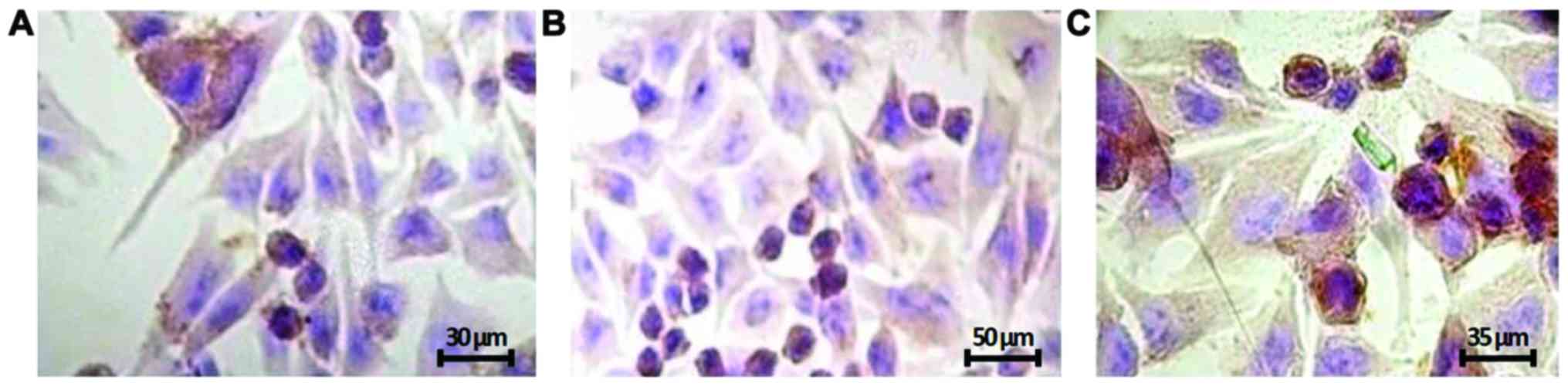
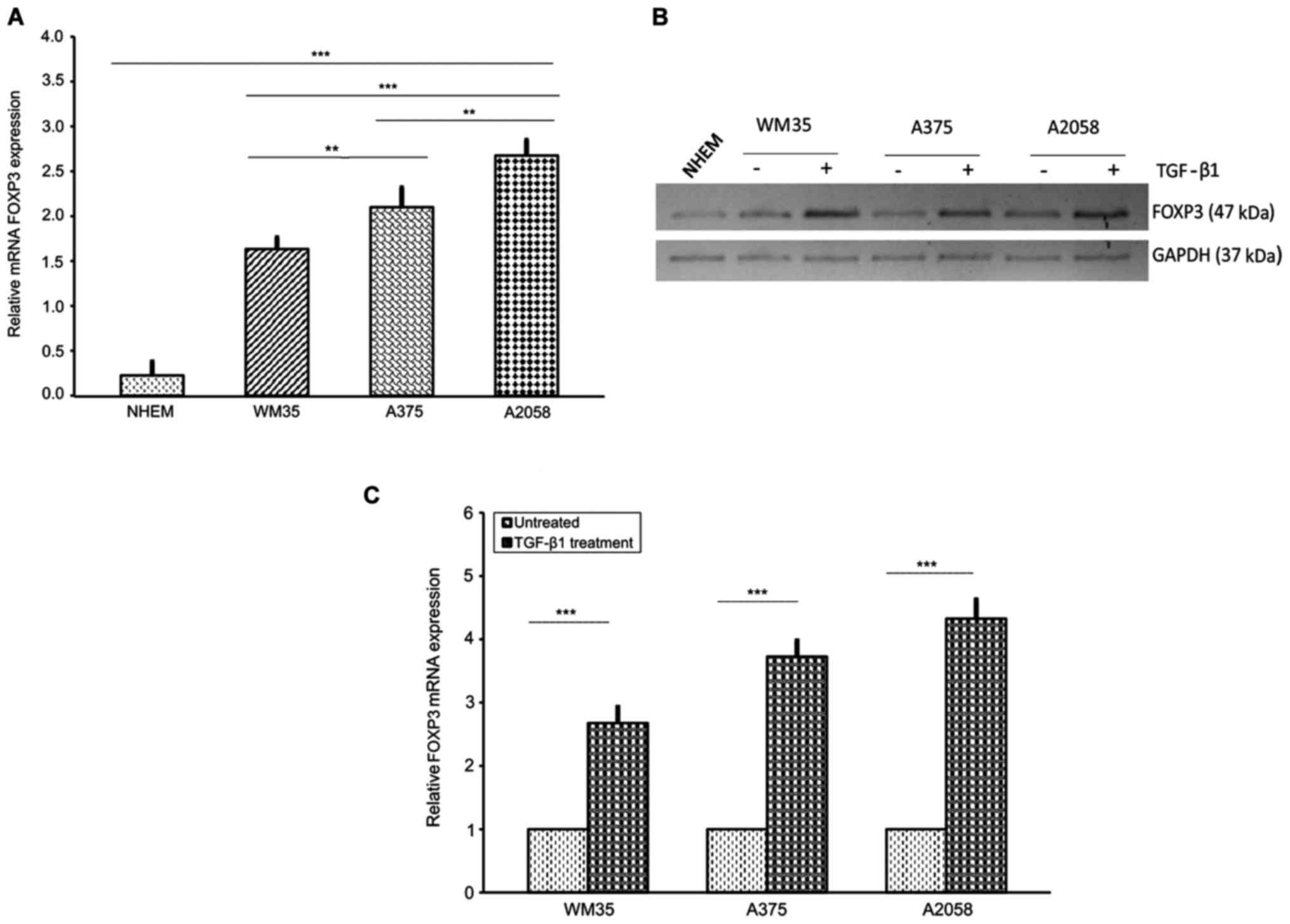
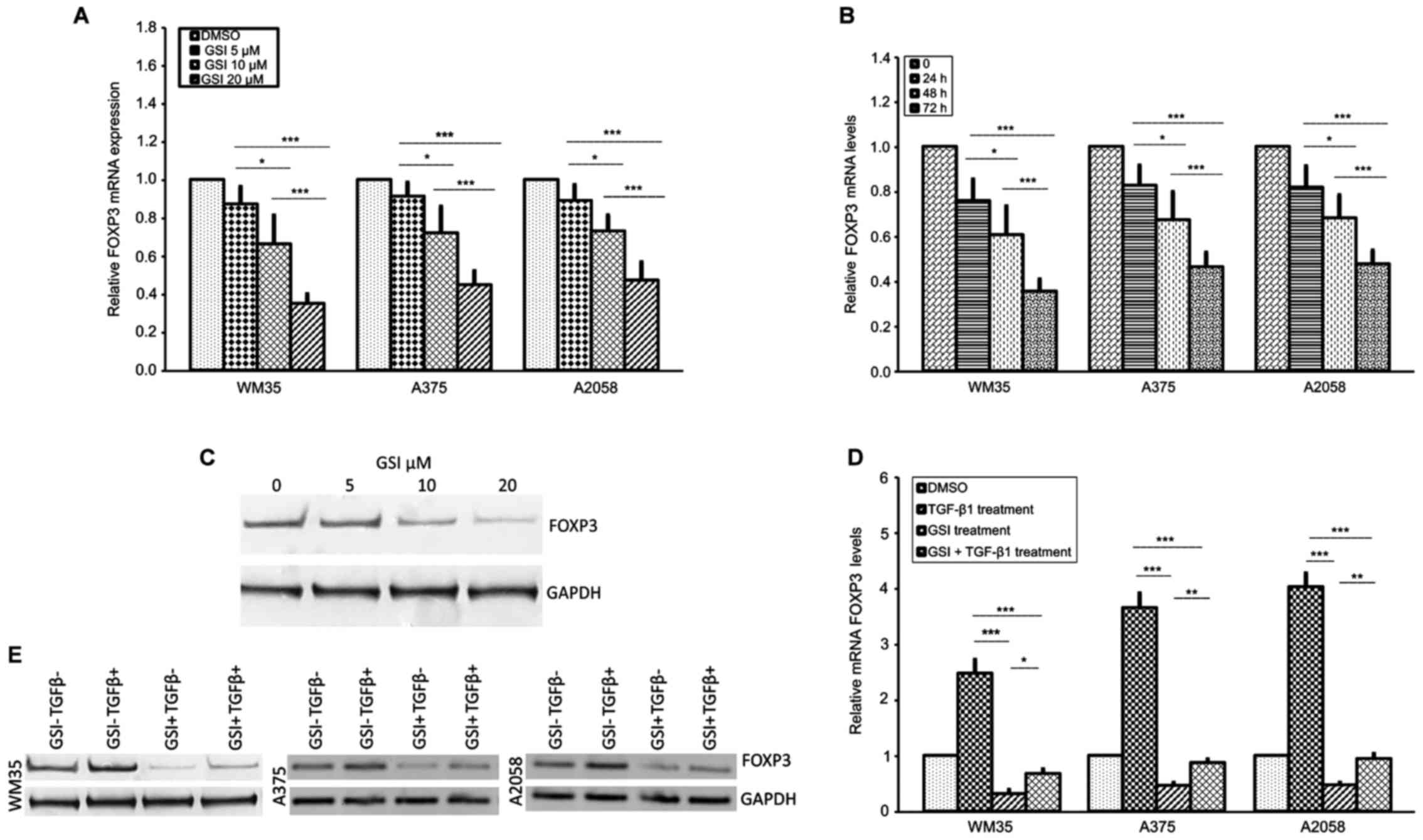
