Introduction
A total of ~2-10% of women suffer from endometriosis
and 50% of them have infertility issues (1), mainly of the estrogen-dependent
inflammatory type (2,3). The glandular epithelium and basement
membrane of human endometrium undergo periodic changes in gene
expression, which must be synchronized to allow the opening of the
implantation window. Any abnormality in gene expression could lead
to a small or absent window. The abnormal inflammatory state caused
by endometriosis impairs endometrial receptivity (4).
Long non-coding RNAs (lncRNAs) are non-coding RNAs
with a length of >200 nucleotides and a wide range of biological
functions. A number of studies reported various lncRNAs to be
associated with endometriosis and compromised fertility (5-8) A
total of two genome-wide screening studies showed that there were
1,000 lncRNA (9) and 1,300 mRNA
(10) transcripts that were
abnormally expressed between eutopic and ectopic endometrial
tissues of patients with endometriosis. Another study revealed that
1,277 lncRNAs and 1,216 mRNAs were differentially expressed between
the eutopic and ectopic endometrium of patients with endometriosis
during the late secretory phase (11). Taken together, these studies
highlight the important regulatory roles of lncRNAs in
endometriosis.
In the present study, the difficulty of obtaining
clinical specimens of the endometrium during the implantation
window was taken into consideration. Therefore, autologous
endometrial transplantation was used to establish a model of
endometriosis in Sprague-Dawley rats. The present study aimed to
determine the changes in lncRNAs and mRNAs in the uterine tissues
of those rats during the implantation window, using microarrays.
The present study hypothesized that the abnormalities in lncRNA and
mRNA profiles have significant effects on the implantation ability
of uterine tissues in rats with endometriosis during the
implantation window, which could be an important cause of
endometrial receptivity impairment in endometriosis.
Materials and methods
Ethics statement
The present study was approved by the Experimental
Animal Center of the Peking Union Medical College Hospital and
Chinese Academy of Medical Sciences (permit no. XHDW-2016-000). All
experiments were performed in accordance with the principles of
experimental animal management and protection. The rats were
humanely cared for and sacrificed to prevent suffering.
Experimental animals and collection of
specimens
Clean, sexually mature and non-pregnant
Sprague-Dawley female (n=35) and male (n=10) rats were selected.
The age of the female rats was 60-70 days and their weight was
200±10 g. The weight of the male rats was 380±10 g (age, 60-70
days). All rats were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd., and they were kept in the specific
pathogen-free facility of the Laboratory Animal Center of Peking
Union Medical College Hospital. Room temperature was 20-25°C and
relative humidity was 45-60%. The rats were housed five per cage.
All rats were kept under 14-h light/10-h dark cycle. They received
sterilized feed and water ad libitum. They were adaptively
fed for 2-3 days before experiments. The vaginal smear method was
used to determine the estrous cycle of each female rat, which was
usually 4-5 days.
The 35 female rats were divided into three groups:
Endometriosis group (n=13), adipose tissue control group (n=8) and
blank control group (n=14). After the observation of three estrous
cycles, the autologous transendocardial transplantation method
described by Vernon and Wilson (12) was performed, but with slight
improvements. In the endometriosis group, a segment of autologous
uterine tissue was collected and transplanted onto the abdominal
wall in the fourth estrus cycle. Pentobarbital sodium (5%, 50
mg/kg, i.p.) was used for anesthesia. The rats were placed in a
supine position and the abdominal skin was prepared. A longitudinal
abdominal incision (5 cm) was made to open the abdominal cavity and
to find the Y-shaped uterus. The left uterine horn was lifted and
uterine tissue (1 cm) was collected and quickly placed in a culture
dish with saline water. The extra adipose tissues outside the
serous layer were removed. A longitudinal incision of the uterine
cavity was made and two pieces (5×5 mm) were sutured on both sides
of the abdominal wall at sites rich in blood vessels. Then the
abdominal incision was sutured layer by layer.
In the adipose tissue control group, eight female
rats were randomly selected. They underwent the same surgery as in
the endometriosis group, but the left uterine horn was ligated and
two pieces of adipose tissues (5×5 mm) were collected from the
abdomen and sutured onto both sides of the abdominal wall. In the
blank control group, no surgery was performed.
A total of 28 days after operating, the rats in the
endometriosis and adipose tissue groups were re-operated to observe
the growth, invasion and adhesion of the transplanted endometrium.
Growth of ectopic endometrium (the transplanted tissues were 5×5
mm), tissue edema (as a sign of inflammation) and vesicle formation
were observed by the naked eye. The models were considered
successful if the transplanted endometrium has grown by at least
2-fold. The model success rate was calculated as the number of
successful models divided by the total number of animals that
underwent modeling. The scoring criteria of the ectopic uterine
tissue growth were: Score 0, no epithelium; score 1, poorly
preserved epithelium; score 2, moderately preserved epithelium; and
score 3, well preserved epithelium (13,14). The abdominal incision was sutured
layer by layer. All rats survived and were prepared for mating.
A total of 10 male rats were kept in different
cages. The female rats were placed in the cages with one male rat
at 18:00 every night and a tray was placed under each cage. At
8:00-9:00 the next morning, the trays were checked for vaginal
plugs and vaginal smear examinations were carried out. If a vaginal
plug and sperm were observed under the microscope at the same time,
it was considered to be the 1st day of pregnancy (D1) (15), and mating was not conducted
anymore. On the 5th day of pregnancy (D5) [i.e., during the short
implanting window; implantation occurs on day 5 in rats (15)], female rats were sacrificed to
collect the uterine tissues. The tissues were divided into two
parts. One part was put in 10% formalin at 4°C for at least 24 h
for subsequent hematoxylin and eosin (H&E) staining and
immunohistochemistry. The other part was quickly put into liquid
nitrogen at -80°C for gene chip analysis and reverse
transcription-quantitative (RT-q)PCR. All experiments were
performed using the samples from the same rats in each group.
RNA extraction
According to the manufacturer's protocol, TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract
the total RNA from the uterine tissues (100 mg). RNA was purified
using the mirVana miRNA Isolation kit (Ambion; Thermo Fisher
Scientific, Inc.). RNA concentration and purity were determined by
spectrophotometry (NanoDrop ND-1000; Thermo Fisher Scientific,
Inc.) to measure optical density at 260/280 nm. The RNA 6000 Nano
Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies,
Inc.) were used and RNA integrity was verified by electrophoresis
on formaldehyde (1%) gels. The tissues with RNA integrity >6
were selected for the subsequent experiments.
Microarray imaging and data analysis
Complementary(c) DNA was synthesized from 5
µl total RNA. The GeeDom® biochip (CapitalBio
Corp.) labeling kit was used. The Nucleospin® Extract II
kit (Macherey-Nagel, GmbH) was used to purify the labeled products.
The rat lncRNA+mRNA Array V1.0 (8×60K format, the sequence included
4,974 control probes; Agilent Technologies, Inc.) was used for
hybridization. The probes in each sequence represented 22,020 rat
lncRNAs and 30,254 rat mRNAs. These lncRNA and mRNA probes
sequences referred to multiple databases including NCBI RefSeq
(https://www.ncbi.nlm.nih.gov/refseq/), Ensembl
(http://www.ensembl.org/index.html),
UCSC genome browser (http://genome.ucsc.edu/), NONCODE V4.0 (http://www.noncode.org) and UCR (https://users.soe.ucsc.edu/~jill/ultra.html) (16). Each RNA is detected by the probe
at least 1-2 times and each microarray includes 4,974 probes.
After hybridization, the chip was washed in the
GeeDom® Slide Washer 8. The Agilent chip scanner
(G2565CA) was used to scan the chip. The Agilent Feature Extraction
(v10.7) software (Agilent Technologies, Inc.) was used to analyze
the images. The GeneSpring software V13.0 (Agilent Technologies,
Inc.) was used for the summary, standardization and quality control
of chip data. Values of ≥2 and ≤-2 fold-change, as well as
P<0.05 were used for screening for differentially expressed
genes. CLUSTER 3.0 software (Stanford University) with data
adjustment functions was used to perform the log2 conversion and
median calculation of chip data. Finally, the Java Treeview v1.8
(Stanford University School of Medicine) was used for cluster
analysis. The samples were tested in triplicates.
RT-qPCR validation of differentially
expressed lncRNAs and mRNAs
The criteria for selecting the candidate
differentially expressed genes for validation were: i) Fold-change
was ≥2; ii) the biological repeat of each group was ≥3; and iii)
P≤0.05. For an upregulated gene, the number of samples with the
detected flag of the experimental group was required to be >60%
of the total number of samples in the group. For a downregulated
gene, the number of samples with the detected flag of the control
group was required to be >60% of the total number of samples in
the group.
Therefore, according to bioinformatics, the
relationship between the targeted regulation of the selected five
lncRNAs and the mRNA microarray results (abnormal expression folds
in the endometriosis group and the blank control group) were
selected for four mRNAs [ADAM metallopeptidase with thrombospondin
type 1 motif 7 (Adamts7), tumor protein p53 (Tp53), distal-less
homeobox 3 (Dlx3) and pyrimidinergic receptor P2Y6 (P2ry6)] for
qPCR validation.
The primers are shown in Table I. The FastQuant RT kit [Tiangen
Biotech (Beijing) Co., Ltd.] was used for the reverse transcription
of total RNA: 1 µg total RNA and 20 µl reaction
system (10X Fast-RT Buffer 2 µl, FQ-RT Primer Mix 2
µl, RT Enzyme Mix 1 µl, RNase-Free water 5 and 10
µl buffer) were kept at 42°C for 15 min, 95°C for 3 min,
cooled on ice and reverse transcribed. The same amount of RNA was
used for all samples. The qPCR reaction system Power
SYBR®-Green PCR Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.; 10 µl) containing Power
SYBR®-Green PCR Master Mix (2X) 5 µl, cDNA sample
0.5 µl, forward primer (10 µM, 0.25 µl),
reverse primer (10 µM, 0.25 µl) and nuclease-free
water (4 µl) were used in a MicroAmp Optical 96-Well
Reaction Plate (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Denaturation was carried out at 95°C for 10 min in a
QuantStudio™ 7 Flex RealTime PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Then, 40 cycles (95°C for 15 sec and 60°C
for 1 min) and a final extension at 60°C for 10 min were performed,
before climbing up to 95°C for melting curve analysis (temperature
ramp of 2%). All RT-qPCR reactions were performed in triplicate.
