Introduction
Rheumatoid arthritis (RA), a systemic and autoimmune
disease, is characterized by chronic inflammation, synovial
hyperplasia, joint swelling and tenderness. This disease mainly
occurs in individuals aged between 20 and 50 years old and its
morbidity rate in women is 2-3 times higher than that in men,
particularly in older women. The pathological changes of RA result
in functional limitations, working disability and poor quality of
life (1-4). One of the important therapeutic
approaches for RA in western medicinal therapy is to alleviate
pain. This control strategy is given priority mainly for
symptomatic treatment. However, this control strategy is not
satisfactory in the clinic, and side effects frequently occur
(5). Therefore, there is an
urgent need to develop novel treatment approaches for RA.
Investigations involving traditional Chinese medicine (TCM) for RA
have been developing for thousands of years. These ancient
therapies remain in use at present as they are empirically
effective and have stood the test of time (6,7).
Astragalus membranaceous (Fisch.), also known
as Huang Qi, is one of the most commonly used herbal medicines in
TCM (8). Astragalus and
its compounds have been widely used for the treatment of RA for
thousands of years in China (9-11).
Astragalosides (AST) are the effective elements extracted from
Astragalus, which mainly contains AST-I-VIII. Among these,
AST-IV is its main active ingredient. Previous studies have
demonstrated that AST has numerous biological effects, including
anti-inflammatory, anti-oxidation, immunological regulation and
anti-aging effects (12-15).
Accumulating evidence has suggested that, rather
than being transcriptional noise, a number of non-coding RNAs
(ncRNAs) serve as master regulators that affect the expression
levels of dozens or even hundreds of target genes (16,17). Among them, long non-coding RNAs
(lncRNAs), a recently discovered class of non-coding RNAs, are
mRNA-like transcripts >200 nucleotides in length that have no or
little protein-coding capacity and serve important roles in a
variety of biological processes, including transcription, splicing
and translation (18). In
addition, lncRNAs are known to be associated with the pathogenesis
of different diseases, including RA (19-21). However, to the best of our
knowledge, there has been minimal focus on the differentially
expressed lncRNAs in RA following AST treatment.
In the present study, in order to observe the effect
of AST on the differentially expressed lncRNAs in adjuvant
arthritis (AA) rats, and to evaluate the possible mechanism of AST,
the Arraystar Rat lncRNA microarray was used. The results will
advance current understanding of the possible mechanism underlying
AST in treatment for RA from the perspective of lncRNAs and provide
novel lncRNAs as potential treatment targets.
Materials and methods
Animal experiments
The protocol was approved by the Committee on the
Ethics of Animal Experiments of Anhui University of Chinese
Medicine (Hefei, China). A total of 40 male Sprague Dawley rats
(age, 6-8 weeks; SPF-grade; weight, 200±20 g) were obtained from
the Experimental Animal Center of Anhui Medical University (Hefei,
China). All rats were healthy and had never received drugs
previously. The animals were housed in the animal facilities of
Anhui University of Chinese Medicine. The animals were housed
individually under a set temperature (18-22°C) and humidity
(40-60%) with a 12-h light/dark cycle, free access to standard
laboratory food and water, and environmental noise kept to a
minimum range. All efforts were made to minimize animal suffering
and the number of animals used.
Drugs and reagents
AST was purchased from Nanjing Zhi Bai Cui Biology
Technology Co. Ltd. (Nanjing, China; purity >98.5%), mainly
containing AST-I-VIII. AST-IV was purchased from Nanjing Zelang
Medical Technology Co. Ltd. (Nanjing, China, purity >98%).
Freund's complete adjuvant (FCA) was purchased from Sigma; Merck
KGaA (Darmstadt, Germany). Sodium pentobarbital was obtained from
Shanghai Chemical Reagent Company (Shanghai, China). TRIzol reagent
was obtained from Invitrogen; Thermo Fisher Scientific, Inc.
(Waltham, MA, USA). The C-reactive protein (CRP) kit was obtained
from Beijing Lidman Co., Ltd. (Beijing, China). The white blood
cell (WBC), red blood cell (RBC), hemoglobin (HGB) and hematocrit
(HCT) kits were obtained from Sysmex Corporation (Shenhu, Japan).
The ELISA kits of interleukin (IL)-1β (cat. no. 20180622), IL-4
(cat. no. 20180526), IL-10 (cat. no. 20180615) and tumor necrosis
factor (TNF)-α (cat. no. 20180702) were obtained from Shanghai
Elisa Biotech Co., Ltd. (Shanghai, China).
Animal experiments
The rats were adaptively fed for 1 week. All rats
were divided randomly into a vehicle group, animal model group, AST
treatment (control) group and AST-IV group (n=10). The animal model
group and control group were established with a single
intracutaneous injection of 0.1 ml of FCA into the right hind
metatarsal footpad (22). The
vehicle group was administered with liquid paraffin for comparison
at the same time. From day 12 after injection of FCA, AST (200
mg/kg) and AST-IV (80 mg/kg) were administered by gavage once a day
for 12 days in the control group. The rats in the vehicle and
animal model groups were given 0.9% saline.
Hind paw swelling and polyarthritis
index
From day 16, swelling of the left hind paw was
measured and the poly-arthritis index was assessed once every 4
days until the animals were sacrificed. Swelling of the paw was
determined by water volume (23):
Swelling (∆ml)=paw thickness (ml) tested value-paw thickness (ml)
original value. The severity of arthritis was graded on a scale of
0-4 with the following criteria (24): 0=no edema or swelling, 1=slight
edema and limited erythema, 2=slight edema and erythema from the
ankle to the tarsal bone, 3=moderate edema and erythema from the
ankle to the tarsal bone, and 4=edema and erythema from the ankle
to the entire leg. The arthritis score for each rat was the sum of
the severity in all four limbs (maximum 16 points per rat).
Synovial tissue samples
On day 28 following immunization, the rats were
anesthetized by intraperitoneal injection with sodium pentobarbital
(60 mg/kg of body weight). Three rats were randomly selected from
each group. Synovial tissue samples were harvested from all rats
and stored at −80°C until use.
Histological examination
The ankle joints and synovial tissue were removed
and fixed in 10% neutral formalin and 2.5% glutaraldehyde
stationary liquid, respectively. Histological changes were
subsequently observed by H&E staining and transmission electron
microscopy (TEM).
Measurements of CRP, WBC, RBC, HGB and
HCT
The content of CRP in serum was measured using an
automatic biochemical analytic instrument; the levels of CRP, WBCs,
RBCs, HGB and HCT in the whole blood were measured using a complete
blood cell counter.
Measurements of IL-1β, TNF-α, IL-4 and
IL-10
The serum contents of IL-1β, TNF-α, IL-4 and IL-10
were determined using the respective ELISA kits, according to the
manufacturer's instructions.
Microarray and data analysis
RNA extraction
Total RNAs were extracted from the three groups
using TRIzol® reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RNA quantity and quality
were measured using a NanoDrop ND-1000 instrument. RNA integrity
was assessed by 2% standard denaturing agarose gel
electrophoresis.
Microarray
The rat lncRNA array was designed for profiling the
lncRNAs and mRNAs. Approximately 9,000 lncRNAs were selected from
the databases of NCBI RefSeq (https://www.ncbi.nlm.nih.gov/refseq/), UCSC
(http://genome.ucsc.edu/), all_mrna records and
orthologs of mouse lncRNAs. The probes for coding genes were
printed once, whereas the probes for lncRNAs were printed three
times. The probes for housekeeping genes and negative probes were
printed multiple times to ensure hybridization quality.
RNA labeling and array
hybridization
Sample labeling and array hybridization were
performed according to the Agilent One-Color Microarray-Based Gene
Expression Analysis protocol (Agilent Technology) with minor
modifications. Briefly, mRNA was purified from total RNA following
the removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation kit,
Epicentre, Illumina, Inc.). Each sample was then amplified in a
circulating water bath at 40°C for 2 h with a Quick Amp Labeling
kit, One-Color (Agilent Technologies, Inc.; cat. no. 5190-0442),
according to the manufacturer's protocol. Subsequently, each sample
was transcribed into fluorescent cRNA along the entire length of
the transcripts without 3′ bias utilizing a mixture of oligo (dT)
and random primers (Arraystar Flash RNA Labeling kit, Arraystar).
The labeled cRNAs were purified using an RNeasy Mini kit (Qiagen,
Inc.). The concentration and specific activity of the labeled cRNAs
(pmol Cy3/µg cRNA) were measured using the NanoDrop ND-1000
instrument. Subsequently, 1 µg of each labeled cRNA was
fragmented by adding 5 µl 10X blocking agent and 1 µl
25X fragmentation buffer, heating the mixture at 60°C for 30 min,
and finally adding 25 µl 2X GE hybridization buffer to
dilute the labeled cRNA. The hybridization solution (50 µl)
was dispensed into the gasket slide and assembled to the lncRNA
expression microarray slide. The slides were incubated for 17 h at
65°C in an Agilent hybridization oven. The hybrid-ized arrays were
washed The hybridized arrays were washed with Gene Expression Wash
Buffer 1 (cat. no. 5188-5325; Agilent Technologies) and Gene
Expression Wash Buffer 2 (cat. no. 5188-5326; Agilent
Technologies), and scanned using the Agilent DNA Microarray Scanner
(model G2505C) (25).
Data analysis
Agilent Feature Extraction software (version
11.0.1.1) was used to analyze the acquired array images.
