hilden, Germany) was added to the exosomes prior to RNA
isolation.
qPCR assays were performed as follows. Briefly, 20
ng cDNA was applied to each qPCR reaction containing 2X SYBR-Green
PCR Master Mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and
500 nM forward and reverse primers in a MyiQ Single-Color RT-PCR
Detection system (Bio-Rad Laboratories, Inc.). RT-PCR for miR-155
was performed for 40 cycles (95°C for 10 sec; 60°C for 30 sec)
following a initial 3 min incubation at 95°C. RT-PCR for eNOS was
performed via the blow procedure (95°C for 3 min; 94°C for 10 sec;
60°C for 30 sec, 72°C for 30 sec; 40 cycles). The primers used are
listed in Table II. The data
obtained from three independent experiments were analyzed for
relative gene expression using the 2−ΔΔCq method
(27).
 | Table IIOligonucleotide primer sequences for
quantitative polymerase chain reaction. |
Table II
Oligonucleotide primer sequences for
quantitative polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| miR-155 |
CGTTAATGCTAATCGTGATAGG | GCAGGGTCCGAGGT |
| miR-39 TCACC | GGGTGTAAATCAG | GCAGGGTCCGAGGT |
| U6 | CTCGCTTCGGCAGCACA
AAC |
GCTTCACGAATTTGCGT |
| eNOS |
CCCTTCAGTGGCTGGTACAT CAC |
GATGGTGACTTTGGCTA |
| 18s rRNA |
CGGCTACCACATCCAAGGAA |
CTGGAATTACCGCGGCT |
NO detection
NO levels were detected in the exosome-free culture
medium of HUVECs treated with serum exosomes for 24 h. NO
concentration was measured using the NO detection kit (S0021;
Beyotime Institute of Biotechnology, Shanghai, China) according to
the manufacturer's protocol.
Statistical analysis
Normally distributed data were presented as the