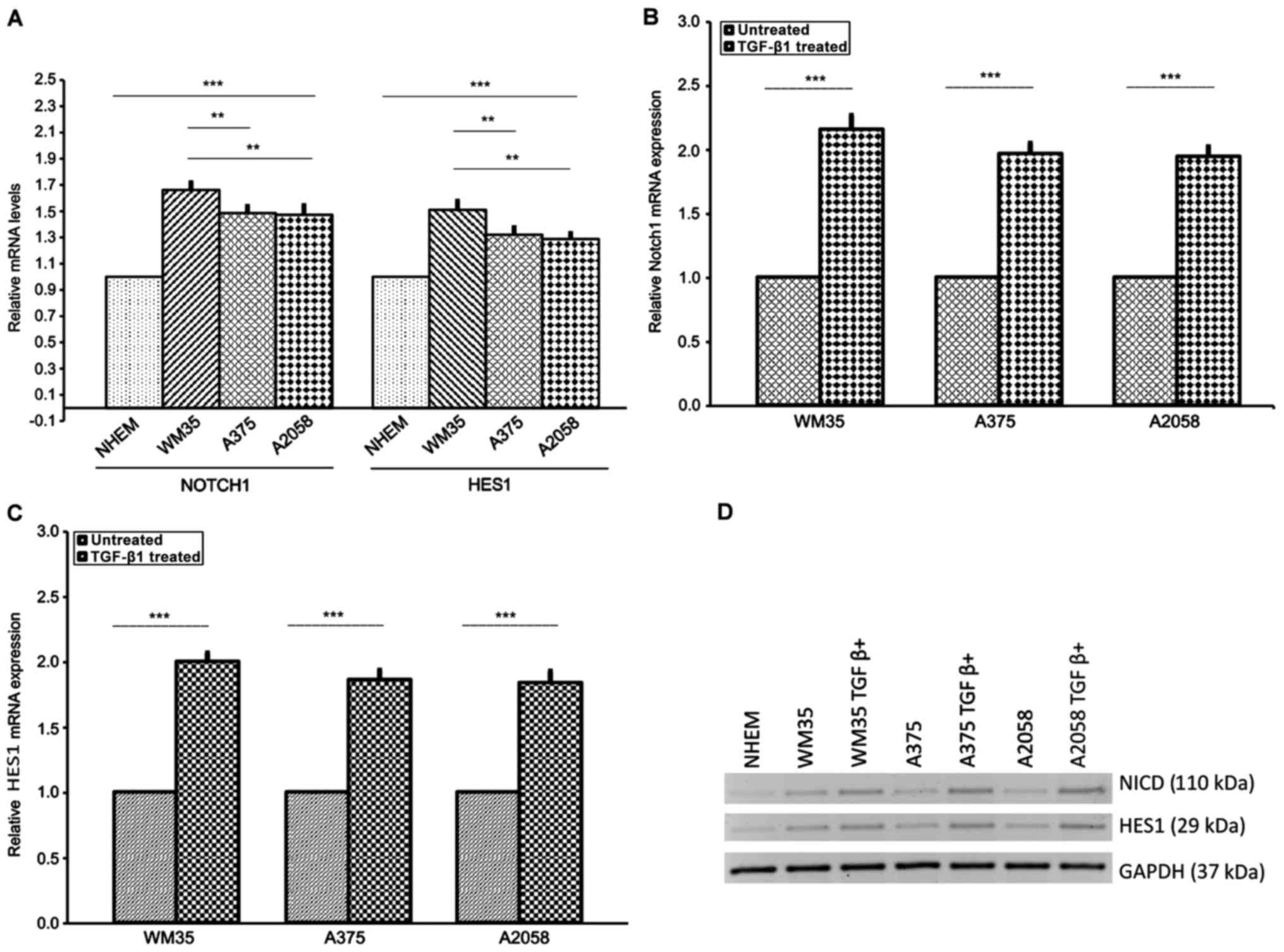
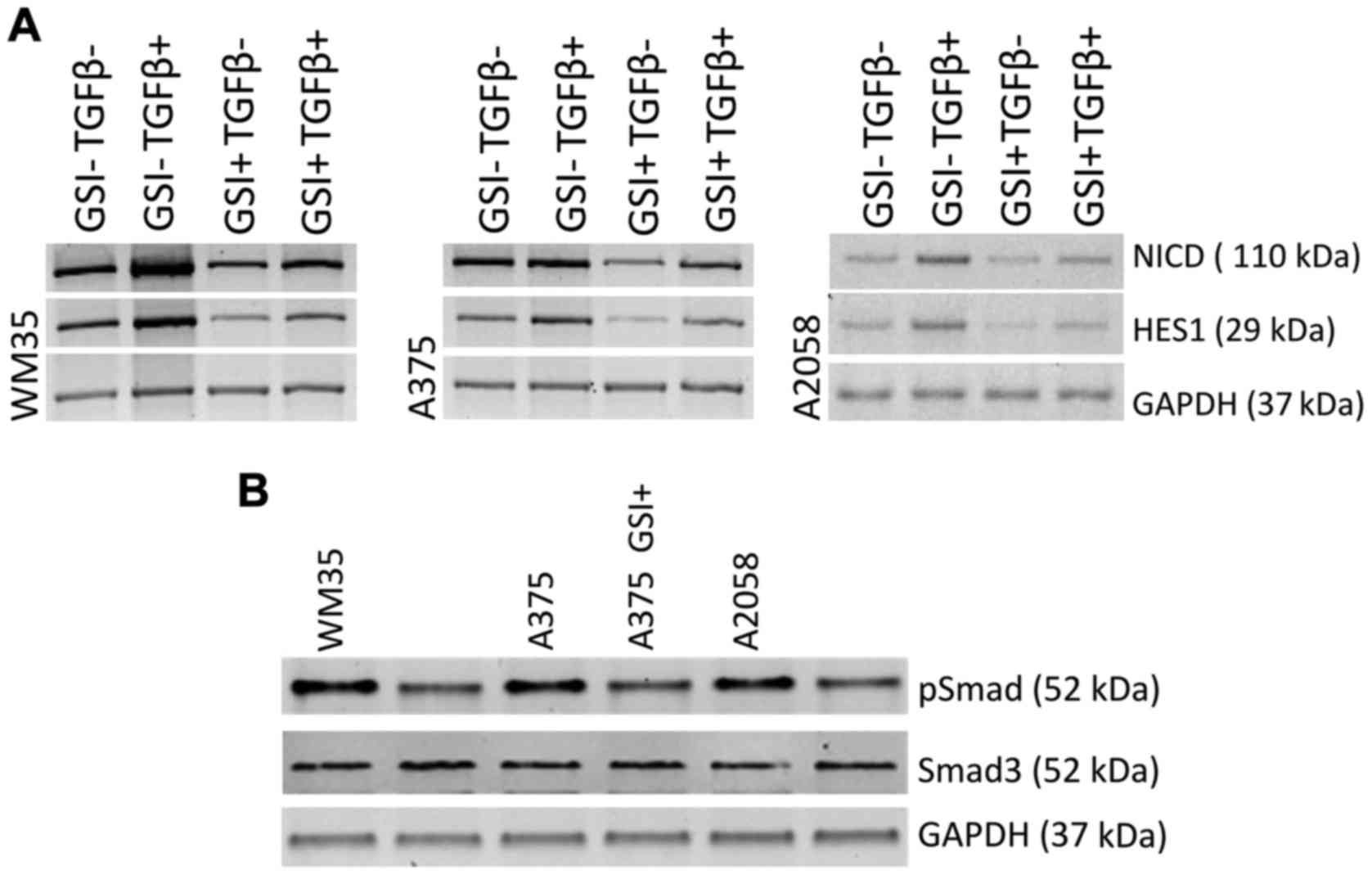
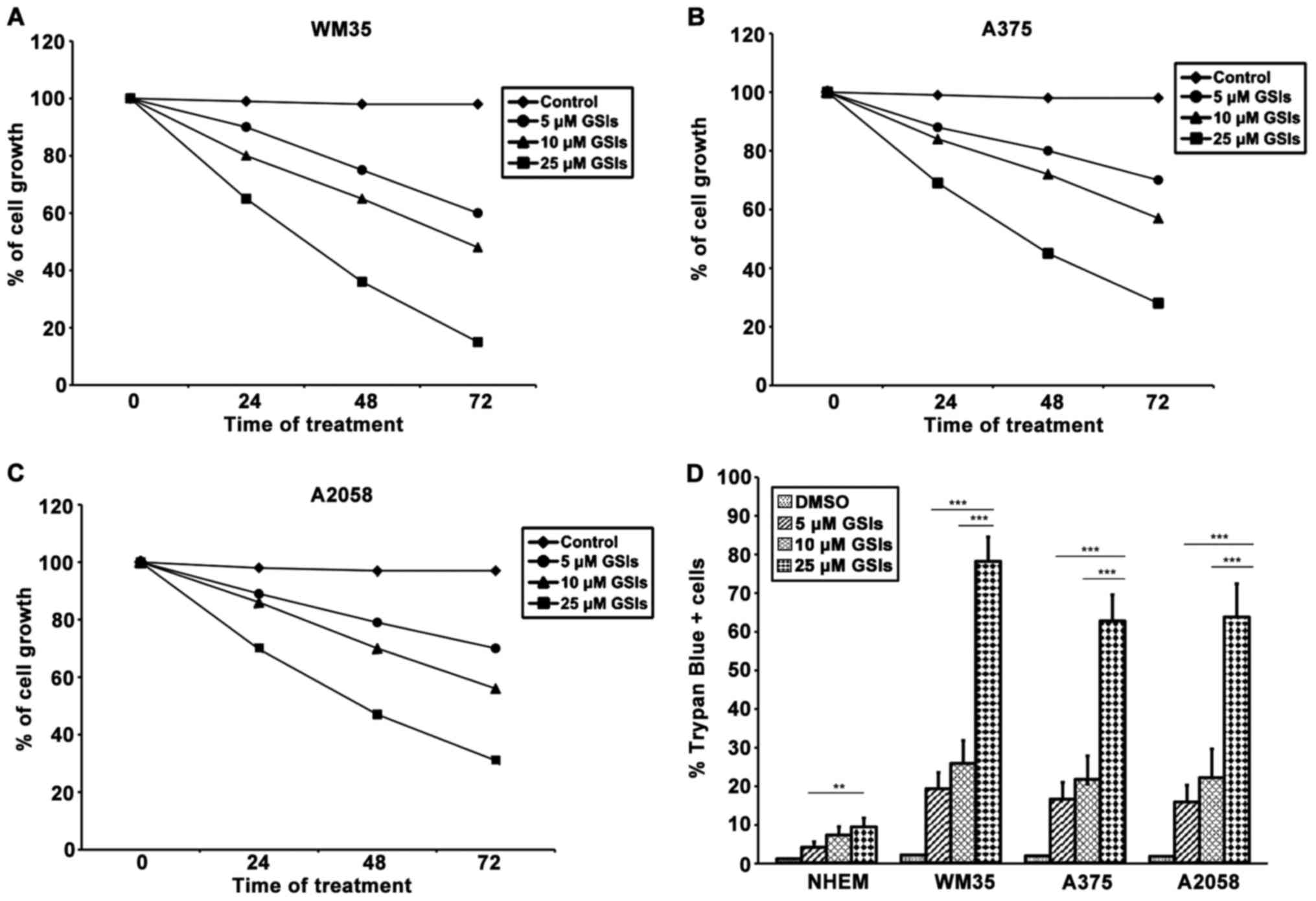
|
1
|
Buzaid AC: Management of metastatic
cutaneous melanoma. Oncology (Williston Park). 18:1443–1450;
discussion 1457–1459. 2004.
