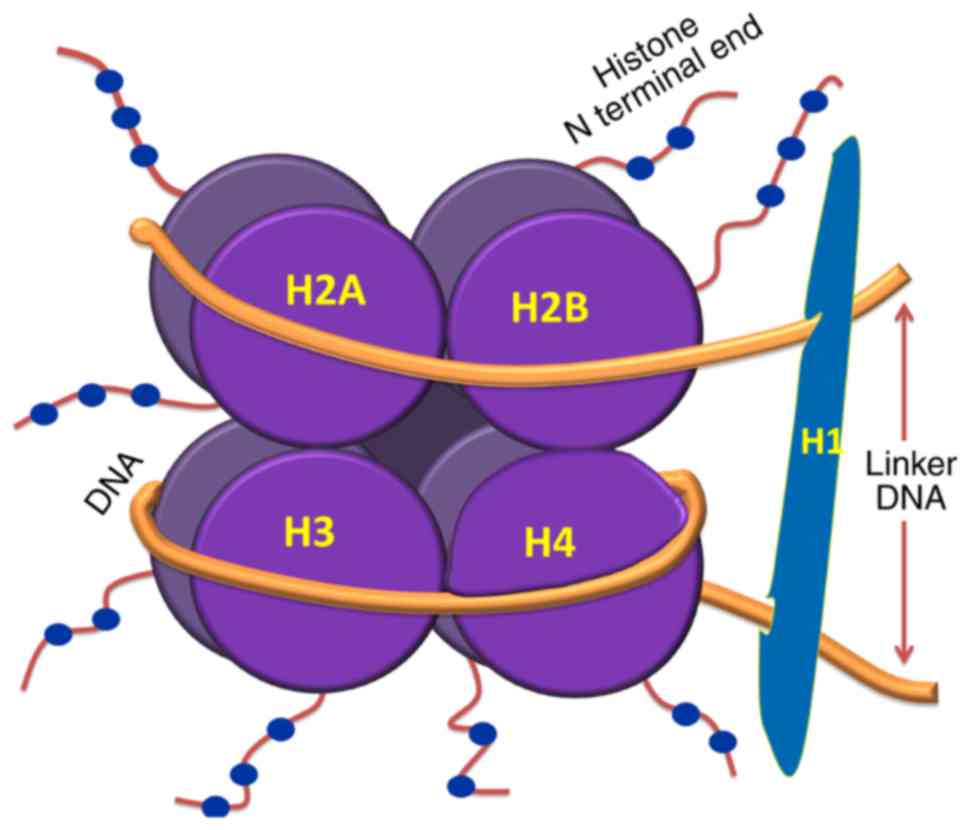
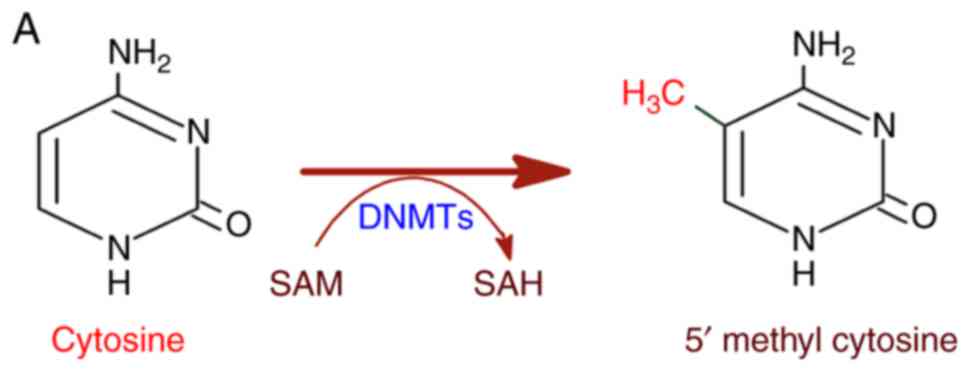
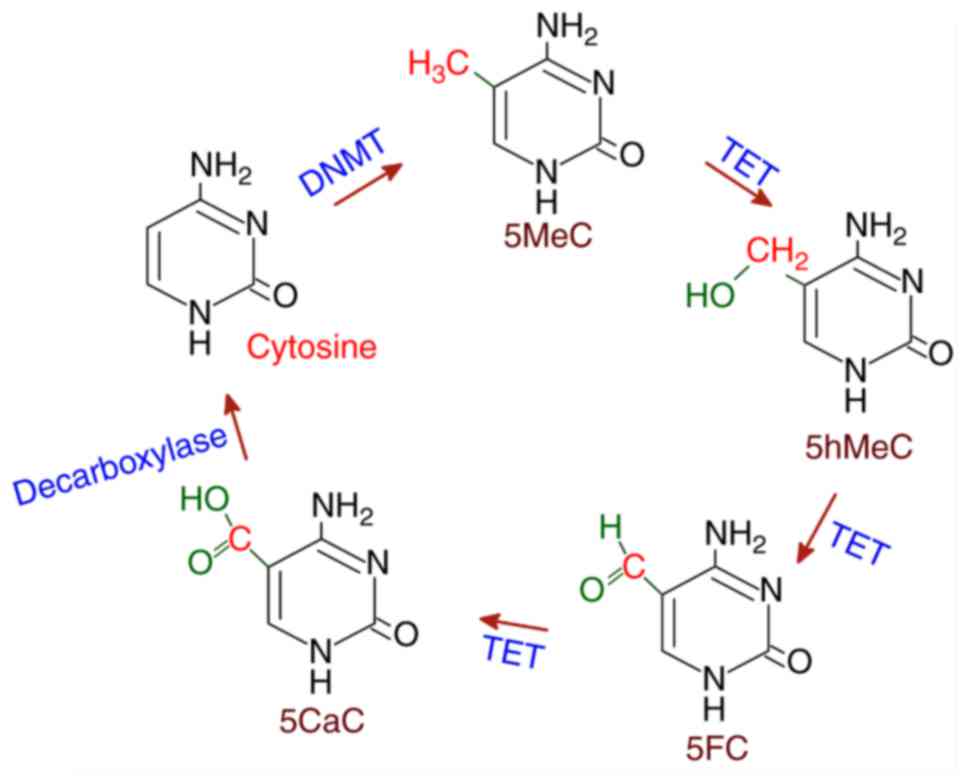
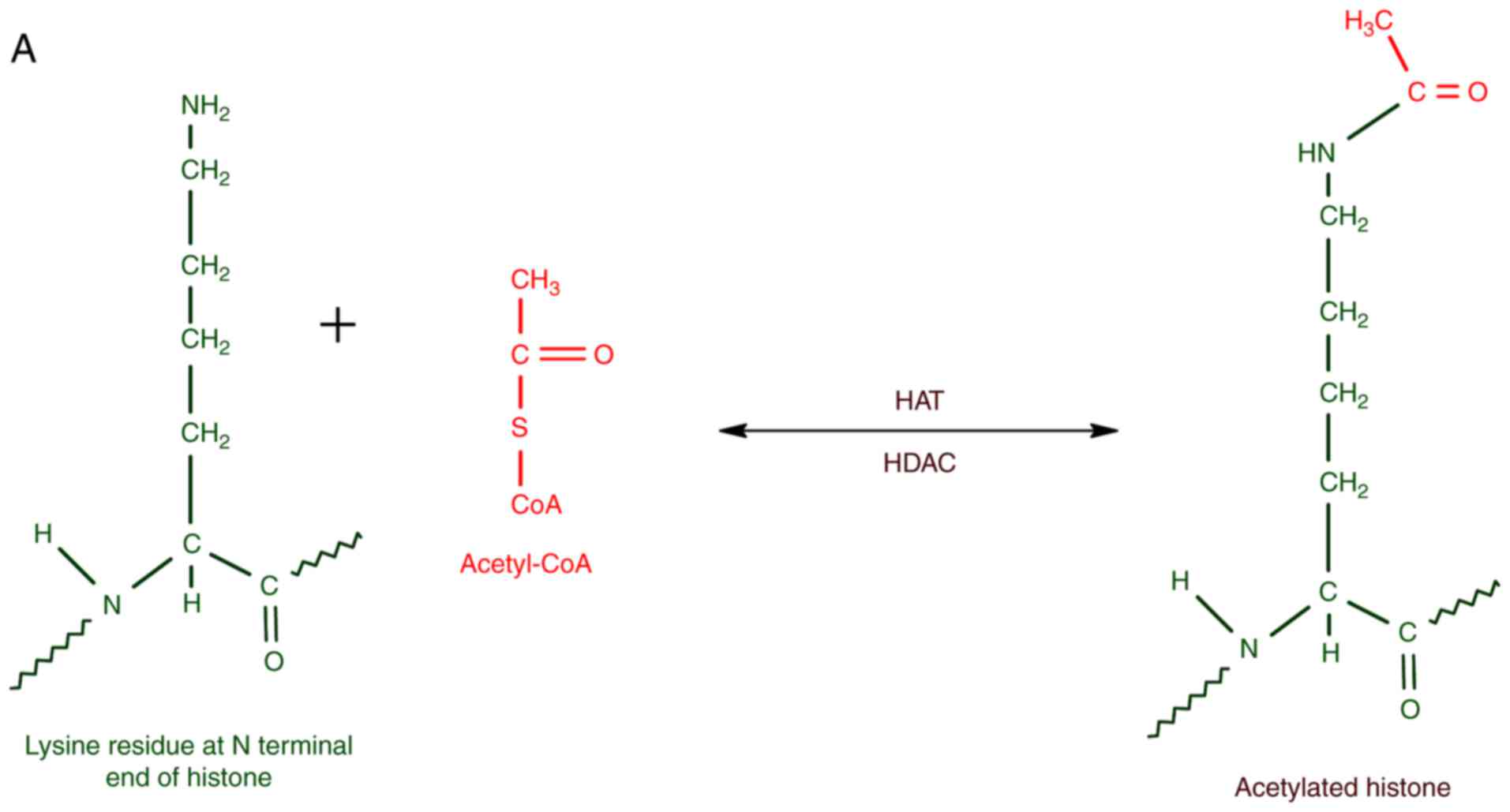
|
1
|
Thankam FG, Dilisio MF, Dougherty KA,
Dietz NE and Agrawal DK: Triggering receptor expressed on myeloid
cells and 5'adenosine monophosphate-activated protein kinase in the
inflammatory response: A potential therapeutic target. Expert Rev
Clin Immunol. 12:1239–1249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thankam FG, Dilisio MF and Agrawal DK:
Immunobiological factors aggravating the fatty infiltration on
tendons and muscles in rotator cuff lesions. Mol Cell Biochem.
417:17–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Page P: Shoulder muscle imbalance and
subacromial impingement syndrome in overhead athletes. Int J Sports
Phys Ther. 6:51–58. 2011.PubMed/NCBI
|
|
4
|
Waddington CH: Towards a theoretical
biology. Nature. 218:525–527. 1968. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kornberg RD: Chromatin structure: A
repeating unit of histones and DNA. Science. 184:868–871. 1974.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weintraub H and Groudine M: Chromosomal
subunits in active genes have an altered conformation. Science.
193:848–856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Narlikar GJ, Fan HY and Kingston RE:
Cooperation between complexes that regulate chromatin structure and
transcription. Cell. 108:475–487. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Felsenfeld G and Groudine M: Controlling
the double helix. Nature. 421:448–453. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Surani MA, Hayashi K and Hajkova P:
Genetic and epigenetic regulators of pluripotency. Cell.
