Introduction
The liver serves a complex and vital role in
metabolism, synthesis, storage and redistribution of nutrients,
carbohydrates, fats and vitamins in the body (1-3).
It is one of the few organs that has retained a high regenerative
potential, allowing the recovery of >50% of its total mass
following damage or loss excretion (1,2,4,5).
The functional units of the liver are organized into lobules, which
are subsequently organized into larger lobes (1). Lobules consist of multiple hepatic
sinusoids, where the flow of blood from the portal triad to the
central vein contributes to a zonation based on decreasing oxygen
tension, which affects both the parenchymal (hepatocytes) and
non-parenchymal cells (NPCs) (1).
The hepatocytes account for 60% of the cell population and are
responsible for the biological functions of the liver, while the
NPCs comprise the remaining 40% of the cell population and serve an
important role in maintaining tissue architecture, mediating
responses to metabolic and toxic stimuli, and in supporting
hepatocyte functions (1). NPCs
contain multiple cell types, including liver sinusoidal endothelial
cells, Kupffer cells, hepatic stellate cells, and pit cells
(natural killer cells) (1). Given
the vital roles of the liver, severe liver damage under
pathological conditions may lead to high morbidity and mortality,
and liver diseases represent a growing global health burden.
The liver is also the major organ responsible for
detoxifying drugs, chemical wastes and xenobiotics through
biotransformation (1,2,4,5).
As a result, the liver is the most important target for
drug-induced toxicity (6).
Drug-induced liver injury (DILI) is a significant leading cause of
acute, chronic liver disease and an important safety issue when
developing new drugs (1,3,6-11).
For example, in the United States of America ~2,000 cases of acute
liver failure occur annually; DILI accounts for >50% of these,
among which 37% are caused by acetaminophen and 13% are
idiosyncratic reactions caused by other medications (11). Furthermore, DILI accounts for 2-5%
of the patients hospitalized with jaundice and ~10% of all cases of
acute hepatitis (11). Therefore,
it is essential to develop valid models to assess and/or predict
drug hepatotoxicity.
In previous decades, numerous in vitro and
in vivo models have been developed to assess drug-induced
hepatotoxicity, particularly for novel drug development (1,3,6-10,12). In vivo animal models have
been used to assess hepatotoxicity, although such models are
usually expensive and time-consuming. Furthermore, animal models
are not always good predictors of human-relevant DILI, owing to
significant species-specific differences in drug metabolism
pathways (3,13,14), although big data approaches may
improve the concordance of the toxicity of pharmaceuticals in
animals and humans (15).
Conversely, the majority of in vitro assay models involve
the use of primary hepatocytes, established liver cell lines, liver
slices, microsomes, perifusion culture systems, co-culture systems,
bioreactors, liver 'organ-on-chip', and/or liver organoids, each of
which has its own advantages and disadvantages (1,3,6-10,12,14,16-24). An ideal in vitro hepatic
function assessment model should retain the normal architecture of
hepatocytes and NPCs, and be easy to construct. (1,3,6-10,12,14,16-24).
The present study aimed to establish a highly
simplified yet effective 3-dimensional (3D) mouse liver microsphere
tissue culture (LMTC) model to assess hepatic functions that may be
impaired by hepatotoxins through a comparison of primary
hepatocytes and a mouse model. By freshly preparing perfused mouse
liver tissue with 80-mesh sift strainer, it was demonstrated that
the liver microsphere tissue exhibited normal hepatic functions for
up to 2 weeks and exhibited normal hepatic functions; however,
apparent tissue degradation and debris release, along with
diminished hepatic functions, were observed in the 2-week
microsphere culture. It was also demonstrated that the microsphere
tissue was responsive to bone morphogenic protein 9 (BMP9)
stimulation with the upregulation of numerous downstream target
genes of BMP9 signaling. Furthermore, it was revealed that 3
commonly used drugs, levofloxacin, azithromycin and paracetamol,
effectively inhibited hepatic indocyanine green (ICG) uptake and
induced higher expression levels of hepatotoxicity-associated genes
compared with that of the animals treated with these drugs in
vivo, suggesting that the LMTC model may be more sensitive in
detecting and predicting drug-associated hepatotoxicity. Therefore,
the simplified LMTC model should be useful for drug hepatotoxicity
and hepatocyte-based singling studies.
Materials and methods
Cell culture and chemicals
Mouse primary hepatocytes (MPH) were obtained from
4-week-old C57 mice using a previously described type I collagenase
liver perfusion protocol (19,25-30). 293-Derived 293pTP cells were
previously described (25,31).
The human colon cancer HCT116 cell line were obtained from the
American Type Culture Collection. The cells were maintained in
complete Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% FBS (Lonsa Science SRL), 100 U penicillin and 100
μg streptomycin at 37°C in 5% CO2. Levofloxacin,
azithromycin and paracetamol were purchased from Sigma-Aldrich;
Merck KGaA. Unless indicated otherwise, all other reagents were
purchased from Thermo Fisher Scientific, Inc. or Sigma-Aldrich;
Merck KGaA.
Mouse perfusion and liver tissue
recovery, and the establishment of mouse LMTC model
The use and care of animals in the present study was
approved by the Research and Experimental Animal Use Ethics
Committee of Chongqing Medical University (Chongqing, China; permit
no. SCXK(YU)20070001). All animal experiments were performed in
accordance with US National Institutes of Health Guide for the Care
and Use of Laboratory Animals (32). The 4-week-old C57BL/6 male mice
were obtained from the Animal Resource Center of Chongqing Medical
University.
Anesthesia was performed using intraperitoneal
injection of 3% sodium pentobarbital at a dose of 50 mg/kg.
Harvesting of mouse liver tissue was performed according to a
modified liver perfusion protocol (33-37). Specifically, following anesthesia
of the mice, following aseptic techniques, an incision was made in
upper-middle abdomen across the abdominal and chest cavities to
expose the liver and heart. Following the blockade of the right
heart circulation, perfusion of ~15 ml cold sterile PBS was rapidly
performed from the left ventricular until the liver turned pale.
Concomitantly, the rapid intra-cardiac perfusion led to acute
cardiac arrest and mortality of the mice, which was additionally
confirmed by cervical dislocation. Mice in the control
non-perfusion group were sacrificed with CO2, followed
by cervical dislocation.
The liver was resected and rinsed in PBS twice in
10-cm cell culture dishes, and then cut into small tissue pieces
with ophthalmic scissors, followed by passing the minced liver
tissue through 80-mesh (sift80) or 200-mesh (sift200) cell
strainer/filter. The recovered microsphere tissue pieces were
washed with sterile PBS by low speed centrifugation [500 × g for 5
min at room temperature (RT)], and immediately used for the in
vitro culture assays as described subsequently.
To establish the LMTC model, the recovered liver
microsphere tissue pieces were cultured in 24-well plates with
various concentrations of FBS and/or bovine calf serum (BCS;
Sijiqing; Zhejiang Tianhang Biotechnology Co., Ltd.) containing 100
U penicillin and 100 μg streptomycin at 37°C in 5%
CO2. Medium was changed daily for the first 3 days, and
then changed every other day.
Amplification and titering of recombinant
adenoviruses expressing BMP9 or green fluorescent protein
(GFP)
Recombinant adenoviruses were generated by using the
AdEasy technology and amplification as described previously
(38-40). The Recombinant adenovirus Ad-BMP9
was previously characterized (41-45). Ad-BMP9 also co-expresses enhanced
GFP. An analogous adenovirus expressing GFP (Ad-GFP) was used as a
mock virus control. For all adenoviral infections, polybrene (4-8
μg/ml) was added to potentiate infection efficiency as
described previously (46).
Preparation of BMP9-conditioned medium
from HCT116 cells
The preparations for BMP9 conditioned medium were
performed as previously described (45). Briefly, subconfluent HCT116 cells
were infected with the optimal titer (MOI =50) of Ad-BMP9. At 24 h
after infection, the culture was changed to serum-free Opti-MEM
media (Thermo Fisher Scientific, Inc.). The media were collected
every 12 h for 4 times consecutively. The pooled BMP9 conditioned
medium was centrifuged at 500 × g for 10 min at RT to remove any
cell debris, aliquoted and stored at −80°C. The control conditioned
medium was also prepared in the same fashion from Ad-GFP infected
HCT116 cells (45,47).
Hematoxylin and eosin (H&E) staining
and Hoechst33258 staining
The recovered liver tissue was rinsed with PBS,
fixed with 4% paraformaldehyde for 30 min at RT, and subjected to
H&E staining as described previously (48-50). The fixed tissue was also stained
with Hoechst33258 (10 μg/ml Hoechst 33258 in PBS) for 5 min
at RT, examined and recorded under a fluorescence microscope
(magnification, ×200).
Immunohistochemical (IHC) and
immunofluorescence (IF) staining on liver microsphere tissue
The IHC and IF staining of the liver microsphere
tissue were performed as described previously (51-53). The recovered liver tissue was
fixed with 4% paraformaldehyde for 30 min at RT, followed by
antigen retrieval and immunostaining with anti-proliferating cell
nuclear antigen (PCNA; 1:100-1:200; cat. no. 13110; Cell Signaling
Technology, Inc.) or anti-Myc proto-oncogene protein (c-Myc;
1:100-1:200; cat. no. ab39688; Abcam) antibodies. Control rabbit
IgG (1:200; cat. no. 011-000-003; Jackson ImmunoResearch
Laboratories, Inc.) was used as a negative control.
ICG uptake/release assay
The ICG uptake/release assay was performed as
described previously (30).
Briefly, recovered liver tissue and cells were washed with PBS and
incubated with ICG (1 mg/ml in complete DMEM) at 37°C for 30 min,
followed by two washes with PBS. For ICG release detection, the
ICG-containing medium was replaced with 2% BCS DMEM, and the tissue
and cells were incubated for an additional 3 h at 37°C in a cell
culture incubator. ICG uptake/release was observed and recorded
under a bright field microscope (magnification, ×200).
Periodic acid-Schiff (PAS) staining
PAS staining was performed as described previously
(19,27,28,30). Briefly, recovered liver tissue was
fixed with 4% paraformaldehyde for 30 min at RT, followed by
staining with 0.5% periodic acid solution for 5 min at RT.
