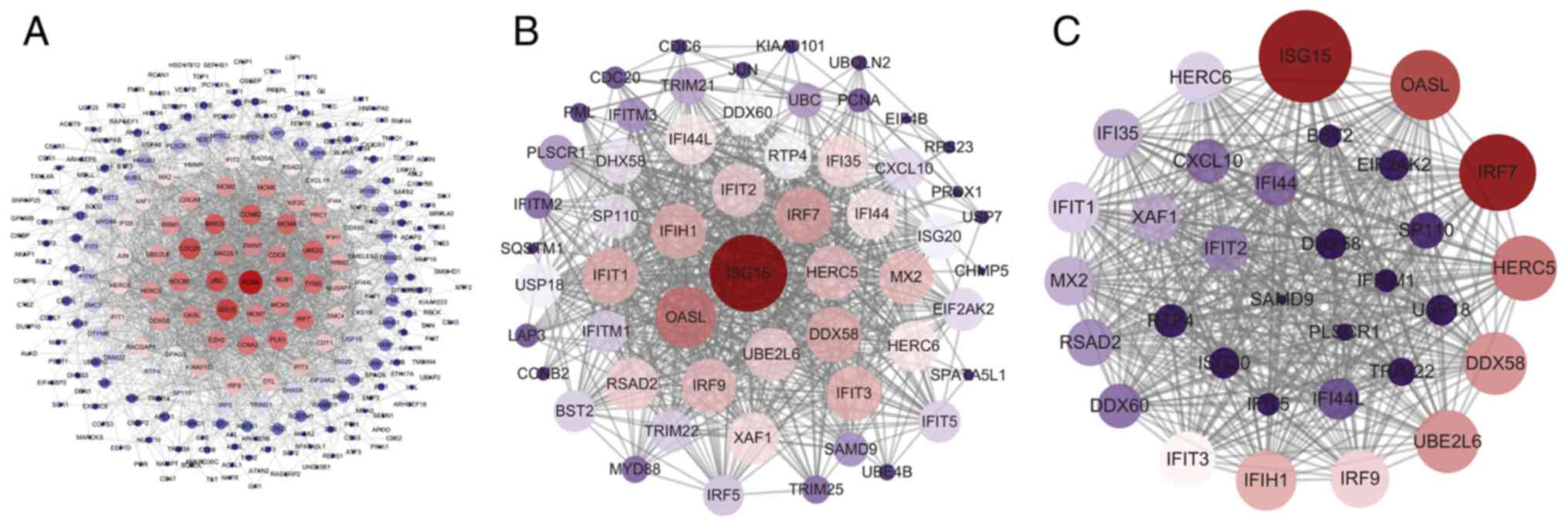
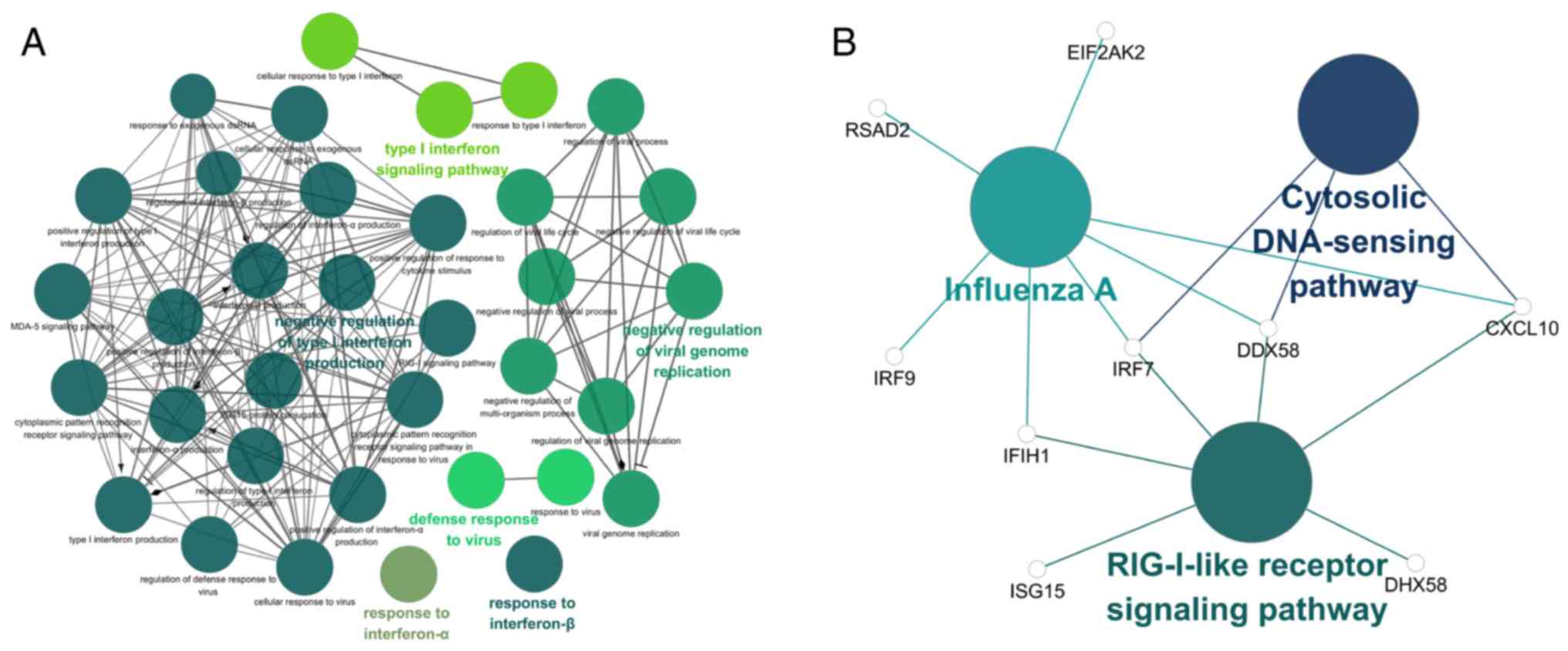
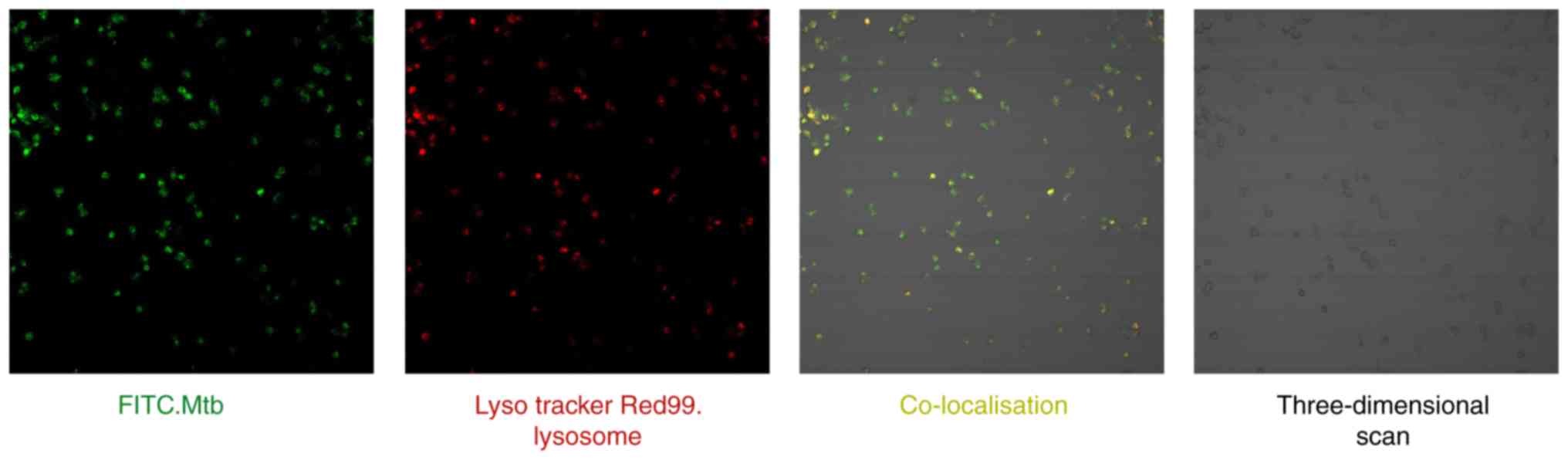
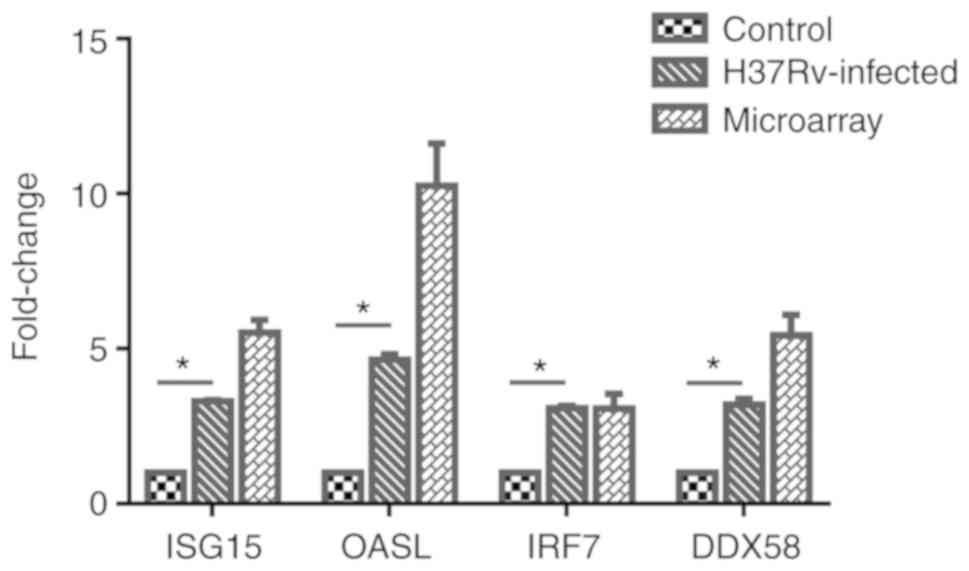
|
1
|
Goldberg MF, Saini NK and Porcelli SA:
Evasion of innate and adaptive immunity by Mycobacterium
tuberculosis. Microbiol Spectr. 5:2014.
|
|
2
|
Comas I, Coscolla M, Luo T, Borrell S,
Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, et
al: Out-of-Africa migration and Neolithic coexpansion of
Mycobacterium tuberculosis with modern humans. Nat Genet.
45:1176–1182. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Organization WH: Global Tuberculosis
Report. 2018, https://www.who.int/tb/publications/global_report/en/.
Accessed September 18, 2018.
|
|
4
|
Aderem A and Underhill DM: Mechanisms of
phagocytosis in macrophages. Annu Rev Immunol. 17:593–623. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weiss G and Schaible UE: Macrophage
defense mechanisms against intracellular bacteria. Immunol Rev.
264:182–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhatt K and Salgame P: Host innate immune
