Introduction
Chronic lymphocytic leukemia (CLL) is a type of
blood cancer that is heterogeneous at the clinical and cellular
levels (1,2), and which arises due to uncontrolled
proliferation of lymphocytes that accumulate in the blood and bone
marrow. These immature cells can permeate other organs, including
the liver, kidney and central nervous system, which can ultimately
result in fatality (2,3). Patients with acute myeloid leukemia
(AML) often exhibit characteristic mutations and dysregulated gene
expression, both of which contribute to the generally poor
prognosis of the disease (4).
Despite significant advances in our understanding of the biology of
leukemia, an optimal treatment regime for CLL is lacking (5-7).
Current treatment regimens rely on chemotherapy, which involves
systemic drug administration. However, the efficacy of chemotherapy
is limited due to immediate clearance of the drugs from blood
circulation. In addition, the development of resistance to
chemotherapy drugs is a major reason for the poor prognosis of
leukemia (8). The development of
resistance is attributed to the mechanism of action of
chemotherapeutic drugs, which activate intrinsic apoptosis pathways
(9). In the case of leukemia,
however, apoptosis pathways are inhibited by Abl expression and a
lack of FAS receptors. Generally, these can be reversed by
administration of high doses of anticancer drugs, but this is
accompanied by marked systemic toxic effects in healthy tissues
(10). Therefore, there is a
requirement to identify non-chemotherapeutic treatments for
leukemia with improved efficacy and reduced side effects.
MicroRNAs (miRNAs) are short noncoding RNAs that are
involved in regulating the expression of their target genes
(11). Certain miRNAs have been
reported to be directly responsible for leukemogenesis and are
involved in the prognosis of cancer. miRNAs regulate the expression
of genes at the posttranscriptional level by altering messenger RNA
(mRNA), thereby modifying associated biological processes or
pathways (12,13). Thus, miRNAs can act as oncogenes
or tumor inhibitors. For example, silencing of miR-15a/16-1 in an
animal model was reported to result in the development of an
indolent form of leukemia (14).
Bcl-2 is an miRNA target, and its interaction with miRNA eliminates
the expression of Bcl-2, resulting in cell death. The anticancer
drug, venetoclax, which inhibits Bcl-2, was recently approved for
use in CLL (15). Other miRNAs
have also been demonstrated to be useful in the regulation of
leukemia, with miRNA-34a reported to downregulate 17p-CLL and halt
disease progression (16,17). These studies lay the groundwork
for the development of miRNA-based therapies. The present study was
performed to examine the effects of miRNA-26a on leukemia
cells.
One of the most important concerns in miRNA-based
therapy is the delivery strategy. Due to their very low stability
in the body, miRNAs require a carrier to elicit their
pharmacological therapeutic effects. The development of nanoscale
biomaterials is a popular topic of interest for targeted treatment
of cancer (18). An ideal
delivery system would deliver the maximum amount of drug to the
target site without being toxic itself (19). Liposomes are ideal carriers for
systemic applications and have been studied in detail in clinical
trials (20). Liposomes are
biocompatible and of an appropriate size for accumulation in tumor
tissue. The long half-life of liposomes in the circulation allows
for accumulation of the carrier in leukemia cells (21).
The aim of the present study was to design an
miRNA-loaded delivery system for the effective treatment of
leukemia. miRNA-26a was physically loaded into liposomes and the
various biological properties, including cell viability, apoptosis
and morphological changes, were studied in vitro.
Materials and methods
Materials
Cholesterol, 1,2-dioleyl-3-trimethylam
monium-propane (DODAP), 1,2-distearoyl-sn-glycero-3-phosphocholine
(DSPC), and N-palmitoyl-sphingosine-1-[succinyl(polyethylene
glycol)]2000 (DSPE-PEG) were purchased from Avanti Polar Lipids
(Alabaster, AL, USA). All other chemicals were of reagent grade and
were used without further purification.
Preparation of miRNA-loaded
liposomes
Liposomes were prepared using the thin-film
hydration method. In brief, DODAP:CHOL:DSPC:DSPE-PEG were dissolved
at a molar ratio of 25:50:23:2 in a mixture of 1 ml of chloroform
(100%) and methanol (100%) (4:1). The organic solvent was agitated
such that all lipids were dissolved, and removed from the rotary
evaporator at 60°C. Thereafter, the lipid film was hydrated using
distilled water at 60°C for 15 min and extruded using a
mini-extruder for 21 cycles through a polycarbonate membrane with a
pore size of 100 nm. The liposomes thus formed were purified by
dialysis for 24 h. Liposomes were stored in glass vials, and 10 µg
miRNA-26a (3-UCG GAU AGG ACC UAA UGA ACU U-5) was added and
incubated for 12 h under constant agitation. The miRNA-loaded
liposomes were stored at 4°C until further use. The miRNA was
complexed with Lipofectamine 2000 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) as a transfection agent and incubated with the
cancer cells.
Characterization of liposomes
The miRNA-loaded liposomes were evaluated in terms
of particle size and particle-size distribution using a Zetasizer
Nano ZS (Malvern Instruments, Worcestershire, UK). The liposomes
(dispersed in 0.1X PBS, density, 1 kg/m3) were diluted
appropriately in ultra-pure water (density, 997 kg/m3)
and experiments were performed in triplicate at room temperature.
The morphology of the liposomes was examined by transmission
electron microscopy (TEM; Tecnai G2 12 TWIN TEM; FEI; Thermo Fisher
Scientific, Inc.). The samples were mixed with 2% phosphotungistic
acid as a counterstain solution for 4 min and placed in a drop onto
a copper grid prior to drying. The samples were then analyzed by
TEM (×10,000, magnification).
