Ovarian cancer (OC) is the 5th most common cancer
and the most lethal gynaecological malignancy in European women
(1). The International Federation
of Gynaecology and Obstetrics characterises four major stages of
OC, with stages I and II constituting tumours localised and mainly
confined to the ovaries, which are associated with a good prognosis
[5-year overall survival for stage I, 87.0-89.5% (2)], and the late stages III and IV, with
confirmed spread to the peritoneum and/or distant metastasis, and
poorer outcome [5-year overall survival for stage IV, 13.2-17.9%
(2)]. Early-stage OC presents
with non‑specific symptoms [including pelvic or abdominal pain,
loss of appetite, fatigue and unexplained weight loss (3,4)]
commonly associated with other diseases or ailments. Additionally,
OC is a relatively rare disease, meaning that general practitioners
will encounter a small number of OC cases throughout their career
(5). Combined, this means that
most patients with malignant growth in the pelvic region are
diagnosed in the late stages of OC.
OC is commonly divided into two major groups,
epithelial and non-epithelial. Epithelial OC (EOC) comprises four
main subtypes, based on the tissue of origin: Serous adenocarcinoma
[high-grade (HGSC) and low-grade]; endometrioid adeno-carcinoma;
ovarian clear cell adenocarcinoma (OCCC); and mucinous
adenocarcinoma. Non-epithelial OC is subdivided into germ cell and
sex chord/stromal OC. Overall, ~86% of OC cases are epithelial, and
of these, 76% are serous histological subtype, with HGSC counting
83% (2). The characteristics of
particularly the four main EOC types differ markedly in origin
tissue, gene and microRNA (miRNA) expression, and morphology, and
there is emerging consensus that they should be recognised as four
distinct diseases (6-8).
This review will present the advances in applying
next-generation sequencing (NGS) in large cohort studies in the
search for genetic variants that act as susceptibility loci and/or
driver mutations for OC.
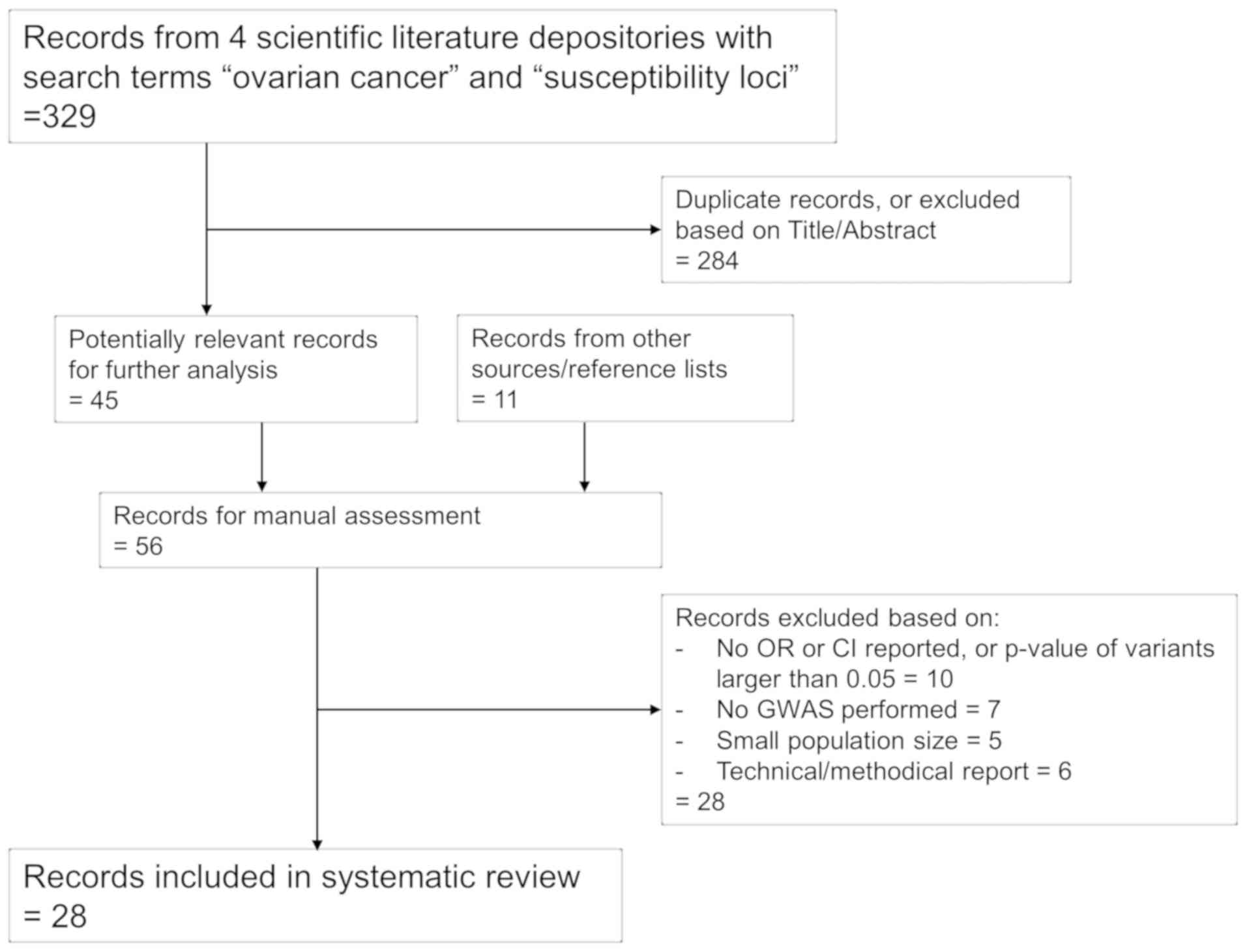
This review was carried out according to the
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
guidelines (21). Studies were
selected based on the search criteria ‘ovarian cancer' and
‘susceptibility loci' in the biomedical databases Medline, EMBASE
and Scopus. Studies reporting genome-wide association studies
(GWAS) in large cohorts were preferred. The aim was to cover as
much of the published literature as possible; however, studies
reporting variants associated with low malignant potential
(borderline) OC subtypes were omitted, and only studies reported in
English were included. In total, 108 susceptibility loci from 28
studies published from 2008 to 2018 were included (Fig. 1).
