Introduction
Lung development is a highly conserved process that
can be broken down into four phases: Origination of the lung
anlage; formation of the trachea; morphogenesis of branches; and
finally, alveologenesis and reduction in cell proliferation with
the emergence of more differentiated cell types (1). In the late stage of lung
development, alveolar airspaces are formed. These are the principal
units involved in gas exchange (2). Lung alveolar epithelial cells are
comprised of alveolar type (AT) 1 and AT2. AT2 cells serve as
progenitors for AT1 cells during development and cell damage
(3-5). This late stage of lung development
requires highly coordinated regulation; any slight disturbance in
alveolarization may lead to severe diseases or developmental
disorders (6).
Small ubiquitin-like modifier (SUMO) conjugation to
proteins is a reversible post-translational modification that
participates in various cellular and developmental processes
(7). In mammals, there are three
main SUMO protein subtypes: SUMO1, SUMO2 and SUMO3. Due to their
high sequence homology, SUMO2 and SUMO3 are usually referred as
SUMO2/3. SUMO1 is conjugated to proteins even under normal
physiological conditions (8).
SUMO2/3 are usually found unconjugated, but rapidly bind to
proteins in several stress conditions (9). Covalent attachment of SUMO to
proteins is assisted by the E1 activating enzyme, E2 conjugating
enzyme (Ubc9) and E3 ligase. Deconjugation from targets is
regulated by SUMO specific proteases (SENPs). In addition to
deSUMOylation, SENPs also catalyze the maturation of SUMO
precursor. Thus, SENPs are a key regulatory mechanism for
maintaining a SUMO balance. Six SENPs (SENP1, 2, 3, 5, 6 and 7) are
present in humans and mice. Out of these six proteins, SENP1 has
been the most widely studied. SENP1 is involved in various
pathophysiological processes, including transcriptional regulation,
cell proliferation, differentiation and development (10-12). Current studies indicate that
disorder in either SUMO conjugation or deconjugation can contribute
to embryonic lethality, chromosomal defects, abnormal nuclear
morphology and promote the occurrence of diseases (13,14). Ubc9 deficiency in mice leads to
embryonic lethality at the early developmental stage (15). Mutation of SENP1 in mice causes an
increase in SUMO1 conjugation and phenotypic defects in the
placenta (16). Experimental data
of Ubc9 deficiency and SENP1 mutation indicate that the maintenance
of the SUMO cycle is essential for normal growth and
development.
In the respiratory system, it has been previously
reported that the CCAAT/enhancer-binding protein α (C/EBPα) is
modified by SUMO1 to mediate lung growth and differentiation
(17). SENP1 enhances the
proliferative ability of pulmonary artery smooth muscle cells
(18). In addition, during the
pathogenesis of lung disease, SUMO1 may participate in the
modulation of hypoxia-inducible factor-1α through SUMOylation in
hypoxic pulmonary hypertension (19). Hypoxia triggers deSUMOylation of
Kruppel-like factor 15 by SENP1 and transcriptional regulation of
arginase 2 in lung endothelium (20). SENP1 was reported to be
overexpressed in lung cancer tissues, while modulation of SENP1
expression was demonstrated to significantly affect the
proliferation of lung cancer cells (21). However, little attention has been
devoted to the role of SENP1 in the process of lung
differentiation. To clarify this process, the expression levels of
SUMO1 and SENP1 were examined in rat lung tissue in the current
study. The results indicated that SENP1 regulates deSUMOylation of
SUMO1-modified proteins during the alveolar development period. It
was also demonstrated that SENP1 is required for the
differentiation of AT2 cells. Together, these results reveal that
SENP1 is involved in lung development and differentiation.
Materials and methods
Animals and tissue preparation
A total of 32 Sprague-Dawley neonatal rats (weight
6.2±0.3 g) were purchased from the Animal Center of Jiangsu
University (Zhenjiang, China). The rats were randomly assigned to 4
groups with 8 rats in each group. Each group of neonatal rats was
fed by a 3-month-old female rat. Rats were kept on a 12-h
light/dark cycle at a room temperature of 23±2°C and supplied with
sufficient food and water. Neonatal rats were sacrificed via
cervical dislocation, followed by a sternotomy. Phosphate buffered
saline (PBS) was gently perfused into the trachea with an injector
to clear the lung tissue. Lungs were collected and frozen
immediately for further study at postnatal day 1, 4, 7 and 14 (P1,
P4, P7 and P14). All animal research was approved by the Animal
Center of Jiangsu University.
Histological analysis
Tissues were fixed with 4% paraformaldehyde for 24 h
at 4°C and washed with PBS. Subsequently, samples were dehydrated
using an alcohol gradient (75% alcohol, 1.5 h; 95% alcohol, 1.5 h;
100% alcohol, 1.5 h; 100% alcohol, 1 h; two xylene washes, 0.5 h
each) and embedded in paraffin. Sections were sliced at 3
µm, followed by conventional dewaxing in water. Antigen
retrieval was performed in 10 mM citrate buffer (pH 6.0) and boiled
for 20 min. The tissue sections were stained with hematoxylin and
eosin (Solarbio Science & Technology Co., Ltd., Beijing, China)
2-3 min each for histological analysis. All steps were performed at
a room temperature. The sections were acquired by confocal light
microscopy (Olympus Corporation, Tokyo, Japan) at magnification of
×400.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) for lung tissue
RT-qPCR was used to detect the mRNA expression of
SUMO1 and SENP1 at each time-point. Total RNA was isolated from the
tissue samples using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). For RT, cDNA was synthesized using the
Prime Script RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) according to the manufacturerֺ’s instructions. qPCR
was performed using the SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd., Dalian, China). The reaction conditions were as follows:
95°C for 30 sec, and 40 cycles of 95°C for 5 sec and 60°C for 30
sec. Target sequences of SUMO1, SENP1 and β-actin were as follows:
SUMO1, 5′-AAG TTA TTG GAC AGG ACA GCA-3′ and 5′-CAT TCC CAG TTC TTT
TGG AG-3′; SENP1, 5′-CGC CAG ATT GAA GAG CAG A-3′ and 5′-AGA GGA
ACA CGA AGG TGG AG-3′; β-actin, 5′-TGT CAC CAA CTG GGA CGA TA-3′
and 5′-GGG GTG TTG AAG GTC TCA AA-3′. The specificity of the PCR
product was ensured by melting curve analysis using the
LightCycler® 96 (Roche Diagnostics, Basel, Switzerland).
All calculations of the relative expression of the target gene were
performed using the 2−ΔΔCq method (22).
Cell culture and grouping
For the experiments, human primary type II alveolar
epithelial cells (AT2; cat. no. HUM-iCELL-a002; purchased from
iCell Bioscience, Inc., Shanghai, China) were cultured in
Dulbeccoֺ’s modified Eagleֺ’s medium/F12 (DME/F-12; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA), supplemented with 10%
fetal bovine serum (Wisent, Inc., St. Bruno, QC, Canada), in a 5%
CO2 at 37°C. For AT2 differentiation, retinoic acid (RA)
was added to medium at 1 µM (23). RA powder was purchased from Target
Molecule Corp. (Boston, MA, USA) and dissolved in dimethyl
sulfoxide. To minimize the effect of AT2 cells transforming into
AT1 in vitro and ensure stable growth, the primary AT2 cells
were passaged for three generations prior used for differentiation.
After reaching 80-90% confluency, the cells were divided into
normal control group (NC group), RA group (with 1 µM RA in
the medium), si-NS group, si-SENP1 group, RA + si-NS group, and RA
+ si-SENP1 group. The si-SENP1 cells and si-NS cells were
transfected with SENP1 small interfering RNA (siRNA) or
non-specific (NS) siRNA.
Protein extraction and western
blotting
All protein extraction handling was performed on
ice. Free SUMO1 and SUMOylated proteins were analyzed. As described
by Sharma et al (24),
individual lung tissue protein lysates were prepared either using
4% sodium dodecyl sulfate (SDS) or 1% Nonident P40 (NP40). SDS
denatures the action of SENPs and preserves conjugated SUMO.
Therefore, the measured free SUMO1 is the naturally existing free
unconjugated SUMO1 protein. NP40 separates SUMO1 from the target.
Thus, the measured free SUMO1 represents total SUMO1 including
unconjugated and separated SUMO1 in lung tissue. Free SUMO1 and
SUMOylated proteins were extracted by 4% SDS, unless otherwise
indicated. Protein extraction for SENP1 detection was performed as
described. AT2 cells were harvested using the
radioimmunoprecipitation assay buffer containing protease inhibitor
phenylmethanesulfonyl fluoride (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for cell lysis. The extract was centrifuged at
12,000 × g, 4°C for 15 min and the supernatant was collected. The
protein concentration was detected using a bicinchoninic acid kit
(Beyotime Institute of Biotechnology, Haimen, China). Protein
extracts (10 µl) were fractionated by 8-16% SDS-PAGE
(GenScript, Piscataway, NJ, USA) and transferred to polyvinylidene
difluoride membranes. The membranes were blocked in 5%
milk-Tris-buffered saline (TBS) buffer containing 0.1% Tween-20 at
37°C for 1 h, then incubated overnight at 4°C with primary
antibodies. Primary antibodies were applied at dilutions of 1:200
for SENP1 (cat. no. HPA011765; Sigma-Aldrich; Merck KGaA), 1:1,000
for surfactant protein C (SP-C; cat. no. sc-13979; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), 1:1,000 for aquaporin-5
(AQP5; cat. no. sc-514022; Santa Cruz Biotechnology, Inc.), 1:1,000
for SUMO1 and 1:1,000 for β-actin (cat. no. 4903 and cat. no. 3700
from Cell Signaling Technology, Inc., Danvers, MA, USA). Following
washing with TBS buffer-0.1% Tween-20, horseradish
peroxidase-coupled secondary antibodies (1:2,000; cat. nos.
