Introduction
Extracellular vesicles (EVs), particularly exosomes,
have attracted considerable interest in cancer research owing to
the discovery of their role in inter-cellular communications.
Exosomes are small endosome-derived lipid nanoparticles (50-100
nm), actively secreted by exocytosis in the majority of living
cells. Exosome release occurs either constitutively or upon
induction, under both normal and pathological conditions; this
dynamically regulated process is functionally relevant, such that
the molecular composition of exosomes reflects the features of the
parent cell. Exosomes transport a distinct molecular cargo of
proteins and nucleic acids (mRNA, miRNA and genomic DNA) in
peripheral blood (PB) and other bio-fluids (e.g., urine and
saliva), and are recognized as relevant for the diagnosis and
prognosis of certain pathologies, particularly solid tumors.
Several studies have demonstrated that the proteins and nucleic
acids contained within the exosomes of cancer patients can be
isolated and identified as exosomal markers associated with cancer
evolution (1-5).
Emerging evidence has also suggested that
membrane-bound carriers (EVs and exosomes) released by cancer cells
can mediate cell-cell communication via the delivery of their
contents (6). When considering
leukemic cells, membrane-bound carriers can potentially alter the
physiological equilibrium of extra-medullary sites (7), an interesting aspect in
hematological malignancies. Innovative approaches, based on the
selection of different sub-populations of tumor-associated exosomes
(8), appear to be powerful and
more informative than conventional isolation methods for diagnostic
and prognostic purposes (9-13).
This is grounded in the evidence that exosomes released by tumor
cells present specific and pan-cancer antigens at the membrane
surface (14-16), enabling the possibility to
determine the nature of the cell of origin, and to isolate the
exosome fraction using the appropriate antibodies. Exosome
enrichment has already been explored in the solid tumors field,
with successful results (17-20).
Currently, at least to the best of our knowledge,
there are no studies available reporting the possibility of
enriching the tumor-derived exosome fraction in patients affected
by different types of leukemia. However, certain groups have
revealed that exosomes isolated from cell line cultures modulate
the crosstalk between leukemia cells and the bone marrow (BM)
microenvironment, and that they also carry molecular tumor markers
(21-24). Moreover, recent studies have
indicated that leukemia-derived exosomes can be utilized as
prognostic (25), diagnostic
(26) and therapeutic biomarkers
(27) for individuals suffering
from different hematologic malignancies, such as acute myeloid
leukemia (28-30), multiple myeloma (31) and chronic lymphocytic leukemia
(32).
Corrado et al (33) reported that chronic myeloid
leukemia (CML) cells may release exosomes, and that the addition of
these vesicles to vascular endothelial cells, as well as to BM
stromal cells cultures, affects both in vitro and in
vivo tumor progression. CML is a clonal myeloproliferative
disease characterized by a reciprocal translocation between
chromosomes 9 and 22 [t(9;22)], resulting in Philadelphia
chromosome-positive (Ph+) CML and the formation of a new
fusion genes encoding for the chimeric breakpoint cluster
region-proto-oncogene 1 tyrosine-protein kinase (BCR-ABL1) p210
oncoprotein (34). BCR-ABL1
exhibits a constitutively high tyrosine kinase activity and is
considered the hallmark of CML Ph+, as it plays a
pivotal role in the pathogenesis (35) and progression of the disease.
CML is characterized by three distinct disease
phases: The chronic phase (CP), the accelerated phase and the
blastic phase. Indeed, BCR-ABL1 reduction or ablation is necessary
to avoid progression to the advanced blastic phase of disease
(36). Currently, treatment with
tyrosine kinase inhibitors (TKIs) targeting the BCR-ABL1 p210
protein is the only treatment able to successfully attenuate the
progression of CML to the blastic phase, inducing a significant
reduction in the expression of the BCR-ABL1 transcript,
namely the major or deep molecular response (DMR), in 80-90% of
patients. The detection and quantification of the BCR-ABL1
transcript in PB cells by reverse transcription-quantitative PCR
(RT-qPCR), normalized to a housekeeping gene, is recognized as the
international standardized method for determining minimal residual
disease (MRD), and plays a key role in the management of patients
with CML (37,38). As aforementioned, the MRD level is
distinguished as a major molecular response (MMR), with BCR-ABL1
≤0.1% and ABL1 >10.000 copies; or a DMR if BCR-ABL1 ≤0.01%, or
ABL >10.000 copies when BCR-ABL1 is undetectable (37). The achievement of the DMR is
associated with survival and the opportunity for treatment-free
remission (TFR).
Nevertheless, numerous studies have demonstrated the
persistence of CML leukemic cells in the BM niche following
treatment, even in patients with undetectable levels of the
BCR-ABL1 transcript by RT-qPCR. The persistence of these
cells in patients with CML has also been confirmed in clinical
practice, where molecular relapse is experienced in ~50% of
patients undergoing TKI discontinuation programs (39-42). Residual leukemic cells may be
quiescent stem cells, detectable only by CD26 recognition methods
(43), or active CML cells.
Residual BM CML cells, which indicate the activation of the
BCR-ABL1 pathways, may re-enter the proliferative
cycle and may be responsible for molecular relapse. In this
scenario, the persistence of the residual active leukemic cells
pool, surviving indefinitely into tumor-specific niches of the BM,
is not evaluated by the standardized MRD monitoring system.
Therefore, a deep and undetectable MR measured on PB cells may not
be sufficient to detect the persistence of BM-active CML leukemic
cells.
Collectively, these biological findings render
Ph+ CML a suitable model with which to explore the
feasibility of tumor-exosome enrichment in hematological
malignancies, and consequently, to investigate new possibilities
for the detection and evaluation of residual tumor-cell activity.
At present, limited data concerning exosome evaluation in patients
with CML, and the identification of the BCR-ABL1 transcript
in these vesicles have been reported. To the best of our knowledge,
only Kang et al (44)
demonstrated the possibility of quantifying the BCR-ABL1
transcript in the total exosome pool, conventionally isolated from
the PB of patients with CML in the advanced disease phases.
Conversely, negative results were obtained when detecting the
transcript by nested PCR in TKI-treated patients with CML in the
CP.
Supported by the above-mentioned considerations, CML
was identified as the most suitable hematological neoplasia for a
feasibility study on neoplastic exosome enrichment. The present
study indicates the results of an explorative feasibility study
based on the quantification of the BCR-ABL1 transcript by
digital PCR (dPCR), a more specific and accurate tool for the
detection of the MRD in patients with CML (45,46), on a tumor-derived exosome fraction
enriched in the PB of patients in the CP, who are also receiving
TKI treatment.
Materials and methods
Patients and controls
A total of 10 patients with Ph+ CML in
the CP, treated with TKIs [imatinib (IM) or dasatinib (DAS)] and in
DMR as confirmed by conventional RT-qPCR; 10 healthy subjects; and
4 patients affected by Ph negative (Ph−) hemato-logical
malignancies, were enrolled in the present study. For CML case no.
1, 5 additional samples from diagnosis to the DMR achievement were
retrospectively analyzed. Healthy subjects and Ph−
patients served as healthy and pathological Ph−
controls, respectively. The 10 healthy subjects were selected based
on having no previous onco-hematological diseases, no relatives
affected by hematological or solid tumors, not undergoing
therapeutic treatment, and having no comorbidities at the time of
sampling. They were 3 males and 7 females, with a median age of
31.5 years (range, 27-49 years). The pathological Ph−
control group included 1 case of acute myeloid leukemia, 1 case of
multiple myeloma, 1 case of myelofibrosis and 1 of acute
lymphoblastic leukemia B. The absence of chromosomal rearrangement
involving BCR or ABL genes was evaluated by FISH and confirmed by
RT-qPCR. The characteristics of the patients and the control
cohorts are presented in Tables I
and II, respectively.
 | Table IThe clinical characteristics of the
CML patient cohort. |
Table I
The clinical characteristics of the
CML patient cohort.
| Variables | Median (range) |
|---|
| No. of
Ph+ CML patients | 10 |
| Sex (M/F) | 6/4 |
| Age at study
(median and range in years) | 70 (33-85) |
| BCR-ABL transcript
at diagnosis | |
| B3A2 | 7 |
| B2A2 | 3 |
| TKI current
treatment | |
| IM | 7 |
| DAS | 3 |
| DMR duration
(months) | 57 (33-81) |
| MR at study | |
| MR4.0
total/undetectable | 1/0 |
| MR4.5
total/undetectable | 3/1 |
| MR5.0
total/undetectable | 6/5 |
 | Table IIClinical characteristics of
Philadelphia-negative patients included in the study. |
Table II
Clinical characteristics of
Philadelphia-negative patients included in the study.
| Ph− 1
| Ph− 2
| Ph− 3
| Ph− 4
|
|---|
| Sex | M | F | F | M |
|---|
| Diagnosis | Acute lymphoblastic
leukemia B | Acute myeloid
leukemia | Myelofibrosis | Multiple
myeloma |
| Age (years) | 38 | 67 | 64 | 49 |
| Disease phase | Diagnosis | CR after
consolidation therapy | CR at 3 months post
allo-HSCT | Relapse |
The present explorative study was approved by the
Ethics Committee of Brescia (local study no. NP2370 approved in
May, 2016) and performed according to good clinical practice and
Declaration of Helsinki. All patients and subjects gave their
written informed consent for the enrollment in the study, the use
of their samples for research purposes and for the publication of
the encompassed data.