means ± standard deviation using SPSS 17.0 software. The data
obtained from three independent experiments were analyzed.
Differences between means were analyzed using two-tailed Student's
t-test, and one-way analysis of variance was used to detect
differences among more than two groups. P<0.05 was considered to
indicate a statistically significant difference. The correlation
between miR-155 expression in placenta-associated serum exosomal
from patients with PE and eNOS protein expression in HUVECs treated
with placenta-associated serum exosomes was analyzed by GraphPad
Prism 5.
Results
Characteristics of placenta-associated
exosomes derived from the sera of patients with PE and normal
pregnant women
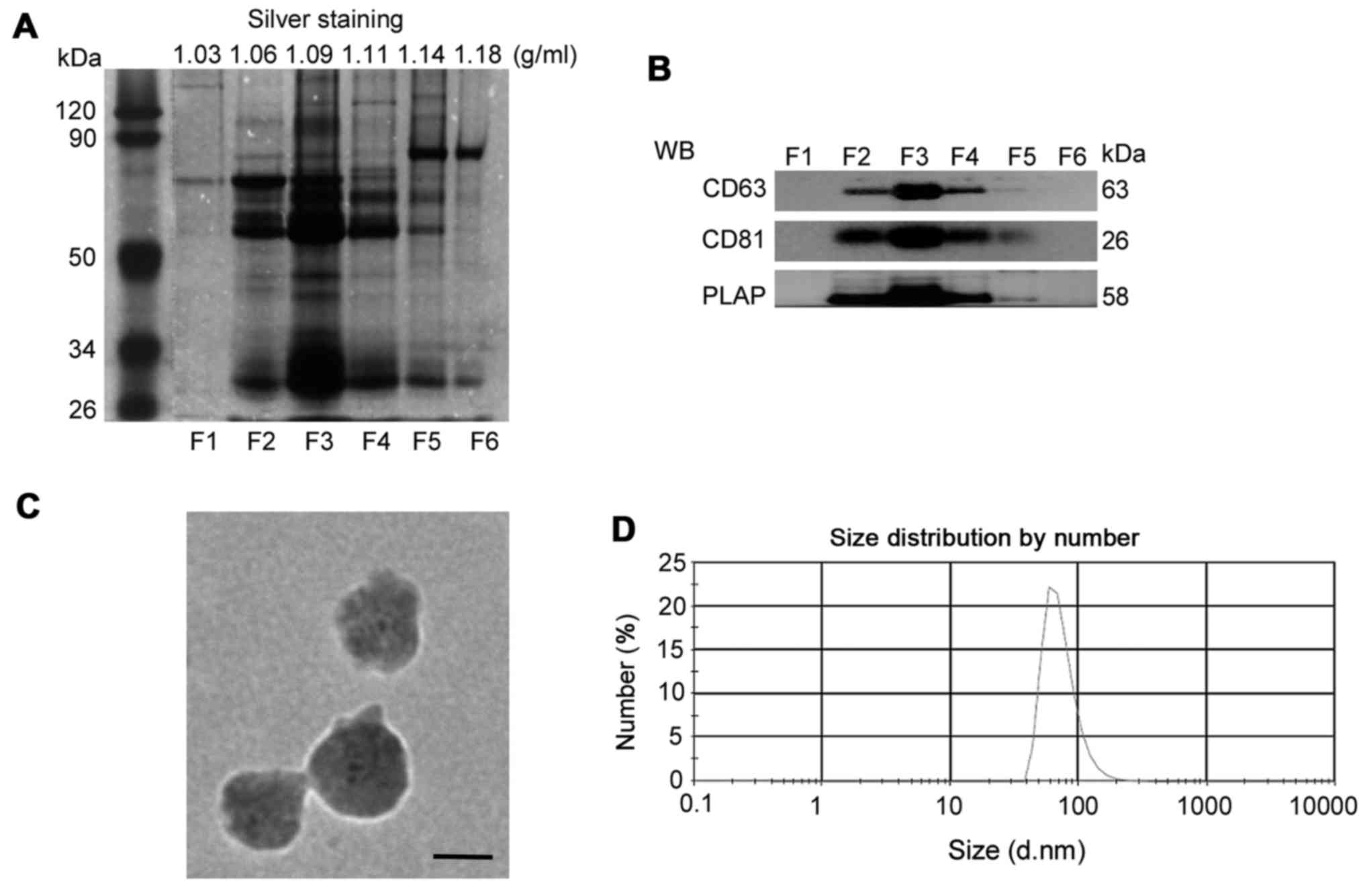
Using sucrose gradients, isolated pelleted vesicles
from the sera of patients with PE were separated into six fractions
(F1-F6) of different densities. As shown in Fig. 1A, the pelleted vesicles in F3
(1.09 g/ml) contained more bands than the other fractions, as
determined by silver staining. Since tetraspanins are known to be
expressed in exosomes (28), the
present study examined CD63 and CD81 expression in the isolated
serum fractions. As shown in Fig.
1B, CD63 and CD81 were detected in the pelleted vesicles in F2,
F3 and F4; the expression of these molecules was highest in F3.
Using PLAP as a marker for placental-derived exosomes (29), the present study confirmed that
the vesicles in F3 contained the highest expression of PLAP. In
addition, the present study measured the size of serum exosomes in
F3 via TEM and DLS analysis. As presented in Fig. 1C, the diameter of isolated
