Introduction
Lung cancer is the second most common cancer, which
also accounts for the most common cause of cancer-associated
mortalities in males and the second most common cause in females.
Approximately 14% of new cancer cases were lung cancer from
2005–2014 in USA (1). There are
two main types of lung cancer, small-cell lung cancer (SCLC) and
non-SCLC (NSCLC), and 80-85% lung cancer cases were classified as
NSCLC globally in 2008, which include a number of subtypes,
including adenocarcinoma (~40%), squamous cell carcinoma (25–30%),
undifferentiated carcinoma or large cell carcinoma (10–15%) and a
few other subtypes, such as sarcomatoid carcinoma (2). Patients with NSCLC are frequently
diagnosed at advanced stages of disease (1). The mean 5-year survival rate for
patients with NSCLC in 2015 is only 18% globally (1), due to its known heterogeneity,
including the complex molecular and tumor microenvironmental
factors, and the limited therapy options (2). Recently, studies regarding NSCLC
therapeutics have been conducted, but the treatment methods remain
limited due to the poor prognosis (3–6).
Therefore, understanding the molecular mechanism responsible for
the pathogenesis of NSCLC remains urgently required.
MicroRNAs (miRNAs) are a large group of small
non-coding RNAs that generally negatively regulate gene expression
at the post-transcriptional level by binding to the 3′untranslated
region (3′UTR) of the transcripts (7). They serve fundamental effects in the
regulation of different intracellular processes, including cell
proliferation, migration, invasion, metastasis, cell cycle and
apoptosis (8–10). Numerous studies revealed that
dysregulation of miRNAs is associated with the initiation and
development of cancer, and miRNAs act as oncogenes or suppressor
genes (11–15).
miR-505 is located in chromosome X of humans, and a
number of studies reported its functional roles in different
diseases, including osteosarcoma, cervical cancer and hepatoma cell
cancer (16–19). For example Yang et al
(17), demonstrated that the
level of miR-505 in plasma was significantly elevated in
hypertensive patients, and miR-505 overexpression impaired the
migration and tube formation of endothelial cells by targeting
fibroblast growth factor 18. Escate et al (18), reported that miR-505 was
significantly downregulated in monocyte-derived macrophages of
patients with familiar hypercholesterolemia (FH), and regulated the
chemokine receptor C-C motif chemokine receptor 3 (CCR3), CCR4 and
C-X-C motif chemokine receptor 1 expression via the transcription
factor runt related transcription factor 1, indicating that miR-505
is involved in the chronic inflammatory condition in FH innate
immunity cells. In human osteosarcoma (19) and hepatomacarcinoma (20), miR-505 was downregulated in cancer
specimens and cell lines. Functional experiments revealed that
miR-505 suppressed proliferation and invasion by directly targeting
high mobility group box 1 (20).
Similarly, miR-505 acts as a tumor inhibitor in cervical cancer
(21) and endometrial cancer
(22) by directly targeting
frizzled class receptor 4 and transforming growth factor-α,
respectively. However, there is limited knowledge regarding the
molecular function and mechanism of miR-505 in NSCLC.
In the present study, it was demonstrated that
miR-505 was downregulated in NSCLC tissues and cell lines, compared
with their paired normal groups and the normal lung epithelial
cells, respectively. Functionally, it was indicated that miR-505
inhibited cell proliferation, tumor growth and
epithelial-mesenchymal transition (EMT) processes in NSCLC cells.
Mechanically, mitogen-activated protein kinase kinase kinase 3
(MAP3K3) was identified as a direct target of miR-505 and it was
demonstrated to mediate the suppressor role of miR-505 by
inhibiting AKT/nuclear factor-κB (NFκB) activation in NSCLC cells.
The data provide a novel insight into the pathogenesis of NSCLC and
the associated molecules may serve as novel biomarkers for the
diagnosis or therapeutic applications for NSCLC in the future.
Materials and methods
Clinical specimen and cell lines
A total of 21 pairs of human NSCLC tissues were
obtained from the surgical resection of patients with NSCLC in
Qingdao Municipal Hospital (Qingdao, China) from December 2017 to
February 2018. Written informed consent was obtained from all
enrolled patients, and all relevant investigations were performed
according to the principles of the Declaration of Helsinki. All
samples were confirmed through the Department of Pathology in
Qingdao Municipal Hospital. The clinical specimens used in the
present study were pathologically based on the eighth edition of
the Tumor-Node-Metastasis Staging Classification for Urologic
Cancers (23). Pathologists from
the Qiagdao Municipal Hospital who were blind to the study
confirmed the pathology of the specimens. All tissue samples were
frozen in liquid nitrogen immediately following resection and
stored at −80°C until further use. Ethical approval for the study
was obtained from the Ethics Committee of Qingdao Municipal
Hospital (approval no. QMH20170233). The human lung cancer cell
lines A549, H1066, H460, H522 and SPC-A1, and the human normal lung
epithelial cell BEAS-2B were purchased from ATCC (American Type
Culture Collection, Manassas, VA, USA), and cultured in RPMI-1640
or Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) and 1% penicillin-streptomycin. Cell transfection was
performed by Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. A549 cells or H460 cells (4×105) were planted
in 6-well plates overnight at 37°C to ensure that cell confluence
could reach 60-80% at the time of transfection. Following 6 h
transfection at 37°C, the serum-free Opti-MEM medium (Gibco; Thermo
Fisher Scientific, Inc.) was replaced with Dulbecco's modified
Eagle's medium containing 10% FBS. They were transiently
transfected with 3 µg pri-miR-505 or pcDNA3 and 20
µmol antisense oligonucleotide (ASO)-miR-505 or ASO-negative
control (NC) in a well of 6-well plate. The sequences were as
follows: pri-miR-505-sense, 5′-CGCGGATCCCAGACTCCCAGCA ATCAC-3′,
pri-miR-505-antisense, 5′-CCGGAATTCGCAGT ATTCCCACCATTT-3′;
ASO-miR-505, 5′-AGGAAACCAG CAAGUGUUGACG-3′, ASO-NC,
5′-UCAUCGUAUCAGC UAUAUCGCA-3′ (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China). Cells were collected for reverse
transcription-quantitative PCR (RT-qPCR), proliferation, migration,
invasion, apoptosis and cell cycle assay analyses at 24 h. At 48 h
post-transfection, cells were collected for western blot analysis,
immunofluorescence staining and the enhanced green fluorescent
protein fluorescent reporter assay.
RNA extraction and (RT-qPCR)
Total RNA was extracted from A549, H1066, H460,
H522, SPC-A1 and BEAS-2B cells and frozen NSCLC tissues using
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. Reverse transcription
was then performed. Gene-specific primers were used to synthesize
miR-505 or MAP3K3 cDNA from total RNA. U6 small nuclear RNA (snRNA)
and β-actin were used as internal controls for miR-505 and MAP3K3
detection, respectively. The expression levels were determined by
qPCR with a SYBR-Green PCR Master Mix kit (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The qPCR conditions were as follows: 95°C
pre-denaturation for 5 min, followed by 33 cycles of denaturation
at 94°C for 30 sec, annealing and synthesis at 58°C for 30 sec and
70°C for 30 sec, and a final extension step at 72°C for 5 min,
using an Applied Biosystems Prism 7900HT Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer
sequences were listed in Table I.
The relative levels of mature miR-505 and MAP3K3 mRNA were
calculated by the 2-ΔΔCq method (24) and normalized to U6 snRNA or
β-actin mRNA levels, respectively.
 | Table IPrimers for the present study. |
Table I
Primers for the present study.
| RT-qPCR
primers | Primer sequence
(5′–3′) |
|---|
| miR-505-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGGAAAC |
|
miR-505-qPCR-forward |
TGCGGCGTCAACACTTGCT |
| Oligo-dT |
TTTTTTTTTTTTTTTTTT |
| U6-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC |
| U6-forward |
TGCGGGTGCTCGCTTCGGCAGC |
| miRNA-universal
reverse |
CCAGTGCAGGGTCCGAGGT |
|
MAP3K3-qPCR-forward |
CAGCGGCGAGGGCTATGGAA |
|
MAP3K3-qPCR-reverse |
CAGCCTTGCCTGGGAGAA |
|
β-actin-qPCR-forward |
CGTGACATTAAGGAGAAGCTG |
|
β-actin-qPCR-reverse |
CTAGAAGCATTTGCGGTGGAC |
Proliferation assays
The cell proliferation was measured in A549 and H460
cells using a Cell Counting Kit-8 (CCK-8; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). A total of 5×103 cells/well
of A549 and H460 cells were transfected with 0.3 µg
pri-miR-505 (overexpression), 20 nM ASO-miR-505 (knockdown) or
different combinations (pcDNA3+MAP3K3 or pri-miR-505+MAP3K3) of 0.3
µg MAP3K3 overexpression vectors (Vigene Biosciences Inc.,
Rockville, MD, USA), and their respective controls were plated in
96-well plates at a proportionate density (5×103
cells/well for all groups) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols and incubated at 37°C. Subsequently, cell
viability at 24, 48 and 72 h post-transfection was determined with
a CCK-8 assay. Briefly, 10 µl CCK-8 was added to each well
and the plates were incubated for 4 h at 37°C. The optical density
(OD) at a wave length of 450 nm was determined using a microplate
reader (Hitachi, Ltd., Tokyo, Japan). The OD values reflect the
relative number of viable cells.