Only a decade ago, sequencing the genome of a single
individual took months, if not years. Subsequent advances in
micro-array and sequencing technologies prompted by large-scale
sequencing efforts such as the Human Genome Project (22) and the 1,000 Genomes Project
(23) have revolutionised the
field of genomic research, and today this can be accomplished over
~1 week using high-throughput sequencing (24). Targeted sequencing of only parts
of the genome, such as transcriptome or whole exome sequencing, or
sequencing of a subset of genes known to be involved in
tumorigenesis, have enabled scientists and clinicians to develop
and tailor research and treatment to the individual patient, a
fundamental premise for precision medicine initiatives, and the
overall goal for the treatment of patients with OC (25).
As high-throughput sequencing evolved into NGS (also
known by the more appropriate term, massively-parallel sequencing),
several hundred thousand genetic variants in thousands of patients
can now be investigated in only a fraction of the time (26). Naturally, this has spawned large
cohort studies, often with participation of clinics across the
world, as well as the invention of specific arrays or chips focused
on variants or genes suspected to be the cause of specific
diseases. The Cancer Genome Atlas (TCGA) Research Network
investigated 33 different cancer forms using high-throughput single
nucleotide polymorphism (SNP), exome and genome sequencing, as well
as gene expression, copy number variation, DNA methylation and
miRNA profiling; these findings were recently summarised (27). OC was one of the three cancer
types selected for the pilot project, and a cohort of 489 patients
with HGSC were selected for analysis. Among other findings, the
researchers found TP53 to be mutated in almost all cases,
and were able to classify tumours into several subtypes depending
on transcription, miRNA and methylation profiles (28).
NGS studies of OC have been reported in the last
five years, mainly stemming from two large global initiatives with
significant overlaps: The US‑based OncoArray Network and its
eponymous genotyping array chip (29); and the mega-consortium
Collaborative Oncological Gene-environment Study (COGS) with the
iCOGS array, and updated OncoChip (30). Established in 2005, the Ovarian
Cancer Association Consortium (OCAC) is a major collaboration, with
contributors from the United States, United Kingdom, Australia, The
Netherlands, Denmark, Poland, Germany and numerous other countries,
and consists of 25,509 population-based EOC cases and 40,941
controls (31). The consortium
was included in COGS together with Breast Cancer Association
Consortium (BCAC), Prostate Cancer Association Group to Investigate
Cancer-Associated Alterations in the Genome and The Consortium of
Investigators of Modifiers of BRCA1/2 (CIMBA), with the aim of
studying the genetics and risk factors of these three
hormone-related cancers [summarised in (30)]. For this collaboration, a custom
genotyping array chip called iCOGS capable of genotyping
>211,000 SNPs was developed and used on >250,000 subjects
(30). Like COGS, the OncoArray
Network's research and the OncoArray chip capable of genotyping
570,000 SNPs have resulted in numerous articles on glioblastoma,
breast, ovarian, prostate and lung cancers, using, among others,
the OCAC and BCAC cohorts (31-35).
In total, 108 susceptibility loci for OC were
identified following a systematic literature search (summarised in
Table I and Table SI for variants with
P<5.0×10−8 and P>5.0×10−8,
respectively). These loci were mainly found via GWAS, in which
genetic variations in a cohort of patients are compared to a cohort
of healthy controls to isolate variants that may contribute to
developing the disease. Variants are given an odds ratio (OR)
score, depending on whether the variant is found predominantly in
the patient cohorts (OR >1) or in the healthy controls (OR
<1).
In total, >50% of the OC susceptibility loci were
found to be involved in HGSC (59/108), which was perhaps expected,
as this is by far the most prevalent subtype of OC and thus the one
most frequently encountered. Certain variants have been reported in
>1 subtype, most notably rs757210, which seems to be linked with
poor prognosis in HGSC (odds ratio 1.12), but predicts superior
outcomes in OCCC (OR 0.80), demonstrating the importance of
stratifying GWAS findings by OC subtype (36-38). rs757210 sits in the promoter
region of HFN1B, which is known to be overexpressed in OCCC
(39) and downregulated in serous
OC (36), as well as being a
susceptibility gene for diabetes type II (40), prostate cancer (41,42), uterine corpus cancer (43) and endometrial cancer (44,45). Shen et al (36) hypothesised that the difference in
expression levels could be due to promoter methylation of
HNF1B in proximity to this variant, which was later
confirmed (46).