exosomes was ~100 nm. DLS analysis consistently demonstrated that
the diameter of serum exosomes in F3 was 73.74±27.56 nm (Fig. 1D). The characteristics of
placenta-associated serum exosomes from normal pregnant women are
shown in Fig. 2.
Collectively, these data indicated that isolated
vesicles in F3 derived from the peripheral sera of patients with PE
and normal pregnant women contained a considerable amount of
placenta-associated exosomes. The isolated vesicles in F3 were used
for subsequent experiments.
Placenta-associated serum exosomes from
patients with PE decrease NO production and eNOS expression in
HUVECs
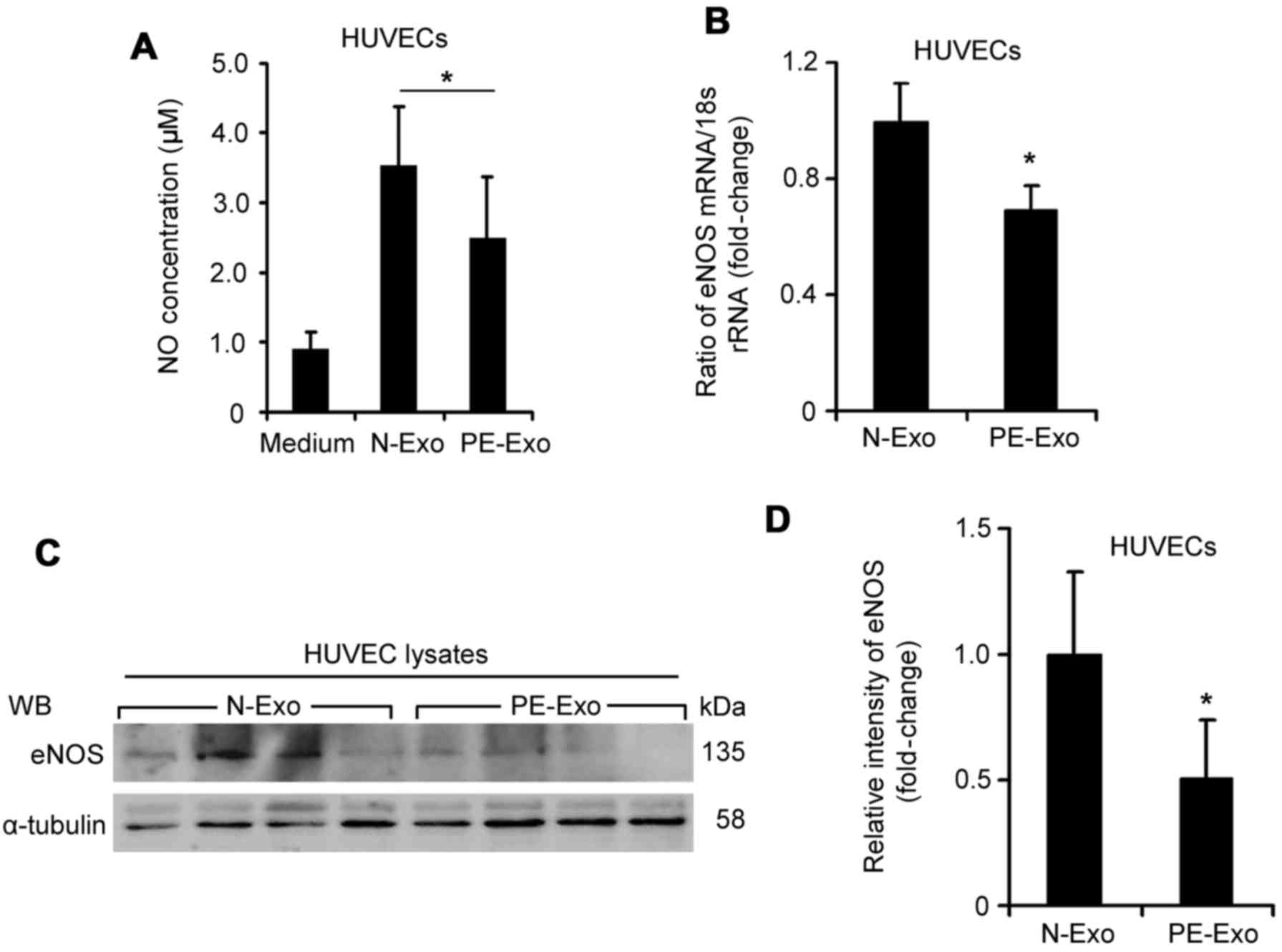
The present study investigated the effects of serum
exosomes on NO production and eNOS expression in HUVECs. Although
the morphology of serum exosomes from normal pregnant women was
similar to that of exosomes from patients with PE, the NO
concentration in HUVECs treated with placenta-associated serum
exosomes from patients with PE was significantly decreased
(2.48±0.89 µM vs. 3.53±0.88 µM, Fig. 3A; P<0.05) compared with those
from normal pregnant women. The mRNA expression levels of eNOS in
HUVECs incubated with placenta-associated serum exosomes from
patients with PE were reduced by 31.04% (Fig. 3B; P<0.05). In addition, the
protein expression levels of eNOS were significantly downregulated
in HUVECs incubated with placenta-associated serum exosomes from
patients with PE (Fig. 3C and D;
P<0.05). These data indicated that placenta-associated serum
exosomes from patients with PE may inhibit the expression of eNOS
and NO production in endothelial cells.