Colony formation assay
The differently-treated A549 and H460 cells (500
cell/well) were seeded into 12-well plates and cultured in
Dulbecco's modified Eagle's medium containing 10% FBS for 2 weeks
at 37°C for colony formation. The culture medium was changed every
3 days. The colonies were then fixed in 100% methanol for 30 min at
room temperature, stained with crystal violet for 30 min at room
temperature and the numbers of macroscopically observable colonies
were recorded.
Cell cycle analysis by flow
cytometry
At post-transfection 48 h, the A549 and H460 cells
were trypsinized. Following washing by PBS three times,
1×106 cells were fixed in 100% thanol for 10 min at room
temperature and incubated with 50 µg/ml Propidium Iodide
(Cell Cycle Staining kit; Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) for 10 min at room temperature in the dark and
analyzed within 20 min using BD FACS Calibur (BD Biosciences,
Franklin Lakes, NJ, USA). Flow cytometry was used to detect cell
apoptosis and analyzed using FlowJo v.7.6.1 (FlowJo, LLC, Ashland,
OR, USA).
Transwell migration and invasion
assays
The migration and invasion were determined by
Transwell assays individually. Briefly, 6×104 cells A549
and H460 cells were seeded into 8 µm cell culture inserts
(BD Biosciences), according to the manufacturer's protocols and
placed in 24-well cell culture plates. Additionally, the upper
chamber was coated with 100 µl diluted Matrigel (2 mg/ml;
Sigma-Aldrich; Merck KGaA) for the invasion assay. The lower
chamber was filled with 600 µl 20% FBS (Gibco; Thermo Fisher
Scientific, Inc.) medium. A549 and H460 cells (6×104) in
200 µl serum-free Dulbecco's modified Eagle's medium were
gently loaded onto each filter insert (upper chamber) and then
incubated at 37°C for 48 h. The filter inserts were removed from
the chambers, fixed with 95% methanol (Sigma-Aldrich; Merck KGaA)
for 10 min at room temperature and stained with Harris' hematoxylin
(Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. The
samples were subsequently washed with purified water three times,
dried and mounted onto slides. Cell migration and invasion were
assessed using a Transwell system (Corning Incorporated, Corning,
NY, USA). The number of migrated and invaded cells was counted
under a microscope (at ×200).
Western blot analysis
Total A549 cell lysates from different experiments
(migration and invasion assay, EGFP reporter, NFκB pathway and
xenograft model assays) were obtained by lysing the cells in
radio-immunoprecipitation assay (RIPA) buffer (Wanlei Co., Ltd.,
Shanghai, China). The nuclear protein extraction from cells was
obtained with a CelLytic™ NuCLEAR™ Extraction kit (Sigma-Aldrich;
Merck KGaA), according to the manufacturer's protocols. For tissues
protein extraction, the tissues were ground up with liquid
nitrogen, and then RIPA buffer was added, followed by ice bath
cracking for 20 min at 4°C. Subsequently, the cells were
centrifuged at 4°C at 10,000×g for 15 min. Protein concentrations
were quantified using the bicinchoninic acid protein assay kit
(Beyotime Institute of Biotechnology, Shanghai, China), according
to the manufacturer's protocols. A total of 30 µg protein
was resolved by 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). Following
blocking with 5% non-fat milk for 2 h at room temperature, the
membranes were incubated with primary antibodies overnight at 4°C,
followed by incubation with a horseradish peroxidase-conjugated
goat-anti-rabbit (cat. no. 4414S; 1:2,000) or goat-anti-mouse
secondary antibody (cat. no. 4410S, 1:3,000) (both from Cell
Signaling Technology, Inc., Danvers, MA, USA) at room temperature
for 1 h. Membranes were stripped and then re-incubated with a
primary antibody against GAPDH (for total lysate or cytoplasm
fraction; cat. no. ab9485; 1:5,000; Abcam, Cambridge, UK) or
centromere protein A (for nuclear fraction; cat. no. ab65678;
1:2,000; Abcam) for normalization aim at room temperature for 4 h,
according to a previous study (25). Specific bands were visualized
using enhanced chemiluminescence detection (Thermo Fisher
Scientific, Inc.). The signal intensity was determined with ImageJ
software v.1.48 (National Institutes of Health, Bethesda, MD, USA).
The antibodies used in this study were listed in Table II.
 | Table IIAntibodies used in the present
study. |
Table II
Antibodies used in the present
study.
| Name | Company | Cat. no. | Dilution |
|---|
| E-cadherin | Wanlei Co., Ltd.,
Shanghai, China | WL01482 | 1:2,000 |
| Vimentin | Wanlei Co.,
Ltd. | WL00742 | 1:1,000 |
| GAPDH | Abcam, Cambridge,
MA, USA | ab9485 | 1:5,000 |
| Bcl2 | Abcam | ab196495 | 1:3,000 |
| Caspase 3 | Wanlei Co.,
Ltd. | WL02117 | 1:1,500 |
| PARP | Wanlei Co.,
Ltd. | WL01932 | 1:1,500 |
| MAP3K3 | Abcam | ab154362 | 1:2,000 |
| IKKα | Abcam | ab32041 | 1:2,000 |
| IKKβ | Abcam | ab32135 | 1:3,000 |
| pAKT | Abcam | ab38449 | 1:1,000 |
| AKT | Abcam | ab8805 | 1:2,000 |
| p50 | Wanlei Co.,
Ltd. | WL01917 | 1:500 |
| p65 | Wanlei Co.,
Ltd. | WL01980 | 1:1,000 |
| CENPA | Abcam | ab45694 | 1:1,500 |
Prediction of miRNA targets
The hypothetical target of miR-505 was predicted
using miRDB (www.mirdb.org), microRNA.org (www.microrna.org) and TargetScan human 7.2 (www.targetscan.org), which revealed that the 3′UTR of
MAP3K3 may be complementarily paired with the seed sequences of
miR-505.
EGFP reporter assay
The MAP3K3 3′UTR was cloned into a pcDNA3/EGFP
vector (Shanghai GeneChem Co., Ltd., Shanghai, China) and mutations
were introduced at potential miR-505 binding sites. The constructed
pcDNA3/EGFP-MAP3K3-3′UTR (0.5 µg) or its mutated form
pcDNA3/EGFP- MAP3K3-3′UTR-mut (0.5 µg) were co-transfected
with pri-miR-505 (0.5 µg; pri-miR-505-sense, 5′-CGCGGATCCCA
GACTCCCAGCAATCAC-3′, pri-miR-505-antisense, 5′-CCG
GAATTCGCAGTATTCCCACCATTT-3′), ASO-miR-505 (20 nM; ASO-miR-505,
5′-AGGAAACCAGCAAGUGUU GACG-3′) or their respective control vector
(pcDNA3; Vigene Biosciences Inc.) or oligos (ASO-NC, 5′-UCAUCGUAUCA
GCUAUAUCGCA-3′) into A549 and H460 cells, and pDsRed2-1 vector
(Clontech Laboratories, Inc., Mountainview, CA, USA) also
co-transfected for normalization aim in 48-well plates using
Lipofectamine 2000 (DNA:Lipofectamine 2000, 1:1). At 48 h
post-transfection, the total proteins were extracted by RIPA buffer
and the fluorescence intensities of GFP and red fluorescent protein
(RFP) were measured. The EGFP activity was measured using a
spectrophotometer (cat. no. F4500; Hitachi, Ltd.) at 528 nm.
RFP-expressing plasmid was integrated as a transfection efficiency
control.
Immunofluorescence staining
4×104 A549 cells transfected with
specific plasmids (1 µg pcDNA3, 1 µg pri-miR-505, 1
µg MAP3K3 or 0.5 µg pri-miR-505+0.5 µg MAP3K3)
using Lipofectamine® 2000, according to the
manufacturer's protocols were seeded with Dulbecco's modified
Eagle's medium containing 10% FBS in 24-well plates for 48 h and
then for immunofluorescence staining. The cells were washed in PBS
three times and fixed with 4% paraformaldehyde for 30 min at room
temperature. Following cells being washed with PBS three times, the
cells were permeabilized using 0.1% Triton-X-100 for 10 min at room
temperature and blocked in 10% donkey serum (Beyotime Institute of
Biotechnology) for 30 min at room temperature. The cells were
subsequently incubated with primary antibodies against p50 (cat.
no. WL01917, 1:50) and p65 (cat. no. WL01980; 1:50) (both from
Wanlei Co., Ltd.) overnight at 4°C. The following day, cells were
washed in PBS three times and then incubated at room temperature
for 1 h with a goat anti-rabbit IgG H&L Alexa Fluor®
488 (cat. no. ab150077; 1:100; Abcam), followed by incubation with
DAPI (1:1,000; Sigma-Aldrich; Merck KGaA) at room temperature.
Images were captured under a confocal microscope (at ×1,000).