128:747–762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Human Genome Structural Variation Working
Group; Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks
LD, Carter NP, Church DM, Felsenfeld A, Guyer M, et al: Completing
the map of human genetic variation. Nature. 447:161–165. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sebat J: Major changes in our DNA lead to
major changes in our thinking. Nat Genet. 39(Suppl 7): S3–S5. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin B, Li Y and Robertson KD: DNA
Methylation: Superior or subordinate in the epigenetic hierarchy.
Genes Cancer. 2:607–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Voelter-Mahlknecht S: Epigenetic
associations in relation to cardiovascular prevention and
therapeutics. Clin Epigenetics. 8:42016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shea JM, Serra RW, Carone BR, Shulha HP,
Kucukural A, Ziller MJ, Vallaster MP, Gu H, Tapper AR, Gardner PD,
et al: Genetic and epigenetic variation, but not diet, shape the
sperm methylome. Dev Cell. 35:750–758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lachner M and Jenuwein T: The many faces
of histone lysine methylation. Curr Opin Cell Biol. 14:286–298.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pradhan S, Bacolla A, Wells RD and Roberts
RJ: Recombinant human DNA (cytosine-5) methyltransferase. I.
Expression, purification, and comparison of de novo and maintenance
methylation. J Biol Chem. 274:33002–33010. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Leonhardt H, Page AW, Weier HU and Bestor
TH: A targeting sequence directs DNA methyltransferase to sites of
DNA replication in mammalian nuclei. Cell. 71:865–873. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Borgel J, Guibert S, Li Y, Chiba H,
Schübeler D, Sasaki H, Forné T and Weber M: Targets and dynamics of
promoter DNA methylation during early mouse development. Nat Genet.
42:1093–1100. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu H, Coskun V, Tao J, Xie W, Ge W,
Yoshikawa K, Li E, Zhang Y and Sun YE: Dnmt3a-dependent nonpromoter
DNA methylation facilitates transcription of neurogenic genes.
Science. 329:444–448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ooi SK, Qiu C, Bernstein E, Li K, Jia D,
Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al:
DNMT3L connects unmethylated lysine 4 of histone H3 to de novo
methylation of DNA. Nature. 448:714–717. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramsahoye BH, Biniszkiewicz D, Lyko F,
Clark V, Bird AP and Jaenisch R: Non-CpG methylation is prevalent
in embryonic stem cells and may be mediated by DNA
methyltransferase 3a. Proc Natl Acad Sci USA. 97:5237–5242. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pinney S: Mammalian Non-CpG methylation:
Stem cells and beyond. Biology (Basel). 3. pp. 739–751. 2014
|
|
25
|
Suzuki MM and Bird A: DNA methylation
landscapes: Provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ehrlich M, Gama-Sosa MA, Huang LH, Midgett
RM, Kuo KC, McCune RA and Gehrke C: Amount and distribution of
5-methylcytosine in human DNA from different types of tissues of
cells. Nucleic Acids Res. 10:2709–2721. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reik W, Dean W and Walter J: Epigenetic
reprogramming in mammalian development. Science. 293:1089–1093.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen L, Kondo Y, Guo Y, Zhang J, Zhang L,
Ahmed S, Shu J, Chen X, Waterland RA and Issa JP: Genome-wide
profiling of DNA methylation reveals a class of normally methylated
CpG island promoters. PLoS Genet. 3:2023–2036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Henderson IR and Jacobsen SE: Tandem
repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA
methylation and initiate siRNA spreading. Genes Dev. 22:1597–1606.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lienert F, Wirbelauer C, Som I, Dean A,
Mohn F and Schübeler D: Identification of genetic elements that
autonomously determine DNA methylation states. Nat Genet.