Following rinsing in distilled water for 3 min, the tissues were
incubated in the Schiff's reagent for 15 min, followed by thorough
rinsing with tap water. Cell staining was recorded under a bright
field microscope (magnification, ×200).
Total RNA isolation and touchdown reverse
transcription- quantitative polymerase chain reaction (TqPCR)
analysis
Total RNA from both tissue and cells was isolated by
using TRIzol reagent (CoWin Biosciences) according to the
manufacturer's protocol. The perfused mouse liver tissues from
normal 6-week-old C57BL/6 mice (n=5 males/group) and drug or
PBS-treated 6-week-old C57BL/6 mice (n=5 males/group) were
dissected out and homogenized in the TRIzol reagent. The recovered
liver tissue and cells were lysed in TRIzol reagent. Total RNA was
extracted and subjected to reverse transcription reactions with
hexamer and M-MuLV reverse transcriptase (New England Biolabs,
Inc.). The cDNA products were additionally diluted and used as PCR
templates. The gene-specific PCR primers (Table SI) were designed using Primer3
Plus (http://www.bioinformatics.nl/cgi-bin/primer-3plus/primer3plus.cgi).
TqPCR was performed using SYBR Green-based TqPCR analysis on a
CFX-Connectunit system (Bio-Rad Laboratories, Inc.), as described
previously (54). TqPCR reactions
were performed in triplicate. GAPDH was used as the
reference gene. Quantification of gene expression was performed
using the 2−ΔΔCq method (55).
WST-1 cell proliferation assay
Cell proliferation was assessed using the Premix
WST-1 Cell Proliferation Assay System (Clontech Laboratories,
Inc.), as described previously (56-59). The MPH seeded in 96-well plates at
6,000 cells/well were treated with levofloxacin (at 1, 5 and 25
μM), azithromycin (at 25, 125 and 625 nM), paracetamol (at
20, 100 and 500 μM) or DMSO for 24, 48 or 72 h. The Premix
WST-1 Reagent was added to each well, followed by incubation at
37°C for 60 min and reading at 440 nm using the EL800 microplate
reader (BioTek Instruments, Inc.). Each assay was performed in
triplicate.
Statistical analysis
All quantitative experiments were performed in
triplicate and/or repeated 3 times. Data are presented as mean ±
standard deviation. Significant differences between groups were
determined using a one-way analysis of variance followed by a Least
Significant Difference post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Optimization of the culturing conditions
for the 3D LMTC model using mouse liver tissue
Liver tissue is rich in blood cells and lipofuscin,
pyridine (NADPH) and flavin coenzymes, which are common causes of
high auto-fluorescence. To minimize the cytotoxicity and
auto-fluorescence caused by blood cell disintegration, left
ventricular perfusion was performed to remove intrahepatic blood in
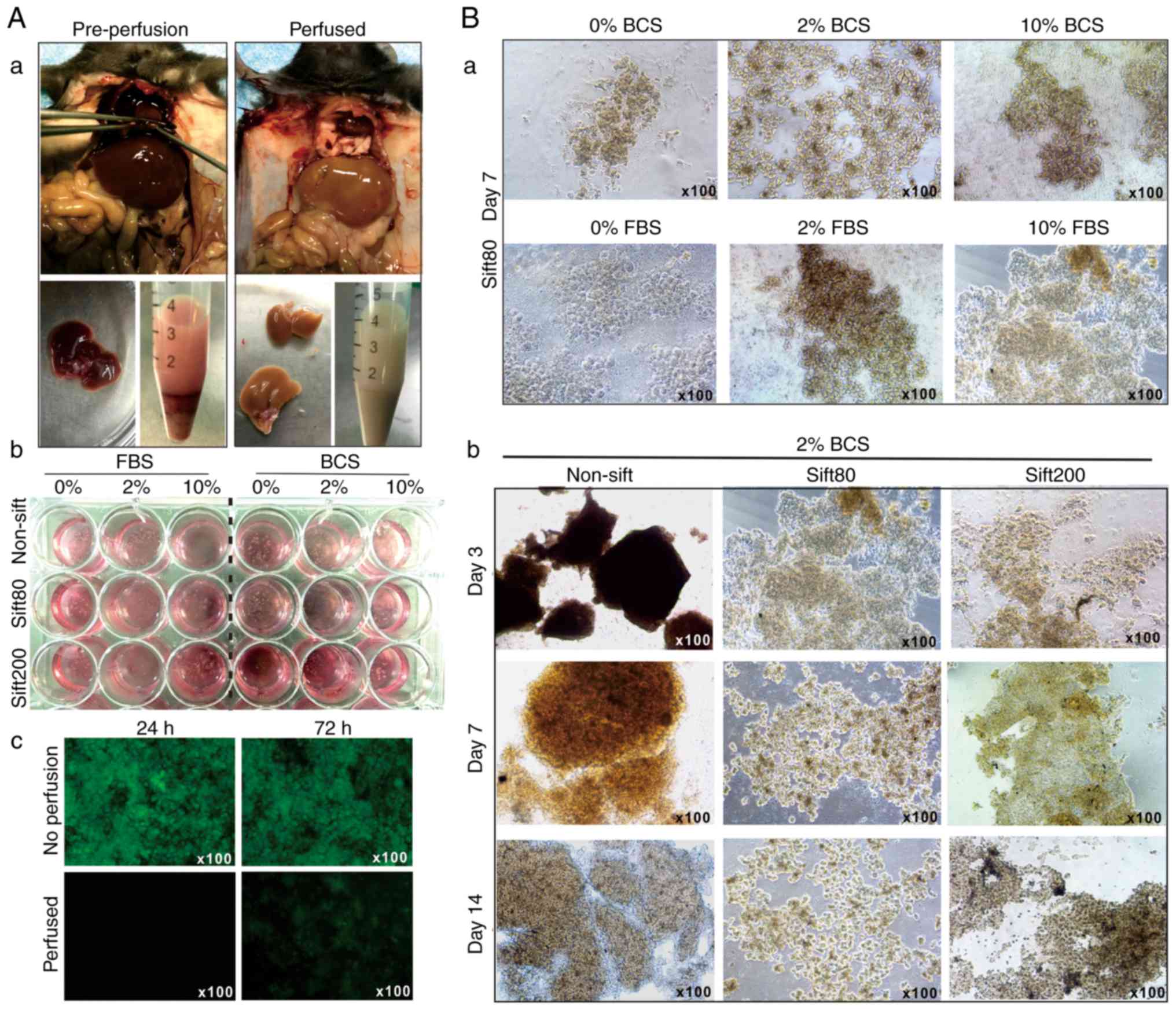
the anesthetized mice (Fig.
1A-a). The liver tissue was prepared in different sizes by
sifting through sift80 or sift200 filter strainers, and the
microsphere tissue preparations were cultured in different
concentrations of FBS or BCS-containing DMEM (Fig. 1A-b). The perfused liver tissue
culture exhibited significantly decreased auto-fluorescence at 24
and 72 h after culturing, compared with culture established with
tissue from the non-perfusion group (Fig. 1A-c).
When the liver microsphere tissue sifted with the
sift80 strainer was cultured with 0, 2 or 10% FBS or BCS DMEM
medium, followed by medium change every day for the first 3 days,
and then every other day for the rest of the study period, it was
identified that the microsphere tissue was in the healthiest state
in 2% BCS medium (Fig. 1B-a).
It was also identified that non-sifted liver tissue
pieces were mostly suspended in 2% BCS medium, and the sift80
tissue was mostly attached to the bottom of the culture plates and
grew healthily for up to 14 days in 2% BCS medium (Fig. 1B-b). Notably, the sift200
microsphere tissue was demonstrated to grow somewhere in between
that of the non-sifted tissue and sift80 microsphere tissue at day
7, but significantly deteriorated at day 14 (Fig. 1B-b). It is noteworthy that,
although the LMTC culture could be passaged 1 to 2 times, the
percentage of viable cells that survived the passages was rather
low, at <10% (data not shown). Collectively, these results
indicated that the optimal LMTC culturing conditions should include
preparation of the mouse liver tissue with 80-mesh sift strainer
and culturing of the sift80 microsphere tissue in 2% BCS/DMEM
medium with daily medium change for the first 72 h, and then once
every 2 days thereafter.