In vivo xenograft model
Animal protocols were approved by Qingdao Municipal
Hospital Animal Care and Use Committee (approval no. QMH20170331A;
Qingdao, China). The methods were conducted in accordance with the
approved guidelines. For the in vivo study, 6-week-old
female BALB/c athymic nude mice (Institute of Zoology, Chinese
Academy of Sciences, Shanghai, China) were used [n=8; divided into
2 groups; weight, 20–30 g; maintenance conditions: Temperature,
18-29°C; relative humidity, 50–60%; free access to clean food and
water; and lighting for 10 h (lights turned on at 8:00 every day
and turned off at 18:00)]. A total number of 1×107
stably transfected (Lenti-control or Lenti-miR-505) A549 cells were
implanted subcutaneously into the armpit of nude mice. For stable
transfections, A549 cells were plated in a 6-well plate
(3×104 cells/ml). After 24 h, a mixture of 3 µg
miR-505 (Lenti-miR-505, 5′-CGUCAACACUUGCUGGUUUCCU-3′) or Control
(Lenti-control, 5′-UCAUCGUAUCAGCUA UAU CGCA-3′) (Guangzhou RiboBio
Co., Ltd.), Lipofectamine 2000 (DNA:Lipofectamine 2000, 1:1), and
Opti-MEM was added to the cells for 6 h. Cells with stable
expression were obtained following culturing in Dulbecco's modified
Eagle's medium for 12 days. These vectors contain a Neomycin
resistance cassette, which was used for mammalian cell clone
selection and maintenance. Independent clones were picked and grown
individually. RNA was isolated using a mirVana miRNA Isolation kit
(Ambion; Thermo Fisher Scientific, Inc.), following the
manufacturer's protocols. The expression levels of miR-505 were
determined by qPCR with a SYBR®-Green PCR Master Mix kit
according to the manufacturer's protocols. Tumor size was measured
every 5 days with calipers following injection, and the tumor
volume was calculated based on formula: 0.5 × (greatest length ×
greatest width2). Tumor weight was measured using an
electronic scale, and the Student's t-test was used to compare
tumor growth among groups. All mice were euthanized with
CO2 at the end of 30 days following implantation. All
animals received human care according to the Institutional Animal
Care and Treatment Committee of Qingdao Municipal Hospital.
Statistics analysis
Statistical analysis was performed using GraphPad
Prism v.5.0 (GraphPad Software, Inc., San Diego, CA, USA). All data
are presented as the mean ± standard deviation, and the experiments
were repeated three times independently. Differences were analyzed
with Student's t-test between two paired groups. For comparisons of
three or more groups, one-way analysis of variance was followed by
the Bonferroni post hoc test for comparison of two selected
treatment groups; the Dunnett's post-hoc test was used for
comparisons of the other treatment groups with the corresponding
controls. Associations between miR-505 expression and
clinicopathological characteristics were assessed using the
χ2 test. P<0.05 was considered to indicate a
statistically significant difference.
Results
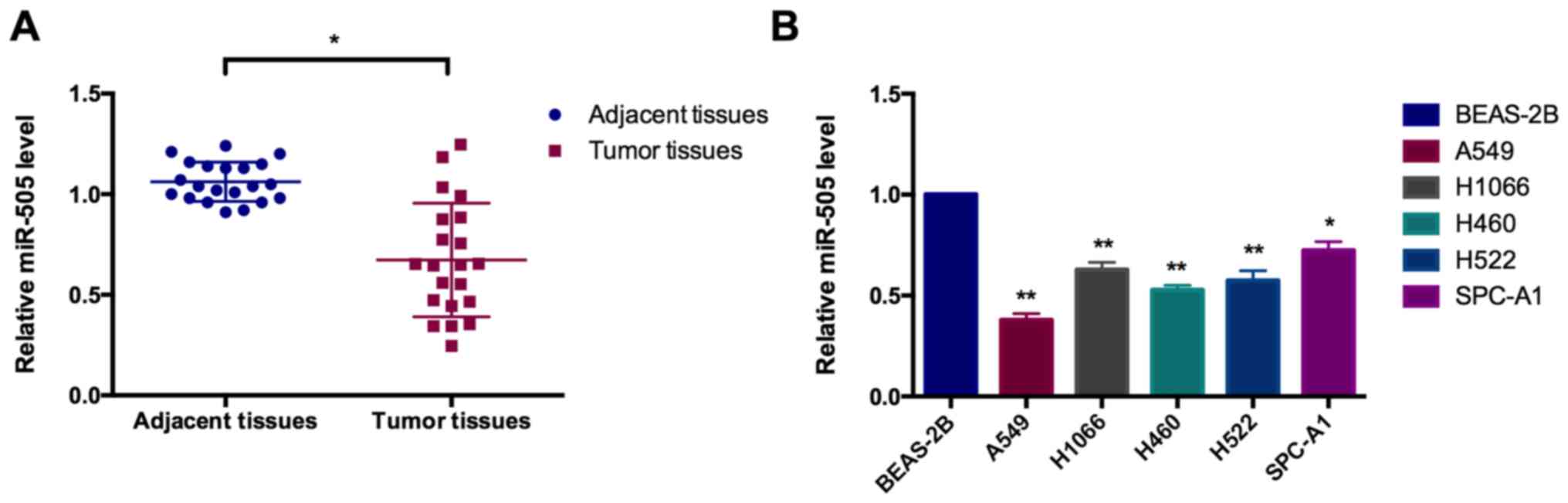
miR-505 is downregulated in NSCLC tissues
and cell lines
The level of miR-505 was determined in 21 paired
NSCLC tissues as well as 6 cell lines using a RT-qPCR assay. As
depicted in Fig. 1A, miR-505 was
significantly reduced in NSCLC tissues, compared with the matched
adjacent non-cancerous controls, which is significantly negatively
associated with large tumor size, TNM stage and distant metastasis
in patients with NSCLC (Table
III). Compared with the human normal lung epithelial cells
BEAS-2B, the level of miR-505 in different lung cancer cell lines
(including A549, H1066, H460, H522 and SPC-A1) was significantly
reduced (Fig. 1B).
 | Table IIIAssociation of miR-505 with the
clinicopathological features of patients. |
Table III
Association of miR-505 with the
clinicopathological features of patients.
| Variables | No. of cases
(n=21) | miR-505 expression
| P-value |
|---|
| Low (n=13) | High (n=8) |
|---|
| Age, years | |
| <60 | 12 | 7 | 5 | 0.681 |
| ≥60 | 9 | 6 | 3 | |
| Sex | |
| Male | 15 | 10 | 5 | 0.477 |
| Female | 6 | 3 | 3 | |
| Tumor size, cm | |
| ≥5 | 13 | 10 | 3 | 0.049a |
| <5 | 8 | 3 | 5 | |
| TNM stage (23) | |
| I-II | 11 | 4 | 7 | 0.011a |
| III-IV | 10 | 9 | 1 | |
| Distant
metastasis | |
| No | 10 | 3 | 7 | 0.004a |
| Yes | 11 | 10 | 1 | |
| Histological type
(23) | |
| Squamous | 9 | 6 | 3 | 0.697 |
|
Adenocarcinoma | 12 | 7 | 5 | |
miR-505 serves as a suppressor gene in
NSCLC cells
To investigate the role of miR-505 in NSCLC cells,
gain- and loss-of-function experiments were performed to analyze
proliferation, cell cycle, migration, invasion, apoptosis and EMT
in A549 and H460 cells. miR-505 overexpression (pri-miR-505) and
knockdown vectors (ASO-miR-505) was constructed and the
transfection efficiencies were validated with a RT-qPCR assay. The
relative miR-505 level was significantly increased by pri-miR-505
transfection, compared with the pcDNA3 control. However, the
relative miR-505 level was significantly decreased upon ASO-miR-505
treatment, compared with ASO-NC treatment in A549 and H460 cells
(Fig. 2A).