Quantitative normalization and subsequent data processing were
performed using the GeneSpring GX v12.1 software package (Agilent
Technologies). Following quantitative normalization of the raw
data, lncRNAs and mRNAs that had at least three of nine samples
have flags in Present or Marginal ('All Targets Value') were
selected for further data analysis. Differentially expressed
lncRNAs and mRNAs were identified through P-value and fold change
filtering. Hierarchical clustering was performed using homemade
scripts.
Functional group analysis
To assess functional enrichment, Gene Ontology (GO)
biological process (BP) term and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses of the differentially expressed
mRNAs were performed using the Database for Annotation,
Visualization and Integration Discovery (https://david.ncifcrf.gov/; v6.8).
Co-expression network
It is well known that lncRNAs perform their function
by positively or negatively regulating gene expression at the
transcriptional and post-transcriptional level. Therefore, it was
inferred that the function of each lncRNA was based on the function
of connected mRNAs. Correlation between the expression of lncRNAs
and mRNAs was evaluated using Pearson's correlation coefficient
(PCC). The lncRNA-mRNA pairs with PCC ≥0.94 and P<0.05 were
selected to construct the co-expression network and was visualized
using Cytoscape 3.2.8 software (http://cytoscape.github.io/).
Key lncRNAs
The selection of key lncRNAs was based on two
aspects: i) node degree: It is well known that hub nodes serve
critical roles in the biological network, therefore all node
degrees of the lncRNA-mRNA network were calculated. ii) Correlation
between bioinformatics analysis and RA: GO and pathway annotations
were performed for those lncRNAs with the top 10 node degrees.
According to the node degree and bioinformatics analysis, four
lncRNAs were selected as potential key lncRNAs.
Key lncRNA-mRNA sub-networks
Every key lncRNA, its linked lncRNAs and mRNAs in
the co-expression network were extracted and used to reconstruct
new sub-networks in Cytoscape software. For further analysis, GO
and pathway annotations were performed for the key lncRNAs by using
their first mRNA neighbors in the key lncRNA-mRNA sub-networks.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
The total RNA was extracted using TRIzol reagent and
then cDNA was synthesized with PrimeScript™ RT reagent kit (Takara
Bio, Inc.) at 65°C for 5 min and 42°C for 60 min. RT-qPCR analysis
was performed using a LightCycler 480 II (Roche, Germany) with TB
Green™ Advantage® qPCR Premix (Takara Bio, Inc.). The
thermocycling conditions were as follows: Pre-denaturation at 95°C
for 5 min, 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The
expression of each lncRNA was determined as the fold change using
the 2−ΔΔCq method (26). The primers were designed and shown
in Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene name | Primer sequence
(5′-3′)
| Product length,
bp |
|---|
| Forward | Reverse |
|---|
| β-actin |
5′-CGAGTACAACCTTCTTGCAGC-3′ |
5′-ACCCATACCCACCATCACAC-3′ | 202 |
| MRAK012530 |
5′-TGATAACAGCATGAGACCCAA-3′ |
5′-AAGCCAAGAACAAGAGCACAA-3′ | 160 |
| MRAK132628 |
5′-TTCCAGAGGACCTTCTCAATG-3′ |
5′-GACAAATCCCACCCTTACCTAC-3′ | 171 |
| MRAK003448 |
5′-TTTTGTGAGTAAGGGAGGGTGA-3′ |
5′-GGGGATTAAGAAGCCATGCTAT-3′ | 157 |
| XR_006457 |
5′-CAGCAAGCAGGAGTACGATGA-3′ |
5′-AGTGACAGTCCGCCTAGAAGC-3′ | 71 |
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 statistical software package (SPSS, Inc., Chicago, IL,
USA). One-way analysis of variance was used to perform statistical
analysis with Tukey's post hoc test for multiple comparisons.
Numerical data are presented as the mean ± standard error.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Hind paw swelling and polyarthritis
index
Secondary inflammation was observed in the right
hind paws from day 12, which indicated successful establishment of
the AA animal model. The results showed that the hind paw swelling
in the AA rats was markedly increased compared with that in the
vehicle group and, between days 20 and 28, swelling of the right
hind paws was significantly attenuated by AST treatment (Table II). Compared with that in the
vehicle rats, the polyarthritis index of the AA rats was
significantly increased. Administration with AST resulted in a
marked decrease in the polyarthritis index compared with that in
the AA rats between days 20 and 28 (Table III).
 | Table IIEffect of astragalosides on hind paw
swelling in adjuvant arthritis rats. |
Table II
Effect of astragalosides on hind paw
swelling in adjuvant arthritis rats.
| Group | Dose, mg/kg | Day 16, ∆ml | Day 20, ∆ml | Day 24, ∆ml | Day 28, Δml |
|---|
| Vehicle | - | 0.203±0.070 | 0.235±0.077 | 0.258±0.066 | 0.288±0.068 |
| Animal model | - | 0.573±0.172a | 0.750±0.172a | 0.883±0.118a | 0.782±0.104a |
| Control | 200 | 0.526±0.138 | 0.605±0.129b | 0.705±0.138c | 0.632±0.112c |
 | Table IIIEffect of astragalosides on the
polyarthritis index in adjuvant arthritis rats. |
Table III
Effect of astragalosides on the
polyarthritis index in adjuvant arthritis rats.
| Group | Dose, mg/kg | Day 16 | Day 20 | Day 24 | Day 28 |
|---|
| Vehicle | - | 0.40±0.52 | 0.30±0.67 | 0.20±0.42 | 0.30±0.48 |
| Animal model | - | 6.90±2.47a | 8.50±2.12a | 9.80±1.93a | 8.00±1.76a |
| Control | 200 | 5.50±1.58 | 6.40±2.59b | 7.60±2.64b |
5.8±1.81c |
Levels of CRP, WBCs, RBCs, HGB and
HGB
As shown in Table
IV, compared with the vehicle group, the levels of CRP and WBCs
were significantly elevated in the animal model group. Compared
with those in the animal model group, the levels of CRP and WBCs
were decreased following treatment with AST. However, there were no
significant changes in the levels of RBCs, HGB or HCT among the
three groups.
 | Table IVEffect of astragalosides on
biomarkers of inflammation in adjuvant arthritis rats. |
Table IV
Effect of astragalosides on
biomarkers of inflammation in adjuvant arthritis rats.
| Group | Dose, mg/kg | CRP, ng/l | WBC,
109/l | RBC,
1012/l | HGB, g/l | HCT, % |
|---|
| Vehicle | - | 4.99±1.23 | 8.73±1.44 | 7.16±0.75 | 151.48±12.48 | 44.69±4.65 |
| Animal model | - | 13.45±2.58a | 12.85±1.47a | 7.53±0.66 | 142.2±12.25 | 43.04±4.15 |
| Control | 200 | 10.27±2.86b | 10.69±1.69b | 7.26±0.77 | 145.23±14.93 | 44.07±4.89 |
Levels of IL-1β, TNF-α, IL-4 and
IL-10
Compared with the vehicle group, the animal model
group exhibited higher levels of IL-1β and TNF-α, and lower levels
of IL-4 and IL-10. Following AST intervention, compared with those
in the animal model group, the levels of IL-1β and TNF-α were
markedly decreased, and those of IL-4 and IL-10 were significantly
increased in the control group (Table
V).
 | Table VEffect of astragalosides on the
levels of IL-1β, TNF-α, IL-4 and IL-10 in adjuvant arthritis
rats. |
Table V
Effect of astragalosides on the
levels of IL-1β, TNF-α, IL-4 and IL-10 in adjuvant arthritis
rats.
| Group | Dose, mg/kg | IL-1β, ng/l | TNF-α, ng/l | IL-4, ng/l | IL-10, ng/l |
|---|
| Vehicle | - | 35.39±10.68 | 162.65±25.4 | 94.23±15.11 | 197.66±21.1 |
| Animal model | - | 73.99±13.18a | 284.2±41.28a | 61.11±13.51a | 125.8±28.96a |
| Control | 200 | 57.90±12.05b |
223.78±36.47c | 78.47±9.38b |
157.39±14.14b |
Histopathology in rats with AA
H&E staining of samples from the vehicle group
showed normal arrangement of syno-vial cells, without synovial
hyperplasia or inflammatory cell infiltration. However, samples
from the animal model group exhibited local thickening of the
surrounding synovium tissue and inflammatory cell infiltration
accompanied by spalling pannus formation. In the control group,
less pathological tissue injury was present compared with that in
the animal model group (Fig.
1A).