FMS-MS01, FMS-Rb01 and FMS-Gt01) from Fcmacs Biotech Co., Ltd.
(Nanjing, China) were added for 1 h at 37°C. The signals were
visualized using FluorChem FC3 chemiluminescence (ProteinSimple,
San Jose, CA, USA). Due to the significant difference in the
protein expression quantity of free SUMO1 and conjugated SUMO1, the
exposure time of conjugated SUMO1 (20 sec) was shorter than that of
free SUMO1 (50 sec). Thus, they are typically shown separately in
immunoblot bands (24,25). Semi-quantitative analysis was
performed using LANE 1D software (Beijing Sage Creation Science
Co., Ltd., Beijing, China). The target protein and β-actin were
analyzed, and the expression of the target protein at each
time-point was compared with with the corresponding internal
reference. Statistical analysis was conducted to estimate the
relative expression of the target proteins.
Cell differentiation and
immunofluorescence
Retinoic acid (RA) is a major bioactive metabolite
of vitamin A. It regulates normal lung development and maturation,
and preserves alveolar formation in a caloric restriction model
(26,27). To investigate the effect of RA on
AT2 differentiation, 6×105 cells were seeded in 2 ml
medium. At a density of 1×107 cells per 6-cm culture
dish, AT2 cells were randomly divided into negative control (NC)
group and RA group (with 1 µM RA added to the medium). Cells
were harvested at 24, 48 and 72 h. Double immunofluorescent
staining was used to observe the expression of SP-C and AQP5. AT2
cells were seeded in 24-well plates at a density of
1×104 cells/well. The cells were separated into NC group
and RA group. Cells were allowed to grow for 24, 48 and 72 h, then
fixed with 4% paraformalde-hyde at room temperature for 20 min and
blocked in 3% fetal calf serum at 37°C for 1 h. Primary antibodies
against SP-C (1:200; cat. no. sc-13979) and AQP5 (1:200, cat. no.
sc-514022; Santa Cruz Biotechnology, Inc.) were incubated overnight
at 4°C. Anti-SP-C was detected using a secondary fluorescein
isothiocyanate (FITC)-conjugated antibody (goat anti-rabbit IgG;
cat. no. A22120; 1:200; Abbkine Scientific Co., Ltd., Wuhan, China)
and anti-AQP5 was detected using a secondary Dylight 594-conjugated
antibody (goat anti-mouse IgG; cat. no. ab96873; 1:200; Abcam,
Cambridge, MA, USA) with incubated for 1 h at 37°C. Cell nuclei
were stained with DAPI (1:1,000) for 3 min at a room temperature of
23±2°C. Fluorescent images were obtained using a fluorescent
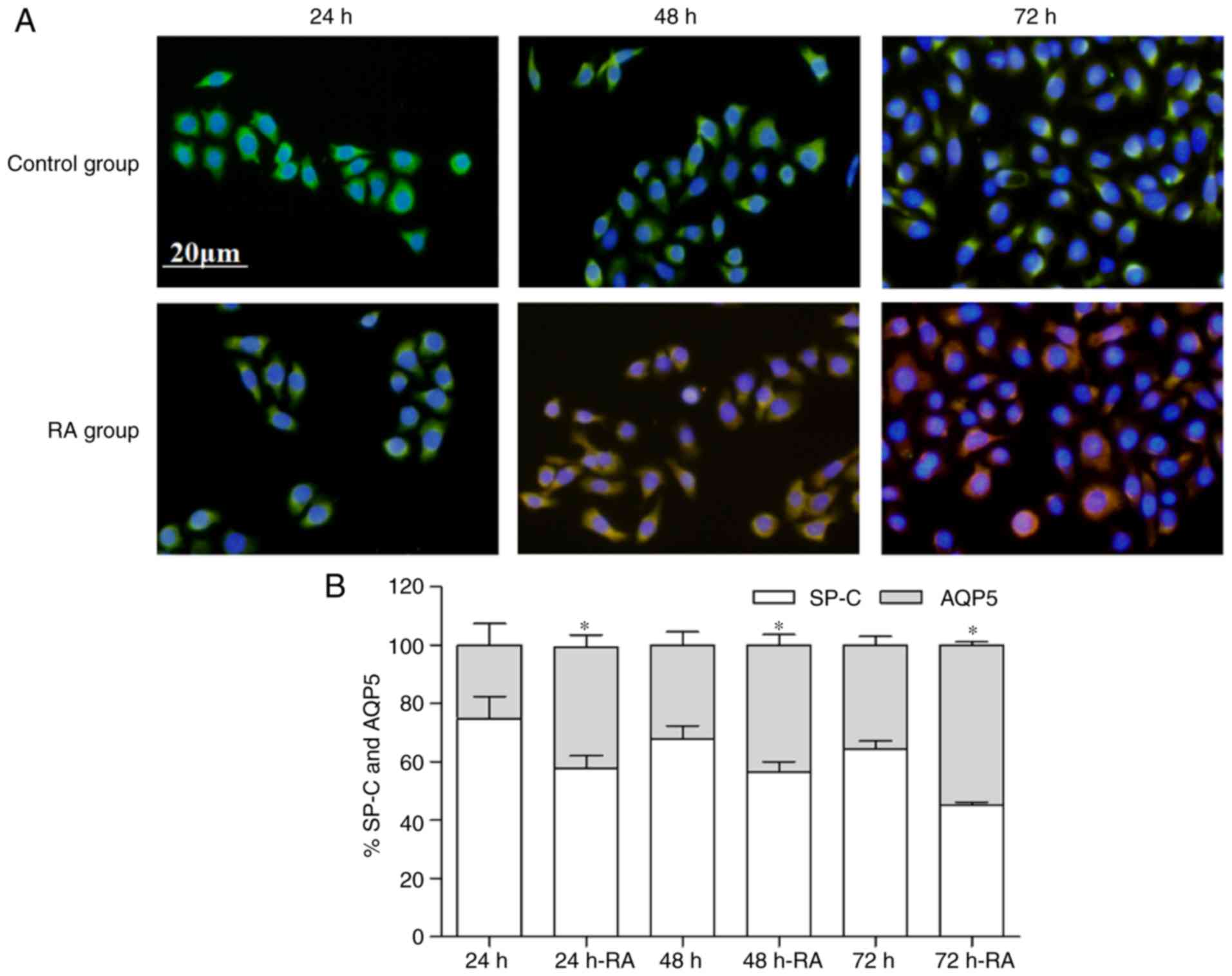
microscope (Olympus Corporation). Quantification of expression
levels of SP-C and AQP5 in fluorescent images was performed using
Image-Pro Plus III (Media Cybernetics, Inc., Rockville, MD, USA).
The percentage of SP-C or AQP5 is the expression level of SP-C or
AQP5/(SP-C+AQP5 expression level).
Cell transient transfection and
transfection based on RA
Transfections were performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), in accordance with the manufacturerֺ’s
instructions, when the cells reached 33% confluency. To verify the
transfection efficiency, AT2 cells were divided into si-NS group
and si-SENP1 group with no RA added in medium. To investigate the
suppression impact of SENP1 on differentiation, cells were
transfected with a SENP1 siRNA or NS siRNA with RA added in the
medium. The concentration of each siRNA used for transfection was
50 nM. Cells were harvested after transfection for 24, 48 and 72 h.
The transfection efficiency of SENP1 was linked to the resulting
mRNA and protein expression levels of conjugated SUMO1. The
suppressive influence of SENP1 with RA added was detected by
RT-qPCR and western blot analysis. Protein and gene expression
levels of SP-C and AQP5 were measured to analyze the effects on
differentiation. The siRNA sequences were as follows: SENP1 siRNA,
5′-GCC UGA CCA UUA CAC GCA ATT-3′ and 5′-UUG CGU GUA AUG GUC AGG
CTT-3′; NS siRNA: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and 5′-ACG UGA
CAC GUU CGG AGA ATT-3′. Primer sequences for detecting SP-C, AQP5
and β-actin were as follows: SP-C, 5′-TTA CCA CTG CCA CCT TCT CC-3′
and 5′-TCA AGA CTG GGG ATG CTC TC-3′; AQP5, 5′-ACT GGG TTT TCT GGG
TAG GG-3′ and 5′-GTG GTC AGC TCC ATG GTC TT-3′; β-actin, 5′-UGA CCU
CAA CUA CAU GGU UTT-3′ and 5′-AAC CAU GUA GUU GAG GUC ATT-3′. The
RT-qPCR processes were performed as described.
Cell proliferation
Cell counting kit-8 (CCK-8; Biosharp, Hefei, China)
was used to analyze cell proliferation. AT2 cells were randomly
into si-NS group and si-SENP1 group. They were then cultured in
DME/F-12 for 24, 48 and 72 h. The cells were then seeded in a
96-well plate at a density of 5,000 cells/well in 100 µl
medium and 10 µl CCK-8; four duplicate wells were set for
each sample. After 2 h in the cell incubator, and ELISA reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to measure
absorbance at 450 nm.