response to Mycobacterium tuberculosis. J Clin Immunol. 27:347–362.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hmama Z, Peña-Díaz S, Joseph S and Av-Gay
Y: Immunoevasion and immunosuppression of the macrophage by
Mycobacterium tuberculosis. Immunol Rev. 264:220–232. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meena LS and Rajni: Survival mechanisms of
pathogenic Mycobacterium tuberculosis H37Rv. FEBS J. 277:2416–2427.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
Large-scale meta-analysis of cancer microarray data identifies
common transcriptional profiles of neoplastic transformation and
progression. Proc Natl Acad Sci USA. 101:9309–9314. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rung J and Brazma A: Reuse of public
genome-wide gene expression data. Nat Rev Genet. 14:89–99. 2013.
View Article : Google Scholar
|
|
12
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smid M, Dorssers LC and Jenster G: Venn
mapping: Clustering of heterologous microarray data based on the
number of co-occurring differentially expressed genes.
Bioinformatics. 19:2065–2071. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
DeConde RP, Hawley S, Falcon S, Clegg N,
Knudsen B and Etzioni R: Combining results of microarray
experiments: A rank aggregation approach. Stat Appl Genet Mol Biol.
5:Article152006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia J, Fjell CD, Mayer ML, Pena OM,
Wishart DS and Hancock RE: INMEX-a web-based tool for integrative
meta-analysis of expression data. Nucleic Acids Res. 41:W63–W70.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia J, Gill EE and Hancock RE:
NetworkAnalyst for statistical, visual and network-based
meta-analysis of gene expression data. Nat Protoc. 10:823–844.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang R, Cai Y, Zhang B and Wu Z: A 16-gene
expression signature to distinguish stage I from stage II lung
squamous carcinoma. Int J Mol Med. 41:1377–1384. 2018.
|
|
18
|
Shao K, Shen LS, Li HH, Huang S and Zhang
Y: Systematic-analysis of mRNA expression profiles in skeletal
muscle of patients with type II diabetes: The glucocorticoid was
central in pathogenesis. J Cell Physiol. 233:4068–4076. 2018.
