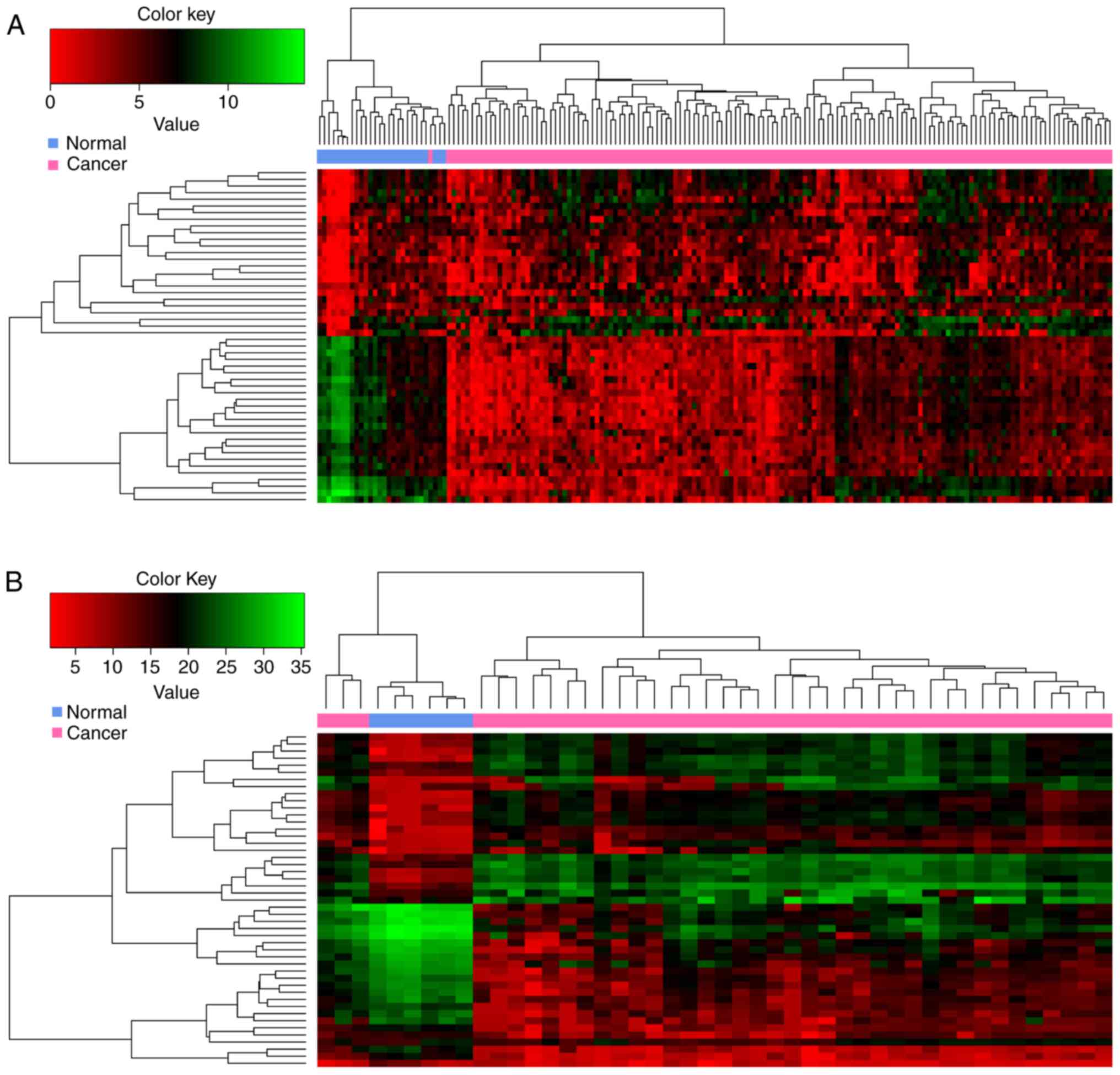
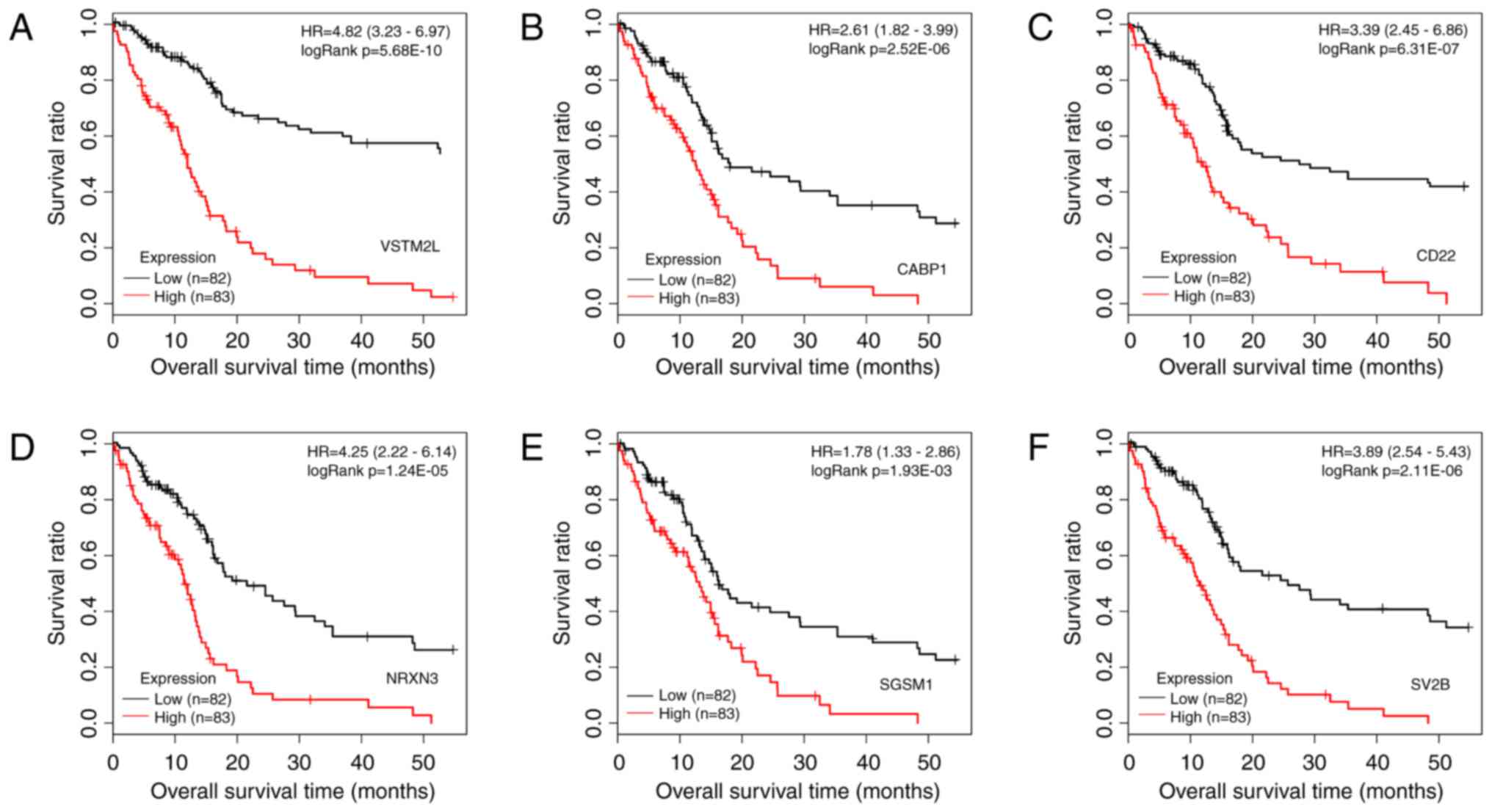
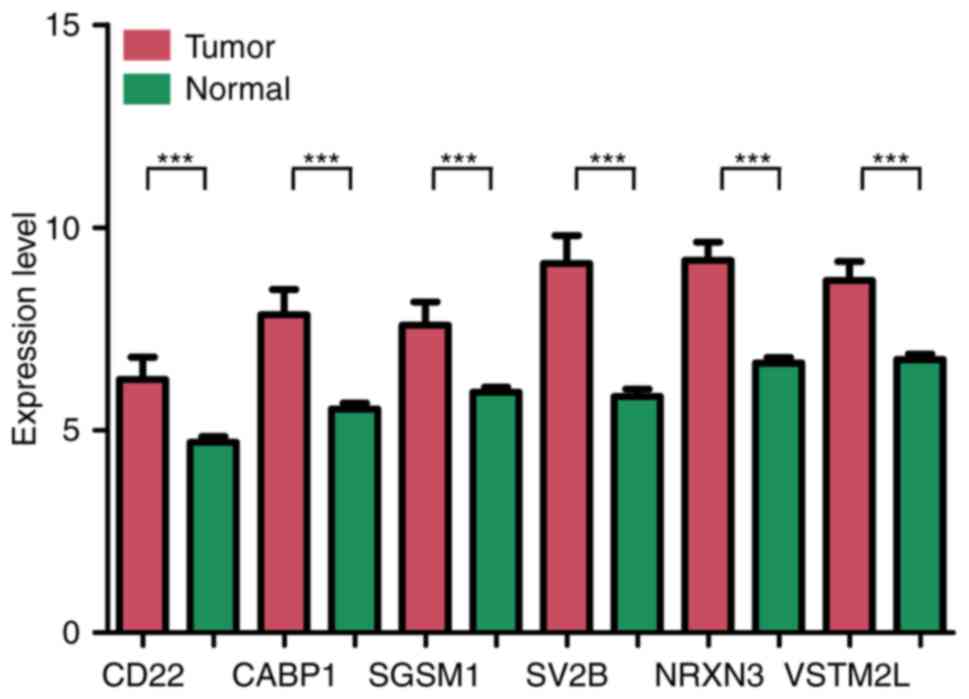