Mutation hotspots are a common feature in cancer
genomics, and some of the identified susceptibility loci were
situated in or near genes that are frequently altered in cancer
cells. As such, Pooley et al and Bojesen et al
(47,48) investigated the telomerase gene
TERT, which maintains chromosome telomeres. Somatic
mutations, especially in the promoter region of TERT, have
been found in cancers of the brain, thyroid gland, bladder and skin
(49). Bojesen et al
(48) reported a locus associated
with HGSC (rs10069690) in intron 4 with the minor allele conferring
increased risk of disease and creates an alternative splice site
that results in a truncated protein and impaired telomerase
function. This reinforces the hypothesis that shorter telomeres
increase cancer risk.
GWAS analyses have become the golden standard for
finding disease susceptibility loci, but there are certain
limitations as well. With hundreds of thousands of variants
examined on a single chip, the risk of false positives increases
dramatically, and stringent data processing must be employed. In
general, GWAS studies favour common variants in the population,
meaning that fine‑mapping and additional filtering are required to
discover variants with a minor allele frequency (MAF) of <5%
(58). Genetic variation occurs
semi-randomly and is widespread throughout the genome, and large
cohort sizes in the thousands are required to obtain statistically
significant and reliable results. There is an ongoing debate
regarding which P-value threshold should be the standard for GWAS,
or whether Bayesian approaches should be employed instead (59). The generally accepted P‑value is
P≤5.0×10−8 for common variants, as first introduced by
The International HapMap Project (60) and subsequently by Pe'er et
al (61), and which has been
recently evaluated and confirmed (62). This latest study concluded that
this threshold is too relaxed for rare variants (MAF ≤0.5%), and
cut‑offs for these should be: 3×10−8 for MAF ≥1%,
2×10−8 for MAF ≥0.5% and 1×10−8 for MAF ≥0.1%
(conditions: Whole-genome sequencing studies in European
populations with all variants having an LD r2>0.8). For the
present review, and contrary to two recent reviews of GWAS
susceptibility loci (63,64), it was determined that all variants
reported by the original articles would be included, with a more
relaxed cut-off of P≤0.05. Variants meeting the threshold criteria
discussed above (P≤5.0×10−8) are presented in Table I; the remaining variants are
included in Table SI. For
simplicity, for articles fine‑mapping a susceptibility loci region
and finding additional SNPs with lower P-values, but in strong LD
with the index SNP (48,54), only the novel variant with the
strongest association was included.
Genetic variants are not randomly distributed in the
genome, but often aggregate in the same populations (23). Genomic research has taken
advantage of this, by examining only those variants or
polymorphisms already reported in large population studies.
Nevertheless, the number of genetic variants in the human genome
amounts to tens of millions, which is not feasible to investigate
in a research setting on a large number of patients. Instead, an
array of representative or index SNPs are frequently used to cover
all variants in a genomic region, utilising the fact that
neighbouring SNPs are often in tight LD and thus inherited together
(65,66). Data from The 1,000 Genomes Project
estimates that any given trait-associated variant in the National
Human Genome Research Institute GWAS database will have 56
neighbouring variants in LD with r2≥0.5 (28). It follows that fine‑mapping of the
region is required to determine if the index SNP is indeed the
causal variant, or merely a proxy for other SNPs in the region.
Three examples of this have been described in the subsection
‘Genome-wide association studies identify numerous susceptibility
loci for ovarian cancer': Bojesen et al fine-mapped the
TERT locus and SNPs in LD with rs10069690 and rs7705526
(48); Lawrenson et al
examined the ABHD8/ANKLE1 locus and rs4808075
(54); and Shen et al
investigated the HNF1B region and rs7405776 and rs11651755
(36). Following the initial
findings of the COGS initiative, Earp et al (67) analysed 11 known susceptibility
regions and found novel associated variants with more robust
P-values and ORs than those previously reported (Table I) (20,37,48,50,68-71).
Several studies over the last few years have
fine‑mapped the 9p22.2 region by rs3814113 first reported by Song
et al in 2009 (68). eQTL
analyses concluded the nearby zinc finger protein basonuclin-2
(BNC2), which has been implicated in oocyte differentiation
(72), to be the most likely
causal candidate gene (69,73). Additional SNPs were found to be
associated with abnormal ovarian ultrasound results (74) and to modify OC risk in
BRCA1/2 mutation carriers (75). BNC2 was reported to
contribute to a HOX-centric network of transcription factors
associated with serous OC risk (55), and Carter et al (76) found a significant association
between germline rs3814113 and tumour formation in OC. Finally, a
recent study by Buckley et al (73) reported additional SNPs in LD with
rs3814113, as well as SNPs located in the regulatory regions of
BNC2, including some in this gene's scaffold/matrix
attachment region, suggesting that they influence chromosomal
three-dimensional organisational optimization for transcription in
an allele‑specific manner.
The potential impact of a genetic variant is
associated with its location in the gene. Only 1% of the human
genome codes for proteins; the remaining regions are
intra/intergenic, promoters, enhancers and long stretches of ‘gene
deserts', where genes are tens or hundreds of kilobases apart
(79). It follows that a variant
within a protein-coding region is potentially more detrimental to
the cell than one located in a gene desert. In the present study,
21 of the 108 identified variants alter amino acid sequences
(Table II). Two algorithms have
been developed to evaluate the potential damage caused by these
changes: SIFT (80) and
PolyPhen-2 (81) scores. Both
have values between 0 and 1, but the values have reciprocal
interpretation. A variant with a SIFT score approaching 0 is
considered deleterious, while one with a PolyPhen-2 score
approaching 1 is considered damaging. Several variants in Table II are located in notable genes
from an OC perspective: ANKLE1 and BRCA2, as
discussed earlier in this review; BTD, which has shown
promise as a biomarker for breast (82) and cervical (83) cancers; ZFHX3, which is a
tumour suppressor gene frequently mutated in prostate (84) and endometrial (85) cancer; and LEKR, which,
although the variant rs62273959 is considered benign, was found to
be in tight LD (r2=0.90) with rs7651446 in the aforementioned
TIPARP gene (77).