Proliferative and apoptotic
characteristics of the 3DLMTC model
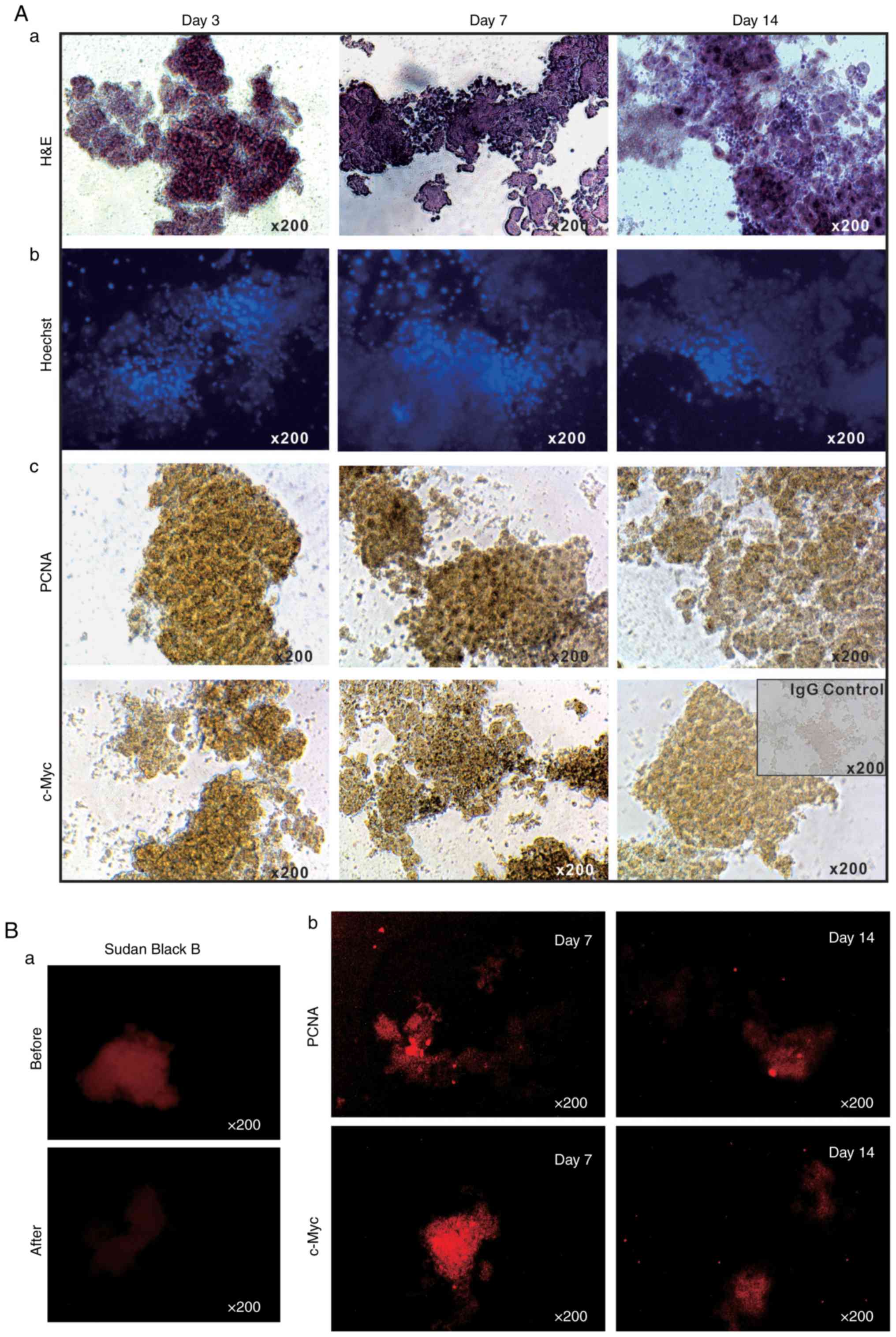
The long-term survival of the LMTC model was
additionally analyzed by culturing sift80 microspheres in 2%
BCS/DMEM for up to 14 days. H&E staining of the sift80
microsphere tissue in 2% BCS/DMEM was performed and it was
identified that the cellularity was preserved at the 3 time-points
examined (days 3, 7 and 14), although the microsphere tissue
scaffolds became loose and disintegrated to a certain extent
(Fig. 2A-a). Accordingly, the
Hoechst 33258 staining assay indicated that the majority of the
cell nuclei were readily visible without any apparent fragmentation
(Fig. 2A-b).
It was also identified that the sift80 microsphere
tissue exhibited strong positive staining for PCNA at days 3 and 7,
with a decreased level of staining at day 14 (Fig. 2A-c, top row). Similarly, high
levels of c-Myc expression were observed at days 3 and 7, although
the levels had significantly decreased by day 14 (Fig. 2A-c, bottom row). These IHC
staining results were further confirmed by IF staining. As liver
tissue exhibits auto-fluorescence, Sudan Black B was employed to
effectively suppress background fluorescence (Fig. 2B-a). IF staining with PCNA and
c-Myc antibodies indicated that the sift80 microspheres markedly
expressed both PCNA and c-Myc at day 7, but that the levels were
rather weak at day 14. Taken together, these results demonstrate
that the sift80 microspheres may be healthily maintained for at
least 7 days, and potentially for up to 14 days.
Hepatic functional characteristics of the
3D LMTC model
To determine whether the cultured microsphere tissue
exhibited normal liver functions, ICG uptake/release and Periodic
Acid-Schiff (PAS) staining of glycogen storage assays were
performed, which are liver-specific functions (19,28,60). It was identified that the sift80
microsphere tissue cultured at days 3 and 7, and to a lesser extent
at day 14, was able to uptake ICG effectively (Fig. 3A-a). Conversely, it was
demonstrated that the ICG was effectively released from the
cultured sift80 microsphere tissue at the 3 time points examined
(Fig. 3A-b). Furthermore, marked
PAS staining of the cultured sift80 microsphere tissues indicated
that the hepatocytes were viable and healthy, particularly at days
3 and 7 (Fig. 3B). These results
suggested that the sift80 microsphere tissue may retain normal
hepatic functions under the in vitro culture conditions for
up to 14 days.
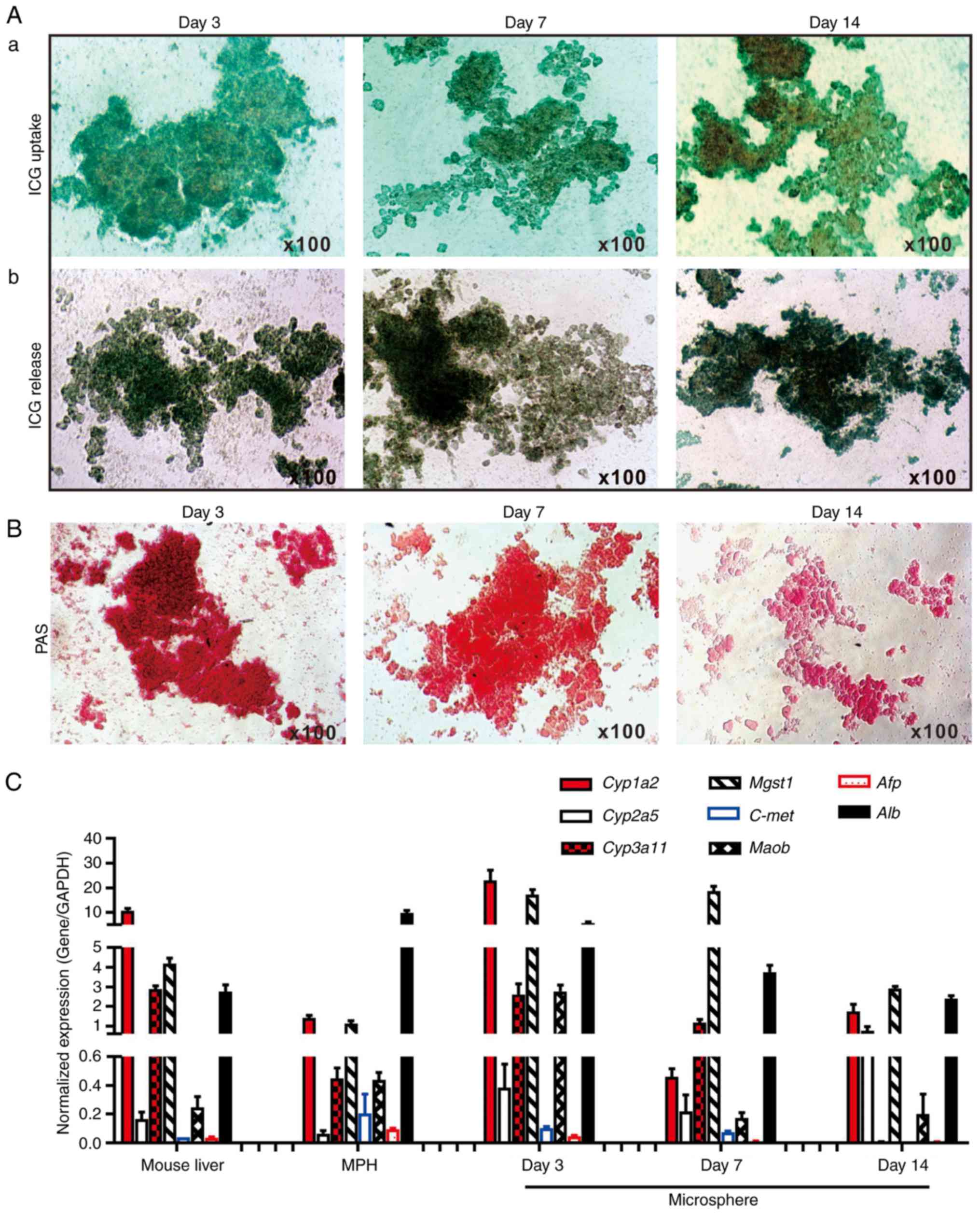 | Figure 3Hepatic functional characterization
of 3-dimensional LMTC model. (A) ICG uptake and release assay. (a)
The freshly recovered mouse liver sift80 microsphere tissue was
incubated with ICG (1 mg/ml in 2% BCS/DMEM) at 37°C for 30 min. ICG
uptake was recorded under a bright field microscope. (b) The
microsphere tissue was then incubated in 2% BCS/DMEM for an
additional 3 h, and the ICG release was recorded under a bright
field microscope. Each assay condition was performed in triplicate.
Representative results are presented. (B) PAS staining for hepatic
glycogen storage. At the indicated time points, the cultured sift80
microsphere tissue was subjected to PAS staining, and the PAS
staining was recorded under a bright field microscope. Each assay
condition was performed in triplicate. Representative results are
presented. (C) Expression profile of hepatocyte-specific genes in
freshly isolated mouse liver tissues, the MPH and the LMTC
microsphere tissue cultures at different time points. Total RNA was
isolated from the 6-week old C57BL mouse liver, the MPH, and
microsphere tissue cultured for 3, 7 and 14 days, and subjected to
touchdown quantitative polymerase chain reaction analysis to detect
the expression of hepatic genes. All samples were normalized with
respective GAPDH expression levels. Each assay condition was
performed in triplicate. LMTC, liver microsphere tissue culture;
ICG, indocyanine green; DMEM, Dulbecco's modified Eagle's medium;
PAS, Periodic acid-Schiff; MPH, mouse primary hepatocytes; Cyp1a2,
cytochrome P450 family 1 subfamily Amember; Cyp2a5, cytochrome P450
2A5; Cyp3a11, cytochrome P450 family 3 subfamily A member 4; Mgst1,
microsomal glutathione S-transferase 1; C-Met, MET proto-oncogene,
receptor tyrosine kinase; Maob, monoamine oxidase B; Afp, α
fetoprotein; Alb, albumin. |
To additionally determine whether the in
vitro culture would affect hepatic gene expression pattern, the
expression profiles of a panel of eight hepatocyte-specific genes
were compared (19,27,28,30,61-63) in the cultured microspheres and
that of the freshly-isolated mouse liver tissue and the MPH. Using
TqPCR analysis, it was identified that cytochrome P450 family 1
subfamily Amember 2, cytochrome P450 family 3 subfamily
Amember 4, microsomal glutathione S-transferase 1 and
albumin (Alb), and to a lesser extent cytochrome P450 2A5
and Maob, were highly expressed in the sift80 microsphere
tissue cultured at days 3, 7 and 14, and in the freshly-prepared
mouse liver tissue and in the MPH, although their expression in the
sift80 microsphere tissue samples decreased over time (Fig. 3C). Conversely, the expression
levels of α fetoprotein and MET proto-oncogene, receptor tyrosine
kinase were low or undetectable in all samples (Fig. 3C). The hepatic gene expression in
the microsphere tissue samples appeared to decrease over time,
particularly at day 14, which may be indicative of lower viability
of the microsphere tissue after culture for 2 weeks. Collectively,
these results demonstrated that the LMTC samples exhibited a normal
hepatic gene expression profile under the in vitro culture
conditions for at least 7 days, and potentially up to 14 days,
although the 2-week microsphere culture exhibited apparent
morphological changes with signs of tissue degradation and tissue
debris (data not shown). Therefore, considering the morphological
changes and the diminished hepatic functions and gene expression
profile of the 2-week microsphere culture, the LMTC microspheres'
viability may be limited to no longer than 2 weeks under the
culture conditions of the present study.