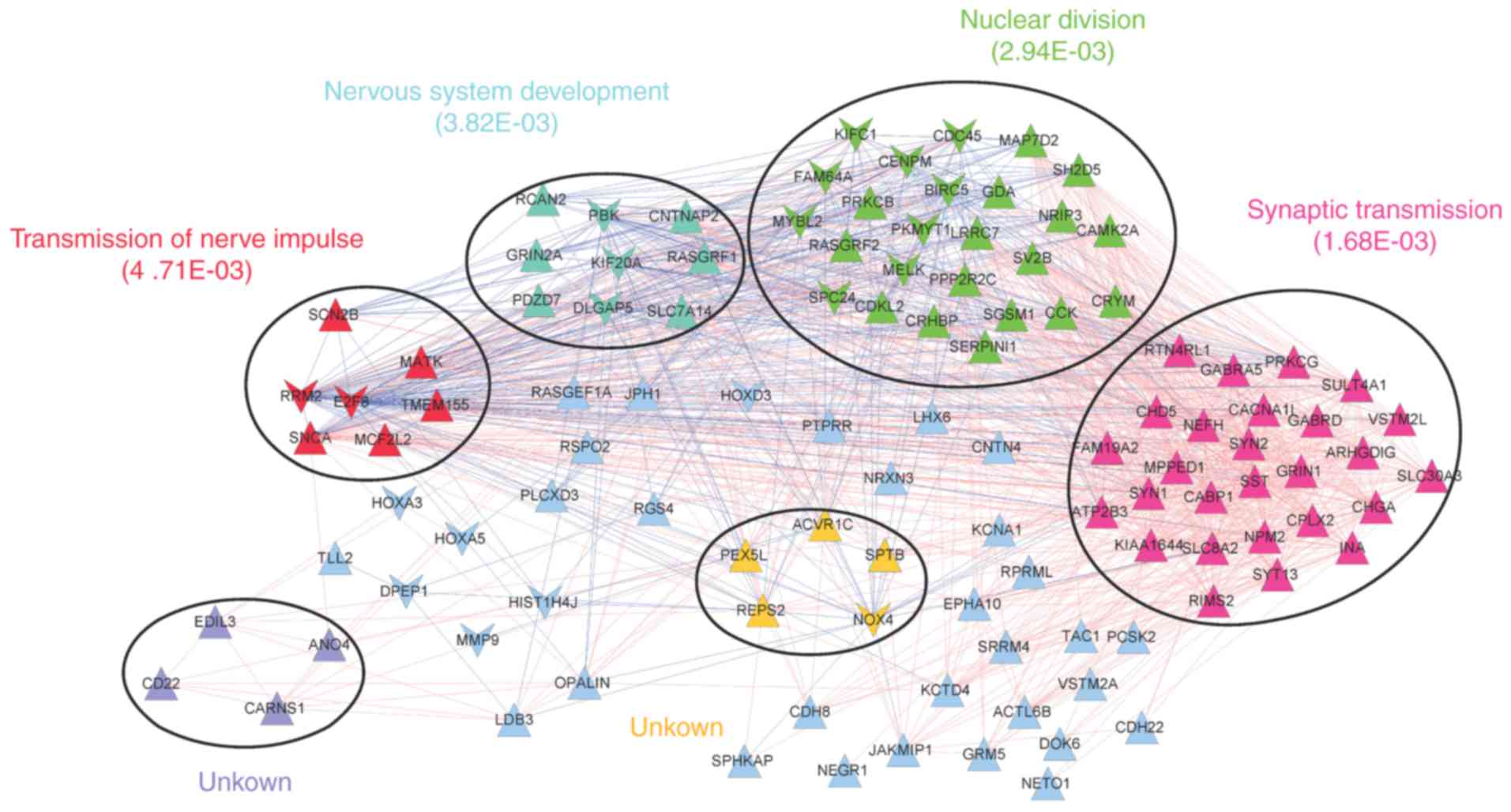
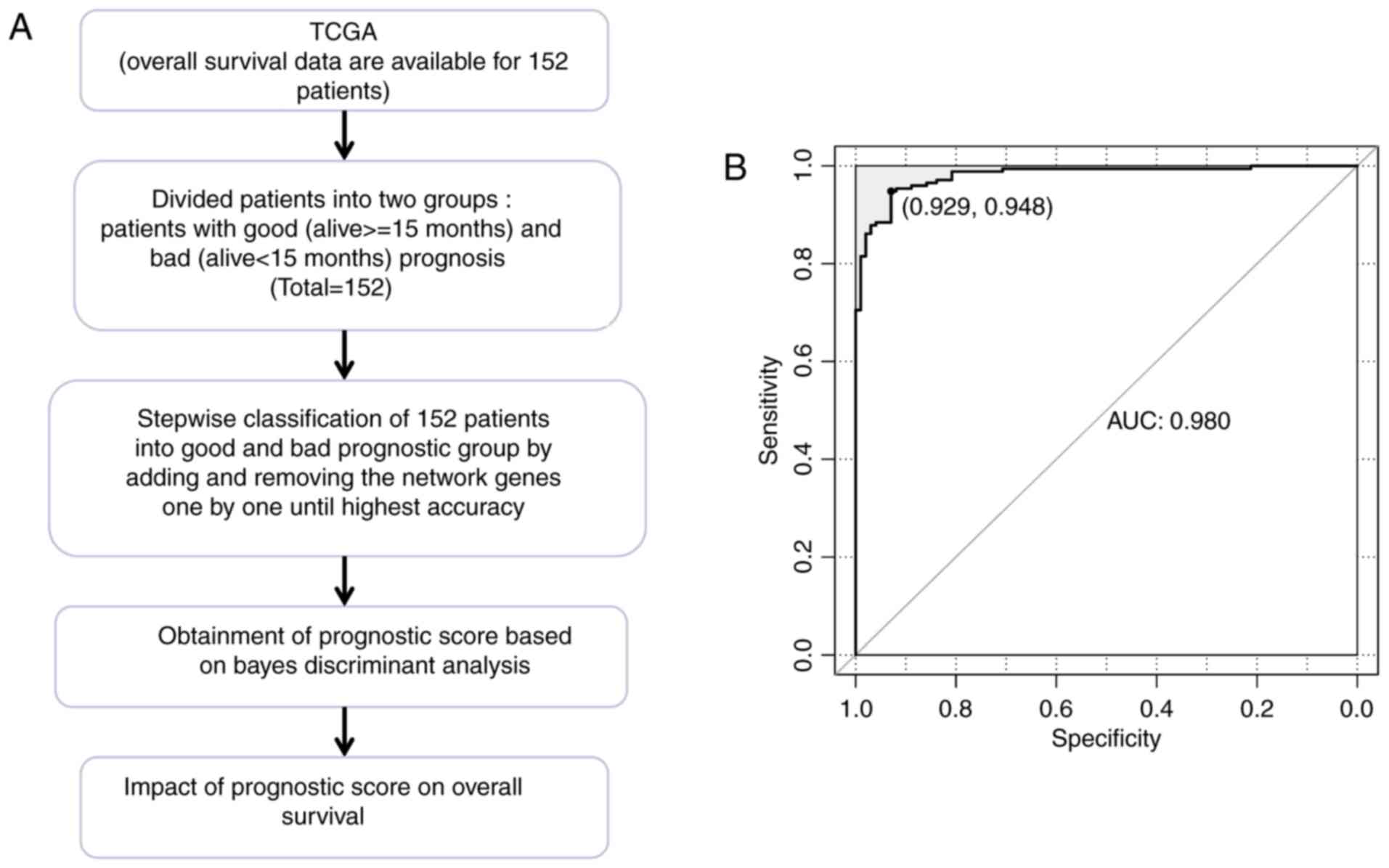
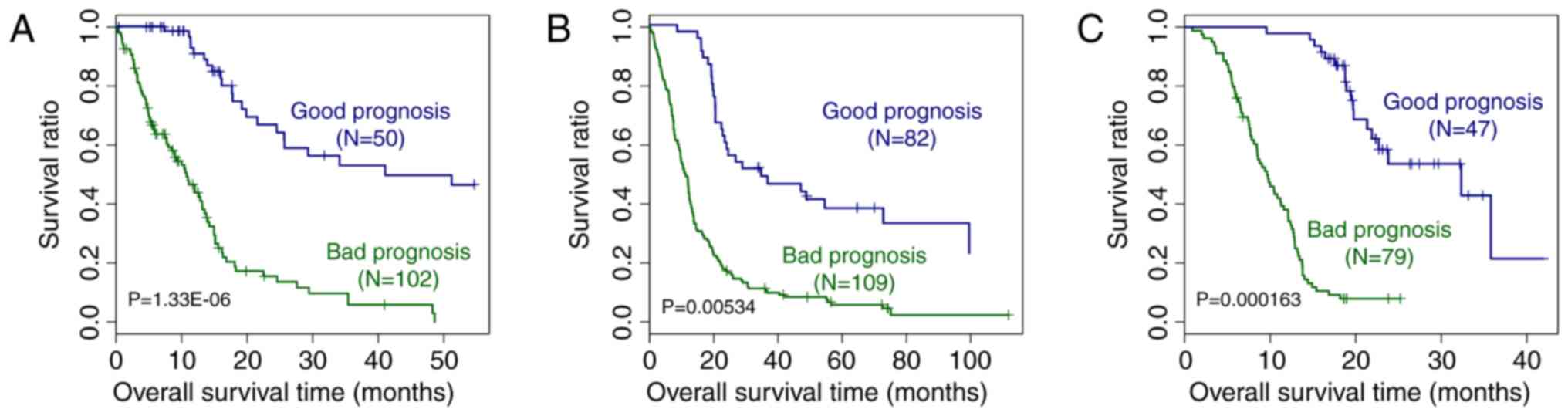
