Introduction
Hyperlipidemia is a major contributing factor to the
pathogenesis of β-cell dysfunction in subjects with type 2 diabetes
(1). Hypercholesterolemia leads
to the accumulation of cholesterol in islets and reduces insulin
secretion, while a reduction in cholesterol levels restores insulin
secretion. Excessive cholesterol accumulation in pancreatic β-cells
not only affects the expression of insulin genes but also impairs
the formation of insulin granules and their trafficking and fusion
with the plasma membrane (2-5).
However, the detailed mechanism by which high intracellular
cholesterol levels affect insulin biosynthesis and secretion is not
completely understood.
MicroRNAs (miRNAs/miR) are a class of short,
nonprotein-coding gene products that target the 3′ untranslated
regions (3′UTRs) of mRNAs by repressing translation. Islet-specific
miRNAs have been identified in mice and humans, and some of the
dysregulated miRNAs are involved in the processes of insulin
biosynthesis and exocytosis (6-9).
miR-24 is a miRNA that is expressed at high levels in pancreatic
β-cells and is shown to be further increased in islets from obese
or high-fat diet-fed mice. Additionally, miR-24 regulates the
expression of insulin gene transcriptional activators neuronal
differentiation 1 (Neurod1) and Hnf1a, through which it negatively
regulates insulin biosynthesis (10). Notably, miR-24 is the miRNA
expressed at high levels in islets from db/db mice, leading to
obesity, hyperglycemia, a reduction in insulin biosynthesis and an
impairment in glucose-stimulated insulin secretion (GSIS) (10). However, the specific role of
miR-24 in insulin secretion has not been explored.
Insulin secretion is a highly complex and concerted
process that is orchestrated by a cascade of regulatory factors.
Secretagogin (Scgn), an EF-hand Ca2+-binding protein
that is expressed at high levels in pancreatic β-cells, has been
shown to be a positive regulator of insulin exocytosis through
effects on actin dynamics and focal adhesion (11-13). Sp1, an experimentally verified
transcriptional regulator of Scgn, which is involved in the control
of GSIS (11), was predicted to
harbor a putative miR-24 binding site in its 3′UTR. Therefore, it
was speculated that cholesterol accumulation decreases insulin
secretion in pancreatic β-cells and may be related to miR-24
overexpression. The present study also postulated that
cholesterol-mediated Sp1 downregulation inhibits the expression of
Scgn and its downstream focal adhesion molecules, subsequently
reducing the docking and fusion of insulin granules with the cell
membrane. The present study sought to confirm the proposed
molecular mechanisms of insulin secretion by employing MIN6
insulinoma cells as a model system.
Materials and methods
Cell culture
Mouse insulinoma-derived MIN6 cells (Fuheng Cell
Center) were used for the functional and imaging experiments in the
current study because of their stable response to glucose
stimulation and low autofluorescence. Moreover, the MIN6 mouse
β-cell line has been widely employed to assess the effect of
cholesterol on pancreatic β-cells. The cells were maintained in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 16.7 mM glucose and supplemented with
10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 2 mM gluta-mine, 100 U/ml penicillin and 0.1
mg/ml streptomycin. The culture medium was changed every 3-4
days.
Oil red O staining
MIN6 cells were seeded in 6-well plates at a density
of 1×106 cells/well and grown overnight at 37°C, and
then incubated with water-soluble non-esterified cholesterol
(Sigma-Aldrich; Merck KGaA) at 37°C for 12 h. Then the cells were
washed three times with PBS and fixed with 4% paraformaldehyde for
30 min at room temperature. A freshly diluted oil red O solution
(Sigma-Aldrich; Merck KGaA) was added to each well and incubated
for 15 min at room temperature, followed by decolorization with a
70% ethanol solution for 15 sec and staining with a hematoxylin
solution for 30 sec at room temperature. After two rinses with
distilled water, intracellular cholesterol droplets were observed
and imaged under a fluorescence microscope at the light level
(Leica Microsystems GmbH).
Determination of the intracellular
cholesterol concentration
MIN6 cells were seeded in 12-well plates
(4×105 cells/well), cultured overnight and subsequently
treated with soluble cholesterol (Sigma-Aldrich; Merck KGaA). A
total of 12 h after treatment, cells were harvested and the
cholesterol content was quantified using a cholesterol assay kit
(cat. no. MAK043; Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. Briefly, 1×106 cells were lysed
in 200 μl of a mixture of chloroform: Isopropanol: NP-40 (7:11:0.1)
with a microhomogenizer and then the supernatant was collected and
dried in a vacuum. The dried lipids were dissolved in 200 μl of
cholesterol assay buffer by sonication (at a frequency of 20 kHz
with an on/off cycle of 10/10 sec repeated for 2 min at 4°C) and
vortexing until the solution appeared cloudy. Next, the reactions
containing the cholesterol probe, esterase, enzyme mix, assay
buffer and samples or standards were processed at 37°C for 1 h. The
absorbance was measured at 570 nm in a microplate reader and the
intracellular cholesterol concentrations were adjusted for the DNA
content.
Quantitation of the cell viability with
the Cell Counting Kit-8 (CCK-8)
MIN6 cells were cultivated in 96-well plates (5,000
cells/well) overnight and subsequently treated with different
concentrations of cholesterol (0, 2.5, 5 or 10 mM) for 12 h. After
removing the cell culture medium, 10 μl CCK-8 solution (Dojindo
Molecular Technologies, Inc.) were added to each well of the plate
and then incubated at 37°C for 2 h. Finally, the absorbance was
measured at 450 nm using a microplate reader.
miRNA transfection and mithramycin A
(MMA) treatment
For the transfection assay, MIN6 cells were seeded
in 24-well plates at a density of 2×105 cells/well and
grown overnight, followed by treatment with 5 mM cholesterol alone
or in combination with 40 nM miR-24 mimic (cat. no.
miR10000219-1-5; sequences commercially unavailable)/80 nM miR-24
inhibitor (cat. no. miR20000219-1-5; sequences commercially
unavailable) for 48 h using the riboFECT™CP reagent according to
the manufacturer's protocol (Guangzhou RiboBio Co., Ltd.).
Meanwhile, a scrambled random miRNA sequence
(5′-UUCUCCGAACGUGUCACGUTT-3′; 40 nM), which was commercially
synthesized by Guangzhou RiboBio Co., Ltd., was used to treat the
control cells. The expression level of miR-24 in these cells was
identified at 48 h after transfection. In addition, MIN6 cells were
pretreated with 10 μM MMA for 2 h and then subjected to the GSIS
assay to identify the effect of direct inhibition of Sp1 expression
on insulin secretion.