Gel electrophoresis
Physical entrapment of miRNA in liposomes was
evaluated by electrophoresis through 2% agarose gels in
Tris-acetate-ethylenediaminetetraacetic acid (EDTA) buffer
containing 0.5 µg/ml of GelRED (Biotium, Inc., Fremont, CA, USA).
The free miRNA and miRNA-loaded liposomes were mixed with 10%
glycerin, 1% bromothymol blue and 2% SDS, subjected to
electrophoresis at 120 V for 20 min, and then photographed using a
gel imaging system (ChemiDocTM; Bio-Rad Laboratories, Inc.).
Cytotoxicity assay
CLL cells (American Type Culture Collection; ATCC;
Manassas, VA, USA) were maintained in RPMI medium supplemented with
10% FBS (Lonza Group, Ltd., Basel, Switzerland) and 1%
penicillin-streptomycin antibiotic mixture in an atmosphere of 65%
humidity with 5% CO2 and 37°C. The cells were seeded in
96-well plates at a density of 1.2×104 cells per well
and incubated for 24 h. The cells were then treated with
miRNA-loaded liposomes or blank liposomes (0.1, 1, 10, 50 and 100
µM) and incubated for a further 24 h. The cells were treated for 24
h with increasing concentrations of miRNA-26a (25, 50 and 100 µM)
to study concentration-dependent cytotoxicity against leukemia
cells. Untreated cells were maintained throughout the study period.
The cells were treated with 100 µl 1.25 mg/ml MTT solution and
incubated for 4 h at 37°C, after which the formazan crystals were
dissolved in DMSO. The absorbance was determined at 570 nm using a
microplate reader (Infinite M200 reader; Tecan, Männedorf,
Switzerland). For comparison, NIH-3T3 cells (ATCC) was purchased
and grown in RPMI medium supplemented with 10% FBS (Lonza Group,
Ltd.) and 1% penicillin-streptomycin antibiotic mixture in a
humidified atmosphere (65%) with 5% CO2 and 37°C. The
same protocol was followed for MTT assay of these cells.
Hoechst 33342 assay
The cells were seeded in 6-well plates at a density
of 3×105 cells per well and incubated for 24 h. The
cells were then treated with miRNA-loaded liposomes (MRL) or blank
liposomes (25, 50 and 100 µM), and incubated for a further
24 h. The cells were treated for 24 h with increasing
concentrations of the miRNA-26a (25, 50 and 100 µM) to
examine concentration-dependent effects on apoptosis. The following
day, the cells were washed carefully with ultrapure water and
stained with 10 µg/ml Hoechst 22242 for 15 min at 37°C. The
cells were then fixed with 4% paraformaldehyde and washed again
with PBS. Apoptosis was qualitatively assessed by fluorescence
microscopy.
Apoptosis
The cells were seeded in 12-well plates at a density
of 2×105 cells per well and incubated for 24 h, then
treated with miRNA-loaded liposomes or blank liposomes (25, 50 and
100 µM), and incubated for a further 24 h. The cells were
treated with 200 µM MRL and incubated for an additional 24
h. The following day, the cells were washed carefully with PBS and
centrifuged at 1,300 x g at 4°C. The cell pellets were resus-pended
in binding buffer (BD Biosciences, Franklin Lakes, NJ, USA) and
incubated with 2.5 µl Annexin V and 2.5 µl propidium
iodide (PI) (BD Biosciences) for 15 min, followed by flow
cytometric analysis (FACSCalibur; BD Biosciences, Franklin Lakes,
NJ, USA).
Western blot analysis
Cells were treated with blank liposome and
miR-26a-loaded liposomes (25, 50 and 100 µM), harvested
after 24 h and lysed using radioimmunoprecipitation assay lysis
buffer for 15 min at 24°C. The lysed cells were centrifuged at
12,000 x g for 15 min at 4°C. The supernatant was collected and the
protein concentration was determined using a BCA protein assay kit
(Thermo Fischer Scientific, Inc.). A total of 25 µg protein
per lane was separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (Bio-Rad Laboratories, Hercules,
CA, USA). The membranes were then blocked with 5% skimmed milk for
1 h, followed by incubation with the following primary antibodies
at 4°C overnight: Anti-cyclin-dependent kinase 6 (CDK6; cat. no.
3136; 1:1,000), anti-MCL1 (cat. no. 4572; 1:1,000), BCL2 family
apoptosis regulator (BCL2 cat. no. 2870; 1:1,000), anti-poly
(ADP-Ribose) polymerase (PARP; cat. no. 9542; 1:1,000) and
anti-GAPDH (dilution, 1:1,000; cat. no. 2118; all from Cell
Signaling Technology, Inc., Danvers, MA, USA). The following day,
the membranes were incubated with horseradish peroxidase-conjugated
anti-mouse IgG secondary antibodies (cat. no. 7076; 1:3,000).