Cell apoptosis
An Annexin V/propidium iodide (PI)-FITC apoptosis
detection kit I (BD Biosciences, Franklin Lakes, NJ, USA) was used
to analyze cell apoptosis by flow cytometry. AT2 cells were plated
at a density of 6×105 cells per 6-cm culture dish, and
divided into si-NS and si-SENP1 groups. Cells were harvested after
transfection for 24, 48 and 72 h. Cells and medium were collected
in a 15 ml centrifuge tube. Trypsinization without EDTA was used to
suspend cells. Samples were handled according to the
manufacturerֺ’s instructions of the apoptosis detection kit. The
apoptosis rate of cells in each group was analyzed using an Accuri
C6 flow cytometer (CANTO 10C; BD Bioscience).
Statistical analysis
Data are expressed as the mean ± standard deviation
and analyzed using SPSS 17.0 statistical software (SPSS, Inc.,
Chicago, IL, USA). One-way analysis of variance was used to analyze
data differences among multiple groups, and then comparisons
between groups were assessed using a Student-Newman-Keuls test.
Student’s t-test was used to analyze the difference between groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Histomorphological variations of lung
morphogenesis at different stages
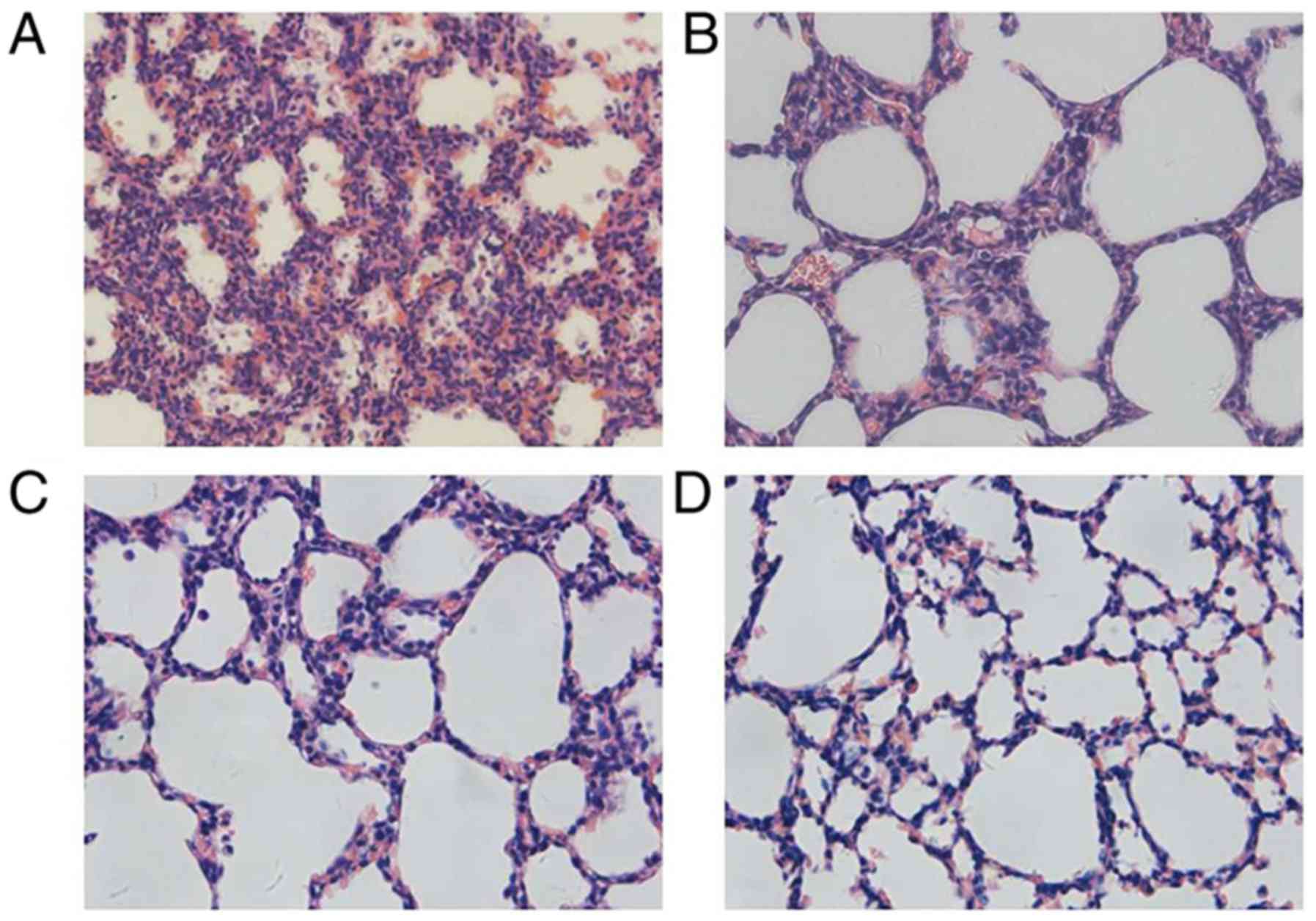
As presented in Fig.
1, irregularity in the structures could be observed in the
primary alveoli at P1. At P4, the late canalicular stage, an
increasing number of alveolar septa were formed with some ridges
protruding into the alveolar space. At P7, the size of air sacs
tended to be homogeneous and the interstitial tissues thinned out.
By P14, mature alveoli were the basic unit in the lung, appeared
uniformly sized and separated by a thin septum.
Lung development proceeds with changes in
free SUMO1, but no changes in total SUMO1
To investigate the role of SUMOylation in lung
development, whether the process involves changes in the levels of
SUMO1 conjugation was determined. Neonatal rats 14 days after birth
were used. At this age, their lungs are in the saccular and
alveolar period, an important phase of lung maturation. Initially,
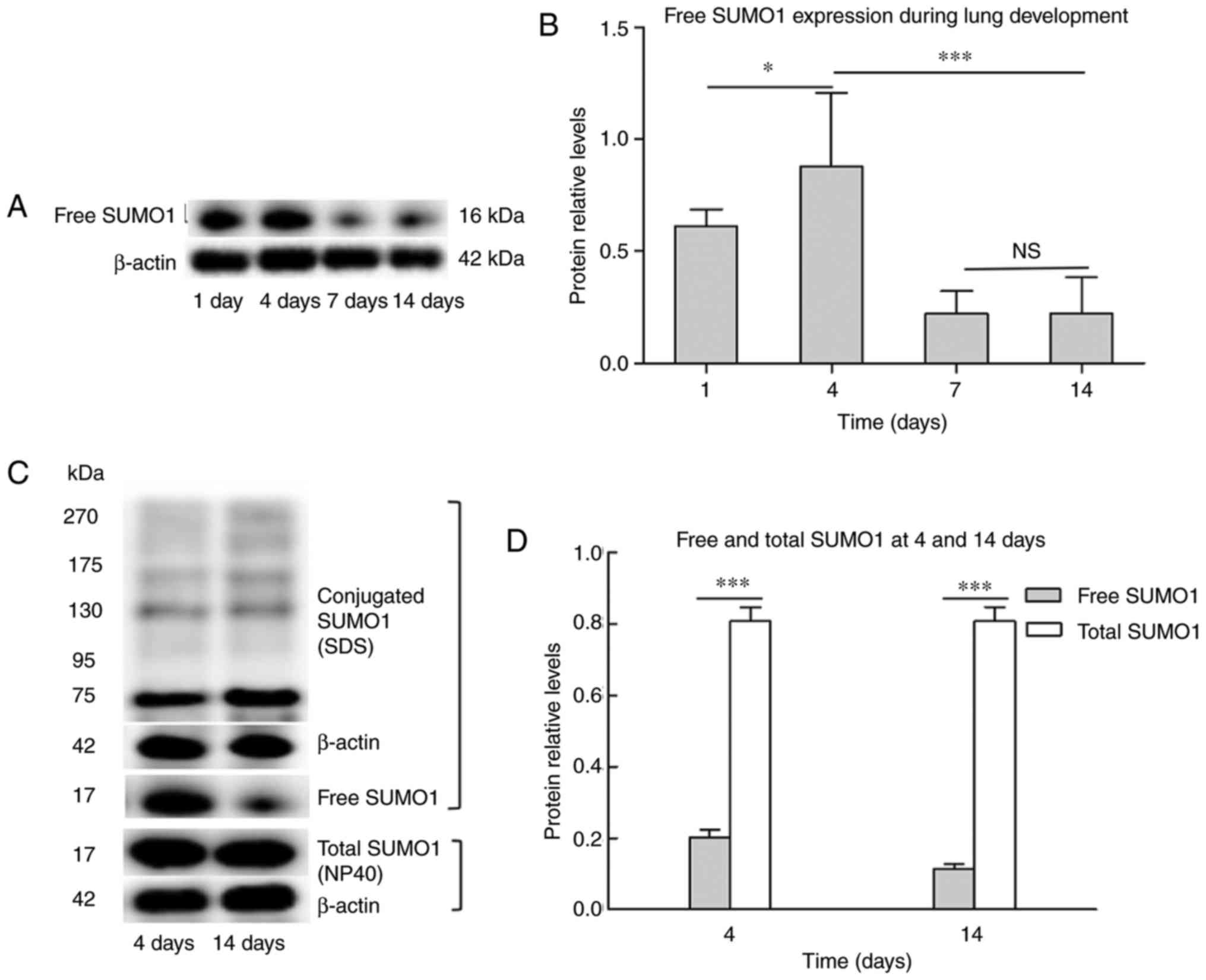
the protein levels of naturally existing free SUMO1 were detected.