View Article : Google Scholar
|
|
19
|
Tuo Y, An N and Zhang M: Feature genes in
metastatic breast cancer identified by MetaDE and SVM classifier
methods. Mol Med Rep. 17:4281–4290. 2018.PubMed/NCBI
|
|
20
|
Tseng GC, Ghosh D and Feingold E:
Comprehensive literature review and statistical considerations for
microarray meta-analysis. Nucleic Acids Res. 40:3785–3799. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Spandidos A, Wang X, Wang H and Seed B:
PrimerBank: A resource of human and mouse PCR primer pairs for gene
expression detection and quantification. Nucleic Acids Res.
38:D792–D799. 2010. View Article : Google Scholar :
|
|
25
|
Stanley SA, Raghavan S, Hwang WW and Cox
JS: Acute infection and macrophage subversion by Mycobacterium
tuberculosis require a specialized secretion system. Proc Natl Acad
Sci USA. 100:13001–13006. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsu T, Hingley-Wilson SM, Chen B, Chen M,
Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et
al: The primary mechanism of attenuation of bacillus
Calmette-Guerin is a loss of secreted lytic function required for
invasion of lung interstitial tissue. Proc Natl Acad Sci USA.
100:12420–12425. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guinn KM, Hickey MJ, Mathur SK, Zakel KL,
Grotzke JE, Lewinsohn DM, Smith S and Sherman DR: Individual
RD1-region genes are required for export of ESAT-6/CFP-10 and for
virulence of Mycobacterium tuberculosis. Mol Microbiol. 51:359–370.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stanley SA, Johndrow JE, Manzanillo P and
Cox JS: The type I IFN response to infection with Mycobacterium
tuberculosis requires ESX-1-mediated secretion and contributes to
pathogenesis. J Immunol. 178:3143–3152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Veer MJ, Holko M, Frevel M, Walker E,
Der S, Paranjape JM, Silverman RH and Williams BR: Functional
classification of interferon-stimulated genes identified using
microarrays. J Leukoc Biol. 69:912–920. 2001.PubMed/NCBI
|
|
30
|
Taylor KE and Mossman KL: Recent advances
in understanding viral evasion of type I interferon. Immunology.
138:190–197. 2013. View Article : Google Scholar :
|
|
31
|
Manca C, Tsenova L, Bergtold A, Freeman S,
Tovey M, Musser JM, Barry CE III, Freedman VH and Kaplan G:
Virulence of a Mycobacterium tuberculosis clinical isolate in mice
is determined by failure to induce Th1 type immunity and is
associated with induction of IFN-alpha/beta. Proc Natl Acad Sci
USA. 98:5752–5757. 2001. View Article : Google Scholar
|
|
32
|
Flynn JL and Chan J: What's good for the
host is good for the bug. Trends Microbiol. 13:98–102. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ordway D, Henao-Tamayo M, Harton M,
Palanisamy G, Troudt J, Shanley C, Basaraba RJ and Orme IM: The
hypervirulent Mycobacterium tuberculosis strain HN878 induces a
potent TH1 response followed by rapid down-regulation. J Immunol.
179:522–531. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Antonelli LR, Gigliotti Rothfuchs A,
Gonçalves R, Roffê E, Cheever AW, Bafica A, Salazar AM, Feng CG and
Sher A: Intranasal Poly-IC treatment exacerbates tuberculosis in
mice through the pulmonary recruitment of a pathogen-permissive
monocyte/macrophage population. J Clin Invest. 120:1674–1682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Berry MP, Graham CM, McNab FW, Xu Z, Bloch
SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et
al: An interferon-inducible neutrophil-driven blood transcriptional
signature in human tuberculosis. Nature. 466:973–977. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Desvignes L, Wolf AJ and Ernst JD: Dynamic
roles of type I and type II IFNs in early infection with
Mycobacterium tuberculosis. J Immunol. 188:6205–6215. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McNab FW, Ewbank J, Rajsbaum R,
Stavropoulos E, Martirosyan A, Redford PS, Wu X, Graham CM, Saraiva
M, Tsichlis P, et al: TPL-2-ERK1/2 signaling promotes host
resistance against intracellular bacterial infection by negative
regulation of type I IFN production. J Immunol. 191:1732–1743.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dorhoi A, Yeremeev V, Nouailles G, Weiner
J III, Jörg S, Heinemann E, Oberbeck-Müller D, Knaul JK, Vogelzang
A, Reece ST, et al: Type I IFN signaling triggers immunopathology
in tuberculosis-susceptible mice by modulating lung phagocyte
dynamics. Eur J Immunol. 44:2380–2393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Der SD, Zhou A, Williams BR and Silverman