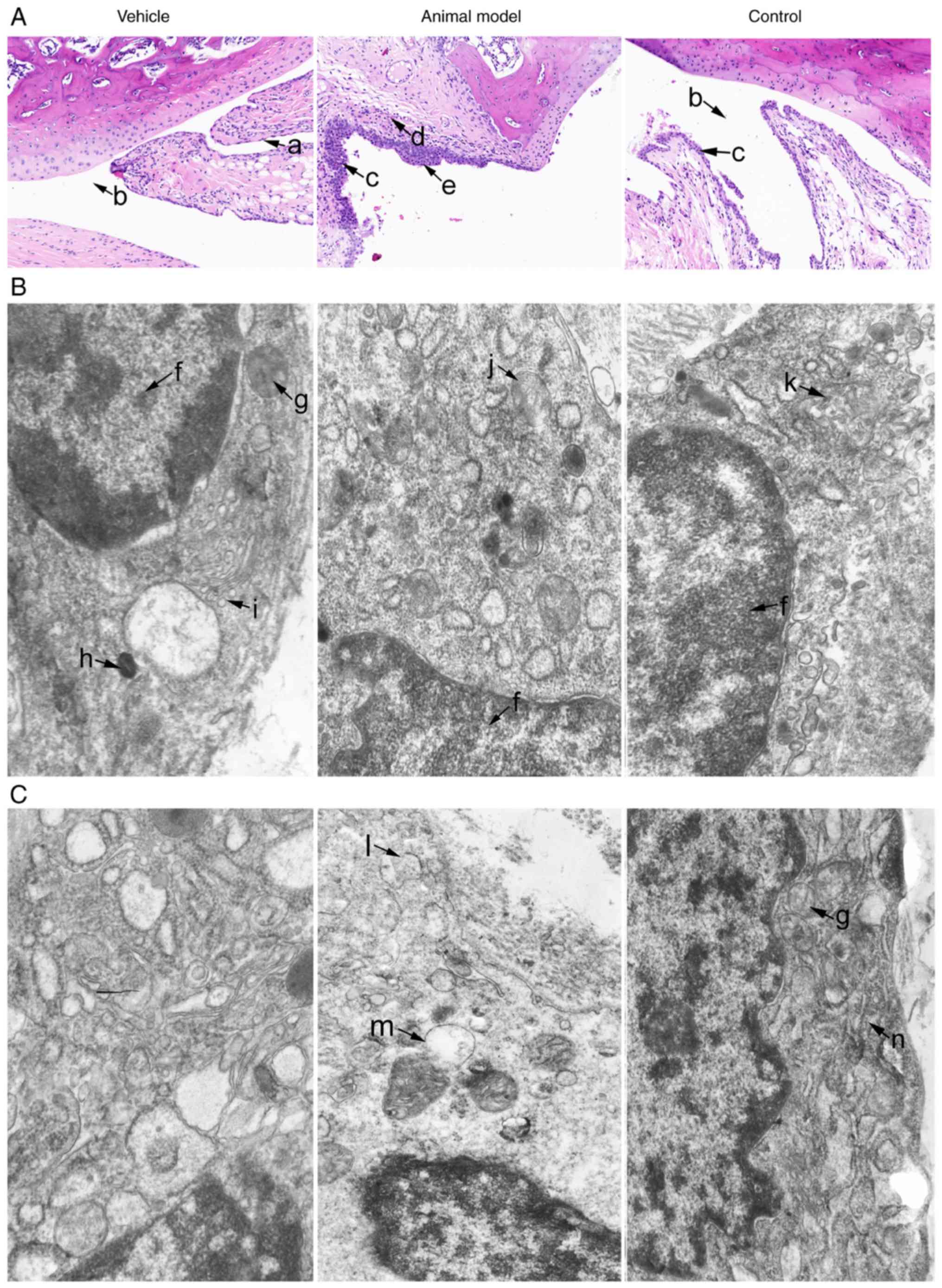 | Figure 1Effect of AST on the histopathology
of ankle joints and synovial tissues in the vehicle, animal model
and control groups. (A) Effect of AST on the histopathology of
ankle joints by H&E staining (magnification, ×200). TEM images
show the effects of AST on the histopathology of (B) type A
synoviocytes and (C) type B synoviocytes in synovial tissues, as
shown by TEM (magnification, ×20,000). a, normal synoviocytes; b,
articular cavity; c, incrassation of synoviocytes; d, formation of
pannus tissue; e, inflammatory cell infiltration; f, nucleus; g,
mitochondrion; h, lysosome; i, golgi complex; j, mitochondrial
swelling; k, alleviation of swollen mitochondria; l, rough
endoplasmic reticulum expansion failure; m, mitochondrial vacuolar
and degeneration; n, rough endoplasmic reticulum. AST,
astragalosides; TEM, transmission electron microscopy. |
TEM revealed that two basic types of synoviocyte in
the vehicle group. Type A synoviocytes were similar to macrophages
with numerous mitochondria, which possessed the main
characteristics of the phase of phagocytosis with multitudinous
vacuoles, lysosomes, parts of rough endoplasmic reticulum and
ribosomes dispersed in the cytoplasm. Type B synoviocytes were
similar to fibroblasts with the main characteristics of the phase
of protein secretion. Chromatin was loose and was distributed
around the nucleus with a fine granular appearance. As shown in
Fig. 1B and C, the vehicle group
exhibited abundant rough endoplasmic reticulum, which were ranged
in stratiform and without vacuoles. In the animal model group,
mitochondria and vacuoles were significantly increased, and
mitochondria were swollen in the type A synoviocytes; rough
endoplasmic reticulum and dense bodies were significantly
increased. In addition, the endoplasmic reticulum cistern was
swollen and there was destruction of the structure in the type B
synoviocytes. Following AST intervention, the vacuoles were reduced
and there was alleviation of mitochondrial swelling in the type A
synoviocytes. Furthermore, the rough endoplasmic reticulum was
reduced and chromatin in the cell nucleus exhibited pyknosis. The
volume of the cell was also reduced in the type B synoviocytes.
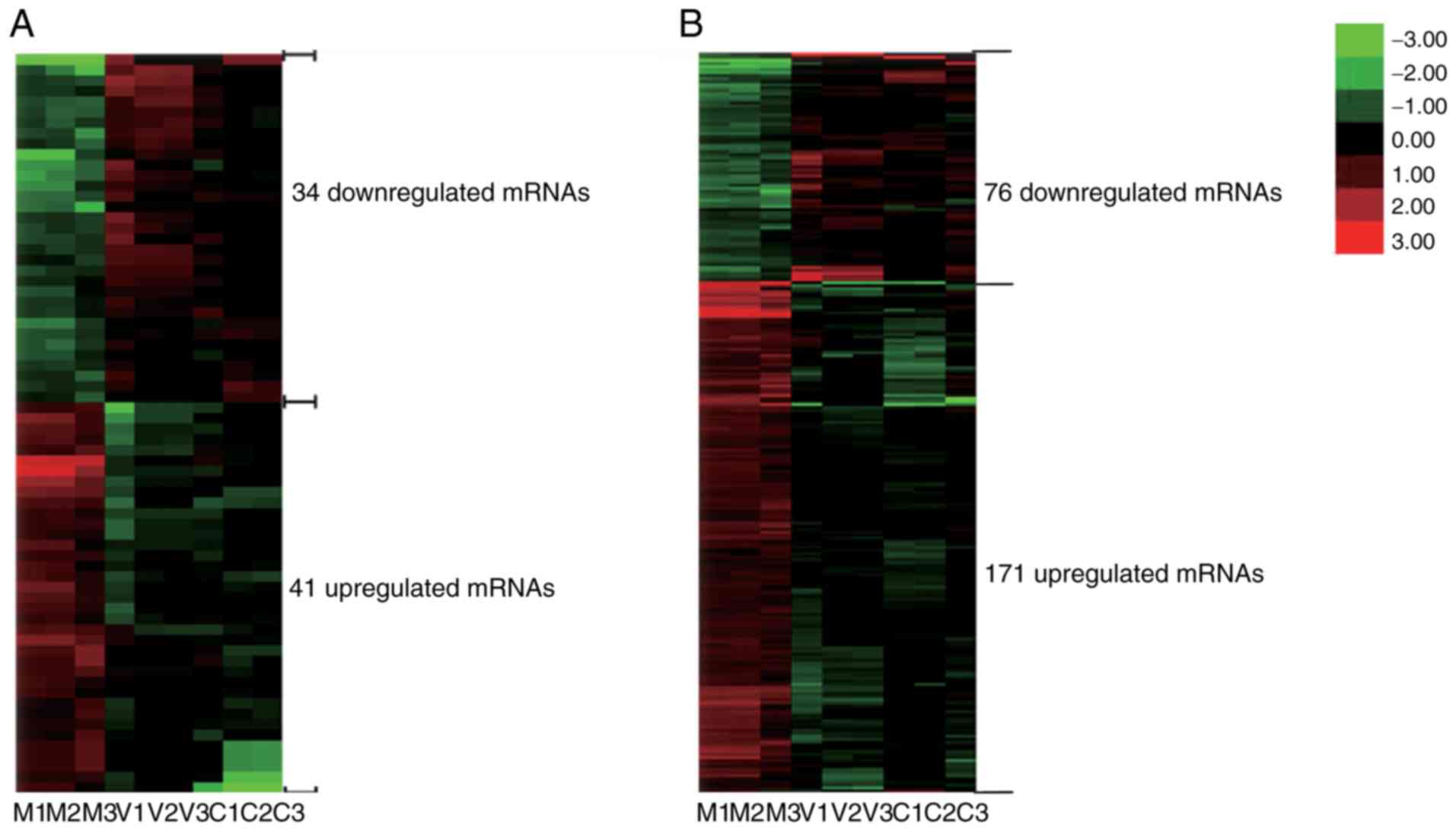
Analysis of differentially expressed
lncRNAs
Differentially expressed lncRNAs were distinguished
among the vehicle, animal model and control groups according to the
filtering criteria (P<0.05, fold change >1.5). It was found
that there were 260 significantly differentially expressed lncRNAs
between the animal model group and the vehicle group, and the
expression levels of 915 lncRNAs were significantly altered in the
control group compared with those in the animal model group. There
were 75 differentially expressed lncRNAs, which were co-expressed
differentially in the three groups. A hierarchical clustering
analysis was applied to the differentially expressed lncRNAs based
on their expression levels among samples, the results are shown in
Fig. 2A.
Analysis of differentially expressed
mRNAs
According to the filtering criteria P<0.05 and
fold change >1.5, 675 differentially expressed mRNAs were found
between the vehicle and AA animal model groups, and the expression
of 1,717 mRNAs were significantly altered between the animal model
and control groups. Subsequently, 247 differentially expressed
mRNAs were found among the three groups. Their distinct expression
patterns are also shown in Fig.
2B through hierarchical clustering analysis.