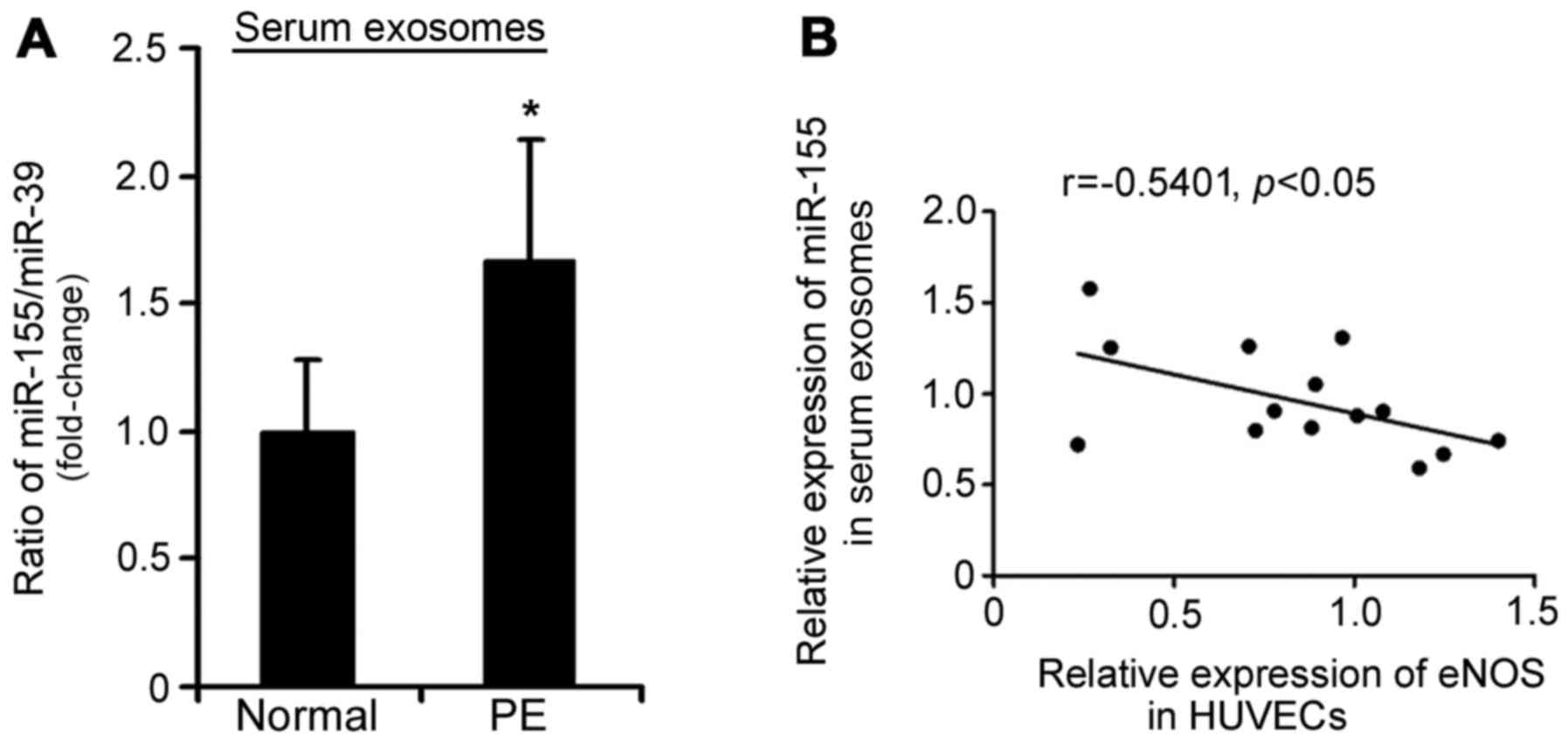
miR-155 expression is increased in
placenta-associated serum exosomes from patients with PE
Our previous study indicated that miR-155 is
upregulated in the placentas of patients with PE (19). Therefore, the present study aimed
to determine whether increased miR-155 expression was transferred
via exosomes from the placenta into maternal circulation during
pregnancy. Using RT-qPCR, it was demonstrated that miR-155
expression was increased in placenta-associated serum exosomes from
patients with PE compared with in those from normal pregnant women
(1.58-fold, P<0.05; Fig. 4A).
Furthermore, an inverse correlation existed between miR-155
expression in placenta-associated serum exosomes from patients with
PE and eNOS protein expression in HUVECs treated with
placenta-associated serum exosomes (r=−0.5401, P<0.05; Fig. 4B).
miR-155-overexpressed exosomes from
trophoblast cell medium inhibit eNOS expression in HUVECs in
vitro
BeWo cells were infected with either Ad-miR-155 or
Ad-LacZ, and exosomes were isolated from the culture medium. The
characteristics of cell medium exosomes were identified, as
follows: CD63, CD81 and PLAP were expressed in the isolated
exosomes from BeWo cell medium, as determined by western blotting;
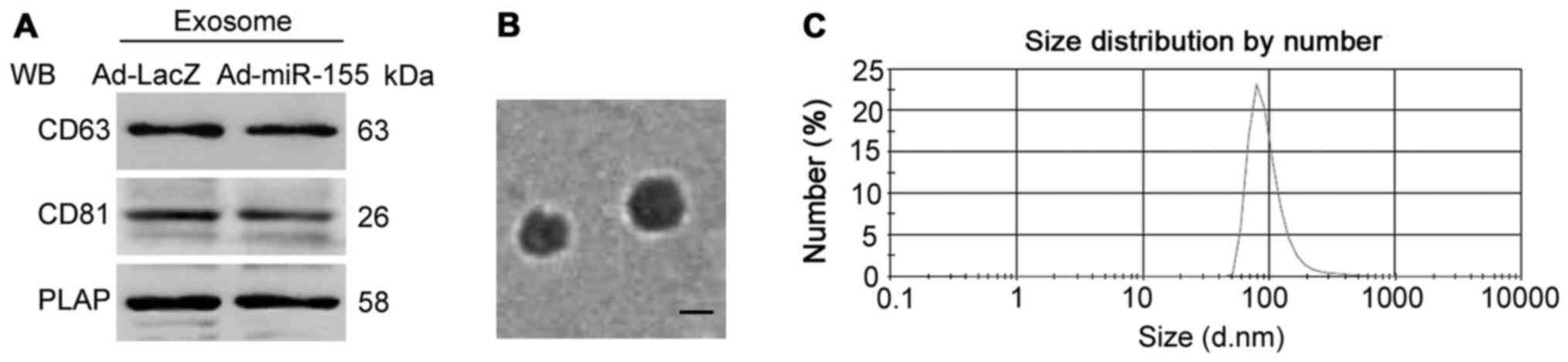
and the diameter of the isolated exosomes was ~100 nm (Fig. 5). Compared with exosomes from
Ad-LacZ-infected cell medium, miR-155 expression was increased by
2.41-fold in exosomes from Ad-miR-155-infected cell medium
(Fig. 6A; P<0.05). The ability
of exosomes to transfer to HUVECs was determined by examining the
uptake of PKH67-labeled exosomes. Fluorescence microscopy
demonstrated that PKH67-labeled BeWo cell exosomes were taken up by
HUVECs and were distributed within the perinuclear compartments
(Fig. 6B). As depicted in
Fig. 6C, the expression levels of
miR-155 were 4.93-fold higher (P<0.05) in HUVECs treated with
exosomes isolated from Ad-miR-155-infected BeWo cells compared with
in those treated with Ad-LacZ-infected BeWo cell exosomes.