GSIS assay and determination of the
insulin content
Insulin secretion and the insulin content in MIN6
cells were measured using an ELISA and normalized to the DNA
content. Briefly, MIN6 cells were cultured in 24-well plates
(2×105 cells/well) and grown overnight, and then
incubated with cholesterol or in combination with the miR-24
mimic/miR-24 inhibitor for 48 h, the cells were subsequently
incubated with KRB buffer supplemented with 3.3 mM glucose [5 mM
KCl, 120 mM NaCl, 24 mM NaHCO3, 15 mM Hepes (pH 7.4), 1
mM MgCl2, 2 mM CaCl2, 1 mg/ml ovalbumin] for
1 h. Next, cells were incubated with KRB buffer containing a
stimulatory concentration of glucose (16.7 mM) for 30 min and the
supernatant was collected to measure insulin concentration. After
the GSIS assay, the cells were lysed in 200 μl of M-PER Mammalian
Protein Extraction Buffer (Thermo Fisher Scientific, Inc.) and then
centrifuged at 4,200 × g for 5 min at 4°C, and the supernatants
were harvested for the insulin content assay using a Mouse High
Range Insulin ELISA kit (cat. no. 80-INSMSH-E01; ALPCO). The
insulin concentrations were calculated and normalized to the DNA
content, as determined using a Quant-iT™ dsDNA assay kit (Thermo
Fisher Scientific, Inc.).
Immunofluorescence staining
Immediately after the GSIS assay, the insulin
content in MIN6 cells (2×105 cells/well) was also
detected using immunofluorescence staining. Briefly, cells were
fixed with a freshly prepared solution of 4% paraformaldehyde for
20 min at room temperature, followed by permeabilization with 0.1%
Triton X-100 for 10 min. After blocking with PBS-Tween-3% bovine
serum albumin (Sigma-Aldrich; Merck KGaA) for 1 h, cells were
incubated with a primary antibody against insulin (cat. no. I2018;
1:200; Sigma-Aldrich; Merck KGaA) overnight at 4°C and then
incubated with a fluorescein isothiocyanate-conjugated secondary
antibody (cat. no. 711-545-1500; 1:200; Jackson ImmunoResearch
Laboratories, Inc.) for 1 h, followed by nuclear staining with
DAPI. Images of the immunofluorescence staining were captured with
a fluorescence microscope (Carl Zeiss AG).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted using the TRIzol reagent
(Thermo Fisher Scientific, Inc.) and reverse transcribed with MMLV
Reverse Transcription Reagents at 37°C for 1 h (Promega
Corporation). Relative qPCR was performed using SYBR-Green
detection chemistry (Qiagen, Inc.) on a PRISM 7500HT Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling reaction consisted of the following conditions: 1
cycle at 95°C for 15 min, followed by 40 cycles of a denaturation
step at 95°C for 15 sec, an annealing step at 60°C for 30 sec, and
an extension step at 70°C for 30 sec. The expression levels of
miR-24, Ins1, Ins2 and Scgn were validated
using RT-qPCR. The U6 small nuclear RNA and Gapdh were used
as endogenous controls for miRNA and mRNAs, respectively, and the
relative gene expression data were analyzed using the
2−ΔΔCq method (14).
The primers used for RT-qPCR are described in Table I.
 | Table IPrimers sequences used for
miR-24, Ins1, Ins2 and Scgn
amplification. |
Table I
Primers sequences used for
miR-24, Ins1, Ins2 and Scgn
amplification.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| miR-24 |
TGGCTCAGTTCAGCAGGAACAG | Unique
quantitative-PCR reverse primer from the cDNA Synthesis kit |
| Ins1 |
CTTGCCCTCTGGGAGCCCA |
TGAAGGTCCCCGGGGCTTC |
| Ins2 |
CCACAAGTGGCACAACTGGA |
CTACAATGCCACGCTTCTGC |
| Scgn |
CCCAGAAGTGGATGGATTTG |
GTTGGGGATCAGGGGTTTAT |
Bioinformatics analysis and luciferase
reporter assays
The miR-24 sequences were obtained from miRBase
database and its target genes were predicted using online websites,
TargetScan and miRanda. The minimum free energy of the
hybridization between Sp1 3′UTR and miR-24 was analyzed with
RNAhybrid. A total of transcription factor prediction databases,
PROMO, Tfsitescan and DBD, were used to predict whether there is a
direct interaction between Sp1 and the Ins1 promoter. The
detailed websites of these databases are listed in the
supplementary file: Table
SI.
The 3′UTR of Sp1, which was predicted to
contain the putative target site for miR-24, was amplified by
RT-PCR from total RNA extracted from MIN6 cells with primers
containing XhoI and NotI sites. For comparison,
site-directed mutagenesis was used to introduce the seed region of
the predicted miR-24 binding sites within the 3′UTR of Sp1
using a Multisite-Quick Change kit (Agilent Technologies, Inc.).
Wild-type and mutant inserts were cloned into a psiCHECK™-2 vector
(Promega Corporation). Each recombinant plasmid was confirmed by
sequencing (Takara Biotechnology Co., Ltd.) and then cotrans-fected
with a miR-24 mimic into 293T cells (Fuheng Cell Center).
Luciferase activity was measured using the Dual-Glo Luciferase
Assay System (Promega Corporation) at 48 h post-transfection. The
relative luciferase activity of each group was reported as the
percentage of Renilla luciferase activity normalized to the
corresponding firefly luciferase activity.
Western blotting analysis
At 30 min after stimulation with high glucose (16.7
mM), MIN6 cells were washed twice with PBS and cellular proteins
were extracted with M-PER Mammalian Protein Extraction Buffer
(Thermo Fisher Scientific, Inc.). Protein concentration was
measured using a BCA protein quantitation kit (Beyotime Institute
of Biotechnology). Total cellular proteins (30 μg) were
fractionated on 10% SDS-PAGE gels. Proteins were transferred to
PVDF membranes and then western blotting was performed according to
standard procedures described in a previous study (15). The primary antibodies used were as
follows: Anti-Scgn antibody (cat. no. ab137017; 1:2,000; Abcam),
anti-Sp1 antibody (cat. no. ab227383; 1:1,000), anti-focal adhesion
kinase (FAK) antibody (cat. no. 3285; 1:1,000; Cell Signaling
Technology, Inc.), anti-phospho-FAK (Tyr397) antibody
(cat. no. 3283; 1:1,000; Cell Signaling Technology, Inc.),
anti-paxillin antibody (cat. no. 2542; 1:1,000; Cell Signaling
Technology, Inc.), anti-phospho-paxillin (Tyr118)
antibody (cat. no. 2541; 1:1,000; Cell Signaling Technology, Inc.)
and β-actin antibody (cat. no. 3700; 1:2,000; Cell Signaling
Technology, Inc.). Accordingly, a horseradish peroxidase-conjugated
goat anti-rabbit immunoglobulin G secondary antibody (cat. no.
A0208; 1:2,000; Beyotime Institute of Biotechnology) and goat
anti-mouse antibody (cat. no. A0216; 1:2,000; Beyotime Institute of
Biotechnology) were used to detect the proteins.