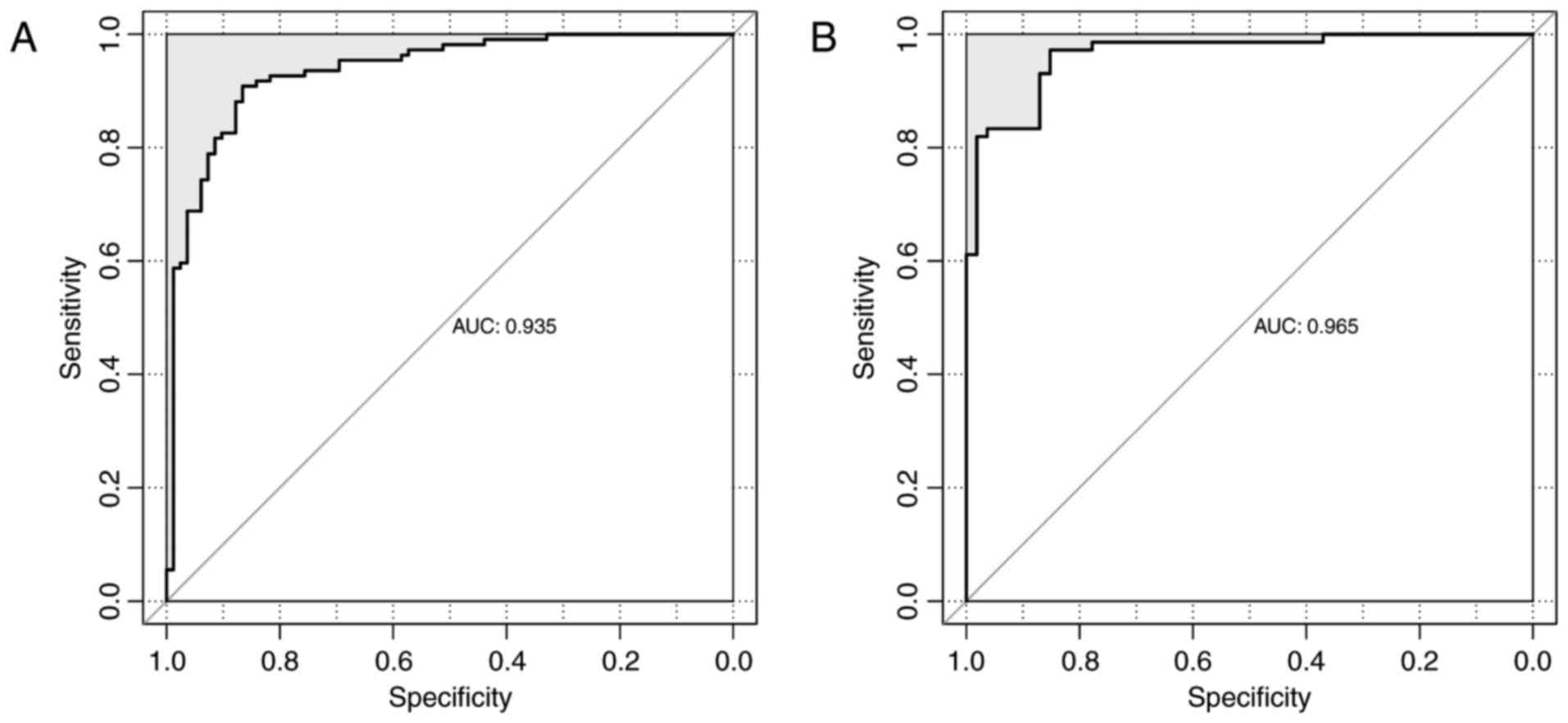
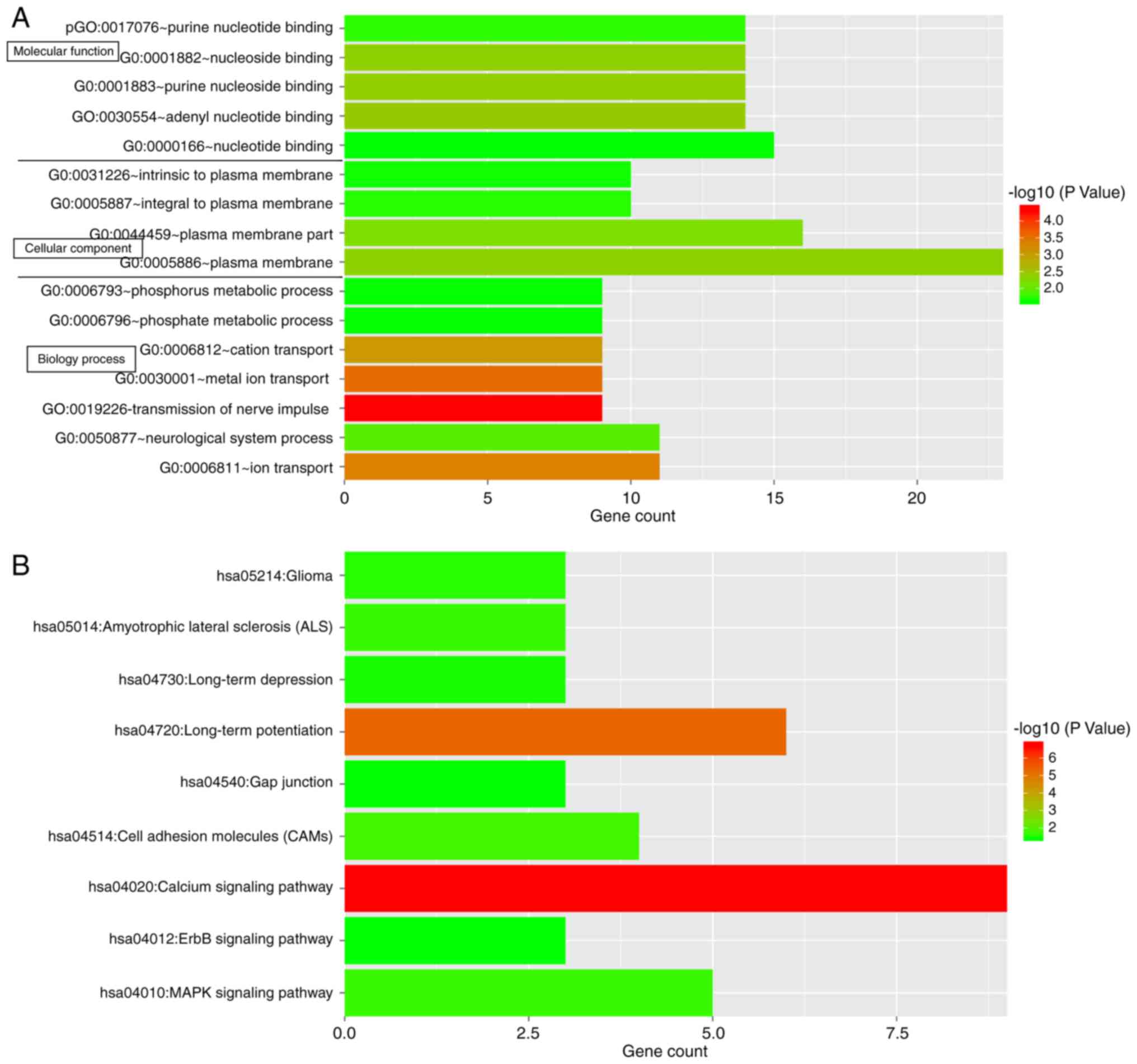
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Catt S, Chalmers A and Fallowfield L:
Psychosocial and supportive-careneeds in high-grade glioma. Lancet
Oncol. 9:884–891. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smoll NR, Schaller K and Gautschi OP:
Long-term survival of patients with glioblastoma multiforme (GBM).