Images of the blots were obtained using an Odyssey infrared imaging
system (LI-COR Biosciences, Lincoln, NE, USA) and quantified using
ImageJ software (version 7.0; National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
The data are presented as the mean ± standard
deviation. All analyses were performed with SPSS software (version
17; SPSS, Inc., Chicago, IL, USA). Comparisons between groups were
assessed by one-way analysis of variance. In instances of multiple
comparisons, analysis of variance was performed, followed by the
Scheffé post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results and Discussion
Preparation and characterization of
miRNA-loaded liposomes
Despite significant advances in our understanding of
the biology of leukemia, there remains no optimal treatment for
CLL. Current treatment options involve chemotherapy, however, they
have poor efficacy due to the immediate clearance of drugs from the
circulation and the development of drug resistance. Although miRNAs
serve important roles in the pathogenesis of cancer, they can also
function as tumor suppressors (22). The more established small
interfering RNAs (siRNAs) silence the expression of a single gene,
while miRNAs can silence multiple genes simultaneously (23). Therefore, miRNA-based therapy has
potential in the treatment of various types of cancer. In the
present study, miRNA-26a was selected for the treatment of leukemia
cells. Delivery strategy remains an important concern in
miRNA-based therapy, as the stability of miRNA in vivo is
low. Liposomes are among the most well-studied carrier systems in
