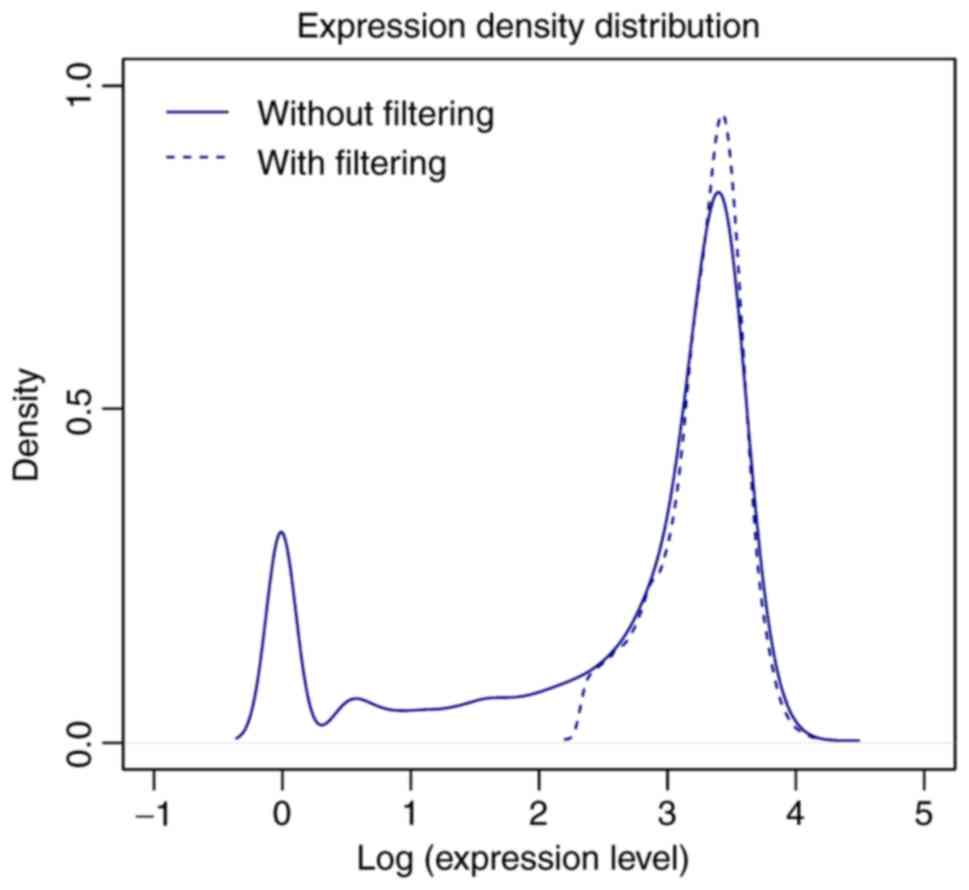
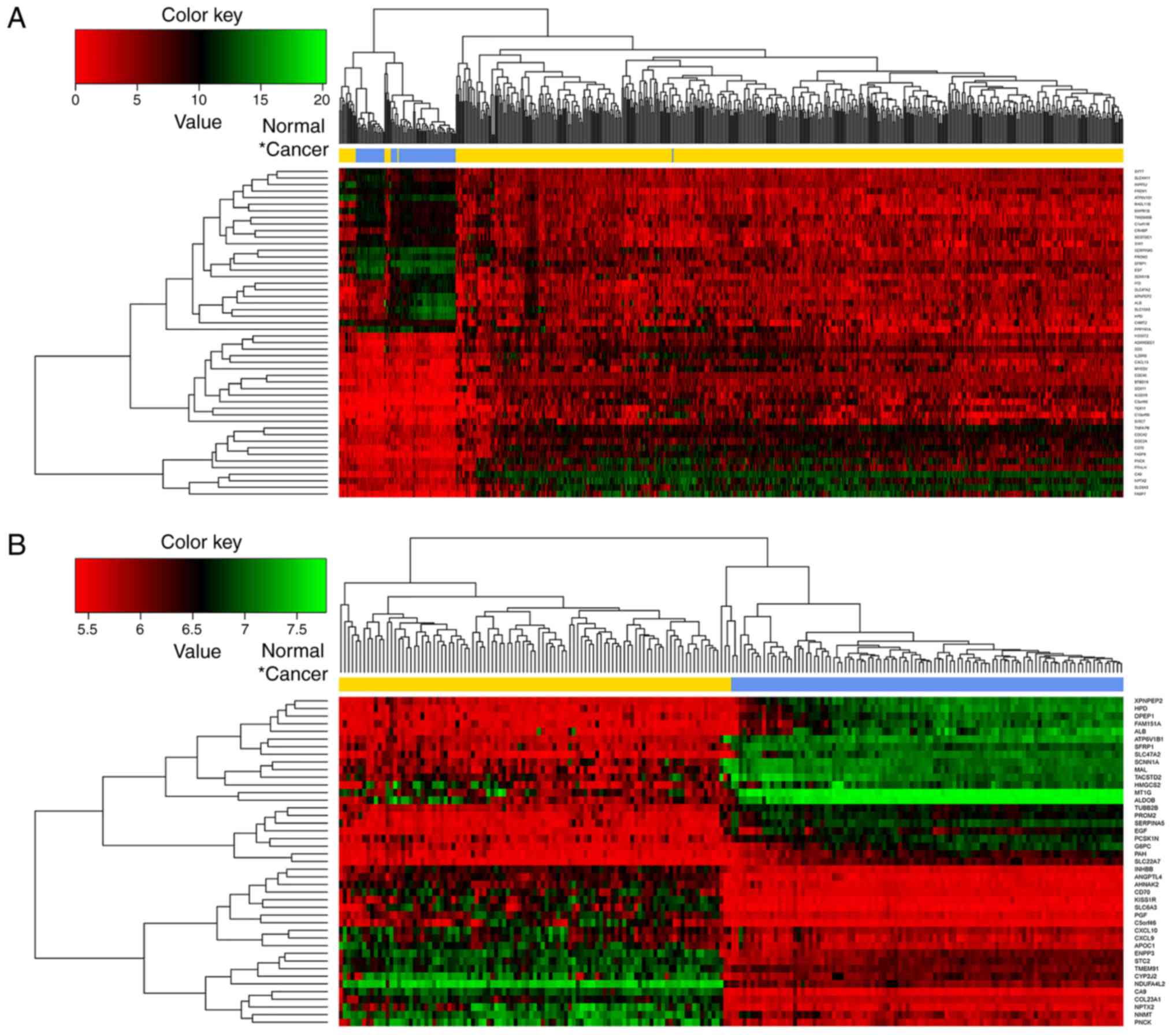
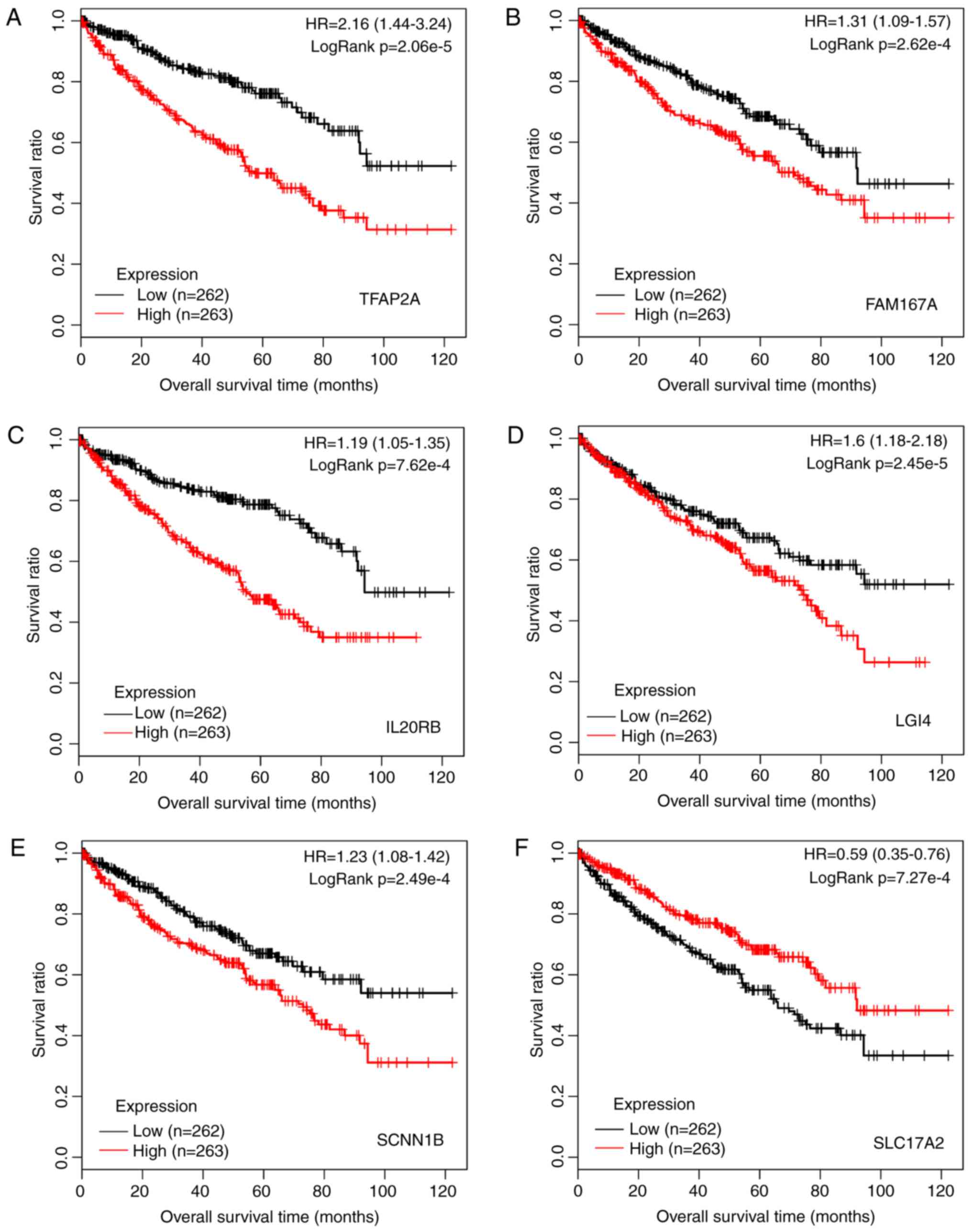
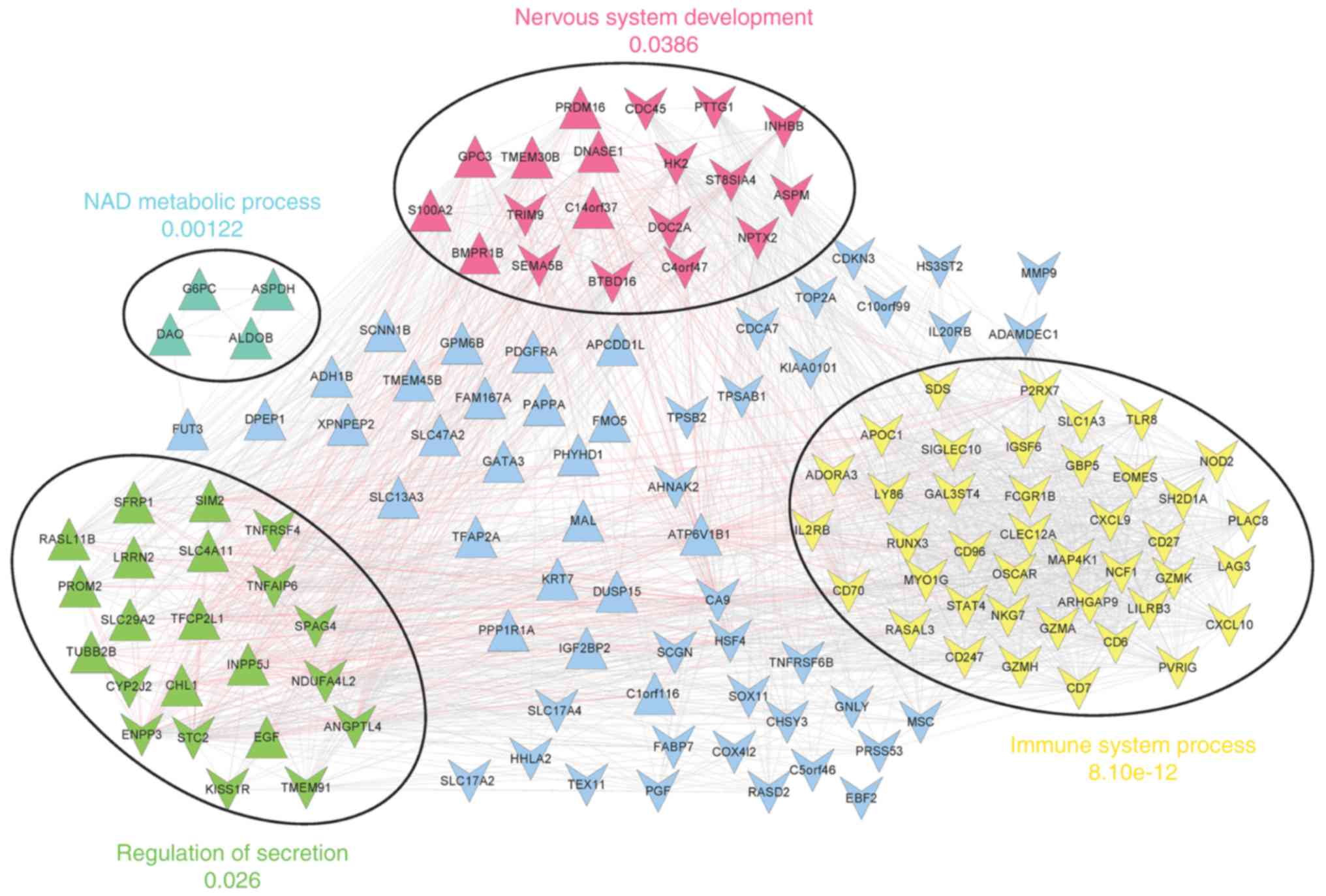
|
1
|
Cohen HT and Mcgovern FJ: Renalcell
carcinoma. N Engl Med. 353:2477–2490. 2005. View Article : Google Scholar
|
|
2
|
Jonasch E, Gao J and Rathmell WK: Renal
cell carcinoma. BMJ. 349:g47972014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberg B, Cowan NC, Hanbury DC, Hora M,
Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC;
European Association of Urology Guideline Group: EAU guidelines on