3D LMTC model for hepatocyte-based cell
signaling investigation
Whether the LMTC model could be used to study the
cell-based signaling pathways that may be involved in the effective
delivery of transgenes into the microsphere tissue or exposure to
secreted growth factors was examined. Whether the mouse liver
microsphere tissue could be effectively transduced with recombinant
adenoviruses, which represent one of the most effective gene
delivery approaches, was first investigated. Considering the fact
that the liver tissue may exhibit different proliferative potential
at different ages, sift80 microsphere tissues from newborn,
14-day-old and 28-day-old mouse liver tissue were prepared, and
then infected with the same titer of adenovirus Ad-GFP. Only
sparsely infected GFP+ hepatocytes were detected in the
sift80 microsphere tissue prepared from newborn mice at days 3 and
7, while GFP+ cells were very few or undetectable in the
microsphere tissue from 14-day-old and 28-day-old mouse liver
tissue; the Ad-GFP-infected MPH cells were used for the control
(Fig. 4A). In fact, the
adenovirus-mediated transgene delivery to whole mouse liver tissue
was analyzed in vitro at 0, 3, 7, 10 and 14 days after
birth, and only sparsely distributed, focal Ad-GFP infection was
observed (data not shown). These results suggest that mouse liver
tissues, whilst viable in culture, may have limited intrinsic
proliferative potential for transgene expression in
vitro.
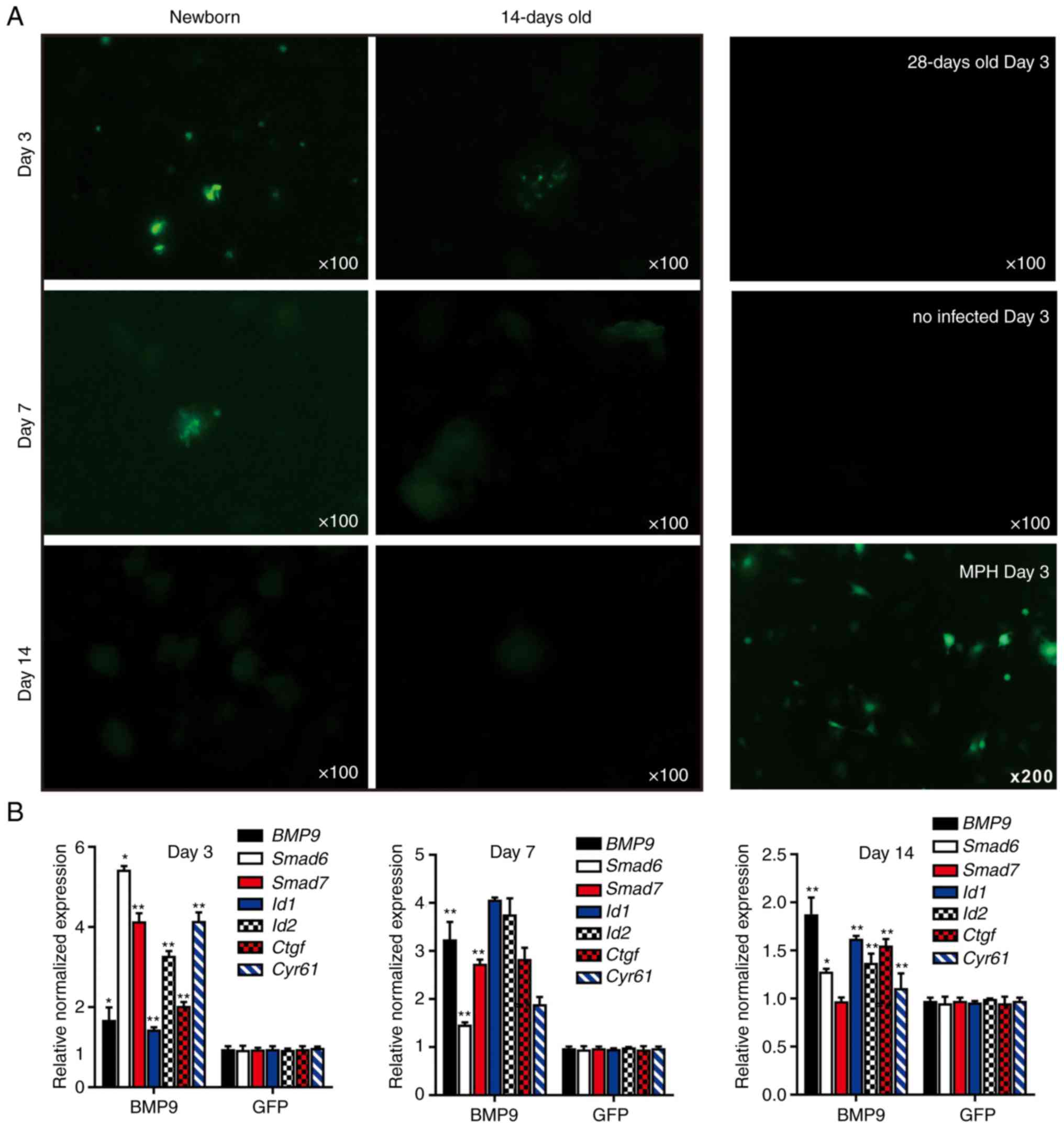 | Figure 43-Dimensional liver microsphere
tissue culture model for hepatic exogenous investigation. (A)
Cultured liver tissue was relatively refractory to Ad-mediated
transgene delivery. The microsphere tissue samples were prepared
from newborn, 14- and 28-day-old C57BL mouse liver samples, and
were infected with the same titer of Ad-GFP. The GFP signal was
recorded at 3, 7 and 14 days after infection. The MPH were also
infected with Ad-GFP as a control, and the GFP signal at day 14 is
presented. (B) Stimulation of the sift80 tissue microspheres with
BMP9-conditioned medium to examine cell signaling and TqPCR
analysis. Total RNA was isolated from the sift80 tissue
microspheres treated with BMP9 or GFP conditioned medium at the
indicated time points, and subjected to TqPCR analysis of BMP9
downstream target genes. All samples were normalized with
respective GAPDH expression levels. *P<0.05
and **P<0.01 vs. GFP groups. Each assay condition was
done in triplicate. Ad, adenovirus; GFP, green fluorescent protein;
MPH, mouse primary hepatocytes; BMP9, bone morphogenic protein 9;
TqPCR, touchdown quantitative polymerase chain reaction analysis;
Id1, inhibitor of DNA binding 1, HLH protein; Id2, inhibitor of DNA
binding 2; Ctgf, connective tissue growth factor; Cyr61, cellular
communication network factor 1. |
Whether liver microsphere tissues were responsive to
signal molecule stimulation was then examined. It has been
previously demonstrated that BMP9 is a highly expressed secreted
protein in fetal mouse liver tissues, and that it exerts
pleiotropic effects on stem cell proliferation and differentiation
(41-43,64-68). The BMP9-containing conditioned
medium was first prepared by infecting HCT116 cells with Ad-BMP9,
as described previously (45,47,69), while the Ad-GFP-infected HCT116
cells were used to prepare for the control conditioned medium
(Fig. S1). A high level of BMP9
expression in the Ad-BMP9-infected HCT116, but not in the Ad-GFP
infected cells, was confirmed by qPCR (Fig. S1). The BMP9-conditioned medium
and GFP control medium were used to stimulate the freshly prepared
sift80 microsphere tissue (Fig.
4B). At 3, 7 and 14 days post-BMP9 stimulation, total RNA was
isolated and TqPCR analysis of BMP9 downstream target genes was
performed as described previously (65,70-73). It was identified that the BMP9
downstream early responsive genes, Smad6 and Smad7,
were significantly upregulated at day 3, while other target genes
including inhibitor of DNA binding 1, HLH protein, inhibitor
of DNA binding 2, connective tissue growth factor and
cellular communication network factor 1 were significantly
upregulated at days 3 and 7. However, the expression of the
majority of the target genes significantly decreased to basal level
at day 14 of culturing. Taken together, these results suggested
that mouse liver microsphere tissues, whilst not serving as an
ideal recipient for transgene delivery, may be used as an effective
in vitro 3D model system for cytokine or growth factor-based
cell signaling studies.
3D LMTC model for drug-induced
hepatotoxicity assays
As DILI remained the leading cause of acute liver
failure and post-market drug withdrawals (5), whether the liver microspheres could
be used to assess the hepatotoxicity induced by liver injury drugs
was assessed. A total of 3 commonly-used hepatotoxic drugs,
levofloxacin, azithromycin and paracetamol, were selected. Using a
WST-1 cell proliferation assay, it was identified that the 3 drugs
inhibited cell proliferation rates of the MPH in a dose- and
time-dependent manner (Fig. 5A).
Therefore, the following optimal doses were selected for subsequent
in vitro studies: Levofloxacin at 5 μM (Fig. 5A-a), azithromycin at 125 nM
(Fig. 5A-b) and paracetamol at
100 μM (Fig. 5A-c).