Electrophoresis (agarose gel, 2.0%) was used to detect the
amplification specificity of the products. GAPDH was selected as
the internal reference gene. Expression was measured using the
2−ΔΔCq method (17).
 | Table IPrimers for reverse
transcription-quantitative PCR. |
Table I
Primers for reverse
transcription-quantitative PCR.
| Gene | Primers
(5′-3′) | Length |
|---|
| NONRATT003997 | F:
CAGGGTCGGGAAACTTTGAGA | 22 bp |
| R:
CATCCAGTGTCCAGTGAGGG | |
|
gi|672027621|ref|XR_592747.1| | F:
CACGGAATGACAAGTGTGGG | 104 bp |
| R:
GGTGACTAATTGCCCCCGTA | |
|
gi|672045999|ref|XR_591544.1| | F:
CAGCTGGATCTGCTTTCGGA | 127 bp |
| R:
AGGAGACCCCAGATAAGCCT | |
| NONRATT006252 | F:
ACATAGCATTGGGCCCTGTC | 134 bp |
| R:
TCTAAAAGTCTCAGAGCATTCTCAA | |
|
gi|672033904|ref|XR_589853.1| | F:
AAGGGTTGGGAAGACCATGAC | 120 bp |
| R:
GAGACTCCCTGCTCACTTACG | |
| Adamts7 | F:
TCAATTTCTGTGAGACGCTGC | 147 bp |
| R:
ATGGTGCGTGGGTTTATCGT | |
| Tp53 | F:
CCCCACCGCCTGTAAGATTC | 110 bp |
| R:
GAGGGGTGGGGGATGGATA | |
| Dlx3 | F:
TCAATCTCAATGGGCTCGCA | 90 bp |
| R:
GGTACGCTCCATACTGTCGG | |
| P2ry6 | F:
GGTAGCTTGAGGCTGAGAGG | 126 bp |
| R:
TGACAGAAGTGTGTACGGCAT | |
| GAPDH | F:
CCTCAAGATCATCAGCAAT | 141 bp |
| R:
CCATCCACAGTCTTCTGGGT | |
Immunohistochemistry
The SP method was used for immunohistochemistry
(18). All specimens were fixed
in neutral formalin (10%) for at least 24 h at room temperature,
paraffin-embedded, sectioned (4 µm) and dewaxed at 60°C
overnight. Then, they were incubated in xylene for 45 min, hydrated
in decreasing ethanol series and washed with distilled water for 5
min. After antigen retrieval, the slides were placed in PBS (pH
7.4), shaken and washed three times, 15 min each time. The sections
were placed in hydrogen peroxide (3%) to block endogenous
peroxidase and incubated at room temperature for 25 min. The slides
were placed in PBS (pH 7.4), shaken and washed three times, 15 min
each time. Bovine serum albumin (3%) (Wuhan Servicebio Technology,
Co., Ltd.) was added and incubated at room temperature for 30 min.
After adding antibodies against Adamts7 (1:200; cat. no. 250456;
Abbiotec, Inc.), P2ry6 (1:50; cat. no. GTX16829; GeneTex, Inc.),
Dlx3 (1:100; cat. no. 27-520; ProSci, Inc.) and Tp53 (1:200; cat.
no. 70-527; ProSci, Inc.), the slides were incubated at 4°C in a
wet box for the night. The DAKO K5007 rat/rabbit secondary antibody
(cat. no. K5007; ready-to-use preparation; Dako; Agilent
Technologies, Inc.) was added and incubated at room temperature for
50 min. DAB coloration (cat. no. G1211; Wuhan Servicebio
Technology, Co., Ltd.) was performed and stained with hematoxylin
for 3 min at room temperature, before observation under a light
microscope.
Protein extraction and western
blotting
The frozen uterine tissues were homogenized in 5X
protein loading buffer (G2013; Wuhan Servicebio Technology, Co.,
Ltd.) with phosphorylation protease inhibitor (G2007; Wuhan
Servicebio Technology, Co., Ltd.), cooled on ice for 30 min and
centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was
kept. The bicinchoninic kit (Wuhan Servicebio Technology, Co.,
Ltd.) was used to determine protein concentration. The protein
solution [mixed 4:1 with 5X protein sample buffer (Wuhan Servicebio
Technology, Co., Ltd.)] was boiled for 15 min and stored at -20°C.
Proteins (30 µg) were separated using 10% SDS-PAGE. Proteins
were transferred to polyvinylidene difluoride membranes, which were
blocked with 5% skimmed milk for 1 h at room temperature. The
primary antibodies against P2ry6 (1:1,000), Adamts7 (1:1,000), Dlx3
(1:1,000) and Tp53 (1:1,000; cat. no. orb319621; Biorbyt, Ltd.)
were incubated at 4°C overnight. The membranes were washed in TBST
(Tween-20, 0.05%) three times at room temperature. The secondary
antibody HRP-labeled goat anti rabbit (1:3,000; cat. no. GB23303;
Wuhan Servicebio Technology, Co., Ltd.) was incubated at room
temperature for 30 min. The membranes were washed in TBST and
enhanced chemiluminescence reagents A and B (cat. no. G2014; Wuhan
Servicebio Technology, Co., Ltd.) was carried out. Adobe PhotoShop
CS5 (Adobe Inc.) was used for image analysis. β-actin (1:3,000;
cat. no. GB12001; Wuhan Servicebio Technology, Co., Ltd.) was
selected as internal reference. Grey value analysis (alpha-EaseFC
v4.0; Alpha Innotech Corporation) was carried out.
Functional prediction of lncRNAs and
co-expression with mRNAs
As in a previous study (19), the Pearson correlation coefficient
(PCC) between each abnormally expressed lncRNA and its co-expressed
mRNA was calculated. PCC >0.8 or <0.8 and P<0.01
represented significantly related lncRNA/mRNA. The hypergeometric
cumulative distribution function was used to conduct functional
enrichment analysis for co-expressed mRNA. The function of lncRNA
was predicted through gene ontology (GO) (http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/). On this basis, protein
regulatory genes could be predicted using cis and trans (20,21). On the basis of the screening
results of lncRNA and mRNA co-expression (correlation >0.99 or
<-0.99 and P<0.05), cis-prediction looked for lncRNA-mRNA
pair whose genomic location was within 10 kb and trans-prediction
used Blat (http://hgdownload.cse.ucsc.edu/admin/exe/) to compare
lncRNA and mRNA (3′UTR) sequences, and selected lncRNA-mRNA pair
that had similar sequence and beyond 100 kb distance.
Statistics analysis
The tiff image data after hybrid scanning of the
Gem®lncRNA+mRNA expression spectrum chip was
preprocessed by the Feature Extraction software v10.7 (Agilent
Technologies, Inc.). The GeneSpring GX software v13.0 (Agilent
Technologies, Inc.) was used to calculate the differences of gene
expression and statistical significance (P-value). Continuous data
(such as the RT-qPCR data) were expressed as the mean ± standard
deviation and analyzed using analysis of variance with post hoc
Tukey's test. The Fisher's exact test was used to analyze GO and
KEGG data. The PCC was used to calculate the co-expression
relationship between lncRNA and mRNA. SPSS 19.0 (IBM, Corp.) was
used to conduct statistical analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Endometriosis modeling was
successful
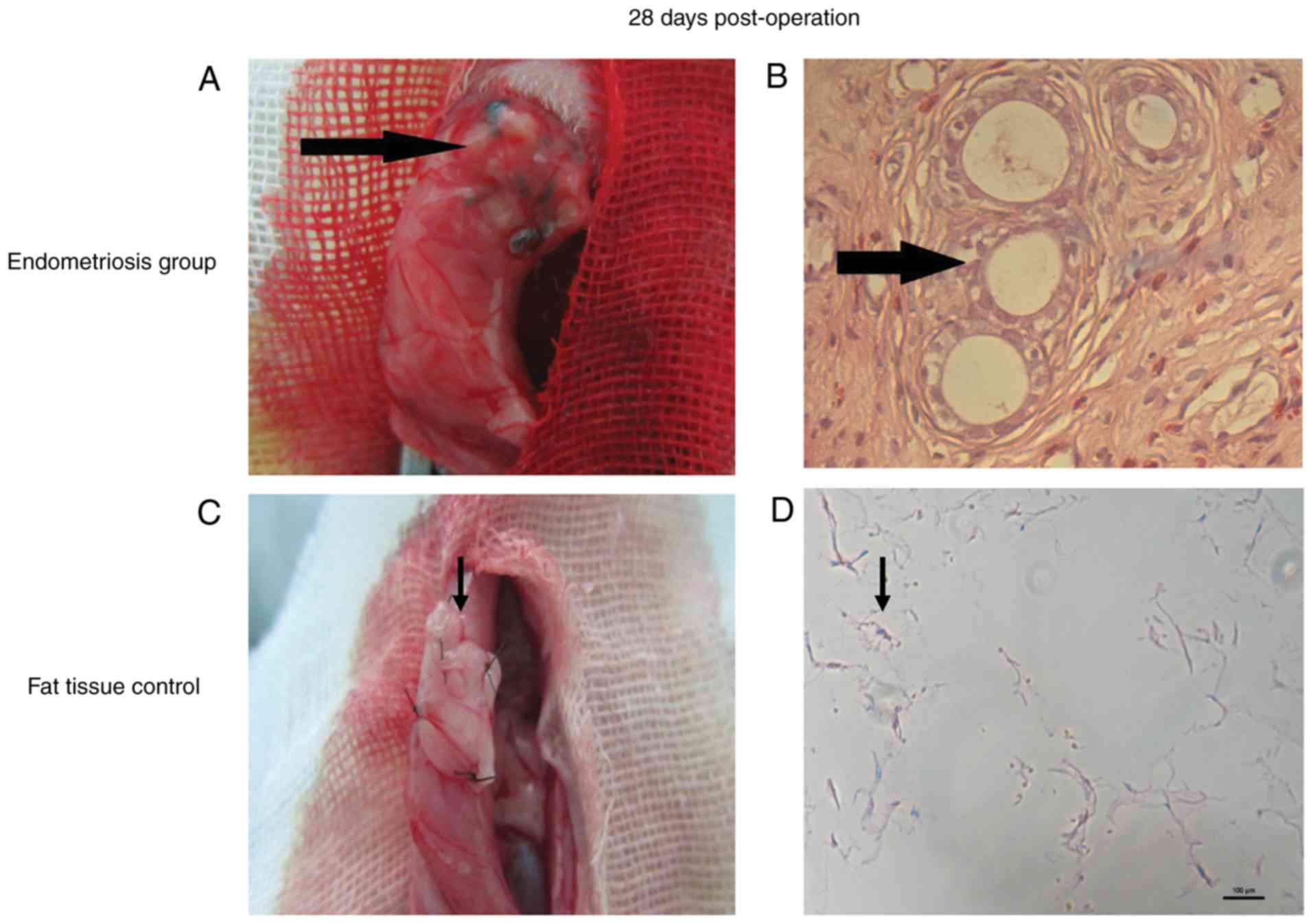
On the 28th day after surgery, growth of ectopic
endometrium of the transplanted tissues was 5×5 mm, tissue edema (a
sign of inflammation) and vesicle formation (Fig. 1A) were observed. All rats in the
endometriosis group were scored 3 with well-preserved epithelium
H&E staining showed that endometrial glands developed (Fig. 1B). In the adipose tissue control
group, there was no change in the transplant (Fig. 1C) and H&E staining showed no
endometrial glands (Fig. 1D). The
abdominal walls of rats in the blank control group were normal.
According to observation and H&E staining, the success rate of
the endometrial transplantation modeling was 100%.