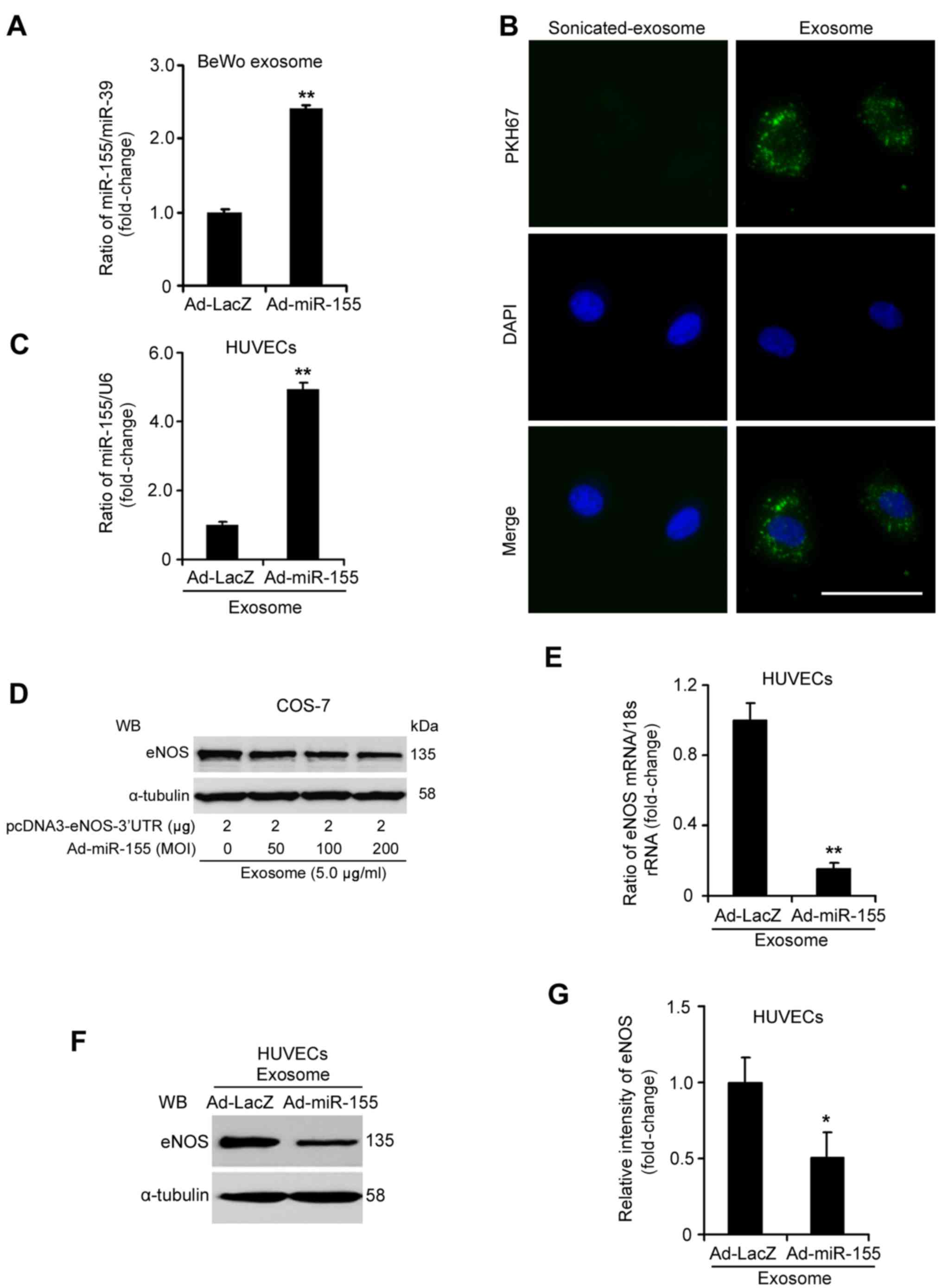 | Figure 6miR-155-overexpressed exosomes from
BeWo cell medium suppress eNOS expression in HUVECs. (A) RT-qPCR
analysis of miR-155 in exosomes from BeWo cells infected with
Ad-miR-155 or Ad-LacZ. (B) HUVECs were incubated for 6 h with PKH67
fluorescently-labeled exosomes and were observed under a
fluorescence microscope (PKH67, green; DAPI, blue). Scale bar, 25
µm. (C) Expression of miR-155 was increased in HUVECs
treated with exosomes from Ad-miR-155-infected BeWo cells. HUVECs
were treated with exosomes (5.0 µg/ml) for 24 h. (D)
miR-155-overexpressed exosomes from BeWo cells inhibited eNOS
expression by targeting its 3′-UTR in COS-7 cells. The pcDNA3-eNOS
plasmid containing the 3′-UTR of the eNOS gene was transfected into
COS-7 cells for 24 h, then miR-155-overexpressed exosomes from BeWo
cells (5.0 µg/ml) were added to the transfected cells for 24
h. (E) eNOS mRNA and (F and G) protein levels were decreased in
HUVECs treated with exosomes from Ad-miR-155-infected BeWo cells.
eNOS expression was detected by RT-qPCR and WB. The relative
intensity of eNOS protein expression was assessed using ImageJ
software. These data were from three independent experiments.
*P<0.05, **P<0.01 vs. Ad-LacZ group.
3′-UTR, 3′-untranslated region; Ad, adenovirus; eNOS, endothelial
nitric oxide synthase; HUVECs, human umbilical vein endothelial
cells; miR-155, microRNA-155; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; WB, western
blotting. |
To examine whether exosomal miR-155 could suppress
eNOS expression via direct 3′-UTR interaction, the present study
investigated eNOS expression in pcDNA3-eNOS-3′-UTR-transfected
COS-7 cells treated with exosomes from Ad-miR-155-infected BeWo
cells (18). As shown in Fig. 6D, miR-155-overexpressed exosomes
inhibited eNOS protein expression in a dose-dependent manner.
Subsequently, the present study investigated eNOS expression in
HUVECs treated with exosomes from Ad-miR-155-infected BeWo cells.
The mRNA expression levels of eNOS were reduced by ~84.73% in