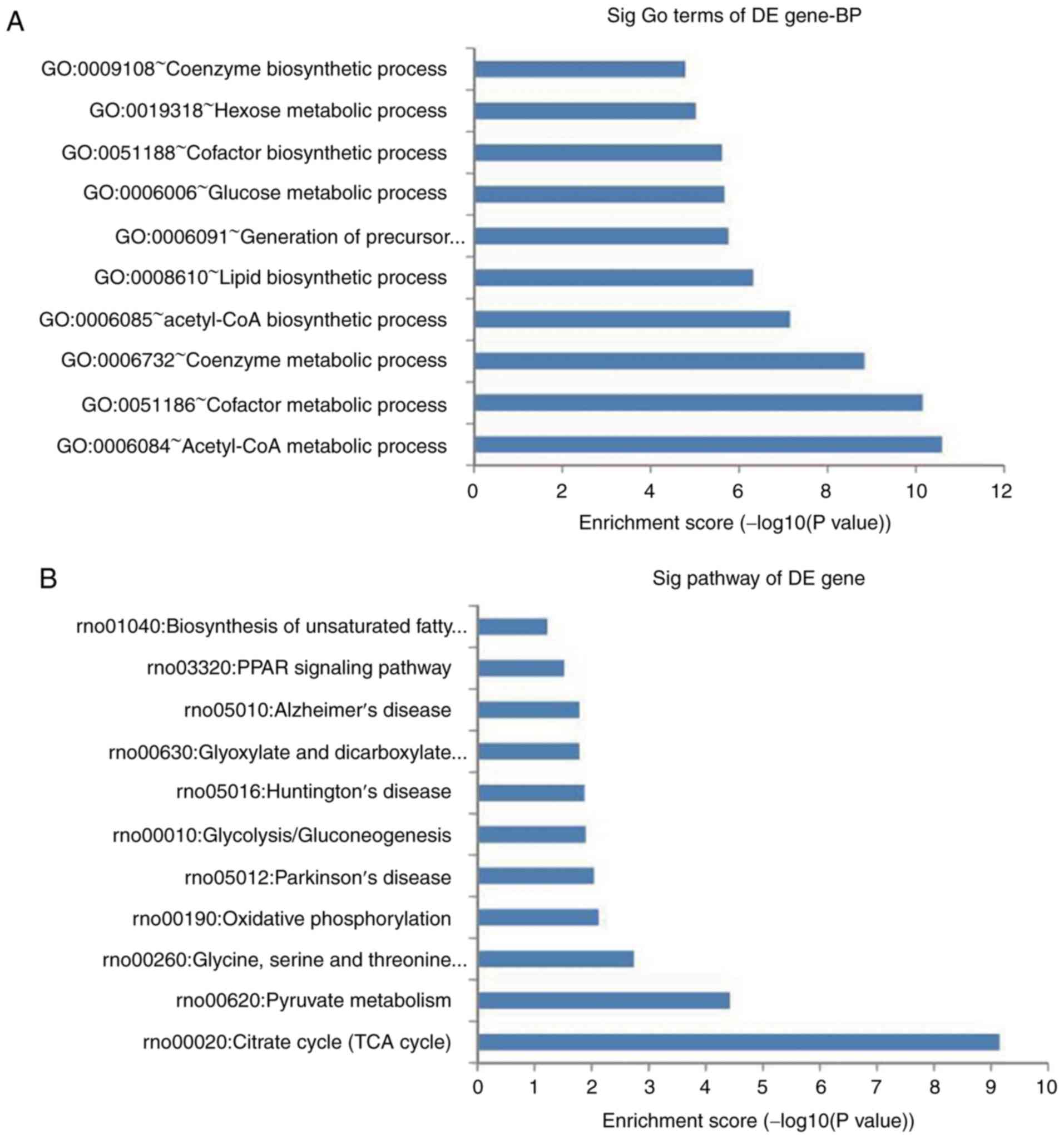
GO and pathway analysis
A total of 135 GO terms were found to be
significantly altered, including acetyl-CoA metabolic process,
cofactor metabolic process and coenzyme metabolic process. The top
10 significantly regulated BPs are shown in Fig. 3A. Pathway analysis demonstrated
that 17 pathways were significantly enriched among the upregulated
and downregulated genes, which were mainly associated with the TCA
cycle, pyruvate metabolism, and oxidative phosphorylation. The top
10 significant pathways are shown in Fig. 3B. According to the functional
annotation and fold change, 15 upregulated and five downregulated
mRNAs were selected (Table VI)
to clarify the functions of the differentially expressed lncRNAs.
The results revealed that the upregulated genes were mainly
associated with regulation of the inflammatory response, the immune
response and various metabolic processeses, and the downregulated
genes were mainly associated with various types of signaling
pathways.
 | Table VIFunctional annotation of 20
differentially expressed genes. |
Table VI
Functional annotation of 20
differentially expressed genes.
| Number | Gene | Trend | Fold change | P-value | Gene
annotation |
|---|
| 1 | Dio3 | Up | 5.77 |
9.93×10−4 | Oxidation-reduction
process |
| 2 | Mlph | Up | 7.31 |
5.38×10−5 | Intracellular
protein transport |
| 3 | Tm6sf2 | Up | 2.99 |
2.47×10−3 | Regulation of lipid
metabolic process |
| 4 | Gys2 | Up | 6.44 |
5.75×10−3 | Glycogen
biosynthetic process, response to glucose |
| 5 | Mogat2 | Up | 7.34 | 0.011 | Lipid metabolic
process |
| 6 | Hgd | Up | 5.32 |
2.14×10−3 | Cellular amino acid
metabolic process, oxidation-reduction process |
| 7 | Dlat | Up | 3.96 |
1.73×10−3 | Acetyl-coa
biosynthetic process from pyruvate, glucose metabolic |
| 8 | Fmo1 | Up | 1.95 | 0.013 | Organic acid
metabolic process, NADPH oxidation, drug metabolic |
| 9 | Gcgr | Up | 6.81 |
8.30×10−5 | G-protein coupled
receptor signaling pathway, regulation of glycogen metabolic
process |
| 10 | Me1 | Up | 5.71 |
2.33×10−3 | Oxidation-reduction
process, regulation of NADP metabolic process |
| 11 | L2hgdh | Up | 2.66 | 0.021 | Cellular protein
metabolic process, oxidation-reduction process |
| 12 | Pdhb | Up | 2.38 | 0.024 | Glucose metabolic
process, acetyl-coa biosynthetic process |
| 13 | Aco1 | Up | 2.71 |
2.55×10−3 | Tricarboxylic acid
cycle |
| 14 | Acly | Up | 5.43 |
1.03×10−4 | Fatty acid
biosynthetic process |
| 15 | Acy1 | Up | 2.02 |
3.65×10−3 | Cellular amino acid
metabolic process |
| 16 | Angptl4 D | own | 4.76 |
4.25×10−5 | Cell
differentiation, negative regulation of apoptotic process |
| 17 | C1qtnf3 D | own | 3.20 |
3.81×10−4 | Fat cell
differentiation, positive regulation of cytokine secretion |
| 18 | Dhx58 D | own | 1.82 |
4.27×10−4 | Immune system
process, regulation of innate immune response |
| 19 | Cblc | Down | 2.62 | 0.017 | Negative regulation
of MAP kinase activity, cell surface receptor signaling
pathway |
| 20 | Npr3 D | own | 5.60 |
2.13×10−3 | Osteoclast
proliferation, phospholipase C-activating G-protein coupled
receptor signaling pathway |
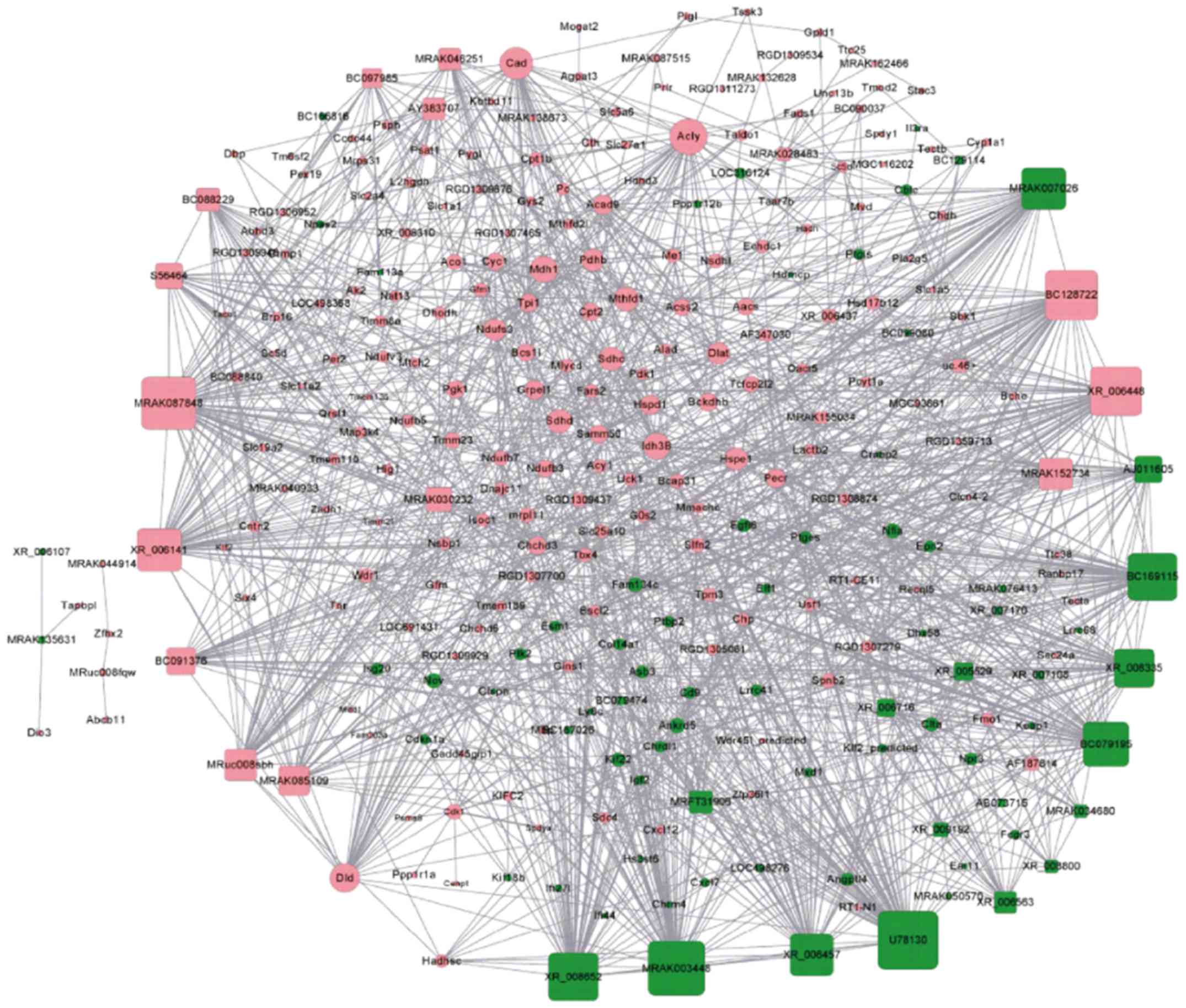
lncRNA-mRNA co-expression network
The lncRNA-mRNA co-expression network among the
vehicle, animal model and control groups with differentially
expressed lncRNAs and mRNAs, which had PCC values ≥0.94, were
constructed using the Cytoscape program and then visualized. As
shown in Fig. 4, the lncRNA-mRNA
co-expression network was composed of 58 lncRNA nodes, 229 mRNA
nodes and 1,617 edges. Based on the node degree and correlation
between bioinformatics analysis and RA, four significant lncRNAs
were selected, two of which were upregulated (MRAK012530 and
MRAK132628) and two of which were downregulated (MRAK003448 and
XR_006457) in the animal model group.