Statistical analysis
All data are presented as the mean ± standard
deviation and significant differences were assessed using one-way
ANOVA followed by Student-Newman-Keuls test. SPSS statistical
software (version 18.0; IBM Corp.) was used to conduct the data
analysis. P<0.05 was considered to indicate a statistically
significant difference. Independent experiments were performed at
least three times.
Results
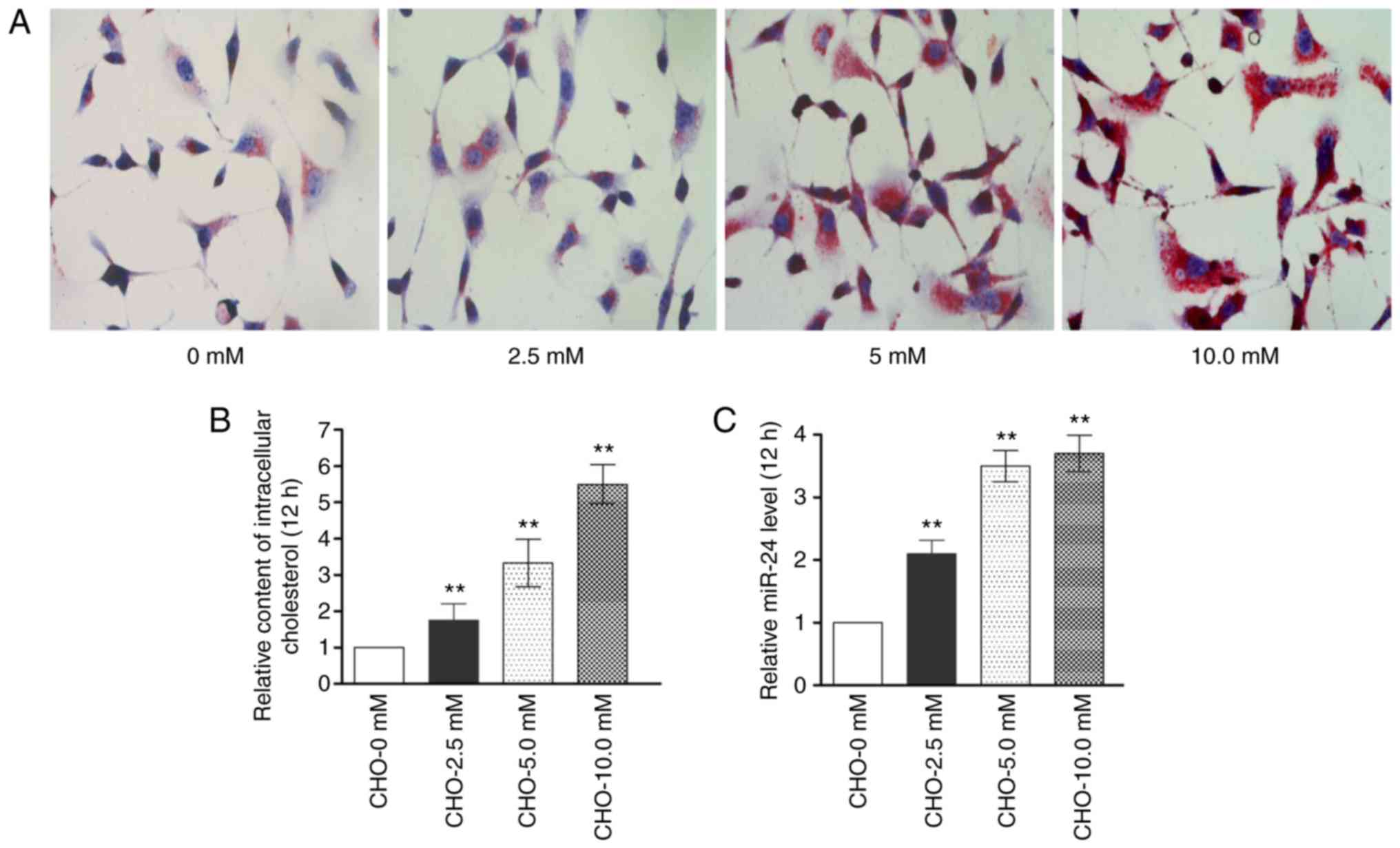
Accumulation of cholesterol in MIN6 cells
induces miR-24 expression
A total of 4 groups of cells treated with increasing
concentrations of cholesterol (0, 2.5, 5.0 or 10.0 mM for 12 h)
were evaluated using RT-qPCR to determine the changes in miR-24
expression in cholesterol-treated MIN6 cells. Cholesterol induced a
dose-dependent increase in the intracellular cholesterol content in
MIN6 cells (Fig. 1A and B).
Meanwhile, miR-24 expression was significantly increased as the
cholesterol concentration increased from 0-5 mM (P<0.01) and an
increase, although to a lesser extent, was observed as the
cholesterol concentration further increased from 5-10 mM (Fig. 1C). Since this study focuses on
insulin biosynthesis and secretion in β-cells, a 5 mM soluble
cholesterol concentration which is greater than the concentration
in normal culture medium was demonstrated to have little impact on
cell viability (Fig. S1A);
therefore, this concentration was used in the subsequent
experiments.