Endometriosis is associated with
differentially expressed lncRNAs
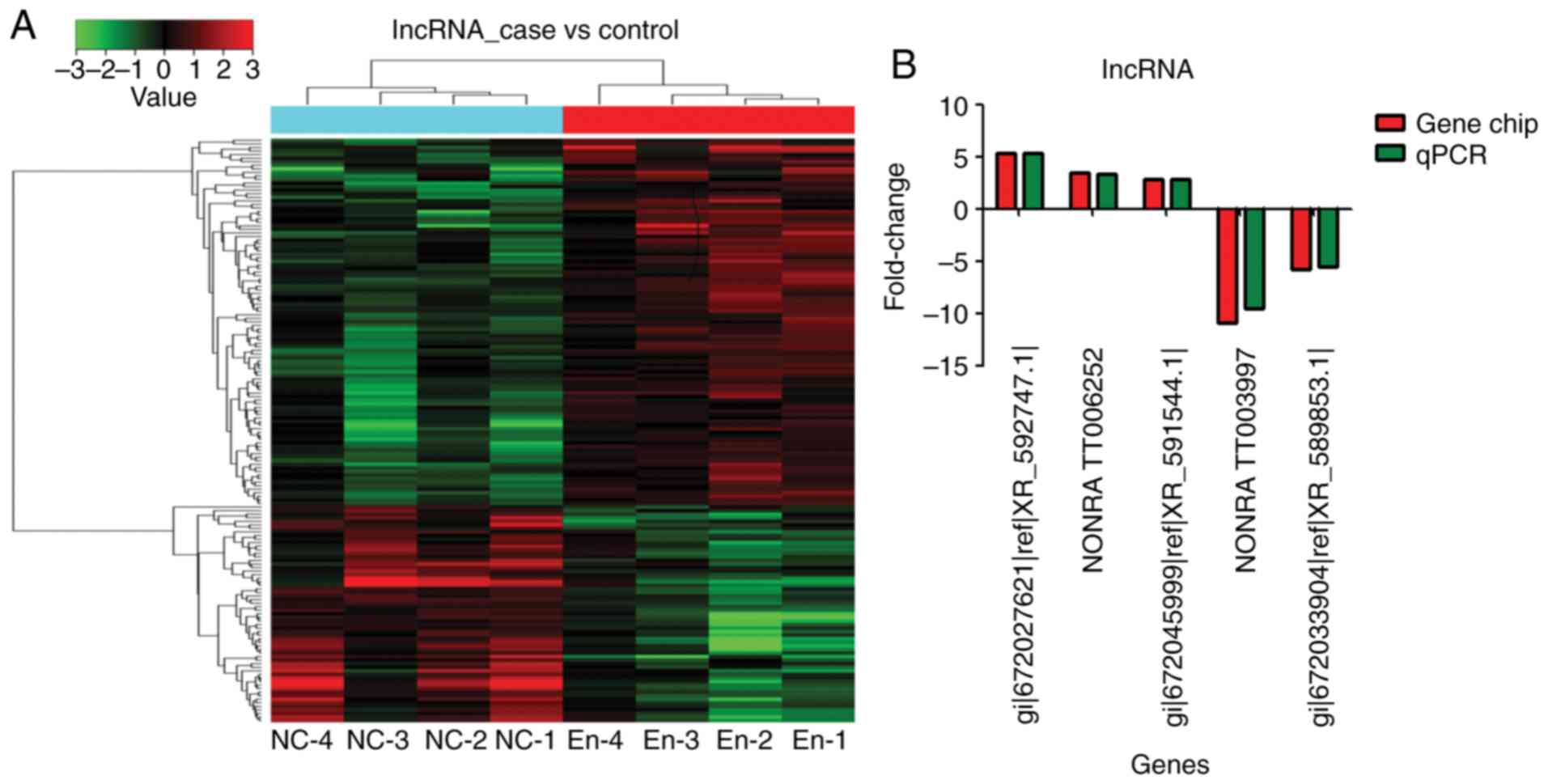
Compared with the blank control group, the
endometriosis model group showed 166 differentially expressed
lncRNAs: 115 upregulated and 51 downregulated (fold-change >2)
(Fig. 2A and Tables II and III).
 | Table IITop 20 upregulated lncRNA. |
Table II
Top 20 upregulated lncRNA.
| lncRNA (probe
name) | FC (abs) | P-value | lncRNA (name) |
|---|
|
gi|672066614|ref|XR_594547.1| | 5.34641 | 0.00327 | LOC103693263 |
|
gi|672076298|ref|XR_595802.1| | 4.895411 | 0.02629 | LOC103693752 |
|
gi|672079914|ref|XR_360377.2| | 4.506577 | 0.032437 | LOC102548312 |
|
gi|672036138|ref|XR_590129.1| | 3.46012 | 0.001325 | LOC103691084 |
| NONRATT018493 | 3.433898 | 0.008362 | |
| NONRATT006252 | 3.42446 | 0.01579 | |
| NONRATT020400 | 3.267124 | 0.019514 | |
| NONRATT021467 | 3.102348 | 0.007103 | |
| NONRATT025682 | 3.051757 | 0.00634 | |
| NONRATT030954 | 3.017166 | 0.011163 | |
| NONRATT011228 | 3.015996 | 0.002754 | |
| NONRATT027460 | 2.978006 | 0.018386 | |
|
gi|672045999|ref|XR_591544.1| | 2.87815 | 0.01452 | LOC103691820 |
| NONRATT012251 | 2.869275 | 0.01117 | |
| NONRATT016194 | 2.819013 | 0.038048 | |
| NONRATT010960 | 2.785293 | 0.022526 | |
|
gi|672052626|ref|XR_592505.1| | 2.765093 | 0.006744 | LOC103692352 |
|
gi|672020316|ref|XR_346989.2| | 2.726432 | 0.001031 | LOC102548283 |
|
gi|672037438|ref|XR_590367.1| | 2.619791 | 0.00196 | |
| NONRATT028550 | 2.596784 | 0.019415 | |
 | Table IIITop 20 downregulated lncRNA. |
Table III
Top 20 downregulated lncRNA.
| lncRNA (probe
name) | FC (abs) | P-value | lncRNA (name) |
|---|
| NONRATT003997 | 10.9511 | 0.00237 | |
| NONRATT024485 | 10.62082 | 0.018243 | |
| NONRATT009394 | 9.938666 | 0.044736 | |
|
gi|672086757|ref|XR_597443.1| | 8.693425 | 0.014136 | LOC103694443 |
| NONRATT016350 | 8.277449 | 0.007646 | |
| NONRATT001658 | 7.982786 | 0.002575 | |
| NONRATT005792 | 6.014078 | 0.00994 | |
|
gi|672033904|ref|XR_589853.1| | 5.79144 | 0.01007 | LOC102546604 |
| NONRATT014091 | 5.124274 | 0.014968 | |
|
gi|672040874|ref|XR_590738.1| | 4.626765 | 0.018434 | LOC103691378 |
| uc.340 | 3.967024 | 0.00138 | |
| NONRATT011360 | 3.901989 | 0.002246 | |
|
gi|672030085|ref|XR_598174.1| | 3.857016 | 0.017871 | LOC102553701 |
|
gi|672060549|ref|XR_355981.2| | 3.720889 | 0.007524 | LOC102547634 |
| NONRATT003965 | 3.631949 | 0.001985 | |
|
gi|672027547|ref|XR_592687.1| | 3.62932 | 0.002161 | LOC102547023 |
|
gi|672034604|ref|XR_589986.1| | 3.453335 | 0.00527 | LOC103690992 |
| NONRATT003634 | 3.397446 | 0.014216 | |
| NONRATT018412 | 3.134341 | 0.015503 | |
|
gi|672027788|ref|XR_340220.2| | 3.128051 | 0.010927 | LOC102551595 |
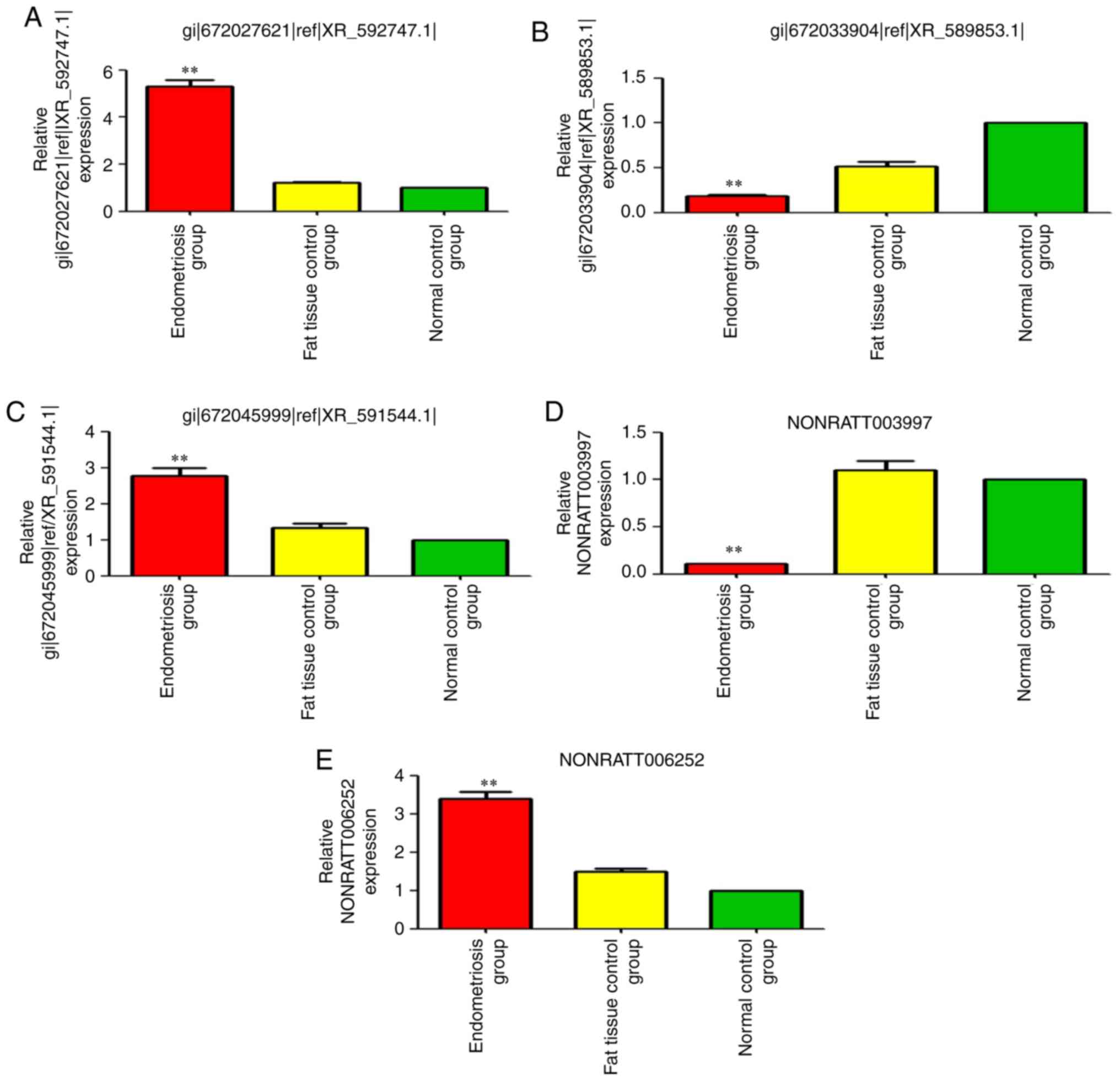
In order to confirm the results of the gene chip
analysis, three lncRNAs with upregulated expression
(gi|672027621|ref|XR_592747.1|, NONRATT006252 and
gi|672045999|ref|XR_591544.1|) and two lncRNAs with downregulated
expression were selected for RT-qPCR validation. The expression
trend and amplitude were consistent with the results of the gene
chip analysis (Fig. 2B).
gi|672066614|ref|XR_594547.1 (LOC103693263) was the
most upregulated lncRNA [fold-change (FC)=5.35; P<0.01]
(Tables II and III, and Fig. 3). NONRATT006252 (FC=3.42;
P<0.01) and gi|672045999|ref|XR_591544.1| (FC=2.88, P<0.01)
were also significantly upregulated (Tables II and III, and Fig. 3).