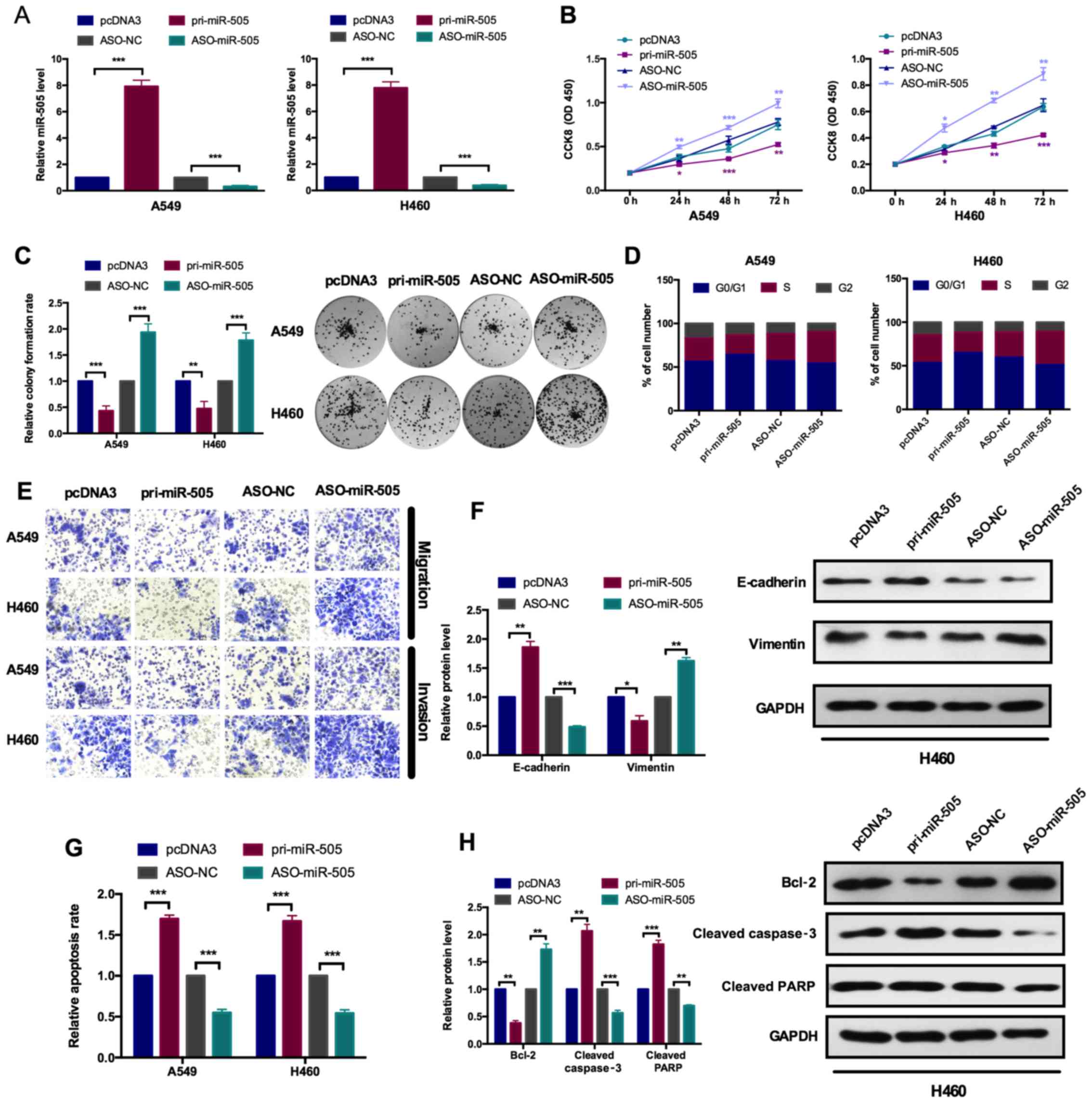 | Figure 2miR-505 inhibits cell proliferation
and the epithelial-mesenchymal transition process. (A) A reverse
transcription-quantitative polymerase chain reaction assay
demonstrated the efficiency of miR-505 overexpression and knockdown
plasmids in A549 and H460 cells. Cells were transfected with the
pri-miR-505 vector or ASO-miR-505 vector and the control groups,
respectively. ***P<0.001 vs. pcDNA3 or ASO-NC group.
(B) The role of miR-505 on A549 and H460 cellular viabilities was
determined with a Cell Counting Kit-8 assay. Overexpression of
miR-505 inhibited cell viability and knockdown of miR-505 promoted
cell viability. *P<0.05, **P<0.01 and
***P<0.001 vs. pcDNA3 for pri-miR-505 or ASO-NC for
ASO-miR-505. (C) Relative colony formation rates of A549 and H460
cells with the indicated transfection were determined with a colony
formation assay. Overexpression of miR-505 inhibited colony
formation ability and knockdown of miR-505 promoted colony
formation ability. **P<0.01 and
***P<0.001 vs. pcDNA3 or ASO-NC group. (D) Flow
cytometry cell cycle assay demonstrated the role of miR-505 on the
cell cycle in A549 and H460 cells. Overexpression of miR-505
decreased the number of A549 and H460 cells in the S and G2 phases,
and decreased the number of cells in the G1 phase. Knockdown of
miR-505 increased the number of HeLa and SiHa cells in the S and G2
phases, and decreased the number of cells in the G1 phase. (E)
Transwell migration and Matrigel invasion assays indicated that
overexpression of miR-505 suppressed cell migration and invasion
ability in A549 and H460 cells. (F) Western blot assays
demonstrated the protein levels of E-cadherin and vimentin
following transfection with pri-miR-505 or ASO-miR-505 vectors in
A549 and H460 cells. *P<0.05, **P<0.01
and ***P<0.001 vs. pcDNA3 or ASO-NC group. (G) Flow
cytometry apoptosis assay indicated the role of miR-505 on
apoptosis in A549 and H460 cells transfected with pri-miR-505
vector or ASO-miR-505 vector and the control group.
***P<0.001 vs. pcDNA3 or ASO-NC group. (H) Western
blot assays demonstrated the protein levels of Bcl2, and cleaved
caspase-3 and PARP following transfection with pri-miR-505 or
ASO-miR-505 in A549 and H460 cells. **P<0.01 and
***P<0.001 vs. pcDNA3 or ASO-NC group. Data are
presented as mean ± standard deviation. The experiments were
repeated three times. miR, microRNA; ASO, antisense
oligonucleotide; NC, negative control; Bcl2, B-cell lymphoma 2;
PARP, poly (ADP) ribose polymerase. |
To determine the effects of miR-505 on cell
proliferation, CCK-8 and colony formation assays were performed. In
CCK-8 analysis, the cell viability was measured at 0, 24, 48 and 72
h post-transfection, presented as the OD450 value in Fig. 2B. Pri-miR-505 significantly
inhibited the cell viability in A549 and H460 cells, compared with
the pcDNA3 group, while ASO-miR-505 significantly increased the
cell viability in these cell lines, compared with the ASO-NC group
(Fig. 2B). Colony formation
analyses provided the similar results. As indicated in Fig. 2C, overexpression of miR-505
significantly decreased the colony formation ability, compared with
the pcDNA3 group, while inhibition of miR-505 could significantly
increase the colony formation ability, compared with the ASO-NC
group (Fig. 2C).
Cell cycle analyses by flow cytometry revealed that
miR-505 overexpression caused an increase in cells at the G0/G1
phase and a decrease in cells at the S phase, while miR-505
knockdown induced a decrease in cells at the G0/G1 phase and the
increase in cells at the S phase (Fig. 2D). Transwell migration and
Matrigel invasion assays were performed to investigate the effects
of miR-505 on cell migration and invasion, respectively. As
indicated in Fig. 2E, it was
observed that the migration and invasion capacities was decreased
by overexpression of miR-505, and the migration and invasion
abilities were enhanced by miR-505 inhibition in A549 and H460
cells (Fig. 2E). Additionally,
the EMT markers were detected by western blot analysis. As
expected, when miR-505 was overexpressed, the epithelial marker
E-cadherin exhibited an increased expression, while the mesenchymal
marker vimentin exhibited a decreased expression in both cell
lines. However, when miR-505 was inhibited, the epithelial marker
E-cadherin decreased and the mesenchymal marker vimentin increased,
compared with their respective controls (Fig. 2F).
As depicted in Fig.
2G, overexpression of miR-505 significantly increased
apoptosis, while inhibition of miR-505 significantly decreased the
cell apoptosis in A549 and H460 cells (Fig. 2G). The expression of apoptosis
inhibitor B-cell lymphoma 2 (Bcl2) was decreased upon miR-505
overexpression and increased while inhibiting miR-505. Furthermore,
it was also demonstrated that the increased expression of cleaved
caspase 3 and cleaved poly (ADP) ribose polymerase (PARP) by
overexpression of miR-505, and the opposite results obtained from
the inhibition of miR-505 (Fig.
2H).
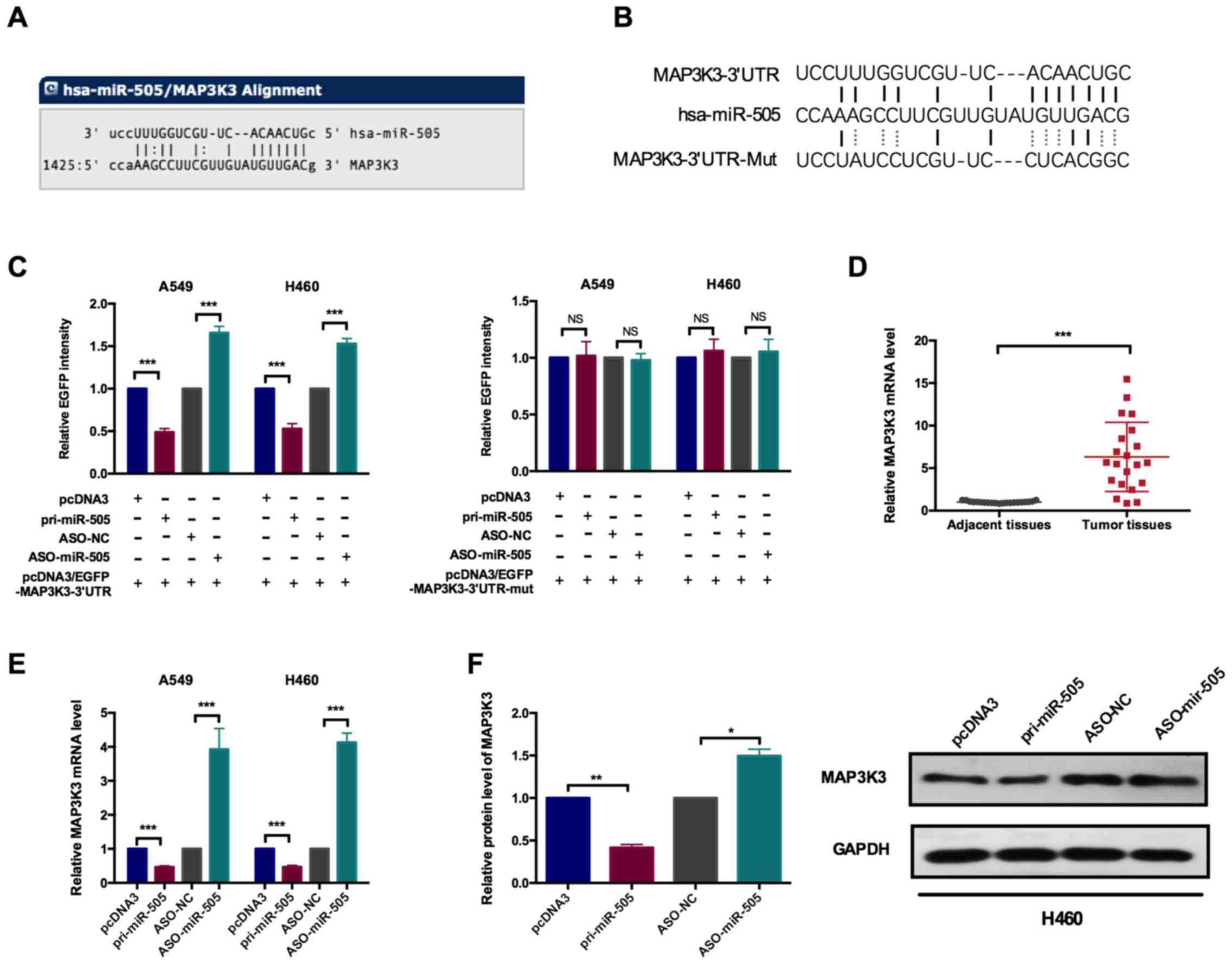
miR-505 directly targets MAP3K3 in NSCLC
cells
By bioinformatics analysis, the putative targets of
miR-505 were analyzed and MAP3K3 was selected for further
validation. Fig. 3A depicts the
alignment of miR-505 binding to the 3′UTR of MAP3K3. The EGFP
reporter vector was contrasted to verify the direct binding of
miR-505 and MAP3K3. By annealing oligos and ligation to the vector,
the putative miR-505 binding site sequences was closed downstream
of the EGFP to construct the wild type and mutated form of the EGFP
reporter vectors, as indicated in Fig. 3B. By co-transfection with
pri-miR-505 or ASO-miR-505 and MAP3K3 3′UTR, the EGFP intensities
were measured. As depicted in Fig.