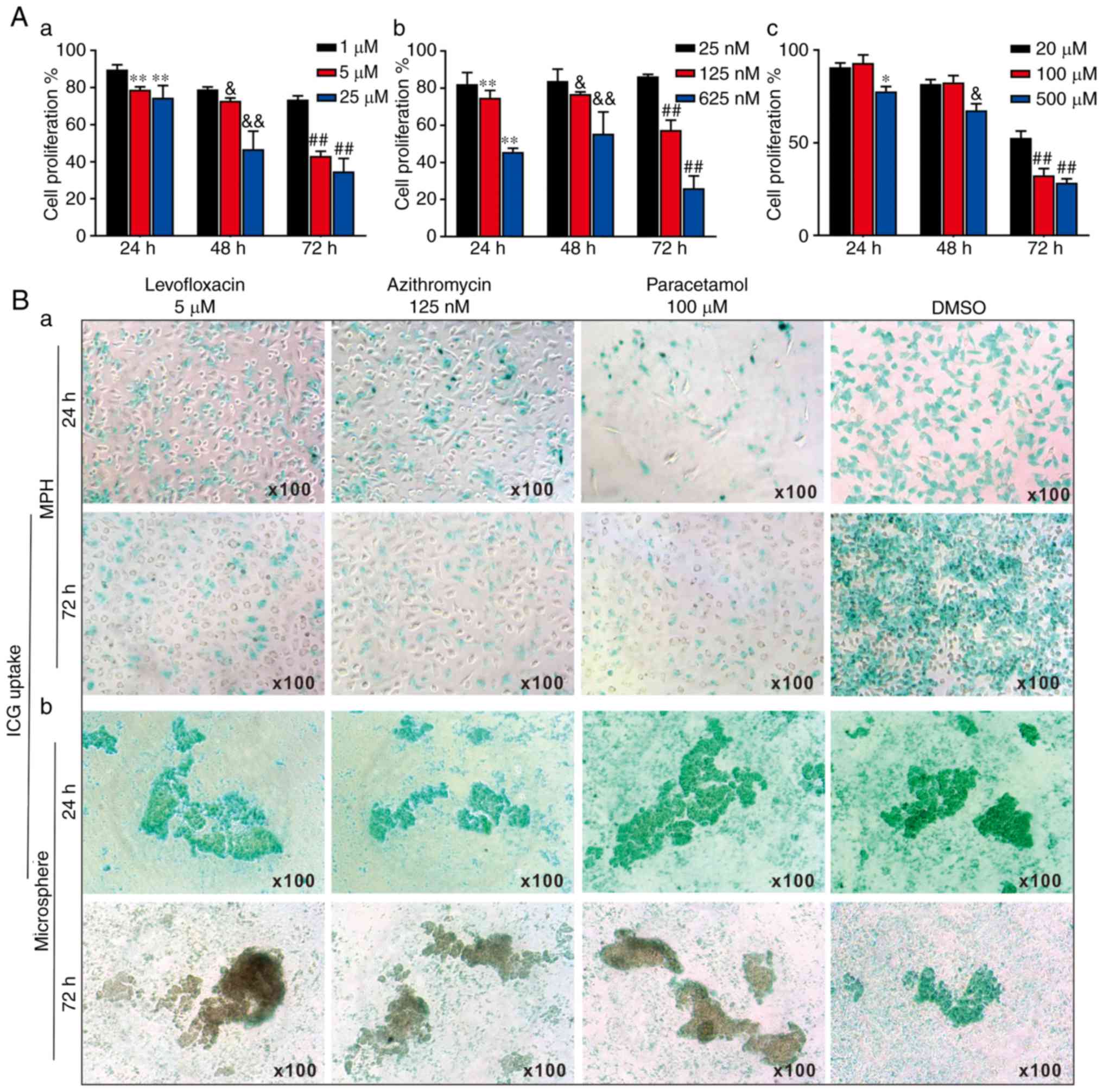 | Figure 53-Dimensional liver microsphere
tissue culture model for assessing the hepatotoxicity of
liver-injury drugs. (A) The effect of 3 liver injury-inducing drugs
on hepatic cell proliferation. Subconfluent MPH were seeded in
96-well plates and treated with (a) levofloxacin, (b) azithromycin,
(c) paracetamol, or DMSO at the indicated concentrations. At 24, 48
and 72 h post-treatment, WST-1 reagent was added to the plates and
incubated for 1 h prior to absorbance measurement. (a)
**P<0.01 vs. 1 μM at 24 h;
&P<0.05 and &&P<0.01 vs. 1
μM at 48 h; ##P<0.01 vs. 1 μM at 72 h.
(b) **P<0.01 vs. 25 nM at 24 h;
&P<0.05 and &&P<0.01 vs. 25
nM at 48 h; ##P<0.01 vs. 25 nM at 72 h. (c)
*P<0.05 vs. 20 μM at 24 h;
&P<0.05 vs. 20 μM at 48 h;
##P<0.01 vs. 20 μM at 72 h. (B) ICG uptake
assay. (a) Subconfluent MPH and (b) sift80 microsphere tissues were
treated with levofloxacin (5 μM), azithromycin (125 nM),
paracetamol (100 μM) or DMSO for up to 72 h. The cells and
microsphere tissue were incubated with ICG (1 mg/ml in 2% BCS/DMEM)
at 37°C for 30 min, and then the ICG-DMEM was changed to complete
DMEM at 24 and 72 h. ICG uptake was recorded under a bright field
microscope. Each assay condition was performed in triplicate.
Representative images are presented. ICG, indocyanine green; MPH,
mouse primary hepatocytes; DMEM, Dulbecco's modified Eagle's
medium. |
The effects of the 3 drugs on ICG uptake in the MPH
and the freshly prepared sift80 microspheres were analyzed. When
subconfluent MPH cells were treated with levofloxacin (5
μM), azithromycin (125 nM), paracetamol (100 μM) or
DMSO control, the ICG uptake was remarkably inhibited by all 3
drugs at both 24 h and 72 h, compared with that of the DMSO control
group (Fig. 5B-a). However, when
the sift80 microsphere tissue was treated with levofloxacin (5
μM), azithromycin (125 nM), paracetamol (100 μM) or
DMSO control, the ICG uptake was not significantly inhibited by the
3 drugs at 24 h, but was markedly inhibited at 72 h, compared with
the DMSO control group (Fig.
5B-b). Therefore, these results demonstrate that the mouse
liver microsphere tissue culture model may be used as an effective
in vitro surrogate system to assess the hepatic functional
abnormality caused by hepatotoxic drugs.
Whether the LMTC model was able to predict the
hepatotoxicity that was closely correlated with the hepatotoxic
effects obtained from in vivo animal studies was also
examined. The mice were treated with the levofloxacin,
azithromycin, paracetamol or DMSO control, and mouse liver tissue
was collected at 24 h or 72 h after treatment. Concomitantly, the
subconfluent MPH and freshly-prepared sift80 microsphere samples
were treated with levofloxacin, azithromycin, paracetamol or DMSO.
Total RNA was isolated from the drug-treated mouse liver tissues,
the MPH and the sift80 microspheres for quantitative analysis of
the expression of a panel of 5 hepatotoxicity-associated genes,
including ATP binding cassette subfamily B member 11
(Abcb11), cytochrome P450 family 2 subfamily E member
1 (Cyp2e1), cytochrome P450 family 7 subfamily
Amember 1, nuclear receptor subfamily 0 group B member 2
(Nr0b2) and nuclear receptor subfamily 1 group H member 4
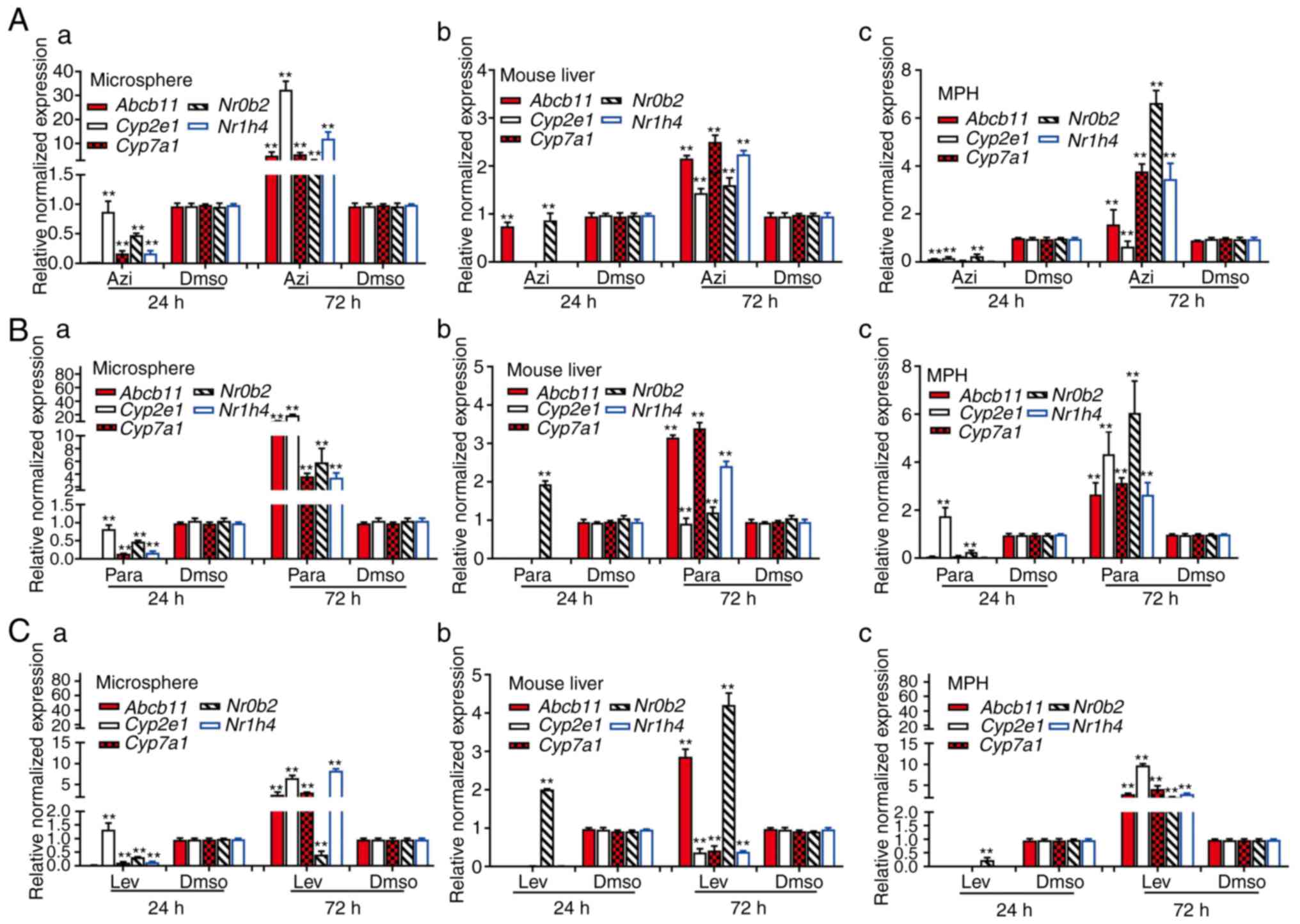
(74-76). In the levofloxacin-treated mouse
liver tissue, MPH and sift80 microsphere tissue groups, while the
majority of the hepatotoxicity-associated genes (with the exception
of Nr0b2 in the mouse liver group) were not significantly
upregulated at 24 h post treatment, 2/5 genes (Abcb11 and
Nr0b2) in the mouse liver group, all 5 genes in the MPH
group, and 4/5 genes in the sift80 microsphere tissue group were
significantly upregulated at 72 h after treatment (Fig. 6A). In the azithromycin treatment
groups, while all 5 genes were repressed at 24 h, all 5 genes were
highly upregulated in all three groups (with the exception of
Cyp2e1 in the MPH group) at 72 h after treatment (Fig. 6B). Similar results were observed
in the paracetamol treatment groups, and all 5 genes were highly
upregulated in all three groups (with the exception of
Cyp2e1 in the mouse liver group) at 72 h after treatment
(Fig. 6C). Notably, for the 3
drugs examined, the magnitudes of gene expression upregulation were
increased in the sift80 microsphere tissue groups compared with
that in the mouse liver groups at the 72 h treatment time point,
suggesting that the LMTC model may be more sensitive in predicting
drug-associated hepatotoxicity.