renal cell carcinoma: the 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Znaor A, Lortet-Tieulent J, Laversanne M,
Jemal A and Bray F: International variations and trends in renal
cell carcinoma incidence and mortality. Eur Urol. 67:519–530. 2015.
View Article : Google Scholar
|
|
5
|
Young JR, Margolis D, Sauk S, Pantuck AJ,
Sayre J and Raman SS: Clear cell renal cell carcinoma:
Discrimination from other renal cell carcinoma subtypes and
oncocytoma at multi-phasic multidetector CT. Radiology.
267:444–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar
|
|
7
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klatte T, Seligson DB, Riggs SB, Leppert
JT, Berkman MK, Kleid MD, Yu H, Kabbinavar FF, Pantuck AJ and
Belldegrun AS: Hypoxia-inducible factor 1α in clear cell renal cell
carcinoma. Clin Cancer Res. 13:7388–7393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Xu Y, Zhu L, Zou Y, Kong W, Dong
B, Huang J, Chen Y, Xue W, Huang Y and Zhang J: Cannabinoid
receptor 2 as a novel target for promotion of renal cell carcinoma
prognosis and progression. J Cancer Res Clin Oncol. 144:39–52.
2018. View Article : Google Scholar
|
|
10
|
Mizutani Y, Nakanishi H, Li YN, Matsubara
H, Yamamoto K, Sato N, Shiraishi T, Nakamura T, Mikami K, Okihara
K, et al: Overexpression of XIAP expression in renal cell carcinoma
predicts a worse prognosis. Int J Oncol. 30:919–925.
2007.PubMed/NCBI
|
|
11
|
Yamada T, Horinaka M, Shinnoh M, Yoshioka
T, Miki T and Sakai T: A novel HDAC inhibitor OBP-801 and a PI3K
inhibitor LY294002 synergistically induce apoptosis via the
suppression of survivin and XIAP in renal cell carcinoma. Int J
Oncol. 43:1080–1086. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tostain J, Li G, Gentilperret A and
Gigante M: Carbonic anhydrase 9 in clear cell renal cell carcinoma:
A marker for diagnosis, prognosis and treatment. Eur J Cancer.
