Introduction
Bradyarrhythmia, including bradycardia, sick sinus
syndrome and atrioventricular block, is a group of diseases
characterized by cardiac pacing or conduction dysfunction. Although
drug therapy and electronic pacemakers have saved the lives of many
patients with bradyarrhythmia, long-term medication cannot solve
the underlying problem and there may be serious side effects,
including wire break, electromagnetic interference, pocket
infection, limited battery life and the lack of an independent
reaction. With the progress of genetic engineering and molecular
biology, biological pacemakers have emerged (1-3).
Studies investigating biological pacemakers have primarily focused
on the following two aspects: The restoration of cardiac pacing and
conduction function. Studies have been performed to introduce
specific genes into tissues with damaged autonomic rhythms or a
damaged conduction system (4-9),
in addition to transplanting pacemaker cells or pacemaker-like
cells to repair or replace the damaged tissues (10-12).
To avoid problems with aberration and ethics,
numerous studies have attempted to initiate automaticity in the
atria and ventricles by using gene- and cell-based hybrid
approaches (13-15). Stem cells have the potential for
proliferation and pluripotent differentiation, and thus stem
cell-derived pacemaker cells are the most common cell source.
Adipose tissue-derived stem cells (ADSCs) are promising candidate
cells for biological pacemakers, the advantages of which include
accessibility, high harvesting efficiency and myocardial
differentiation potential (16,17).
Insulin gene enhancer binding protein-1 (ISL-1), a
subtype of the LIM homologous domain transcription factors, marks
undifferentiated cardiac progenitor cells in the second heart field
and is important in promoting cardiac development and
differentiation (18,19). ISL-1-positive progenitor cells are
able to differentiate into a variety of cell types in the heart and
have persistent expression from the embryonic stage to adulthood in
the murine sinoatrial node (SAN), compared with the transient
expression of other components in the second heart field, which is
consistent with results observed in the human SAN (20). At 9.5 embryonic days,
ISL-1+ progenitor cells are primarily expressed in the
venous sinus, namely the primitive pacemaker region, in early
embryonic development (21). Sun
et al (22) reported that
ISL‑1 is a specific marker for a subset of pacemaker cells at the
developmental stages examined. ISL-1-expressing cells, organized as
a ring-shaped structure around the venous pole, exhibit pacemaker
functions and are able to rescue short stature homeobox 2-mediated
bradycardia in the adult zebrafish heart (23,24). Vedantham et al (25) used RNA sequencing to confirm that
ISL‑1 has an upstream regulatory role in the development of
bradycardia in the mouse SAN. Dorn et al (26) also confirmed that the
overexpression of ISL‑1 in embryonic stem cells or Xenopus
embryos can lead to the upregulation of SAN‑specific genes and the
downregulation of working myocardial genes. Liang et al
(27) reported that ISL-1 is a
necessary condition for development of the SAN, and influences the
survival, reproduction and function of pacemaker cells.
Therefore, numerous studies have demonstrated that
ISL-1 is located upstream of a variety of transcription factors and
ion channels, and regulates the expression of SAN‑specific genes.
The present study examined whether ISL-1 was able to successfully
direct the differentiation of ADSCs into pacemaker cells in order
to provide a novel breakthrough for building an extracorporeal
biological pacemaker by combining gene therapy with cell
therapy.
Materials and methods
Animals
Adult male Sprague-Dawley (SD) rats (n=6; age, 3-4
weeks; weight, 40-80 g) and newborn SD rats (n=60; age, 1-3 days;
weight, 5-10 g) were purchased from the Center for Disease Control
and Prevention of Hubei Province (Hubei, China). All animals were
housed in micro-isolators under specific pathogen‑free conditions
at 24°C in a 12 h light/dark cycle, with free access to food and
water. Animals received care in accordance with the guidelines for
animal care published by the United States National Institutes of
Health (Guide for the Care and Use of Laboratory Animals,
Department of Health and Human Services, NIH Publication no. 86-23,
revised 1985). The present study was approved by the Experimental
Animal Committee of Wuhan University (Hubei, China; no.
WDRM20171015).
Isolation and culture of ADSCs
All experimental procedures were conducted in
accordance with the Institutional Guidelines for the Care and Use
of Laboratory Animals at Wuhan University and conformed to the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals. The adult male SD rats were anesthetized with
an intraperitoneal injection of 2% pentobarbital sodium (40 mg/kg)
and then sacrificed by cervical dislocation. ADSCs were obtained
using a previously described method with modifications (28). Briefly, SD rat inguinal adipose
tissue was digested in 5 ml Dulbecco’s modified Eagle’s medium
(DMEM)/F-12 medium (cat. no. SH30023, HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) containing 0.1% (w/v) collagenase type I
(cat. no. C0130, Sigma; Merck KGaA) at 37°C for 45 min with gentle
agitation. Following filtering and centrifugation at 1,000 x g for
10 min at room temperature, the floating top layer was discarded.
The pellet was both washed and resuspended in DMEM/F-12
supplemented with 10% fetal bovine serum (FBS; cat. no. 16000‑044;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1%
penicillin/streptomycin (cat. no. 15070063, Invitrogen; Thermo
Fisher Scientific, Inc.). The cells were seeded in 6-well plates
(Corning, Inc., Corning, NY, USA) and incubated at 37°C with a 5%
CO2 atmosphere. The medium was replaced every 2 days.
When the cells reached 80‑90% confluence, they were passaged using
0.25% trypsin (cat. no. GNM25200; Genom, Hangzhou, China). Cells at
passages 3-5 were used for all subsequent experiments.
Construction of the human ISL‑1
lentiviral vector and ISL‑1 infection
The lentiviral vector expressing ISL-1
(Ubi-MCS-ISL-1-3FLAG-SV40-mCherry) was constructed by inserting the
human ISL-1 gene (positive clone sequence:
ATGGGAGACATGGGCGATCCACCAAAAAAAAAACGTCTGATTTCCCTGTGTGTTGGTTGCGGCAATCAAATTCACGACCAGTATATTCTGAGGGTTTCTCCGGATTTGGAGTGGCATGCAGCATGTTTGAAATGTGCGGAGTGTAATCAGTATTTGGACGAAAGCTGTACGTGCTTTGTTAGGGATGGGAAAACCTACTGTAAAAGAGATTATATCAGGTTGTACGGGATCAAATGCGCCAAGTGCAGCATAGGCTTCAGCAAGAACGACTTCGTGATGCGCGCCCGCTCTAAGGTGTACCACATCGAGTGTTTCCGCTGTGTAGCCTGCAGCCGACAGCTCATCCCGGGAGACGAATTCGCCCTGCGGGAGGATGGGCTTTTCTGCCGTGCAGACCACGATGTGGTGGAGAGAGCCAGCCTGGGAGCTGGAGACCCTCTCAGTCCCTTGCATCCAGCGCGGCCTCTGCAAATGGCAGCCGAACCCATCTCGGCTAGGCAGCCAGCTCTGCGGCCGCACGTCCACAAGCAGCCGGAGAAGACCACCCGAGTGCGGACTGTGCTCAACGAGAAGCAGCTGCACACCTTGCGGACCTGCTATGCCGCCAACCCTCGGCCAGATGCGCTCATGAAGGAGCAACTAGTGGAGATGACGGGCCTCAGTCCCAGAGTCATCCGAGTGTGGTTTCAAAACAAGCGGTGCAAGGACAAGAAACGCAGCATCATGATGAAGCAGCTCCAGCAGCAGCAACCCAACGACAAAACTAATATCCAGGGGATGACAGGAACTCCCATGGTGGCTGCTAGTCCGGAGAGACATGATGGTGGTTTACAGGCTAACCCAGTAGAGGTGCAAAGTTACCAGCCGCCCTGGAAAGTACTGAGTGACTTCGCCTTGCAAAGCGACATAGATCAGCCTGCTTTTCAGCAACTGGTCAATTTTTCAGAAGGAGGACCAGGCTCTAATTCTACTGGCAGTGAAGTAGCATCGATGTCCTCGCAGCTCCCAGATACACCCAACAGCATGGTAGCCAGTCCTATTGAGGCA)
into the Ubi-MCS-3FLAG-SV40-Cherry vector (GeneChem Co., Ltd.,
Shanghai, China) using BamHI (cat. no. FD0054) and
AgeI (cat. no. CON181) restriction sites, all obtained from
GeneChem Co., Ltd. The ADSCs at passages 3-5 were removed from the
culture dishes by digestion. A cell suspension was prepared and
then inoculated onto 6-well plates. For infection, the cells were
randomly divided into two groups: In group A, a lentivirus
overexpressing mCherry or ISL-1-mCherry in DMEM/F-12 was added to
the cells at different multiplicity of infection (MOI) values
(MOI=0, 20, 50, 80 and 100) when cell confluence reached 30‑50%; in
group B, a lentivirus overexpressing mCherry or ISL-1-mCherry, 5-10
μg/ml polybrene (cat. no. H9268; Sigma; Merck KGaA) and
enhanced infection solution (cat. no. REVG0002; GeneChem Co., Ltd.)
were mixed together and added to the culture medium of cells at
different MOI values (MOI=0, 20, 50, 80 and 100) when cell
confluence reached 30-50%. The culture medium was replaced
following culture at 37°C in 5% CO2 for 12-24 h, with
the transfection time determined by the state of the cells.