Plasma collection, exosome isolation and
RNA extraction
To isolate exosomes, 4.9 ml of EDTA-treated PB from
patients and healthy donors was centrifuged at 2,000 x g for 10 min
in EDTA-K2 gel S-monovette® tubes (Sarsted Inc.). To
avoid cellular contamination, the plasma was aspirated up to 1 cm
of the gel front, and stored at -80°C until analysis. Upon thawing,
2 ml of plasma from each sample was clarified by centrifugation
(1,200 x g for 20 min, 10°C) to eliminate residual red blood cells
and cellular debris.
Following tumor exosome isolation from the plasma,
exosomal RNA purification was performed using the SoRTEV™ EV-RNA
Low Volume Enrichment kit (Exosomics S.p.A), according to the
manufacturer's protocol, and the total exosomal RNA was eluted in a
volume of 15 µl. A total of 12 µl extracted RNA was
reverse-transcribed using the RNAUsScript Reverse Transcriptase kit
(LeGene Biosciences) following the manufacturer's instructions. The
final reaction volume was 20 µl, and 5 µl was used
for each dPCR quantification. The isolation of the total exosome
fraction from the plasma of 5 healthy controls (cases healthy
controls nos. 6, 7, 8, 9 and 10) and of 10 CML samples (case nos.
5, 6, 7, 8, 9 and 1a-e) was performed using the Total Exosome
Isolation kit and Total Exosome RNA and Protein Isolation kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions.
RT-qPCR and dPCR analyses RT-qPCR
analysis
Conventional RT-qPCR was carried out at the
Reference Laboratory (Spedali Civili of Brescia, member of LabNet,
Italy), according to European Leukemia Net (ELN) Guidelines
(37). A total of 10 ml of PB was
obtained and used for RT-qPCR analysis for all subjects enrolled in
the present study.
Molecular responses (MRs) by RT-qPCR were defined
according to the latest laboratory recommendations and using
ABL1 as a reference gene. Measurable MRs were assigned
following the international scale (IS), and scored MR4.0
if BCR-ABL1 %IS was ≤0.01%, MR4.5 if
BCR-ABL1 %IS was ≤0.0032%, and MR5.0 if
BCR-ABL1 %IS was ≤0.001. The minimum sum of the ABL1
reference gene transcripts, irrespective of BCR-ABL1
detection, was 10,000, 32,000 and 100,000 for MR4.0,
MR4.5 and MR5.0, respectively (37). The quantification values of
BCR-ABL1 and ABL1 performed by the Reference Laboratory are
presented in Table III.
 | Table IIIResults of the quantification of
BCR-ABL1/ABL1 by RT-qPCR aligned with IS and of
BCR-ABL1 and ABL1 transcript by RT-qPCR in CML
patients, before the normalization and the calculation of the
ratio. |
Table III
Results of the quantification of
BCR-ABL1/ABL1 by RT-qPCR aligned with IS and of
BCR-ABL1 and ABL1 transcript by RT-qPCR in CML
patients, before the normalization and the calculation of the
ratio.
| RT-qPCR
BCR-ABL1/ABL1
| | |
|---|
| Case no. | MR | IS | BCR-ABL1
transcript copies | ABL1
transcript copies |
|---|
| 1 | 4.5 | 0.0013 | 3 | 94,484 |
| 2 | 4.5 | 0 | 0 | 34,853 |
| 3 | 4.5 | 0.0031 | 4 | 96,732 |
| 4 | 5.0 | 0 | 0 | 102,813 |
| 5 | 5.0 | 0 | 0 | 109,406 |
| 6 | 5.0 | 0 | 0 | 101,818 |
| 7 | 5.0 | 0 | 0 | 110,005 |
| 8 | 5.0 | 0 | 0 | 105,513 |
| 9 | 5.0 | 0.0008 | 3 | 143,946 |
| 10 | 4.0 | 0.0053 | 8 | 148,729 |
| 1a | 0.0 | 19.528 | 4,480 | 23,198 |
| 1b | 2.0 | 0.1021 | 142 | 136,406 |
| 1c | 3.0 | 0.0112 | 5 | 48,072 |
| 1d | 3.0 | 0.0707 | 70 | 105,953 |
| 1e | 4.0 | 0.0054 | 8 | 148,921 |
dPCR analysis
Since dPCR has been demonstrated to be more accurate
and sensitive than conventional RT-qPCR for the quantification of
the MRD in patients with CML (by detecting the BCR-ABL1
transcript), in order to compare the results obtained from the
tumor exosomes, BCR-ABL1 dPCR analysis was first performed
on cDNA from PB cells, as previously described (45,46). To reduce bias, the cDNA samples
were the same as those used during the RT-qPCR analysis.
A 16 µl reaction mixture was prepared,
containing 8 µl 2X QuantStudio 3D Digital PCR Master Mix
(Thermo Fisher Scientific, Inc.), 0.8 µl 20X
TaqMan-MGB-FAM-probe assay for BCR-ABL1, 1.1 µl cDNA
(50 ng/µl) and 6.1 µl nuclease-free water (Qiagen,
Inc.). The negative control reaction mix contained 8 µl 2X
QuantStudio 3D Digital PCR Master Mix, 0.8 µl 20X
TaqMan-MGB-FAM-probe assay for BCR-ABL1 and 7.2 µl
nuclease-free water; one negative control was loaded for each round
of thermal cycling, containing samples prepared with the same mix.
The reverse transcription negative control reaction mixture
contained 8 µl 2X QuantStudio 3D Digital PCR Master Mix, 0.8
µl 20X TaqMan-MGB-FAM-probe assay for BCR-ABL1, 1.2
µl reverse transcription blank and 6.1 µl
nuclease-free water. dPCR analysis was performed on cDNA extracted
from exosomes, and was set up and optimized considering the low
quantity of RNA within vesicles.
To quantify the RNA extracted from the vesicles, the
Y4 transcript was used as an internal reference gene
(47,48), and quantified using a 20X
TaqMan-MGB-FAM-probe assay for Y4 (5' Y4 RNA)
(49). The conventional
ABL1 gene was excluded since it is not considered a reliable
internal reference gene for exosomes (47,48,50-52).
A 16 µl of reaction mixture containing 8
µl 2X QuantStudio 3D Digital PCR Master Mix (Thermo Fisher
Scientific, Inc.), 0.8 µl 20X TaqMan-MGB-FAM-probe assay for
BCR-ABL1, 5 µl diluted cDNA and 2.2 µl
nuclease-free water (Qiagen, Inc.). The negative control reaction
contained no cDNA and was made up to a total of 16 µl with
7.2 µl nuclease-free water; a negative control sample was
used for each round of thermocycling (as aforementioned). The
reverse transcription negative control contained 8 µl 2X
QuantStudio 3D Digital PCR Master Mix, 0.8 µl 20X
TaqMan-MGB-FAM-probe assay for BCR-ABL1, 5 µl reverse
transcription blank and 2.2 µl nuclease-free water. The same
analysis parameters were replicated using the 20X
TaqMan-MGB-FAM-probe assay for Y4.
For each sample of the cellular and exosomal cDNA
groups, 15 µl of each reaction mixtures was loaded onto a
QuantStudio 3D Digital PCR 20K Chip (Thermo Fisher Scientific,
Inc.) using the automatic chip loader, and the signal was amplified
using the following thermocycling profile: 95°C for 8 min, 45
cycles at 95°C for 15 sec and 60°C for 1 min, with a final
extension step at 60°C for 2 min. Amplification was followed by
chip imaging, and secondary analysis was performed using the
QuantStudio 3D Analysis Suite Cloud Software (version 3.1.5). All
samples were independently analyzed by two different operators, and
quantification was performed in a blinded manner; in the case of a
discrepancy, a third dPCR analysis was performed. The final results
were expressed as the means of the number of BCR-ABL1 and
Y4 copies/µl of reaction, and the emission threshold
was fixed at 4,000 relative fluorescence units.