|
|
2
|
La Porta CA: Mechanism of drug sensitivity
and resistance in melanoma. Curr Cancer Drug Targets. 9:391–397.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fava P, Astrua C, Chiarugi A, Crocetti E,
Pimpinelli N, Fargnoli MC, Maurichi A, Rubegni P, Manganoni AM,
Bottoni U, et al: Differences in clinicopathological features and
distribution of risk factors in Italian melanoma patients.
Dermatology. 230:256–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maio M: Melanoma as a model tumour for
immune-oncology. Ann Oncol. 23(Suppl 8): viii10–14. 2012.
View Article : Google Scholar
|
|
5
|
Shrayer DP, Bogaars H, Wolf SF, Hearing VJ
and Wanebo HJ: A new mouse model of experimental melanoma for
vaccine and lymphokine therapy. Int J Oncol. 13:361–374.
1998.PubMed/NCBI
|
|
6
|
Nakai N, Katoh N, Kitagawa T, Ueda E,
Takenaka H and Kishimoto S: Immunoregulatory T cells in the
peripheral blood of melanoma patients treated with melanoma
antigen-pulsed mature monocyte-derived dendritic cell vaccination.
J Dermatol Sci. 54:31–37. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Russo A, Ficili B, Candido S, Pezzino FM,
Guarneri C, Biondi A, Travali S, McCubrey JA, Spandidos DA and
Libra M: Emerging targeted therapies for melanoma treatment
(Review). Int J Oncol. 45:516–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakamura K and Okuyama R: Immunotherapy
for advanced melanoma: Current knowledge and future directions. J
Dermatol Sci. 83:87–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Slingluff CL Jr, Chianese-Bullock KA,
Bullock TN, Grosh WW, Mullins DW, Nichols L, Olson W, Petroni G,
Smolkin M and Engelhard VH: Immunity to melanoma antigens: From
self-tolerance to immunotherapy. Adv Immunol. 90:243–295. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hori S, Nomura T and Sakaguchi S: Control
of regulatory T cell development by the transcription factor Foxp3.
Science. 299:1057–1061. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramsdell F: Foxp3 and natural regulatory T
cells: Key to a cell lineage. Immunity. 19:165–168. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakaguchi S: Naturally arising
CD4+ regulatory t cells for immunologic self-tolerance
and negative control of immune responses. Annu Rev Immunol.
22:531–562. 2004. View Article : Google Scholar
|
|
13
|
Takeuchi Y and Nishikawa H: Roles of
regulatory T cells in cancer immunity. Int Immunol. 28:401–409.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu R, Li S, Yang WH and Wang L: IPEX
syndrome, FOXP3 and cancer. J Syndr. 1:72013.PubMed/NCBI
|
|
15
|
Martin F, Ladoire S, Mignot G, Apetoh L
and Ghiringhelli F: Human FOXP3 and cancer. Oncogene. 29:4121–4129.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coffer PJ and Burgering BM: Forkhead-box
transcription factors and their role in the immune system. Nat Rev
Immunol. 4:889–899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen Z, Chen L, Hao F and Wu J:
Transcriptional regulation of Foxp3 gene: Multiple signal pathways
on the road. Med Res Rev. 29:742–766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu H: FOXP3 expression and prognosis: Role
of both the tumor and T cells. J Clin Oncol. 27:1735–1736. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hinz S, Pagerols-Raluy L, Oberg HH,
Ammerpohl O, Grüssel S, Sipos B, Grützmann R, Pilarsky C,
Ungefroren H, Saeger HD, et al: Foxp3 expression in pancreatic
carcinoma cells as a novel mechanism of immune evasion in cancer.
Cancer Res. 67:8344–8350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karanikas V, Speletas M, Zamanakou M,
Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI and
Germenis AE: Foxp3 expression in human cancer cells. J Transl Med.
6:192008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang WH, Jiang CL, Yan W, Zhang YH, Yang
JT, Zhang C, Yan B, Zhang W, Han W, Wang JZ and Zhang YQ: FOXP3
expression and clinical characteristics of hepatocellular
carcinoma. World J Gastroenterol. 16:5502–5509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu HY, Li C, Yang W, Gai XD, Jia T, Lei YM
and Li Y: FOXP3 and TLR4 protein expression are correlated in
non-small cell lung cancer: Implications for tumor progression and
escape. Acta Histochem. 115:151–157. 2013. View Article : Google Scholar
|
|
23
|
Kim M, Grimmig T, Grimm M, Lazariotou M,
Meier E, Rosenwald A, Tsaur I, Blaheta R, Heemann U, Germer CT, et
al: Expression of Foxp3 in colorectal cancer but not in Treg cells
correlates with disease progression in patients with colorectal
cancer. PLoS One. 8:e536302013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Merlo A, Casalini P, Carcangiu ML,
Malventano C, Triulzi T, Mènard S, Tagliabue E and Balsari A: FOXP3
expression and overall survival in breast cancer. J Clin Oncol.