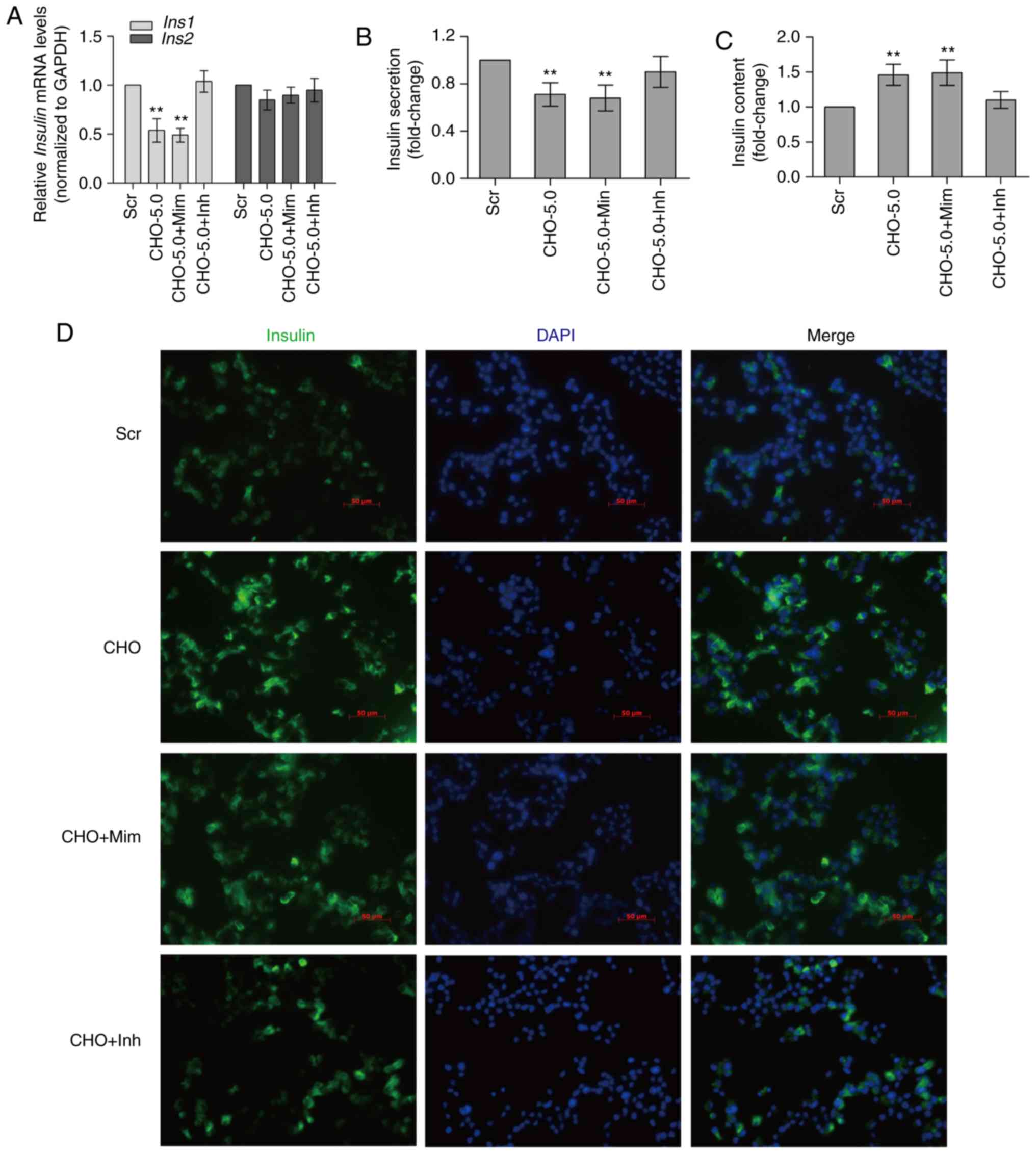
Cholesterol accumulation in MIN6 cells
inhibits Ins1 expression and insulin secretion
The effects of cholesterol accumulation in MIN6
cells on insulin mRNA expression and insulin secretion were
determined by exposing cells to 5 mM cholesterol for 12 h. Based on
the results of the RT-qPCR assays, the expression of the
Ins1 mRNA was significantly reduced in MIN6 cells treated
with cholesterol alone or in combination with 40 nM miR-24 mimic
(P<0.01), whereas the expression of Ins2 was not
decreased (Fig. 2A). Meanwhile,
GSIS was significantly reduced in cells treated with cholesterol
alone or in combination with the miR-24 mimic (P<0.01; Fig. 2B). In general, the intracellular
insulin content increased by ~50% in β-cells treated with
cholesterol alone or in combination with the miR-24 mimic,
indicating that cholesterol alone or in combination with the miR-24
mimic inhibited insulin secretion to a greater extent than insulin
transcription. Notably, the miR-24 inhibitor rescued the reductions
in insulin transcription and secretion, particularly insulin
secretion, induced by the treatment with cholesterol alone or in
combination with the miR-24 mimic, which subsequently decreased the
insulin content in MIN6 cells (Fig.
2C-D). Therefore, the cholesterol-induced decrease in insulin
secretion is associated with increased miR-24 expression.
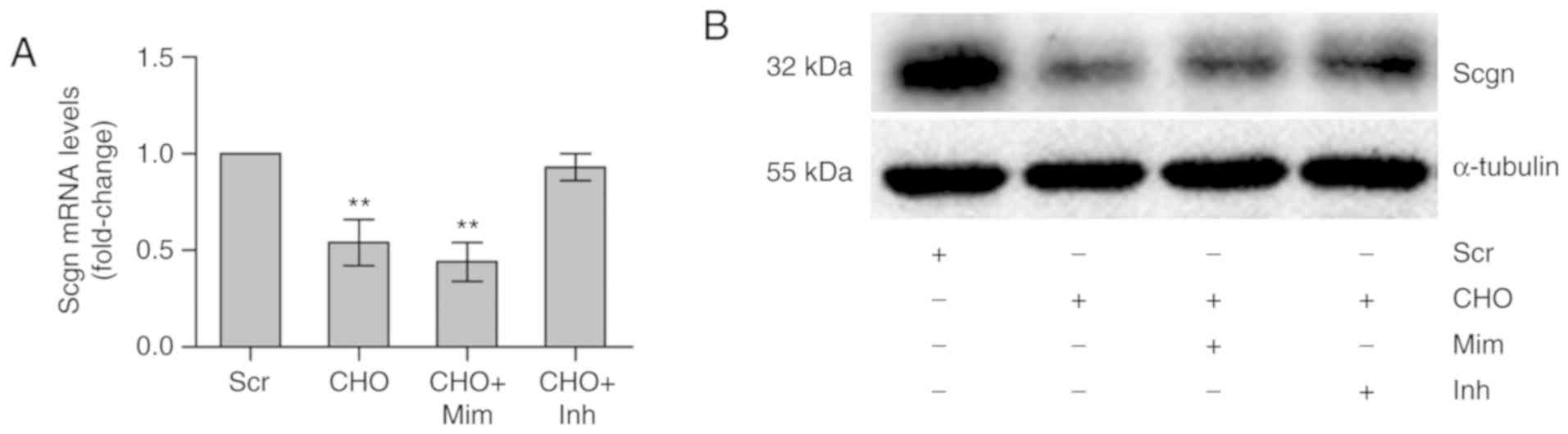
Cholesterol inhibits the expression of
Scgn at both the mRNA and protein levels
The expression of Scgn was evaluated in MIN6
cells treated with cholesterol alone or in combination with a
miR-24 mimic/miR-24 inhibitor to investigate the mechanism
underlying the effect of cholesterol on insulin secretion. After
confirming the successful modulation of miR-24 expression (Fig. S1B), cholesterol alone or in
combination with a miR-24 mimic significantly reduced in Scgn
expression at both the mRNA and protein levels (P<0.01), whereas
the reduction was markedly rescued after co-incubation with a
miR-24 inhibitor for 12 h (Fig.
3). These results indicate a regulatory relationship between
miR-24 and Scgn.