Fluorescent microscope analysis and flow
cytometric analysis
The expression of mCherry 48 h following infection
of the cells was observed and recorded under a fluorescent
microscope (BX51 system; Olympus Corporation, Tokyo, Japan). The
transfected cells were digested with 0.25% trypsin at 72-96 h
post-transfection and centrifuged at 1,000 x g for 5 min at room
temperature. The cell density was 1-2×106/ml.
Non-transfected cells served as a negative control. The percentage
of red fluorescent protein‑positive cells was detected by flow
cytometric analysis (BD Biosciences, Franklin Lakes, NJ, USA).
Isolation and culture of NRVMs and
co‑culture systems
Primary NRVMs were isolated from newborn SD rats in
1-3 days according to a previously described method with
modifications (29). Briefly, the
neonatal rat heart tissue was derived from newborn SD rats 1-3 days
following sacrifice via cervical dislocation and was digested with
0.125% trypsin (cat. no. C0201, Beyotime Institute of
Biotechnology, Shanghai, China) at 37°C for 10 min. The
precipitation were then repeatedly digested with mixed liquor
containing 0.125% trypsin and 0.08% collagenase II (cat. no. C6885;
Sigma; Merck KGaA) 5‑8 times at 37°C for 5 min. Following being
filtered and centrifuged at 1,000 x g for 10 min at room
temperature, the floating top layer was discarded. The pellets were
resuspended and seeded in 6-well plates with fresh DMEM/F-12
supplemented with 15% FBS and 1% penicillin/streptomycin. The
harvested NRVMs were purified by differential adhesion time to
isolate the cardiomyocytes from the fibroblasts and 0.1 mmol/l
bromodeoxyuridine (cat. no. B-5002, Sigma; Merck KGaA) to inhibit
the mitosis of fibroblasts. For co‑culture experiments, the ADSCs
and NRVMs were mixed and plated at a ratio of 1:10 onto the 6-well
plates (30,31). The complete culture medium was
replaced every 2 days, and the cell morphology and beating rate
were observed.
Reverse transcription‑quantitative
polymerase chain reaction (RT‑qPCR) analysis
Total cellular RNA was extracted from the co-culture
systems after 5-7 days using TRIzol reagent (cat. no. 15596-026,
Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was produced
using the PrimeScript™ RT reagent kit with gDNA Eraser (cat. no.
RR047A, Takara Bio, Inc., Otsu, Japan) in a 15-μl mixture.
All primers (Table I) for PCR
amplification were synthesized by Invitrogen; Thermo Fisher
Scientific, Inc. Reactions were carried out in a 20 μl
reaction volume (containing 4 μl cDNA, 4 μl forward
primer, 0.4 μl reverse primer, 10 μl SYBR Green
Master mix, 0.4 μl 50X ROX reference dye 2 and 4.8 μl
H2O). RT-qPCR analysis was performed with standard
SYBR® Premix Ex Taq™ (cat. no. RR420A, Takara Bio, Inc.)
on a StepOne™ Real-Time PCR (Thermo Fisher Scientific, Inc.)
instrument as follows: Pre‑denaturation at 95°C for 5 min,
denaturation at 95°C for 30 sec, annealing at 60°C for 20 sec, and
a final extension at 60°C, a total of 40 cycles. The dissolution
curve was from 60‑95°C and the temperature was raised by 1°C per 20
sec. Relative gene expression was calculated using the
2-ΔΔCq method (32)
following normalization to glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) expression. To ensure accuracy, all results were repeated
at least three times.
 | Table IPolymerase chain reaction primers
used in the present study. |
Table I
Polymerase chain reaction primers
used in the present study.
| Gene | | Primer sequence
(5′-3′) | Reaction
conditions | Product size
(bp) |
|---|
| H‑ISL‑1 | Forward |
5′‑GGCAATCAAATTCACGACCA3′ | 50°C 2 min, 95°C 10
min; 95°C | 124 |
| Reverse |
5′-CCCTAACAAAGCACGTACAGCT-3′ | 30 sec, 60°C 30
sec, 40 cycles | |
| R‑HCN4 | Forward |
5′‑CACTAAGGGCAACAAGGAGACC‑3′ | 50°C 2 min, 95°C 10
min; 95°C | 281 |
| Reverse |
5′‑GGTAGTTGAAGACGCCTGAGTTG‑3′ | 30 sec, 60°C 30
sec, 40 cycles | |
| R‑Cx43 | Forward |
5′‑GCTGGTGGTGTCCTTGGTGT‑3′ | 50°C 2 min, 95°C 10
min; 95°C | 213 |
| Reverse |
5′‑GGAGGAGACATAGGCGAGAGTG‑3′ | 30 sec, 60°C 30
sec, 40 cycles | |
| R‑Cx45 | Forward |
5′‑CCCAGGCTATAACATTGCTGTC‑3′ | 50°C 2 min, 95°C 10
min; 95°C | 198 |
| Reverse |
5′‑ATTGCTAGATCCAAGCGTTCC‑3′ | 30 sec, 60°C 30
sec, 40 cycles | |
| R‑GAPDH | Forward |
5′‑CGCTAACATCAAATGGGGTG‑3′ | 50°C 2 min, 95°C 10
min; 95°C | 201 |
| Reverse |
5′‑TTGCTGACAATCTTGAGGGAG‑3′ | 30 sec, 60°C 30
sec, 40 cycles | |
Western blot analysis
The cells were harvested using RIPA lysis buffer
(cat. no. AS1004, Aspen Biological, Wuhan, China). Then the total
protein concentrations in the sample were measured with a
bicinchoninic acid kit (cat. no. AS1086; Aspen Biological)
according to the manufacturer’s protocol. The protein samples (40
μg protein each well) were mixed with 5X SDS-PAGE buffer
(cat. no. AS1012; Aspen Biological) in a water bath at 95‑100°C for
5 min and transferred onto a polyvinylidene fluoride membranes. The
membranes were incubated with primary antibodies against
hyperpolarization-activated cyclic nucleotide-gated cation channel
(HCN)4 (cat. no. ab32675), connexin (Cx)43 (cat. no. ab11370; both
Abcam, Cambridge, MA, USA) and Cx45 (cat. no. F5108; Affinity
Biosciences. OH, USA) overnight at 4°C. Following at least three
washes in Tris‑buffered saline with Tween-20, the membranes were
incubated for 30 min at room temperature with corresponding
secondary antibodies: HRP-goat anti rat, (cat. no. AS1093) or
HRP-goat anti rabbit (cat. no. AS1107; Aspen Biological), raised in
the appropriate species. Specific bands of target proteins were
then visualized using an enhanced chemiluminescence detection kit
(cat. no. AS1059; Aspen Biological) according to the manufacturer’s
recommendations. The level of GAPDH was used to normalize the
signal intensities. The image collection and densitometry analyses
were performed with the Quantity One analysis software version 4.0
(AlphaEaseFC, Protein Simple, San Jose, CA, USA). The experiments
were performed at least three times to verify results.