NONRATT003997 was the most significantly
downregulated (absolute FC=10.95; P<0.01) (Tables II and III, and Fig. 3). gi|672033904|ref|XR_589853.1|
was also significantly downregulated (absolute FC=5.79; P<0.01)
(Tables II and III, and Fig. 3).
Endometriosis is associated with
differentially expressed mRNAs
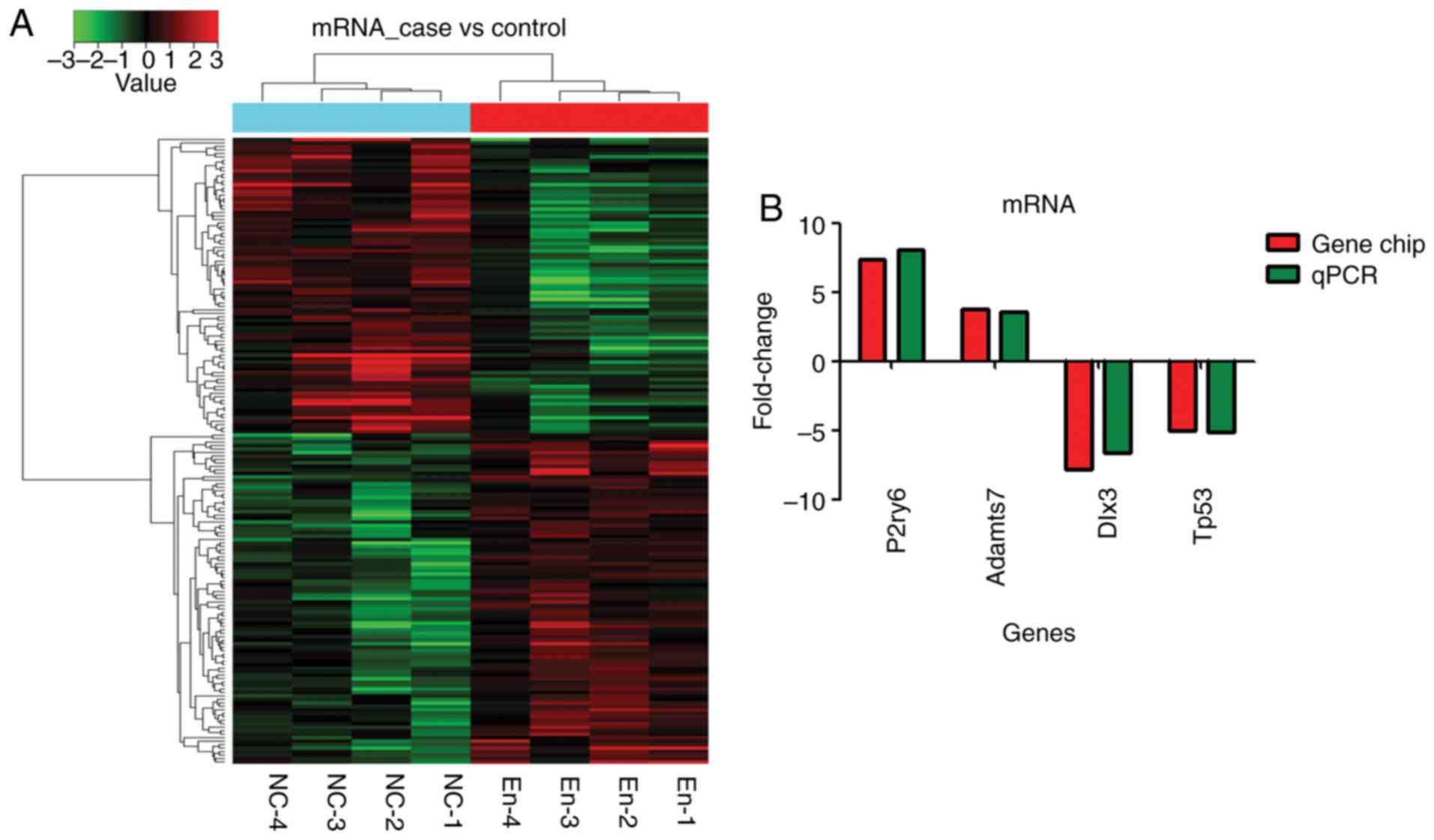
Overall, 182 mRNA transcripts were differentially
expressed: 97 were upregulated and 85 were downregulated
(fold-change >2) (Fig. 4A).
According to bioinformatics, four mRNAs (Adamts7, Tp53, Dlx3 and
P2ry6) were selected in the prediction of target regulation
relationship with five lncRNAs and gene chip results. RT-qPCR
validation showed that the expression of those mRNA was consistent
with the gene chip analysis (Fig.
4B).
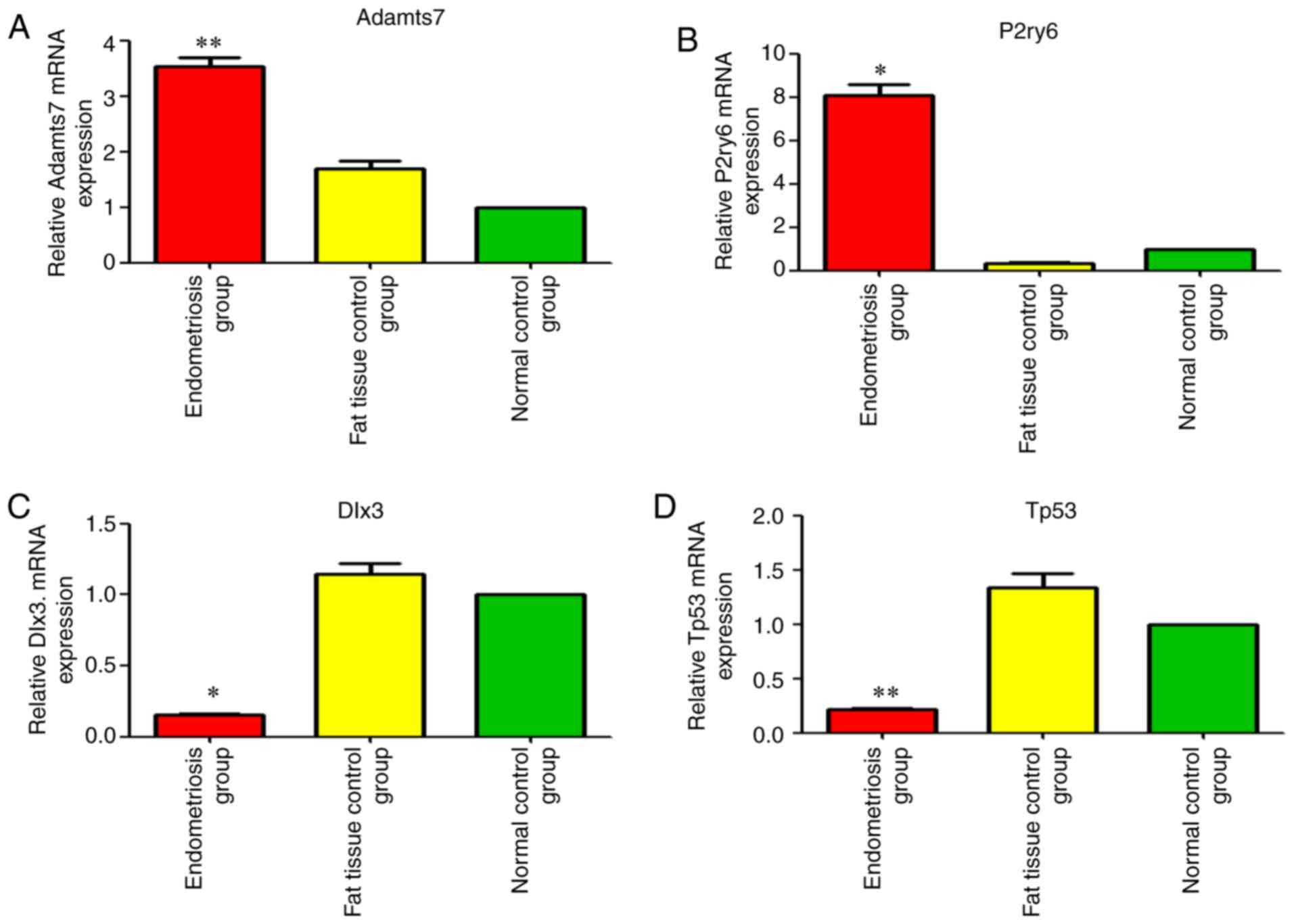
Adamts7 (A_44_P964460; FC=3.75, P<0.01) and P2ry6
(A_64_P042495; FC=7.37; P<0.05) were among the first 20
upregulated mRNAs (Tables IV and
V and Fig. 5). Dlx3 was the second
downregulated mRNA [FC absolute (ABS)=7.79; P<0.05] Tables IV and V, and Fig.
5). Tp53 was among the first 20 downregulated mRNAs [FC
(ABS)=5.01, P<0.01] (Tables
IV and V and Fig. 5).
 | Table IVTop 20 upregulated mRNA. |
Table IV
Top 20 upregulated mRNA.
| Probe name | mRNA | FC (abs) | P-value |
|---|
| A_64_P366031 | Cbln1 | 7.464442 | 0.002725 |
| A_64_P042495 | P2ry6 | 7.37147 | 0.02699 |
| A_64_P066251 | Ms4a14 | 5.004323 | 0.026015 |
| A_64_P021845 | Tac3 | 4.34276 | 0.024744 |
| A_44_P241448 | LOC102550973 | 3.948064 | 0.022094 |
| A_64_P057276 | LOC102549073 | 3.933694 | 0.023946 |
| A_42_P817214 | Efcab3 | 3.786581 | 0.004338 |
| A_44_P964460 | Adamts7 | 3.75427 | 0.01493 |
| A_44_P506980 | Pou3f4 | 3.655452 | 0.008571 |
| A_64_P044628 | Agtr1b | 3.09492 | 0.017631 |
| A_64_P012957 | Dntt | 3.094423 | 0.014797 |
| A_64_P016978 | Zfp488 | 3.061541 | 0.003415 |
| A_64_P087831 | Galnt13 | 3.038959 | 0.005398 |
| A_44_P135990 | Cd3g | 2.947655 | 0.002446 |
| A_64_P071732 | Lrrc4c | 2.944548 | 0.01782 |
| A_44_P375658 | Nxph1 | 2.943451 | 0.002413 |
| A_44_P1071620 | Sit1 | 2.608929 | 0.006022 |
| A_64_P059820 | Xkr6 | 2.532615 | 0.013807 |
| A_64_P003997 | Igsf1 | 2.527809 | 0.023865 |
| A_64_P113401 | Fam183b | 2.519156 | 0.00364 |
 | Table VTop 20 downregulated mRNA. |
Table V
Top 20 downregulated mRNA.