Finally, it is worth noting that rs587778134 causes a frame shift
mutation in the DNA repair gene BRIP1 (also known as
FANCJ), which is a well-known OC susceptibility gene that
interacts with BRCA1 (19,86).
Although it is not found in the SIFT/PolyPhen-2 databases, it does
have an entry for OC susceptibility (RCV000409984.1) in the ClinVar
database of potentially clinically relevant genetic variants and is
designated as ‘Likely pathogenic' (Table II) (87).
OC is suspected to be a hormonal disease and related
to breast and prostate cancers (88,89). Three new susceptibility loci were
found by investigating a cohort of patients with breast or ovarian
cancer harbouring mutations in BRCA1 (90). As part of the COGS initiative, the
three cancers were examined in individual GWAS projects (20,32,91), whereas Kar et al (92) combined the results from the three
projects in a single three-cancer meta-analysis, as well as
one-by-one comparisons. The findings showed clear pleiotropy among
the diseases, with three susceptibility loci identified in all
three cancers (rs17041869, rs7937840 and rs1469713), and four loci
shared between breast and ovarian cancers (rs635634, rs11571833,
rs200182588 and rs8037137). No shared loci were found for prostate
and ovarian cancer alone.
Surprisingly few of the susceptibility loci were
found in genes commonly associated with OC, such as TP53
(93), BRCA2 (92) or HNF1B (36,37). This may be explained by the fact
that a GWAS only detects susceptibility loci of a single or few
nucleotides, often conferring subtle differences in gene
expression, whereas some mutations in the classic causal genes are
large deletions or inactivating mutations that are detrimental to
normal protein function. Additionally, most of the variants have
relatively low ORs (<2.0), meaning that they only have moderate
effects on OC risk. Incidentally, the variant with the
second-highest OR (rs587778134, OR=8.13) was located in
BRIP1, and mutations in this gene have been established to
confer a moderate to high risk of developing OC (19,86).
Much attention has been focused on finding isolated
susceptibility loci on a genome-wide scale over the past decade. A
large number of the identified variants were situated in intergenic
regions far from the genes they potentially affect, and while
several GWAS have been performed and analysed, few studies have
fine‑mapped and functionally validated any of the findings. With
most loci situated in genes not previously associated with OC,
including hits in long noncoding RNAs (94), there is an urgent requirement and
potential for examining these further, particularly those that are
near or in genes implicated in oocyte and ovary development, or
tumour progression.
The focus must be on finding candidate causal genes
for OC. Promising studies have been released in recent years,
including the transcriptome-wide association study by Lu et
al (95). In this study, they
performed a ‘reverse GWAS' by cross‑matching existing OC‑specific
gene expression profiles with all known susceptibility loci and
candidate SNPs, and reported the Frizzled gene FZD4 as a
novel candidate causal gene. This is an area that complements and
overlaps well with the search for novel susceptibility loci.
We are currently performing whole exome sequencing
of patients with HGSC and OCCC, to identify variants that are
subtype‑ and survival‑specific. Combined with the published
variants summarised in this review, a screen of a large number of
patients with OC will be performed to identify potential biomarkers
for the early detection of OC that may decrease the mortality rates
for patients.
This study was supported by the Danish Mermaid III
project, who was not involved in the decision to write the
paper.
All data generated or analyzed during this study are
included in this published article.
All authors designed the study. MKC drafted and
edited the manuscript, EH supervised and edited, and CH supervised.
All authors contributed to and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Stewart BW and Wild CP: World Cancer
Report 2014. 2014.
|
|
2
|
Danish Gynecologic Cancer Group: Annual
Report of the Danish Gynecologic Cancer Database 2016-17. Danish
Gynecol Cancer Database. 2017.
|
|
3
|
Goff BA, Mandel LS, Drescher CW, Urban N,
Gough S, Schurman KM, Patras J, Mahony BS and Andersen MR:
Development of an ovarian cancer symptom index: Possibilities for
earlier detection. Cancer. 109:221–227. 2007. View Article : Google Scholar
|
|
4
|
Hamilton W, Peters TJ, Bankhead C and
Sharp D: Risk of ovarian cancer in women with symptoms in primary
care: Population based case-control study. BMJ. 339:b29982009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sundar S, Neal RD and Kehoe S: Diagnosis
of ovarian cancer. BMJ. 351:h44432015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Köbel M, Kalloger SE, Boyd N, McKinney S,
Mehl E, Palmer C, Leung S, Bowen NJ, Ionescu DN, Rajput A, et al:
Ovarian carcinoma subtypes are different diseases: Implications for
biomarker studies. PLoS Med. 5:e2322008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prat J: Ovarian carcinomas: Five distinct
diseases with different origins, genetic alterations, and
clinicopathological features. Virchows Arch. 460:237–249. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bast RC Jr, Klug TL, St John E, Jenison E,
Niloff JM, Lazarus H, Berkowitz RS, Leavitt T, Griffiths CT, Parker
L, et al: A radioim-munoassay using a monoclonal antibody to
monitor the course of epithelial ovarian cancer. N Engl J Med.