46:3141–3148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choueiri TK, Regan MM, Rosenberg JE, Oh
WK, Clement J, Amato AM, McDermott D, Cho DC, Atkins MB and
Signoretti S: Carbonic anhydrase IX and pathological features as
predictors of outcome in patients with metastatic clear-cell renal
cell carcinoma receiving vascular endothelial growth
factor-targeted therapy. Bju Int. 106:772–778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang W, Huang P, Zhao M and Lau YL:
Biomarker identification for early tumor detection aided by
bioinformatics gene expression analysis. International Conference
on Biomedical Engineering and Informatics. Sanya; Hainan, China:
pp. 469–473. 2008
|
|
15
|
Fonseca AL, da Silva VL, da Fonsêca MM,
Meira ITJ, da Silva TE, Kroll JE, Ribeiro-dos-Santos AM, Freitas
CR, Furtado R, de Souza JE, et al: Bioinformatics analysis of the
human surfaceome reveals new targets for a variety of tumor types.
Int J Genomics. 2016.1–7. 2016. View Article : Google Scholar
|
|
16
|
Smyth GK: Limma: Linear Models for
Microarray Data. Springer; New York: 2005
|
|
17
|
Wang Y, Zeigler MM, Lam GK, Hunter MG,
Eubank TD, Khramtsov VV, Tridandapani S, Sen CK and Marsh CB: The
role of the NADPH oxidase complex, p38 MAPK, and Akt in regulating
human monocyte/macrophage survival. Am J Respir Cell Mol Biol.
36:68–77. 2007. View Article : Google Scholar
|
|
18
|
Bruford EA, Lush MJ, Wright MW, Sneddon
TP, Sue P and Ewan B: The HGNC Database in 2008: A resource for the
human genome. Nucleic Acids Res. 36:445–448. 2008. View Article : Google Scholar
|
|
19
|
Pollard KS, Dudoit S and van der Laan MJ:
Multiple testing procedures: The multtest package and applications
to genomics. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Du ST: Springer; New York: pp. 465. 2005
|
|
20
|
Therneau TM: Survival analysis. R package
survival, version 2. 39–5. 2016.
|
|
21
|
Kleinbaum DG and Klein M: Kaplan-meier
survival curves and the log-rank test. Statistics for Biology and
Health. Springer; New York: pp. 45–82. 2005
|
|
22
|
Porcher R: CORR Insights(®): Kaplan-meier
survival analysis overestimates the risk of revision arthroplasty:
A meta-analysis. Clin Orthop Relat Res. 473:3443–3445. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nowicka-Zagrajek J and Weron R: COR:
MATLAB function to compute the correlation coefficients. Hsc
software. 2008.
|
|
24
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar
|
|
25
|
Yu G, Wang LG, Han Y and He QY: Cluster
profiler: An R package for comparing biological themes among gene
clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:169–175. 2007. View Article : Google Scholar
|
|
27
|
Reimand J, Tooming L, Peterson H, Adler P
and Vilo J: GraphWeb: Mining heterogeneous biological networks for
gene modules with functional significance. Nucleic Acids Res.
36:452–459. 2008. View Article : Google Scholar
|
|
28
|
Dimitriadou E, Hornik K, Leisch F, Meyer D
and Weingesse A: The e1071 package. Ethnos J Anthropol. 23:55–56.
2006.
|
|
29
|
Wuttig D, Zastrow S, Füssel S, Toma MI,
Meinhardt M, Kalman K, Junker K, Sanjmyatav J, Boll K, Hackermüller
J, et al: CD31, EDNRB and TSPAN7 are promising prognostic markers
in clear-cell renal cell carcinoma revealed by genomewide
expression analyses of primary tumors and metastases. Int J Cancer.