HUVECs treated with exosomes from Ad-miR-155-infected BeWo cells
(Fig. 6E; P<0.05).
Consistently, the protein expression levels of eNOS were markedly
decreased in HUVECs incubated with miR-155-overexpressed exosomes
compared with in those treated with exosomes from Ad-LacZ-infected
BeWo cells (Fig. 6F and G;
P<0.05). Taken together, these results indicated that exosomal
miR-155 derived from trophoblasts may suppress eNOS expression in
HUVECs.
Discussion
During human pregnancy, placenta-associated serum
exosomes are involved in cell-cell communication between the
placenta and peripheral vascular endothelial cells (29). The present study isolated serum
exosomes with PLAP-positive expression by PEG precipitation and
ultracentrifugation with a sucrose gradient, which led to the
isolation of placenta-associated exosomes with high purity and
yield. The present study demonstrated that placenta-associated
serum exosomes from patients with PE reduced NO production and eNOS
expression in endothelial cells; this effect may be partly due to
the increased expression of miR-155 in placenta-associated serum
exosomes.
Previous studies have accumulated evidence regarding
the physiological function of serum exosomal miRNAs. Squadrito
et al demonstrated that serum/plasma exosomal miR-122 and
miR-155 are predo minantly associated with inflammatory liver
injury (30). Another study
indicated that serum exosomal miR-21 is positively correlated with
tumor progression and aggressiveness (31). In addition, miRNAs in plasma
exosomes have been reported to be stable under various storage
conditions (32). These data have
led to the recognition that serum exosomal miRNAs serve important
roles in pathophysiological alterations and the pathogenesis of
diseases. Placenta exosomal miRNAs may mediate crosstalk between
the fetoplacental unit and the maternal system during pregnancy
(33,34). The present study initially
detected the upregulation of miR-155 in placenta-associated serum
exosomes from patients with PE compared with in those from normal
pregnant women.