Immunofluorescence
The cell cultures were washed three times with
phosphate-buffered saline (PBS; cat. no. GNB20012; Genom) and fixed
with 4% paraformaldehyde (cat. no. AS1018; Aspen Biological) for 15
min at room temperature. Following washing in PBS, the cell were
treated with 0.2% Triton (cat. no. T8787; Sigma; Merck KGaA) for 15
min. The cells then were incubated with the primary antibody
anti-cardiac troponin T (cTnT, cat. no. ab19615, Abcam) or HCN4
(cat. no. ab8295; Abcam) overnight at 4°C and incubated with goat
anti-mouse or goat anti-rat secondary antibody Alexa Fluor 647
(cat. no. A0473; Beyotime Institute of biotechnology) or Alexa
Fluor 647 (cat. no. ab150167; Abcam) for 50 min at room
temperature. Nuclei stained with 4′,6-diamidino-2-phenylindole was
used as a location control. The fluorescent images were obtained
with a Leica-LCS-SP8-STED confocal laser-scanning microscope (Leica
Microsystems GmbH, Solms, Germany). Images were captured randomly
of three visual fields in three samples to observe the positive
rates of cTnT and HCN4 detection.
Electrophysiological recordings
Single, funny current (If) measurements
were obtained by using a standard micro-electrode whole-cell,
patch-clamp technique with an Axon patch-clamp amplifier 700B
(Molecular Devices LLC, Sunnyvale, CA, USA). A digital 700AD/DA
converter and 6.0.4 pClamp (both Axon Instruments, Union City, CA,
USA) were used for recording and analyzing the data. Following
co-culture for 5-7 days, the cells were incubated with 180
μmol/l 2-aminoethoxydiphenyl borate (cat. no. D9754; Sigma;
Merck KGaA) for 15 min to block intercellular electrical
conduction. The cells were perfused with a normal Tyrode’s solution
containing (mmol/l): NaCl 135, KCl 5.4, CaCl2 1.8,
MgCl2 1.0, glucose 10, Bacl2 2.0 and HEPES
5.5 (pH 7.4) with NaOH. Pipette solution contained (mmol/L): KCl
120, Cacl2 5.0, MgCl2 5.0, HEPES 10 and EGTA
10 (pH 7.3) with KOH. The impedance of the fluid filled electrode
was 3‑4 MΩ. The Clampex program was applied to the sample. The
sampling frequency was 10 kHz, and the filtering rate was 5 kHz.
The holding potential was set at -30mV, and a family of voltage
steps from -140 to -40 mV for 1.5 sec with 20 mV increments was
applied to elicit If. CsCl (4 mmol/l) was administered
to detect the change in If.
Statistical analysis
All data are represented as the mean ± standard
error of the mean. The statistical significance of the differences
between two groups were analyzed using Student’s t-test and
comparisons among multiple groups were analyzed by one-way analysis
of variance with SPSS 19.0 software (IBM Corp., Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Culture of ADSCs and optimum MOI value
for transfection
When initially isolated, the ADSCs appeared
primarily round in shape. The ADSCs began to adhere at 4-5 h; a
small number of cells became adherent with an irregular shape at 24
h; the largest number of cells became adherent and spindle-shaped
with a relatively flat morphology, among which were cells with a
high oval individual visible refractive index, at 48 h. Following
culture for 5-7 days, the cell density reached ~90%. The sizes of
the cultured cells were marginally different, based on their
spindle-like morphology. The adjacent cells grew in a certain
direction. As the number of passages increased, the sub‑cultured
cells were increasingly purified, homogenous and spindle‑like. The
cells reached 80‑90% confluency in a fascicular-like or
whirlpool-like manner following culture for 2‑3 days (Fig. 1A). The rate of proliferation was
significantly increased following propagation in culture.
Sub-cultured cells at passage 3-5 were used in the present
study.
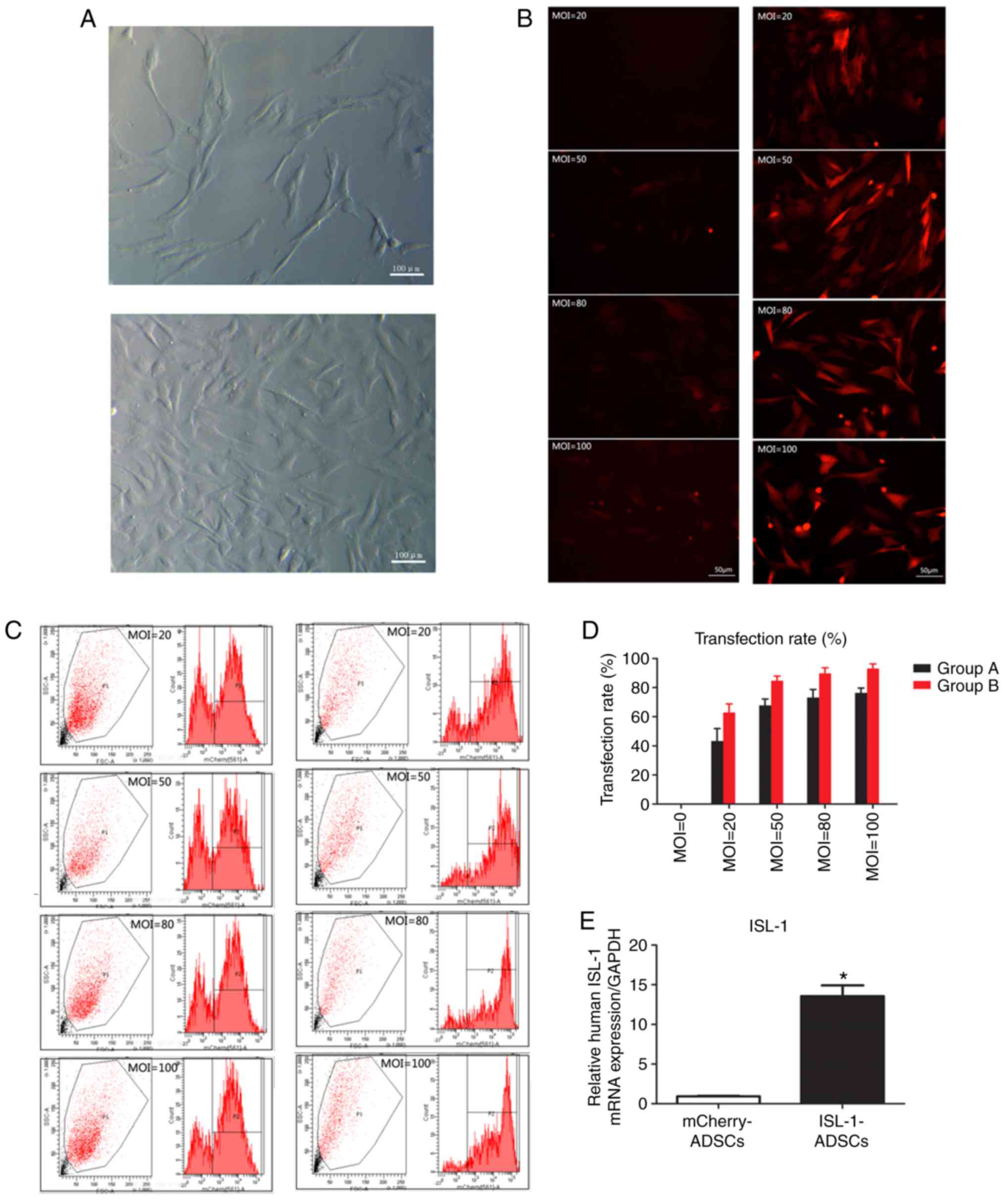 | Figure 1Transfection rate and the expression
of ISL‑1. (A) ADSCs observed following culture for 48 h and 5‑7
days under a light microscope (magnification, ×100). (B)
ISL‑1‑ADSCs observed under a fluorescence microscope at different
MOI values (magnification, ×200). (C) Numbers of positive cells at
MOI values detected by flow cytometric analysis. (D) Transfection
rate at different MOI values. In group A, cells were transfected
with lentivirus overexpressing mCherry or ISL-1-mCherry. In group
B, cells were transfected with a mix of lentivirus overexpressing
mCherry or ISL-1-mCherry, 5-10 μg/ml polybrene and enhanced
infection solution. (E) mRNA expression of human ISL-1 in the
transfected groups. *P<0.05, vs. mCherry-ADSCs.