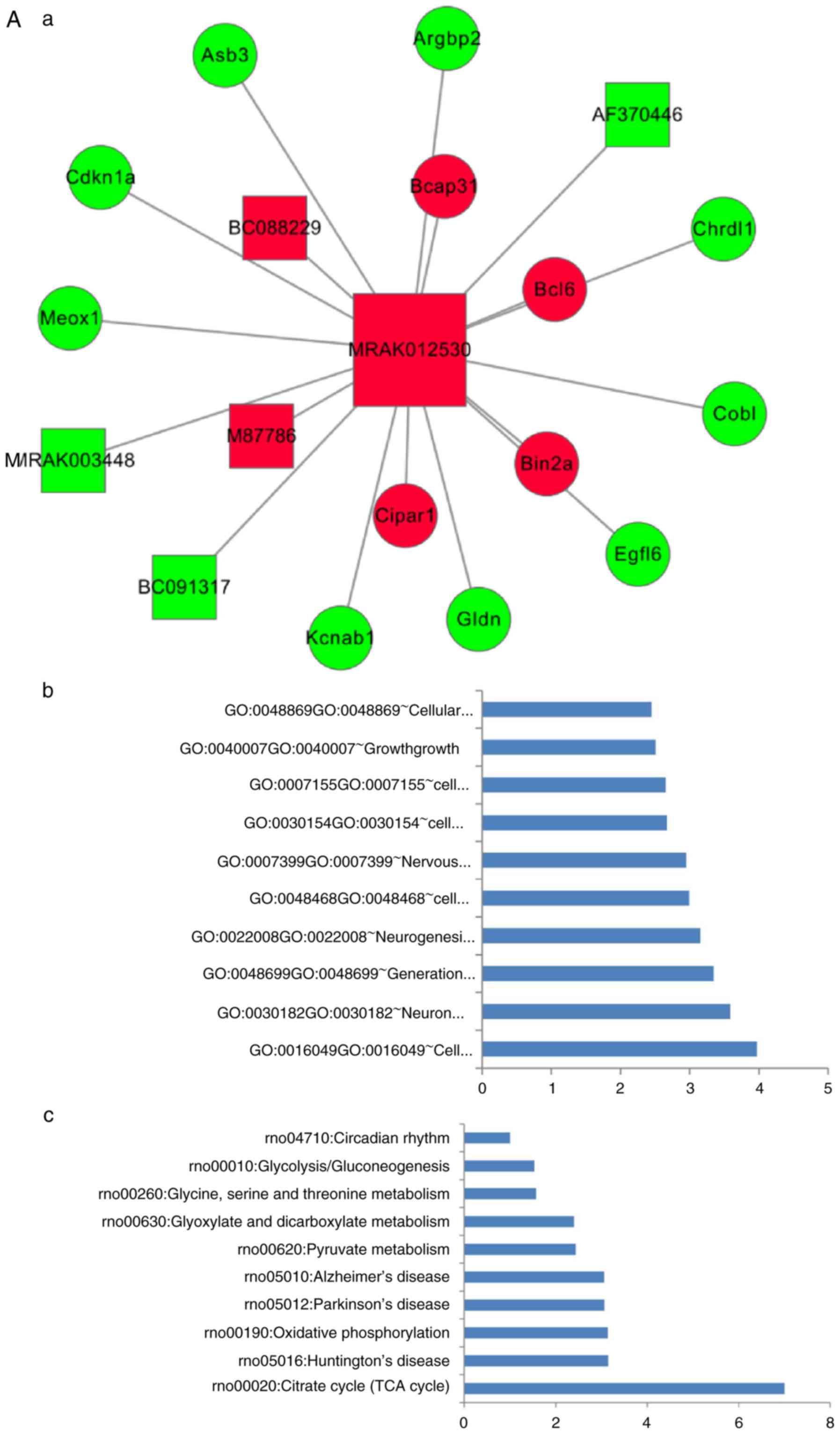
Key lncRNAs-mRNA sub-networks
The new key lncRNA-mRNA sub-networks were
constructed by extracting the key lncRNAs and their linked mRNAs
from the co-expression network. As shown in Fig. 5, there were six lncRNA nodes, 13
mRNA nodes and 19 edges (Fig.
5A-a), seven lncRNA nodes, 14 mRNA nodes and 21 edges (Fig. 5B-a), eight lncRNA nodes, 30 mRNA
nodes and 38 edges (Fig. 5C-a),
and 12 lncRNA nodes, 39 mRNA nodes and 51 edges (Fig. 5D-a) in the lncRNA MRAK012530,
MRAK132628, MRAK003448 and XR_006457-mRNA sub-networks,
respectively. The top 10 significantly regulated BP and pathways
for the key lncRNAs, determined using their first mRNAs neighbors
in the key lncRNA-mRNA sub-networks, are shown in Fig. 5A-b and -c, B-b and -c, C-b and -c and
D-b and -c.
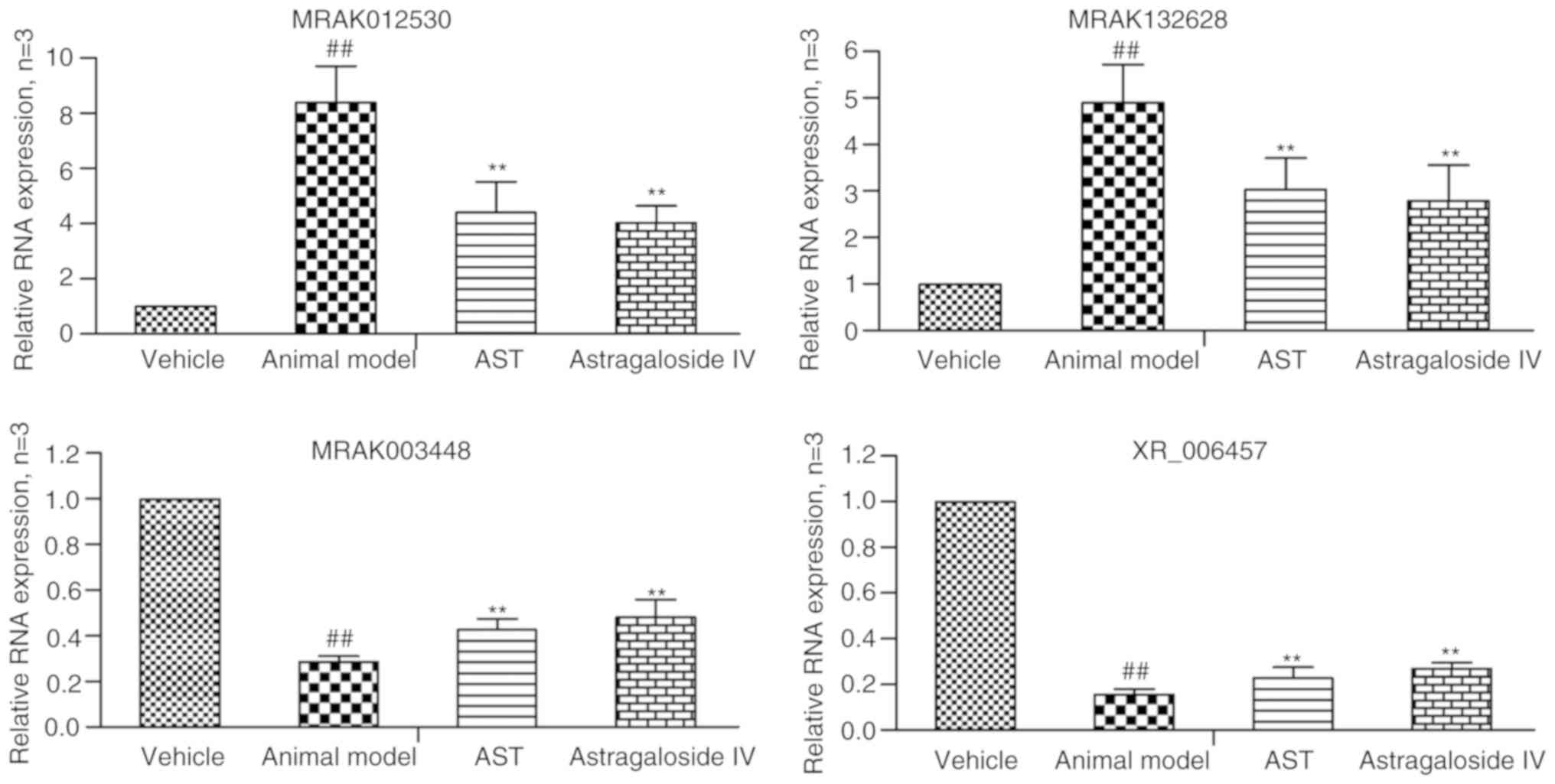
RT-qPCR validation
In order to verify the reliability of the microarray
data, a number of notable candidate lncRNAs (MRAK012530,
MRAK132628, MRAK003448 and XR_006457) were selected and analyzed
using RT-qPCR analysis. As shown in Fig. 6, the results demonstrated that
lncRNAs MRAK012530 and MRAK132628 were upregu-lated and lncRNAs
MRAK003448 and XR_006457 were downregulated in the animal model
relative to their levels in the vehicle group. Following AST and
AST-IV intervention, the expression levels of lncRNAs MRAK012530
and MRAK132628 were significantly downregulated and those of
lncRNAs MRAK003448 and XR_006457 were significantly upregulated.