3C, it was observed that the EGFP intensities following
co-transfection with the wild-type MAP3K3 3′UTR reporter vector and
pri-miR-505 or ASO-miR-505were significantly decreased and
increased, respectively, compared with their respective controls
(Fig. 3C). When the
co-transfection with the mutated form of the reporter vector was
performed, there were no significant changes in the EGFP
intensities by modulating the expression of miR-505.
Subsequently, the expression levels of MAP3K3 were
determined in clinical samples. As depicted in Fig. 3D, there was significantly
upregulated expression of MAP3K3 in lung cancer tissues, compared
with the matched adjacent normal tissues (Fig. 3D). The expression levels of
miR-505 were also modulated in A549 and H460 cells, and the MAP3K3
level at the mRNA and protein levels were detected by RT-qPCR and
western blot analysis, respectively. RT-qPCR assays revealed the
significantly decreased MAP3K3 mRNA level upon pri-miR-505
transfection and significantly increased MAP3K3 level by inhibition
miR-505 in A549 and H460 cells (Fig.
3E). The similar results of MAP3K3 protein levels were
indicated in Fig. 3F.
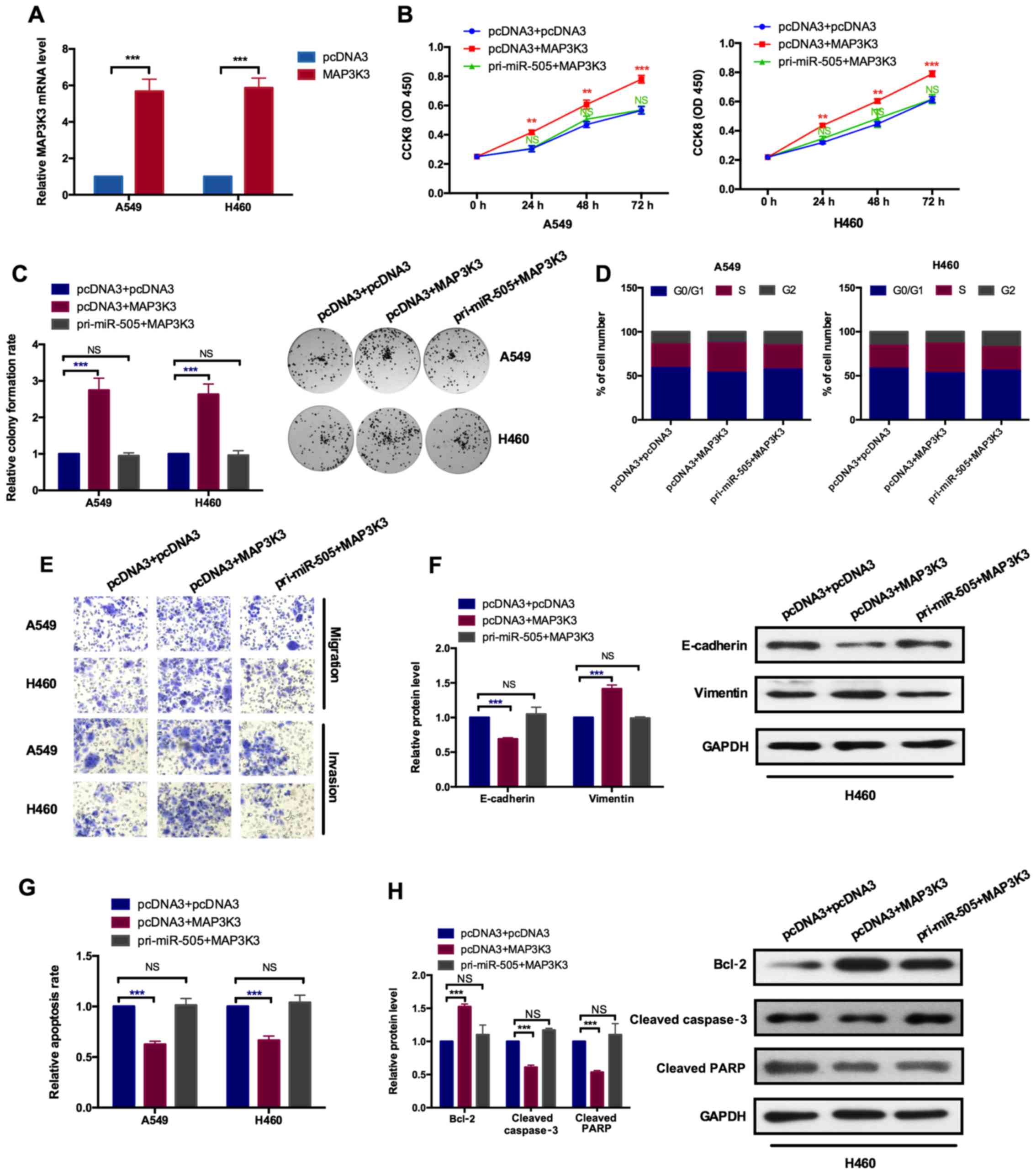
MAP3K3 serves an oncogenic role and
miR-505 neutralizes its effects in NSCLC cells
To further confirm the effects of miR-505 on cell
proliferation, cell cycle, apoptosis, migration, invasion and EMT
in NSCLC cells were mediated by MAP3K3, rescue experiments were
performed. The transfection efficiency of MAP3K3 expression vectors
was determined. As depicted in Fig.
4A, the mRNA level of MAP3K3 was significantly increased upon
overexpression of MAP3K3, compared with the empty vector control
pcDNA3, in A549 and H460 cells. Subsequently, co-transfection
experiments with MAP3K3 and miR-505 expression vectors were
performed and the combined effects of MAP3K3 and miR-505 together
were detected. As indicated in Fig.
4B and C, overexpression of MAP3K3 increased cell
proliferation, while overexpression of MAP3K3 and miR-505 together
decreased the enhanced role induced by MAP3K3, as assessed by CCK-8
and colony formation assays individually. The cell cycle analysis
revealed that MAP3K3 induced the increase of cells in the S phase
and the decrease of cells in the G0/G1 phase, while combination of
miR-505 and MAP3K3 eliminated the oncogenic roles caused by MAP3K3
(Fig. 4D). Transwell migration
and Matrigel invasion assays, and western blot analysis was used to
detect the EMT markers, which demonstrated the increased migration
and invasion capabilities, and decreased expression of E-cadherin
and increased expression of vimentin upon overexpressing MAP3K3.
Furthermore, cells treated with MAP3K3 and miR-505 together had
increased effects on migration, invasion and EMT, caused by MAP3K3,
reduced back to the normal control level (Fig. 4E and F). Similarly, the apoptosis
detected by flow cytometry analysis and western blot assays
indicated the inhibition effects induced by MAP3K3, which was
opposite to the effects induced by miR-505, while combination of
MAP3K3 and miR-505 overexpression abolished the inhibition effects
induced by MAP3K3, as indicated by the relative apoptosis rates
assessed by flow cytometry (Fig.