ADSCs, adipose tissue-derived stem cells; ISL-1, insulin gene
enhancer binding protein 1; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; MOI, multiplicity of infection. |
At 2 days post‑transfection, the red fluorescence of
the infected ADSCs was detected by fluorescence microscopy in at
least five randomly selected fields. The transfection efficiency
was >80% at an MOI value of ≥50. At an MOI value of 80, certain
fluorescent cells appeared as roundish cells with vacuoles in the
cytoplasm, and gradually began to float. This phenomenon was more
marked at an MOI of 100. Compared with group A, group B exhibited
higher fluorescence intensity and clearer cell morphological
features (Fig. 1B). The flow
cytometric analysis demonstrated that the transfection efficiency
of the ADSCs in group B reached 84.7±2.3% at an MOI value of 50
(Fig. 1C). With the increase in
MOI, the fluorescence intensity and cell toxicity increased
gradually (Fig. 1D). Through
fluorescence microscopy and flow cytometric analyses, it was
ascertained that the optimum MOI value for transfection was 50
using the transfection protocol of group B, and this was adopted
for the following experiments. The mRNA expression of ISL-1 in the
ISL‑1‑ADSCs group was significantly higher compared with that in
the control group at day 5 post-transfection (Fig. 1E). The results demonstrated that
ISL-1 achieved steady overexpression in the ADSCs.
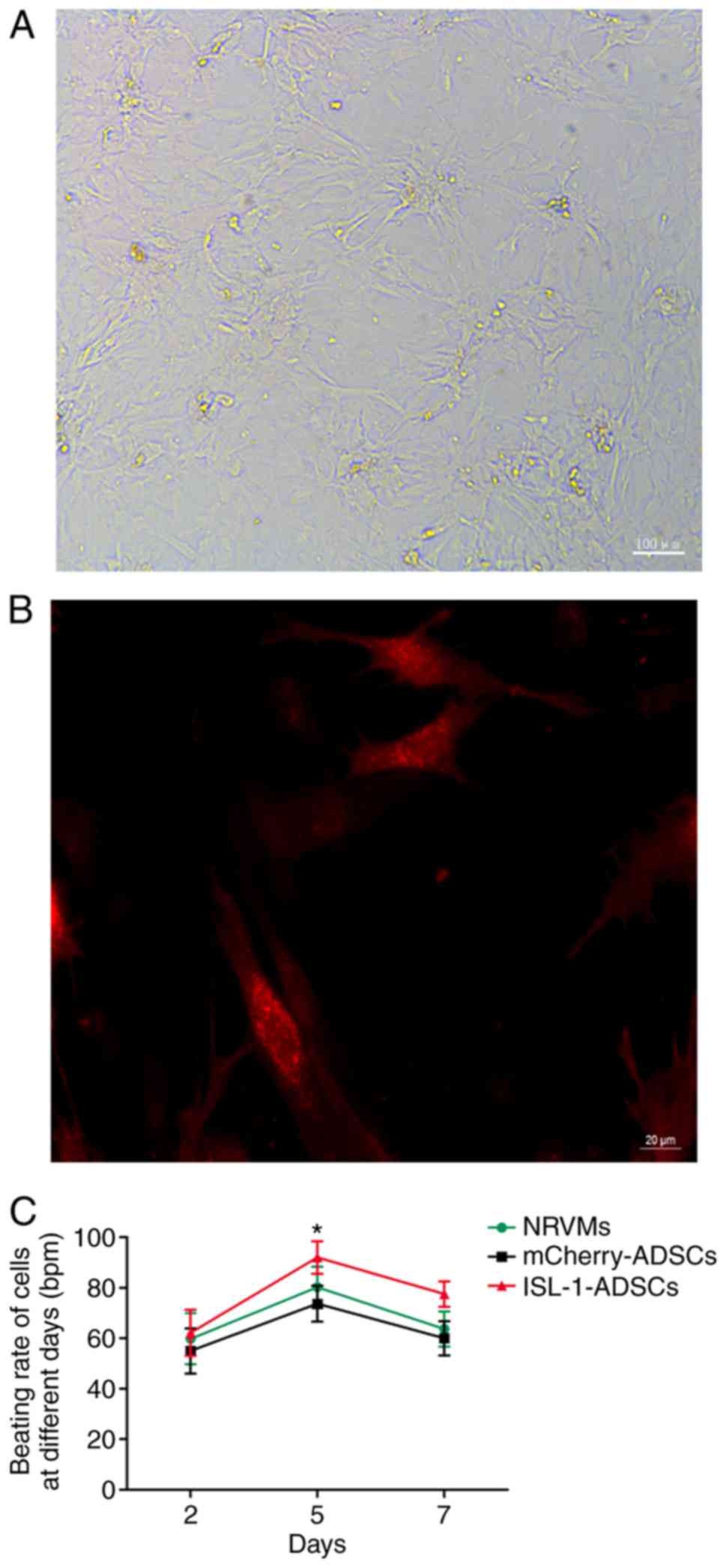
Cell morphology and beating rate
The proliferation rate decreased and the morphology
diversified following transfection, with the morphology dominated
by the long spindle shape, in the ISL-1-ADSCs group compared with
the mCherry-ADSCs group. Following co-culture for 2 days, the ADSCs
and NRVMs had begun to form connections and cell clusters; the
majority of cells were connected in a network and exhibited
synchronous pulsation within a cluster following co-culture for 5
days; all cells had formed mutual a connection with a low but
stable pulsation frequency following co-culture for 7 days
(Fig. 2A). The morphology of the
ADSCs was more irregular following co-culture with NRVMs, as
determined by fluorescence microscopy, with three principal forms:
Fusiform, triangular and quadrilateral (Fig. 2B).
The cells of the ISL-1-ADSCs group and mCherry-ADSCs
group were not observed to be beating cells. Following
co‑culture with NRVMs, red‑fluorescing ISL‑1‑transfected ADSCs were
clearly observed to be spontaneously beating; however,
red‑fluorescing mCherry‑transfected ADSCs were not spontaneously
beating, although they exhibited synchronous pulsation driven by
the surrounding cardiomyocytes, as determined by fluorescence
microscopy. Furthermore, ADSCs transfected with ISL-1 had a higher
pulsation frequency compared with the ADSCs transfected with
mCherry and the NRVMs without ADSCs. The beating rate of the cells
reached ~92±6.42 bpm (n=15) in the ISL-1-ADSCs+NRVM group at day 5.
By comparison, the beating rate of the cells in the
mCherry-ADSCs+NRVM group was ~73.67±7.09 bpm (n=15) at day 5; the
beating rate of the NRVMs without ADSCs was ~80.17±8.13 bpm (n=15)
at day 5 (Fig. 2C).
Expression of associated proteins and
genes assessed by western blotting and RT‑qPCR
To evaluate the role of ISL-1 in the differentiation
of the SAN, the expression status of a number of genes that are
known to be important for SAN formation and function was
investigated. The genes detected in the present study included:
HCN4, a crucial marker of pacemaker cells; Cx45, which prevents the
formation of areas of conductivity inside the SAN; and Cx43, a
marker of the working myocardium. As presented in Fig. 3A-C, the mRNA and protein
expression levels of HCN4 and Cx45 were significantly increased in
the ADSCs transfected with ISL-1 lentivirus (ISL-1-ADSCs group)
compared with the empty lentivirus group (mCherry-ADSCs group), and
the difference was statistically increased when co-cultured with
NRVMs (P<0.05). By contrast, the mRNA and protein expression
levels of CX43 were significantly increased in the group
co‑cultured with NRVMs, and downregulated in the ISL-1-ADSCs+NRVMs
group compared with the mCherry-ADSCs+NRVMs group (P<0.05).