| Probe name | mRNA | FC (abs) | P-value |
|---|
| A_64_P009530 | LOC687483 | 8.781071 | 0.003707 |
| A_44_P575055 | Dlx3 | 7.794150 | 0.014970 |
| A_64_P142988 | Fosb | 7.026777 | 0.029057 |
| A_44_P295042 | Cxcl5 | 6.987926 | 0.012344 |
| A_64_P094855 | Slc46a2 | 5.017081 | 0.002092 |
| A_64_P121136 | Tp53 | 5.013210 | 0.008090 |
| A_43_P11576 | Hspa1b | 4.539404 | 0.024537 |
| A_64_P043411 | Elf3 | 4.520617 | 0.006673 |
| A_42_P576446 | Krt17 | 4.418501 | 0.019014 |
| A_64_P062413 | Calml3 | 4.182893 | 0.004898 |
| A_64_P017515 | Ppp1r35 | 3.717559 | 0.014440 |
| A_64_P014090 | Grp | 3.703025 | 0.028939 |
| A_44_P271658 | LOC100912449 | 3.625274 | 0.008212 |
| A_64_P072638 | Jsrp1 | 3.393673 | 0.006873 |
| A_64_P124621 | LOC102555023 | 3.319351 | 0.006541 |
| A_64_P135713 | Mbnl3 | 3.274797 | 0.010593 |
| A_64_P113635 | RGD1563753 | 3.230319 | 0.005243 |
| A_64_P031497 | Murc | 3.213777 | 0.013858 |
| A_44_P107653 | Lrrc26 | 2.927319 | 0.027704 |
| A_64_P002809 | Tubb4a | 2.853953 | 0.008596 |
Adamts7 and P2ry6 proteins are
differentially expressed in endometriosis, but not Dlx3 and
Tp53
Validation was performed using samples from 13 rats
in the endometriosis group, eight in the adipose tissue control
group and 14 in the blank control group. Fig. 6A shows that Adamts7 was expressed
in the uterine tissues of all three groups during the implantation
stage, mainly in the endometrial stroma, glands and uterine cavity
epithelium. The expression in the endometriosis group was
upregulated significantly in comparison with the two control groups
(adipose tissue and blank controls) in the three types of tissues
(P<0.05). The Adamts7 expression was significantly increased in
the adipose tissue control group compared with in the blank control
group in uterine stroma and glandular epithelium (P<0.05;
Fig. 6B).
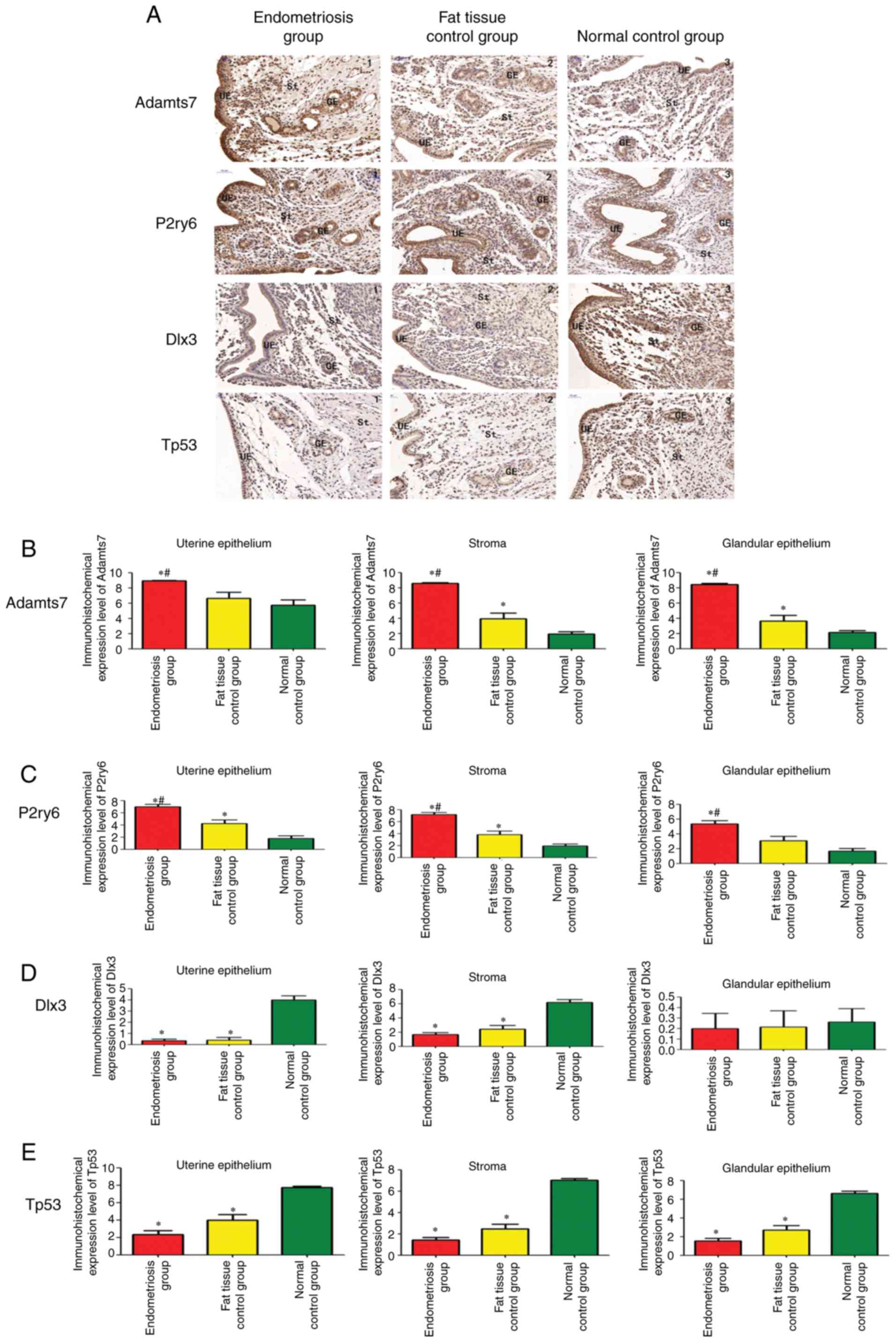 | Figure 6Immunohistochemistry of uterine
tissue in rats. (A) Expression of Adamts7, P2ry6, Dlx3 and Tp53 in
the endometriosis group (n=13), adipose tissue control group (n=8)
and blank control group (n=14). (magnification, x200).
Quantification of the immunohistochemistry results for (B) Adamts7,
(C) P2ry6, (D) Dlx3 and (E) Tp53. The samples were tested in
triplicates. *P<0.05 vs. blank controls.
#P<0.05 vs. adipose tissue controls. Adamts7, ADAM
metallopep-tidase with thrombospondin type 1 motif 7; Tp53, tumor
protein p53; Dlx3, distal-less homeobox 3; P2ry6, pyrimidinergic
receptor P2Y6; GE, glandular epithelium; St, endometrial stroma;
UE, uterine epithelium. |
P2ry6 was expressed in the uterine tissues of all
three groups of rats during the implantation stage, mainly in the
endometrial stroma, glands and uterine cavity epithelium. The
expression of P2ry6 was significantly increased in the
endometriosis group compared with in the two control groups in the
three tissue types (P<0.05). The expression of P2ry6 was also
significantly increased in the adipose tissue control group
compared with in the blank control group in uterine epithelium and
stroma (P<0.05; Fig. 6C).
Dlx3 was expressed in the uterine tissues of all
three groups of rats, mainly in the endometrial stroma and uterine
cavity epithelium. The expression in the glands was relatively
weak. In the endometrial stroma and uterine epithelium, there was
no significant difference in Dlx3 expression between the
endometriosis and adipose tissue control groups (P=0.291 and
P=0.98), and the expression in the endometriosis and adipose tissue
control groups was significantly downregulated in comparison with
the blank control group (P<0.05). There were no significant
differences in the expression of Dlx3 in the uterine glandular
epithelium among the three groups (P=0.80; Fig. 6D).
Tp53 was expressed in the uterine tissues of all
three groups of rats, mainly in the endometrial stroma and uterine
cavity epithelium. In all three tissue types, there was no
significant difference in Tp53 expression between the endometriosis
and adipose tissue control groups (P=0.089) and the expression the
endometriosis and adipose tissue control groups was significantly
downregulated in comparison with the blank control group
(P<0.05; Fig. 6E).
Taken together, those results suggest that
endometriosis is associated with increased protein expression of
Adamts7 and P2ry6, while Dlx3 and Tp53 are not changed by
endometriosis. Changes in Dlx3 and Tp53 protein expression could be
due to the surgery.
Western blot analysis
Adamts7 was significantly upregulated in the
endometriosis group in comparison with the adipose tissue control
group (P=0.002) and blank control group (P=0.003), and its
expression in the adipose tissue control group was significantly
upregulated compared with the blank control group (P=0.004;
Fig. 7A and B). The expression of
P2ry6 in the endometriosis group was significantly upregulated
compared with the adipose tissue control group (P=0.001) and blank
control group (P=0.001), and its expression in the adipose tissue
control group was significantly increased compared with in the
blank control group (P=0.004; Fig. 7A
and C). The expression of TP53 in the adipose tissue control
group was significantly down-regulated. compared with the blank
control group (P=0.004; Fig. 7A and
E). The expression of Dlx3 in the endometriosis group was
significantly downregulated compared with the adipose tissue
control group (P= 0.002) and blank control group (P= 0.002); its
expression in the adipose tissue control group was significantly
downregulated compared with the blank control group (P= 0.002;
Fig. 7A and D). Taken together,
those results show that the Adamts7 and P2ry6 proteins are
upregulated by endometriosis, while Dlx3 and Tp53 are downregulated
(Fig. 7).