27:1746–1752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wolf D, Wolf AM, Rumpold H, Fiegl H,
Zeimet AG, Muller-Holzner E, Deibl M, Gastl G, Gunsilius E and
Marth C: The expression of the regulatory T cell-specific forkhead
box transcription factor FoxP3 is associated with poor prognosis in
ovarian cancer. Clin Cancer Res. 11:8326–8331. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kiniwa Y, Miyahara Y, Wang HY, Peng W,
Peng G, Wheeler TM, Thompson TC, Old LJ and Wang RF:
CD8+ Foxp3+ regulatory T cells mediate
immunosuppression in prostate cancer. Clin Cancer Res.
13:6947–6958. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Niu J, Jiang C, Li C, Liu L, Li K, Jian Z
and Gao T: Foxp3 expression in melanoma cells as a possible
mechanism of resistance to immune destruction. Cancer Immunol
Immunother. 60:1109–1118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ebert LM, Tan BS, Browning J, Svobodova S,
Russell SE, Kirkpatrick N, Gedye C, Moss D, Ng SP, MacGregor D, et
al: The regulatory T cell-associated transcription factor FoxP3 is
expressed by tumor cells. Cancer Res. 68:3001–3009. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Douglass S, Ali S, Meeson AP, Browell D
and Kirby JA: The role of FOXP3 in the development and metastatic
spread of breast cancer. Cancer Metastasis Rev. 31:843–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng C, Yao Y, Jie W, Zhang M, Hu X, Zhao
Y, Wang S, Yin J and Song Y: Up-regulation of Foxp3 participates in
progression of cervical cancer. Cancer Immunol Immunother.
62:481–487. 2013. View Article : Google Scholar
|
|
31
|
Triulzi T, Tagliabue E, Balsari A and
Casalini P: FOXP3 expression in tumor cells and implications for
cancer progression. J Cell Physiol. 228:30–35. 2013. View Article : Google Scholar
|
|
32
|
Quaglino P, Osella-Abate S, Marenco F,
Nardò T, Gado C, Novelli M, Savoia P and Bernengo MG: FoxP3
expression on melanoma cells is related to early visceral spreading
in melanoma patients treated by electrochemotherapy. Pigment Cell
Melanoma Res. 24:734–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gerber AL, Münst A, Schlapbach C, Shafighi
M, Kiermeir D, Hüsler R and Hunger RE: High expression of FOXP3 in
primary melanoma is associated with tumour progression. Br J
Dermatol. 170:103–109. 2014. View Article : Google Scholar
|
|
34
|
Viguier M, Lemaître F, Verola O, Cho MS,
Gorochov G, Dubertret L, Bachelez H, Kourilsky P and Ferradini L:
Foxp3 expressing CD4+CD25(high) regulatory T cells are
overrepresented in human metastatic melanoma lymph nodes and
inhibit the function of infiltrating T cells. J Immunol.
173:1444–1453. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Knol AC, Nguyen JM, Quéreux G, Brocard A,
Khammari A and Dréno B: Prognostic value of tumor-infiltrating
Foxp3+ T-cell subpopulations in metastatic melanoma. Exp
Dermatol. 20:430–434. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jandus C, Bioley G, Speiser DE and Romero
P: Selective accumulation of differentiated FOXP3(+) CD4 (+) T
cells in metastatic tumor lesions from melanoma patients compared
to peripheral blood. Cancer Immunol Immunother. 57:1795–1805. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L and Zhao Y: The regulation of
Foxp3 expression in regulatory CD4(+)CD25(+)T cells: Multiple
pathways on the road. J Cell Physiol. 211:590–597. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Liu Y, Dai L, Liu Q, Jia L, Wang
H, An L, Jing X, Liu M, Li P and Cheng Z: Foxp3 downregulation in
NSCLC mediates epithelial-mesenchymal transition via NF-κB
signaling. Oncol Rep. 36:2282–2288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ou-Yang HF, Zhang HW, Wu CG, Zhang P,
Zhang J, Li JC, Hou LH, He F, Ti XY, Song LQ, et al: Notch
signaling regulates the FOXP3 promoter through RBP-J- and
Hes1-dependent mechanisms. Mol Cell Biochem. 320:109–114. 2009.
View Article : Google Scholar
|
|
40
|
Maruyama T, Konkel JE, Zamarron BF and
Chen W: The molecular mechanisms of Foxp3 gene regulation. Semin
Immunol. 23:418–423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uzdensky AB, Demyanenko SV and Bibov MY:
Signal transduction in human cutaneous melanoma and target drugs.
Curr Cancer Drug Targets. 13:843–866. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu J, Sato C, Cerletti M and Wagers A:
Notch signaling in the regulation of stem cell self-renewal and
differentiation. Curr Top Dev Biol. 92:367–409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bray SJ: Notch signalling: A simple
pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Radtke F and Raj K: The role of Notch in
tumorigenesis: Oncogene or tumour suppressor. Nat Rev Cancer.
3:756–767. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Roy M, Pear WS and Aster JC: The
multifaceted role of Notch in cancer. Curr Opin Genet Dev.
17:52–59. 2007. View Article : Google Scholar
|
|
47
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fortini ME: Notch signaling: The core
pathway and its posttranslational regulation. Dev Cell. 16:633–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schroeter EH, Kisslinger JA and Kopan R:
Notch-1 signalling requires ligand-induced proteolytic release of
intracellular domain. Nature. 393:382–386. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Koch U and Radtke F: Notch signaling in
solid tumors. Curr Top Dev Biol. 92:411–455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao J, Dong Y, Zhang B, Xiong Y, Xu W,
Cheng Y, Dai M, Yu Z, Xu H and Zheng G: Notch1 activation
contributes to tumor cell growth and proliferation in human
hepatocellular carcinoma HepG2 and SMMC7721 cells. Int J Oncol.
41:1773–1781. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bolós V, Mira E, Martínez-Poveda B, Luxán
G, Cañamero M, Martínez-A C, Mañes S and de la Pompa JL: Notch
activation stimulates migration of breast cancer cells and promotes
tumor growth. Breast Cancer Res. 15:R542013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Reedijk M, Odorcic S, Zhang H, Chetty R,
Tennert C, Dickson BC, Lockwood G, Gallinger S and Egan SE:
Activation of Notch signaling in human colon adenocarcinoma. Int J
Oncol. 33:1223–1229. 2008.PubMed/NCBI
|
|
54
|
Yuan X, Wu H, Xu H, Han N, Chu Q, Yu S,
Chen Y and Wu K: Meta-analysis reveals the correlation of Notch
signaling with non-small cell lung cancer progression and
prognosis. Sci Rep. 5:103382015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hijioka H, Setoguchi T, Miyawaki A, Gao H,
Ishida T, Komiya S and Nakamura N: Upregulation of Notch pathway
molecules in oral squamous cell carcinoma. Int J Oncol. 36:817–822.