As shown in Fig. 2A, free SUMO1
increased at day 4 compared with day 1, visibly decreased at day 7
compared with day 4, and then expressed at a similar level until
day 14. Day 4 and day 14 were selected as time-points for detecting
the degree of SUMO conjugation. In western blot analysis, free
SUMO1 and bands of higher molecular mass (>70 kDa) were detected
indicating SUMOylated proteins (Fig.
2B). Free SUMO1 prepared using 4% SDS (which represented
naturally existing free SUMO1) was compared with free SUMO1
prepared using 1% NP40 (which represented total SUMO1). Naturally
existing free SUMO1 at day 14 exhibited a sharp decrease compared
with day 4, while no significant changes were observed in total
SUMO1 (Fig. 2B). The results
indicate that the total SUMO1 was steadily expressed and the
SUMOylation and deSUMOylation were maintained in a dynamic
balance.
SUMO1 mRNA expression is stable, while
SENP1 is differentially expressed during lung development
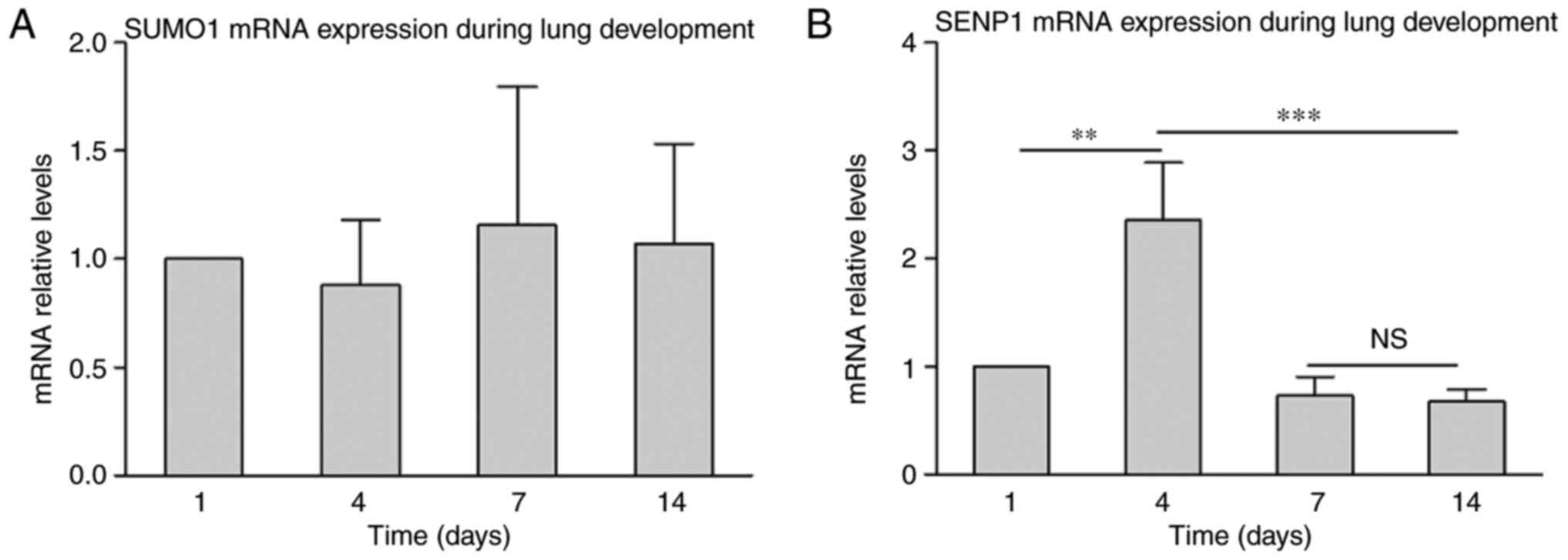
Gene expression analysis was performed to further
analyze the role of SUMO1 in lung development. RT-qPCR results
indicated that SUMO1 mRNA was not significantly changed during lung
development from P1 to P4 (Fig.
3A). This indicates that mRNA expression of SUMO1 was
maintained in at a stable level. Subsequently, gene expression
analysis of SENP1 was performed (Fig.
3B). The results demonstrated an increase in SENP1 mRNA at P4
compared with P1, which decreased at P7 and was maintained a steady
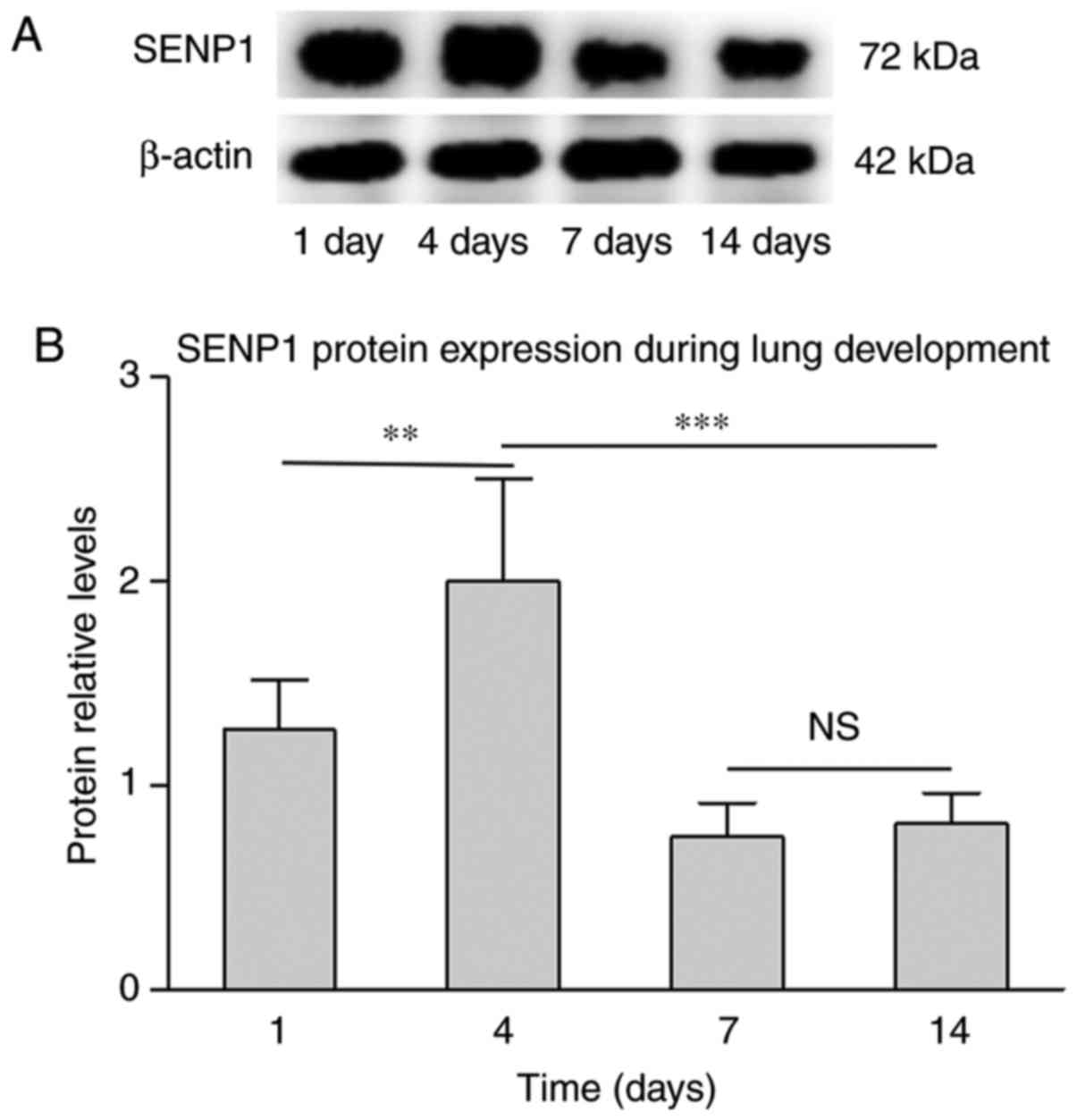
level from P7 to P14. To confirm if varied gene expression is
consistent with protein expression, SENP1 was also detected using
western blot analysis. As presented in Fig. 4, the gene and protein levels of
SENP1 exhibited a similar trend during lung development. However,
compared with the induction of SENP1 mRNA at P4, the change in
protein expression level was marginal. This difference may be
associated with the post-translational modifications, and that
protein translation is slightly delayed compared with mRNA
transcription. This confirmed that SENP1 may impact lung
development.
Retinoic acid promotes AT2
differentiation into AT1 with changes in SENP1
In a previous study, tritium [3H] was
injected into pregnant rats and the fetal rats were examined
(28). This experiment revealed
that AT2 cells could convert to AT1 cells during lung development.
According to the previous study in rats, during the 14 days after
birth, the expression of SP-C was gradually reduced, whereas the
expression of AQP5 increased (17,29). SP-C and AQP5 are specific markers
of AT2 and AT1 cells, respectively (30,31). This indicated that AT2
differentiation to AT1 may occur during this period. To determine
whether SENP1 affects lung development by participating in the
differentiation of AT2, AT2 cells were cultured in vitro in
the current study and RA was used to promote differentiation.