J Clin Neurosci. 20:670–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Drappatz J, Norden AD and Wen PY:
Therapeutic strategies for inhibiting invasion in glioblastoma.
Expert Rev Neurother. 9:519–534. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thakkar JP, Dolecek TA, Horbinski C,
Ostrom QT, Lightner DD, Barnholtz-Sloan JS and Villano JL:
Epidemiologic and molecular prognostic review of glioblastoma.
Cancer Epidemiol Biomarkers Prev. 23:1985–1996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gan HK, Kaye AH and Luwor RB: The EGFRvIII
variant in glioblastoma multiforme. J Clin Neurosci. 16:748–754.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Srividya MR, Thota B, Shailaja BC,
Arivazhagan A, Thennarasu K, Chandramouli BA, Hegde AS and Santosh
V: Homozygous 10q23/PTEN deletion and its impact on outcome in
glioblastoma: A prospective translational study on a uniformly
treated cohort of adult patients. Neuropathology. 31:376–383. 2011.
View Article : Google Scholar
|
|
8
|
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ,
Yan W, Liu YW, Zhang W, Chen L and Jiang T: Prognostic value of a
nine-gene signature in glioma patients based on mRNA expression
profiling. CNS Neurosci Ther. 20:112–118. 2014. View Article : Google Scholar
|
|
9
|
Sun Y, Zhang W, Chen D, Lv Y, Zheng J,
Lilljebjörn H, Ran L, Bao Z, Soneson C, Sjögren HO, et al: A glioma
classification scheme based on coexpression modules of EGFR and
PDGFRA. Proc Natl Acad Sci USA. 111:3538–3543. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen PY and Huse JT: 2016 World Health
Organization classification of central nervous system tumors.
Continuum (Minneap Minn). 23:1531–1547. 2017.
|
|
11
|
Li H, Yu B, Li J, Su L, Yan M, Zhang J, Li
C, Zhu Z and Liu B: Characterization of differentially expressed
genes involved in pathways associated with gastric cancer. PloS
One. 10:e01250132015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu C and Shen T: cDNA microarray and
bioinformatic analysis for the identification of key genes in
Alzheimer’s disease. Int J Mol Med. 33:457–461. 2014. View Article : Google Scholar
|
|
13
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
14
|
Reimand J, Tooming L, Peterson H, Adler P
and Vilo J: GraphWeb: Mining heterogeneous biological networks for
gene modules with functional significance. Nucleic Acids Res.
36(Web Server issue): W452–W459. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Joutsijoki H, Haponen M, Rasku J,
Aalto-Setälä K and Juhola M: Error-correcting output codes in
classification of human induced pluripotent stem cell colony
images. Biomed Res Int. 2016:30250572016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Voigt AP, Eidenschink Brodersen L, Pardo
L, Meshinchi S and Loken MR: Consistent quantitative gene product
expression: #1. Automated identification of regenerating bone
marrow cell populations using support vector machines. Cytometry A.
89:978–986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
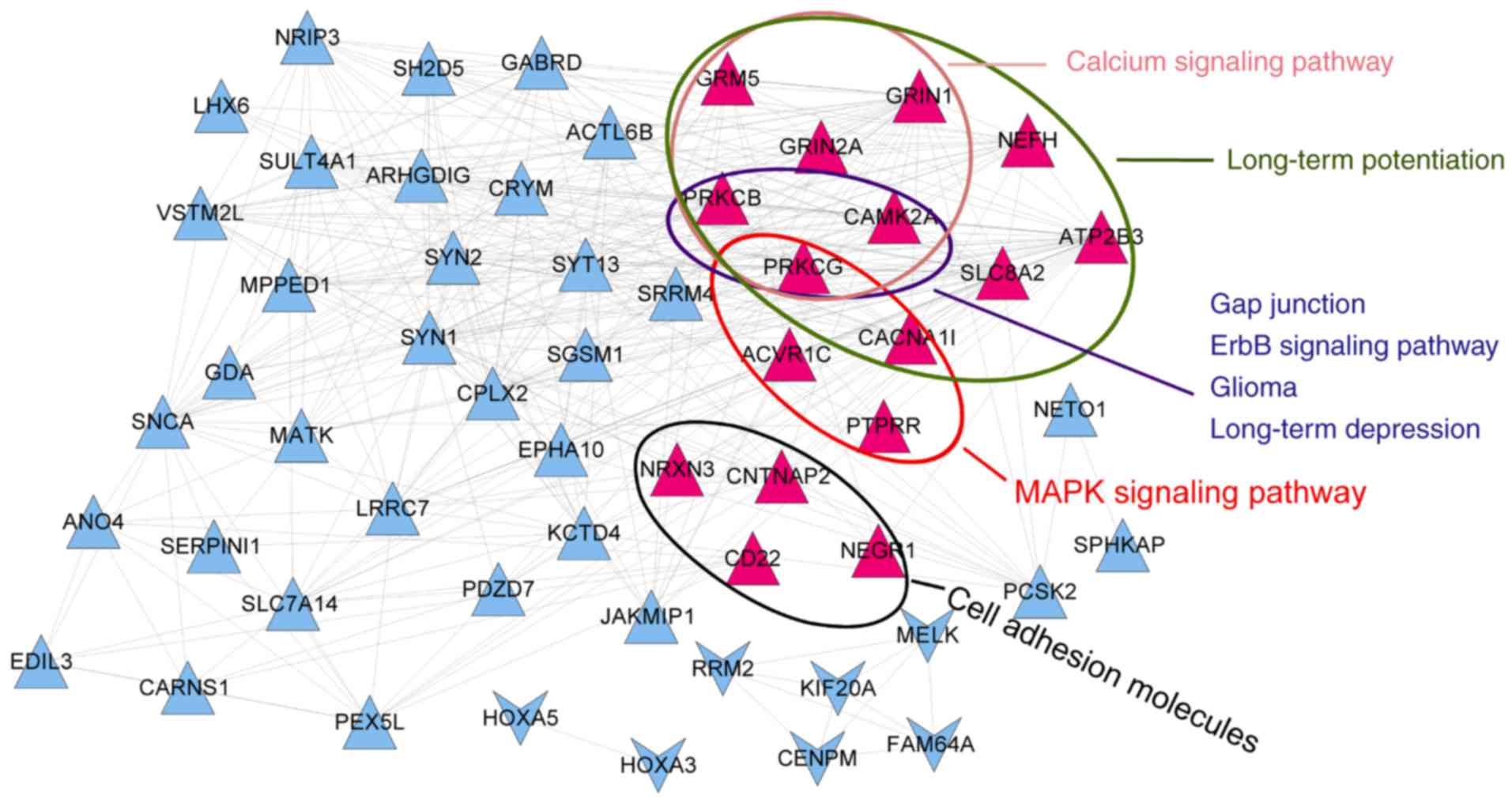
Park P, Sanderson TM, Amici M, Choi SL,
Bortolotto ZA, Zhuo M, Kaang BK and Collingridge GL:
Calcium-permeable AMPA receptors mediate the induction of the
protein kinase a-dependent component of long-term potentiation in
the hippo-campus. J Neurosci. 36:622–631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schroeder JI and Hagiwara S: Cytosolic
calcium regulates ion channels in the plasma membrane of Vicia faba
guard cells. Nature. 338:427–430. 1989. View Article : Google Scholar
|
|
20
|
Leclerc C, Haeich J, Aulestia FJ,
Kilhoffer MC, Miller AL, Néant I, Webb SE, Schaeffer E, Junier MP,
Chneiweiss H and Moreau M: Calcium signaling orchestrates
glioblastoma development: Facts and conjunctures. Biochim Biophys
Acta. 1863:1447–1459. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zohrabian VM, Forzani B, Chau Z, Murali R
and Jhanwar-Uniyal M: Rho/ROCK and MAPK signaling pathways are
involved in glioblastoma cell migration and proliferation.
Anticancer Res. 29:119–123. 2009.PubMed/NCBI
|
|
22
|
Sangpairoj K, Vivithanaporn P,
Apisawetakan S, Chongthammakun S, Sobhon P and Chaithirayanon K:
RUNX1 regulates migration, invasion, and angiogenesis via p38 MAPK
pathway in human glioblastoma. Cell Mol Neurobiol. 37:1243–1255.
2017. View Article : Google Scholar
|
|
23
|
Schlaepfer IR, Clegg HV, Corley RP,
Crowley TJ, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH,
Stallings MC, et al: The human protein kinase C gamma gene (PRKCG)
as a susceptibility locus for behavioral disinhibition. Addict
Biol. 12:200–209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geiges D, Meyer T, Marte B, Vanek M,
Weissgerber G, Stabel S, Pfeilschifter J, Fabbro D and Huwiler A:
Activation of protein kinase C subtypes alpha, gamma, delta,
epsilon, zeta and eta by tumor-promoting and nontumor-promoting
agents. Biochem Pharmacol. 53:865–875. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Louhimo R, Aittomäki V, Faisal A, Laakso
M, Chen P, Ovaska K, Valo E, Lahti L, Rogojin V, Kaski S and
Hautaniemi S: Systematic use of computational methods allows
stratifying treatment responders in glioblastoma multiforme.
Proceedings of the CAMDA Conference. Critical Assessment of Massive
Data Analysis, 2011. Systems Biomedicine. 1:130–136. 2013.
View Article : Google Scholar
|
|
26
|
Verbeek DS, Goedhart J, Bruinsma L, Sinke
RJ and Reits EA: PKC gamma mutations in spinocerebellar ataxia type
14 affect C1 domain accessibility and kinase activity leading to
aberrant MAPK signaling. J Cell Sci. 121:2339–2349. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thomas SL, Alam R, Lemke N, Schultz LR,
Gutiérrez JA and Rempel SA: PTEN augments SPARC suppression of
proliferation and inhibits SPARC-induced migration by suppressing
SHC-RAF-ERK and AKT signaling. Neuro Oncol. 12:941–955. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dhillon AS, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 3279–3290.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eleveld TF, Schild L, Ebus ME, van Sluis
PG, Westerhout EM, Caron HN, Koster JJB, Versteeg R and Molenaar
JJ: Abstract A31: Activation of the RAS-MAPK pathway in primary
neuroblastoma tumors is associated with poor prognosis. Cancer Res.