4G) and presented by the increased expression of Bcl2 as well
as the decreased expressions of cleaved caspase 3 and cleaved PARP
(Fig. 4H). These results
demonstrated that MAP3K3 mediated the role of miR-505 on cell
proliferation, cell cycle arrest, apoptosis, migration, invasion as
well as EMT progress in NSCLC cells.
miR-505 regulates the AKT/NFκB pathway
through MAP3K3
MAP3K3, as a member of mitogen-activated protein
kinase (MAPK) family, could interact with AKT to active the NFκB
signaling pathway (26). It was
determined whether miR-505 was also involved in AKT/NFκB pathways
via MAP3K3 by western blot analysis to analyze the expression level
of AKT (total AKT and phosphorylated form of AKT) and its
downstream target IκB kinase α (IKKα) and IKKβ as well as the
activation of the NFκB pathway.
Firstly, the level of miR-505 in NSCLC cells was
modulated by gain- and loss-of-function experiments and the
expression levels of the indicated molecules were detected. As
indicated in Fig. 5A,
overexpression of miR-505 significantly inhibited the
phosphorylation of AKT (pAKT) but not the total AKT, and the
expression of its downstream targets IKKα and IKKβ was also
significantly decreased by miR-505 overexpression (Fig. 5A). To further evaluate the effects
of miR-505 on NFκB pathway activation, the nuclear and cytoplasmic
cell lysates was isolated and the expression level of p50 and p65
was determined by western blot analysis. It was demonstrated that
nuclear p50 and p65 expression was significantly decreased while
cytoplasmic p50 and p65 expression was significantly increased upon
overexpression of miR-505 in cells (Fig. 5B), indicating the activation of
the NFκB pathway was inhibited by miR-505 overexpression. However,
miR-505 knockdown resulted in the activation of AKT (increased
expression of pAKT), increased expression of IKKα and IKKβ as well
as the elevated expression of nuclear p50 and p65, and decreased
expression of cytoplasmic p50 and p65 (Fig. 5A and B).
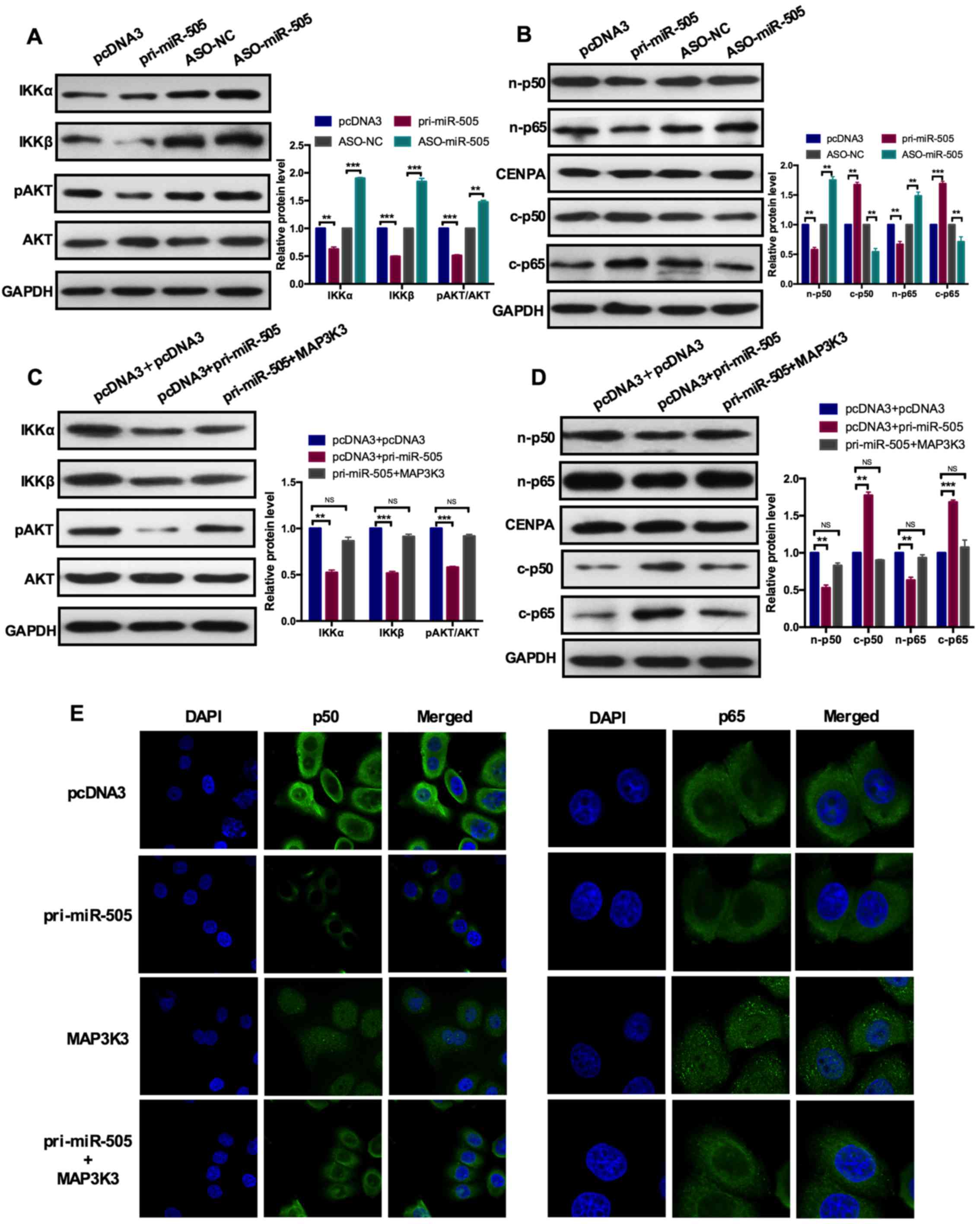 | Figure 5miR-505 inhibits the AKT-nuclear
factor-κB pathway through MAP3K3. (A) A western blot assay
demonstrated the protein expression levels of IKKa, IKKβ, pAKT, AKT
with pri-miR-505 or ASO-miR-505 and the control group in A549
cells. miR-505 overexpression inhibited IKKa, IKKβ and pAKT
expression but not the total AKT expression. **P<0.01
and ***P<0.001 vs. pcDNA3 or ASO-NC group. (B) A
western blot assay indicated the protein expression levels of
n-p50, c-p50, n-p65 and c-p65 with pri-miR-505 or ASO-miR-505 and
the control group in A549 cells. miR-505 overexpression inhibited
n-p50 and n-p65 expression. **P<0.01 and
***P<0.001 vs. pcDNA3 or ASO-NC group. (C) A western
blot assay demonstrated the protein expression levels of IKKa,
IKKβ, pAKT, AKT with pcDNA3 and pcDNA3 or pcDNA3 and pri-miR-505 or
pri-miR-505 and MAP3K3 in A549 cells. MAP3K3 overexpression rescued
the role of miR-505 induced on the expression level of the
indicated markers. NS vs. pri-miR-505 and MAP3K3;
**P<0.01 and ***P<0.001 vs. pcDNA3 and
pri-miR-505 group. (D) A western blot assay indicated the protein
expression levels of n-p50, c-p50, n-p65 and c-p65 with pcDNA3 and
pcDNA3 or pcDNA3 and pri-miR-505 or pri-miR-505 and MAP3K3 in A549
cells. NS vs. pri-miR-505 and MAP3K3; **P<0.01 and
***P<0.001 vs. pcDNA3 and pri-miR-505 group. (E) An
immunofluorescence assay indicated the distribution of p50 and p65
transfected with the indicated plasmids in A549 cells. miR-505
inhibited but MAP3K3 promoted the nuclear distribution of p50 and
p65. Data are presented as mean ± standard deviation. The
experiments were repeated three times. pAKT, phosphorylated AKT;
n-p50, nuclear p50; c-p50, cytoplasmic p50; miR, microRNA; ASO,
antisense oligonucleotide; NC, negative control; MAP3K3;
mitogen-activated protein kinase kinase kinase 3; IKKβ, IκB kinase
β; CENPA, centromere protein A. |
Subsequently, co-transfection with miR-505 and
MAP3K3 in NSCLC cells was performed to re-evaluate the effects of
miR-505 on the AKT/NFκB pathway activation. As depicted in Fig. 5C and D, overexpression of miR-505
significantly inhibited AKT/NFκB pathway activation, presented as
decreased expression of pAKT, IKKα, IKKβ, and nuclear p50 and p65,
and increased expression of cytoplasmic p50 and p65. However,
ectopic expression of MAP3K3 in the presence of miR-505
counteracted the effects induced by miR-505 to the normal control
level, presented as increased expression levels of pAKT, IKKα,
IKKβ, and nuclear p50 and p65, and decreased expression of
cytoplasmic p50 and p65, compared with only miR-505 overex-pression
(Fig. 5C and D). Additionally, an
immunofluorescence assay in A549 cells demonstrated the nuclear
distribution of p50 and p65 transfected with miR-505 was reduced,
compared with the pcDNA3 group. The nuclear distribution of p50 and
p65 transfected with MAP3K3 was increased, compared with the pcDNA3
group. Furthermore, co-transfection with miR-505 and MAP3K3 rescued
the role of miR-505 and MAP3K3 induced in cells (Fig. 5E). These results provided the
direct evidence that miR-505 inhibited the AKT/NFκB pathway by
downregulating MAP3K3 expression in NSCLC cells.