Initially, the differentiation efficiency of RA was examined. AT2
cells were exposed to 1 µM RA and the expression levels of
SP-C and AQP5 were then detected using western blot analysis. As
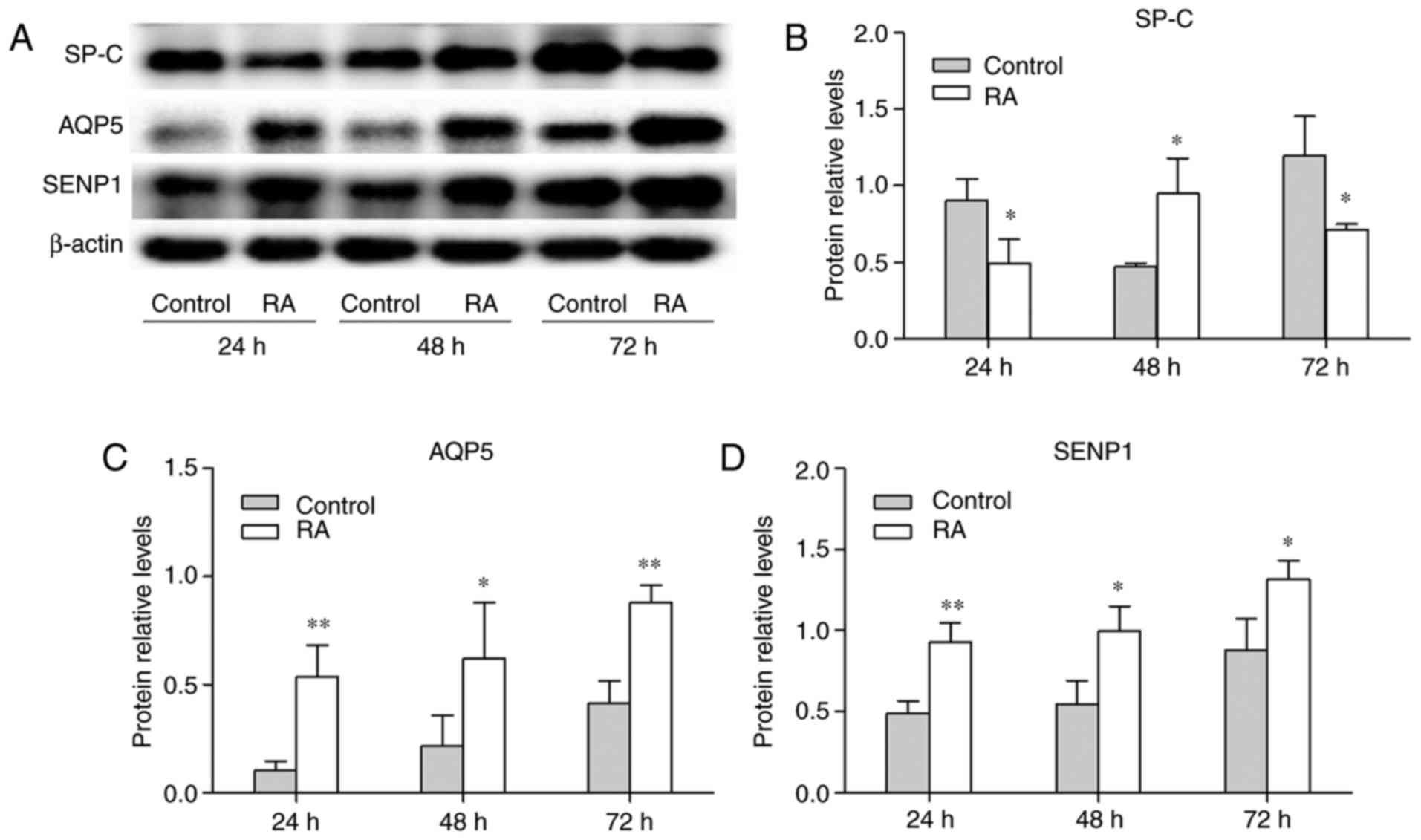
shown in Fig. 5A-C, the
expression of SP-C was decreased at 24 and 72 h following treatment
with RA. However, it increased was 48 h, which may have been caused
by cell proliferation. RA enhanced the expression of AQP5 at 24, 48
and 72 h compared with untreated cells (Fig. 5C). As shown in Fig. 5A and D, SENP1 protein was
increased over time in the control and RA groups. Additionally, the
expression of SENP1 in the RA group was increased compared with the
control group at 24, 48 and 72 h. To validate the changes in SP-C
and AQP5, the expression were also measured by immunofluorescence
(Fig. 6A and B). These results
also demonstrated that the decrease in SP-C was accompanied by an
increase in AQP5. The experiments demonstrated that RA promoted the
differentiation of AT2. Taken together, these results suggest that
SENP1 may participate in the differentiation of AT2.
Inhibition of SENP1 results in conjugated
SUMO1 changes and impairs AT2 differentiation
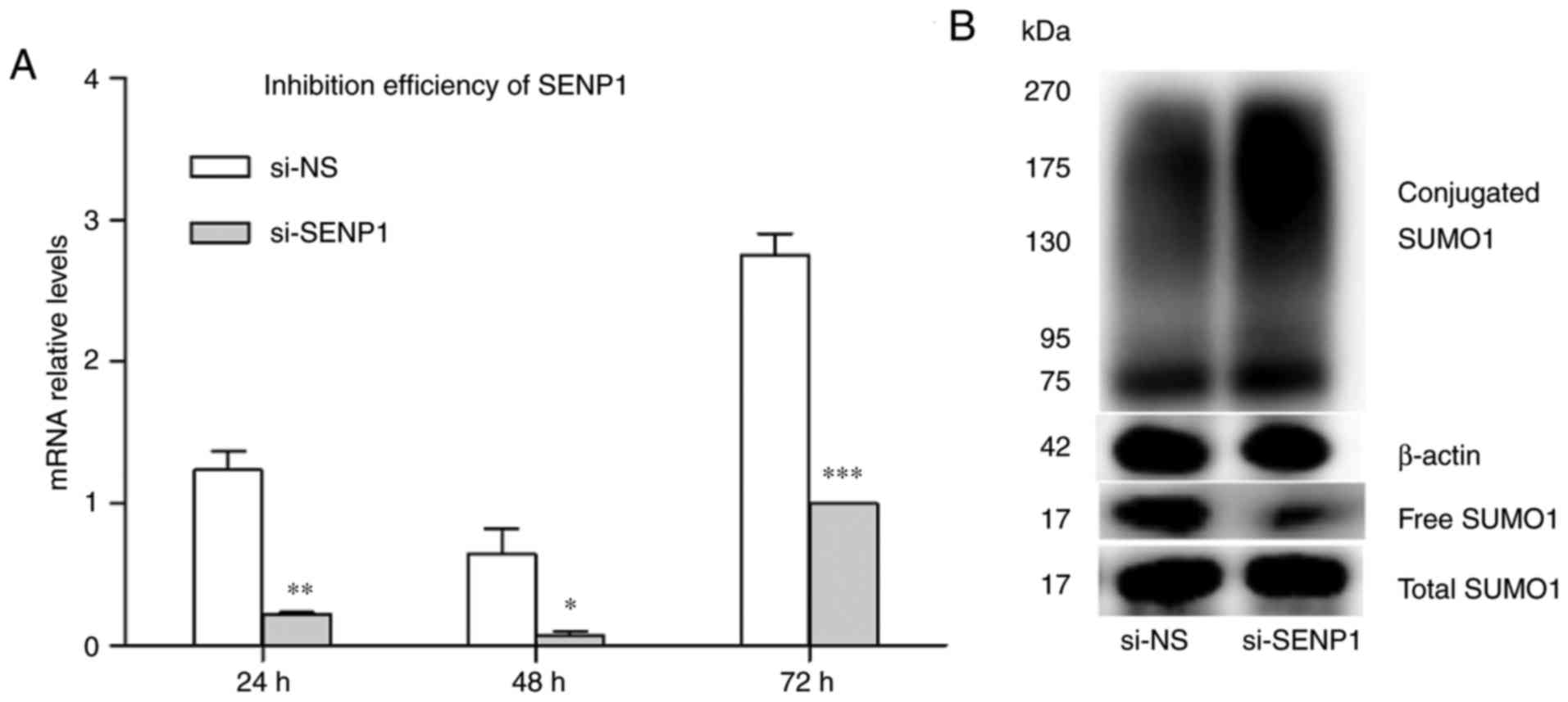
siRNA was used to inhibit SENP1 expression in AT2.
Cells were transfected with si-SENP1 and non-specific RNA (si-NS).