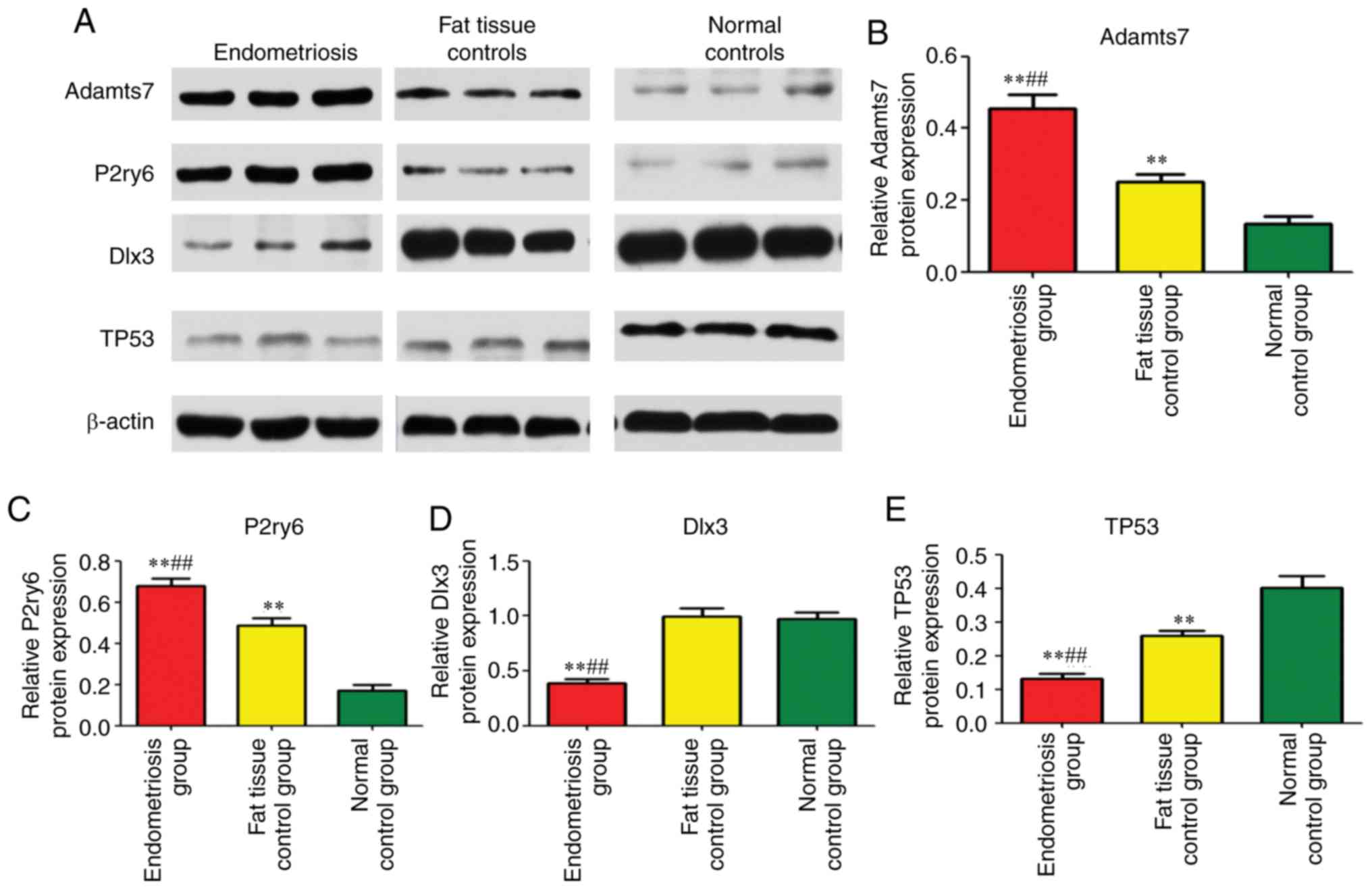 | Figure 7Western blot analysis of uterine
tissue in rats. (A) Expression of Adamts7, P2ry6, Dlx3 and Tp53 in
the endometriosis group (n=13), adipose tissue control group (n=8)
and blank control group (n=14). Quantification of the western
blotting results for (B) Adamts7, (C) P2ry6, (D) Dlx3 and (E) Tp53.
The samples were tested in triplicates. β-actin was used to
normalize the data. **P<0.01 vs. normal control,
##P<0.01 vs. adipose tissue controls. Adamts7, ADAM
metallopeptidase with thrombospondin type 1 motif 7; Tp53, tumor
protein p53; Dlx3, distal-less homeobox 3; P2ry6, pyrimidinergic
receptor P2Y6. |
GO and KEGG analysis
In the GO pathway analysis, the mRNAs that were
abnormally expressed were mainly involved in cell differentiation,
response to oxygen-containing compounds, ureteric bud
morphogenesis, ureter maturation, embryonic organ morphogenesis,
endocrine hormone secretion, female pregnancy, the immune response,
response to steroid hormones, the ER-nucleus signaling pathway,
parturition, response to oxidative stress, reproductive processes,
response to estrogen, regulation of reproductive processes, in
utero embryonic development and placenta development (Table VI).
 | Table VIGO analysis of mRNA. |
Table VI
GO analysis of mRNA.
| Term | ID | Background
number | P-value |
|---|
| Reproduction | GO:0000003 | 715 | 0.934613717 |
| Steroid metabolic
process | GO:0008202 | 205 | 0.907802417 |
| Embryo development
ending in birth or egg hatching | GO:0009792 | 519 | 0.907802417 |
| Sexual
reproduction | GO:0019953 | 543 | 0.828851221 |
| Placenta
development | GO:0001890 | 127 | 0.809271211 |
| Steroid
biosynthetic process | GO:0006694 | 115 | 0.785775079 |
| Embryonic
epithelial tube formation | GO:0001838 | 105 | 0.767818727 |
| In utero
embryonic development | GO:0001701 | 321 | 0.720657678 |
| Regulation of
hormone secretion | GO:0046883 | 196 | 0.706512715 |
| Blastocyst
development | GO:0001824 | 66 | 0.655993287 |
| Embryonic heart
tube development | GO:0035050 | 64 | 0.648745020 |
| Maternal process
involved in female pregnancy | GO:0060135 | 59 | 0.632235095 |
| Embryo
development | GO:0009790 | 780 | 0.605305219 |
| Response to
estrogen | GO:0043627 | 231 | 0.572555551 |
| Multicellular
organismal reproductive behavior | GO:0033057 | 25 | 0.453156618 |
| Hormone
transport | GO:0009914 | 253 | 0.437730680 |
| Chronic
inflammatory response | GO:0002544 | 22 | 0.432311123 |
| Regulation of
inflammatory response to antigenic stimulus | GO:0002861 | 20 | 0.411517366 |
| Parturition | GO:0007567 | 17 | 0.389834482 |
| Uterus
development | GO:0060065 | 16 | 0.383439641 |
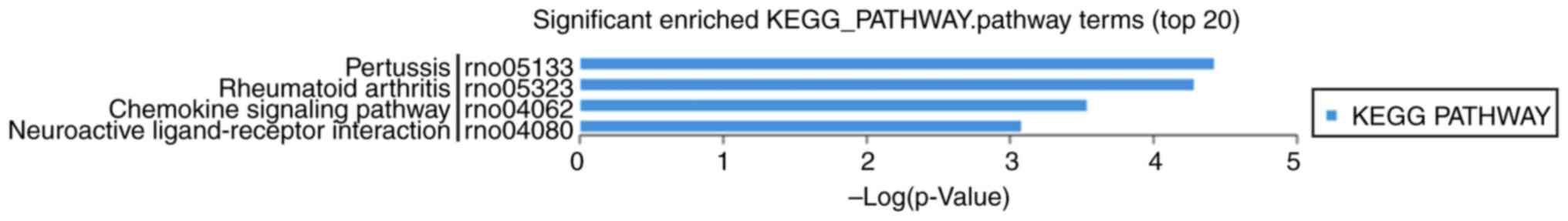
In the KEGG analysis (Fig. 8), differentially expressed mRNAs
were mainly involved in estrogen signaling pathway, GnRH signaling
pathway, inflammatory mediator regulation of TRP channels,
endometrial cancer, ovarian steroidogenesis, steroid hormone
biosynthesis, apoptosis signaling pathway, insulin/IGF
pathway-mitogen activated protein kinase/MAP kinase cascade,
inflammation mediated by chemokine and cytokine signaling
pathway.
In unsupervised hierarchical clustering analysis,
the differentially expressed lncRNA (Fig. 2A) and mRNA (Fig. 4A) were used to generate heat maps,
and they segregated into the endometriosis group and the normal
control group clusters.
lncRNA and mRNA co-expression profiles
and lncRNA function prediction
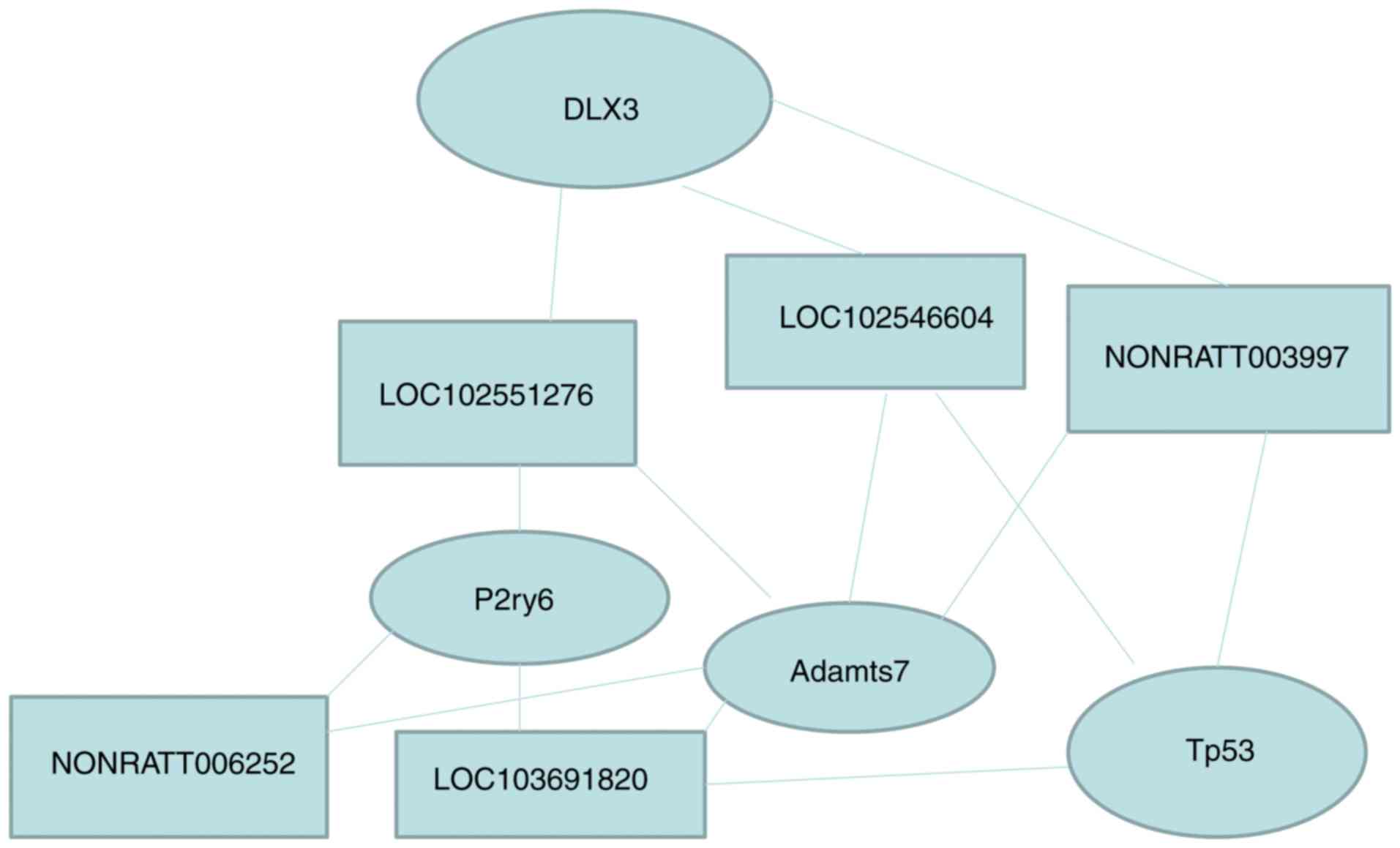
Numerous lncRNA bind thousands of mRNA to achieve
their biological functions. One lncRNA can bind to multiple mRNAs
and one mRNA might be the target gene for multiple lncRNAs. For
example, as shown in this study, Dlx3 was targeted by 32 lncRNA,
TP53 was targeted by 35 lncRNA, P2ry6 was targeted by 33 lncRNA,
Adamts7 was targeted by 40 lncRNA, NONRATT003997 targeted 21 mRNA,
gi|672027621|ref|XR_592747.1| targeted 23 mRNA,
gi|672045999|ref|XR_591544.1| targeted 29 mRNA, NONRATT006252
targeted 24 mRNA, and gi|672033904|ref|XR_589853.1| targeted 32
mRNA (Tables VI and V). Taken together, these results
indicate that the effect of endometriosis on uterine lncRNA and
mRNA is complex (Table VII and
Fig. 9).