2010.PubMed/NCBI
|
|
56
|
Ai Q, Ma X, Huang Q, Liu S, Shi T, Zhang
C, Zhu M, Zhang Y, Wang B, Ni D, et al: High-level expression of
Notch1 increased the risk of metastasis in T1 stage clear cell
renal cell carcinoma. PLoS One. 7:e350222012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pinnix CC, Lee JT, Liu ZJ, McDaid R,
Balint K, Beverly LJ, Brafford PA, Xiao M, Himes B, Zabierowski SE,
et al: Active Notch1 confers a transformed phenotype to primary
human melanocytes. Cancer Res. 69:5312–5320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Howard JD, Moriarty WF, Park J, Riedy K,
Panova IP, Chung CH, Suh KY, Levchenko A and Alani RM: Notch
signaling mediates melanoma-endothelial cell communication and
melanoma cell migration. Pigment Cell Melanoma Res. 26:697–707.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Müller CS: Notch signaling and malignant
melanoma. Adv Exp Med Biol. 727:258–264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu ZJ, Xiao M, Balint K, Smalley KS,
Brafford P, Qiu R, Pinnix CC, Li X and Herlyn M: Notch1 signaling
promotes primary melanoma progression by activating
mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt
pathways and up-regulating N-cadherin expression. Cancer Res.
66:4182–4190. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Balint K, Xiao M, Pinnix CC, Soma A, Veres
I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M and Liu ZJ:
Activation of Notch1 signaling is required for
beta-catenin-mediated human primary melanoma progression. J Clin
Invest. 115:3166–3176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Massi D, Tarantini F, Franchi A,
Paglierani M, Di Serio C, Pellerito S, Leoncini G, Cirino G,
Geppetti P and Santucci M: Evidence for differential expression of
Notch receptors and their ligands in melanocytic nevi and cutaneous
malignant melanoma. Mod Pathol. 19:246–254. 2006. View Article : Google Scholar
|
|
63
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer - a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001.PubMed/NCBI
|
|
64
|
Trapani JA: The dual adverse effects of
TGF-beta secretion on tumor progression. Cancer Cell. 8:349–350.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Massagué J: TGFbeta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li MO, Wan YY, Sanjabi S, Robertson AK and
Flavell RA: Transforming growth factor-beta regulation of immune
responses. Annu Rev Immunol. 24:99–146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huber S, Schramm C, Lehr HA, Mann A,
Schmitt S, Becker C, Protschka M, Galle PR, Neurath MF and Blessing
M: Cutting edge: TGF-beta signaling is required for the in vivo
expansion and immunosuppressive capacity of regulatory
CD4+CD25+ T cells. J Immunol. 173:6526–6531.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen W, Jin W, Hardegen N, Lei KJ, Li L,
Marinos N, McGrady G and Wahl SM: Conversion of peripheral
CD4+CD25− naive T cells to
CD4+CD25+ regulatory T cells by TGF-beta
induction of transcription factor Foxp3. J Exp Med. 198:1875–1886.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Pyzik M and Piccirillo CA: TGF-beta1
modulates Foxp3 expression and regulatory activity in distinct
CD4+ T cell subsets. J Leukoc Biol. 82:335–346. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Oft M, Heider KH and Beug H: TGFbeta
signaling is necessary for carcinoma cell invasiveness and
metastasis. Curr Biol. 8:1243–1252. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang HJ, Wang HY, Zhang HT, Su JM, Zhu J,
Wang HB, Zhou WY, Zhang H, Zhao MC, Zhang L and Chen XF:
Transforming growth factor-β1 promotes lung adenocarcinoma invasion
and metastasis by epithelial-to-mesenchymal transition. Mol Cell
Biochem. 355:309–314. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lee D, Chung YH, Kim JA and Lee YS, Lee D,
Jang MK, Kim KM, Lim YS, Lee HC and Lee YS: Transforming growth
factor beta 1 overexpression is closely related to invasiveness of
hepatocellular carcinoma. Oncology. 82:11–18. 2012. View Article : Google Scholar
|
|
73
|
Teraoka H, Sawada T, Yamashita Y, Nakata
B, Ohira M, Ishikawa T, Nishino H and Hirakawa K: TGF-β1 promotes
liver metastasis of pancreatic cancer by modulating the capacity of
cellular invasion. Int J Oncol. 19:709–715. 2001.PubMed/NCBI
|
|
74
|
Malaponte G, Zacchia A, Bevelacqua Y,
Marconi A, Perrotta R, Mazzarino MC, Cardile V and Stivala F:
Co-regulated expression of matrix metalloproteinase-2 and
transforming growth factor-β in melanoma development and
progression. Oncol Rep. 24:81–87. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ostroukhova M, Qi Z, Oriss TB,
Dixon-McCarthy B, Ray P and Ray A: Treg-mediated immunosuppression
involves activation of the Notch-HES1 axis by membrane-bound
TGF-beta. J Clin Invest. 116:996–1004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Samon JB, Champhekar A, Minter LM, Telfer
JC, Miele L, Fauq A, Das P, Golde TE and Osborne BA: Notch1 and
TGFbeta1 cooperatively regulate Foxp3 expression and the
maintenance of peripheral regulatory T cells. Blood. 112:1813–1821.
2008. View Article : Google Scholar
|
|
77
|
Zhou J, Jain S, Azad AK, Xu X, Yu HC, Xu
Z, Godbout R and Fu Y: Notch and TGFβ form a positive regulatory
loop and regulate EMT in epithelial ovarian cancer cells. Cell
Signal. 28:838–849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zavadil J, Cermak L, Soto-Nieves N and
Böttinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial-to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Blokzijl A, Dahlqvist C, Reissmann E, Falk
A, Moliner A, Lendahl U and Ibáñez CF: Cross-talk between the Notch
and TGF-beta signaling pathways mediated by interaction of the
Notch intracellular domain with Smad3. J Cell Biol. 163:723–728.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Klüppel M and Wrana JL: Turning it up a
Notch: Cross-talk between TGF beta and Notch signaling. BioEssays.
27:115–118. 2005. View Article : Google Scholar
|
|
81
|
Barbarulo A, Grazioli P, Campese AF,
Bellavia D, Di Mario G, Pelullo M, Ciuffetta A, Colantoni S, Vacca
A, Frati L, et al: Notch3 and canonical NF-kappaB signaling
pathways cooperatively regulate Foxp3 transcription. J Immunol.
186:6199–6206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Burghardt S, Claass B, Erhardt A, Karimi K
and Tiegs G: Hepatocytes induce Foxp3+ regulatory T
cells by Notch signaling. J Leukoc Biol. 96:571–577. 2014.