Transfection efficiency was demonstrated by RT-qPCR and western
blot analysis. The results indicated that the gene expression of
SENP1 was decreased at 24, 48 and 72 h after transfection compared
with si-NS. SENP1 was decreased 8.8-fold at 48 h compared with the
corresponding control group, indicating efficient inhibition
(Fig. 7A). As SENP1 is a
SUMO-specific protease, mediating protein deSUMOylation is its main
role. Changes in SUMOylated proteins were detected when SENP1 was
depleted using siRNA. The level of conjugated SUMO1 increased
sharply, while free SUMO1 was decreased by SENP1 knockdown compared
with si-NS (Fig. 7B). This result
indicates that the inhibition of SENP1 expression interfered with
deSUMOlation. The effect of SENP1 inhibition on cell
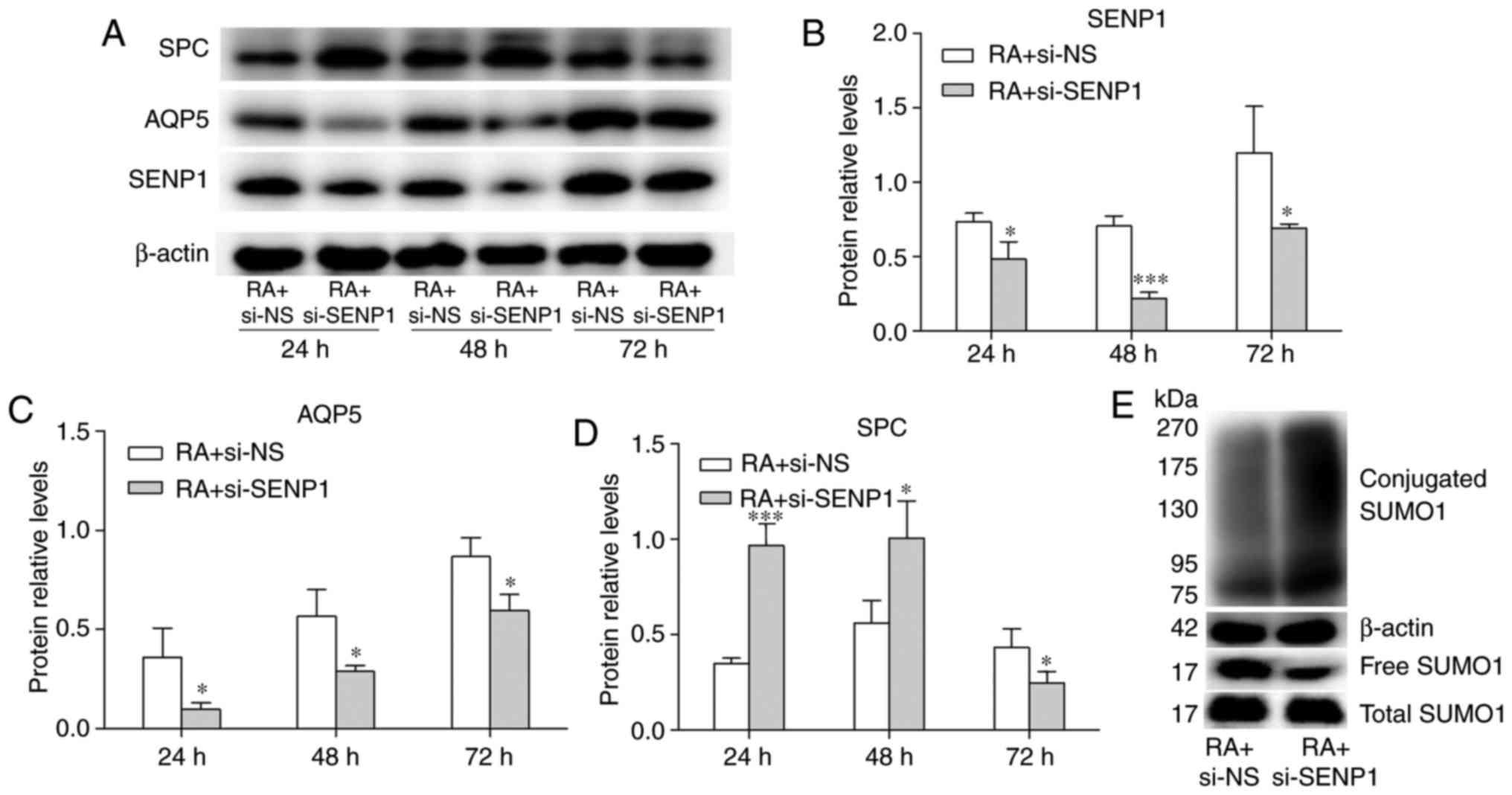
differentiation was investigated further. AT2 cells were cultured
in medium containing RA, then transfected with si-NS (RA + si-NS
group) or si-SENP1 (RA + si-SENP1 group). Western blot analysis
demonstrated that the expression of SENP1 protein decreased
significantly in the RA + si-SENP1 group compared to the RA + si-NS
group (Fig. 8A and B), with a
1.5-fold decrease in protein levels at 24 h, 3.2-fold at 48 h, and
1.6-fold at 72 h. The inhibition efficiency was the most pronounced
at 48 h, in accordance with the changes in mRNA expression. The
protein expression levels of SP-C and AQP5 were also assessed using
western blot analysis. As shown in Fig. 8C and D, compared to the control
group, the inhibition of SENP1 decreased the protein level of AQP5
expression at 24, 48 and 72 h. The protein expression of SP-C was
increased at 24 and 48 h, but decreased at 72 h. The decrease of
SP-C may have been caused by growth inhibition. The change in
SUMOylated proteins was also detected. As shown in Fig. 8E, the conjugated SUMO1 was
increased by si-SENP1 and free SUMO1 was decreased, which was
similar to the result with no RA was added (Fig. 7B). The expression level of free
and total SUMO1 under these two conditions revealed no
statistically significant difference. These results indicate that
the inhibition efficiency of SENP1 on conjugated and free SUMO1 has
not been affected by the effect of RA on the differentiation of AT2
cells. To determine the effect of SENP1 on cell differentiation,
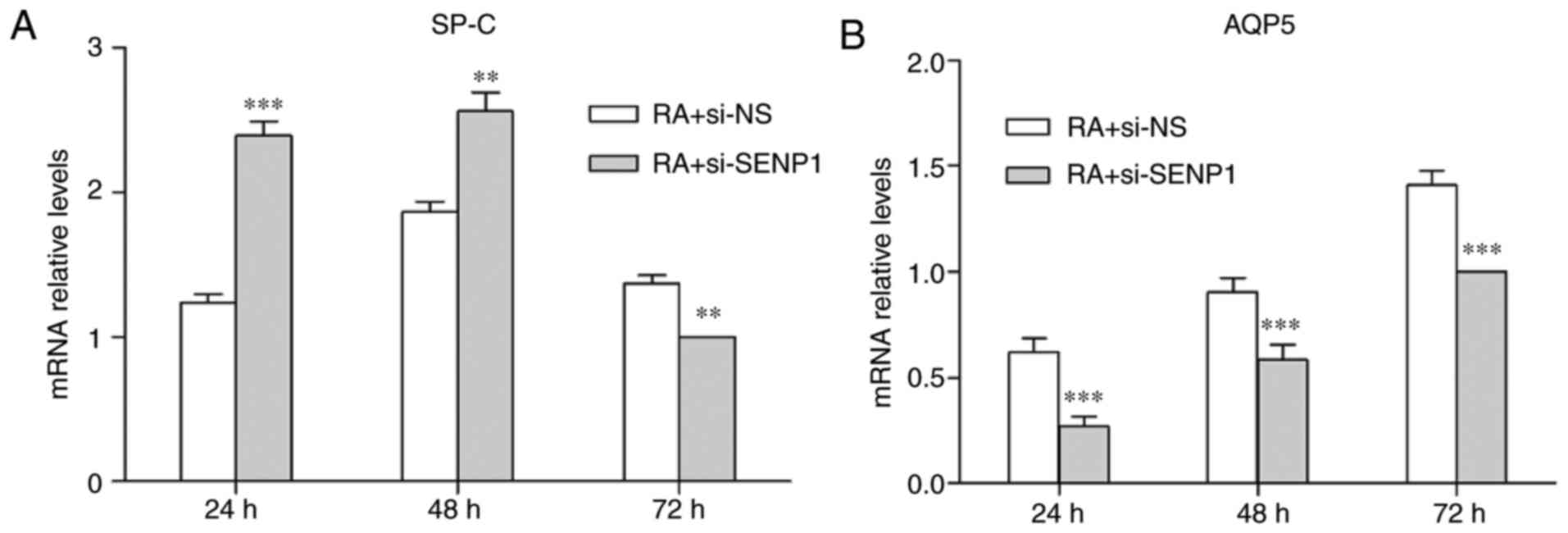
SP-C and AQP5 levels were also analyzed by RT-qPCR. SP-C and AQP5
mRNA expression levels exhibited a similar expression pattern to
the relevant protein levels following SENP1 knockdown (Fig. 9). These results indicate that
inhibition of SENP1 hinders the differentiation of AT2, and that
SENP1 is an important regulator of the differentiation process.