309:883–887. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hellström I, Raycraft J, Hayden-Ledbetter
M, Ledbetter JA, Schummer M, McIntosh M, Drescher C, Urban N and
Hellström KE: The HE4 (WFDC2) protein is a biomarker for ovarian
carcinoma. Cancer Res. 63:3695–3700. 2003.PubMed/NCBI
|
|
11
|
Karlsen MA, Fagö-Olsen C, Høgdall E,
Schnack TH, Christensen IJ, Nedergaard L, Lundvall L, Lydolph MC,
Engelholm SA and Høgdall C: A novel index for preoperative,
non-invasive prediction of macro-radical primary surgery in
patients with stage IIIC-IV ovarian cancer-a part of the Danish
prospective pelvic mass study. Tumor Biol. 37:12619–12626. 2016.
View Article : Google Scholar
|
|
12
|
Jacobs I, Oram D, Fairbanks J, Turner J,
Frost C and Grudzinskas JG: A risk of malignancy index
incorporating CA 125, ultrasound and menopausal status for the
accurate preoperative diagnosis of ovarian cancer. Br J Obstet
Gynaecol. 97:922–929. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moore RG, McMeekin DS, Brown AK,
DiSilvestro P, Miller MC, Allard WJ, Gajewski W, Kurman R, Bast RC
Jr and Skates SJ: A novel multiple marker bioassay utilizing HE4
and CA125 for the prediction of ovarian cancer in patients with a
pelvic mass. Gynecol Oncol. 112:40–46. 2009. View Article : Google Scholar
|
|
14
|
Skates SJ: Ovarian cancer screening:
Development of the risk of ovarian cancer algorithm (ROCA) and ROCA
screening trials. Int J Gynecol Cancer. 22(Suppl 1): S24–S26. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ueland FR, Desimone CP, Seamon LG, Miller
RA, Goodrich S, Podzielinski I, Sokoll L, Smith A, van Nagell JR Jr
and Zhang Z: Effectiveness of a multivariate index assay in the
preoperative assessment of ovarian tumors. Obstet Gynecol.
117:1289–1297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karlsen MA, Sandhu N, Høgdall C,
Christensen IJ, Nedergaard L, Lundvall L, Engelholm SA, Pedersen
AT, Hartwell D, Lydolph M, et al: Evaluation of HE4, CA125, risk of
ovarian malignancy algorithm (ROMA) and risk of malignancy index
(RMI) as diagnostic tools of epithelial ovarian cancer in patients
with a pelvic mass. Gynecol Oncol. 127:379–383. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jacobs IJ and Menon U: Progress and
challenges in screening for early detection of ovarian cancer. Mol
Cell Proteomics. 3:355–366. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Norquist BM, Harrell MI, Brady MF, Walsh
T, Lee MK, Gulsuner S, Bernards SS, Casadei S, Yi Q, Burger RA, et
al: Inherited mutations in women with ovarian carcinoma. JAMA
Oncol. 2:482–490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ramus SJ, Song H, Dicks E, Tyrer JP,
Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward
J, Philpott S, et al: Germline mutations in the BRIP1, BARD1,
PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer
Inst. 107:djv2142015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee
A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM,
et al: Identification of six new susceptibility loci for invasive
epithelial ovarian cancer. Nat Genet. 47:164–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moher D, Liberati A, Tetzlaff J and Altman
DG; PRISMA Group: Preferred reporting items for systematic reviews
and meta-analyses: The PRISMA statement. PLoS Med. 6:e10000972009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
International Human Genome Sequencing
Consortium: Finishing the euchromatic sequence of the human genome.
Nature. 431:931–945. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
The 1000 Genomes Project Consortium; Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar
|
|
25
|
Ashley EA: Towards precision medicine. Nat
Rev Genet. 17:507–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goodwin S, McPherson JD and McCombie WR:
Coming of age: Ten years of next-generation sequencing
technologies. Nat Rev Genet. 17:333–351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blum A, Wang P and Zenklusen JC: SnapShot:
TCGA-analyzed tumors. Cell. 173:5302018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cancer Genome Atlas Research Network:
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Amos CI, Dennis J, Wang Z, Byun J,
Schumacher FR, Gayther SA, Casey G, Hunter DJ, Sellers TA, Gruber
SB, et al: The oncoarray consortium: A network for understanding
the genetic architecture of common cancers. Cancer Epidemiol
Biomarkers Prev. 26:126–135. 2017. View Article : Google Scholar :
|
|
30
|
Sakoda LC, Jorgenson E and Witte JS:
Turning of COGS moves forward findings for hormonally mediated
cancers. Nat Genet. 45:345–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar
SP, Lawrenson K, Winham SJ, Dennis J, Pirie A, Riggan MJ, Chornokur
G, et al: Identification of 12 new susceptibility loci for
different histotypes of epithelial ovarian cancer. Nat Genet.
49:680–691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Michailidou K, Beesley J, Lindstrom S,
Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah
M, et al: Genome-wide association analysis of more than 120,000
individuals identifies 15 new susceptibility loci for breast
cancer. Nat Genet. 47:373–380. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McKay JD, Hung RJ, Han Y, Zong X,
Carreras-Torres R, Christiani DC, Caporaso NE, Johansson M, Xiao X,
Li Y, et al: Large-scale association analysis identifies new lung
cancer susceptibility loci and heterogeneity in genetic
susceptibility across histological subtypes. Nat Genet.