Endothelial cell functions can be affected by
exosomes via various mechanisms (9,35).
Since vasomotor dysfunction is one of the pathological alterations
associated with PE, the present study focused on vasomotor
function, and demonstrated that placenta-associated serum exosomes
from patients with PE exhibited downregulated eNOS expression and
NO production in HUVECs. The expression of eNOS can be modulated at
numerous levels, and post-transcriptional regulation has an
important role in the modulation. The present study revealed the
inhibitory effect of exosomal miR-155 on eNOS expression using
pcDNA3-eNOS-3′-UTR-overexpressing COS-7 cells, which do not possess
endogenous eNOS expression. In addition, miR-155-overexpressed
exosomes isolated from trophoblast cell medium were able to
suppress the expression of eNOS in HUVECs. Therefore,
placenta-associated serum exosomal miR-155 may suppress eNOS
expression through direct 3′-UTR interaction. These findings
provided evidence to suggest that serum placenta-associated
exosomes may regulate endothelial cell functions during PE
development.
In conclusion, the present study identified that
placenta-associated serum exosomes from patients with PE suppressed
NO production and eNOS expression in endothelial cells. In
addition, increased miR-155 expression in placenta-associated serum
exosomes may be associated with the effects of serum exosomes. Our
findings indicated that serum exosomal miR-155 may regulate
endothelial cell functions and contribute to the development of
PE.
Acknowledgments
The authors would like to thank Dr Zhiqun Wang and
Dr Yimin Dai (both from Nanjing Drum Tower Hospital) for collection
of human serum samples. The study was funded in part by grants from
the National Natural Science Foundation (grant nos. 81370724,
81571462 and 81701472), the Jiangsu Biobank of Clinical Resources
(grant no. BM2015004) and the Jiangsu Key Laboratory for Molecular
Medicine (grant no. BM2007208), Project of Nanjing Clinical
Medicine.
References
|
1
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
James JL, Whitley GS and Cartwright JE:
Preeclampsia: fitting together the placental, immune and
cardiovascular pieces. J Pathol. 221:363–378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Redman CW, Tannetta DS, Dragovic RA,
Gardiner C, Southcombe JH, Collett GP and Sargent IL: Review: does
size matter? Placental debris and the pathophysiology of
preeclampsia. Placenta. 33(Suppl): S48–S54. 2012. View Article : Google Scholar
|
|
4
|
Redman CW and Sargent IL: Placental
debris, oxidative stress and preeclampsia. Placenta. 21:597–602.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Keller S, Sanderson MP, Stoeck A and
Altevogt P: Exosomes: from biogenesis and secretion to biological
function. Immunol Lett. 107:102–108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nazimek K, Ptak W, Nowak B, Ptak M,
Askenase PW and Bryniarski K: Macrophages play an essential role in
antigen-specific immune suppression mediated by T CD8+
cell-derived exosomes. Immunology. 146:23–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee HD, Kim YH and Kim DS: Exosomes
derived from human macrophages suppress endothelial cell migration
by controlling integrin trafficking. Eur J Immunol. 44:1156–1169.
2014. View Article : Google Scholar
|
|
9
|
Chen DB and Wang W: Human placental
microRNAs and preeclampsia. Biol Reprod. 88:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hromadnikova I, Kotlabova K, Hympanova L
and Krofta L: Cardiovascular and cerebrovascular disease associated
microRNAs are dysregulated in placental tissues affected with
gestational hypertension, preeclampsia and intrauterine growth
restriction. PLoS One. 10:e01383832015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo SS, Ishibashi O, Ishikawa G, Ishikawa
T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A,
et al: Human villous trophoblasts express and secrete
placenta-specific microRNAs into maternal circulation via exosomes.
Biol Reprod. 81:717–729. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pankratz F, Bemtgen X, Zeiser R, Leonhardt
F, Kreuzaler S, Hilgendorf I, Smolka C, Helbing T, Hoefer I, Esser
JS, et al: Micro RNA-155 exerts cell-specific antiangiogenic but
proarteriogenic effects during adaptive neovascularization.
Circulation. 131:1575–1589. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bala S, Csak T, Saha B, Zatsiorsky J,
Kodys K, Catalano D, Satishchandran A and Szabo G: The
pro-inflammatory effects of miR-155 promote liver fibrosis and
alcohol-induced steatohepatitis. J Hepatol. 64:1378–1387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murphy MS, Casselman RC, Tayade C and
Smith GN: Differential expression of plasma microRNA in
preeclamptic patients at delivery and 1 year postpartum. Am J
Obstet Gynecol. 213:367.e1–367.e9. 2015. View Article : Google Scholar
|
|
15
|
Zhang Y, Diao Z, Su L, Sun H, Li R, Cui H
and Hu Y: Micro-RNA-155 contributes to preeclampsia by
down-regulating CYR61. Am J Obstet Gynecol. 202:466.e1–466.e7.