Depletion of SENP1 inhibits proliferation
and promotes apoptosis of AT2 cells
SENP1 has an important role in regulating the
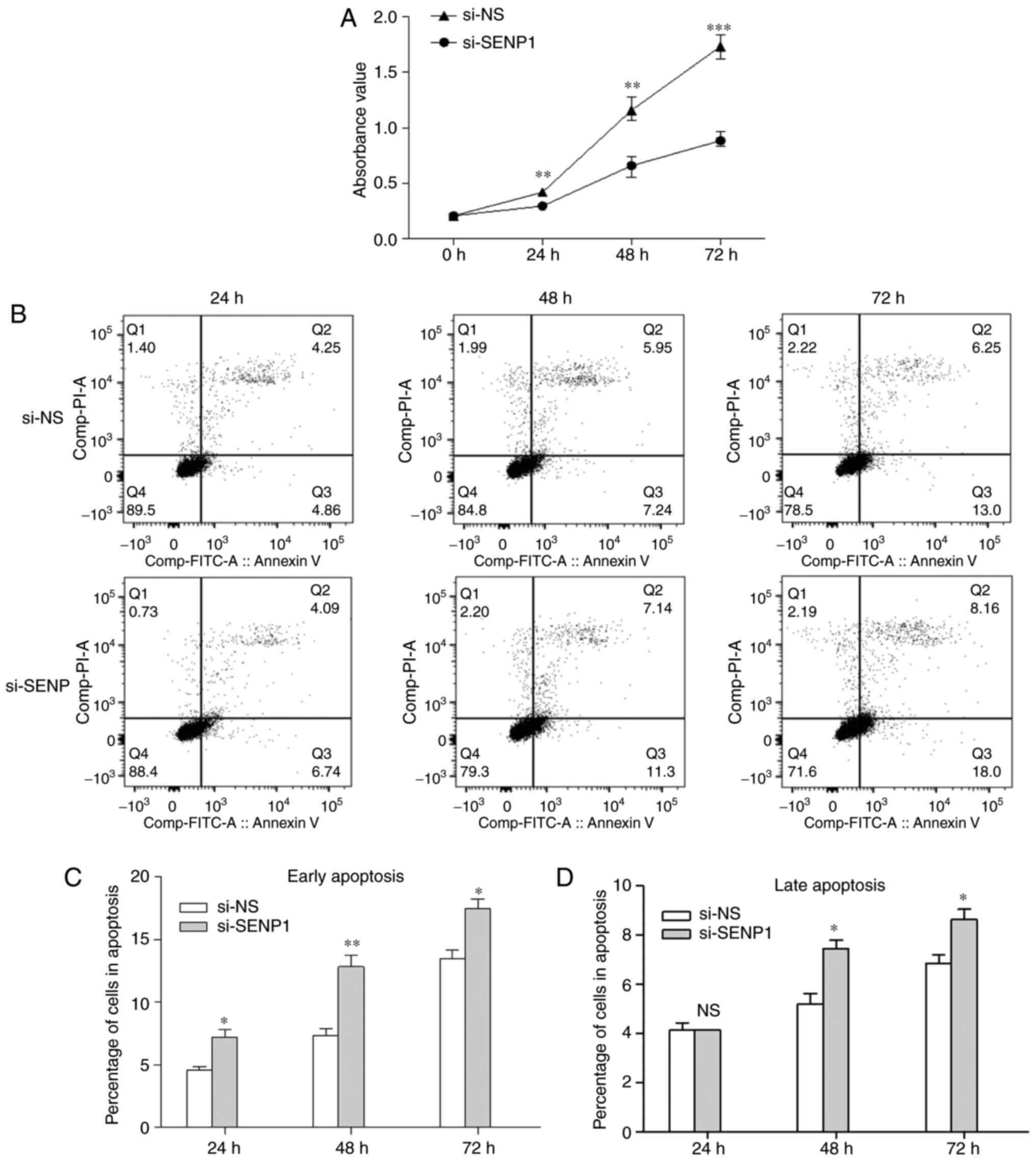
proliferation and apoptosis of cells. CCK-8 was used to evaluate
the AT2 proliferation in cells transfected with SENP1 siRNA for 24,
48 and 72 h. As shown in Fig.
10A, cell proliferation was reduced in the si-SENP1 group
compared with si-NS (P<0.05). To examine whether SENP1 knockdown
also affects apoptosis, flow cytometry analysis and an Annexin
V-FITC/PI apoptosis detection kit was used. Early apoptosis rate
(Q3) and late apoptosis rate (Q2) were combined as the total
apoptosis rate. SENP1 inhibition promoted cell apoptosis when AT2
cells were transfected with si-SENP1 for 24, 48 and 72 h (Fig. 10B). Following transfection with
si-SENP1 for 24 h, the percentage of cells in the early apoptosis
subgroup was significantly higher than in the si-NS group
(7.230±0.816 vs. 4.573±0.398%); however, SENP1 inhibition had no
influence on the late apoptosis. The percentage of cells in both
the early and late apoptosis subgroups were higher compared with
the si-NS group (12.833±1.266 vs. 7.317±0.831%; and 7.453±0.479 vs.
5.183±0.629%, respectively) when SENP1 inhibition was increased to
48 h. Furthermore, the percentage of cells in the early and late
apoptosis subgroups were higher compared with the si-NS group
(17.500±1.080 vs. 13.467±1.034%; and 8.637±0.585 vs. 6.860±0.483%,
respectively) at 72 h after transfection. The significant decrease
in cell apoptosis confirmed that the suppression of SENP1 caused
AT2 cell death. Overall, the data indicate that SENP1 inhibition
can reduce the proliferation of AT2 cells and promote
apoptosis.
Discussion
Similar to human lung development, the lung
development of rats is divided into five stages: Embryonic period,
pseudoglandular period, canalicular period, saccular period and
alveolar stage. At 1-4 days after birth, the rats are in the
saccular stage of lung development. At days 5-14 (equivalent to 36
weeks of fetal lung development in human pregnancy), the rat lungs
are in the alveolar phase (32).
Therefore, neonatal rats can be used to investigate the association
between the expression of SUMO1 and SENP1, and lung
development.
Although free SUMO1 protein is differentially
expressed during the lung development of neonatal rats, the mRNA
levels of SUMO1 remain is stable in lung tissue at this stage. The
protein levels of free and conjugated SUMO1 were also analyzed;
with the total SUMO1 protein levels remaining unchanged. However,
the expression level of free SUMO1 fluctuated and reached the
highest level at post-natal day 4. The level of conjugated SUMO1
was also variable, although its level was higher at day 14 than at
day 4. This data suggests that the observed changes in free SUMO
are associated with the degree of protein SUMOylation. Sharma et
al (24) reported that SENP1
is a major mediator of SUMO1 deconjugation and has a limited role
in deSUMOylating SUMO2/3-modified proteins. On the basis of SUMO1
overexpression in Ca Ski cells, Yuasa and Saitoh (33) labeled SUMO1 protein with GFP in Ca
Ski cells then added SENP1 catalytic domain into cell culture
medium. The study revealed that the labeled SUMO1 was decreased
significantly through the function of the SENP1 catalytic domain;
the deSUMOylation of GFP directly demonstrated the effect of SENP1
on SUMO1 modification. In the current study, the expression of
SENP1 was determined and revealing that the expression trend of
SENP1 in at the gene and protein levels was consistent with that of
free SUMO1 protein. Tissue morphological data indicated that that
P4 is the most obvious period of alveolar formation. The alveolar
morphology began to stabilize at P7-14. Consistent with these
results, the expression of SENP1 decreased at P7 compared with P4,
and expression was stable at P7-14. This indicates that SENP1 may
regulate SUMO1 deconjugation to maintain the dynamic balance of
protein SUMOylation and have an important role in lung
development.
To further investigate the effect of SENP1 on
protein SUMOylation and lung development in the present study,
SENP1 was silenced in AT2 cells. AT2 is considered to be a stem
cell of the alveolar epithelium (3,5).
In the process of normal cell renewal and repair, AT2 cells can
differentiate into AT1 cells, or produce progeny AT2 via mitosis to
maintain the cell population (34). SUMO1-conjugation was markedly
increased in cells with SENP silencing compared with the control
cells, indicating that depletion of SENP1 leads to disorder in
SUMOylation and deSUMOylation. Previous studies have demonstrated
that SUMOylation imbalance can lead to tumorigenesis, inflammatory
diseases, DNA damage and impair cell differentiation (10,14). Bronchopulmonary dysplasia (BPD) is
a common serious respiratory disease in preterm infants. Compared
with normal infants, the expression of free SUMO1 in the peripheral
blood mononuclear cells of children with BPD is increased, while
the expression of NAD-dependent protein deacetylase sirtuin-1
(SIRT1) and SUMOylated SIRT1 are decreased (35). These results suggest that the
increase in free SUMO1 and decrease in SUMOylated SIRT1 may be
associated with the occurrence of BPD. A previous study reported
that the differentiation of multipotent stem cells into neurons was
inhibited by overexpression of SUMO1 (25). It was speculated that SENP1 may
have a function in the differentiation of AT2; thus, this was
investigated by culturing AT2 cells in vitro. Compared with
the control group, the SENP1 expression level was increased by
adding RA to the medium to promote the differentiation of AT2. This
confirmed that SENP1 was involved in the differentiation.
Subsequently, RA was used to promote differentiation and expression
of SENP1 in AT2 cells was inhibited using siRNA. Compared with the
RA + si-NS group, the expression levels of AQP5 in the RA +
si-SENP1 group were suppressed at the protein and mRNA level. The
expression of SP-C was increased at 24 and 48 h after transfection,
but decreased at 72 h. These results demonstrate that despite the
differentiation promoting effect of RA, the differentiation of AT2
was inhibited when SENP1 was suppressed. The inhibition of SENP1 in
cells was confirmed to affect cell growth. The increase in SP-C was
caused by inhibition of cell differentiation, while the decrease
may be influenced by cell death. Further analysis demonstrated that
silencing SENP1 in AT2 cells impairs cell proliferation and
promotes apoptosis. This effect was most marked at 72 h. The above
results indicate that SENP1 is important for the differentiation of
AT2s and can affect their growth. Numerous target proteins are
regulated by the SUMOylation and deSUMOylation balance. Our
previous study (17) demonstrated
that SUMOylated C/EBPα was gradually decreased during lung
differentiation and was negatively correlated with pulmonary
surfactant secretion. This suggested that SUMO modification may be
involved in C/EBPα-mediated lung growth and differentiation.