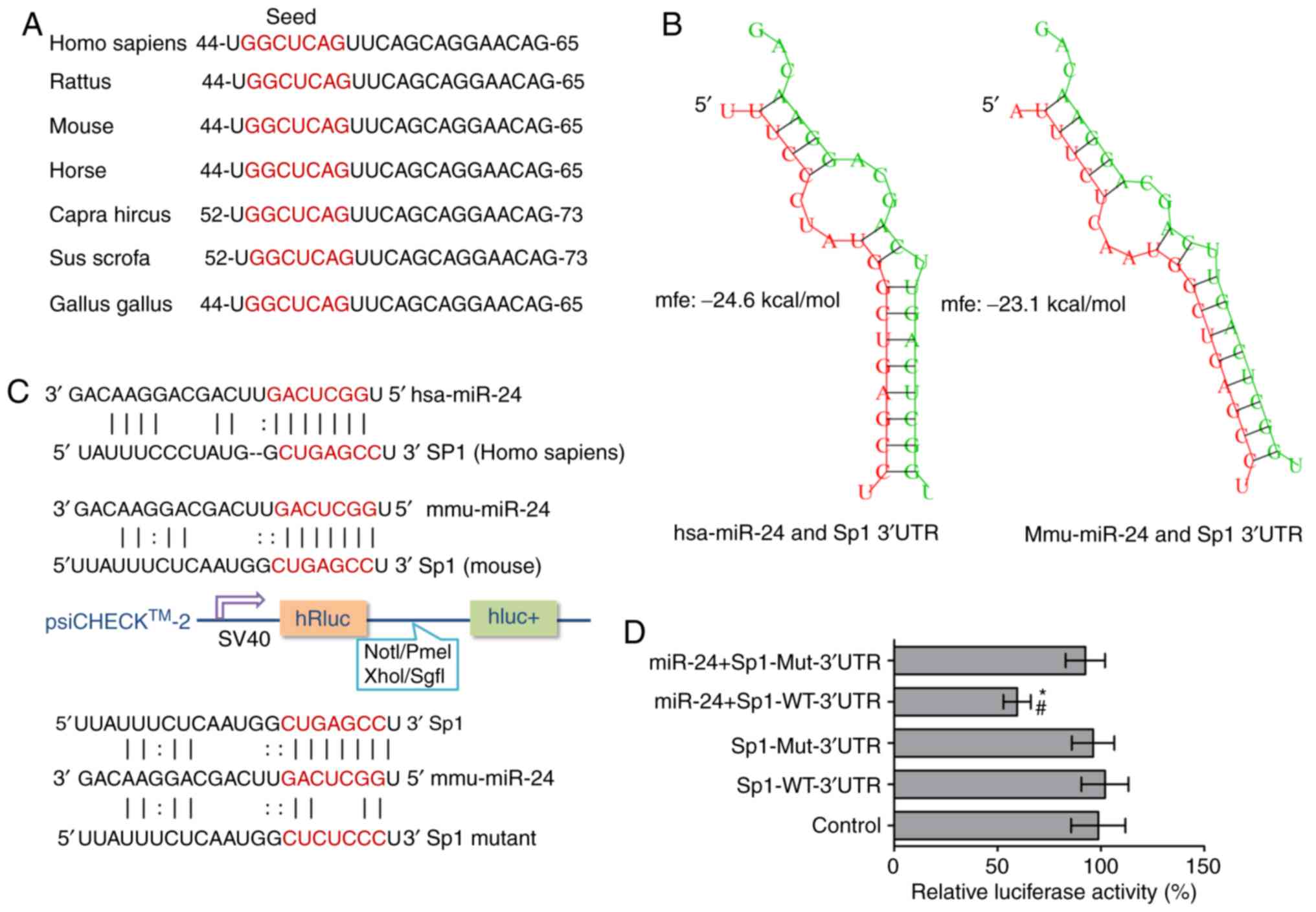
Sp1 is a direct target gene of
miR-24
Notably, miR-24 is broadly conserved among diverse
species, indicating that it has biological functions under
physiological and/or pathological conditions (Fig. 4A). Although miR-24 expression was
significantly increased in cholesterol-treated MIN6 cells, its
function in insulin secretion is largely unknown. Potential target
genes of miR-24 were first searched for using the online analysis
programs mentioned in the methods to further investigate the
regulatory relationship between miR-24 and Scgn, but
putative miR-24 binding sites within the 3′UTR of the Scgn
mRNA were not identified. However, Sp1, an experimentally confirmed
transcriptional activator of the Scgn gene (11), contains a putative miR-24 binding
site within its 3′UTR. Interestingly, low free energy scores (in
RNAhybrid) for the hybridization of miR-24 and the Sp1 mRNA
were observed in mice and humans, suggesting that miR-24 most
likely directly targets the 3′UTR of the Sp1 mRNA and
represses its expression (Fig.
4B). Furthermore, in silico promoter analysis excluded a
potential direct interaction between Sp1 and the Ins1
promoter (data not shown).
Next, a luciferase reporter assay was performed in
293T cells to verify the direct targeting effect of miR-24 on the
3′UTR of the Sp1 mRNA. psi-CHECK™-2 luciferase vectors were
constructed harboring the wild-type (Sp1-WT) or mutant (Sp1-Mut)
sequence of the mouse Sp1 3′UTR, in which the putative
miR-24 binding site 5′-CUGAGCC-3′ was mutated to 5′-CUCUCCC-3′
(Fig. 4C). Meanwhile, a vector
containing a nonrelated cDNA fragment was constructed as a control.
Following the cotransfection of the miR-24 mimic with Sp1-WT,
Sp1-Mut or a control, the luciferase activity in the Sp1-WT
transfected cells was significantly reduced by the miR-24 mimic
(P<0.05), whereas it was not affected in the Sp1-Mut and control
groups (Fig. 4D). Based on these
results, miR-24 plays a crucial role in the posttranscriptional
repression of Sp1 expression via directly targeting its
3′UTR.
miR-24 regulates the Scgn/FAK/paxillin
pathway in cholesterol-treated MIN6 cell by targeting Sp1
The miR-24 mimic or miR-24 inhibitor was transfected
into cholesterol-treated MIN6 cells and the levels of Sp1, Scgn,
FAK, phosphorylated (p)-FAK, Paxillin and p-paxillin were measured
using western blotting to confirm whether miR-24 inhibited insulin
secretion by directly regulating Sp1 expression and the GSIS
pathway. Cholesterol-induced over-expression of miR-24 decreased
the levels of Sp1, Scgn and its downstream focal adhesion
molecules, p-FAK and p-paxillin, during GSIS. In contrast, the
miR-24 inhibitor significantly rescued the reduction in Sp1, Scgn,
p-FAK and p-paxillin levels during the GSIS process (P<0.01;
Figs. 3B and 5).