2010. View Article : Google Scholar
|
|
16
|
Johal T, Lees CC, Everett TR and Wilkinson
IB: The nitric oxide pathway and possible therapeutic options in
preeclampsia. Br J Clin Pharmacol. 78:244–257. 2014. View Article : Google Scholar
|
|
17
|
Kim YJ, Park HS, Lee HY, Ha EH, Suh SH, Oh
SK and Yoo HS: Reduced L-arginine level and decreased placental
eNOS activity in preeclampsia. Placenta. 27:438–444. 2006.
View Article : Google Scholar
|
|
18
|
Sun HX, Zeng DY, Li RT, Pang RP, Yang H,
Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, et al: Essential role
of microRNA-155 in regulating endothelium-dependent vasorelaxation
by targeting endothelial nitric oxide synthase. Hypertension.
60:1407–1414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cunningham FG, Leveno KJ, Bloom SL, Spong
CY, Dashe JS, Hoffman BL, Casey BM and Sheffield JS: Hypertensive
disorders. Williams Obstetrics. 24th edition. McGraw-Hill
Education; New York, NY: pp. 729–731. 2014
|
|
20
|
Baudin B, Bruneel A, Bosselut N and
Vaubourdolle M: A protocol for isolation and culture of human
umbilical vein endothelial cells. Nat Protoc. 2:481–485. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vlassov A, Li M, Zeringer E and Conrad R:
Methods and compositions for exosome isolation. US Patent 8,901,284
B2. Filed February 26, 2014 issued December 2. 2014
|
|
22
|
Rider MA, Hurwitz SN and Meckes DG Jr:
ExtraPEG: a polyethylene glycol-based method for enrichment of
extracellular vesicles. Sci Rep. 6:239782016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H,
Liu J, Pan T, Chen J, Wu M, et al: Exosomes mediate the
cell-to-cell transmission of IFN-α-induced antiviral activity. Nat
Immunol. 14:793–803. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Putz U, Howitt J, Doan A, Goh CP, Low LH,
Silke J and Tan SS: The tumor suppressor PTEN is exported in
exosomes and has phosphatase activity in recipient cells. Sci
Signal. 5:ra702012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pegtel DM, Cosmopoulos K, Thorley-Lawson
DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD,
Würdinger T and Middeldorp JM: Functional delivery of viral miRNAs
via exosomes. Proc Natl Acad Sci USA. 107:6328–6333. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hnasko TS and Hnasko RM: The western bot.
Methods Mol Bio. 318:87–96. 2015. View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Rana S, Yue S, Stadel D and Zöller M:
Toward tailored exosomes: the exosomal tetraspanin web contributes
to target cell selection. Int J Biochem Cell Biol. 44:1574–1584.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salomon C, Torres MJ, Kobayashi M,
Scholz-Romero K, Sobrevia L, Dobierzewska A, Illanes SE, Mitchell
MD and Rice GE: A gestational profile of placental exosomes in
maternal plasma and their effects on endothelial cell migration.
PLoS One. 9:e986672014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Squadrito ML, Baer C, Burdet F, Maderna C,
Gilfillan GD, Lyle R, Ibberson M and De Palma M: Endogenous RNAs
modulate microRNA sorting to exosomes and transfer to acceptor
cells. Cell Reports. 8:1432–1446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar
|
|
32
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1568–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ouyang Y, Mouillet JF, Coyne CB and
Sadovsky Y: Review: placenta-specific microRNAs in exosomes - good
things come in nano-packages. Placenta. 35(Suppl): S69–S73. 2014.
View Article : Google Scholar
|
|
34
|
Kambe S, Yoshitake H, Yuge K, Ishida Y,
Ali MM, Takizawa T, Kuwata T, Ohkuchi A, Matsubara S, Suzuki M, et
al: Human exosomal placenta-associated miR-517a-3p modulates the
expression of PRKG1 mRNA in Jurkat cells. Biol Reprod. 91:1292014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gambim MH, do Carmo AO, Marti L,
Veríssimo-Filho S, Lopes LR and Janiszewski M: Platelet-derived
exosomes induce endothelial cell apoptosis through peroxynitrite
generation: experimental evidence for a novel mechanism of septic
vascular dysfunction. Crit Care. 11:R1072007. View Article : Google Scholar : PubMed/NCBI
|