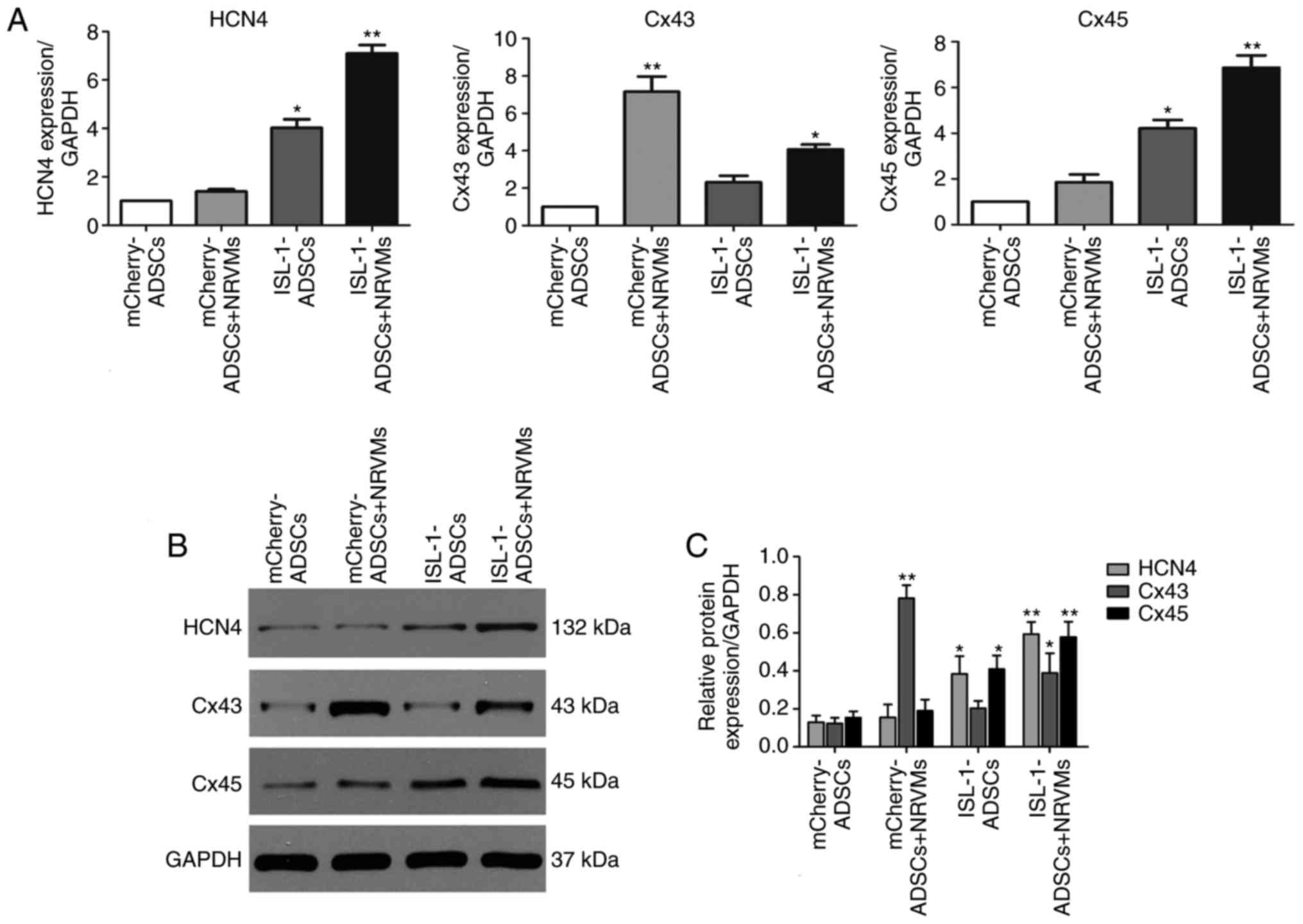 | Figure 3Expression of related genes by
western blot and RT-qPCR analyses following co-culture for 5-7
days. (A) Gene expression levels of HCN4, Cx45 and Cx43 were
examined by RT-qPCR analysis. (B) Protein expression of HCN4, Cx45
and Cx43 was examined using western blotting. (C) Quantitative
assessment of protein levels of HCN4, Cx45 and Cx43 by integrated
optical density analyses. Similar results were obtained in three
independent experiments. GAPDH was used as the protein control.
*P<0.05 vs. other groups, **P<0.01, vs.
other groups. HCN4, hyperpolarization-activated cyclic
nucleotide-gated cation channel 4; Cx45, connexin 45; Cx43,
connexin 43; GAPDH, glyceraldehyde 3-phosphate dehydrogenase;
ADSCs, adipose tissue-derived stem cells; NRVMs, neonatal rat
ventricular cardiomyocytes; ISL-1, insulin gene enhancer binding
protein 1; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
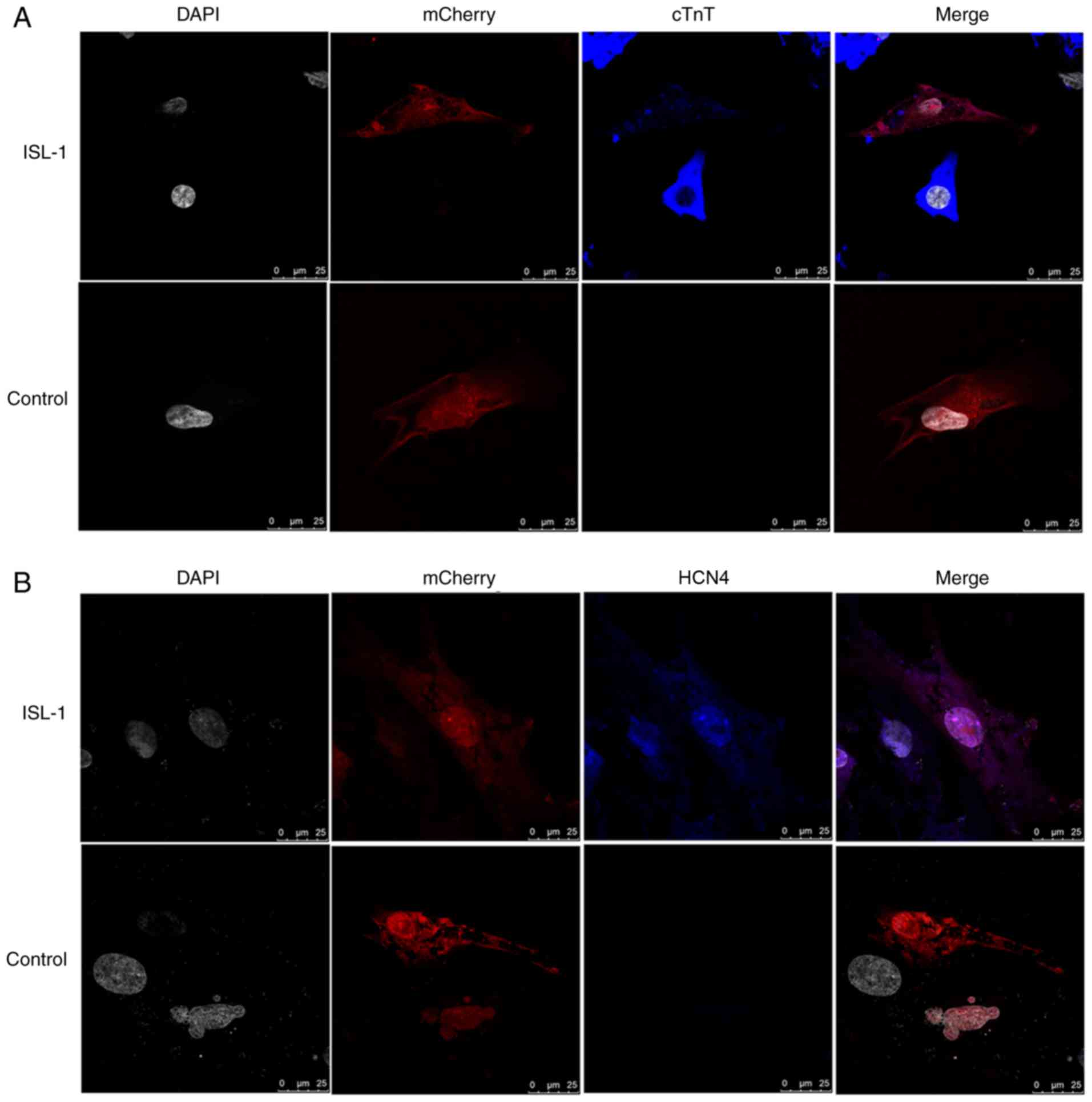
Localization of associated gene
expression in co‑culture systems, as assessed by
immunofluorescence
As demonstrated in Fig. 4, cTnT, a myocardium‑specific
marker, and HCN4, a marker of SAN function, were detected by
immunofluorescence following co-culture for 5-7 days. The ADSCs
were randomly distributed in the culture and a large number existed
in a plane below the NRVMs, with some interspersed between the
NRVMs. The immunofluorescence staining results revealed that the
NRVMs and cardiac muscle-like cells were all positive for the
expression of cTnT, and that the ADSCs transfected with ISL-1 had
abundant positive staining for HCN4 protein. However, in the
mCherry-transfected group, cTnT and HCN4 protein expression was
barely detectable. The ADSCs transfected with ISL-1 also exhibited
a high percentage of cells positive for the specific proteins,
compared with negative control cells.