43:1091–1097. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meissner A, Mikkelsen TS, Gu H, Wernig M,
Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB,
et al: Genome-scale DNA methylation maps of pluripotent and
differentiated cells. Nature. 454:766–770. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Smith ZD, Chan MM, Mikkelsen TS, Gu H,
Gnirke A, Regev A and Meissner A: A unique regulatory phase of DNA
methylation in the early mammalian embryo. Nature. 484:339–344.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aran D, Sabato S and Hellman A: DNA
methylation of distal regulatory sites characterizes dysregulation
of cancer genes. Genome Biol. 14:R212013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharif O and Knapp S: From expression to
signaling: Roles of TREM-1 and TREM-2 in innate immunity and
bacterial infection. Immunobiology. 213:701–713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maunakea AK, Nagarajan RP, Bilenky M,
Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C,
Zhao Y, et al: Conserved role of intragenic DNA methylation in
regulating alternative promoters. Nature. 466:253–257. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Laurent L, Wong E, Li G, Huynh T, Tsirigos
A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J and Wei CL:
Dynamic changes in the human methylome during differentiation.
Genome Res. 20:320–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mayer W, Niveleau A, Walter J, Fundele R
and Haaf T: Demethylation of the zygotic paternal genome. Nature.
403:501–502. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Inoue A and Zhang Y: Replication-dependent
loss of 5- hydroxymethylcytosine in mouse preimplantation embryos.
Science. 334:1942011. View Article : Google Scholar
|
|
40
|
Guo JU, Su Y, Zhong C, Ming GL and Song H:
Hydroxylation of 5-methylcytosine by TET1 promotes active DNA
demethylation in the adult brain. Cell. 145:423–434. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang H, Lin H, Xu H, Zhang L, Cheng L, Wen
B, Shou J, Guan K, Xiong Y and Ye D: TET-catalyzed 5-methylcytosine
hydroxylation is dynamically regulated by metabolites. Cell Res.
24:1017–1020. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu H and Zhang Y: Mechanisms and functions
of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev.
25:2436–2452. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gill G: SUMO and ubiquitin in the nucleus:
Different functions, similar mechanisms. Genes Dev. 18:2046–2059.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tsankova N, Renthal W, Kumar A and Nestler
EJ: Epigenetic regulation in psychiatric disorders. Nat Rev
Neurosci. 8:355–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ragunathan K, Jih G and Moazed D:
Epigenetic inheritance uncoupled from sequence-specific
recruitment. Science. 348:12586992015. View Article : Google Scholar :
|
|
47
|
Covic M, Hassa PO, Saccani S, Buerki C,
Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G and Hottiger
MO: Arginine methyltransferase CARM1 is a promoter-specific
regulator of NF-kappaB-dependent gene expression. EMBO J. 24:85–96.
2005. View Article : Google Scholar
|
|
48
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H and Shinkai Y:
G9a histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Botuyan MV, Lee J, Ward IM, Kim JE,
Thompson JR, Chen J and Mer G: Structural basis for the methylation
state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in
DNA repair. Cell. 127:1361–1373. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Techima M: Inquiring for new approach to
nursing education. III. A case in clinical practicum (2). Kango
Kenkyu. 24:366–372. 1991.In Japanese.
|
|
51
|
Fischle W, Tseng BS, Dormann HL,
Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H and
Allis CD: Regulation of HP1-chromatin binding by histone H3
methylation and phosphorylation. Nature. 438:1116–1122. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Margueron R, Trojer P and Reinberg D: The
key to development: Interpreting the histone code. Curr Opin Genet
Dev. 15:163–176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wysocka J, Swigut T, Xiao H, Milne TA,
Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et
al: A PHD finger of NURF couples histone H3 lysine 4 trimethylation
with chromatin remodelling. Nature. 442:86–90. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Andrews FH, Gatchalian J, Krajewski K,
Strahl BD and Kutateladze TG: Regulation of methyllysine readers
through phosphorylation. ACS Chem Biol. 11:547–553. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Couture JF, Collazo E and Trievel RC:
Molecular recognition of histone H3 by the WD40 protein WDR5. Nat
Struct Mol Biol. 13:698–703. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kadota S and Nagata K: pp32, an INHAT
component, is a transcription machinery recruiter for maximal
induction of IFN-stimulated genes. J Cell Sci. 124:892–899. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shao Z, Raible F, Mollaaghababa R, Guyon
JR, Wu CT, Bender W and Kingston RE: Stabilization of chromatin
structure by PRC1, a Polycomb complex. Cell. 98:37–46. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chandrasekharan MB, Huang F and Sun ZW:
Ubiquitination of histone H2B regulates chromatin dynamics by
enhancing nucleosome stability. Proc Natl Acad Sci USA.