clinical trials, are highly biocompatible and of a size appropriate
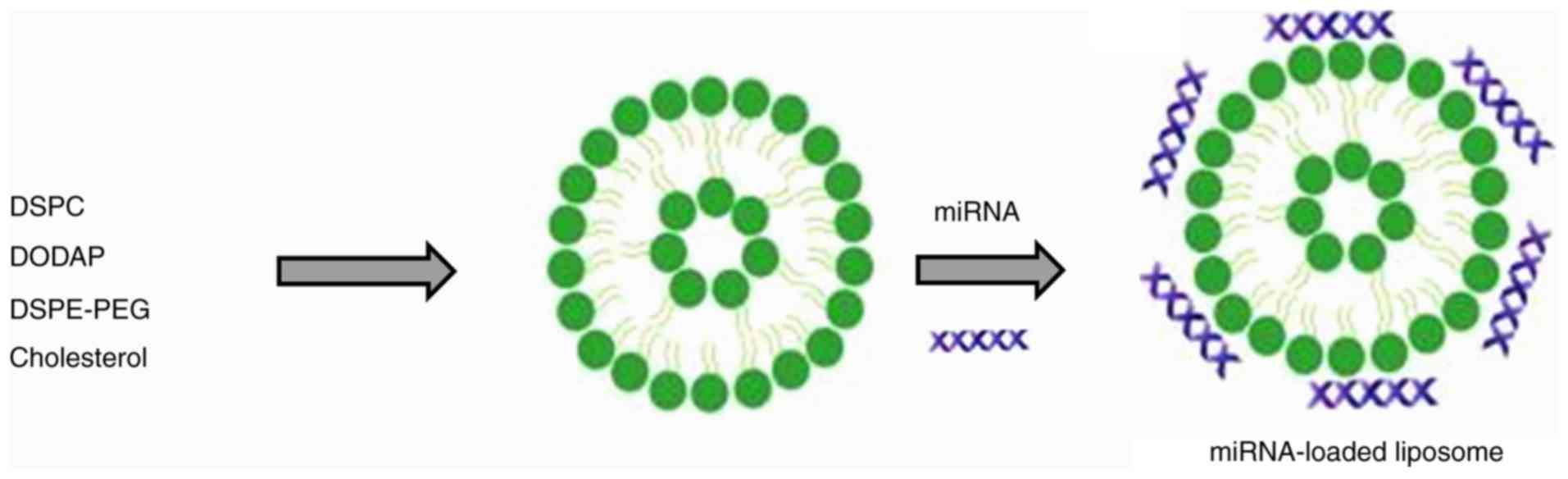
for accumulation in tumor tissue (24). The negatively charged miR-26a
forms an electrostatic complex with the positively charged surfaces
of the liposomes (Fig. 1)
(25).
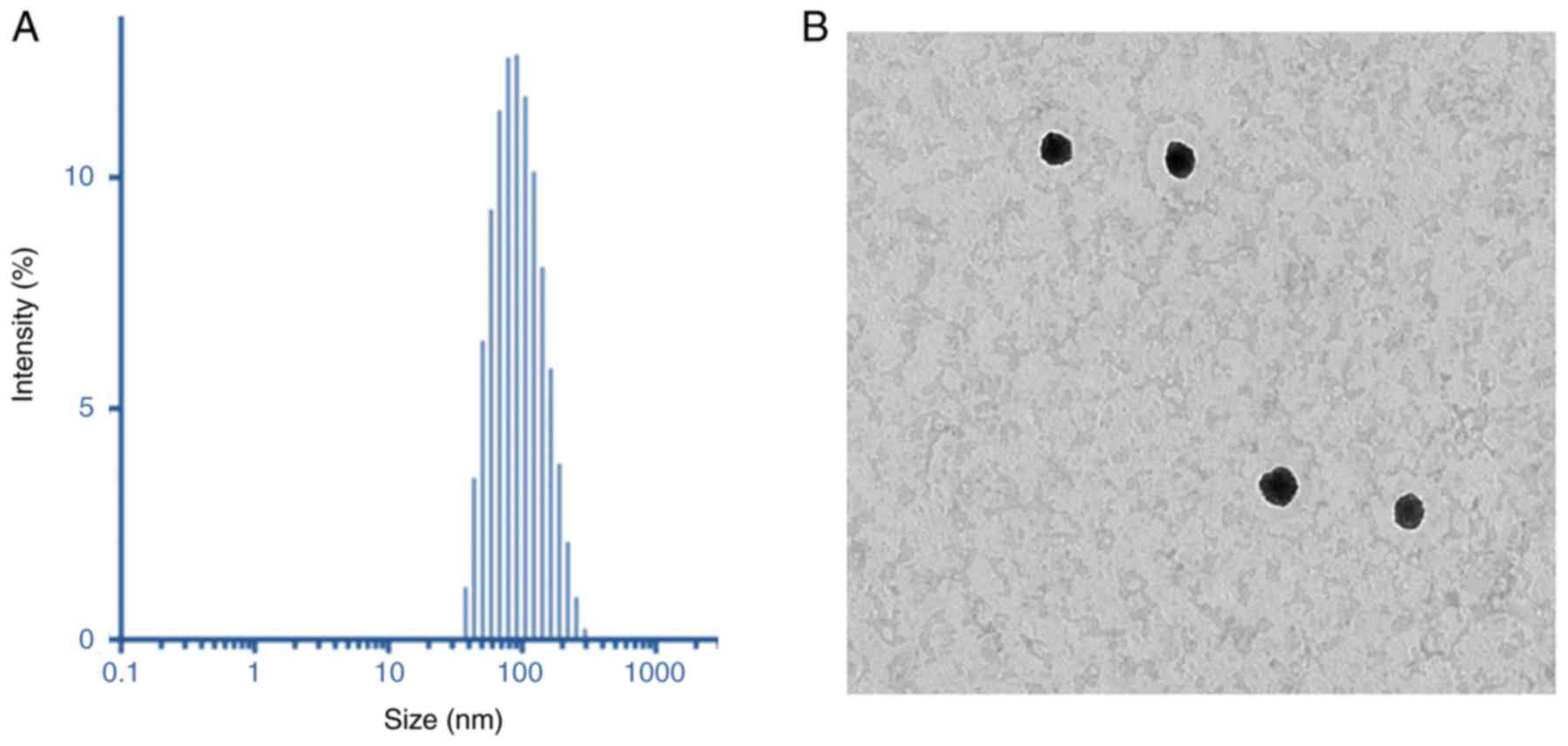
Dynamic light scattering (DLS) analysis was
performed to characterize the final particle size and size
distributions of the liposome preparations. As demonstrated in
Fig. 3A, the particles were 110
nm in size, with a uniform dispersity index of 0.15. The particle
size of MRL was small enough for cancer-targeting applications
(26). The surface charge of MRL
was +21.5±1.25 mV, indicating the presence of cationic charged
liposomes. The dried particles were spherical in shape and
dispersed. The particle size observed from TEM was consistent with
that determined by DLS analysis (Fig.
3A). The positively charged liposomes were predicted to be
internalized into the cancer cells, thereby further enhancing the
efficacy of cancer treatment. The morphology of the particles was
analyzed by TEM (Fig. 3B).
Gel electrophoresis
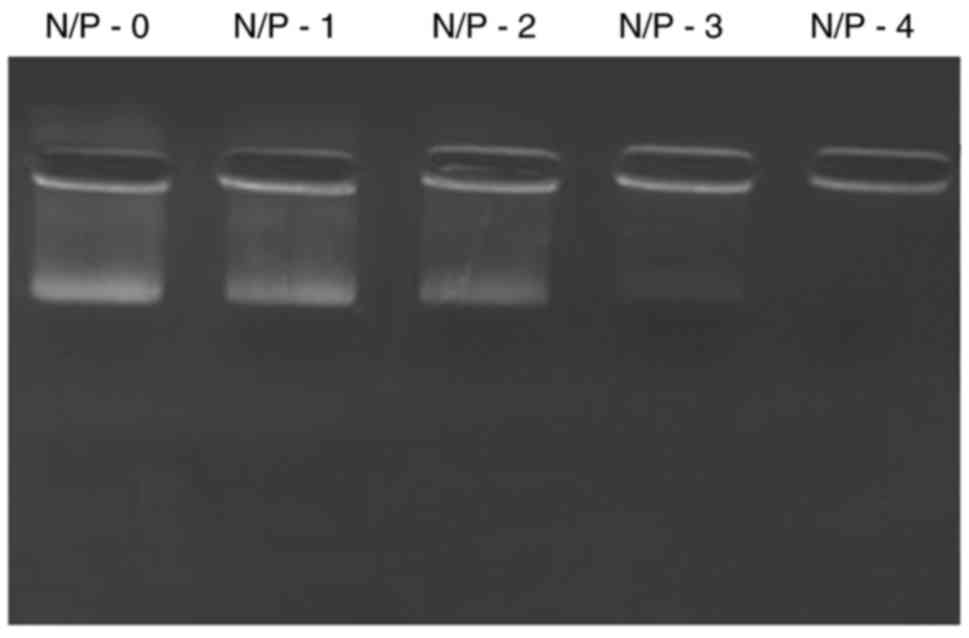
The loading of miR-26a into liposomes was confirmed
by gel electrophoresis (Fig. 2).
Free miRNA was electrophoresed to the opposite end of the gel,
while loading of miRNA into liposomes prevented its release and
migration in a concentration-dependent manner. The results
indicated the ability of liposomes to withhold the encapsulated
miRNA and thereby improve its stability and therapeutic
efficacy.