The RT-qPCR results were consistent with those of the microarray
analysis, indicating that the false positive results of the gene
microarray caused by individual differences and tissue
heterogeneity could be excluded, that these four key lncRNAs may be
used as potential critical therapeutic targets of AST, and that
AST-IV was the main active ingredient of AST, which serves a major
role in regulating differentially expressed lncRNAs.
Discussion
RA is a progressive and debilitating autoimmune
disease (27). Synovial tissue is
principally composed of fibroblast-like synovial cells, which are
closely associated with the pathogenesis and development of RA,
particularly joint damage (28).
lncRNAs, a recently identified type of ncRNA, are involved in
numerous biological processes, including cellular differentiation,
cell growth and cell cycle regulation (29). Evidence from previous research
indicates the aberrant expression of lncRNAs in the pathogenesis of
RA (30,31). In our previous study, it was also
confirmed that up to 260 lncRNAs (170 upregulated and 90
downregulated lncRNAs) were found to be differentially expressed in
the synovium between an AA animal model and normal rats (25). However, fewer studies have
examined the expression profile of lncRNAs in RA following AST
treatment. In the present study, 75 differentially expressed
lncRNAs and 247 differentially expressed mRNAs were found in
synovial tissues of the vehicle, animal model and control groups.
Using the node degree and correlation between bioinformatics
analysis and RA, the results suggested that four lncRNAs
(MRAK012530, MRAK132628, MRAK003448 and XR_006457) could be
selected as critical therapeutic targets of AST. In order to
exclude the false positive results of the gene microarray caused by
individual differences and tissue heterogeneity, RT-qPCR was used
to verify the four lncRNAs. The results showed consistency between
the RT-qPCR and microarray data, which indicated that these four
lncRNAs have increased reliability and repeatability and can be
used as potential treatment targets of AST.
To investigate the potential regulatory roles of
lncRNAs, GO and pathway analyses were used to reveal the biological
functions enriched among the differentially expressed coding-genes
(32). In the present study, the
results of the GO analysis revealed that differentially expressed
genes were mainly involved in acetyl-CoA metabolic process,
cofactor metabolic process and coenzyme metabolic process. The
pathway analysis showed that 17 pathways were enriched and
primarily involved in the citrate cycle (TCA cycle), glyoxylate and
dicarboxylate metabolism, pyruvate metabolism, oxidative
phosphorylation and PPAR signaling pathway, which may have
essential roles in the occurrence and development of RA (33-37).
Although thousands of lncRNAs have been identified
and recorded in several public databases, the functional
characterization of lncRNAs remains in its infancy. Until now, only
a small number of lncRNAs have been well functionally annotated
(38,39). Improved knowledge has suggested
that lncRNAs, owing to their increased length, can regulate
microRNA (miRNA) abundance by binding and sequestering them, acting
as so-called miRNA sponges. In this manner, lncRNAs can regulate
the expression of target mRNAs (40). Therefore, it has been shown that
an efficient way to infer the potential function of lncRNAs is by
examining related mRNAs, whose functions have been annotated
(41-43). In the present study, according to
the fold change and functional annotation, 20 mRNAs were selected
to examine the possible mechanism underlying the effect of AST
treatment in RA.
Deiodinase, iodothyronine type III (Dio3), is
involved in the oxidation-reduction process and positive regulation
of multicellular organism growth (44). The oxidation-reduction reaction is
an integrated aspect of cell metabolism and dynamic equilibrium.
This reaction occurs between oxygen metabolites and certain amino
acids, proteins or enzymes, which have important physiological
functions of reducing its activity and macromolecular substances.
Those products mainly mediate endothelial cells and tissue injury
and aggravate the injury of inflammation-stimulating factors
(45). In this study, AST not
only inhibited the expression of Dio3, but also downregulated
several other oxidation-reduction reaction-related genes, including
homogentisate 1,2-dioxygenase, flavin containing dimethylaniline
monoxygenase 1, malic enzyme 1 and L-2-hydroxyglutarate
dehydrogenase, which indicated that AST regulated the
oxidation-reduction reaction.
In our previous study, using gas chromatography
time-of-flight mass spectrometry and a variety of multivariate
statistical methods, including PCA, PLS-DA and OPLS-DA, it was
confirmed that L-dopa, 1,4-dihydroxy-2-naphthoic acid and tartronic
acid, which are primarily associated with amino acid, glycogen
metabolism and the TCA cycle, were markedly altered (46,47). In the present study, it was found
that certain genes were significantly altered in the animal model
group, including transmembrane 6 superfamily member 2,
monoacylglycerol O-acyltransferase, ATP citrate lyase and
angiopoietin-like 4, which are related to lipid metabolism
(48,49), glycogen synthase 2,
dihydrolipoamide S-acetyltransferase, glucagon receptor and
pyruvate dehydrogenase E1 beta subunit, which are important in
several biological functions including glycogen metabolic process
and response to glucose (50,51), and aconitase 1, which is involved
in TAC (52,53), indicating there were some
metabolic disorders during the development of RA, which is
consistent with our previous studies (46,47). Following the administration of
AST, the expression of these genes returned to normal levels,
indicating that AST can regulate disorders of metabolism.
C1q and tumor necrosis factor-related protein 3
(C1qtnf3) is involved in the negative regulation of interleukin-6
(IL-6) secretion (54). IL-6 is
involved in inflammatory immune cell multiplication and aggravates
the development of inflammation in RA (55,56). DEXH (Asp-Glu-X-His) box
polypeptide 58 (Dhx58) is involved in the RIG-I-like receptor
signaling pathway, which can negatively regulate the innate immune
response (57). Casitas B-lineage
lymphoma c (Cblc) is involved in the JAK-STAT signaling pathway and
is an important negative regulator in the T-cell and B-cell antigen
receptor signaling pathway (58,59). Several experiments have
demonstrated that the T cell receptor signaling pathway and
JAK-STAT signaling pathway are involved in RA (60-63). Following AST treatment, the
expression levels of C1qtnf3, Dhx58 and Cblc were increased. The
levels of CRP, WBCs, IL-1β and TNF-α were decreased and the levels
of IL-4 and IL-10 were significantly increased following treatment
with AST. These results suggested that AST inhibited inflammation
in RA.
The hyperplasia of synoviocytes is one of the major
morphologic features of RA. It has been identified that
synoviocytes can cause severe joint destruction by secreting
various cytokines, inflammatory mediators and proteinases (64,65). Natriuretic peptide receptor 3
(Npr3) negatively regulates cell proliferation (66). In the present study, the level of
Npr3 was significantly increased following AST treatment compared
with that in the AA animal model group. These results suggested
that AST inhibited hyperplasia of the synoviocytes.
In conclusion, the present study advances current
understanding of the possible mechanism of AST in the treatment of
RA from the perspective of lncRNAs, and provides novel lncRNAs as
potential critical therapeutic targets of AST. In addition, AST-IV
was the main active ingredient of AST, which serves a major role in
regulating differentially expressed lncRNAs.
Abbreviations:
|
RA
|
rheumatoid arthritis
|
|
lncRNAs
|
long non-coding RNAs
|
|
AST
|
astragalosides
|
|
AST-IV
|
astragaloside IV
|
|
AA
|
adjuvant arthritis
|
|
GO
|
Gene Ontology
|
|
DAVID
|
Database for Annotation, Visualization
and Integration Discovery
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TCM
|
traditional Chinese medicine
|
|
ncRNAs
|
non-coding RNAs
|
|
TEM
|
transmission electron microscopy
|
|
CRP
|
C-reactive protein
|
|
WBC
|
white blood cell
|
|
RBC
|
red blood cell
|
|
HGB
|
hemoglobin
|
|
HCT
|
hematocrit
|
|
IL
|
interleukin
|
|
TNF-α
|
tumor necrosis factor-α
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PCC
|
Pearson's correlation coefficient
|
|
BP
|
biological process
|
Acknowledgments
The authors thank Mr. Hui Wang (KangChen Biotech
Co., Ltd., Shanghai, China), and Mr. Qiang Fan (Ao Ji Biotech Co.,
Ltd., Shanghai, China) for providing assistance with data
analysis.