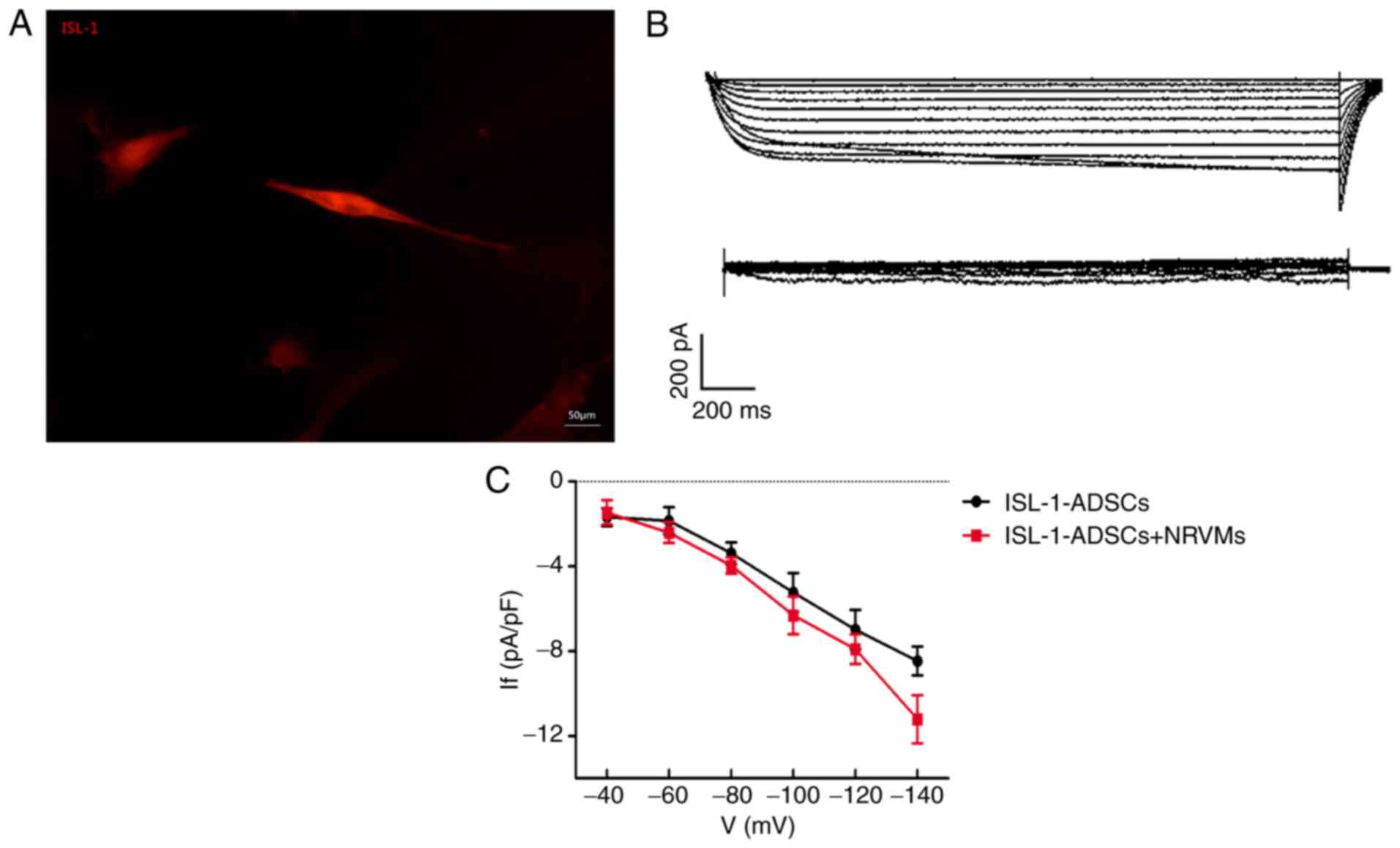
Electrophysiological recording in
pacemaker‑like cells
The red fluorescent cells were used for detecting
intracellular electrical activity via the patch clamp method
(Fig. 5A). The If, a
key contributor to spontaneous phase four depolarization, was
significantly detected in the transfected‑ISL‑1‑ADSCs and
ISL-1-ADSCs+NRVMs, although not in the control group or NRVMs.
Following co-culture with NRVMs, transfected-ISL-1-ADSCs recorded a
greater inward current. The ISL-1-ADSCs had an inward current
(~1/20 cells) that was activated by the hyperpolarization intervals
from -40 mV. The current had clear dependent characteristics of
time and voltage and was sensitive to CsCl. The current was
completely inhibited following the addition of CsCl blocker (4
mmol/l) to the extracellular fluid (Fig. 5B). When CsCl was eluted from the
extracellular fluid, the inward current resumed rapidly. With the
increase in density-voltage and time, the inward current gradually
increased (Fig. 5C). Therefore,
the current was markedly voltage- and time-dependent.
Discussion
It is well known that ISL-1 is essential to the
proper formation and development of the heart. ISL1-positive cells,
isolated from embryonic stem cells (ESCs), induced pluripotent stem
cells, bone mesenchymal stem cells and embryonic or postnatal heart
tissues, have the potential to give rise to diverse cell types in
the cardiovascular system, including cardiomyocytes, smooth muscle
cells, endothelial cells and pacemaker cells (33-35). ISL-1-overexpressing cardiomyocytes
exhibit higher beating frequencies in mouse ESC-derived regions and
Xenopus hearts, which are associated with the upregulation
of nodal‑specific genes and the downregulation of genes associated
with the working myocardium (26). However, ADSCs have high harvesting
efficiency and pluripotency potential and, for these reasons, offer
promise for biological pacing therapy in the future. At present, to
the best of our knowledge, no investigations of biological
pacemakers have provided direct evidence as to whether or not ADSCs
may be induced to become pacemaker cells via the single ISL-1
transcription factor. Therefore, it is particularly important to
examine the role of the single ISL-1 transcription factor in
building a biological pacemaker. In the present study, ISL-1 was
overexpressed via lentiviral vectors to identify whether a single
transcription factor may be sufficient to induce the
differentiation of ADSCs into pacemaker-like cells. To identify
whether the cells transfected with ISL-1 had the specific
phenotypes and functions of pacemaker‑like cells, the outcomes were
evaluated through morphological detection, RT-qPCR analysis,
western blotting, immunofluorescence and electrophysiological
monitoring.
There are a number of principal methods for inducing
cardiomyocyte differentiation in stem cells, including biochemical
induction, cardiac microenvironment induction and genetic
modification (26). Co-culturing
with NRVMs has been demonstrated to generate a model mimicking the
physiological microenvironment of the heart following genetic
modification by cellular factors, chemical substances, electrical
activity and mechanical stretching (36). Studies have demonstrated that
direct contact, rather than indirect contact, is more essential for
adult stem cells to successfully differentiate into myocardial
cells (30,31). Therefore, the transfected ADSCs
were directly co-cultured with NRVMs at a 1:10 ratio, in order to
induce differentiation of the stem cells in the present study. It
was found that the morphology of the ADSCs altered following
transfection with ISL-1 and was more irregular following co-culture
with NRVMs. The expression of ISL-1 markedly increased the
percentage of spontaneously beating ADSCs.
The above observation was accompanied by the altered
expression of a number of genes essential for SAN formation. HCN
channels are expressed throughout cardiac development (37). HCN4 exhibits the highest
expression among the four HCN genes (HCN1-4) and is maintained
until adulthood in the SAN (38).
Studies on numerous species have demonstrated that HCN4 is key in
the generation of the SAN, although a low level of HCN4 has be
observed in human and mouse atrial and ventricular myocytes
(39-42). Therefore, HCN4 may be considered a
specific marker of the SAN region. In our previous study, it was
demonstrated that the overexpression of ISL-1 in ADSCs was able to
upregulate the expression level of HCN4 in vitro, and its
level was significantly increased by co-culture induction. Four
principal connexins have been identified to be expressed in
different cardiac tissues, including Cx40, Cx30.2, Cx43 and Cx45
(43). To prevent the SAN from
over-suppressing the more hyperpolarized non-pacing atrial muscle
surrounding the SAN, electrical coupling should be weak in the
center of the SAN. Furthermore, to ensure that the SAN is able to
drive the atrial muscle, electrical coupling must be strong in the
periphery of the SAN. Consistent with this, Cx43 is the dominant
connexin in working myocytes and Cx45 is primarily expressed in the
center of the SAN, which directly connects nodal cells with atrial
myocytes; cardiomyoblast cells express a combination of Cx43 and
Cx45 in the periphery of the SAN (44,45). In the present study, Cx45 was also
upregulated in ISL-1-transfected ADSCs, and the differences became
more marked when the cells were co-cultured with NRVMs. Cx43 was
downregulated in the ISL-1-transfected ADSCs but was upregulated
following co-culture, possibly due to the addition of NRVMs.