76(5 Suppl)2016. View Article : Google Scholar
|
|
30
|
Martiny-Baron G and Fabbro D: Classical
PKC isoforms in cancer. Pharmacol Res. 55:477–486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang K and Klionsky DJ: Mitochondria
removal by autophagy. Autophagy. 7:297–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bononi A, Agnoletto C, De Marchi E, Marchi
S, Patergnani S, Bonora M, Giorgi C, Missiroli S, Poletti F,
Rimessi A and Pinton P: Protein kinases and phosphatases in the
control of cell fate. Enzyme Res. 2011:3290982011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roessler J, Ammerpohl O, Gutwein J,
Steinemann D, Schlegelberger B, Weyer V, Sariyar M, Geffers R,
Arnold N, Schmutzler R, et al: The CpG island methylator phenotype
in breast cancer is associated with the lobular subtype.
Epigenomics. 7:187–199. 2015. View Article : Google Scholar
|
|
34
|
Hwang E, Yoo KC, Kang SG, Kim RK, Cui YH,
Lee HJ, Kim MJ, Lee JS, Kim IG, Suh Y and Lee SJ: PKCδ activated by
c-MET enhances infiltration of human glioblastoma cells through
NOTCH2 signaling. Oncotarget. 7:4890–4902. 2016. View Article : Google Scholar
|
|
35
|
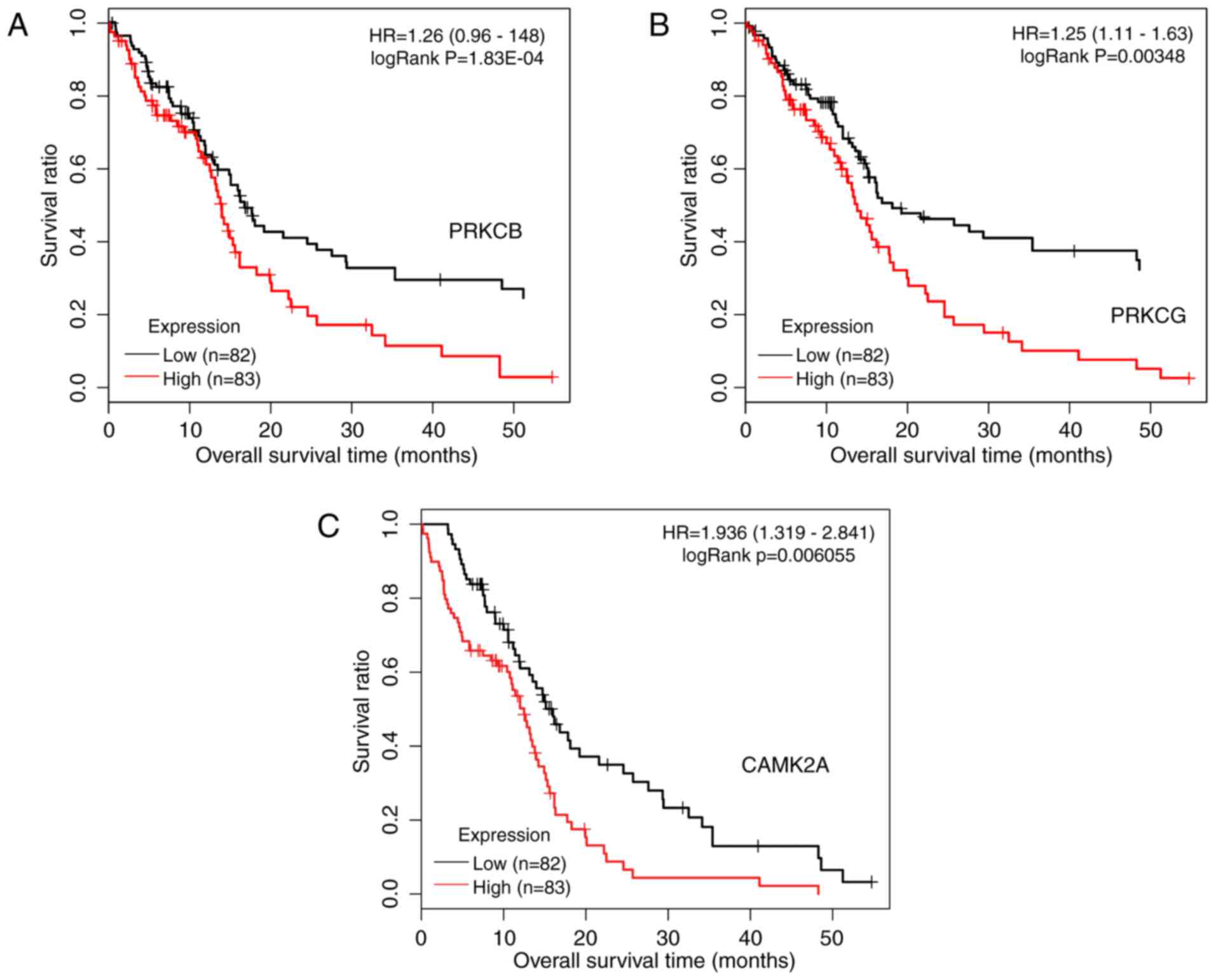
Rodriguez-Mora O, LaHair MM, Howe CJ,
McCubrey JA and Franklin RA: Calcium/calmodulin-dependent protein
kinases as potential targets in cancer therapy. Expert Opin Ther
Targets. 9:791–808. 2005. View Article : Google Scholar : PubMed/NCBI
|