The final results for the exosomes samples were
reported as ratio of BCR-ABL1/Y4 transcript, and normalized
to the total amount of plasma used for tumor exosome isolation and
enrichment. The quantification results are presented in Table IV. Y4 was also quantified
using dPCR on the total exosome fraction of the healthy controls
(case nos. 6, 7, 8, 9 and 10) and in CML samples (nos. 5, 6, 7, 8,
9 and 1a-e), isolated using the Total Exosome Isolation kit and
Total Exosome RNA and Protein Isolation kit (Thermo Fisher
Scientific, Inc.).
 | Table IVResults of the quantification of
BCR-ABL1/ABL1 by RT-qPCR aligned with IS and of
BCR-ABL1 transcript by dPCR in CML patients. |
Table IV
Results of the quantification of
BCR-ABL1/ABL1 by RT-qPCR aligned with IS and of
BCR-ABL1 transcript by dPCR in CML patients.
| RT-qPCR
BCR-ABL1/ ABL1
| dPCR PB cells
BCR-ABL1
| dPCR EXO
BCR-ABL1
| dPCR EXO Y4
|
|---|
| Case no. | MR | IS | DOTS | Copies/µl of
reaction | DOTS | Copies/µl of
reaction | Copies/ml C
plasma | DOTS | Copies/µl of
reaction | Copies/ml
plasma |
|---|
| 1 | 4.5 | 0.0013 | 2 | 0.161 | 3 | 0.256 | 6.67 | 294 | 28.239 | 1,058.9 |
| 2 | 4.5 | 0 | 1 | 0.0857 | 1 | 0.091 | 4.74 | 100 | 10.579 | 661.18 |
| 3 | 4.5 | 0.0031 | 3 | 0.226 | 2 | 0.158 | 8.23 | 251 | 22.711 | 1,419.44 |
| 4 | 5.0 | 0 | 5 | 0.439 | 4 | 0.324 | 20.25 | 2 | 0.299 | 37.37 |
| 5 | 5.0 | 0 | 0 | 0 | 2 | 0.226 | 14.12 | 11 | 0.848 | 35.3 |
| 6 | 5.0 | 0 | 2 | 0.188 | 3 | 0.219 | 13.69 | 6 | 0.432 | 54.0 |
| 7 | 5.0 | 0 | 1 | 0.0773 | 1 | 0.072 | 4.5 | 30 | 2.39 | 298.75 |
| 8 | 5.0 | 0 | 1 | 0.0781 | 5 | 0.407 | 12.72 | 11 | 0.915 | 57.19 |
| 9 | 5.0 | 0.0008 | 1 | 0.0853 | 4 | 0.329 | 10.28 | 8 | 0.613 | 38.31 |
| 10 | 4.0 | 0.0053 | 5 | 0.441 | 5 | 0.378 | 14.76 | 76 | 5.945 | 928.9 |
| 1a | 0.0 | 19.528 | 49 | 3.014 | 8 | 0.645 | 80.625 | 97 | 9.146 | 1,143.25 |
| 1b | 2.0 | 0.1021 | 11 | 0.905 | 5 | 0.381 | 47.625 | 58 | 4.973 | 621.625 |
| 1c | 3.0 | 0.0112 | 3 | 0.250 | 5 | 0.377 | 47.125 | 18 | 1.288 | 161 |
| 1d | 3.0 | 0.0707 | 3 | 0.234 | 3 | 0.241 | 15.0625 | 24 | 1.734 | 108.375 |
| 1e | 4.0 | 0.0054 | 1 | 0.0725 | 3 | 0.228 | 14.25 | 11 | 0.909 | 56.813 |
Results
The present study aimed to evaluate the feasibility
of enriching the tumorexosome fraction in patients with
Ph+ CML in the CP and undergoing treatment with TKIs by
detecting the BCR-ABL1 transcript.
The SoRTEV™ EV-RNA Low Volume Enrichment kit
(EXOSOMICS Spa, Siena, Italy) is an innovative tool to selectively
isolate tumor-enriched exosomes from biological fluids. Exosomal
purification is achieved using immunoaffinity beads coated with
proprietary antibodies against exosome pan-tumor surface antigens,
and allows the selective isolation of tumor-derived nucleic acids
from tumor-enriched exosomes.
Following exosome enrichment, BCR-ABL1 was
quantified in PB cells by RT-qPCR, following the MRD monitoring
gold standards. The BCR-ABL1 transcript was also quantified
by dPCR on PB cell extracts and cDNA was extracted from
tumor-derived exosomes, since it has been described as a more
accurate and sensitive approach for the detection of deeper and
more stable molecular responses (45). dPCR has been considered the most
effective tool for the quantification of the target transcript in
exosome cDNA, due to the low amount of nucleic acid in these
vesicles. The results obtained from tumor-derived exosomes were
subsequently compared with those obtained by dPCR from PB
cells.
RT-qPCR BCR-ABL1 quantification
RT-qPCR was performed on the control samples
following the international ELN Guidelines; the BCR-ABL1
transcript was not detected; thus, these subjects were confirmed as
controls.
The quantification of the BCR-ABL1 transcript
in the CML patient samples was also performed following the
international ELN Guidelines and the MR classes were assigned
according to the IS (37). The
replicates were 3 for BCR-ABL1 quantification and 2 for ABL1
quantification, for all samples. At enrollment, 1/10 (10%) CML
patients resulted in MR4.0, 3/10 (20%) resulted in
MR4.5 and 6/10 (70%) resulted in MR5.0; 1/3
(33%) of patients with MR4.5 and 5/6 (83%) of patients
in MR5.0 presented with undetectable levels of
BCR-ABL1 transcript, respectively (Tables I and III).
dPCR BCR-ABL1 quantification in PB
cells
The cDNA obtained from PB cells of both the patients
with CML and the controls was deemed suitable for the
quantification of BCR-ABL1 by dPCR. No BCR-ABL1
transcript was detected in the healthy controls or in the
Ph− pathological controls, while the fusion gene
transcript was quantifiable in all but one of the CML patient
samples; this sample (case no. 5) also possessed an
MR5.0 (undetectable by RT-qPCR) following the IS. The
median quantification value of the fusion-gene transcript in the
CML patient samples was 0.123 BCR-ABL1 copies/µl
(range 0-0.441), and the raw dPCR data are presented in Table IV.
dPCR BCR-ABL1/Y4 quantification in the
tumor-exosome enriched fraction
The BCR-ABL1 expression levels were
quantified by dPCR from cDNA isolated from the exosomes of the
patients with CML and the control subjects. In order to estimate
the total amount of the exosomal RNA, Y4 was quantified. The
raw quantification data, expressed as both copies/ml of reaction
and copies/ml of plasma, are presented in Table IV.
In the healthy control subjects, dPCR analysis
revealed an auto-fluorescence smeared signal at the
6-carboxy-fluorescein (FAM) wavelength, for both in Y4
(Fig. 1A) and BCR-ABL1
(Fig. 1B). The signal was not
superimposable with FAM-labeled probe signal, and there was no
separation in terms of fluorescence intensity between the negative
and auto-fluorescent microparticles. Nevertheless, the identified
signal was under the emission threshold of all samples, and was
therefore assumed to be background noise.