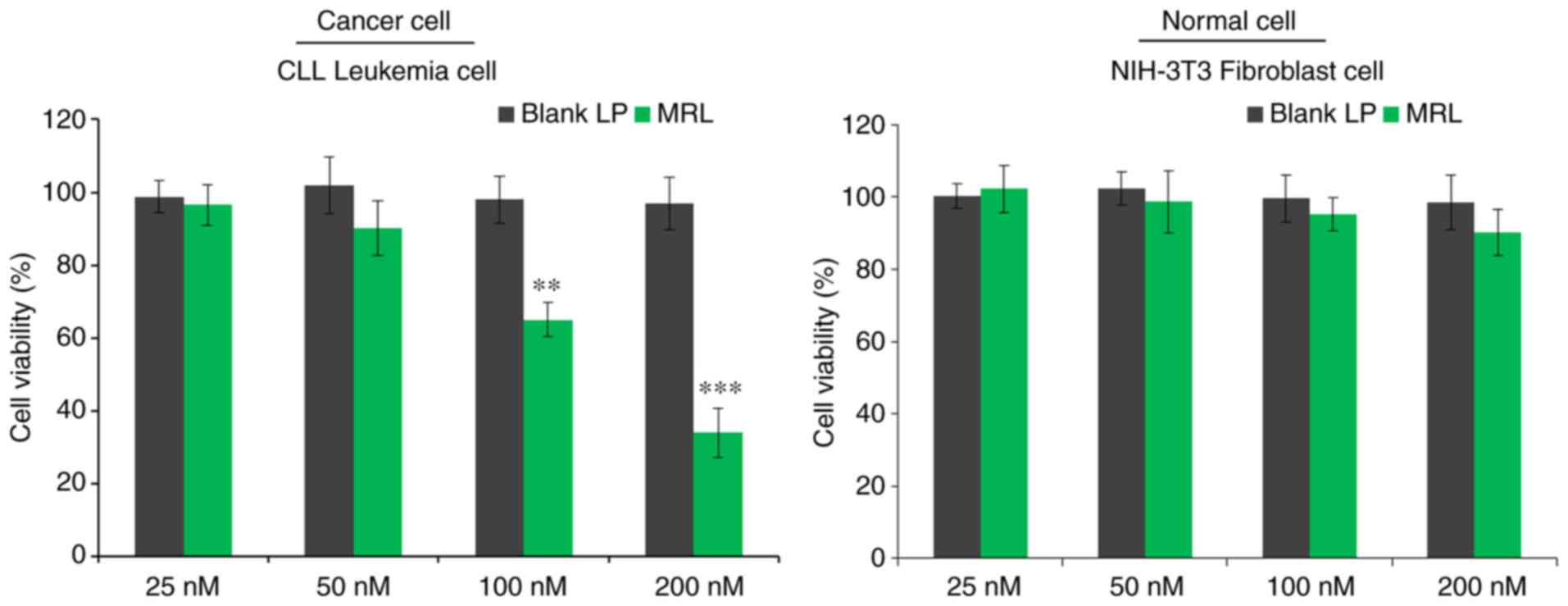
In vitro cell viability
The viability of CLL cells in vitro was
evaluated by MTT assay (Fig. 4).
Briefly, cells were treated with blank liposomes or miRNA-loaded
liposomes and incubated for 24 h. The control blank liposomes had
no effect on the viability of cancer cells, indicating that the
liposomes were non-toxic and biocompatible vectors. As expected,
miRNA-26a-loaded liposomes decreased the viability of cancer cells
in a concentration-dependent manner (P<0.001), with >60%
cells killed when treated with the highest concentration (200 nM).
The results indicated the anticancer effect of miRNA-26a against
leukemia cells.
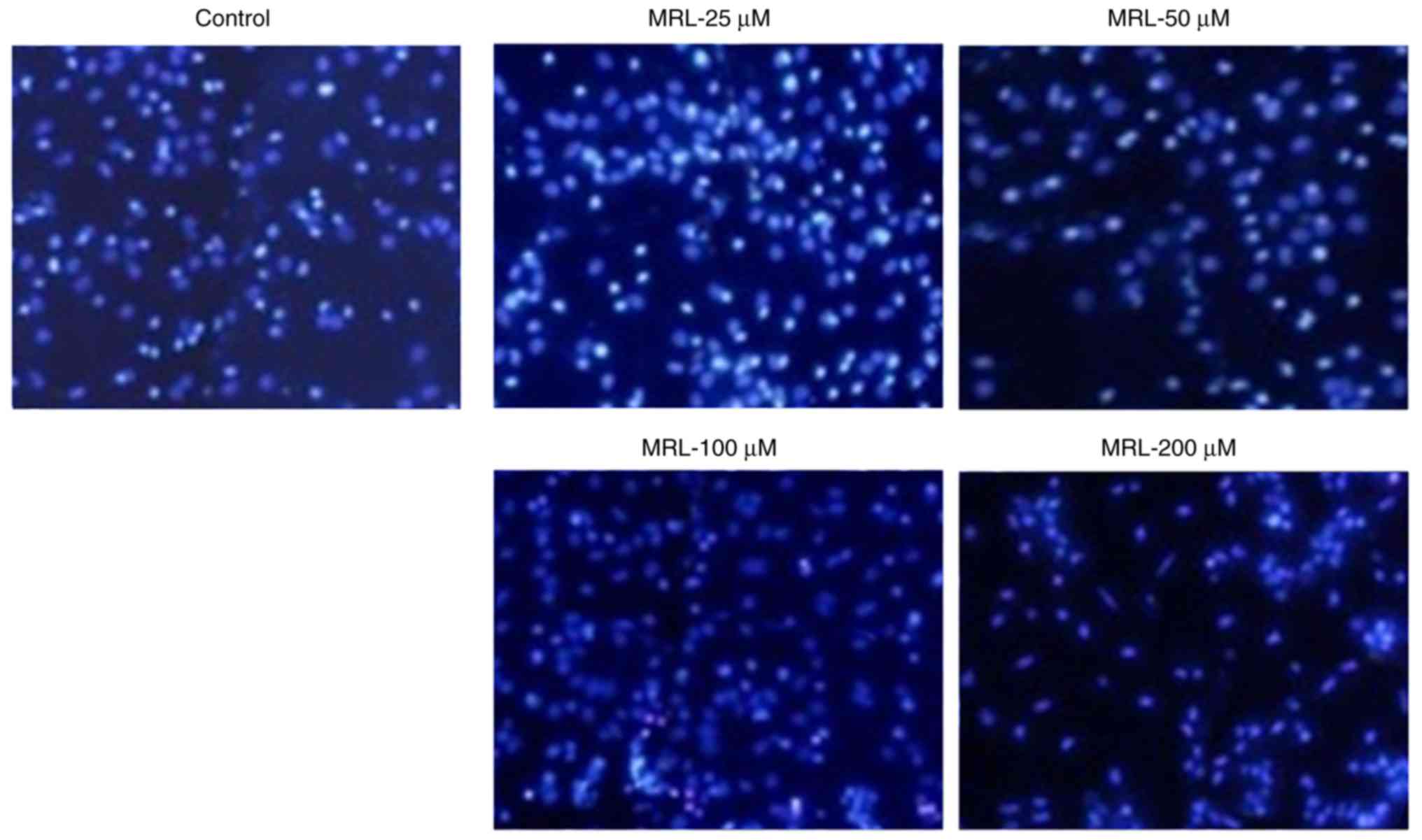
Hoechst 33342 assay
The anticancer effect of miRNA-26a was further
evaluated by Hoechst 33342 staining (Fig. 5). Untreated cells maintained their