Therefore, it was suggested that ISL-1 may promote pacemaker
differentiation by enhancing the expression of HCN4 and Cx45 in
vitro and reducing the expression of Cx43 in the working
myocardium.
Cardiac pacemaker activity originates from the SAN.
The automaticity of the SAN is due to slow diastolic
depolarization. There are two clocks that act interdependently and
synergistically to initiate the heartbeat, modulated by autonomic
neurons, including the voltage clock generated by HCN channels,
referred to as If, the funny current; and the calcium
clock generated by rhythmic Ca2+ release from the
sarcoplasmic reticulum (46-48). In the present study, the typical
If current demonstrated that the overexpression of ISL-1
in ADSCs resulted in normal electrophysiologically functional
cells, which were enriched in the nodal subtype at the expense of
the ventricular lineage. Following initial heart tube formation,
the heart tube elongates via the addition of ISL-1- and NK2
homeobox 5 (Nkx2.5)-positive progenitor cells, which maintain their
expression until cardiac differentiation (21). The regulatory association between
ISL-1 and Nkx2.5 may also exhibit opposite effects (26) Nkx2.5 is able to induce
differentiation in the working myocardium, resulting in the
downregulation of HCN4 and Tbx3, and the ectopic expression of Cx40
and natriuretic peptide A in the SAN region (49). By contrast, ISL-1 is able to
upregulate nodal‑specific genes and downregulate transcripts
associated with the working myocardium to induce differentiation of
the SAN. The results of the present study demonstrated that the
morphology, biochemical characteristics and electrophysiological
characteristics of ADSCs altered considerably following co-culture
with NRVMs. This indicated that the cardiac microenvironment is
important in inducing stem cells to differentiate into
pacemaker-like cells. Furthermore, it suggested that the in
vivo environment may improve the quality of pacemaker-like
cells.
In conclusion, the present results suggested that
ISL-1-transfected ADSCs had been successfully transformed into
spontaneously beating cells that exhibited behaviors similar to
those of pacemaker cells. The following lines of evidence support
this result: i) The finding of transfected cells with morphological
features distinctive to pacemaker-like cells; ii) characteristic
mRNA and protein alterations, including the upregulation of HCN4
and Cx45, and the suppression of Cx43; iii) the localization of the
expression of HCN4 and cTnT, as demonstrated by immunofluorescence;
and iv) If, the current specific to the SAN, was
recorded in ISL-1-transfected cells. However, the present study had
a number of limitations. No further animal experiments were
performed owing to the low efficiency of lentivirus transfection.
In addition, the Na+ ion currents, K+ ion
currents, and action potential of the cells was not examined due to
technical and instrumental limitations. The present findings are
not intended to be used as a direct precursor to the long-term
application of biological pacing tools, as further investigation in
in vivo animal models is required to evaluate safety and
validity prior to application in patients with sinus dysfunction.
However, the results of the present study may provide insights into
the progression of pacemaker cell development, as these cells are
reflected in the ADSC system, and this may offer a new perspective
for the examination of a novel approach in the generation of
biological pacemakers.
Funding
The present study was supported by the Fundamental
Research Funds for the Central Universities of China (grant no.
2042015kf0229).
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors’ contributions
JZ made substantial contributions to the conception
and design of the study, performed the experiments and wrote the
paper. MY, AKY and YTC assisted in the development of experiments
and analyzed the data. XW, YHT, QYZ and TW participated in research
design and coordinated the study. CXH revised the manuscript and
gave final approval of the version to be published. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was provided by the Ethics Committee
of Wuhan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors are grateful to Wuhan University School
of Basic Medical Science and the Medical Research Center for
Structural Biology for their assistance in conducting the
experiments on the fluorescent images.
References
|
1
|
Cho HC and Marbán E: Biological therapies
for cardiac arrhythmias: Can genes and cells replace drugs and
devices. Circ Res. 106:674–685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boink GJ, Christoffels VM, Robinson RB and
Tan HL: The past, present, and future of pacemaker therapies.
Trends Cardiovasc Med. 25:661–673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cingolani E, Goldhaber JI and Marban E:
Next-generation pacemakers: From small devices to biological
pacemakers. Nat Rev Cardiol. 15:139–150. 2018. View Article : Google Scholar :
|
|
4
|
Edelberg JM, Aird WC and Rosenberg RD:
Enhancement of murine cardiac chronotropy by the molecular transfer
of the human beta2 adrenergic receptor cDNA. J Clin Invest.
101:337–343. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boink GJ, Nearing BD, Shlapakova IN, Duan
L, Kryukova Y, Bobkov Y, Tan HL, Cohen IS, Danilo P Jr, Robinson
RB, et al: Ca(2+)-stimulated adenylyl cyclase AC1 generates
efficient biological pacing as single gene therapy and in
combination with HCN2. Circulation. 126:528–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miake J, Marbán E and Nuss HB: Biological
pacemaker created by gene transfer. Nature. 419:132–133. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qu J, Plotnikov AN, Danilo P Jr,
Shlapakova I, Cohen IS, Robinson RB and Rosen MR: Expression and
function of a biological pacemaker in canine heart. Circulation.
107:1106–1109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bucchi A, Plotnikov AN, Shlapakova I,
Danilo P Jr, Kryukova Y, Qu J, Lu Z, Liu H, Pan Z, Potapova I, et
al: Wild-type and mutant HCN channels in a tandem
biological-electronic cardiac pacemaker. Circulation. 114:992–999.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boink GJ, Duan L, Nearing BD, Shlapakova
IN, Sosunov EA, Anyukhovsky EP, Bobkov E, Kryukova Y, Ozgen N,
Danilo P Jr, et al: HCN2/SkM1 gene transfer into canine left bundle
branch induces stable, autonomically responsive biological pacing
at physiological heart rates. J Am Coll Cardiol. 61:1192–1201.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruhparwar A, Tebbenjohanns J, Niehaus M,
Mengel M, Irtel T, Kofidis T, Pichlmaier AM and Haverich A:
Transplanted fetal cardiomyocytes as cardiac pacemaker. Eur J
Cardiothorac Surg. 21:853–857. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kehat I, Khimovich L, Caspi O, Gepstein A,
Shofti R, Arbel G, Huber I, Satin J, Itskovitz-Eldor J and Gepstein
L: Electromechanical integration of cardiomyocytes derived from
human embryonic stem cells. Nat Biotechnol. 22:1282–1289. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xue T, Cho HC, Akar FG, Tsang SY, Jones
SP, Marbán E, Tomaselli GF and Li RA: Functional integration of
electrically active cardiac derivatives from genetically engineered
human embryonic stem cells with quiescent recipient ventricular
cardiomyocytes: Insights into the development of cell-based
pacemakers. Circulation. 111:11–20. 2005. View Article : Google Scholar
|
|
13
|
Plotnikov AN, Shlapakova I, Szabolcs MJ,
Danilo P Jr, Lorell BH, Potapova IA, Lu Z, Rosen AB, Mathias RT,
Brink PR, et al: Xenografted adult human mesenchymal stem cells
provide a platform for sustained biological pacemaker function in
canine heart. Circulation. 116:706–713. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shlapakova IN, Nearing BD, Lau DH, Boink
GJ, Danilo P Jr, Kryukova Y, Robinson RB, Cohen IS, Rosen MR and
Verrier RL: Biological pacemakers in canines exhibit positive
chronotropic response to emotional arousal. Heart Rhythm.
7:1835–1840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chauveau S, Anyukhovsky EP, Ben-Ari M,
Naor S, Jiang YP, Danilo P Jr, Rahim T, Burke S, Qiu X, Potapova
IA, et al: Induced pluripotent stem cell-derived cardiomyocytes
provide in vivo biological pacemaker function. Circ Arrhythm
Electrophysiol. 10:e0045082017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dmitrieva RI, Minullina IR, Bilibina AA,
Tarasova OV, Anisimov SV and Zaritskey AY: Bone marrow- and
subcutaneous adipose tissue-derived mesenchymal stem cells:
Differences and similarities. Cell Cycle. 11:377–383. 2012.