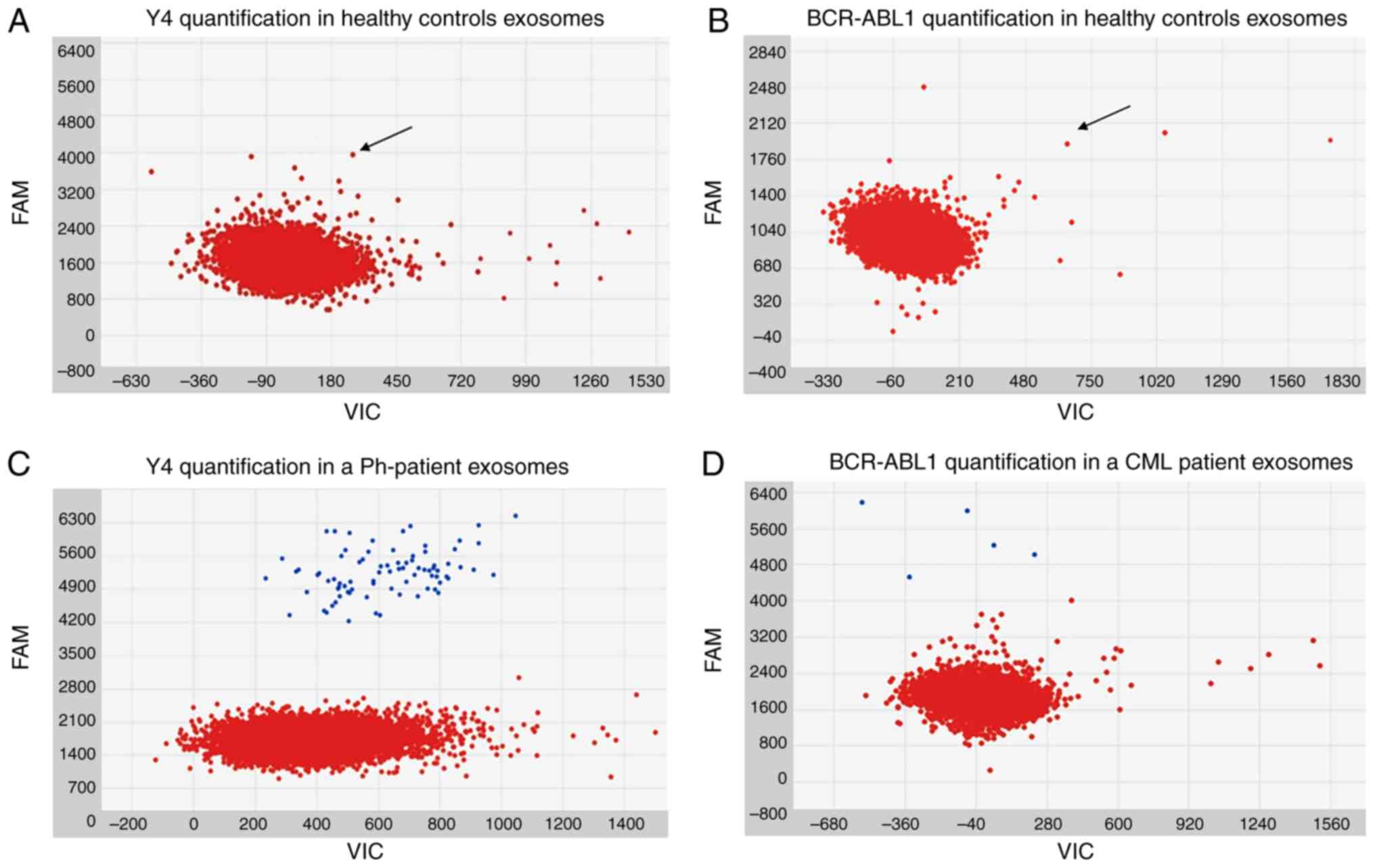 | Figure 1Images obtained during the analysis
of dPCR chips by Analysis Suite software. In all the figures, the
x-axis corresponds to the VIC wavelength channel. Although no
VIC-labeled probe was present in the preparation and no signal
resulted emitted in VIC wavelength channel, a baseline signal is
naturally present in all the samples and correspond to the
background of emission. VIC and FAM are the two wavelengths
automatically analyzed by Analysis Suite software and are reported
in all the graphs generated by this software. (A) The loaded sample
was tumor-enriched exosome cDNA from healthy control number 2. Red
dots represent the negative micro-reactions. The y-axis corresponds
to FAM (BCR-ABL1 probe label) wavelength emission intensity. In the
image, a smear of negative micro-reactions in FAM channel is
appreciable (arrow). (B) The loaded sample was tumor-enriched
exosome cDNA from healthy control number 2. Red dots represent the
negative micro-reactions. Y-axis corresponds to FAM (Y4 probe
label) wavelength emission intensity. In the image, a smear of
negative micro-reactions in FAM channel is appreciable (arrow). (C)
The loaded sample was tumor-enriched exosome cDNA from
Ph− control number 4. Red and blue dots represent the
negative and the Y4 positive micro-reactions, respectively. Y-axis
corresponds to FAM (Y4 probe label) wavelength emission intensity.
(D) The loaded sample was tumor-enriched exosome cDNA from CML
patient number 10. Red and blue dots represent the negative and the
BCR-ABL1 positive micro-reactions, respectively. The y-axis
corresponds to FAM (BCR-ABL1 probe label) wavelength emission
intensity. dPCR, digital PCR; BCR-ABL1, breakpoint cluster
region-proto-oncogene 1 tyrosine-protein kinase; CML, chronic
myeloid leukemia; Ph−, Philadelphia
chromosome-negative. |
BCR-ABL1 was not amplified by dPCR in the
Ph− controls, with the exception of the multiple myeloma
patient sample, where 2 copies were identified. On the contrary, a
positive FAM signal was detectable in all the samples tested for
the internal reference gene. The median quantification for the
Y4 transcript was 58.93 (range, 14.31-365.31), which was
normal-ized for plasma volume. The Ph− 3 sample
possessed the lowest quantity of Y4, while Ph− 4
presented the highest internal reference gene transcript. A
representative example of Ph− control quantification of
Y4 is displayed in Fig.
1C. The results of BCR-ABL1 and Y4 quantification in the
control samples are presented in Table V.
 | Table VResults of the quantification of
BCR-ABL1 by dPCR in Ph− controls and heathy
controls. |
Table V
Results of the quantification of
BCR-ABL1 by dPCR in Ph− controls and heathy
controls.
| dPCR PB cells
BCR-ABL1
| dPCR EXO
BCR-ABL1
| dPCR EXO Y4
|
|---|
| Case no. | DOTS | Copies/µl of
reaction | DOTS | Copies/µl of
reaction | Copies/µl
plasma | DOTS | Copies/µl of
reaction | Copies/ml
plasma |
|---|
| Ph−
1 | 0 | 0 | 0 | 0 | 0 | 5 | 0.398 | 34.11 |
| Ph−
2 | 0 | 0 | 0 | 0 | 0 | 17 | 1.34 | 83.75 |
| Ph−
3 | 0 | 0 | 0 | 0 | 0 | 3 | 0.229 | 14.31 |
| Ph−
4 | 0 | 0 | 2 | 0.164 | 6.58 | 79 | 5.845 | 365.31 |
| Healthy 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Healthy 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
For the patients with CML, dPCR amplified the
BCR-ABL1 transcript in all exosome samples (median
BCR-ABL1 copies/ml of plasma, 11.50; range, 4.5-20.25;
normalized for plasma volume). The resulting signals were
superimposable with those detected in the PB cells of patients with
CML, considering both the fluorescence intensity and the absence of
background noise (Fig. 1D). The
median quantification value of the Y4 transcript was 177.97
copies/ml of plasma (range 35.3-1,419.44; normal-ized for plasma
volume), and the results of BCR-ABL1 and Y4
transcripts quantification in CML patient samples are presented in
Table IV.