typical morphology and dispersal on the plate, whereas treatment
with miRNA-26a induced apoptosis in a concentration-dependent
manner. Apoptosis of cancer cells was more evident with increasing
concentrations of miRNA-26a (Fig.
5). For example, cells treated with 200 µM miRNA
exhibited typical apoptotic morphology, including condensation of
chromatin, breakdown of the nuclear membrane, and apoptotic body
formation. Taken together, these observations indicated the
anticancer potential of miRNA-26a loaded within a stable
nanocarrier.
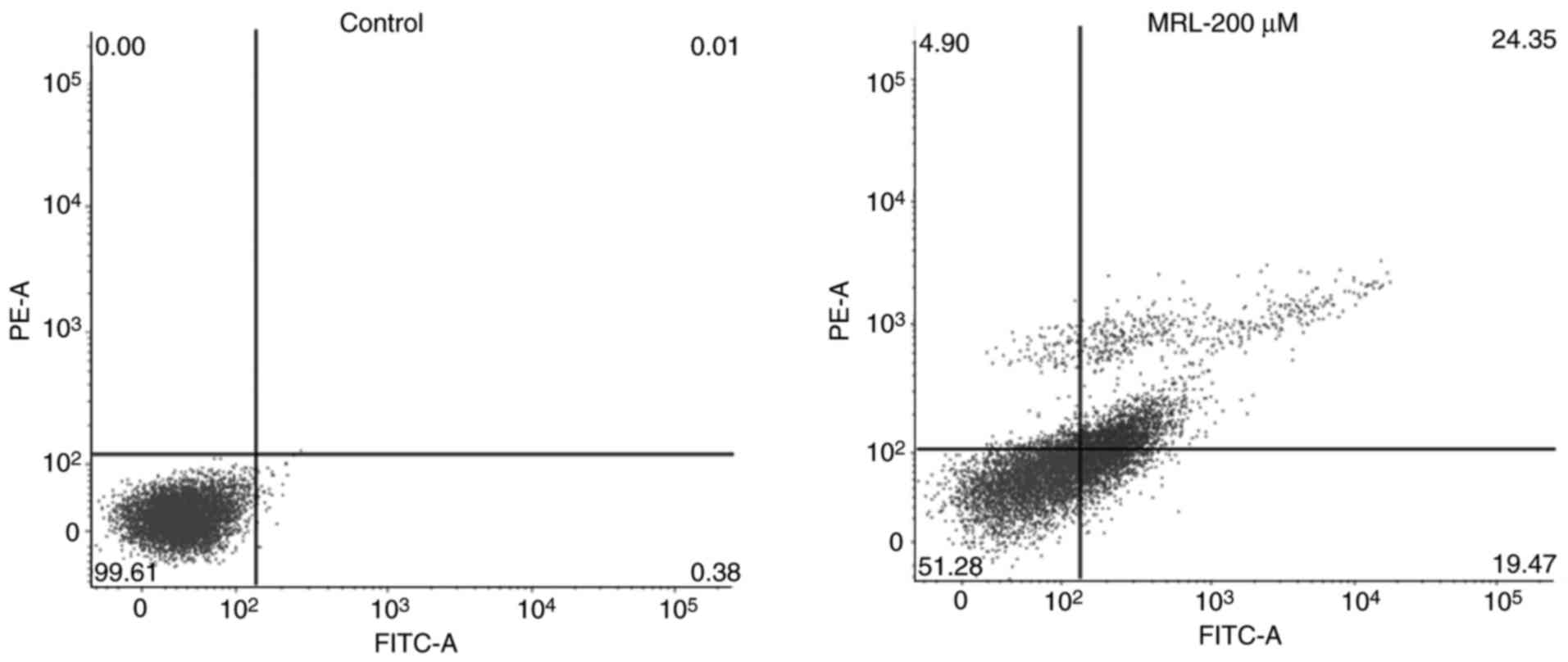
Apoptotic rate, determined by flow
cytometry
Quantitative analysis of apoptosis was performed by
Annexin V/PI staining and flow cytometry (Fig. 6). Cancer cells were treated with
200 µM miRNA for 24 h. A significant (P<0.01) increase in
apoptosis was observed compared with the control group.