RH: Identification of genes differentially regulated by interferon
alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad
Sci USA. 95:15623–15628. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chaussabel D, Semnani RT, McDowell MA,
Sacks D, Sher A and Nutman TB: Unique gene expression profiles of
human macrophages and dendritic cells to phylogenetically distinct
parasites. Blood. 102:672–681. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hermann M and Bogunovic D: ISG15: In
sickness and in health. Trends Immunol. 38:79–93. 2017. View Article : Google Scholar
|
|
42
|
Chua PK, McCown MF, Rajyaguru S, Kular S,
Varma R, Symons J, Chiu SS, Cammack N and Nájera I: Modulation of
alpha interferon anti-hepatitis C virus activity by ISG15. J Gen
Virol. 90:2929–2939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang D and Zhang DE:
Interferon-stimulated gene 15 and the protein ISGylation system. J
Interferon Cytokine Res. 31:119–130. 2011. View Article : Google Scholar :
|
|
44
|
Speer SD, Li Z, Buta S, Payelle-Brogard B,
Qian L, Vigant F, Rubino E, Gardner TJ, Wedeking T, Hermann M, et
al: ISG15 deficiency and increased viral resistance in humans but
not mice. Nat Commun. 7:114962016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sooryanarain H, Rogers AJ, Cao D, Haac
MER, Karpe YA and Meng XJ: ISG15 modulates type I interferon
signaling and the antiviral response during Hepatitis E virus
replication. J Virol. 91:e00621–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kimmey JM, Campbell JA, Weiss LA, Monte
KJ, Lenschow DJ and Stallings CL: The impact of ISGylation during
Mycobacterium tuberculosis infection in mice. Microbes Infect.
19:249–258. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Keller C, Hoffmann R, Lang R, Brandau S,
Hermann C and Ehlers S: Genetically determined susceptibility to
tuberculosis in mice causally involves accelerated and enhanced
recruitment of granulocytes. Infect Immun. 74:4295–4309. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sambarey A, Devaprasad A, Mohan A, Ahmed
A, Nayak S, Swaminathan S, D'Souza G, Jesuraj A, Dhar C, Babu S, et
al: Unbiased identification of blood-based biomarkers for pulmonary
tuberculosis by modeling and mining molecular interaction networks.
EBioMedicine. 15:112–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sadler AJ and Williams BR:
Interferon-inducible antiviral effectors. Nat Rev Immunol.
8:559–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hancks DC, Hartley MK, Hagan C, Clark NL
and Elde NC: Overlapping patterns of rapid evolution in the nucleic
acid sensors cGAS and OAS1 suggest a common mechanism of pathogen
antagonism and escape. PLoS Genet. 11:e10052032015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Choi UY, Kang JS, Hwang YS and Kim YJ:
Oligoadenylate synthase-like (OASL) proteins: Dual functions and
associations with diseases. Exp Mol Med. 47:e1442015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee MS, Kim B, Oh GT and Kim YJ: OASL1
inhibits translation of the type I interferon-regulating
transcription factor IRF7. Nat Immunol. 14:346–355. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee MS, Park CH, Jeong YH, Kim YJ and Ha
SJ: Negative regulation of type I IFN expression by OASL1 permits
chronic viral infection and CD8(+) T-cell exhaustion. PLoS Pathog.
9:e10034782013. View Article : Google Scholar
|
|
54
|
de Toledo-Pinto TG, Ferreira AB,
Ribeiro-Alves M, Rodrigues LS, Batista-Silva LR, Silva BJ, Lemes
RM, Martinez AN, Sandoval FG, Alvarado-Arnez LE, et al:
STING-dependent 2′-5′ oligoadenylate synthetase-like production is
required for intracellular Mycobacterium leprae survival. J Infect
Dis. 214:311–320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Leisching G, Pietersen RD, van Heerden C,
van Helden P, Wiid I and Baker B: RNAseq reveals
hypervirulence-specific host responses to M. tuberculosis
infection. Virulence. 8:848–858. 2017. View Article : Google Scholar :
|
|
56
|
Cheng Y and Schorey JS: Mycobacterium
tuberculosis-induced IFN-β production requires cytosolic DNA and
RNA sensing pathways. J Exp Med. 215:2919–2935. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lin Y, Duan Z, Xu F, Zhang J, Shulgina MV
and Li F: Construction and analysis of the transcription
factor-microRNA co-regulatory network response to Mycobacterium
tuberculosis: A view from the blood. Am J Transl Res. 9:1962–1976.
2017.
|