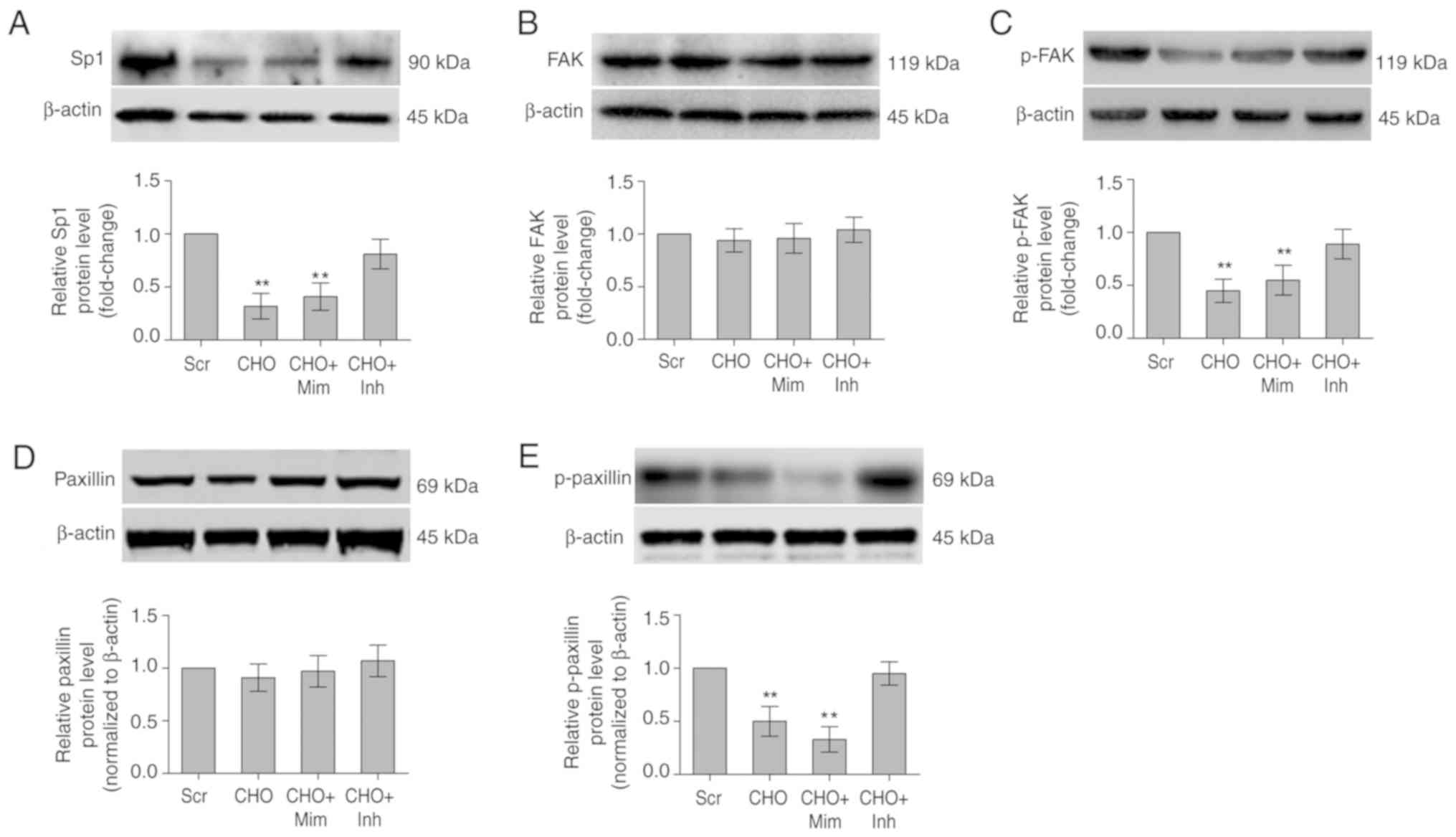 | Figure 5CHO affects the levels of proteins
involved in the Sp1/Scgn-FAK signaling pathway mediating insulin
trafficking. Changes in the levels of the Sp1, FAK, p-FAK, paxillin
and p-paxillin proteins in MIN6 cells treated with 5.0 mM CHO alone
or in combination with 40 nM miR-24 mimic/80 nM miR-24 inhibitor
for 12 h were measured using western blotting assay. β-Actin served
as the loading control. (A) Sp1, (B) FAK, (C) p-FAK (D) paxillin
and (E) p-paxillin. **P<0.01 vs. the Scr-treated
group. Scr, scrambled miRNA sequence; CHO, cholesterol; Mim, miR-24
mimic; Inh, miR-24 inhibitor; Scgn, secretagogin; p-FAK,
phosphorylated-focal adhesion kinase; miR, microRNA. |
In parallel, an experiment designed to identify the
effect of direct inhibition of Sp1 function on insulin secretion in
MIN6 cells was conducted. The inhibition of Sp1 function by a
pretreatment with the Sp1 inhibitor MMA (10 mM for 2 h) decreased
the levels of the Scgn, p-FAK and p-paxillin proteins and
attenuated GSIS (Fig. S2).
Furthermore, online analysis of the Ins1 promoter sequence
did not identify a Sp1-binding site, which excludes a direct
interaction between the Sp1 protein and this mRNA. Therefore,
cholesterol-induced overexpression of miR-24 inhibits GSIS by
negatively regulating Sp1 expression and the
Scgn/FAK/paxillin pathway.
Discussion
Type 2 diabetes is often associated with
hypercholesterolemia. Previous evidence has shown that excessive
accumulation of palmitate in β-cells could lead to β-cell
dysfunction and the development of diabetes (10,16,17). In addition to the uptake of
palmitate, β-cells also take up cholesterol through their
high-affinity low density lipid receptors (5). Cholesterol accumulation in the
islets may contribute to insulin-producing β-cell dysfunction and
the loss of GSIS. However, a role for hypercholesterolemia in the
pathogenesis of type 2 diabetes is not well established. In the
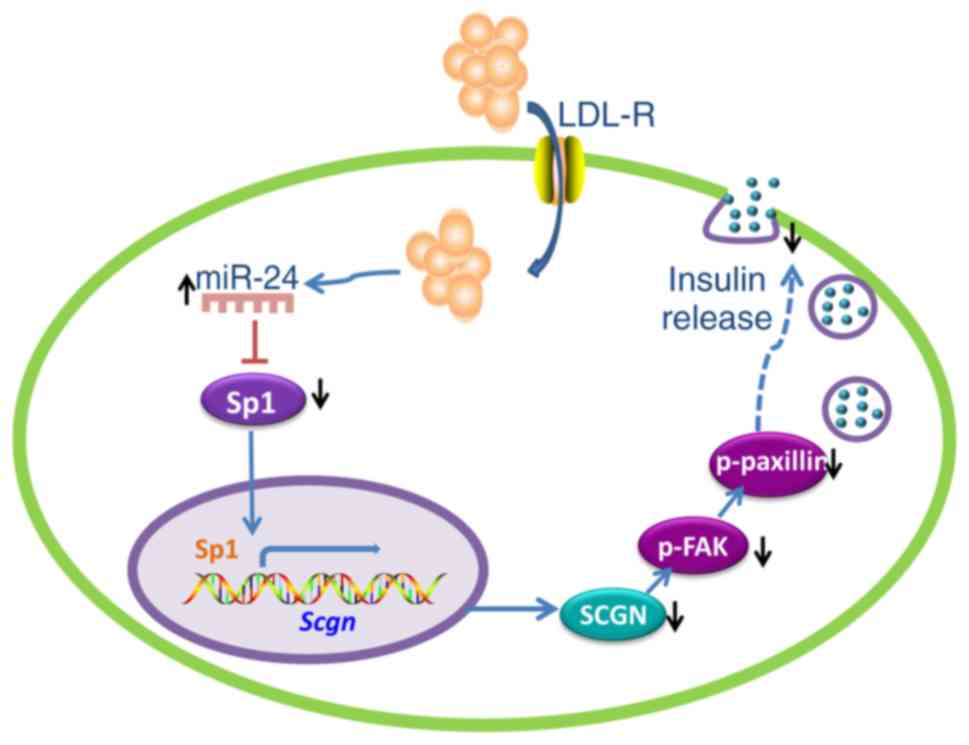
present study, a novel molecular mechanism was investigated by
which cholesterol inhibits insulin secretion in MIN6 cells. As
shown in Fig. 6, cholesterol
accumulation in MIN6 cells increased the expression of miR-24 and
miR-24 overexpression silenced the expression of Sp1 by
directly targeting its 3′UTR. As a transcriptional activator of
Scgn, downregulation of Sp1 resulted in a concomitant
reduction in Scgn expression, followed by a decrease in the levels
of the downstream proteins p-FAK and p-paxillin, which regulate the
focal adhesion process in insulin granules. GSIS was subsequently
impaired.