 | Table VIIExpression of lncRNA and mRNA. |
Table VII
Expression of lncRNA and mRNA.
| Source | Target | Correlation | P-value | Source gene
symbol | Target gene
symbol |
|---|
|
gi|672033904|ref|XR_589853.1| | A_64_P135713 | 0.992091 | 0.000001 | gi|672033904 | Dlx3 |
| NONRATT003997 | A_64_P135713 | 0.928748 | 0.000857 | - | Dlx3 |
|
gi|672027621|ref|XR_592747.1| | A_64_P135713 | −0.92396 | 0.001037 | gi|672027621 | Dlx3 |
|
gi|672033904|ref|XR_589853.1| | A_64_P113635 | 0.961171 | 0.000142 | gi|672033904 | Tp53 |
| NONRATT003997 | A_64_P113635 | 0.96782 | 0.000081 | - | Tp53 |
|
gi|672045999|ref|XR_591544.1| | A_64_P113635 | −0.91168 | 0.00161 | gi|672045999 | Tp53 |
| NONRATT006252 | A_42_P814235 | 0.957719 | 0.000183 | - | P2ry6 |
|
gi|672027621|ref|XR_592747.1| | A_42_P814235 | 0.919696 | 0.001218 | gi|672027621 | P2ry6 |
|
gi|672045999|ref|XR_591544.1| | A_42_P814235 | 0.919036 | 0.001248 | gi|672045999 | P2ry6 |
|
gi|672033904|ref|XR_589853.1| | A_44_P178519 | −0.95751 | 0.000186 | gi|672033904 | Adamts7 |
| NONRATT006252 | A_44_P178519 | 0.912129 | 0.001586 | - | Adamts7 |
| NONRATT003997 | A_44_P178519 | −0.9136 | 0.00151 | - | Adamts7 |
|
gi|672027621|ref|XR_592747.1| | A_44_P178519 | 0.921091 | 0.001157 | gi|672027621 | Adamts7 |
|
gi|672045999|ref|XR_591544.1| | A_44_P178519 | 0.944424 | 0.000411 | gi|672045999 | Adamts7 |
Discussion
The present study showed that there were 115
upregulated lncRNAs, 51 downregulated lncRNAs, 97 upregulated mRNAs
and 85 downregulated mRNAs in the uterine tissues of rats with
endometriosis, compared with the control group. The relative
protein expression levels of Adamts7, P2ry6, Dlx3 and TP53 were
different in the endometriosis group. Bioinformatics could predict
the co-expression relationship of the selected five lncRNAs and
four mRNAs. GO and KEGG analyses predicted that Adamts7, P2ry6,
Dlx3, and TP53 were involved in endometriosis-related inflammation
and reproductive pathways. Taken together, the results strongly
suggest that the changes in the expression of lncRNAs, mRNAs and
Adamts7, P2ry6, Dlx3 and TP53 proteins may possibly affect
endometrial receptivity in rats with endometriosis during the
implantation window.
Wang et al (7) found that the co-expression
relationship of lncRNA(HOX)A11-AS1 (HOXA11 antisense RNA) and
homeobox A (HOXA9, HOXA10, HOXA11 and HOXA13) played an important
role in the pathogenesis of abdominal wall endometriosis. Powell
et al (22) served the
association among the allele 1p36.12 of LINC00339, blood CDC42 and
endometriosis. Sun et al (9) found that 948 lncRNA transcripts and
4,088 mRNA transcripts were abnormally expressed in the ectopic
endometrium of patients with endometriosis using gene chip
technology. Ghazal et al (5) confirmed that H19 acted as a
molecular sponge and attenuated the bioactivity of let-7 in the
eutopic endometrium of patients with endometriosis; the expression
of H19 in the endometrium of patients with endometriosis was
significantly downregulated compared with those without
endometriosis. Sigurgeirsson et al (23) found that the expression of 3,297
mRNAs, 516 lncRNAs and 102 small non-coding RNAs in the endometrium
were significantly different between the proliferative phase and
secretory phase of women's normal menstrual cycle (7-9 days after
ovulation), and they speculated that the changes in the expression
level of lncRNAs and mRNAs were probably the reasons for impaired
endometrial receptivity. Shin et al (24) found that in the four-cell stage of
embryo development in female rats, the X chromosome was selectively
silenced and inactivated; during this process, lncRNAXist was
activated by the downregulation of ubiquitin ligase Rnf12/RLIM3-5
in the embryonic cells. It has been shown that lncRNAs play a key
role in human embryo implantation (25). In the present study, a rat
endometriosis model was established by mating to obtain uterine
tissues during the implantation window. This study is the first
study to the best of our knowledge on the expression profile of
lncRNA and mRNA in endometriosis during the implantation
window.
Adamts7 belongs to a group of proteins that have
platelet-associated activity (26). The overexpression of ADAMTS-7
promotes the decomposition of cartilage oligomeric matrix protein
and accelerates the development of osteoarthritis induced by
surgery and collagen-induced arthritis (27). In the present study, Adamts7 was
mainly expressed in the endometrial stroma, glands and uterine
cavity epithelium, the expression in the uterine tissues of rats
with endometriosis was upregulated, and the expression in the
adipose tissue control group was also upregulated in comparison
with that of the blank control group. Therefore, it could be
hypothesized that implantation failure in endometriosis during the
implantation stage could be related to inflammation and the
function of secretory glands.
Giannattasio et al (28) showed that as a G-protein combined
receptor and a uridine diphosphate receptor, P2ry6 has a high
affinity for G-protein combined receptor and uridine-2-phosphate
receptor, which is an important endogenous inhibitor of the
function of T cells in allergic pulmonary inflammation. Hamby et
al (29) showed that
transforming growth factor-β1, lipopolysaccharide, interferon-γ and
other inflammatory factors can cause immune injury in astrocytes
through the expression of P2ry6, leading to nervous system
dysfunction. In the present study, the expression of P2ry6 in the
uterine tissues of rats with endometriosis was upregulated during
the implantation stage and the expression in the operation control
group was upregulated in comparison with that of the blank control
group. Therefore, this study considered that the gene regulation of
P2ry6 in rats with endometriosis during the implantation window
might be related to inflammation and hormone levels, which needs to
be confirmed.
Dlx3 has been widely studied in the course of
pregnancy and plays an important role in the formation of the
placenta (30-32). DLX3 is expressed in the placenta
tissues during human early pregnancy and plays an important
regulatory role in the trophoblastic layer, syncytial layer and the
formation of primary villi (33).
Berghorn et al (34)
showed that DLX3 in the placental tissues of 8.5-day mouse
embryonic development was not detected, but DLX3 could be detected
at 9.5 days and continued to 15.5 days. DLX3 produces
3β-hydroxysteroid dehydrogenase (VI) during the process of the
secretion of progesterone by placental trophoblast cells and the
expression of DLX3 is not detected in the embryonic stem cells
(Rcho-1) of rats (34). In the
present study, the expression of DLX3 in the endometrial stroma in
rats with endometriosis during the implantation window was not
significantly different from that of the operation control group
and they were both decreased compared with in the blank control
group. It was hypothesized that the effect of DLX3 on rats with
endometriosis during the implantation window was mainly reflected
in the endometrial stroma, which might not be related to gland
activity and hormone levels, but more closely related to
inflammation. Nevertheless, changes in the expression of DLX3 could
be due to the surgery (35).
Additional studies are necessary to examine this.
Tp53 acts as a bridge in the exchange process of
pregnant mother and fetus in the early stages. The inactivation of
Tp53 is affected by the preimplantation factor and may cause the
apoptosis of placental cells, indicating that the interaction of
Tp53 and implantation factors in human placental cytotrophoblasts
promotes the survival and growth of embryonic cells (36). The expression profile of TP53
fluctuates during pregnancy (37). In human placental cells,
adiponectin induces caspase activity by increasing the expression
of TP53 and BAX, leading to a decrease in the expression of
adiponectin receptors, and affecting the nutrient transport
function of the placenta (38).
Tp53 was also a downregulated gene in the uterine tissues of rats
with endometriosis during the implantation window, but there was no
significant difference between the endometriosis group and
operation control group, both of them showing downregulated levels
in comparison with those of the blank control group. Tp53 probably
plays an important role in the formation of endometrial receptivity
in rats with endometriosis during the implantation window, which
might be related to the endometriosis-associated inflammatory
characteristics and gland secretion. As for DLX3, changes in Tp53
could also be due to surgery (39). This will have to be examined in
the future.
The present study is not without limitations.
Although the present study could efficiently avoid the effects of
the menstrual cycle in different individuals, it was still
impossible to avoid the disadvantages of using animal experiments.
There were also some differences among animals and the endometrium
could not be separated because the uterine tissues of rats are too
small. Therefore, the gene expression profile in the uterine
tissues was decreased compared with the endometrium. Moreover,
although human lncRNA sequence are highly similar to rats', there
are still some differences (Table
VIII). Therefore, the next step will be to compare the degree
of conservation of human and rat lncRNA sequences. On the basis of
the obtained gene data of lncRNA and mRNA expression profiles in
the uterine tissues of rats with endometriosis during the
implantation window, five lncRNAs were compared with the
conservative sequence of the human lncRNA database, showing that
the lncRNAs were highly consistent. The correlation analyses
indicate a relationship among the lncRNAs and mRNAs, but not the
exact causal nature of the relationships. Finally, no functional
and mechanistic experiments were performed to confirm the GO and
KEGG results. Additional studies are still necessary to confirm the
roles of the differentially expressed lncRNAs and mRNAs identified
in the present study.
 | Table VIIIComparison between rat and human
lncRNA sequences. |
Table VIII
Comparison between rat and human
lncRNA sequences.
| Rat lncRNA | Human lncRNA | % identical | lncRNA ID |
|---|
|
gi|672027621|ref|XR_592747.1| | p17451 | 90.31 | TCONS_00001579 |
| NONRATT022454 | p44032_v4 | 83.45 | uc001cvx.3 |
|
gi|672045999|ref|XR_591544.1| | p10107 | 83.29 |
ENST00000563759.1 |
| NONRATT014258 |
RNA40309|RefSeq_2352_1430 | 90.06 | |
| NONRATT011779 |
RNA38792|RefSeq_733_3290 | 81.9 | |
| NONRATT029450 |
RNA42607|UCSC_176_6901 | 100 | |
| NONRATT028550 | p28817 | 86.47 | LIT3372 |
| NONRATT003997 | p37546_v4 | 84.17 |
ENST00000514073.1 |
| NONRATT027560 | p28817 | 81.42 | LIT3372 |
| NONRATT000719 |
RNA45098|UCSC_3204_2521 | 88 | |
| NONRATT017840 |
RNA43455|UCSC_1198_3829 | 81.71 | |
The results showed that the changes in the
expression of lncRNAs, mRNAs and proteins (Adamts7, P2ry6, Dlx3 and
TP53) may possibly affect the endometrial receptivity in rats with
endometriosis during the implantation window, probably resulting in
implantation failure of the embryo. This study provides new
insights to investigate the pathogenesis of the disease and new
clues about the possible causes of implantation failure in patients
with endometriosis. It could ultimately reveal treatment targets
for endometriosis infertility, but RNA stability is still a major
barrier to the successful development of RNA therapeutics.