When comparing the BCR-ABL1 transcript levels
detected in the PB cells and tumor-derived exosomes of the patients
with CML, no linear association was identified, considering the
absolute quantification and the BCR-ABL1 transcript levels
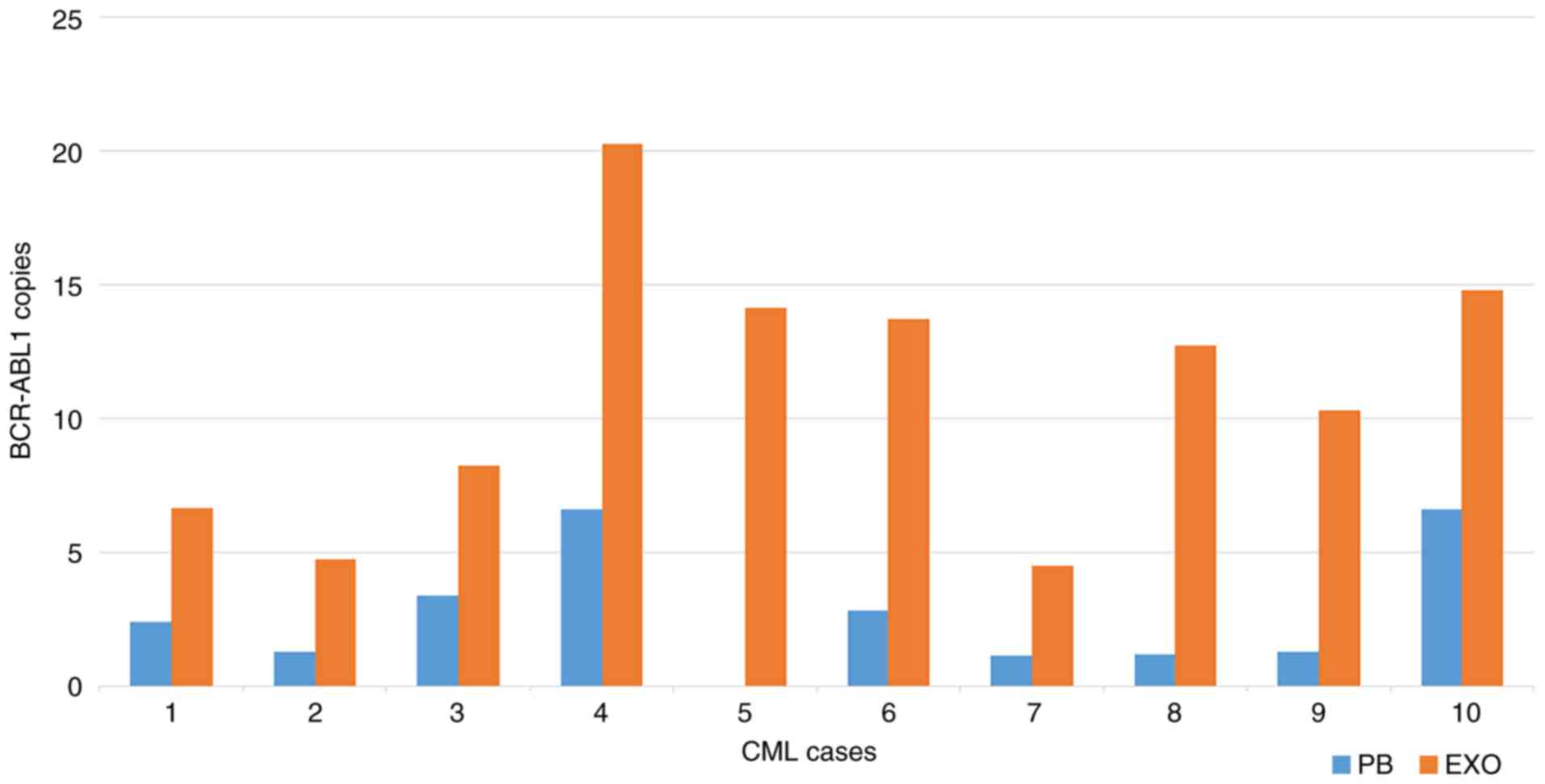
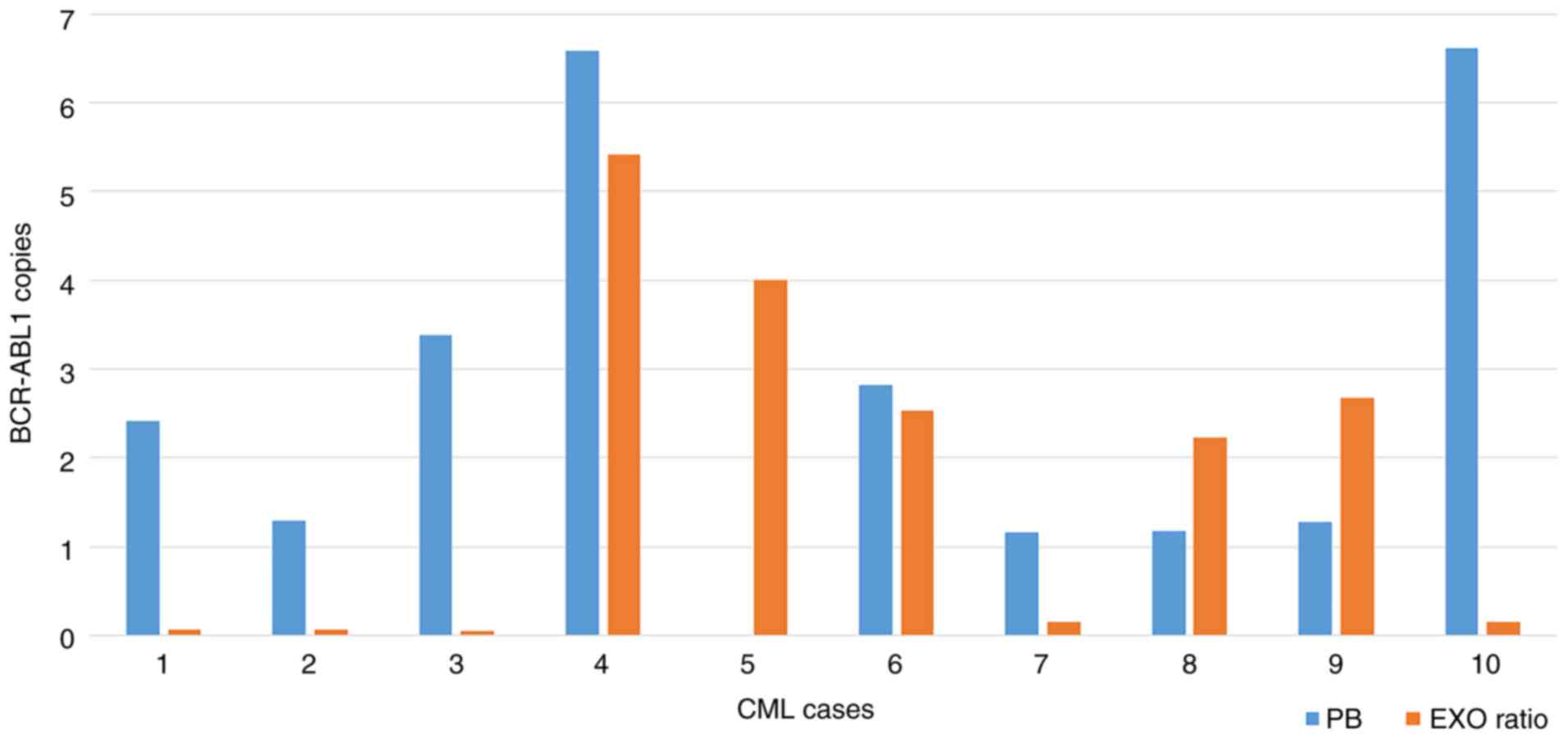
normalized for the plasma volume (Figs. 2 and 3, respectively). Nevertheless, excluding
case no. 5 (where no BCR-ABL1 transcript copies were
identified in PB cells), a trend was identified in the absolute
quantification, since patient nos. 4 and 10 exhibited the highest
amount of transcript in both PB cells and tumor-derived exosomes,
while patient no. 7 possessed the lowest BCR-ABL1 transcript
copy number in both PB cells and tumor-derived exosomes (Fig. 2). This trend was not present when
considering the absolute BCR-ABL1 transcript quantification
value of PB cells (as determined by dPCR) or the ratio
BCR-ABL1/Y4 transcript on exosomal samples (Fig. 3). Notably, patient no. 4 was the
case with the highest level of BCR-ABL1, considering both
the absolute and the relative quantification (Figs. 2 and 3).
dPCR Y4 quantification in the
total-exosome fraction
In order to estimate the total amount of the
exosomal RNA, cDNA obtained from exosomes isolated from the plasma
of CML and healthy controls, using the Total Exosome Isolation kit
and Total Exosome RNA and Protein Isolation kit, were used for
Y4 quantification by dPCR. The raw quantification data,
expressed as both copies/ml of reaction and copies/ml of plasma,
are reported in Table IV. In the
CML samples, the median Y4 transcript quantification value
in the total-exosome fraction was 286.695 Y4 copies/ml of plasma
(range 110.08-2,685.54); while in healthy controls, this was 172.44
Y4 copies/ml of plasma (range 98.33-211) (Table VI).
 | Table VIResults of the dPCR quantification of
Y4 performed on tumor exosomes fraction, obtained by SoRTEV™
EV-RNA Low Volume Enrichment kit, and on total-exosomes fraction
obtained by the Total Exosome Isolation kit and Total Exosome RNA
and Protein Isolation kit. |
Table VI
Results of the dPCR quantification of
Y4 performed on tumor exosomes fraction, obtained by SoRTEV™
EV-RNA Low Volume Enrichment kit, and on total-exosomes fraction
obtained by the Total Exosome Isolation kit and Total Exosome RNA
and Protein Isolation kit.