As shown in recent studies, miRNAs are key factors
involved in the mechanisms regulating insulin biosynthesis, the
trafficking of granules and insulin secretion (18-20). The highest expression of miR-24
was observed in islets isolated from genetically obese or high-fat
diet-fed mice, as well as in islets treated with palmitate
(10). In the study by Zhu et
al (10), miR-24 repressed
the insulin biosynthesis process by targeting two maturity onset
diabetes of the young genes, Hnf1a and Neurod1,
leading to reductions in both basal and stimulated-insulin
secretion. In addition to affecting insulin biosynthesis, further
studies are needed to determine whether miR-24 also regulates the
insulin secretion process. In the present study, dose-dependent
cholesterol accumulation concomitant with miR-24 upregulation in
cholesterol-treated MIN6 cells was revealed. Intriguingly,
following the increase in miR-24 expression, Ins1 mRNA
expression was reduced, whereas the Ins2 mRNA was
unaffected. These results are consistent with existing evidence
that Ins1 expression is significantly decreased and
Ins2 expression is not affected in NeuroD conditional
knockout mice (21), suggesting
that cholesterol-induced miR-24 upregulation in MIN6 may repress
Ins1 expression by targeting NeuroD. The level of
Ins2 mRNA is 9-fold higher than the Ins1 mRNA in MIN6
cells (22), therefore,
miR-24-mediated Ins1 mRNA silencing may have a limited
effect on insulin protein biosynthesis.
Since a significant decrease in insulin secretion
and a marked increase in the insulin content were observed in
cholesterol-treated MIN6 cells after GSIS, changes in the
expression of Scgn, a key regulator of insulin secretion that was
recently identified were subsequently detected (12,13,23,24) and observed a dramatic decrease in
response to the cholesterol treatment. A bioinformatics analysis
was next conducted to predict interactions between miR-24 and
Scgn. Although a direct interaction between miR-24 and
Scgn was not identified, a putative miR-24 binding site
within the Sp1 3′UTR was predicted and experimentally
identified. Following the downregulation of Sp1, the
expression of Scgn, which is transcriptionally regulated by
Sp1 (11), was subsequently
downregulated. It was excluded a potential direct interaction
between Sp1 and the Ins1 promoter, which further supports
that miR-24 decreases insulin secretion rather than insulin
biosynthesis through Sp1.
Scgn is specifically expressed in pancreatic islets
at high levels and further elevated in islets and plasma from
patients and rats with type 2 diabetes mellitus (12,25-27). Scgn is a pivotal regulator of
insulin secretion that functions by activating focal adhesion
complexes, including FAK, paxillin and F-actin, and modulating the
focal adhesion process (13,28). Silencing of focal adhesion
complexes impairs the docking and release of insulin granules
(29,30). Consistent with the results of
these studies, the present study observed decreased levels of p-FAK
and paxillin following the downregulation of Scgn. Notably, the
suppression of the Sp1/Scgn/FAK signaling pathway was rescued by a
miR-24 inhibitor, which further confirmed that a miR-24-to-Scgn
regulatory pathway mediates cholesterol-induced inhibition of
insulin secretion.
Collectively, cholesterol accumulation in MIN6
pancreatic β-cells increases miR-24 expression and miR-24 is a
negative regulator of Sp1. The miR-24-to-Scgn regulatory pathway
was proven to regulate focal adhesion, a critical step in insulin
secretion, in cholesterol-treated MIN6 cells. Moreover, miR-24
inhibition is probably a therapeutic strategy for
cholesterol-induced β-cell dysfunction by regulating Scgn-mediated
insulin secretion. However, understanding of the relationship
between hypercholesterolemia and the miR-24-to-Scgn regulatory
pathway is still at an early stage. Further investigations are
warranted to illuminate how cholesterol accumulation increases the
expression of miR-24 in β-cells and whether miR-24 manipulates
insulin secretion by targeting other genes. Further studies
designed to establish whether the miR-24-to-Scgn pathway is
impaired during insulin exocytosis in patients with
hypercholesterolemia in vivo are needed.
Supplementary Data
Abbreviations:
|
GSIS
|
glucose-stimulated insulin
secretion
|
Acknowledgments
Not applicable.
Funding
The present study was financially support by The
National Natural Sciences Foundation of China (grant no. 81770460),
The Aid Program (grant no. 2017KJ268), The Construction Program of
Innovation Platform (grant no. 2017KJ182) from Science and
Technology Bureau of Hengyang City, and The Third Level of
Chuanshan Talent Project of University of South China (grant no.
2017CST20).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XHX and JHL designed the experiments. JY, YCL, ZBZ,
WL, SMX, LZZ, LLO and ABG performed the experiments. BY and HL
analyzed the data. JY and LLO wrote and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brunham LR, Kruit JK, Verchere CB and
Hayden MR: Cholesterol in islet dysfunction and type 2 diabetes. J
Clin Invest. 118:403–408. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hao M, Head WS, Gunawardana SC, Hasty AH
and Piston DW: Direct effect of cholesterol on insulin secretion: A
novel mechanism for pancreatic beta-cell dysfunction. Diabetes.
56:2328–2338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu Y, Toomre DK, Bogan JS and Hao M:
Excess cholesterol inhibits glucose-stimulated fusion pore dynamics
in insulin exocytosis. J Cell Mol Med. 21:2950–2962. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bogan JS, Xu Y and Hao M: Cholesterol
accumulation increases insulin granule size and impairs membrane
trafficking. Traffic. 13:1466–1480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kruit JK, Kremer PH, Dai L, Tang R, Ruddle
P, de Haan W, Brunham LR, Verchere CB and Hayden MR: Cholesterol
efflux via ATP-binding cassette transporter A1 (ABCA1) and
cholesterol uptake via the LDL receptor influences
cholesterol-induced impairment of beta cell function in mice.