Approximately 25% of the total cells were in the late apoptotic
stage, while 20% were in the early apoptosis stage, indicating the
potent anticancer effect of the MRL formulation.
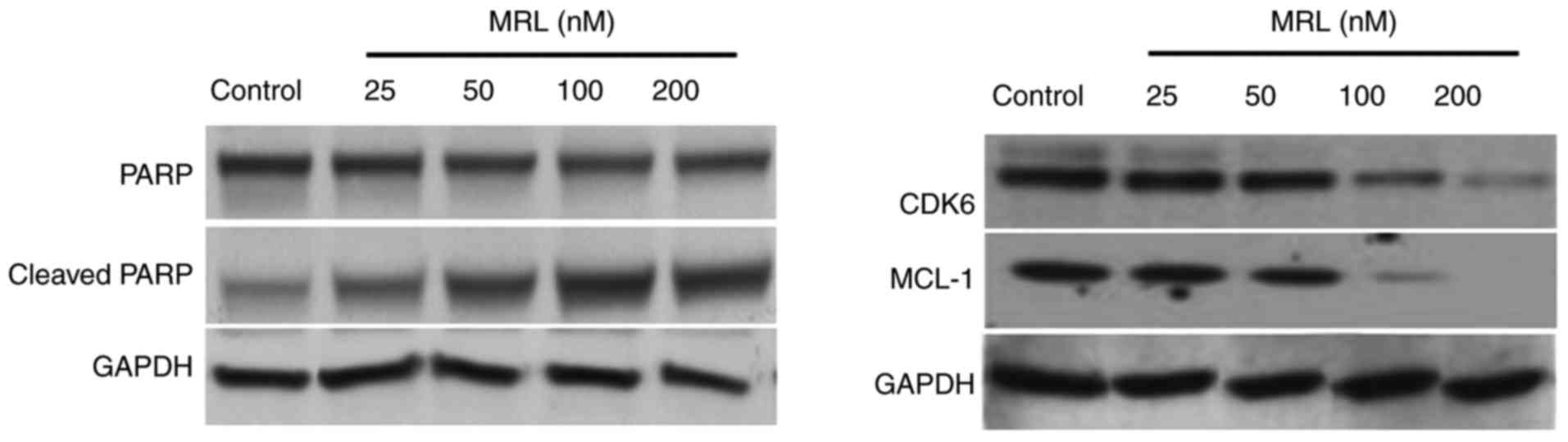
Western blot analysis
The mechanism of action of miRNA-26a was examined by
western blotting (Fig. 7). The
protein expression levels of target genes, cyclin-dependent kinase
6 (Cdk6) and myeloid cell leukemia 1 (Mcl-1), and the marker of
apoptosis, PARP, were analyzed. The results indicated that high
concentrations of miRNA-26a decreased the protein expression level
of PARP and increased that of cleaved PARP, indicating the
apoptosis-inducing potential of miRNA-26a. Seeing as
miRNA-26a-loaded liposomes significantly downregulated the
expression of the target genes, Mcl-1 and Cdk6, this may be the
mechanism of action by which it mediates apoptotic effect. Numerous
studies have demonstrated that miR-26a can target and downregulate
a number of protein-coding gene targets, including CDK6, cyclin D2
and E2, and Mcl-1, in different types of cancer cells (27,28).
In conclusion, miRNA-26a-loaded liposomes
successfully prepared for the effective treatment of leukemia
cells. It was demonstrated that 200 nM miRNA-26a significantly
decreased the viability of CLL cells compared with control. The
miRNA-26a-loaded liposomes exerted a marked apoptosis-inducing
effect, as demonstrated by flow cytometry and Hoechst 33342
staining. Western blot analysis revealed a superior
apoptosis-inducing effect of miRNA-26a-loaded liposomes compared
with free miRNA-26a. miRNA-26a significantly downregulated the
expression of its target genes, Mcl-1 and Cdk6. The results of the
present study indicate that miRNA-26a-loaded liposomes exert
apoptosis-inducing and anticancer effects on leukemia cells,
suggesting their possible utility in future therapies. This
approach has the potential for extrapolation to other types of
neoplasms, including lymphomas and AML.
Funding
The present study was supported by Shandong Blood
Center (Shangdon, China).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and CKS contributed equally to all research. CKS
was responsible for the writing of the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Kwan JM, Fialho AM, Kundu M, Thomas J,
Hong CS, Das Gupta TK and Chakrabarty AM: Bacterial proteins as
potential drugs in the treatment of leukemia. Leuk Res.
33:1392–1399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhojwani D, Yang JJ and Pui CH: Biology of
childhood acute lymphoblastic leukemia. Pediatr Clin North Am.
62:47–60. 2015. View Article : Google Scholar
|
|
3
|
Randhawa JK and Ferrajoli A: A review of
supportive care and recommended preventive approaches for patients
with chronic lymphocytic leukemia. Expert Rev Hematol. 9:235–244.
2016. View Article : Google Scholar
|
|
4
|
Morabito F, Gentile M, Seymour JF and
Polliack A: Ibrutinib, idelalisib and obinutuzumab for the
treatment of patients with chronic lymphocytic leukemia: Three new
arrows aiming at the target. Leuk Lymphoma. 56:3250–3256. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabbri G and Dalla-Favera R: The molecular
pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer.