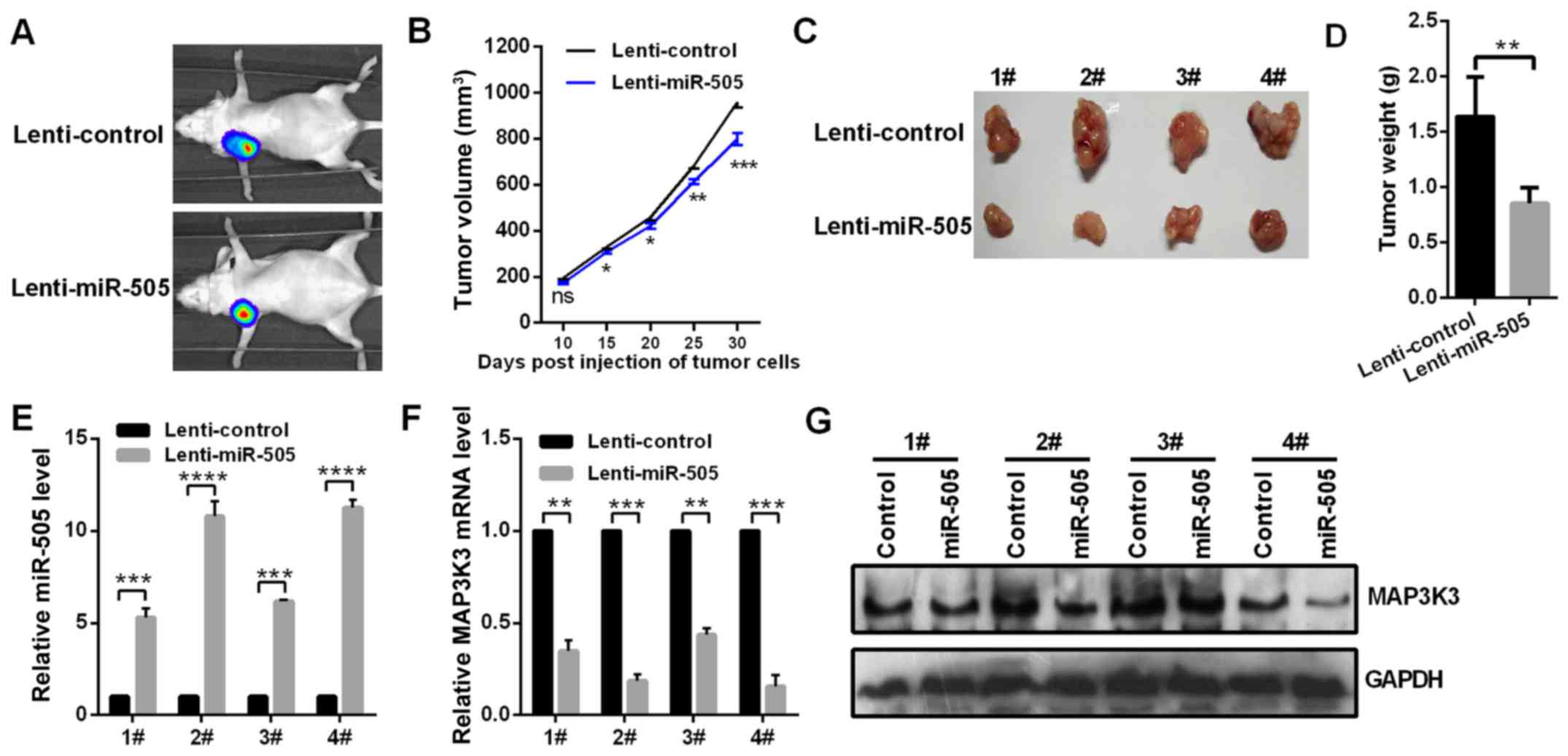
miR-505 inhibits tumor growth in
vivo
To verify whether miR-505 regulates the tumor growth
in vivo, miR-505-over-expressing and control A549 cells were
subcutaneously transplanted into the right flank of mice. After 30
days, it was determined that stable transfection of lenti-miR-505
in A549 cells significantly inhibited the tumor growth (Fig. 6A and B) and significantly
decreased the tumor weight (Fig. 6C
and D). The RT-qPCR assay demonstrated that the level of
miR-505 in the xenograft was upregulated in the miR-505-treated
group (Fig. 6E). Additionally,
the RT-qPCR and western blot assays indicated the mRNA and protein
level of MAP3K3 in the xenograft was significantly downregulated in
the miR-505-treated group (Fig. 6F
and G). These data indicated that miR-505 inhibited tumor
growth in vivo.
Discussion
miRNA expression signature for lung cancer was
characterized by previous studies (27,28). Recently, a number of individual
studies reported the dysregulated miRNAs in NSCLC and the
functional roles of the individual miRNAs were investigated
(29–35). For example, Yang et al
(29) reported that miR-183 has a
decreased expression level in NSCLC tissues and act as a tumor
suppressor by downregulating metastasis associated 1 in NSCLC cells
(29). Other miRNAs, including
miR-30b (30), miR-485-5p
(31), miR-449a (32), miR-1253 (33) and miR-615-3p (34), also exhibit reduced expression
levels in NSCLC tissues, compared with the adjacent normal tissues,
and could inhibit cell proliferation and invasion in NSCLC cells
individually by binding to the 3′UTR of their different targets.
Additionally, Ding et al (35) reported that miR-25 was highly
expressed in NSCLC tissues, and enhanced cell migration and
invasion by inhibiting kruppel like factor 4 in NSCLC cells. These
experimental data, including the present results, provide direct
evidence that miRNAs are involved in the pathogenesis of NSCLC.
MAP3K3, a family member of serine/threonine protein
kinase, acts as an upstream regulator of the MAPK pathway and
activated multiple MAPKs, including extracellular regulated kinase
1 (ERK1)/ERK2, c-Jun N-terminal kinase, p38 and ERK5, regulating
numerous different cellular functions, including proliferation,
migration, immune response and cell cycle (36,37). Additionally, MAP3K3 could active
the IKK/NFκB pathway (38). Apart
from its roles in the immune responses, MAP3K3 was previously
reported to be associated with tumorigenesis. For example, Hasa
et al (39) reported that
MAP3K3 expression was significantly increased in esophageal
dysplasia and esophageal squamous cell carcinoma in comparison with
the normal mucosa, and had the potential to serve as a predictor of
poor disease prognosis (39).
There are 8-20% patients with breast cancer that harbor MAP3K3 gene
amplification, and in vitro and in vivo studies
(40) demonstrated that MAP3K3
contributes to breast carcinogenesis and may endow resistance of
breast cancer cells to cytotoxic chemotherapy, indicating its
potential valuable therapeutic target in patients with
MAP3K3-amplified breast cancer (40). A number of studies also evaluated
the prognostic applications of MAP3K3 in different types of cancer
(41,42). Jia et al (41) reported that MAP3K3 overexpression
was observed in ~60% of ovarian carcinoma cases and was
significantly associated with histological type, grade and
chemotherapy response, indicating that MAP3K3 overexpression may be
an independent poor prognostic indicator in ovarian carcinoma.
Additionally, He et al (42) investigated the associations
between the expression of MAP3K3 and the clinical outcomes in
primary lung cancer. Their data indicated that overexpression of
MAP3K3 associated with the active immune response and the improved
patient survival in patients with lung cancer.
As small regulator RNA molecules, miRNAs serve
important roles in regulating gene expression at the
post-transcriptional level (43).
It had been demonstrated that one gene could be regulated by
different miRNAs and one miRNA could regulate hundreds of different
genes according to the different cell context (43). Previous studies had identified a
number of miRNAs could directly regulate the expression of MAP3K3
(44-46). For example, brain-specific miR-124
is significantly downregulated in LPS-treated BV2 cells and
MPTP-induced model of Parkinson's disease, and miR-124 directly
inhibits the expression of MAP3K3 by binding to its 3′UTR in the
inflammatory pathogenesis of Parkinson's disease (44). Zheng et al (45), reported that miR-188 was
upregulated in aged lineage-negative bone marrow cells, enhanced
cell senescence by regulating MAP3K3 expression and provided a
novel strategy for prevention and treatment of cardiovascular
disease. Recently, Zhao et al (46) reported that miR-188 directly
targeted MAP3K3 in NSCLC and functioned as a tumor suppressor
(46).
In the present study, for the first time, to the
best of our knowledge, it was demonstrated that miR-505 was
down-regulated and MAP3K3 was upregulated in NSCLC tissues, and
MAP3K3 was identified as a direct target of miR-505. Additionally,
the functional roles of miR-505 were assessed by different assays,
and its tumor suppressor functions in NSCLC cells were confirmed
in vitro by inhibiting tumor growth and EMT progress. By
directly binding to the 3′UTR of MAP3K3, miR-505 inhibited MAP3K3
expression and subsequently inactivated the AKT/NFκB pathway,
resulting in the decreased expression levels of IKKα, IKKβ, pAKT,
and nuclear p50 and p65, as well as the accumulation of the
cytoplasmic p50 and p65. By rescue experiments, the tumor
suppressor roles of miR-505 mediated directly by MAP3K3 in NSCLC
cells were confirmed. MAP3K3 subsequently mediated the inhibition
of AKT/NFκB activation induced by overexpression of miR-505, and
constructed an indirect regulation axis between miR-505 and the
AKT/NFκB pathway. The present data provided evidence of miRNAs
involved in the pathogenesis of NSCLC and may serve as valuable
biomarkers for clinical applications.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HT and YH designed the study. WL collated the data,
and designed and developed the database. WS performed the data
analyses and produced the initial draft of the manuscript. HT, QB
and WL obtained the results and validated them. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Qingdao Municipal Hospital (Qingdao, China) and
informed consent was obtained from all patients prior to the
study.