Diabetologia. 53:1110–1119. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Osmai M, Osmai Y, Bang-Berthelsen CH,
Pallesen EM, Vestergaard AL, Novotny GW, Pociot F and
Mandrup-Poulsen T: MicroRNAs as regulators of beta-cell function
and dysfunction. Diabetes Metab Res Rev. 32:334–349. 2016.
View Article : Google Scholar
|
|
8
|
Li Y, Luo T, Wang L, Wu J and Guo S:
MicroRNA-19a-3p enhances the proliferation and insulin secretion,
while it inhibits the apoptosis of pancreatic cells via the
inhibition of SOCS3. Int J Mol Med. 38:1515–1524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tugay K, Guay C, Marques AC, Allagnat F,
Locke JM, Harries LW, Rutter GA and Regazzi R: Role of microRNAs in
the age-associated decline of pancreatic beta cell function in rat
islets. Diabetologia. 59:161–169. 2016. View Article : Google Scholar
|
|
10
|
Zhu Y, You W, Wang H, Li Y, Qiao N, Shi Y,
Zhang C, Bleich D and Han X: MicroRNA-24/MODY gene regulatory
pathway mediates pancreatic beta-cell dysfunction. Diabetes.
62:3194–3206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malenczyk K, Girach F, Szodorai E, Storm
P, Segerstolpe A, Tortoriello G, Schnell R, Mulder J, Romanov RA,
Borok E, et al: A TRPV1-to-secretagogin regulatory axis controls
pancreatic beta-cell survival by modulating protein turnover. EMBO
J. 36:2107–2125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wagner L, Oliyarnyk O, Gartner W, Nowotny
P, Groeger M, Kaserer K, Waldhausl W and Pasternack MS: Cloning and
expression of secretagogin, a novel neuroendocrine- and pancreatic
islet of langerhans-specific Ca2+-binding protein. J Biol Chem.
275:24740–24751. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang SY, Lee JJ, Lee JH, Lee K, Oh SH, Lim
YM, Lee MS and Lee KJ: Secretagogin affects insulin secretion in
pancreatic beta-cells by regulating actin dynamics and focal
adhesion. Biochem J. 473:1791–1803. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Lv YC, Tang YY, Peng J, Zhao GJ, Yang J,
Yao F, Ouyang XP, He PP, Xie W, Tan YL, et al: MicroRNA-19b
promotes macrophage cholesterol accumulation and aortic
atherosclerosis by targeting ATP-binding cassette transporter A1.
Atherosclerosis. 236:215–226. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Staaf J, Ubhayasekera SJ, Sargsyan E,
Chowdhury A, Kristinsson H, Manell H, Bergquist J, Forslund A and
Bergsten P: Initial hyperinsulinemia and subsequent beta-cell
dysfunction is associated with elevated palmitate levels. Pediatr
Res. 80:267–274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barlow J, Jensen VH, Jastroch M and
Affourtit C: Palmitate-induced impairment of glucose-stimulated
insulin secretion precedes mitochondrial dysfunction in mouse
pancreatic islets. Biochem J. 473:487–496. 2016. View Article : Google Scholar
|
|
18
|
Tiwari J, Gupta G, de Jesus Andreoli Pinto
T, Sharma R, Pabreja K, Matta Y, Arora N, Mishra A, Sharma R and
Dua K: Role of microRNAs (miRNAs) in the pathophysiology of
diabetes mellitus. Panminerva Med. 60:25–28. 2018.
|
|
19
|
Hashimoto N and Tanaka T: Role of miRNAs
in the pathogenesis and susceptibility of diabetes mellitus. J Hum
Genet. 62:141–150. 2017. View Article : Google Scholar
|
|
20
|
Calderari S, Diawara MR, Garaud A and
Gauguier D: Biological roles of microRNAs in the control of insulin
secretion and action. Physiol Genomics. 49:1–10. 2017. View Article : Google Scholar
|
|
21
|
Gu C, Stein GH, Pan N, Goebbels S,
Hornberg H, Nave KA, Herrera P, White P, Kaestner KH, Sussel L and
Lee JE: Pancreatic beta cells require NeuroD to achieve and
maintain functional maturity. Cell Metab. 11:298–310. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang ZW, Zhang LQ, Ding L, Wang F, Sun
YJ, An Y, Zhao Y, Li YH and Teng CB: MicroRNA-19b downregulates
insulin 1 through targeting transcription factor neuroD1. FEBS
Lett. 585:2592–2598. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JJ, Yang SY, Park J, Ferrell JE Jr,
Shin DH and Lee KJ: Calcium ion induced structural changes promote
dimerization of secretagogin, which is required for its insulin
secretory function. Sci Rep. 7:69762017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kobayashi M, Yamato E, Tanabe K, Tashiro
F, Miyazaki S and Miyazaki J: Functional analysis of novel
candidate regulators of insulin secretion in the MIN6 mouse
pancreatic beta cell line. PLoS One. 11:e01519272016. View Article : Google Scholar
|
|
25
|
Bazwinsky-Wutschke I, Wolgast S, Muhlbauer
E and Peschke E: Distribution patterns of calcium-binding proteins
in pancreatic tissue of non-diabetic as well as type 2 diabetic
rats and in rat insulinoma beta-cells (INS-1). Histochem Cell Biol.
134:115–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Segerstolpe A, Palasantza A, Eliasson P,
Andersson EM, Andreasson AC, Sun X, Picelli S, Sabirsh A, Clausen
M, Bjursell MK, et al: Single-cell transcriptome profiling of human
pancreatic islets in health and Type 2 diabetes. Cell Metabo.
24:593–607. 2016. View Article : Google Scholar
|
|
27
|
Hansson SF, Zhou AX, Vachet P, Eriksson
JW, Pereira MJ, Skrtic S, Jongsm Wallin H, Ericsson-Dahistrand A,
Karlsson D, Ahnmark A, et al: Secretagogin is increased in plasma
from type 2 diabetes patients and potentially reflects stress and
islet dysfunction. PLoS One. 13:e01966012018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rorsman P and Renstrom E: Insulin granule
dynamics in pancreatic beta cells. Diabetologia. 46:1029–1045.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Heaslip AT, Nelson SR, Lombardo AT, Beck
Previs S, Armstrong J and Warshaw DM: Cytoskeletal dependence of
insulin granule movement dynamics in INS-1 beta-cells in response
to glucose. PLoS One. 9. pp. e1090822014, View Article : Google Scholar
|
|
30
|
Rondas D, Tomas A and Halban PA: Focal
adhesion remodeling is crucial for glucose-stimulated insulin
secretion and involves activation of focal adhesion kinase and
paxillin. Diabetes. 60:1146–1157. 2011. View Article : Google Scholar : PubMed/NCBI
|