Abbreviations:
|
cDNA
|
complementary DNA
|
|
H&E
|
hematoxylin and eosin
|
|
lncRNA
|
long non-coding RNA
|
|
PCC
|
Pearson's correlation coefficient
|
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HC conceived and coordinated the study, designed,
performed and analyzed the experiments, wrote the paper. XZ, ZL,
YZ, JL carried out the data collection, data analysis, and revised
the paper. All authors reviewed the results and approved the final
version of the manuscript.
Ethics approval and consent to
participate
This study was approved by the experimental animal
center of Peking Union Medical College Hospital and Chinese Academy
of Medical Sciences (permit no. XHDW-2016-000). All experiments
were performed in accordance with the principles of experimental
animal management and protection. The rats were humanely sacrificed
as necessary to prevent suffering.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Bedaiwy MA, Alfaraj S, Yong P and Casper
R: New developments in the medical treatment of endometriosis.
Fertil Steril. 107:555–565. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao Y, Gong P, Chen Y, Nwachukwu JC,
Srinivasan S, Ko C, Bagchi MK, Taylor RN, Korach KS, Nettles KW, et
al: Dual suppression of estrogenic and inflammatory activities for
targeting of endometriosis. Sci Transl Med. 7:271ra92015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Ziegler D, Pirtea P, Galliano D,
Cicinelli E and Meldrum D: Optimal uterine anatomy and physiology
necessary for normal implantation and placentation. Fertil Steril.
105:844–854. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lessey BA and Kim JJ: Endometrial
receptivity in the eutopic endometrium of women with endometriosis:
It is affected, and let me show you why. Fertil Steril. 108:19–27.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghazal S, McKinnon B, Zhou J, Mueller M,
Men Y, Yang L, Mueller M, Flannery C, Huang Y and Taylor HS: H19
lncRNA alters stromal cell growth via IGF signaling in the
endometrium of women with endometriosis. EMBO Mol Med. 7:996–1003.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang Z, Chen Y, Zhao Y, Xu C, Zhang A,
Zhang Q, Wang D, He J, Hua W and Duan P: miR-200c suppresses
endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell
Res Ther. 8:2512017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang M, Hao C, Huang X, Bao H, Qu Q, Liu
Z, Dai H, He S and Yan W: Aberrant expression of lncRNA
(HOXA11-AS1) and homeobox A (HOXA9, HOXA10, HOXA11, and HOXA13)
genes in infertile women with endometriosis. Reprod Sci.
25:654–661. 2018. View Article : Google Scholar
|
|
8
|
Sha L, Huang L, Luo X, Bao J, Gao L, Pan
Q, Guo M, Zheng F and Wang H: Long non-coding RNA LINC00261
inhibits cell growth and migration in endometriosis. J Obstet
Gynaecol Res. 43:1563–1569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun PR, Jia SZ, Lin H, Leng JH and Lang
JH: Genome-wide profiling of long noncoding ribonucleic acid
expression patterns in ovarian endometriosis by microarray. Fertil
Steril. 101:1038–1046.e7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rekker K, Saare M, Eriste E, Tasa T,
Kukuškina V, Roost AM, Anderson K, Samuel K, Karro H, Salumets A
and Peters M: High-throughput mRNA sequencing of stromal cells from
endo-metriomas and endometrium. Reproduction. 154:93–100. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Li Y, Yang Z, Liu K and Wang D:
Genome-wide microarray analysis of long non-coding RNAs in eutopic
secretory endometrium with endometriosis. Cell Physiol Biochem.
37:2231–2245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vernon MW and Wilson EA: Studies on the
surgical induction of endometriosis in the rat. Fertil Steril.
44:684–694. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Keenan JA, Williams-Boyce PK, Massey PJ,
Chen TT, Caudle MR and Bukovsky A: Regression of endometrial
explants in a rat model of endometriosis treated with the immune
modulators loxoribine and levamisole. Fertil Steril. 72:135–141.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai H, Zhu XX, Li ZF, Zhu YP and Lang JH:
MicroRNA dysregulation and steroid hormone receptor expression in
uterine tissues of rats with endometriosis during the implantation
window. Chin Med J (Engl). 131:2193–2204. 2018. View Article : Google Scholar
|
|
15
|
Makrigiannakis A, Zoumakis E, Kalantaridou
S, Coutifaris C, Margioris AN, Coukos G, Rice KC, Gravanis A and
Chrousos GP: Corticotropin-releasing hormone promotes blastocyst
implantation and early maternal tolerance. Nat Immunol.
2:1018–1024. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao L, Wu J and Wang JY, Chung HK,
Kalakonda S, Rao JN, Gorospe M and Wang JY: Long noncoding RNA
uc.173 promotes renewal of the intestinal mucosa by inducing
degradation of microRNA 195. Gastroenterology. 154:599–611. 2018.
View Article : Google Scholar :
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Konno R, Yamakawa H, Utsunomiya H, Ito K,
Sato S and Yajima A: Expression of survivin and Bcl-2 in the normal
humanendometrium. Mol Hum Reprod. 6:529–534. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong ES, Schmitt BM, Kazachenka A, Thybert
D, Redmond A, Connor F, Rayner TF, Feig C, Ferguson-Smith AC,
Marioni JC, et al: Interplay of cis and trans mechanisms driving
transcription factor binding and gene expression evolution. Nat
Commun. 8:10922017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Wang N, Cai R, Zhao F, Xiong Y, Li
X, Wang A, Lin P and Jin Y: Genome-wide analysis and functional
prediction of long non-coding RNAs in mouse uterus during the
implantation window. Oncotarget. 8:84360–84372. 2017.PubMed/NCBI
|
|
22
|
Powell JE, Fung JN, Shakhbazov K, Sapkota
Y, Cloonan N, Hemani G, Hillman KM, Kaufmann S, Luong HT, Bowdler
L, et al: Endometriosis risk alleles at 1p36.12 act through inverse
regulation of CDC42 and LINC00339. Hum Mol Genet. 25:5046–5058.
2016.
|
|
23
|
Sigurgeirsson B, Amark H, Jemt A, Ujvari
D, Westgren M, Lundeberg J and Gidlöf S: Comprehensive RNA
sequencing of healthy human endometrium at two time points of the
menstrual cycle. Biol Reprod. 96:24–33. 2017.PubMed/NCBI
|
|
24
|
Shin J, Wallingford MC, Gallant J, Marcho
C, Jiao B, Byron M, Bossenz M, Lawrence JB, Jones SN, Mager J and
Bach I: RLIM is dispensable for X-chromosome inactivation in the
mouse embryonic epiblast. Nature. 511:86–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bouckenheimer J, Assou S, Riquier S, Hou
C, Philippe N, Sansac C, Lavabre-Bertrand T, Commes T, Lemaitre JM,
Boureux A and De Vos J: Long non-coding RNAs in human early
embryonic development and their potential in ART. Hum Reprod
Update. 23:19–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu CJ: The role of ADAMTS-7 and ADAMTS-12
in the pathogenesis of arthritis. Nat Clin Pract Rheumatol.
5:38–45. 2009. View Article : Google Scholar
|
|
27
|
Zhang Y, Wei F and Liu CJ: Overexpression
of ADAMTS-7 leads to accelerated initiation and progression of
collagen-induced arthritis in mice. Mol Cell Biochem. 404:171–179.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Giannattasio G, Ohta S, Boyce JR, Xing W,
Balestrieri B and Boyce JA: The purinergic G protein-coupled
receptor 6 inhibits effector T cell activation in allergic
pulmonary inflammation. J Immunol. 187:1486–1495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hamby ME, Coppola G, Ao Y, Geschwind DH,
Khakh BS and Sofroniew MV: Inflammatory mediators alter the
astrocyte transcriptome and calcium signaling elicited by multiple
G-protein-coupled receptors. J Neurosci. 32:14489–14510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
King JH, Kwan STC, Yan J, Klatt KC, Jiang
X, Roberson MS and Caudill MA: Maternal choline supplementation
alters fetal growth patterns in a mouse model of placental
insufficiency. Nutrients. 9:E7652017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Murthi P, Hiden U, Rajaraman G, Liu H,
Borg AJ, Coombes F, Desoye G, Brennecke SP and Kalionis B: Novel
homeobox genes are differentially expressed in placental
microvascular endo-thelial cells compared with macrovascular cells.
Placenta. 29:624–630. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clark PA, Brown JL, Li S, Woods AK, Han L,
Sones JL, Preston RL, Southard TL, Davisson RL and Roberson MS:
Distal-less 3 haploinsufficiency results in elevated placental
oxidative stress and altered fetal growth kinetics in the mouse.
Placenta. 33:830–838. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chui A, Pathirage NA, Johnson B,
Cocquebert M, Fournier T, Evain-Brion D, Roald B, Manuelpillai U,
Brennecke SP, Kalionis B and Murthi P: Homeobox gene distal-less 3
is expressed in proliferating and differentiating cells of the
human placenta. Placenta. 31:691–697. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berghorn KA, Clark PA, Encarnacion B,
Deregis CJ, Folger JK, Morasso MI, Soares MJ, Wolfe MW and Roberson
MS: Developmental expression of the homeobox protein distal-less 3
and its relationship to progesterone production in mouse placenta.
J Endocrinol. 186:315–323. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhan Y, Li X, Gou X, Yuan G, Fan M and
Yang G: DLX3 inhibits the proliferation of human dental pulp cells
through inactivation of canonical Wnt/β-catenin signaling pathway.
Front Physiol. 9:16372018. View Article : Google Scholar
|
|
36
|
Moindjie H, Santos ED, Gouesse RJ,
Swierkowski-Blanchard N, Serazin V, Barnea ER, Vialard F and
Dieudonné MN: Preimplantation factor is an anti-apoptotic effector
in human trophoblasts involving p53 signaling pathway. Cell Death
Dis. 7:e25042016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kaczmarek MM, Krawczynski K, Najmula J,
Reliszko ZP, Sikora M and Gajewski Z: Differential expression of
genes linked to the leukemia inhibitor factor signaling pathway
during the estrus cycle and early pregnancy in the porcine
endometrium. Reprod Biol. 14:293–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Duval F, Santos ED, Poidatz D, Serazin V,
Gronier H, Vialard F and Dieudonné MN: Adiponectin inhibits
nutrient transporters and promotes apoptosis in human villous
cytotrophoblasts: Involvement in the control of fetal growth. Biol
Reprod. 94:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hausmann R, Nerlich A and Betz P: The
time-related expression of p53 protein in human skin wounds-a
quantitative immunohis-tochemical analysis. Int J Legal Med.
111:169–172. 1998. View Article : Google Scholar
|