49:1126–1132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schumacher FR, Al Olama AA, Berndt SI,
Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D,
Anokian E, Cieza-Borrella C, et al: Association analyses of more
than 140,000 men identify 63 new prostate cancer susceptibility
loci. Nat Genet. 50:928–936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Melin BS, Barnholtz-Sloan JS, Wrensch MR,
Johansen C, Il'yasova D, Kinnersley B, Ostrom QT, Labreche K, Chen
Y, Armstrong G, et al: Genome-wide association study of glioma
subtypes identifies specific differences in genetic susceptibility
to glioblastoma and non-glioblastoma tumors. Nat Genet. 49:789–794.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen H, Fridley BL, Song H, Lawrenson K,
Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, et
al: Epigenetic analysis leads to identification of HNF1B as a
subtype-specific susceptibility gene for ovarian cancer. Nat
Commun. 4:16282013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pharoah PD, Tsai YY, Ramus SJ, Phelan CM,
Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, et
al: GWAS meta‑analysis and replication identifies three new
susceptibility loci for ovarian cancer. Nat Genet. 45:362–370.
2013. View Article : Google Scholar
|
|
38
|
Kelemen LE, Lawrenson K, Tyrer J, Li Q,
Lee JM, Seo JH, Phelan CM, Beesley J, Chen X, Spindler TJ, et al:
Genome-wide significant risk associations for mucinous ovarian
carcinoma. Nat Genet. 47:888–897. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tsuchiya A, Sakamoto M, Yasuda J, Chuma M,
Ohta T, Ohki M, Yasugi T, Taketani Y and Hirohashi S: Expression
profiling in ovarian clear cell carcinoma: Identification of
hepatocyte nuclear factor-1beta as a molecular marker and a
possible molecular target for therapy of ovarian clear cell
carcinoma. Am J Pathol. 163:2503–2512. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gudmundsson J, Sulem P, Steinthorsdottir
V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T,
Gudbjartsson D, Agnarsson BA, Baker A, et al: Two variants on
chromosome 17 confer prostate cancer risk and the one in TCF2
protects against type 2 diabetes. Nat Genet. 39:977–983. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun J, Zheng SL, Wiklund F, Isaacs SD,
Purcell LD, Gao Z, Hsu FC, Kim ST, Liu W, Zhu Y, et al: Evidence
for two independent prostate cancer risk-associated loci in the
HNF1B gene at 17q12. Nat Genet. 40:1153–1155. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thomas G, Jacobs KB, Yeager M, Kraft P,
Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et
al: Multiple loci identified in a genome‑wide association study of
prostate cancer. Nat Genet. 40:310–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Spurdle AB, Thompson DJ, Ahmed S, Ferguson
K, Healey CS, O'Mara T, Walker LC, Montgomery SB, Dermitzakis ET;
Australian National Endometrial Cancer Study Group; et al:
Genome-wide association study identifies a common variant
associated with risk of endometrial cancer. Nat Genet. 43:451–455.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
44
|
Painter JN, O'Mara TA, Batra J, Cheng T,
Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson K, et
al: Fine‑mapping of the HNF1B multicancer locus identifies
candidate variants that mediate endometrial cancer risk. Hum Mol
Genet. 24:1478–1492. 2015. View Article : Google Scholar
|
|
45
|
Setiawan VW, Haessler J, Schumacher F,
Cote ML, Deelman E, Fesinmeyer MD, Henderson BE, Jackson RD,
Vöckler JS, Wilkens LR, et al: HNF1B and endometrial cancer risk:
Results from the PAGE study. PLoS One. 7:e303902012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ross-Adams H, Ball S, Lawrenson K, Halim
S, Russell R, Wells C, Strand SH, Ørntoft TF, Larson M, Armasu S,
et al: HNF1B variants associate with promoter methylation and
regulate gene networks activated in prostate and ovarian cancer.
Oncotarget. 7:74734–74746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pooley KA, Bojesen SE, Weischer M, Nielsen
SF, Thompson D, Amin Al Olama A, Michailidou K, Tyrer JP, Benlloch
S, Brown J, et al: A genome-wide association scan (GWAS) for mean
telomere length within the COGS project: Identified loci show
little association with hormone-related cancer risk. Hum Mol Genet.
22:5056–5064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bojesen SE, Pooley KA, Johnatty SE,
Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen
HC, Smart CE, et al: Multiple independent variants at the TERT
locus are associated with telomere length and risks of breast and
ovarian cancer. Nat Genet. 45:371–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vinagre J, Almeida A, Pópulo H, Batista R,
Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al:
Frequency of TERT promoter mutations in human cancers. Nat Commun.
4:21852013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bolton KL, Tyrer J, Song H, Ramus SJ,
Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY,
et al: Common variants at 19p13 are associated with susceptibility
to ovarian cancer. Nat Genet. 42:880–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shao G, Patterson-Fortin J, Messick TE,
Feng D, Shanbhag N, Wang Y and Greenberg RA: MERIT40 controls
BRCA1-Rap80 complex integrity and recruitment to DNA double-strand
breaks. Genes Dev. 23:740–754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang B, Hurov K, Hofmann K and Elledge SJ:
NBA1, a new player in the Brca1 A complex, is required for DNA
damage resistance and checkpoint control. Genes Dev. 23:729–739.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Feng L, Huang J and Chen J: MERIT40
facilitates BRCA1 localization and DNA damage repair. Genes Dev.
23:719–728. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lawrenson K, Kar S, McCue K, Kuchenbaeker
K, Michailidou K, Tyrer J, Beesley J, Ramus SJ, Li Q, Delgado MK,
et al: Functional mechanisms underlying pleiotropic risk alleles at
the 19p13.1 breast-ovarian cancer susceptibility locus. Nat Commun.