131:E693–E704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Arai E, Ushijima S, Tsuda H, Fujimoto H,
Hosoda F, Shibata T, Kondo T, Imoto I, Inazawa J, Hirohashi S and
Kanai Y: Genetic clustering of clear cell renal cell carcinoma
based on array-comparative genomic hybridization: Its association
with DNA methylation alteration and patient outcome. Clin Cancer
Res. 14:5531–5539. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Verine J, Lehmann-Che J, Soliman H,
Feugeas JP, Vidal JS, Mongiat-Artus P, Belhadj S, Philippe J,
Lesage M, Wittmer E, et al: Determination of angptl4 mRNA as a
diagnostic marker of primary and metastatic clear cell renal-cell
carcinoma. PLoS One. 5:e104212010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Galaup A, Cazes A, Le Jan S, Philippe J,
Connault E, Le Coz E, Mekid H, Mir LM, Opolon P, Corvol P, et al:
Angiopoietin-like 4 prevents metastasis through inhibition of
vascular permeability and tumor cell motility and invasiveness.
Proc Natl Acad Sci USA. 103:18721–18726. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong D, Jia L, Zhou Y, Ren L, Li J and
Zhang J: Serum level of ANGPTL4 as a potential biomarker in renal
cell carcinoma. Urol Oncol. 35:279–285. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xin M, Gu L, Li H, Gao Y, Li X, Shen D,
Gong H, Li S, Niu S, Zhang Y, et al: Hypoxia-induced overexpression
of stannio-calcin-1 is associated with the metastasis of early
stage clear cell renal cell carcinoma. J Transl Med. 13:562015.
View Article : Google Scholar
|
|
35
|
Meyer HA, Tölle A, Jung M, Fritzsche FR,
Haendler B, Kristiansen I, Gaspert A, Johannsen M, Jung K and
Kristiansen G: Identification of stanniocalcin 2 as prognostic
marker in renal cell carcinoma. Eur Urol. 55:669–678. 2009.
View Article : Google Scholar
|
|
36
|
Girgis AH, Iakovlev VV, Beheshti B, Bayani
J, Squire JA, Bui A, Mankaruos M, Youssef Y, Khalil B, Khella H, et
al: Multilevel whole-genome analysis reveals candidate biomarkers
in clear cell renal cell carcinoma. Cancer Res. 72:5273–5284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Knaup KX, Monti J, Hackenbeck T,
Jobst-Schwan T, Klanke B, Schietke RE, Wacker I, Behrens J, Amann
K, Eckardt KU, et al: Hypoxia regulates the sperm associated
antigen 4 (SPAG4) via HIF, which is expressed in renal clear cell
carcinoma and promotes migration and invasion in vitro. Mol
Carcinog. 53:970–978. 2014.
|
|
38
|
Kennedy C, Sebire K, de Kretser DM and
O’Bryan MK: Human sperm associated antigen 4 (SPAG4) is a potential
cancer marker. Cell Tissue Res. 315:279–283. 2004. View Article : Google Scholar
|
|
39
|
Shiraishi T, Terada N, Zeng Y, Mooney S,
Takahashi S, Takaha N, Miki T, Getzenberg R and Kulkarni P: 433
sperm associated antigen 4 is a novel biomarker for renal cell
carcinoma. J Urol. 187:e177-e1782012. View Article : Google Scholar
|
|
40
|
Shoji K, Murayama T, Mimura I, Wada T,
Kume H, Goto A, Ohse T, Tanaka T, Inagi R, van der Hoorn FA, et al:
Sperm-associated antigen 4, a novel hypoxia-inducible factor 1
target, regulates cytokinesis, and its expression correlates with
the prognosis of renal cell carcinoma. Am J Pathol. 182:2191–2203.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jin HA, Heo JH, Kang YH, Kim KH, Han KS
and Hong SJ: Abstract B18: CXCL10 suppresses tumor angiogenesis and
impedes expression of critical angiogenic factors in renal cell
carcinoma. Cancer Res. 76:B182016. View Article : Google Scholar
|
|
42
|
Liu W, Liu Y, Qiang F, Zhou L, Chang Y, Xu
L, Zhang W and Xu Ji: Elevated expression of IFN-inducible CXCR3
ligands predicts poor prognosis in patients with non-metastatic
clear-cell renal cell carcinoma. Oncotarget. 7:139762016.PubMed/NCBI
|
|
43
|
Ruf M, Moch H and Schraml P: Interaction
of tumor cells with infiltrating lymphocytes via CD70 and CD27 in
clear cell renal cell carcinoma. Oncoimmunology. 4:e10498052015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruf M, Mittmann C, Nowicka AM, Hartmann A,
Hermanns T, Poyet C, van den Broek M, Sulser T, Moch H and Schraml
P: pVHL/HIF-regulated CD70 expression is associated with
infiltration of CD27+ lymphocytes and increased serum
levels of soluble CD27 in clear cell renal cell carcinoma. Clin
Cancer Res. 21:889–898. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen F, Bai J, Li W, Mei P, Liu H, Li L,
Pan Z, Wu Y and Zheng J: RUNX3 suppresses migration, invasion and
angiogenesis of human renal cell carcinoma. PLoS One. 8:e562412013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang G, Qin W, Zheng J, Wei M, Zhou X,
Wang H and Wen W: Expressions of EZH2 and RUNX3 in renal cell
carcinoma and their clinical significance. Xi Bao Yu Fen Zi Mian Yi
Xue Za Zhi. 29:82–84. 2013.In Chinese.