Discussion
In the present study, a simple yet effective 3D
microsphere culture system of a mouse liver was successfully
developed. By freshly preparing the perfused mouse liver tissue
with an 80-mesh sift strainer, it was demonstrated that, under the
optimal culture condition of 2% BCS/DMEM, the microspheres remained
viable with marked PCNA and c-Myc expression, and exhibited normal
hepatic functions, including ICG uptake/release and glycogen
synthesis/storage, for up to 2 weeks. However, morphological
analysis of cells after 2 weeks of culture revealed tissue
degradation and observation of tissue debris, together with
diminished hepatic function and gene expression, indicating that
the viability of the microspheres may be limited to <2 weeks. It
was also demonstrated that the cultured microspheres exhibited a
similar expression profile of hepatocyte-specific genes to that of
the freshly isolated mouse liver tissue. While the microspheres
exhibited limited intrinsic proliferative potential for transgene
expression, it was demonstrated that the microspheres were
responsive to BMP9 stimulation, and numerous downstream target
genes of BMP9 signaling were effectively upregulated. Furthermore,
using 3 commonly-used drugs, levofloxacin, azithromycin and
paracetamol, it was revealed that the 3 drugs effectively inhibited
hepatic ICG uptake at 72 h after treatment and induced increased
expression levels of hepatotoxicity-associated genes compared with
that of the animals treated with these drugs in vivo,
suggesting that the LMTC model may be more sensitive in detecting
the expression of hepatotoxicity-associated genes, and therefore
more sensitive in predicting drug-associated hepatotoxicity.
The liver serves a critical role in
biotransformation and disposition of drugs or xenobiotics. As a
result, hepatotoxicity may be caused by a wide range of
pharmaceutical agents, natural products, chemicals, environmental
pollutants or dietary factors (21,77). In fact, hepatotoxicity or DILI is
a major cause for drug withdrawals worldwide (22). Commonly-used techniques to assess
DILI effects include in vivo animal models and various in
vitro models (14,21-23). While the in vivo animal
models may more accurately predict drug hepatotoxicity, they are
more costly and time-consuming to perform (14,23). Overall, the development of in
vitro liver models to study disease and the prediction of
metabolism and drug-induced liver injury in humans remains a
challenge (78). Therefore, in
vivo models are used for the advanced stages of drug
development.
There have been numerous attempts to establish ideal
in vitro hepatotoxicity assessment systems, which include
the uses of primary hepatocytes or established liver cell lines
alone or in co-culture with other cell types in 2D and 3D formats,
including liver slices, microsomes, perifusion culture systems,
co-culture systems, bioreactors, liver 'organ-on-chip' and/or liver
organoids (1,3,6-10,12,14,16-24).
The use of primary hepatocytes alone or in a
co-culture format is largely limited by the inefficient recovery of
primary cells from liver tissue (77,79), which may be easily overcome by the
LMTC microsphere tissue culture system described in the present
study. Liver cancer lines and/or immortalized liver cell lines are
also used as an alternative to primary hepatocytes in these models,
although these cell types may not be able to completely replicate
the biological characteristics of primary hepatocytes (3,10,19,22,24). In these system, protein and urea
synthesis, glucose metabolism and cytochrome (CYP450) activities
were stable over a 2-week culture period, with maximal activities
at the end of the first week in the majority of models (80,81). Nonetheless, the maintenance of
functional primary hepatocytes cultures has been difficult, due to
dedifferentiation and the consequent loss of hepatic function with
limited utility (78).
Compared with the hepatocytes cultured in 2D
format, 3D hepatocyte culture, such as the LMTC system described in
the present study, should result in improved replication of the
morphological structure and growth microenvironment of liver cells
(3,7,10),
as 3D culture models have been demonstrated to be beneficial for
cell viability in other organ systems (50,82,83). In other models, the liver slices
were prepared by using a tissue slice with a 10-mm diameter
motor-driven tissue-coring tool in cold oxygenated (95%
O2 and 5%CO2) (16,18). Precision-cut liver slices have
been used for the investigation of hepatic metabolism,
hepatotoxicity and enzyme induction (8). An advantage of using liver slices is
the potential for examining the toxic effects on hepatocytes that
are mediated by nonparenchymal cells, as the physiological liver
microarchitecture is maintained in cultured slices (8). The liver microsomes were prepared by
homogenizing the liver tissue with a Potter glass homogenizer
equipped with a Teflon pestle followed by ultracentrifugation
(18,20,84). It has been demonstrated that
long-term stable primary hepatic 3D spheroid cultures in chemically
defined conditions may be used to predict drug-induced
hepatotoxicity (85). In future
studies, the liver microphysiological systems, also referred to as
'liver-on-a-chip', present the opportunity to explore system/organ
level effects without using animal experimentation (1,3,78).
However, the complexity of the systems and the requirements of the
equipment make the wider application of a number of these
techniques difficult across the various fields of liver research.
Conversely, the LMTC microsphere tissue culture system described in
the present study is simple, effective and biologically relevant in
terms of replicating hepatic functions in vitro.
In summary, compared with a number of the
aforementioned hepatotoxicity assessment systems, the LMTC model
described in the present study was relatively simple and easy to
prepare, and yet highly effective and reproducible. This
microsphere tissue model system required minimal resources and
could be maintained for up to 2 weeks. Therefore, this system may
be a valuable tool to assess drug-induced hepatotoxicity and
metabolism, and to investigate hepatocyte-based cell signaling
mechanisms.
Supplementary Data
Acknowledgements
Not applicable.
Funding
The present study was supported in part by research
grants from the 2017 Chongqing Postdoctoral Innovation Talent
Support Program (JMF), the China Postdoctoral Research Fund (grant
no. 2018M643426 to JMF) and the National Key Research and
Development Program of China (grant nos. 2016YFC1000803 and
2011CB707906). TCH was also supported by the Mabel Green Myers
Research Endowment Fund and The University of Chicago Orthopaedic
Surgery Alumni Fund.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
YZ, QP, YG, TY and YC performed the experiments.
JF, HW and YL analyzed the data, and contributed to data analysis
and experimental materials. JF, TCH, QS and AH conceptualized the
study design. JF and TH wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The use and care of animals in the present study
was approved by the Research and Experimental Animal Use Ethics
Committee of Chongqing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Beckwitt CH, Clark AM, Wheeler S, Taylor
DL, Stolz DB, Griffith L and Wells A: Liver 'organ on a chip'. Exp
Cell Res. 363:15–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Si-Tayeb K, Lemaigre FP and Duncan SA:
Organogenesis and development of the liver. Dev Cell. 82:175–189.
2010. View Article : Google Scholar
|
|
3
|
Ware BR and Khetani SR: Engineered liver
platforms for different phases of drug development. Trends
Biotechnol. 35:172–183. 2017. View Article : Google Scholar :
|
|
4
|
Thapa BR and Walia A: Liver function tests
and their interpretation. Indian J Pediatr. 74:663–671. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaplowitz N: Idiosyncratic drug
hepatotoxicity. Nat Rev Drug Discov. 4:489–499. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jaeschke H, Gores GJ, Cederbaum AI, Hinson
JA, Pessayre D and Lemasters JJ: Mechanisms of hepatotoxicity.
Toxicol Sci. 65:166–176. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Godoy P, Hewitt NJ, Albrecht U, Andersen
ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger
J, et al: Recent advances in 2D and 3D in vitro systems using
primary hepatocytes, alternative hepatocyte sources and
non-parenchymal liver cells and their use in investigating
mechanisms of hepatotoxicity, cell signaling and ADME. Arch
Toxicol. 87:1315–1530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebhardt R, Hengstler JG, Müller D,
Glöckner R, Buenning P, Laube B, Schmelzer E, Ullrich M, Utesch D,
Hewitt N, et al: New hepatocyte in vitro systems for drug
metabolism: metabolic capacity and recommendations for application
in basic research and drug development, standard operation
procedures. Drug Metab Rev. 35:145–213. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhushan A, Senutovitch N, Bale SS, McCarty
WJ, Hegde M, Jindal R, Golberg I, Berk Usta O, Yarmush ML, Vernetti
L, et al: Towards a three-dimensional microfluidic liver platform
for predicting drug efficacy and toxicity in humans. Stem Cell Res
Ther. 4(Suppl 1): pp. S162013, View Article : Google Scholar
|
|
10
|
Soldatow VY, Lecluyse EL, Griffith LG and
Rusyn I: In vitro models for liver toxicity testing. Toxicol Res
(Camb). 2:23–39. 2013. View Article : Google Scholar
|
|
11
|
Pandit A, Sachdeva T and Bafna P:
Drug-induced hepatotoxicity: A review. J Appl Pharm Sci. 2:233–243.