| dPCR EXO Y4
in tumor-exosomes
| dPCR EXO Y4
in total exosome fraction
|
|---|
| Case no. | DOTS | Copies/µl of
reaction | Copies/ml
plasma | DOTS | Copies/µl of
reaction | Copies/ml
plasma |
|---|
| Healthy 6 | 0 | 0 | 0 | 22 | 1.628 | 203.5 |
| Healthy 7 | 0 | 0 | 0 | 18 | 1.258 | 157.25 |
| Healthy 8 | 0 | 0 | 0 | 23 | 1.688 | 211 |
| Healthy 9 | 0 | 0 | 0 | 19 | 1.379 | 172.44 |
| Healthy 10 | 0 | 0 | 0 | 10 | 0.787 | 98.33 |
| CML 1a | 97 | 9.146 | 1,143.25 | 298 | 21.484 | 2,685.54 |
| CML 1b | 58 | 4.973 | 621.625 | 164 | 15.757 | 1,969.605 |
| CML 1c | 18 | 1.288 | 161 | 76 | 7.639 | 954.855 |
| CML 1d | 24 | 1.734 | 108.375 | 54 | 4.35 | 543.85 |
| CML 1e | 11 | 0.909 | 56.813 | 39 | 2.979 | 372.33 |
| CML 5 | 11 | 0.848 | 35.3 | 11 | 0.881 | 110.08 |
| CML 6 | 6 | 0.432 | 54.0 | 14 | 1.107 | 138.42 |
| CML 7 | 30 | 2.39 | 298.75 | 22 | 1.608 | 201.06 |
| CML 8 | 11 | 0.915 | 57.19 | 21 | 1.546 | 193.29 |
| CML 9 | 8 | 0.613 | 38.31 | 20 | 1.432 | 179.01 |
Discussion
The present explorative study, which was based on a
mono-centric cohort of 10 patients in the CP of CML, with DMR under
TKI treatment, indicated the potential feasibility of tumor-derived
exosome enrichment, in patients with onco-hematological
diseases.
The current interest in exosomes is due to their
ability to robustly protect and carry information to activate
target cells, promoting their epigenetic alteration, transporting
membrane receptors, and proteins of interest. In addition to
cellular communication, exosomes are known to be associated with
immune reactions, cancer viability and invasion, antigen
presentation, and cell migration and differentiation (53). Recently, a number of studies have
demonstrated the role of these EVs in hematological malignancies
(21-23), as well as their involvement at
different diseases phases (33,53).
In recent years, various exosome-isolation
strategies have been developed. The most promising of these methods
for a deep molecular characterization of intra-tumor cell
communication encompass the immune-selective enrichments of tumor
antigens (15,18). These tools, which act
synergistically with innovative molecular biology techniques such
as next generation sequencing and dPCR, may improve the accuracy
and sensitivity of exosomes analysis (45,46). In this context, the CML model is
considered to be particularly suitable, given the presence of
BCR-ABL1 as a hallmark indicator of leukemic cells, and the
established use of RT-qPCR to monitor MRD. The BCR-ABL1
transcript has previously been detected in the exosomes of patients
with CML, but not in those in the CP. Specifically, a previous
study demonstrated the possibility of quantifying the
BCR-ABL1 transcript in exosomes isolated from the PB of
patients with CML in advanced disease phases, such as accelerating
or blast phases. However, they were not be able to detect
BCR-ABL1 by nested PCR in CML patients in the CP (44).
To the best of our knowledge, the present study is
the first to investigate the possibility of isolating
leukemia-derived exosomes from patients with CML, using a
commercial kit developed for solid tumors, and based on a
proprietary antibody affinity method that selectively enriches
tumor-derived exosomes (8). The
evaluation of enrichment specificity was determined by
BCR-ABL1 transcript detection using dPCR, since it has been
demonstrated to be more sensitive and robust than conventional
approaches for BCR-ABL1 quantification in CML cases
(45,46).
In the healthy controls of the present study, the
absence of the BCR-ABL1 and Y4 transcripts confirmed
the specificity of the enrichment, although the absence of the
fusion gene transcript cannot be considered as absence of exosome
isolation. The BCR-ABL1 transcript may not be incorporated
in all tumor-derived exosomes, and some EVs may be isolated by
non-specific immune-recognition. On the other hand, for a lack of
Y4 transcript detection confirmed the power of tumor-derived
exosomes enrichment, as Y4 is considered to be a good
internal reference gene for exosomes evaluation (48,54), and its absence would indicate a
lack of tumor-derived exosomes (as expected in healthy controls).
The smear of a FAM signal under the emission threshold was
considered as background noise, for both low fluorescence intensity
and an abnormal emission pattern (Fig. 1A and 1B). This effect may be due to an
auto-fluorescence property of chemical contaminants present in the
eluate (such as the resin of the SoRTEV™ EV-RNA Low Volume
Enrichment kit); in the absence of target detection, this may in
non-specific reactivity and the production of background noise.
By contrast, in the Ph− control cohort,
Y4 was detectable by dPCR in all samples. This may reflect
the presence of active tumor cells in enrolled patients, and their
subsequent release of exosomes. Considering the quantity of
Y4 in the Ph− patients, a wide range was
appreciated. Even if no correlation analysis between the quantity
of Y4 transcript and disease status was attempted, a trend
may be observed. Notably, the lowest levels of the internal
reference gene (which may reflect the lowest levels of
tumor-derived exosomes) were detected in a Ph− patient
who had undergone allogenic stem cell transplantation (14.31
Y4 copies/ml plasma), and in a Ph− patient in
complete morphological remission (34.11 Y4 copies/ml
plasma). Furthermore, the highest detected levels of Y4
transcript (365.21 Y4 copies/ml plasma) were observed in a
patient with multiple myeloma (Ph− case no. 4) at the
point of relapse, when the activity of the leukemia cells was
potentially increasing or at its most prominent (Table V). In this case, 2 BCR-ABL1
transcript copies were also detected in the cDNA of the
tumor-derived exosome. This result may suggest a lack of
BCR-ABL1 dPCR quantification specificity, but may also be
due to the development of a new Ph+ subclones in a
conventionally Ph− disease. This mechanism of subclone
(or new leukemia clones) development and selection is possible and
has been previously described (55,56). Collectively, these findings
highlight the possibility of improving our understanding of tumor
heterogeneity by selectively isolating the neoplastic exosomes.
Moreover, it is known that the BCR-ABL1 transcript may also
be detected in elderly healthy subjects, which may reflect the
presence of a group of Ph+ cells controlled by the
immune system (57).
Finally, in 10 patients in the CP of CML with a DMR
under TKI treatment (and in 5 samples of CML patient no. 1
monitored from diagnosis to the achievement of the DMR) the exosome
enrichment performed using the SoRTEV™ EV-RNA Low Volume Enrichment
kit highlighted the presence of the BCR-ABL1 transcript in
leukemia-derived EVs. A previous study demonstrated the
detectability of this fusion gene transcript in the total exosomes
pool of patients affected by CML, not enriched for specific
sub-populations; BCR-ABL1 was only detected by conventional
nested PCR in patients in the accelerated phase or in blast crisis
(44). These results contradict
those of the present study, which may be due to the improvements in
the specificity of leukemia-derived exosome enrichment methods
combined with highly sensitive dPCR quantification.
In order to exclude the possibility that the
absence of Y4 RNA reflects an overall lower exosome density in
healthy subject plasma, or in patients with prominent DMRs, the
total-exosome fraction was isolated from 5 healthy controls and
from 10 CML patient samples at different disease phases. A
commercial kit was used considering its performance rating in
published literature (58).
Y4 was quantified to evaluate the expression of the
reference gene, and to indirectly estimate the exosome
concentration in the plasma samples. Samples collected at diagnosis
or during the early phase of the therapy response (samples 1a and
1b) possessed a higher amount of Y4 in the total-exosome
fraction (2,685.54 and 1,969.605 Y4 copies/ml plasma) compared with
those of healthy control samples (median, 172.44 Y4 copies/ml
plasma; range 98.33-211), or the CML samples with MMR (samples 1c
and 1d) or DMR (samples 1e and 5-9; median, 286.695 Y4 copies/ml
plasma; range 110.08-954.855). This result confirms that patients
with clinical evidence of disease exhibit an increased number of
exosomes in the plasma, reflecting the high exosomes-release
activity of tumor and leukemic cells (59). Nevertheless, the application of
tumor-exosome enrichment in healthy subjects, and in those with CML
presenting with MMR or DMR, indicated a notable difference between
these two categories. In fact, as a result of tumor-exosomes
enrichment, no Y4 signal was detectable in the healthy
controls, whilst it was consistent in CML samples derived from
patients with both high and low disease levels.