106:16686–16691. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Canzonetta C, Vernarecci S, Iuliani M,
Marracino C, Belloni C, Ballario P and Filetici P: SAGA DUB-Ubp8
deubiquitylates centromeric histone variant Cse4. G3 (Bethesda).
6:287–298. 2015. View Article : Google Scholar
|
|
60
|
Sun L, Johnston SA and Kodadek T: Physical
association of the APIS complex and general transcription factors.
Biochem Biophys Res Commun. 296:991–999. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Muratani M and Tansey WP: How the
ubiquitin-proteasome system controls transcription. Nat Rev Mol
Cell Biol. 4:192–201. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun ZW and Allis CD: Ubiquitination of
histone H2B regulates H3 methylation and gene silencing in yeast.
Nature. 418:104–108. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Henry KW, Wyce A, Lo WS, Duggan LJ, Emre
NC, Kao CF, Pillus L, Shilatifard A, Osley MA and Berger SL:
Transcriptional activation via sequential histone H2B
ubiquitylation and deubiquitylation, mediated by SAGA-associated
Ubp8. Genes Dev. 17:2648–2663. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gao C, Huang W, Kanasaki K and Xu Y: The
role of ubiquitination and sumoylation in diabetic nephropathy.
Biomed Res Int. 2014.160692:2014.
|
|
65
|
Collins P, Mitxitorena I and Carmody R:
The ubiquitination of NF-κB subunits in the control of
transcription. Cells. 5:pii: E232016. View Article : Google Scholar
|
|
66
|
Moazed D: Small RNAs in transcriptional
gene silencing and genome defence. Nature. 457:413–420. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Matzke M, Matzke AJ and Kooter JM: RNA:
Guiding gene silencing. Science. 293:1080–1083. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Boosani C and Agrawal D: Epigenetic
regulation of innate immunity by microRNAs. Antibodies. 5:82016.
View Article : Google Scholar
|
|
71
|
Castel SE and Martienssen RA: RNA
interference in the nucleus: Roles for small RNAs in transcription,
epigenetics and beyond. Nat Rev Genet. 14:100–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pak J and Fire A: Distinct populations of
primary and secondary effectors during RNAi iC elegans. Science.
315:241–244. 2007. View Article : Google Scholar
|
|
74
|
Halic M and Moazed D: Dicer-independent
primal RNAs trigger RNAi and heterochromatin formation. Cell.
140:504–516. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Motamedi MR, Verdel A, Colmenares SU,
Gerber SA, Gygi SP and Moazed D: Two RNAi complexes, RITS and RDRC,
physically interact and localize to noncoding centromeric RNAs.
Cell. 119:789–802. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bühler M, Verdel A and Moazed D: Tethering
RITS to a nascent transcript initiates RNAi- and
heterochromatin-dependent gene silencing. Cell. 125:873–886. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Holoch D and Moazed D: RNA-mediated
epigenetic regulation of gene expression. Nat Rev Genet. 16:71–84.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bayne EH, White SA, Kagansky A, Bijos DA,
Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J
and Allshire RC: Stc1: A critical link between RNAi and chromatin
modification required for heterochromatin integrity. Cell.