|
|
47
|
Parmar KM, Singla M, Mandal AK,
Bhattacharya S and Singh SK: The expression of RUNX3 gene in renal
cell cancer and its clinical relevance with serum vascular
endothelial growth factor. Int J Mol Immunooncol. 2:732017.
View Article : Google Scholar
|
|
48
|
Chen F, Liu X, Cheng Q, Zhu S, Bai J and
Zheng J: RUNX3 regulates renal cell carcinoma metastasis via
targeting miR-6780a5p/E-cadherin/EMT signaling axis. Oncotarget.
8:101042–101056. 2016.
|
|
49
|
Zhao He, Wang XL, Zhang H, Guo P, Huang C,
Liu C, Yao X, Chen F, Lou YW, et al: RUNX3 mediates suppression of
tumor growth and metastasis of human CCRCC by regulating cyclin
related proteins and TIMP-1. PLoS One. 7:e329612012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bhuvarahamurthy V, Kristiansen GO,
Johannsen M, Loening SA, Schnorr D, Jung K and Staack A: In situ
gene expression and localization of metalloproteinases MMP1, MMP2,
MMP3, MMP9, and their inhibitors TIMP1 and TIMP2 in human renal
cell carcinoma. Oncol Rep. 15:1379–1384. 2006.PubMed/NCBI
|
|
51
|
Cho NH, Shim HS, Rha SY, Kang SH, Hong SH,
Choi YD, Hong SJ and Cho SH: Increased expression of matrix
metallo-proteinase 9 correlates with poor prognostic variables in
renal cell carcinoma. Eur Urol. 44:560–566. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sato A, Nagase H, Obinata D, Fujiwara K,
Fukuda N, Soma M, Yamaguchi K, Kawata N and Takahashi S: Inhibition
of MMP-9 using a pyrrole-imidazole polyamide reduces cell invasion
in renal cell carcinoma. Int J Oncol. 43:1441–1446. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang QB, Ma X, Li HZ, Ai Q, Liu SW, Zhang
Y, Gao Y, Fan Y, Ni D, Wang BJ and Zhang X: Endothelial Delta-like
4 (DLL4) promotes renal cell carcinoma hematogenous metastasis.
Oncotarget. 5:3066–3075. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Struckmann K, Mertz K, Steu S, Storz M,
Staller P, Krek W, Schraml P and Moch H: pVHL co-ordinately
regulates CXCR4/CXCL12 and MMP2/MMP9 expression in human clear-cell
renal cell carcinoma. J Pathol. 214:464–471. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yuan L, Zeng G, Chen L, Wang G and Wang X,
Cao X, Lu M, Liu X, Qian G, Xiao Y and Wang X: Identification of
key genes and pathways in human clear cell renal cell carcinoma
(ccRCC) by co-expression analysis. Int J Biol Sci. 14:266–279.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Feng JY, Diao XW, Fan MQ, Wang PX, Xiao Y,
Zhong X, Wu RH and Huang CB: Screening of feature genes of the
renal cell carcinoma with DNA microarray. Eur Rev Med Pharmacol
Sci. 17:2994–3001. 2013.PubMed/NCBI
|