However, the limited residual quantities of some
samples do not allow additional experiments, such as the
conventional ultracentrifugation. In order to compare the result
obtainable using different isolation technologies, included the
enrichment approach described in this study, further studies are
warranted with this particular aim. Future larger-scale studies
will also allow additional analysis, for example the quantification
of total exosomes isolated by conventional techniques, which were
not possible in this pilot feasibility study. In conclusion, the
present study indicated the feasibility of enriching
leukemia-derived exosomes by immune-affinity recognition, as
reported in solid tumors. Moreover, the detectability of the
BCR-ABL1 transcript in the enriched EVs was demonstrated;
this indicates novel insight into the detectability of molecular
communication between residual leukemic cells resident in the BM,
which are considered to be one of the main contributors to the
relapse of patients undergoing TKI discontinuation. Considering the
biological roles of the exosomes, the presence of the
BCR-ABL1 transcript in leukemia-derived exosomes cannot
currently be considered to have the same significance as the
presence of the fusion gene transcript in PB cells. The
implications of BCR-ABL1-positive exosomes on the disease
progression status of patients with CP-CML, and their subsequent
clinical impact, remain to be fully understood. The present study
may be considered the bases for further larger-scale studies, with
the aim of investigating these challenges, and broadening the
application of tumor-derived exosomes enrichment in the
onco-hematology field, where it currently remains to be
explored.
Acknowledgements
Not applicable.
Funding
This study was supported in part by the European
Leukemia Net (ELN)-European Treatment and Outcome Study (EUTOS) and
Gimema CML-Working Party, Department of Clinical and Experimental
Sciences of University of Brescia funds, and by Cofin 2009.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
DR, SB and CF designed the study; CF, CZ and SB
performed the dPCR analysis; CF and CZ performed the exosome
enrichment and isolation; MM, MF, NP, FR, FC, AT, EM, LG, TZ and
EAB enrolled the patients and collected the clinical data; SB, CF
and DR wrote the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
The present explorative study was approved by the
Ethics Committee of Brescia (local study no. NP2370 approved in
May, 2016) and performed according to good clinical practice and
Declaration of Helsinki. All patients and subjects gave their
written informed consent for the enrollment in the study, the use
of their samples for research purposes and for the publication of
the encompassed data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rahbari M, Pecqueux M, Aust D, Stephan H,
Tiebel O, Chatzigeorgiou A, Tonn T, Baenke F, Rao V, Ziegler N, et
al: Expression of glypican 3 is an independent prognostic biomarker
in primary gastro-esophageal adenocarcinoma and corresponding serum
exosomes. J Clin Med. 8:E6962019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang YL, Liu LC, Hung Y, Chen CJ, Lin YZ,
Wu WR and Wang SC: Long non-coding RNA HOTAIR in circulatory
exosomes is correlated with ErbB2/HER2 positivity in breast cancer.
Breast. 46:64–69. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao A, Guo L, Xu J, Zheng L, Guo Z, Ling
Z, Wang L and Mao W: Identification and validation of circulating
exosomes-based liquid biopsy for esophageal cancer. Cancer Med.
8:3566–3574. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han S, Huo Z, Nguyen K, Zhu F, Underwood
PW, Basso KBG, George TJ and Hughes SJ: The proteome of pancreatic
cancer-derived exosomes reveals signatures rich in key signaling
pathways. Proteomics. 19:e18003942019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grange C, Brossa A and Bussolati B, Grange
C, Brossa A and Bussolati B: Extracellular vesicles and carried
miRNAs in the progression of renal cell carcinoma. Int J Mol Sci.
20:E18322019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wong CH and Chen YC: Clinical significance
of exosomes as potential biomarkers in cancer. World J Clin Cases.
7:171–190. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kinjyo I, Bragin D, Grattan R, Winter SS
and Wilson BS: Leukemia-derived exosomes and cytokines pave the way
for entry into the brain. J Leukoc Biol. 105:741–753. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zarovni N, Corrado A, Guazzi P, Zocco D,
Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J and Chiesi A:
Integrated isolation and quantitative analysis of exosome shuttled
proteins and nucleic acids using immunocapture approaches. Methods.
87:46–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lakkaraju A and Rodriguez-Boulan E:
Itinerant exosomes: Emerging roles in cell and tissue polarity.
Trends Cell Biol. 18:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: A common pathway for a specialized function. J
Biochem. 140:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Février B and Raposo G: Exosomes:
Endosomal-derived vesicles shipping extracellular messages. Curr
Opin Cell Biol. 16:415–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miller IV and Grunewald TG: Tumour-derived
exosomes: Tiny envelopes for big stories. Biol Cell. 107:287–305.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Simpson RJ, Jensen SS and Lim JW:
Proteomic profiling of exosomes: Current perspectives. Proteomics.
8:4083–4099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zocco D, Ferruzzi P, Cappello F, Kuo WP
and Fais S: Extracellular vesicles as shuttles of tumor biomarkers
and anti-tumor drugs. Front Oncol. 4:2672014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
The fifth international meeting of ISEV,
ISEV2016, Rotterdam, the Netherlands, 4-7 May, 2016, OPW 3.8. J
Extracell Vesicles. 5:315522016. View Article : Google Scholar
|
|
17
|
Jiang S, Hu C, Liu P and Lu M:
Tumor-derived exosomes in cancer metastasis risk diagnosis and
metastasis therapy. Clin Transl Oncol. 21:152–159. 2019. View Article : Google Scholar
|
|
18
|
Wen SW, Lima LG, Lobb RJ, Norris EL,
Hastie ML, Krumeich S and Möller A: Breast cancer-derived exosomes
reflect the cell-of-origin phenotype. Proteomics. 19:e18001802019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tatischeff I: Prostate cancer under the
light of tumor cells-derived extracellular vesicles. Cancer Res
Front. 3:83–111. 2017. View Article : Google Scholar
|
|
20
|
Théry C and Witwer K: ISEV2018 abstract
book. J Extracell Vesicles. 7(Suppl 1): 14614502018. View Article : Google Scholar
|
|
21
|
Jafarzadeh N, Safari Z, Pornour M,
Amirizadeh N, Forouzandeh Moghadam M and Sadeghizadeh M: Alteration
of cellular and immune-related properties of bone marrow
mesenchymal stem cells and macrophages by K562 chronic myeloid
leukemia cell derived exosomes. J Cell Physiol. 234:3697–3710.
2019. View Article : Google Scholar
|
|
22
|
Wierz M, Pierson S, Gargiulo E, Guerin C,
Moussay E and Paggetti J: Purification of leukemia-derived exosomes
to study microenvironment modulation. Methods Mol Biol.
1884:231–245. 2019. View Article : Google Scholar
|
|
23
|
Cheng H, Sun G and Cheng T: Hematopoiesis
and microenvironment in hematological malignancies. Cell Regen
(Lond). 7:22–26. 2018. View Article : Google Scholar
|
|
24
|
Bouyssou JM, Liu CJ, Bustoros M,
Sklavenitis-Pistofidis R, Aljawai Y, Manier S, Yosef A, Sacco A,
Kokubun K, Tsukamoto S, et al: Profiling of circulating exosomal
miRNAs in patients with waldenström macroglobulinemia. PLoS One.
13:e02045892018. View Article : Google Scholar
|
|
25
|
Tadokoro H, Umezu T, Ohyashiki K, Hirano T
and Ohyashiki JH: Exosomes derived from hypoxic leukemia cells
enhance tube formation in endothelial cells. J Biol Chem.
288:34343–34351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharifi H, Shafiee A, Molavi G, Razi E,
Mousavi N, Sarvizadeh M and Taghizadeh M: Leukemia-derived
exosomes: Bringing onco-genic signals to blood cells. J Cell
Biochem. 120:16307–16315. 2019.PubMed/NCBI
|
|
27
|
Yao Y, Wang C, Wei W, Shen C, Deng X, Chen
L, Ma L and Hao S: Dendritic cells pulsed with leukemia
cell-derived exosomes more efficiently induce antileukemic
immunities. PLoS One. 9:e914632014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szczepanski MJ, Szajnik M, Welsh A,
Whiteside TL and Boyiadzis M: Blast-derived microvesicles in sera
from patients with acute myeloid leukemia suppress natural killer
cell function via membrane-associated transforming growth
factor-beta1. Haematologica. 96:1302–1309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wojtuszkiewicz A, Schuurhuis GJ, Kessler
FL, Piersma SR, Knol JC, Pham TV, Jansen G, Musters RJ, van Meerloo
J, Assaraf YG, et al: Exosomes secreted by apoptosis-resistant
acute myeloid leukemia (AML) blasts harbor regulatory network
proteins potentially involved in antagonism of apoptosis. Mol Cell
Proteomics. 15:1281–1298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huan J, Hornick NI, Goloviznina NA,
Kamimae-Lanning AN, David LL, Wilmarth PA, Mori T, Chevillet JR,
Narla A, Roberts CT Jr, et al: Coordinate regulation of residual
bone marrow function by paracrine trafficking of AML exosomes.