140:666–677. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gerace EL, Halic M and Moazed D: The
methyltransferase activity of Clr4Suv39h triggers RNAi
independently of histone H3K9 methylation. Mol Cell. 39:360–372.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zaratiegui M, Castel SE, Irvine DV, Kloc
A, Ren J, Li F, de Castro E, Marí L, Chang AY, Goto D, et al: RNAi
promotes heterochromatic silencing through replication-coupled
release of RNA Pol II. Nature. 479:135–138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li F, Martienssen R and Cande WZ:
Coordination of DNA replication and histone modification by the
Rik1-Dos2 complex. Nature. 475:244–248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Millar NL, Murrell GA and McInnes IB:
Inflammatory mechanisms in tendinopathy-towards translation. Nat
Rev Rheumatol. 13:110–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Campbell AL, Smith NC, Reilly JH, Kerr SC,
Leach WJ, Fazzi UG, Rooney BP, Murrell GA and Millar NL: IL-21
receptor expression in human tendinopathy. Mediators Inflamm.
2014.481206:2014.
|
|
84
|
Legerlotz K, Jones ER, Screen HR and Riley
GP: Increased expression of IL-6 family members in tendon
pathology. Rheumatology (Oxford). 51:1161–1165. 2012. View Article : Google Scholar
|
|
85
|
Dean BJ, Gettings P, Dakin SG and Carr AJ:
Are inflammatory cells increased in painful human tendinopathy? A
systematic review. Br J Sports Med. 50:216–220. 2016. View Article : Google Scholar
|
|
86
|
Thankam FG, Dilisio MF, Dietz NE and
Agrawal DK: TREM-1, HMGB1 and RAGE in the shoulder tendon: Dual
mechanisms for inflammation based on the coincidence of
glenohumeral arthritis. PLoS One. 11:e01654922016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Guo J, Zhang JF, Li G and Chan KM: A
Mini-review: MicroRNA in tendon injuries. J Stem Cell Res Ther.
5:3032015.
|
|
88
|
Millar NL, Gilchrist DS, Akbar M, Reilly
JH, Kerr SC, Campbell AL, Murrell GA, Liew FY, Kurowska-Stolarska M
and McInnes IB: MicroRNA29a regulates IL-33-mediated tissue
remodelling in tendon disease. Nat Commun. 6:67742015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Thankam FG, Boosani CS, Dilisio MF, Dietz
NE and Agrawal DK: MicroRNAs associated with shoulder tendon
matrisome disorganization in glenohumeral arthritis. PLoS One.
11:e01680772016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thankam FG, Boosani CS, Dilisio MF and
Agrawal DK: MicroRNAs associated with inflammation in shoulder
tendinopathy and glenohumeral arthritis. Mol Cell Biochem.
437:81–97. 2018. View Article : Google Scholar
|
|
91
|
Medzhitov R and Horng T: Transcriptional
control of the inflammatory response. Nat Rev Immunol. 9:692–703.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wierda RJ, Geutskens SB, Jukema JW, Quax
PH and van den Elsen PJ: Epigenetics in atherosclerosis and
inflammation. J Cell Mol Med. 14:1225–1240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
De Santa F, Totaro MG, Prosperini E,
Notarbartolo S, Testa G and Natoli G: The histone H3 lysine-27
demethylase Jmjd3 links inflammation to inhibition of
polycomb-mediated gene silencing. Cell. 130:1083–1094. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim
BW, Riggs AD, Chu FF and Pfeifer GP: Methylation of polycomb target
genes in intestinal cancer is mediated by inflammation. Cancer Res.
68:10280–10289. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chen X, El Gazzar M, Yoza BK and McCall
CE: The NF-kappaB Factor RelB and Histone H3 lysine
methyltransferase G9a directly interact to generate epigenetic
silencing in endotoxin tolerance. J Biol Chem. 284:27857–27865.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gazzar ME, Yoza BK, Chen X, Hu J, Hawkins
GA and McCall CE: G9a and HP1 couple histone and DNA methylation to
TNF transcription silencing during endotoxin tolerance. J Biol
Chem. 283:32198–32208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Villagra A, Sotomayor EM and Seto E:
Histone deacetylases and the immunological network: Implications in
cancer and inflammation. Oncogene. 29:157–173. 2010. View Article : Google Scholar
|
|
98
|
Bayarsaihan D: Epigenetic Mechanisms in
Inflammation. J Dent Res. 90:9–17. 2011. View Article : Google Scholar :
|
|
99
|
Shuto T, Furuta T, Oba M, Xu H, Li JD,
Cheung J, Gruenert DC, Uehara A, Suico MA, Okiyoneda T and Kai H:
Promoter hypomethylation of Toll-like receptor-2 gene is associated
with increased proinflammatory response toward bacterial
peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB
J. 20:782–784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Takahashi N: Microbial ecosystem in the
oral cavity: Metabolic diversity in an ecological niche and its
relationship with oral diseases. Int Congr Ser. 1284:103–112. 2005.