View Article : Google Scholar
|
|
17
|
Joo HJ, Kim JH and Hong SJ: Adipose
tissue-derived stem cells for myocardial regeneration. Korean Circ
J. 47:151–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brade T, Gessert S, Kuhl M and Pandur P:
The amphibian second heart field: Xenopus islet-1 is required for
cardiovascular development. Dev Biol. 311:297–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bu L, Jiang X, Martin-Puig S, Caron L, Zhu
S, Shao Y, Roberts DJ, Huang PL, Domian IJ and Chien KR: Human ISL1
heart progenitors generate diverse multipotent cardiovascular cell
lineages. Nature. 460:113–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weinberger F, Mehrkens D, Friedrich FW,
Stubbendorff M, Hua X, Müller JC, Schrepfer S, Evans SM, Carrier L
and Eschenhagen T: Localization of Islet-1-positive cells in the
healthy and infarcted adult murine heart. Circ Res. 110:1303–1310.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mommersteeg MT, Dominguez JN, Wiese C,
Norden J, de Gier-de VC, Burch JB, Kispert A, Brown NA, Moorman AF
and Christoffels VM: The sinus venosus progenitors separate and
diversify from the first and second heart fields early in
development. Cardiovasc Res. 87:92–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun Y, Liang X, Najafi N, Cass M, Lin L,
Cai CL, Chen J and Evans SM: Islet 1 is expressed in distinct
cardiovascular lineages, including pacemaker and coronary vascular
cells. Dev Biol. 304:286–296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tessadori F, van Weerd JH, Burkhard SB,
Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM and
Bakkers J: Identification and functional characterization of
cardiac pacemaker cells in zebrafish. PLoS One. 7:e476442012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hoffmann S, Berger IM, Glaser A, Bacon C,
Li L, Gretz N, Steinbeisser H, Rottbauer W, Just S and Rappold G:
Islet1 is a direct transcriptional target of the homeodomain
transcription factor Shox2 and rescues the Shox2-mediated
bradycardia. Basic Res Cardiol. 108:3392013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vedantham V, Galang G, Evangelista M, Deo
RC and Srivastava D: RNA sequencing of mouse sinoatrial node
reveals an upstream regulatory role for Islet-1 in cardiac
pacemaker cells. Circ Res. 116:797–803. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dorn T, Goedel A, Lam JT, Haas J, Tian Q,
Herrmann F, Bundschu K, Dobreva G, Schiemann M, Dirschinger R, et
al: Direct nkx2-5 transcriptional repression of isl1 controls
cardio-myocyte subtype identity. Stem Cells. 33:1113–1129. 2015.
View Article : Google Scholar
|
|
27
|
Liang X, Zhang Q, Cattaneo P, Zhuang S,
Gong X, Spann NJ, Jiang C, Cao X, Zhao X, Zhang X, et al:
Transcription factor ISL1 is essential for pacemaker development
and function. J Clin Invest. 125:3256–3268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bunnell BA, Flaat M, Gagliardi C, Patel B
and Ripoll C: Adipose-derived stem cells: Isolation, expansion and
differentiation. Methods. 45:115–120. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Golden HB, Gollapudi D, Gerilechaogetu F,
Li J, Cristales RJ, Peng X and Dostal DE: Isolation of cardiac
myocytes and fibroblasts from neonatal rat pups. Methods Mol Biol.
843:205–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu Y, Liu T, Song K, Ning R, Ma X and Cui
Z: ADSCs differentiated into cardiomyocytes in cardiac
microenvironment. Mol Cell Biochem. 324:117–129. 2009. View Article : Google Scholar
|
|
31
|
Choi YS, Dusting GJ, Stubbs S,
Arunothayaraj S, Han XL, Collas P, Morrison WA and Dilley RJ:
Differentiation of human adipose-derived stem cells into beating
cardiomyocytes. J Cell Mol Med. 14:878–889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Moretti A, Caron L, Nakano A, Lam JT,
Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et
al: Multipotent embryonic isl1+ progenitor cells lead to cardiac,
smooth muscle, and endothelial cell diversification. Cell.
127:1151–1165. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moretti A, Bellin M, Jung CB, Thies TM,
Takashima Y, Bernshausen A, Schiemann M, Fischer S, Moosmang S,
Smith AG, et al: Mouse and human induced pluripotent stem cells as
a source for multipotent Isl1+ cardiovascular progenitors. FASEB J.
24:700–711. 2010. View Article : Google Scholar
|
|
35
|
Yi Q, Xu H, Yang K, Wang Y, Tan B, Tian J
and Zhu J: Islet-1 induces the differentiation of mesenchymal stem
cells into cardiomyocyte-like cells through the regulation of Gcn5
and DNMT-1. Mol Med Rep. 15:2511–2520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen H, Wang Y, Zhang Z, Yang J, Hu S and
Shen Z: Mesenchymal stem cells for cardiac regenerative therapy:
Optimization of cell differentiation strategy. Stem Cells Int.
2015:5247562015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schuleri KH, Boyle AJ and Hare JM:
Mesenchymal stem cells for cardiac regenerative therapy. Handb Exp
Pharmacol. 195–218. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biel M, Schneider A and Wahl C: Cardiac
HCN channels: Structure, function, and modulation. Trends
Cardiovasc Med. 12:206–212. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi W, Wymore R, Yu H, Wu J, Wymore RT,
Pan Z, Robinson RB, Dixon JE, McKinnon D and Cohen IS: Distribution
and prevalence of hyperpolarization-activated cation channel (HCN)
mRNA expression in cardiac tissues. Circ Res. 85:e1-e61999.
View Article : Google Scholar
|
|
40
|
Liu J, Dobrzynski H, Yanni J, Boyett MR
and Lei M: Organisation of the mouse sinoatrial node: Structure and
expression of HCN channels. Cardiovasc Res. 73:729–738. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zicha S, Fernández-Velasco M, Lonardo G,
L’Heureux N and Nattel S: Sinus node dysfunction and
hyperpolarization-activated (HCN) channel subunit remodeling in a
canine heart failure model. Cardiovasc Res. 66:472–481. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li N, Csepe TA, Hansen BJ, Dobrzynski H,
Higgins RS, Kilic A, Mohler PJ, Janssen PM, Rosen MR, Biesiadecki
BJ and Fedorov VV: Molecular mapping of sinoatrial node HCN channel
expression in the human heart. Circ Arrhythm Electrophysiol.
8:1219–1227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gros DB and Jongsma HJ: Connexins in
mammalian heart function. Bioessays. 18:719–730. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Martinez AD, Hayrapetyan V, Moreno AP and
Beyer EC: Connexin43 and connexin45 form heteromeric gap junction
channels in which individual components determine permeability and
regulation. Circ Res. 90:1100–1107. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Boyett MR, Inada S, Yoo S, Li J, Liu J,
Tellez J, Greener ID, Honjo H, Billeter R, Lei M, et al: Connexins
in the sinoatrial and atrioventricular nodes. Adv Cardiol.
42:175–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lakatta EG, Vinogradova T, Lyashkov A,
Sirenko S, Zhu W, Ruknudin A and Maltsev VA: The integration of
spontaneous intracellular Ca2+ cycling and surface membrane ion
channel activation entrains normal automaticity in cells of the
heart’s pacemaker. Ann NY Acad Sci. 1080:178–206. 2006. View Article : Google Scholar
|
|
47
|
Maltsev VA and Lakatta EG: Dynamic
interactions of an intracellular Ca2+ clock and membrane ion
channel clock underlie robust initiation and regulation of cardiac
pacemaker function. Cardiovasc Res. 77:274–284. 2008. View Article : Google Scholar
|
|
48
|
Lakatta EG, Maltsev VA and Vinogradova TM:
A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane
voltage clocks controls the timekeeping mechanism of the heart’s
pacemaker. Circ Res. 106:659–673. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Espinoza-Lewis RA, Liu H, Sun C, Chen C,
Jiao K and Chen Y: Ectopic expression of Nkx2.5 suppresses the
formation of the sinoatrial node in mice. Dev Biol. 356:359–369.
2011. View Article : Google Scholar : PubMed/NCBI
|