2012.
|
|
12
|
Groneberg DA, Grosse-Siestrup C and
Fischer A: In vitro models to study hepatotoxicity. Toxicol Pathol.
30:394–399. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olson H, Betton G, Robinson D, Thomas K,
Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, et al:
Concordance of the toxicity of pharmaceuticals in humans and in
animals. Regul Toxicol Pharmacol. 32:56–67. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maes M, Vinken M and Jaeschke H:
Experimental models of hepatotoxicity related to acute liver
failure. Toxicol Appl Pharmacol. 290:86–97. 2016. View Article : Google Scholar
|
|
15
|
Clark M and Steger-Hartmann T: A big data
approach to the concordance of the toxicity of pharmaceuticals in
animals and humans. Regul Toxicol Pharmacol. 96:94–105. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Granhall C, Floby E, Nordmark A,
Orzechowski A, Thorne A, Tybring G and Sohlenius-Sternbeck AK:
Characterization of testosterone metabolism and 7-hydroxycoumarin
conjugation by rat and human liver slices after storage in liquid
nitrogen for 1 h up to 6 months. Xenobiotica. 32:985–996. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Omura T and Sato R: The carbon
monoxide-binding pigment of liver microsomes. I. Evidence for its
hemoprotein nature. J Biol Chem. 239:2370–2378. 1964.PubMed/NCBI
|
|
18
|
Houston JB and Carlile DJ: Prediction of
hepatic clearance from microsomes, hepatocytes, and liver slices.
Drug Metab Rev. 29:891–922. 1997. View Article : Google Scholar
|
|
19
|
Bi Y, He Y, Huang J, Su Y, Zhu GH, Wang Y,
Qiao M, Zhang BQ, Zhang H, Wang Z, et al: Functional
characteristics of reversibly immortalized hepatic progenitor cells
derived from mouse embryonic liver. Cell Physiol Biochem.
34:1318–1338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cogger VC, O'Reilly JN, Warren A and Le
Couteur DG: A standardized method for the analysis of liver
sinusoidal endothelial cells and their fenestrations by scanning
electron microscopy. J Vis Exp. 98:pp. e526982015
|
|
21
|
Bale SS, Moore L, Yarmush M and Jindal R:
Emerging in vitro liver technologies for drug metabolism and
inter-organ interactions. Tissue Eng Part B Rev. 22:383–394. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bale SS, Vernetti L, Senutovitch N, Jindal
R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, et
al: In vitro platforms for evaluating liver toxicity. Exp Biol Med
(Maywood). 239:1180–1191. 2014. View Article : Google Scholar
|
|
23
|
Bhakuni GS, Bedi O, Bariwal J, Deshmukh R
and Kumar P: Animal models of hepatotoxicity. Inflamm Res.
65:13–24. 2016. View Article : Google Scholar
|
|
24
|
May JE, Xu J, Morse HR, Avent ND and
Donaldson C: Toxicity testing: The search for an in vitro
alternative to animal testing. Br J Biomed Sci. 66:160–165. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seglen PO: Preparation of isolated rat
liver cells. Methods Cell Biol. 13:29–83. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bi Y, Gong M, Zhang X, Zhang X, Jiang W,
Zhang Y, Chen J, Liu Y, He TC and Li T: Pre-activation of retinoid
signaling facilitates neuronal differentiation of mesenchymal stem
cells. Dev Growth Differ. 52:419–431. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bi Y, Huang J, He Y, Zhu GH, Su Y, He BC,
Luo J, Wang Y, Kang Q, Luo Q, et al: Wnt antagonist SFRP3 inhibits
the differentiation of mouse hepatic progenitor cells. J Cell
Biochem. 108:295–303. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang J, Bi Y, Zhu GH, He Y, Su Y, He BC,
Wang Y, Kang Q, Chen L, Zuo GW, et al: Retinoic acid signalling
induces the differentiation of mouse fetal liver-derived hepatic
progenitor cells. Liver Int. 29:1569–1581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Cui J, Zhang BQ, Zhang H, Bi Y,
Kang Q, Wang N, Bie P, Yang Z, Wang H, et al: Decellularized liver
scaffolds effectively support the proliferation and differentiation
of mouse fetal hepatic progenitors. J Biomed Mater Res A. 102:pp.
1017–1025. 2014, View Article : Google Scholar :
|
|
30
|
Fan J, Wei Q, Liao J, Zou Y, Song D, Xiong
D, Ma C, Hu X, Qu X, Chen L, et al: Noncanonical Wnt signaling
plays an important role in modulating canonical Wnt-regulated
stemness, proliferation and terminal differentiation of hepatic
progenitors. Oncotarget. 8:pp. 27105–27119. 2017, PubMed/NCBI
|
|
31
|
Wu N, Zhang H, Deng F, Li R, Zhang W, Chen
X, Wen S, Wang N, Zhang J, Yin L, et al: Overexpression of Ad5
precursor terminal protein accelerates recombinant adenovirus
packaging and amplification in HEK-293 packaging cells. Gene Ther.
21:629–637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
National Research Council(US): Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals Guide for the Care and Use of Laboratory Animals. National
Academies Press; (US), Washington, DC: pp. 963–965. 2011
|
|
33
|
Cabral F, Miller CM, Kudrna KM, Hass BE,
Daubendiek JG, Kellar BM and Harris EN: Purification of hepatocytes
and sinusoidal endothelial cells from mouse liver perfusion. J Vis
Exp. Feb 12–2018, Epub ahead of print. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Huang X, Werner M, Broering R, Yang
D and Lu M: Advanced method for isolation of mouse hepatocytes,
liver sinusoidal endothelial cells, and kupffer cells. Methods Mol
Biol. 1540:249–258. 2017. View Article : Google Scholar
|
|
35
|
Choi WM, Eun HS, Lee YS, Kim SJ, Kim MH,
Lee JH, Shim YR, Kim HH, Kim YE, Yi HS and Jeong WI: Experimental
applications of in situ liver perfusion machinery for the study of
liver disease. Mol Cells. 42:45–55. 2019.PubMed/NCBI
|
|
36
|
Zelepukin IV, Yaremenko AV, Petersen EV,
Deyev SM, Cherkasov VR, Nikitin PI and Nikitin MP: Magnetometry
based method for investigation of nanoparticle clearance from
circulation in a liver perfusion model. Nanotechnology.
30:1051012019. View Article : Google Scholar
|
|
37
|
Mederacke I, Dapito DH, Affo S, Uchinami H
and Schwabe RF: High-yield and high-purity isolation of hepatic
stellate cells from normal and fibrotic mouse livers. Nat Protoc.
102:305–315. 2015. View Article : Google Scholar
|
|
38
|
He TC, Zhou S, da Costa LT, Yu J, Kinzler
KW and Vogelstein B: A simplified system for generating recombinant
adenoviruses. Proc Natl Acad Sci USA. 955:2509–2514. 1998.
View Article : Google Scholar
|
|
39
|
Luo J, Deng ZL, Luo X, Tang N, Song WX,
Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, et al: A protocol
for rapid generation of recombinant adenoviruses using the AdEasy
system. Nat Protoc. 2:1236–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee CS, Bishop ES, Zhang R, Yu X, Farina
EM, Yan S, Zhao C, Zheng Z, Shu Y, Wu X, et al: Adenovirus-mediated
gene delivery: Potential applications for gene and cell-based
therapies in the new era of personalized medicine. Genes Dis.
4:43–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng H, Jiang W, Phillips FM, Haydon RC,
Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, et al:
Osteogenic activity of the fourteen types of human bone
morphogenetic proteins (BMPs). J Bone Joint Surg Am. 85:1544–1552.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kang Q, Song WX, Luo Q, Tang N, Luo J, Luo
X, Chen J, Bi Y, He BC, Park JK, et al: A comprehensive analysis of
the dual roles of BMPs in regulating adipogenic and osteogenic
differentiation of mesenchymal progenitor cells. Stem Cells Dev.
18:545–559. 2009. View Article : Google Scholar
|
|
43
|
Kang Q, Sun MH, Cheng H, Peng Y, Montag
AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, et al:
Characterization of the distinct orthotopic bone-forming activity
of 14 BMPs using recombinant adenovirus-mediated gene delivery.
Gene Ther. 11:1312–1320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li R, Zhang W, Cui J, Shui W, Yin L, Wang
Y, Zhang H, Wang N, Wu N, Nan G, et al: Targeting BMP9-promoted
human osteosarcoma growth by inactivation of notch signaling. Curr
Cancer Drug Targets. 14:274–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li R, Yan Z, Ye J, Huang H, Wang Z, Wei Q,
Wang J, Zhao L, Lu S, Wang X, et al: The prodomain-containing BMP9
produced from a stable line effectively regulates the
differentiation of mesenchymal stem cells. Int J Med Sci. 13:8–18.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao C, Wu N, Deng F, Zhang H, Wang N,
Zhang W, Chen X, Wen S, Zhang J, Yin L, et al: Adenovirus-mediated
gene transfer in mesenchymal stem cells can be significantly
enhanced by the cationic polymer polybrene. PLoS One. 9:pp.
e929082014, View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kong Y, Zhang H, Chen X, Zhang W, Zhao C,
Wang N, Wu N, He Y, Nan G, Zhang H, et al: Destabilization of
heterologous proteins mediated by the GSK3beta phosphorylation
domain of the β-catenin protein. Cell Physiol Biochem.
32:1187–1199. 2013. View Article : Google Scholar
|
|
48
|
Zhao C, Zeng Z, Qazvini NT, Xu X, Zhang R,
Yan S, Shu Y, Zhu Y, Duan C, Bishop E, et al: Thermoresponsive
citrate-based graphene oxide scaffold enhances bone regeneration
from BMP9-stimulated adipose-derived mesenchymal stem cells. ACS
Biomater Sci Eng. 4:2943–2955. 2018. View Article : Google Scholar
|
|
49
|
Gao Y, Huang E, Zhang H, Wang J, Wu N,
Chen X, Wang N, Wen S, Nan G, Deng F, et al: Crosstalk between
Wnt/beta-catenin and estrogen receptor signaling synergistically
promotes osteogenic differentiation of mesenchymal progenitor
cells. PLoS One. 8:pp. e824362013, View Article : Google Scholar
|
|
50
|
Yan Z, Yin L, Wang Z, Ye J, Zhang Z, Li R,
Denduluri SK, Wang J, Wei Q, Zhao L, et al: A novel organ culture
model of mouse intervertebral disc tissues. Cells Tissues Organs.