7:126752016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kar SP, Tyrer JP, Li Q, Lawrenson K, Aben
KK, Anton-Culver H, Antonenkova N, Chenevix-Trench G; Australian
Cancer Study; Australian Ovarian Cancer Study Group; et al:
Network-based integration of GWAS and gene expression identifies a
HOX-Centric network associated with serous ovarian cancer risk.
Cancer Epidemiol Biomarkers Prev. 24:1574–1584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lawrenson K, Li Q, Kar S, Seo JH, Tyrer J,
Spindler TJ, Lee J, Chen Y, Karst A, Drapkin R, et al: Cis-eQTL
analysis and functional validation of candidate susceptibility
genes for high-grade serous ovarian cancer. Nat Commun. 6:82342015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ghoussaini M, Song H, Koessler T, Al Olama
AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall
E, et al: Multiple loci with different cancer specificities within
the 8q24 gene desert. J Natl Cancer Inst. 100:962–966. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Panagiotou OA, Evangelou E and Ioannidis
JP: Genome-wide significant associations for variants with minor
allele frequency of 5% or less-an overview: A HuGE review. Am J
Epidemiol. 172:869–889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Stephens M and Balding DJ: Bayesian
statistical methods for genetic association studies. Nat Rev Genet.
10:681–690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
International HapMap Consortium: A
haplotype map of the human genome. Nature. 437:1299–1320. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pe'er I, Yelensky R, Altshuler D and Daly
MJ: Estimation of the multiple testing burden for genomewide
association studies of nearly all common variants. Genet Epidemiol.
32:381–385. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fadista J, Manning AK, Florez JC and Groop
L: The (in)famous GWAS P-value threshold revisited and updated for
low-frequency variants. Eur J Hum Genet. 24:1202–1205. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kar SP, Berchuck A, Gayther SA, Goode EL,
Moysich KB, Pearce CL, Ramus SJ, Schildkraut JM, Sellers TA and
Pharoah PDP: Common genetic variation and susceptibility to ovarian
cancer: Current insights and future directions. Cancer Epidemiol
Biomarkers Prev. 27:395–404. 2018. View Article : Google Scholar
|
|
64
|
Jones MR, Kamara D, Karlan BY, Pharoah PDP
and Gayther SA: Genetic epidemiology of ovarian cancer and
prospects for poly-genic risk prediction. Gynecol Oncol.
147:705–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Elmas A, Ou Yang TH, Wang X and
Anastassiou D: Discovering genome-wide tag SNPs based on the mutual
information of the variants. PLoS One. 11:e01679942016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Johnson GC, Esposito L, Barratt BJ, Smith
AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge
F, et al: Haplotype tagging for the identification of common
disease genes. Nat Genet. 29:233–237. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Earp M, Winham SJ, Larson N, Permuth JB,
Sicotte H, Chien J, Anton-Culver H, Bandera EV, Berchuck A, Cook
LS, et al: A targeted genetic association study of epithelial
ovarian cancer susceptibility. Oncotarget. 7:7381–7389. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Song H, Ramus SJ, Tyrer J, Bolton KL,
Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer
DW, DiCioccio R, et al: A genome‑wide association study identifies
a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet.
41:996–1000. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
69
|
Goode EL, Chenevix-Trench G, Song H, Ramus
SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson
MC, Kjaer SK, et al: A genome-wide association study identifies
susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet.
42:874–879. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
70
|
Permuth-Wey J, Lawrenson K, Shen HC,
Velkova A, Tyrer JP, Chen Z, Lin HY, Chen YA, Tsai YY, Qu X, et al:
Identification and molecular characterization of a new ovarian
cancer susceptibility locus at 17q21.31. Nat Commun. 4:16272013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen K, Ma H, Li L, Zang R, Wang C, Song
F, Shi T, Yu D, Yang M, Xue W, et al: Genome-wide association study
identifies new susceptibility loci for epithelial ovarian cancer in
Han Chinese women. Nat Commun. 5:46822014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Romano RA, Li H, Tummala R, Maul R and
Sinha S: Identification of Basonuclin2, a DNA‑binding zinc‑finger
protein expressed in germ tissues and skin keratinocytes. Genomics.