Funding
This study was financially supported by the National
Natural Science Foundation of China (grant no. 81873139) and the
Introducing and Cultivating College Leading Talents Project of the
Education Department of Anhui Province (Excellent Young and
Middle-aged Backbones of College Visiting and Training Abroad in
2016; grant no. gxfxZD2016118).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WPL made substantial contributions to the conception
and design of the study. FRW, XJQ and NNJ performed the
experiments. HJ contributed to data acquisition, and data analysis
and interpretation. JL revised the manuscript critically for
important intellectual content. All authors agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The protocol was approved by the Committee on the
Ethics of Animal Experiments of Anhui University of Chinese
Medicine. All surgical procedures were performed under sodium
pentobarbital anesthesia, and all efforts were made to minimize
animal suffering and the number of animals used.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sarabi ZS, Saeidi MG, Khodashahi M, Rezaie
AE, Hashemzadeh K, Khodashahi R and Heidari H: Evaluation of the
anti-inflammatory effects of atorvastatin on patients with
rheumatoid arthritis: A randomized clinical trial. Electron
Physician. 8:2700–2706. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wechalekar MD, Lester S, Hill CL, Lee A,
Rischmueller M, Smith MD, Walker JG and Proudman SM: Active foot
synovitis in patients with rheumatoid arthritis: Unstable remission
status, radiographic progression, and worse functional outcomes in
patients with foot synovitis in apparent remission. Arthritis Care
Res (Hoboken). 68:1616–1623. 2016. View Article : Google Scholar
|
|
3
|
Wang Wu Q, Wang Y, Yu Q, Wang D, Song Y,
Liu L, Ye Z, Xu X, Cao PH, et al: The bispecific antibody aimed at
the vicious circle of IL-1β and IL-17A, is beneficial for the
collagen-induced rheumatoid arthritis of mice through NF-κB
signaling pathway. Immunol Lett. 179:68–79. 2016. View Article : Google Scholar
|
|
4
|
Burmester GR, Rubbert-Roth A, Cantagrel A,
Hall S, Leszczynski P, Feldman D, Rangaraj MJ, Roane G, Ludivico C,
Bao M, et al: Efficacy and safety of subcutaneous tocilizumab
versus intravenous tocilizumab in combination with traditional
DMARDs in patients with RA at week 97 (SUMMACTA). Ann Rheum Dis.
75:68–74. 2016. View Article : Google Scholar :
|
|
5
|
Rossi D, Modena V, Sciascia S and
Roccatello D: Rheumatoid arthritis: Biological therapy other than
anti-TNF. Int Immunopharmacol. 27:185–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang L, Lv Q, Xie D, Shi T and Wen C:
Deciphering the potential pharmaceutical mechanism of chinese
traditional medicine (Gui-Zhi-Shao-Yao-Zhi-Mu) on rheumatoid
arthritis. Sci Rep. 6:226022016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Mao X, Guo Q, Bai M, Zhang B, Liu
C, Sun Y, Li S and Lin N: Pathway of PPAR-gamma coactivators in
thermogenesis: A pivotal traditional Chinese medicine-associated
target for individualized treatment of rheumatoid arthritis.
Oncotarget. 7:15885–15900. 2016.PubMed/NCBI
|
|
8
|
Lin YC, Chang CW and Wu CR:
Anti-nociceptive, anti-inflammatory and toxicological evaluation of
Fang-Ji-Huang-Qi-Tang in rodents. BMC Complement Altern Med.
15:102015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu XY, Xu L, Wang Y, Li JX, Zhang Y,
Zhang C, Wang SS and Zhang XM: Protective effects of total
flavonoids of Astragalus against adjuvant-induced arthritis in rats
by regulating OPG/RANKL/NF-κB pathway. Int Immunopharmacol.
44:105–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang JB, Qiu JD, Yang LH, He JP, Smith GW
and Li HQ: Therapeutic effects of Astragalus polysaccharides on
inflammation and synovial apoptosis in rats with adjuvant-induced
arthritis. Int J Rheum Dis. 13:396–405. 2010. View Article : Google Scholar
|
|
11
|
Wan L, Liu J, Huang CB, Wang Y, Chen X,
Zhang WD, Wang GZ, Fan HX, Ge Y, Chen RL, et al: Xinfeng capsule
for the treatment of rheumatoid arthritis patients with decreased
pulmonary function -a randomized vehicleled clinical trial. Chin J
Integr Med. 22:168–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Wang DD, Wei T, He SM, Zhang GY
and Wei QL: Effects of astragalosides from Radix Astragali on high
glucose-induced proliferation and extracellular matrix accumulation
in glomerular mesangial cells. Exp Ther Med. 11:2561–2566. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu YY, Wu WY, Gong HL, Li WZ and Yin YY:
Astragalosides attenuate learning and memory impairment in rats
following ischemiareperfusion injury. Mol Med Rep. 9:1319–1324.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu JM, Zhang XB, Jiang W, Wang HD and
Zhang YN: Astragalosides promote angiogenesis via vascular
endothelial growth factor and basic fibroblast growth factor in a
rat animal animal model of myocardial infarction. Mol Med Rep.
12:6718–6726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Q, Lu JT, Zhou AW, Wang B, He GW and
Chen MZ: Antinociceptive effect of astragalosides and its mechanism
of action. Acta Pharmacol Sin. 22:809–812. 2001.PubMed/NCBI
|
|
16
|
Juan L, Wang G, Radovich M, Schneider BP,
Clare SE, Wang Y and Liu Y: Potential roles of microRNAs in
regulating long intergenic noncoding RNAs. BMC Med Genomics.
6(Suppl 1): S72013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye N, Rao S, Du T, Hu H, Liu Z, Shen Y and
Xu Q: Intergenic variants may predispose to major depression
disorder through regulation of long non-coding RNA expression.
Gene. 601:21–26. 2017. View Article : Google Scholar
|
|
18
|
Szcześniak MW and Makałowska I: lncRNA-RNA
interactions across the human transcriptome. PLoS One.
11:e01503532016. View Article : Google Scholar
|
|
19
|
Yang L, Xu L, Wang Q, Wang M and An G:
Dysregulation of long non-coding RNA profiles in human colorectal
cancer and its association with overall survival. Oncol Lett.
12:4068–4074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rühle F and Stoll M: Long non-coding RNA
databases in cardiovascular research. Genomics Proteomics
Bioinformatics. 14:191–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Messemaker TC, Frank-Bertoncelj M, Marques
RB, Adriaans A, Bakker AM, Daha N, Gay S, Huizinga TW, Toes RE,
Mikkers HM and Kurreeman F: A novel long non-coding RNA in the
rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels.
Genes Immun. 17:85–92. 2016. View Article : Google Scholar
|
|
22
|
Li R, Cai L, Hu CM, Wu TN and Li J:
Expression of hedgehog signal pathway in articular cartilage is
associated with the severity of cartilage damage in rats with
adjuvant-induced arthritis. J Inflamm (Lond). 12:242015. View Article : Google Scholar
|
|
23
|
Hao J, Wu X, Setrerrahmane S, Qian K, Hou
Y, Yu L, Lin C, Wu Q and Xu H: Combination therapy of PEG-HM-3 and
methotrexate retards adjuvant-induced arthritis. Int J Mol Sci.
18:E15382017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Wang QW, Zuo J, Chen JW and Li X:
Anti-arthritic activity of ethanol extract of Claoxylon indicum on
Freund's complete adjuvant-induced arthritis in mice. BMC
complement Altern Med. 17:112017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang H, Qin XJ, Li WP, Ma R, Wang T and
Li ZQ: lncRNAs expression in adjuvant-induced arthritis rats
reveals the potential role of lncRNAs contributing to rheumatoid
arthritis pathogenesis. Gene. 593:131–142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Chen D, Liu D, Liu D, He M, Peng A, Xu J,
Lin L, Luo F, Chen L, Huang X, et al: Rheumatoid arthritis
fibroblast-like synoviocyte suppression mediated by PTEN involves
survivin gene silencing. Sci Rep. 7:3672017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang J, Zhao F and Nie J: Anti-rheumatic
effects of Aconitum leucostomum Worosch. on human fibroblast-like
synoviocyte rheumatoid arthritis cells. Exp Ther Med. 14:453–460.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Y, Pan J, Zhang N, Wei W, Yu S and Ai
L: Knockdown of long non-coding RNA H19 inhibits multiple myeloma
cell growth via NF-κB pathway. Sci Rep. 7:180792017. View Article : Google Scholar
|
|
30
|
Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF,
Guan XL, Li A, Wang F, Zhao QF, Wang HY, et al: Long noncoding RNA
expression profile in fibroblast-like synoviocytes from patients
with rheumatoid arthritis. Arthritis Res Ther. 18:2272016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung
CH and Lai NS: Increased expression of long noncoding RNAs
LOC100652951 and LOC100506036 in T cells from patients with
rheumatoid arthritis facilitates the inflammatory responses.
Immunol Res. 64:576–583. 2016. View Article : Google Scholar
|
|
32
|
Luo Q, Li X, Xu C, Zeng L, Ye J, Guo Y,
Huang Z and Li J: Integrative analysis of long non-coding RNAs and
messenger RNA expression profiles in systemic lupus erythematosus.