However, there has been limited research on the effects of protein
SUMOylation and deSUMOylation on lung development and
differentiation. The specifics of which proteins undergo SUMO
modification and have roles in lung development and/or
differentiation still require further clarification.
In conclusion, to the best of our knowledge, the
current study is the first to demonstrate that SENP1 maintains the
dynamic balance between protein SUMOylation and deSUMOylation
during the alveolar period. In vitro experiments revealed
that SENP1 regulates the proliferation and differentiation of AT2
cells via protein SUMOylation. The findings indicated that SENP1 is
a key factor involved in normal lung development; however, whether
SUMOylation of specific target proteins has a key role in the
process requires further investigation.
Acknowledgments
Not applicable.
Funding
This work was financially supported by the National
Natural Science Foundation of China (grant no. 81370746, 81741052)
and the National Natural Science Foundation of Jiangsu Province
(grant no. BK20161356).
Availability of data and materials
The datasets used and/or analyzed in this study are
available from the corresponding author on reasonable request.
Authorsֺ’ contributions
XQW conducted most of the experiments and was a
major contributor in writing the paper. HYL, QXW and HMJ conceived
the concept of the study. JYC and YZ cultivated cells and performed
the PCR assay. HTZ performed the statistical analysis. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Center
of Jiangsu University (Zhenjiang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warburton D and Bellusci S: The molecular
genetics of lung morphogenesis and injury repair. Paediatr Respir
Rev. 5(Suppl A): S283–S287. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Claudio N and Morty RE: MicroRNA in late
lung development and bronchopulmonary dysplasia: The need to
demonstrate causality. Mol Cell Pediatr. 3:192016. View Article : Google Scholar
|
|
3
|
Barkauskas CE, Cronce MJ, Rackley CR,
Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW and Hogan LM:
Type 2 alveolar cells are stem cells in adult lung. J Clin Invest.
123:3025–3036. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Adamson IY and Bowden DH: The type 2 cell
as progenitor of alveolar epithelial regeneration: A cytodynamic
study in mice after exposure to oxygen. Lab Invest. 30:35–42.
1974.PubMed/NCBI
|
|
5
|
Adamson IY and Bowden DH: Derivation of
type 1 epithelium from type 2 cells in the developing rat lung. Lab
Invest. 32:736–745. 1975.PubMed/NCBI
|
|
6
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lomelí H and Vázquez M: Emerging roles of
the SUMO pathway in development. Cell Mol Life Sci. 68:4045–4064.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garciadominguez M and Reyes JC: SUMO
association with repressor complexes, emerging routes for
transcriptional control. Biochim Biophys Acta. 1789:451–459. 2009.
View Article : Google Scholar
|
|
9
|
Tempé D, Piechaczyk M and Bossis G: SUMO
under stress. Biochem Soc Trans. 36:874–878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim JH and Baek SH: Emerging roles of
desumoylating enzymes. Biochim Biophys Acta. 1792:155–162. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saho E, Takuya A, Hiroshi A, Shunsuke K,
Barnabas S, Yusuke Y, Akira M, Shunichi T and Dana B: The SUMO
protease SENP1 is required for cohesion maintenance and mitotic
arrest following spindle poison treatment. Biochem Biophys Res
Commun. 42:310–316. 2012.
|
|
12
|
Chen CH, Chang CC, Lee TH, Luo M, Huang P,
Liao P, Wei S, Li F, Chen R, Zhou XZ, et al: SENP1 deSUMOylates and
regulates Pin1 protein activity and cellular function. Cancer Res.
73:3951–3962. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Flotho A and Melchior F: Sumoylation: A
regulatory protein modification in health and disease. Annu Rev
Biochem. 82:357–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bawa-Khalfe T and Yeh ET: SUMO losing
balance: SUMO proteases disrupt SUMO homeostasis to facilitate
cancer development and progression. Genes Cancer. 1:748–752. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nacerddine K, Lehembre F, Bhaumik M, Artus
J, Cohen- Tannoudji M, Babinet C, Pandolfi PP and Dejean A: The
SUMO pathway is essential for nuclear integrity and chromosome
segregation in mice. Dev Cell. 9:769–779. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamaguchi T, Sharma P, Athanasiou M, Kumar
A, Yamada S and Kuehn MR: Mutation of SENP1/SuPr-2 reveals an
essential role for desumoylation in mouse development. Mol Cell
Biol. 25:5171–5182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen YD, Liu JY, Lu YM, Zhu HT, Tang W,
Wang QX and Lu HY: Functional roles of C/EBPα and SUMO-modification
in lung development. Int J Mol Med. 40:1037–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou F, Dai A, Fu D, Jiang Y, Tan X and
Zhang X: SENP-1 enhances hypoxia-induced proliferation of rat
pulmonary artery smooth muscle cells by regulating
hypoxia-inducible factor-1α. Mol Med Rep. 13:3482–3490. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Y, Wang J, Tian H, Li G, Zhu H, Liu
L, Hu R and Dai A: Increased SUMO-1 expression in response to
hypoxia: Interaction with HIF-1α in hypoxic pulmonary hypertension.
Int J Mol Med. 36:271–281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pandey D, Nomura Y, Rossberg MC, Hori D,
Bhatta A, Keceli G, Leucker T, Santhanam L, Shimoda LA, Berkowitz D
and Romer L: Hypoxia triggers SENP1 (sentrin-specific protease 1)
modulation of KLF15 (Kruppel-like factor 15) and transcriptional
regulation of Arg2 (Arginase 2) in pulmonary endothelium.
Arterioscler Thromb Vasc Biol. 38:913–926. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang RT, Zhi XY, Zhang Y and Zhang J:
Inhibition of SENP1 induces radiosensitization in lung cancer
cells. Exp Ther Med. 6:1054–1058. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Gao RW, Kong XY, Zhu XX, Zhu GQ, Ma JS and
Liu XX: Retinoic acid promotes primary fetal alveolar epithelial
type II cell proliferation and differentiation to alveolar
epithelial type I cells. In Vitro Cell Dev Biol Anim. 51:479–487.
2015. View Article : Google Scholar
|
|
24
|
Sharma P, Yamada S, Lualdi M, Dasso M and
Kuehn MR: SENP1 is essential for desumoylating SUMO1-modified
proteins but dispensable for SUMO2 and SUMO3 deconjugation in the
mouse embryo. Cell Rep. 3:1640–1650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Juarez-Vicente F, Luna-Pelaez N and
Garcia-Dominguez M: The SUMO protease SENP7 is required for proper
neuronal differentiation. Biochim Biophys Acta. 1863:1490–1498.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Besnard V, Nabeyrat E, Henrion-Caude A,
Chadelat K, Perin L, Le Boucn Y and Clement A: Protective role of
retinoic acid from antiproliferative action of TNF-alpha on lung
epithelial cells. Am J Physiol Lung Cell Mol Physiol.
282:L863–L871. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Londhe VA, Maisonet TM, Lopez B, Shin BC,
Huynh J and Devaskar SU: Retinoic acid rescues alveolar hypoplasia
in the calorierestricted developing rat lung. Am J Respir Cell Mol
Biol. 48:179–187. 2013. View Article : Google Scholar :
|
|
28
|
Evans MJ, Cabral LJ, Stephens RJ and
Freeman G: Renewal of alveolar epithelium in the rat following
exposure to NO2. Am J Pathol. 70:175–198.
1973.PubMed/NCBI
|
|
29
|
Lu H, Chang L, Li W, Jiang N, Peng Q, Cai
C and Liu J: Effects of hyperoxia on the dynamic expression of
Aquaporin5 in premature rats lung development. J Huazhong Univ Sci
Technolog Med Sci. 27:318–312. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
George UM, Ashna U, Kumar SS and Nandkumar
AM: Effect of tobacco extract on surfactant synthesis and its
reversal by retinoic acid-role of cell-cell interactions in vitro.
In Vitro Cell Dev Biol Anim. 49:260–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nomura J, Horie I, Seto M, Nagai K,
Hisatsune A, Miyata T and Isohama Y: All-trans retinoic acid
increases expression of aquaporin-5 and plasma membrane water
permeability via transactivation of Sp1 in mouse lung epithelial
cells. Biochem Biophys Res Commun. 351:1048–1053. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schittny JC: Development of the lung. Cell
Tissue Res. 367:427–444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuasa E and Saitoh H: In situ SUMOylation
and DeSUMOylation assays: fluorescent methods to visualize
SUMOylation and DeSUMOylation in permeabilized cells. Methods Mol
Biol. 1475:151–159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Desai TJ, Brownfield DG and Krasnow MA:
Alveolar progenitor and stem cells in lung development, renewal and
cancer. Nature. 507:190–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan F, Dong W, Lei X, Li Q, Kang L, Zhao S
and Zhang C: Attenuated SUMOylation of sirtuin 1 in premature
neonates with bronchopulmonary dysplasia. Mol Med Rep.
17:1283–1288. 2018.
|