16:145–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nimmanapalli R and Bhalla K: Novel
targeted therapies for Bcr-Abl positive acute leukemias: Beyond
STI571. Oncogene. 21:8584–8590. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Soni G and Yadav KS: Applications of
nanoparticles in treatment and diagnosis of leukemia. Mater Sci Eng
C. 47:156–164. 2015. View Article : Google Scholar
|
|
8
|
Terme M, Borg C, Guilhot F, Masurier C,
Flament C, Wagner EF, Caillat-Zucman S, Bernheim A, Turhan AG,
Caignard A and Zitvogel L: BCR/ABL promotes dendritic cell-mediated
natural killer cell activation. Cancer Res. 65:6409–6417. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagata Y and Todokoro K: Requirement of
activation of JNK and p38 for environmental stress-induced
erythroid differentiation and apoptosis and of inhibition of ERK
for apoptosis. Blood. 94:853–863. 1999.PubMed/NCBI
|
|
10
|
Jia L, Patwari Y, Kelsey SM and Newland
AC: Trail-induced apoptosis in Type I leukemic cells is not
enhanced by overexpression of bax. Biochem Biophys Res Commun.
283:1037–1045. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Negrini M, Ferracin M, Sabbioni S and
Croce CM: MicroRNAs in human cancer: From research to therapy. J
Cell Sci. 120:1833–1840. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and downregulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar
|
|
13
|
Klein U, Lia M, Crespo M, Siegel R, Shen
Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G and
Dalla-Favera R: The DLEU2/miR-15a/161 cluster controls B cell
proliferation and its deletion leads to chronic lymphocytic
leukemia. Cancer Cell. 17:28–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roberts AW, Davids MS, Pagel JM, Kahl BS,
Puvvada SD, Gerecitano JF, Kipps TJ, Anderson MA, Brown JR,
Gressick L, et al: Targeting BCL2 with venetoclax in relapsed
chronic lymphocytic leukemia. N Engl J Med. 374:311–322. 2016.
View Article : Google Scholar
|
|
16
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A microRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fabbri M, Bottoni A, Shimizu M, Spizzo R,
Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE,
et al: Association of a microRNA/TP53 feedback circuitry with
pathogenesis and outcome of B-cell chronic lymphocytic leukemia.
JAMA. 305:59–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramasamy T, Ruttala HB, Gupta B, Poudel
BK, Choi HG, Yong CS and Kim JO: Smart chemistry-based nanosized
drug delivery systems for systemic applications: A comprehensive
review. J Control Release. 258:226–253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choi JY, Ramasamy T, Tran TH, Ku SK, Shin
BS, Choi HG, Yong CS and Kim JO: Systemic delivery of axitinib with
nano-hybrid liposomal nanoparticles inhibits hypoxic tumor growth.
J Mater Chem B. 3:408–416. 2015. View Article : Google Scholar
|
|
20
|
Ramasamy T, Ruttala HB, Sundaramoorthy P,
Poudel BK, Youn YS, Ku SK, Choi HG, Yong CS and Kim JO: Multimodal
selenium nanoshell-capped Au@mSiO2 nanoplatform for
NIR-responsive chemophotothermal therapy against metastatic breast
cancer. NPG Asia Mater. 10:197–216. 2018. View Article : Google Scholar
|
|
21
|
Ramasamy T, Haidar ZS, Tran TH, Choi JY,
Choi HG, Jeong JH, Shin BS, Choi HG, Yong CS and Kim JO:
Layer-by-layer assembly of liposomal nanoparticles with PEGylated
polyelectrolytes enhances systemic delivery of multiple anticancer
drugs. Acta Biomater. 10:5116–5127. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Y, Gao DY and Huang L: In vivo
delivery of miRNAs for cancer therapy: Challenges and strategies.
Adv Drug Deliv Rev. 81:128–141. 2015. View Article : Google Scholar
|
|
23
|
Lam JK, Chow MY, Zhang Y and Leung SW:
siRNA versus miRNA as therapeutics for gene silencing. Mol Ther
Nucleic Acids. 4:e2522015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ruttala HB, Ramasamy T, Madeshwaran T,
Hiep TT, Kandasamy U, Oh KT, Choi HG, Yong CS and Kim JO: Emerging
potential of stimulus-responsive nanosized anticancer drug delivery
systems for systemic applications. Arch Pharm Res. 41:111–129.
2018. View Article : Google Scholar
|
|
25
|
Ahmadzada T, Reid G and McKenzie DR:
Fundamentals of siRNA and miRNA therapeutics and a review of
targeted nanoparticle delivery systems in breast cancer. Biophys
Rev. 10:69–86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ganju A, Khan S, Hafeez BB, Behrman SW,
Yallapu MM, Chauhan SC and Jaggi MM: miRNA nanotherapeutics for
cancer. Drug Discov Today. 22:424–432. 2017. View Article : Google Scholar :
|
|
27
|
Yang X, Liang L, Zhang XF, Jia HL, Qin Y,
Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al: MicroRNA-26a
suppresses tumor growth and metastasis of human hepatocellular
carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology.
58:158–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang
JR, Zeng C and Zhuang SM: MicroRNA-26a/b and their host genes
cooperate to inhibit the G1/S transition by activating the pRb
protein. Nucleic Acids Res. 40:4615–4625. 2012. View Article : Google Scholar : PubMed/NCBI
|