View Article : Google Scholar
|
|
101
|
Brand RA: Surgical anatomy of the rotator
cuff and the natural history of degenerative periarthritis. Clin
Orthop Related Res. 466:543–551. 2008. View Article : Google Scholar
|
|
102
|
Claycombe KJ, Brissette CA and Ghribi O:
Epigenetics of inflammation, maternal infection, and nutrition. J
Nutr 145 (Suppl):. 1109S–1115S. 2015.
|
|
103
|
Niwa T, Tsukamoto T, Toyoda T, Mori A,
Tanaka H, Maekita T, Ichinose M, Tatematsu M and Ushijima T:
Inflammatory processes triggered by Helicobacter pylori infection
cause aberrant DNA methylation in gastric epithelial cells. Cancer
Res. 70:1430–1440. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ding W, Mouzaki M, You H, Laird JC, Mato
J, Lu SC and Rountree CB: CD133+ liver cancer stem cells
from methionine adenosyl transferase 1A-deficient mice demonstrate
resistance to transforming growth factor (TGF)-beta-induced
apoptosis. Hepatology. 49:1277–1286. 2009. View Article : Google Scholar
|
|
105
|
Wang YQ, Li YM, Li X, Liu T, Liu XK, Zhang
JQ, Guo JW, Guo LY and Qiao L: Hypermethylation of TGF-β1 gene
promoter in gastric cancer. World J Gastroenterol. 19:5557–5564.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Boosani CS, Dhar K and Agrawal DK:
Down-regulation of hsa-miR-1264 contributes to DNMT1-mediated
silencing of SOCS3. Mol Biol Rep. 42:1365–1376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Cheng C, Huang C, Ma TT, Bian EB, He Y,
Zhang L and Li J: SOCS1 hypermethylation mediated by DNMT1 is
associated with lipopolysaccharide-induced inflammatory cytokines
in macrophages. Toxicol Lett. 225:488–497. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kuo PL, Shen KH, Hung SH and Hsu YL:
CXCL1/GROα increases cell migration and invasion of prostate cancer
by decreasing fibulin-1 expression through NF-κB/HDAC1 epigenetic
regulation. Carcinogenesis. 33:2477–2487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen S, Boosani C and Agrawal DK: Abstract
12818: KPNA4 mediates vitamin D-dependent inhibition of NF-κB
activity in swine epicardial preadipocytes. Circulation.
130:A128182014.
|
|
110
|
Cao B, Yang Y, Pan Y, Jia Y, Brock MV,
Herman JG and Guo M: Epigenetic silencing of CXCL14 induced
colorectal cancer migration and invasion. Discov Med. 16:137–147.
2013.PubMed/NCBI
|
|
111
|
Ramos EA, Grochoski M, Braun-Prado K,
Seniski GG, Cavalli IJ, Ribeiro EM, Camargo AA, Costa FF and
Klassen G: Epigenetic changes of CXCR4 and its ligand CXCL12 as
prognostic factors for sporadic breast cancer. PLoS One.
6:e294612011. View Article : Google Scholar
|
|
112
|
Yasmin R, Siraj S, Hassan A, Khan AR,
Abbasi R and Ahmad N: Epigenetic regulation of inflammatory
cytokines and associated genes in human malignancies. Mediators
Inflamm. 2015.201703:2015.
|