201:38–50. 2016. View Article : Google Scholar :
|
|
51
|
Liao J, Wei Q, Zou Y, Fan J, Song D, Cui
J, Zhang W, Zhu Y, Ma C, Hu X, et al: Notch signaling augments
BMP9-induced bone formation by promoting the
osteogenesis-angiogenesis coupling process in mesenchymal stem
cells (MSCs). Cell Physiol Biochem. 41:1905–1923. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cui J, Zhang W, Huang E, Wang J, Liao J,
Li R, Yu X, Zhao C, Zeng Z, Shu Y, et al: BMP9-induced osteoblastic
differentiation requires functional Notch signaling in mesenchymal
stem cells. Lab Invest. 99:58–71. 2019. View Article : Google Scholar
|
|
53
|
Yu X, Xia Y, Zeng L, Zhang X, Chen L, Yan
S, Zhang R, Zhao C, Zeng Z, Shu Y, et al: A blockade of PI3Kgamma
signaling effectively mitigates angiotensin II-induced renal injury
and fibrosis in a mouse model. Sci Rep. 8:109882018. View Article : Google Scholar
|
|
54
|
Zhang Q, Wang J, Deng F, Yan Z, Xia Y,
Wang Z, Ye J, Deng Y, Zhang Z, Qiao M, et al: TqPCR: A touchdown
qPCR assay with significantly improved detection sensitivity and
amplification efficiency of SYBR Green qPCR. PLoS One. 10:pp.
e01326662016, View Article : Google Scholar
|
|
55
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
56
|
Yu X, Liu F, Zeng L, He F, Zhang R, Yan S,
Zeng Z, Shu Y, Zhao C, Wu X, et al: Niclosamide exhibits potent
anticancer activity and synergizes with sorafenib in human renal
cell cancer cells. Cell Physiol Biochem. 47:957–971. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shu Y, Yang C, Ji X, Zhang L, Bi Y, Yang
K, Gong M, Liu X, Guo Q, Su Y, et al: Reversibly immortalized human
umbilical cord-derived mesenchymal stem cells (UC-MSCs) are
responsive to BMP9-induced osteogenic and adipogenic
differentiation. J Cell Biochem. 119:8872–8886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang N, Zhang W, Cui J, Zhang H, Chen X,
Li R, Wu N, Chen X, Wen S, Zhang J, et al: The piggyBac
transposon-mediated expression of SV40 T antigen efficiently
immortalizes mouse embryonic fibroblasts (MEFs). PLoS One. 9:pp.
e973162014, View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liao Z, Nan G, Yan Z, Zeng L, Deng Y, Ye
J, Zhang Z, Qiao M, Li R, Denduluri S, et al: The anthelmintic drug
niclosamide inhibits the proliferative activity of human
osteosarcoma cells by targeting multiple signal pathways. Curr
Cancer Drug Targets. 15:726–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Desmettre T, Devoisselle JM, Soulie-Begu S
and Mordon S: Fluorescence properties and metabolic features of
indocyanine green (ICG). J Fr Ophtalmol. 22:1003–1016. 1999.In
French. PubMed/NCBI
|
|
61
|
Bi Y, He Y, Huang JY, Xu L, Tang N, He TC
and Feng T: Induced maturation of hepatic progenitor cells in
vitro. Braz J Med Biol Res. 46:559–566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim DS, Ryu JW, Son MY, Oh JH, Chung KS,
Lee S, Lee JJ, Ahn JH, Min JS, Ahn J, et al: A liver-specific gene
expression panel predicts the differentiation status of in vitro
hepatocyte models. Hepatology. 66:1662–1674. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nikoozad Z, Ghorbanian MT and Rezaei A:
Comparison of the liver function and hepatic specific genes
expression in cultured mesenchymal stem cells and hepatocytes. Iran
J Basic Med Sci. 17:27–33. 2014.PubMed/NCBI
|
|
64
|
Luu HH, Song WX, Luo X, Manning D, Luo J,
Deng ZL, Sharff KA, Montag AG, Haydon RC and He TC: Distinct roles
of bone morphogenetic proteins in osteogenic differentiation of
mesenchymal stem cells. J Orthop Res. 25:665–677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lamplot JD, Qin J, Nan G, Wang J, Liu X,
Yin L, Tomal J, Li R, Shui W, Zhang H, et al: BMP9 signaling in
stem cell differentiation and osteogenesis. Am J Stem Cells.
2:1–21. 2013.PubMed/NCBI
|
|
66
|
Luther G, Wagner ER, Zhu G, Kang Q, Luo Q,
Lamplot J, Bi Y, Luo X, Luo J, Teven C, et al: BMP-9 induced
osteogenic differentiation of mesenchymal stem cells: Molecular
mechanism and therapeutic potential. Curr Gene Ther. 11:229–240.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wagner ER, Luther G, Zhu G, Luo Q, Shi Q,
Kim SH, Gao JL, Huang E, Gao Y, Yang K, et al: Defective osteogenic
differentiation in the development of osteosarcoma. Sarcoma.
2011:3252382011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang RN, Green J, Wang Z, Deng Y, Qiao M,
Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al: Bone
Morphogenetic Protein (BMP) signaling in development and human
diseases. Genes Dis. 1:87–105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhou L, An N, Jiang W, Haydon R, Cheng H,
Zhou Q, Breyer B, Feng T and He TC: Fluorescence-based functional
assay for Wnt/beta-catenin signaling activity. Biotechniques.
33:1126–1128. 1130–1132, passim. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Luo Q, Kang Q, Si W, Jiang W, Park JK,
Peng Y, Li X, Luu HH, Luo J, Montag AG, et al: Connective tissue
growth factor (CTGF) is regulated by Wnt and bone morphogenetic
proteins signaling in osteoblast differentiation of mesenchymal
stem cells. J Biol Chem. 279:55958–55968. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng Y, Kang Q, Cheng H, Li X, Sun MH,
Jiang W, Luu HH, Park JY, Haydon RC and He TC: Transcriptional
characterization of bone morphogenetic proteins (BMPs)-mediated
osteogenic signaling. J Cell Biochem. 90:1149–1165. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu
BA, Luu HH, Park JK, Li X, Luo J, et al: Inhibitor of DNA
binding/differentiation helix-loop-helix proteins mediate bone
morphogenetic protein-induced osteoblast differentiation of
mesenchymal stem cells. J Biol Chem. 279:32941–32949. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Si W, Kang Q, Luu HH, Park JK, Luo Q, Song
WX, Jiang W, Luo X, Li X, Yin H, et al: CCN1/Cyr61 is regulated by
the canonical Wnt signal and plays an important role in
Wnt3A-induced osteoblast differentiation of mesenchymal stem cells.
Mol Cell Biol. 26:2955–2964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ramappa V and Aithal GP: Hepatotoxicity
related to anti-tuberculosis drugs: Mechanisms and management. J
Clin Exp Hepatol. 3:37–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Njoku DB: Drug-induced hepatotoxicity:
Metabolic, genetic and immunological basis. Int J Mol Sci.
15:6990–7003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Perwitasari DA, Atthobari J and Wilffert
B: Pharmacogenetics of isoniazid-induced hepatotoxicity. Drug Metab
Rev. 47:222–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bale SS, Golberg I, Jindal R, McCarty WJ,
Luitje M, Hegde M, Bhushan A, Usta OB and Yarmush ML: Long-term
coculture strategies for primary hepatocytes and liver sinusoidal
endothelial cells. Tissue Eng Part C Methods. 21:413–422. 2015.
View Article : Google Scholar :
|
|
78
|
Hughes DJ, Kostrzewski T and Sceats EL:
Opportunities and challenges in the wider adoption of liver and
interconnected microphysiological systems. Exp Biol Med (Maywood).
242:1593–1604. 2017. View Article : Google Scholar
|
|
79
|
Kegel V, Deharde D, Pfeiffer E, Zeilinger
K, Seehofer D and Damm G: Protocol for isolation of primary human
hepatocytes and corresponding major populations of non-parenchymal
liver cells. J Vis Exp. pp. e530692016, PubMed/NCBI
|
|
80
|
Riccalton-Banks L, Liew C, Bhandari R, Fry
J and Shakesheff K: Long-term culture of functional liver tissue:
Three-dimensional coculture of primary hepatocytes and stellate
cells. Tissue Eng. 9:401–410. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Rebelo SP, Costa R, Silva MM, Marcelino P,
Brito C and Alves PM: Three-dimensional co-culture of human
hepatocytes and mesenchymal stem cells: Improved functionality in
long-term bioreactor cultures. J Tissue Eng Regen Med.
11:2034–2045. 2017. View Article : Google Scholar
|
|
82
|
Chen L, Jiang W, Huang J, He BC, Zuo GW,
Zhang W, Luo Q, Shi Q, Zhang BQ, Wagner ER, et al: Insulin-like
growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic
differentiation and bone formation. J Bone Miner Res. 25:2447–2459.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Huang E, Zhu G, Jiang W, Yang K, Gao Y,
Luo Q, Gao JL, Kim SH, Liu X, Li M, et al: Growth hormone
synergizes with BMP9 in osteogenic differentiation by activating
the JAK/STAT/IGF1 pathway in murine multilineage cells. J Bone
Miner Res. 27:1566–1575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Khetani SR and Bhatia SN: Microscale
culture of human liver cells for drug development. Nat Biotechnol.
26:120–126. 2008. View Article : Google Scholar
|
|
85
|
Vorrink SU, Zhou Y, Ingelman-Sundberg M
and Lauschke VM: Prediction of drug-induced hepatotoxicity using
long-term stable primary hepatic 3D spheroid cultures in chemically
defined conditions. Toxicol Sci. 163:655–665. 2018. View Article : Google Scholar : PubMed/NCBI
|