View Article : Google Scholar
|
|
83
|
Mota C, Nunes-Silva V, Pires AR, Matoso P,
Victorino RM, Sousa AE and Caramalho I: Delta-like 1-mediated Notch
signaling enhances the in vitro conversion of human memory CD4 T
cells into FOXP3-expressing regulatory T cells. J Immunol.
193:5854–5862. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Trehanpati N, Shrivastav S, Shivakumar B,
Khosla R, Bhardwaj S, Chaturvedi J, Sukriti, Kumar B, Bose S, Mani
Tripathi D, et al: Analysis of Notch and TGF-β signaling expression
in different stages of disease progression during hepatitis B virus
infection. Clin Transl Gastroenterol. 3:e232012. View Article : Google Scholar
|
|
85
|
Luo X, Tan H, Zhou Y, Xiao T, Wang C and
Li Y: Notch1 signaling is involved in regulating Foxp3 expression
in T-ALL. Cancer Cell Int. 13:342013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Josien H: Recent advances in the
development of gamma-secretase inhibitors. Curr Opin Drug Discov
Devel. 5:513–525. 2002.PubMed/NCBI
|
|
87
|
Cardile V, Frasca G, Libra M, Caggia S,
Umezawa K, Panico A and Malaponte G: Dehydroxymethylepoxyquinomicin
inhibits expression and production of inflammatory mediators in
interleukin-1beta-induced human chondrocytes. Cell Physiol Biochem.
25:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ohnuki H and Tosato G: Notch and TGFβ:
Functional partners facilitating tumor progression. OncoImmunology.
3:e290292014. View Article : Google Scholar
|
|
89
|
Hoek K, Rimm DL, Williams KR, Zhao H,
Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et
al: Expression profiling reveals novel pathways in the
transformation of melanocytes to melanomas. Cancer Res.
64:5270–5282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Perrot CY, Javelaud D and Mauviel A:
Insights into the transforming growth factor-β signaling pathway in
cutaneous melanoma. Ann Dermatol. 25:135–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang J, Wang Y, Li D and Jing S: Notch
and TGF-β/Smad3 pathways are involved in the interaction between
cancer cells and cancer-associated fibroblasts in papillary thyroid
carcinoma. Tumour Biol. 35:379–385. 2014. View Article : Google Scholar
|
|
92
|
Zhang HY and Sun H: Up-regulation of Foxp3
inhibits cell proliferation, migration and invasion in epithelial
ovarian cancer. Cancer Lett. 287:91–97. 2010. View Article : Google Scholar
|
|
93
|
Wang L, Liu R, Li W, Chen C, Katoh H, Chen
GY, McNally B, Lin L, Zhou P, Zuo T, et al: Somatic single hits
inactivate the X-linked tumor suppressor FOXP3 in the prostate.
Cancer Cell. 16:336–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zuo T, Liu R, Zhang H, Chang X and Liu Y,
Wang L, Zheng P and Liu Y: FOXP3 is a novel transcriptional
repressor for the breast cancer oncogene SKP2. J Clin Invest.
117:3765–3773. 2007.PubMed/NCBI
|
|
95
|
Liu Y, Zhang P, Li J, Kulkarni AB,
Perruche S and Chen W: A critical function for TGF-beta signaling
in the development of natural
CD4+CD25+Foxp3+ regulatory T
cells. Nat Immunol. 9:632–640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Brody JR, Costantino CL, Berger AC, Sato
T, Lisanti MP, Yeo CJ, Emmons RV and Witkiewicz AK: Expression of
indoleamine 2,3-dioxygenase in metastatic malignant melanoma
recruits regulatory T cells to avoid immune detection and affects
survival. Cell Cycle. 8:1930–1934. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen C, Rowell EA, Thomas RM, Hancock WW
and Wells AD: Transcriptional regulation by Foxp3 is associated
with direct promoter occupancy and modulation of histone
acetylation. J Biol Chem. 281:36828–36834. 2006. View Article : Google Scholar
|
|
98
|
Dimitrakopoulos FI, Papadaki H,
Antonacopoulou AG, Kottorou A, Gotsis AD, Scopa C, Kalofonos HP and
Mouzaki A: Association of FOXP3 expression with non-small cell lung
cancer. Anticancer Res. 31:1677–1683. 2011.PubMed/NCBI
|
|
99
|
Franco-Molina MA, Miranda-Hernández DF,
Mendoza- Gamboa E, Zapata-Benavides P, Coronado-Cerda EE, Sierra-
Rivera CA, Saavedra-Alonso S, Taméz-Guerra RS and Rodríguez-Padilla
C: Silencing of Foxp3 delays the growth of murine melanomas and
modifies the tumor immunosuppressive environment. OncoTargets Ther.
9:243–253. 2016. View Article : Google Scholar
|
|
100
|
Fantini MC, Becker C, Monteleone G,
Pallone F, Galle PR and Neurath MF: Cutting edge: TGF-beta induces
a regulatory phenotype in CD4+CD25− T cells
through Foxp3 induction and down-regulation of Smad7. J Immunol.
172:5149–5153. 2004. View Article : Google Scholar
|
|
101
|
Selvaraj RK and Geiger TL: A kinetic and
dynamic analysis of Foxp3 induced in T cells by TGF-beta. J
Immunol. 178:7667–7677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Guo X and Wang XF: Signaling cross-talk
between TGF-beta/BMP and other pathways. Cell Res. 19:71–88. 2009.
View Article : Google Scholar
|
|
103
|
Wang Y, Shen RW, Han B, Li Z, Xiong L,
Zhang FY, Cong BB and Zhang B: Notch signaling mediated by
TGF-β/Smad pathway in concanavalin A-induced liver fibrosis in
rats. World J Gastroenterol. 23:2330–2336. 2017. View Article : Google Scholar :
|
|
104
|
Yan XC, Cao J, Liang L, Wang L, Gao F,
Yang ZY, Duan JL, Chang TF, Deng SM, Liu Y, et al: miR-342-5p is a
notch downstream molecule and regulates multiple angiogenic
pathways including notch, vascular endothelial growth factor and
transforming growth factor β signaling. J Am Heart Assoc.
5:e0030422016. View Article : Google Scholar
|
|
105
|
Kared H, Adle-Biassette H, Foïs E, Masson
A, Bach JF, Chatenoud L, Schneider E and Zavala F:
Jagged2-expressing hematopoietic progenitors promote regulatory T
cell expansion in the periphery through notch signaling. Immunity.
25:823–834. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Stockhausen MT, Sjö J and Axelson H:
Regulation of the Notch target gene Hes-1 by TGFalpha induced
Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res.