Patient consent for publication
Consent for publication was obtained from the
participants.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to acknowledge the beneficial
comments on the present study received from reviewers.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abbosh C, Birkbak NJ and Swanton C: Early
stage NSCLC - challenges to implementing ctDNA-based screening and
MRD detection. Nat Rev Clin Oncol. 15:577–586. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frega S, Bonanno L, Guarneri V, Conte P
and Pasello G: Therapeutic perspectives for brain metastases in
non-oncogene addicted non-small cell lung cancer (NSCLC): Towards a
less dismal future? Crit Rev Oncol Hematol. 128:19–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsiara A, Liontos M, Kaparelou M,
Zakopoulou R, Bamias A and Dimopoulos MA: Implementation of
immunotherapy in the treatment of advanced non-small cell lung
cancer (NSCLC). Ann Transl Med. 6:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Economopoulou P and Mountzios G: The
emerging treatment landscape of advanced non-small cell lung
cancer. Ann Transl Med. 6:1382018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan HL, Wang T and Zhang KH: MicroRNAs as
potential biomarkers for diagnosis, therapy and prognosis of
gastric cancer. OncoTargets Ther. 11:3891–3900. 2018. View Article : Google Scholar
|
|
9
|
Han Y and Li H: miRNAs as biomarkers and
for the early detection of non-small cell lung cancer (NSCLC). J
Thorac Dis. 10:3119–3131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lekka E and Hall J: Noncoding RNAs in
disease. FEBS Lett. 592:2884–2900. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
14
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang W, Wan S, Yang Z, Teschendorff AE and
Zou Q: Tumor origin detection with tissue-specific miRNA and DNA
methylation markers. Bioinformatics. 34:398–406. 2018. View Article : Google Scholar
|
|
16
|
Yamamoto Y, Yoshioka Y, Minoura K,
Takahashi RU, Takeshita F, Taya T, Horii R, Fukuoka Y, Kato T,
Kosaka N, et al: An integrative genomic analysis revealed the
relevance of microRNA and gene expression for drug-resistance in
human breast cancer cells. Mol Cancer. 10:1352011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Q, Jia C, Wang P, Xiong M, Cui J, Li
L, Wang W, Wu Q, Chen Y and Zhang T: MicroRNA-505 identified from
patients with essential hypertension impairs endothelial cell
migration and tube formation. Int J Cardiol. 177:925–934. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Escate R, Mata P, Cepeda JM, Padró T and
Badimon L: miR-505-3p controls chemokine receptor up-regulation in
macrophages: Role in familial hypercholesterolemia. FASEB J.
32:601–612. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu YJ, Li W, Chang F, Liu JN, Lin JX and
Chen DX: MicroRNA-505 is downregulated in human osteosarcoma and
regulates cell proliferation, migration and invasion. Oncol Rep.
39:491–500. 2018.
|
|
20
|
Lu L, Qiu C, Li D, Bai G, Liang J and Yang
Q: MicroRNA-505 suppresses proliferation and invasion in hepatoma
cells by directly targeting high-mobility group box 1. Life Sci.
157:12–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma C, Xu B, Husaiyin S, Wang L,
Wusainahong K, Ma J, Zhu K and Niyazi M: MicroRNA-505 predicts
prognosis and acts as tumor inhibitor in cervical carcinoma with
inverse association with FZD4. Biomed Pharmacother. 92:586–594.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Sun KX, Liu BL, Zong ZH and Zhao
Y: MicroRNA-505 functions as a tumor suppressor in endometrial
cancer by targeting TGF-α. Mol Cancer. 15:112016. View Article : Google Scholar
|
|
23
|
Paner GP, Stadler WM, Hansel DE, Montironi
R, Lin DW and Amin MB: Updates in the Eighth Edition of the
Tumor-Node-Metastasis Staging Classification for Urologic Cancers.
Eur Urol. 73:560–569. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Yang Z, Wang XL, Bai R, Liu WY, Li X, Liu
M and Tang H: miR-23a promotes IKKα expression but suppresses ST7L
expression to contribute to the malignancy of epithelial ovarian
cancer cells. Br J Cancer. 115:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Samanta AK, Huang HJ, Le XF, Mao W, Lu KH,
Bast RC Jr and Liao WS: MEKK3 expression correlates with nuclear
factor kappa B activity and with expression of antiapoptotic genes
in serous ovarian carcinoma. Cancer. 115:3897–3908. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Landi MT, Zhao Y, Rotunno M, Koshiol J,
Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola
FM, et al: MicroRNA expression differentiates histology and
predicts survival of lung cancer. Clin Cancer Res. 16:430–441.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du X, Zhang J, Wang J, Lin X and Ding F:
Role of miRNA in lung cancer-potential biomarkers and therapies.
Curr Pharm Des. 23:5997–6010. 2018. View Article : Google Scholar
|
|
29
|
Yang CL, Zheng XL, Ye K, Ge H, Sun YN, Lu
YF and Fan QX: MicroRNA-183 acts as a tumor suppressor in human
non-small cell lung cancer by down-regulating MTA1. Cell. Physiol
Biochem. 46:93–106. 2018.
|
|
30
|
Qi Z, Zhang B, Zhang J, Hu Q, Xu F, Chen B
and Zhu C: MicroRNA-30b inhibits non-small cell lung cancer cell
growth by targeting the epidermal growth factor receptor.
Neoplasma. 65:192–200. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang RS, Zheng YL, Li C, Ding C, Xu C and
Zhao J: MicroRNA-485-5p suppresses growth and metastasis in
non-small cell lung cancer cells by targeting IGF2BP2. Life Sci.
199:104–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu D, Liu J, Chen J, He H, Ma H and Lv X:
MiR-449a suppresses tumor growth, migration and invasion in
non-small cell lung cancer by targeting HMGB1-mediated NF-κB
signaling way. Oncol Res. Mar 21–2018.Epub ahead of print.
|
|
33
|
Liu M, Zhang Y, Zhang J, Cai H, Zhang C,
Yang Z, Niu Y, Wang H, Wei X, Wang W, et al: MicroRNA-1253
suppresses cell proliferation and invasion of non-small-cell lung
carcinoma by targeting WNT5A. Cell Death Dis. 9:1892018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Jia Y, Jia L, Li T, Yang L and
Zhang G: MicroRNA-615-3p inhibits the tumor growth and metastasis
of NSCLC via inhibiting IGF2. Oncol Res. Mar 21–2018.Epub ahead of
print.
|
|
35
|
Ding X, Zhong T, Jiang L, Huang J, Xia Y
and Hu R: miR-25 enhances cell migration and invasion in
non-small-cell lung cancer cells via ERK signaling pathway by
inhibiting KLF4. Mol Med Rep. 17:7005–7016. 2018.PubMed/NCBI
|
|
36
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang J, Lin Y, Guo Z, Cheng J, Huang J,
Deng L, Liao W, Chen Z, Liu Z and Su B: The essential role of MEKK3
in TNF-induced NF-kappaB activation. Nat Immunol. 2:620–624. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hasan R, Sharma R, Saraya A, Chattopadhyay
TK, DattaGupta S, Walfish PG, Chauhan SS and Ralhan R: Mitogen
activated protein kinase kinase kinase 3 (MAP3K3/MEKK3)
overexpression is an early event in esophageal tumorigenesis and is
a predictor of poor disease prognosis. BMC Cancer. 14:1–7. 2014.
View Article : Google Scholar
|
|
40
|
Fan Y, Ge N, Wang X, Sun W, Mao R, Bu W,
Creighton CJ, Zheng P, Vasudevan S, An L, et al: Amplification and
over-expression of MAP3K3 gene in human breast cancer promotes
formation and survival of breast cancer cells. J Pathol. 232:75–86.
2014. View Article : Google Scholar :
|
|
41
|
Jia W, Dong Y, Tao L, Pang L, Ren Y, Liang
W, Jiang J, Cheng G, Zhang WJ, Yuan X, et al: MAP3K3 overexpression
is associated with poor survival in ovarian carcinoma. Hum Pathol.
50:162–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He Y, Wang L, Liu W, Zhong J, Bai S, Wang
Z, Thomas DG, Lin J, Reddy RM, Ramnath N, et al: MAP3K3 expression
in tumor cells and tumor-infiltrating lymphocytes is correlated
with favorable patient survival in lung cancer. Sci Rep.
5:114712015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 18:30065–30067.
2018.
|
|
44
|
Yao L, Ye Y, Mao H, Lu F, He X, Lu G and
Zhang S: MicroRNA-124 regulates the expression of MEKK3 in the
inflammatory pathogenesis of Parkinson's disease. J
Neuroinflammation. 15:132018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zheng Y, Liu H and Kong Y: miR-188
promotes senescence of lineage-negative bone marrow cells by
targeting MAP3K3 expression. FEBS Lett. 591:2290–2298. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao L, Ni X, Zhao L, Zhang Y, Jin D, Yin
W, Wang D and Zhang W: MiroRNA-188 acts as tumor suppressor in
non-small-cell lung cancer by targeting MAP3K3. Mol Pharm.
15:1682–1689. 2018. View Article : Google Scholar : PubMed/NCBI
|