83:821–833. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Buckley MA, Woods NT, Tyrer JP,
Mendoza-Fandiño G, Lawrenson K, Hazelett DJ, Najafabadi HS, Gjyshi
A, Carvalho RS, Lyra PC Jr, et al: Functional analysis and fine
mapping of the 9p22.2 ovarian cancer susceptibility locus. Cancer
Res. 79:467–481. 2019. View Article : Google Scholar
|
|
74
|
Wentzensen N, Black A, Jacobs K, Yang HP,
Berg CD, Caporaso N, Peters U, Ragard L, Buys SS, Chanock S and
Hartge P: Genetic variation on 9p22 is associated with abnormal
ovarian ultrasound results in the prostate, lung, colorectal, and
ovarian cancer screening trial. PLoS One. 6:e217312011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Vigorito E, Kuchenbaecker KB, Beesley J,
Adlard J, Agnarsson BA, Andrulis IL, Arun BK, Barjhoux L, Belotti
M, Benitez J, et al: Fine‑scale mapping at 9p22.2 identifies
candidate causal variants that modify ovarian cancer risk in BRCA1
and BRCA2 mutation carriers. PLoS One. 11:e01588012016. View Article : Google Scholar
|
|
76
|
Carter H, Marty R, Hofree M, Gross AM,
Jensen J, Fisch KM, Wu X, DeBoever C, Van Nostrand EL, Song Y, et
al: Interaction landscape of inherited polymorphisms with somatic
events in cancer. Cancer Discov. 7:410–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Permuth JB, Pirie A, Ann Chen Y, Lin HY,
Reid BM, Chen Z, Monteiro A, Dennis J, Mendoza-Fandino G; AOCS
Study Group; et al: Exome genotyping arrays to identify rare and
low frequency variants associated with epithelial ovarian cancer
risk. Hum Mol Genet. 25:3600–3612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Fong PC, Boss DS, Yap TA, Tutt A, Wu P,
Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et
al: Inhibition of poly(ADP-Ribose) polymerase in tumors from BRCA
mutation carriers. N Engl J Med. 361:123–134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Venter JC, Adams MD, Myers EW, Li PW,
Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al:
The sequence of the human genome. Science. 291:1304–1351. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Vaser R, Adusumalli S, Leng SN, Sikic M
and Ng PC: SIFT missense predictions for genomes. Nat Protoc.
11:1–9. 2016. View Article : Google Scholar
|
|
81
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kang UB, Ahn Y, Lee JW, Kim YH, Kim J, Yu
MH, Noh DY and Lee C: Differential profiling of breast cancer
plasma proteome by isotope‑coded affinity tagging method reveals
biotinidase as a breast cancer biomarker. BMC Cancer. 10:1142010.
View Article : Google Scholar
|
|
83
|
Huang L, Zheng M, Zhou QM, Zhang MY, Jia
WH, Yun JP and Wang HY: Identification of a gene‑expression
signature for predicting lymph node metastasis in patients with
early stage cervical carcinoma. Cancer. 117:3363–3373. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sun X, Frierson HF, Chen C, Li C, Ran Q,
Otto KB, Cantarel BL, Vessella RL, Gao AC, Petros J, et al:
Frequent somatic mutations of the transcription factor ATBF1 in
human prostate cancer. Nat Genet. 37:407–412. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Walker CJ, Miranda MA, O'Hern MJ, McElroy
JP, Coombes KR, Bundschuh R, Cohn DE, Mutch DG and Goodfellow PJ:
Patterns of CTCF and ZFHX3 mutation and associated outcomes in
endo-metrial cancer. J Natl Cancer Inst. 107:djv2492015. View Article : Google Scholar
|
|
86
|
Rafnar T, Gudbjartsson DF, Sulem P,
Jonasdottir A, Sigurdsson A, Jonasdottir A, Besenbacher S, Lundin
P, Stacey SN, Gudmundsson J, et al: Mutations in BRIP1 confer high
risk of ovarian cancer. Nat Genet. 43:1104–1107. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Landrum MJ, Lee JM, Benson M, Brown GR,
Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al:
ClinVar: Improving access to variant interpretations and supporting
evidence. Nucleic Acids Res. 46:D1062–D1067. 2018. View Article : Google Scholar :
|
|
88
|
Henderson BE and Feigelson HS: Hormonal
carcinogenesis. Carcinogenesis. 21:427–433. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Henderson BE, Ross RK, Pike MC and
Casagrande JT: Endogenous hormones as a major factor in human
cancer. Cancer Res. 42:3232–3239. 1982.PubMed/NCBI
|
|
90
|
Couch FJ, Wang X, McGuffog L, Lee A,
Olswold C, Kuchenbaecker KB, Soucy P, Fredericksen Z, Barrowdale D,
Dennis J, et al: Genome-wide association study in BRCA1 mutation
carriers identifies novel loci associated with breast and ovarian
cancer risk. PLoS Genet. 9:e10032122013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Al Olama AA, Kote-Jarai Z, Berndt SI,
Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z,
Saunders E, et al: A meta‑analysis of 87,040 individuals identifies
23 new susceptibility loci for prostate cancer. Nat Genet.
46:1103–1109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kar SP, Beesley J, Amin Al Olama A,
Michailidou K, Tyrer J, Kote-Jarai Z, Lawrenson K, Lindstrom S,
Ramus SJ, Thompson DJ, et al: Genome-wide meta-analyses of breast,
ovarian, and prostate cancer association studies identify multiple
new susceptibility loci shared by at least two cancer types. Cancer
Discov. 6:1052–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Schildkraut JM, Goode EL, Clyde MA,
Iversen ES, Moorman PG, Berchuck A, Marks JR, Lissowska J, Brinton
L, Peplonska B, et al: Single nucleotide polymorphisms in the TP53
region and susceptibility to invasive epithelial ovarian cancer.
Cancer Res. 69:2349–2357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Johnatty SE, Tyrer JP, Kar S, Beesley J,
Lu Y, Gao B, Fasching PA, Hein A, Ekici AB, Beckmann MW, et al:
Genome-wide analysis identifies novel loci associated with ovarian
cancer outcomes: Findings from the ovarian cancer association
consortium. Clin Cancer Res. 21:5264–5276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lu Y, Beeghly-Fadiel A, Wu L, Guo X, Li B,
Schildkraut JM, Im HK, Chen YA, Permuth JB, Reid BM, et al: A
transcrip-tome-wide association study among 97,898 women to
identify candidate susceptibility genes for epithelial ovarian
cancer risk. Cancer Res. 78:5419–5430. 2018. View Article : Google Scholar : PubMed/NCBI
|