Mol Med Rep. 17:3489–3496. 2018.
|
|
33
|
Cheng B, Zheng H, Wu F, Wu J, Liu X, Tang
C, Lu S, Chen Z, Song F, Ruan J, et al: Metabolomics analysis of
Danggui Sini decoction on treatment of collagen-induced arthritis
in rats. J Chromatogr B Analyt Technol Biomed Life Sci.
1061-1062:282–291. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu Y, Lu C, Zha Q, Kong H, Lu X, Lu A and
Xu G: Plasma metabonomics study of rheumatoid arthritis and its
Chinese medicine subtypes by using liquid chromatography and gas
chromatography coupled with mass spectrometry. Mol Biosyst.
8:1535–1543. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huffman KM, Jessee R, Andonian B, Davis
BN, Narowski R, Huebner JL, Kraus VB, McCracken J, Gilmore BF, Tune
KN, et al: Molecular alterations in skeletal muscle in rheumatoid
arthritis are related to disease activity, physical inactivity, and
disability. Arthritis Res Ther. 19:122017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XF, Sun YY, Bao J, Chen X, Li YH, Yang
Y, Zhang L, Huang C, Wu BM, Meng XM and Li J: Functional role of
PPAR-γ on the proliferation and migration of fibroblast-like
synoviocytes in rheumatoid arthritis. Sci Rep. 7:126712017.
View Article : Google Scholar
|
|
37
|
Lin Y and Luo Z: Aberrant methylation
patterns affect the molecular pathogenesis of rheumatoid arthritis.
Int Immunopharmacol. 46:141–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang A, Zhang J, Kaipainen A, Lucas JM
and Yang H: Long non-coding RNA: A newly deciphered 'code' in
prostate cancer. Cancer Lett. 375:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Széll M, Danis J, Bata-Csörgő Z and Kemény
L: PRINS, a primate-specific long non-coding RNA, plays a role in
the keratinocyte stress response and psoriasis pathogenesis.
Pflugers Arch. 468:935–943. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang
AX, Hu JL, Wang YM and Gao YD: Comprehensive analysis of lncRNA and
mRNA expression profiles in lung cancer. Clin Lab. 63:313–320.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang S, Ning Q, Zhang G, Sun H, Wang Z and
Li Y: Construction of differential mRNA-lncRNA crosstalk networks
based on ceRNA hypothesis uncover key roles of lncRNAs implicated
in esophageal squamous cell carcinoma. Oncotarget. 7:85728–85740.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song C, Zhang J, Liu Y, Pan H, Qi HP, Cao
YG, Zhao JM, Li S, Guo J, Sun HL and Li CQ: Construction and
analysis of cardiac hypertrophy-associated lncRNA-mRNA network
based on competitive endogenous RNA reveal functional lncRNAs in
cardiac hypertrophy. Oncotarget. 7:10827–10840. 2016.PubMed/NCBI
|
|
44
|
Molina-Pinelo S, Salinas A, Moreno-Mata N,
Ferrer I, Suarez R, Andrés-León E, Rodríguez-Paredes M, Gutekunst
J, Jantus-Lewintre E, Camps C, et al: Impact of DLK1-DIO3 imprinted
cluster hypomethylation in smoker patients with lung cancer.
Oncotarget. 9:4395–4410. 2016.PubMed/NCBI
|
|
45
|
Moon JS, Nakahira K, Chung KP, DeNicola
GM, Koo MJ, Pabón MA, Rooney KT, Yoon JH, Ryter SW, Stout-Delgado H
and Choi AM: NOX4-dependent fatty acid oxidation promotes NLRP3
inflammasome activation in macrophages. Nat Med. 22:1002–1012.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang H, Liu J, Wang T, Gao JR, Sun Y,
Huang CB, Meng M and Qin XJ: Urinary metabolite profiling provides
potential differentiation to explore the mechanisms of
adjuvant-induced arthritis in rats. Biomed Chromatogr.
30:1397–1405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang H, Liu J, Wang T, Gao JR, Sun Y,
Huang CB, Meng M and Qin XJ: Mechanism of xinfeng capsule on
adjuvant-induced arthritis via analysis of urinary metabolomic
profiles. Autoimmune Dis. 2016:56909352016.PubMed/NCBI
|
|
48
|
Smagris E, Gilyard S, BasuRay S, Cohen JC
and Hobbs HH: Inactivation of Tm6sf2, a gene defective in fatty
liver disease, impairs lipidation but not secretion of very low
density lipoproteins. J Biol Chem. 291:10659–10676. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sookoian S and Pirola CJ: Meta-analysis of
the influence of TM6SF2 E167K variant on plasma concentration of
aminotransferases across different populations and diverse liver
phenotypes. Sci Rep. 6:277182016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ko R and Lee SY: Glycogen synthase kinase
3β in Toll-like receptor signaling. BMB Rep. 49:305–310. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Malaver-Ortega LF, Sumer H, Liu J and
Verma PJ: Inhibition of JAK-STAT ERK/MAPK and glycogen synthase
kinase-3 induces a change in gene expression profile of bovine
induced pluripotent stem cells. Stem Cells Int. 2016:51279842016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sekeli R, Abdullah JO, Namasivayam P, Muda
P, Abu Bakar UK, Yeong WC and Pillai V: RNA interference of
1-aminocyclo-propane-1-carboxylic acid oxidase (ACO1 and ACO2)
genes expression prolongs the shelf life of Eksotika (Carica papaya
L.) papaya fruit. Molecules. 19:8350–8362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu X, Wang A, Zhu S and Zhang L:
Expression of ACO1, ERS1 and ERF1 genes in harvested bananas in
relation to heat-induced defense against Colletotrichum musae. J
Plant Physiol. 168:1634–1640. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang R, Zhong L, Zhou J and Peng Y:
Complement-C1q TNF-related protein 3 alleviates mesangial cell
activation and inflammatory response stimulated by secretory IgA.
Am J Nephrol. 43:460–468. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ruderman EM: Rheumatoid arthritis: IL-6
inhibition in RA-déjà vu all over again? Nat Rev Rheumatol.
11:321–322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sapan HB, Paturusi I, Jusuf I, Patellongi
I, Massi MN, Pusponegoro AD, Arief SK, Labeda I, Islam AA, Rendy L
and Hatta M: Pattern of cytokine (IL-6 and IL-10) level as
inflammation and anti-inflammation mediator of multiple organ
dysfunction syndrome (MODS) in polytrauma. Int J Burns Trauma.
6:37–43. 2016.PubMed/NCBI
|
|
57
|
Li XY, Han CM, Wang Y, Liu HZ, Wu ZF, Gao
QH and Zhao SH: Expression patterns and association analysis of the
porcine DHX58 gene. Anim Genet. 41:537–540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Aranaz P, Hurtado C, Erquiaga I, Miguéliz
I, Ormazábal C, Cristobal I, García-Delgado M, Novo FJ and Vizmanos
JL: CBL mutations in myeloproliferative neoplasms are also found in
the gene's prolinerich domain and in patients with the V617FJAK2.
Haematologica. 97:1234–1241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang F and Gu H: Negative regulation of
lymphocyte development and function by the Cbl family of proteins.
Immunol Rev. 224:229–238. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sakaguchi S, Benham H, Cope AP and Thomas
R: T-cell receptor signaling and the pathogenesis of autoimmune
arthritis: Insights from mouse and man. Immunol Cell Biol.
90:277–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Olasz K, Boldizsar F, Kis-Toth K, Tarjanyi
O, Hegyi A, van Eden W, Rauch TA, Mikecz K and Glant TT: T cell
receptor (TCR) signal strength vehicles arthritis severity in
proteo-glycan-specific TCR transgenic mice. Clin Exp Immunol.
167:346–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang Y, Dong Q and Li R: Matrine induces
the apoptosis of fibroblast-like synoviocytes derived from rats
with collagen-induced arthritis by suppressing the activation of
the JAK/STAT signaling pathway. Int J Mol Med. 39:307–316. 2017.
View Article : Google Scholar :
|
|
63
|
Isomäki P, Junttila I, Vidqvist KL,
Korpela M and Silvennoinen O: The activity of JAK-STAT pathways in
rheumatoid arthritis: Constitutive activation of STAT3 correlates
with interleukin 6 levels. Rheumatology (Oxford). 54:1103–1113.
2015. View Article : Google Scholar
|
|
64
|
Ohshima S, Mima T, Sasai M, Nishioka K,
Shimizu M, Murata N, Yoshikawa H, Nakanishi K, Suemura M, McCloskey
RV, et al: Tumour necrosis factor alpha (TNF-alpha) interferes with
Fas-mediated apoptotic cell death on rheumatoid arthritis (RA)
synovial cells: A possible mechanism of rheumatoid synovial
hyperplasia and a clinical benefit of anti-TNF-alpha therapy for
RA. Cytokine. 12:281–288. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sakurai N, Kuroiwa T, Ikeuchi H, Hiramatsu
N, Maeshima A, Kaneko Y, Hiromura K and Nojima Y: Expression of
IL-19 and its receptors in RA: Potential role for synovial
hyperplasia formation. Rheumatology (Oxford). 47:815–820. 2008.
View Article : Google Scholar
|
|
66
|
Ma N, Ma Y, Nakashima A, Kikkawa U and
Furuyashiki T: The loss of Lam2 and Npr2-Npr3 diminishes the
vacuolar localization of Gtr1-Gtr2 and disinhibits TORC1 activity
in fission yeast. PLoS One. 11:e01562392016. View Article : Google Scholar : PubMed/NCBI
|