310:218–228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Pisklakova A, Grigson E, Ozerova M, Chen
F, Sullivan DM and Nefedova Y: Anti-myeloma effect of
pharmacological inhibition of Notch/gamma-secretase with RO4929097
is mediated by modulation of tumor microenvironment. Cancer Biol
Ther. 17:477–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Tas F, Karabulut S, Yasasever CT and
Duranyildiz D: Serum transforming growth factor-beta 1 (TGF-β1)
levels have diagnostic, predictive, and possible prognostic roles
in patients with melanoma. Tumour Biol. 35:7233–7237. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Takizawa T, Ochiai W, Nakashima K and Taga
T: Enhanced gene activation by Notch and BMP signaling cross-talk.
Nucleic Acids Res. 31:5723–5731. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Asnaghi L, Ebrahimi KB, Schreck KC, Bar
EE, Coonfield ML, Bell WR, Handa J, Merbs SL, Harbour JW and
Eberhart CG: Notch signaling promotes growth and invasion in uveal
melanoma. Clin Cancer Res. 18:654–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sriuranpong V, Borges MW, Ravi RK, Arnold
DR, Nelkin BD, Baylin SB and Ball DW: Notch signaling induces cell
cycle arrest in small cell lung cancer cells. Cancer Res.
61:3200–3205. 2001.PubMed/NCBI
|
|
112
|
Thélu J, Rossio P and Favier B: Notch
signalling is linked to epidermal cell differentiation level in
basal cell carcinoma, psoriasis and wound healing. BMC Dermatol.
2:72002. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Panelos J, Tarantini F, Paglierani M, Di
Serio C, Maio V, Pellerito S, Pimpinelli N, Santucci M and Massi D:
Photoexposition discriminates Notch 1 expression in human cutaneous
squamous cell carcinoma. Mod Pathol. 21:316–325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Banerjee D, Hernandez SL, Garcia A,
Kangsamaksin T, Sbiroli E, Andrews J, Forrester LA, Wei N,
Kadenhe-Chiweshe A, Shawber CJ, et al: Notch suppresses
angiogenesis and progression of hepatic metastases. Cancer Res.
75:1592–1602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Talora C, Cialfi S, Segatto O, Morrone S,
Kim Choi J, Frati L, Paolo Dotto G, Gulino A and Screpanti I:
Constitutively active Notch1 induces growth arrest of HPV-positive
cervical cancer cells via separate signaling pathways. Exp Cell
Res. 305:343–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Takebe N, Nguyen D and Yang SX: Targeting
notch signaling pathway in cancer: Clinical development advances
and challenges. Pharmacol Ther. 141:140–149. 2014. View Article : Google Scholar :
|
|
117
|
Olsauskas-Kuprys R, Zlobin A and Osipo C:
Gamma secretase inhibitors of Notch signaling. Onco Targets Ther.
6:943–955. 2013.PubMed/NCBI
|
|
118
|
Ji X, Wang Z, Geamanu A, Sarkar FH and
Gupta SV: Inhibition of cell growth and induction of apoptosis in
non-small cell lung cancer cells by delta-tocotrienol is associated
with notch-1 down-regulation. J Cell Biochem. 112:2773–2783. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Wang M, Wu L, Wang L and Xin X:
Down-regulation of Notch1 by gamma-secretase inhibition contributes
to cell growth inhibition and apoptosis in ovarian cancer cells
A2780. Biochem Biophys Res Commun. 393:144–149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Hu J, Zhu X and Lu Q: Antiproliferative
effects of γ-secretase inhibitor, a Notch signalling inhibitor, in
multiple myeloma cells and its molecular mechanism of action. J Int
Med Res. 41:1017–1026. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Qi R, An H, Yu Y, Zhang M, Liu S, Xu H,
Guo Z, Cheng T and Cao X: Notch1 signaling inhibits growth of human
hepatocellular carcinoma through induction of cell cycle arrest and
apoptosis. Cancer Res. 63:8323–8329. 2003.PubMed/NCBI
|
|
122
|
Wang L, Qin H, Chen B, Xin X, Li J and Han
H: Overexpressed active Notch1 induces cell growth arrest of HeLa
cervical carcinoma cells. Int J Gynecol Cancer. 17:1283–1292. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Miranda-Hernández DF, Franco-Molina MA,
Mendoza-Gamboa E, Zapata-Benavides P, Sierra-Rivera CA,
Coronado-Cerda EE, Rosas-Taraco AG, Taméz-Guerra RS and
Rodríguez-Padilla C: Expression of Foxp3, CD25 and IL-2 in the
B16F10 cancer cell line and melanoma is correlated with tumor
growth in mice. Oncol Lett. 6:1195–1200. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Del Papa B, Sportoletti P, Cecchini D,
Rosati E, Balucani C, Baldoni S, Fettucciari K, Marconi P, Martelli
MF, Falzetti F and Di Ianni M: Notch1 modulates mesenchymal stem
cells mediated regulatory T-cell induction. Eur J Immunol.
43:182–187. 2013. View Article : Google Scholar
|
|
125
|
Rao P and Kadesch T: The intracellular
form of notch blocks transforming growth factor beta-mediated
growth arrest in Mv1Lu epithelial cells. Mol Cell Biol.
23:6694–6701. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sun XF, Sun XH, Cheng SF, Wang JJ, Feng
YN, Zhao Y, Yin S, Hou ZM, Shen W and Zhang XF: Interaction of the
transforming growth factor-β and Notch signaling pathways in the
regulation of granulosa cell proliferation. Reprod Fertil Dev.
28:1873–1881. 2016. View Article : Google Scholar
|
|
127
|
Masuda S, Kumano K, Shimizu K, Imai Y,
Kurokawa M, Ogawa S, Miyagishi M, Taira K, Hirai H and Chiba S:
Notch1 oncoprotein antagonizes TGF-beta/Smad-mediated cell growth
suppression via sequestration of coactivator p300. Cancer Sci.
96:274–282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Asano N, Watanabe T, Kitani A, Fuss IJ and
Strober W: Notch1 signaling and regulatory T cell function. J
Immunol. 180:2796–2804. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Xu L, Kitani A, Stuelten C, McGrady G,
Fuss I and Strober W: Positive and negative transcriptional
regulation of the Foxp3 gene is mediated by access and binding of
the Smad3 protein to enhancer I. Immunity. 33:313–325. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Tone Y, Furuuchi K, Kojima Y, Tykocinski
ML, Greene MI and Tone M: Smad3 and NFAT cooperate to induce Foxp3
expression through its enhancer. Nat Immunol. 9:194–202. 2008.
View Article : Google Scholar
|