Leukemia. 29:2285–2295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, De Veirman K, Faict S, Frassanito
MA, Ribatti D, Vacca A and Menu E: Multiple myeloma exosomes
establish a favourable bone marrow microenvironment with enhanced
angiogenesis and immunosuppression. J Pathol. 239:162–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yeh YY, Ozer HG, Lehman AM, Maddocks K, Yu
L, Johnson AJ and Byrd JC: Characterization of CLL exosomes reveals
a distinct microRNA signature and enhanced secretion by activation
of BCR signaling. Blood. 125:3297–3305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Corrado C, Saieva L, Raimondo S, Santoro
A, De Leo G and Alessandro R: Chronic myelogenous leukaemia
exosomes modulate bone marrow microenvironment through activation
of epidermal growth factor receptor. J Cell Mol Med. 20:1829–1839.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rowley JD: Letter: A new consistent
chromosomal abnormality in chronic myelogenous leukaemia identified
by quinacrine fluorescence and Giemsa staining. Nature.
243:290–293. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Druker BJ, Sawyers CL, Kantarjian H, Resta
DJ, Reese SF, Ford JM, Capdeville R and Talpaz M: Activity of a
specific inhibitor of the BCR-ABL tyrosine kinase in the blast
crisis of chronic myeloid leukemia and acute lymphoblastic leukemia
with the philadelphia chromosome. N Engl J Med. 344:1038–1042.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sonoyama J, Matsumura I, Ezoe S, Satoh Y,
Zhang X, Kataoka Y, Takai E, Mizuki M, Machii T, Wakao H and
Kanakura Y: Functional cooperation among Ras, STAT5, and
phosphati-dylinositol 3-kinase is required for full oncogenic
activities of BCR/ABL in K562 cells. J Biol Chem. 277:8076–8082.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cross NC, White HE, Colomer D, Ehrencrona
H, Foroni L, Gottardi E, Lange T, Lion T, Machova Polakova K,
Dulucq S, et al: Laboratory recommendations for scoring deep
molecular responses following treatment for chronic myeloid
leukemia. Leukemia. 29:999–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Egan D and Radich J: Monitoring disease
burden in chronic myeloid leukemia: Past, present, and future. Am J
Hematol. 91:742–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mahon FX, Réa D, Guilhot J, Guilhot F,
Huguet F, Nicolini F, Legros L, Charbonnier A, Guerci A, Varet B,
et al: Discontinuation of imatinib in patients with chronic myeloid
leukaemia who have maintained complete molecular remission for at
least 2 years: The prospective, multicentre stop imatinib (STIM)
trial. Lancet Oncol. 11:1029–1035. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Russo D, Malagola M, Skert C, Cancelli V,
Turri D, Pregno P, Bergamaschi M, Fogli M, Testoni N, De Vivo A, et
al: Managing chronic myeloid leukaemia in the elderly with
intermittent imatinib treatment. Blood Cancer J. 5:e3472015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ross DM, Masszi T, Gómez Casares MT,
Hellmann A, Stentoft J, Conneally E, Garcia-Gutierrez V, Gattermann
N, le Coutre PD, Martino B, et al: Durable treatment-free remission
in patients with chronic myeloid leukemia in chronic phase
following frontline nilotinib: 96-week update of the ENESTfreedom
study. J Cancer Res Clin Oncol. 144:945–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cortes J, Rea D and Lipton JH:
Treatment-free remission with first- and second-generation tyrosine
kinase inhibitors. Am J Hematol. 94:346–357. 2019.
|
|
43
|
Bocchia M, Sicuranza A, Abruzzese E, Iurlo
A, Sirianni S, Gozzini A, Galimberti S, Aprile L, Martino B, Pregno
P, et al: Residual peripheral blood CD26+ leukemic stem
cells in chronic myeloid leukemia patients during TKI therapy and
during treatment-free remission. Front Oncol. 8:1942018. View Article : Google Scholar
|
|
44
|
Kang KW, Jung JH, Hur W, Park J, Shin H,
Choi B, Jeong H, Kim DS, Yu ES, Lee SR, et al: The potential of
exosomes derived from chronic myelogenous leukaemia cells as a
biomarker. Anticancer Res. 38:3935–3942. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bernardi S, Ruggieri G, Malagola M,
Cancelli V, Cattina F, Polverelli N, Zanaglio C, Perucca S, Re F,
Montanelli A and Russo D: Digital PCR (Dpcr) a step forward to
detection and quantification of minimal residual disease (MRD) in
Ph+/BCR-ABL1 chronic myeloid leukemia (CML). J Mol Biomark Diagn.
8:3302017. View Article : Google Scholar
|
|
46
|
Bernardi S, Malagola M, Zanaglio C,
Polverelli N, Dereli Eke E, D'Adda M, Farina M, Bucelli C, Scaffidi
L, Toffoletti E, et al: Digital PCR improves the quantitation of
DMR and the selection of CML candidates to TKIs discontinuation.
Cancer Med. 8:2041–2055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tosar JP, Gámbaro F, Sanguinetti J,
Bonilla B, Witwer KW and Cayota A: Assessment of small RNA sorting
into different extracellular fractions revealed by high-throughput
sequencing of breast cell lines. Nucleic Acids Res. 43:5601–5616.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yeri A, Courtright A, Reiman R, Carlson E,
Beecroft T, Janss A, Siniard A, Richholt R, Balak C, Rozowsky J, et
al: Total extracellular small RNA profiles from plasma, saliva, and
urine of healthy subjects. Sci Rep. 7:440612017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li C, Qin F, Hu F, Xu H, Sun G, Han G,
Wang T and Guo M: Characterization and selective incorporation of
small non-coding RNAs in non-small cell lung cancer extracellular
vesicles. Cell Biosci. 8:22018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Slonchak A, Clarke B, Mackenzie J,
Amarilla AA, Setoh YX and Khromykh AA: West Nile virus infection
and interferon alpha treatment alter the spectrum and the levels of
coding and noncoding host RNAs secreted in extracellular vesicles.
BMC Genomics. 20:4742019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bardi GT, Al-Rayan N, Richie JL,
Yaddanapudi K and Hood JL: Detection of inflammation-related
melanoma small extracellular vesicle (sEV) mRNA content using
primary melanocyte sEVs as a reference. Int J Mol Sci.
20:E12352019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen T, Zhang G, Kong L, Xu S, Wang Y and
Dong M: Leukemia-derived exosomes induced IL-8 production in bone
marrow stromal cells to protect the leukemia cells against
chemotherapy. Life Sci. 221:187–195. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Anfossi S, Babayan A, Pantel K and Calin
GA: Clinical utility of circulating non-coding RNAs-an update. Nat
Rev Clin Oncol. 15:541–563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Park SJ, Lee HW, Jeong SH, Park JS, Kim
HC, Seok JY, Kim HJ and Cho SR: Acquisition of a BCR-ABL1
transcript in a patient with disease progression from MDS with
fibrosis to AML with myelodysplasia-related changes. Ann Clin Lab
Sci. 41:379–384. 2011.PubMed/NCBI
|
|
56
|
Miki K, Obara N, Makishima K, Sakamoto T,
Kusakabe M, Kato T, Kurita N, Nishikii H, Yokoyama Y,
Sakata-Yanagimoto M, et al: An unprecedented case of p190 BCR-ABL
chronic myeloid leukemia diagnosed during treatment for multiple
myeloma: A case report and review of the literature. Case Rep
Hematol. 2018:78639432018.
|
|
57
|
Boquett JA, Alves JR and de Oliveira CE:
Analysis of BCR/ABL transcripts in healthy individuals. Genet Mol
Res. 12:4967–4971. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Serrano-Pertierra E, Oliveira-Rodríguez M,
Rivas M, Oliva P, Villafani J, Navarro A, Blanco-López MC and
Cernuda-Morollón E: Characterization of plasma-derived
extracellular vesicles isolated by different methods: A comparison
study. Bioengineering (Basel). 6. pp. E82019, View Article : Google Scholar
|
|
59
|
Navarro-Tableros V, Gomez Y, Camussi G and
Brizzi MF: Extracellular vesicles: New players in ymphomas. Int J
Mol Sci. 20:E412018. View Article : Google Scholar
|