Introduction
Cholangiocarcinoma (CCA) is known as one of the most
aggressive malignancies worldwide. CCA is responsible for 5-15% of
all primary liver cancer cases and is the second most frequent type
of liver cancer (1). CCA is a
highly invasive malignancy, exhibiting early distal organ and lymph
node metastasis and resistance to chemotherapy (2,3).
In recent years, the morbidity and mortality of CCA have increased
worldwide (4). Therefore, it is
crucial to elucidate the mechanism underlying the occurrence and
progression of CCA in order to uncover new cancer treatment
targets.
MicroRNAs (miRNAs/miRs), a class of non-coding RNA
molecules consisting of 18-25 nucleotides, have been verified to
regulate the translation of various target genes and are involved
in cancer occurrence, metastasis, progression and recurrence
(5,6). In CCA, certain miRNAs have been
previously demonstrated to exert stimulatory/suppressive effects on
cancer progression (7). For
example, miR-383 was found to act as a tumor suppressor in CCA and
inhibits tumor development through decreasing the expression of
interferon regulatory factor 1 (8). Through inhibiting aldolase A
translation, miR-122-5p decreased bile duct carcinoma cell
proliferation and promoted cell apoptosis (9). By regulating autophagy-related cell
death, miR-124 was shown to act as tumor suppressor in CCA
(10).
The function and underlying mechanism of action of
miR-137 have been investigated in various types of cancer (11-14). A study on non-small cell lung
cancer revealed that miR-137 may decrease tumor growth and
sensitize cancer cells to chemotherapy via regulating AKT2
(15). Similarly, in colon
cancer, patients with low expression levels of miR-137 have adverse
clinical prognosis (16,17). However, the biological function
and molecular mechanism of action of miR-137 in CCA remain
elusive.
The aim of the present study was to investigate the
expression of miR-137 in CCA tissues and cell lines, and elucidate
whether its effects on the proliferation, migration and invasion
ability of CCA cells were mediated via regulation of Wnt family
member 2B (WNT2B), in order to determine whether miR-137 and its
target gene, WNT2B, may be novel biomarkers for the diagnosis and
clinical treatment of CCA.
Materials and methods
Tissue samples
A total of 29 patients (mean age, 52.3±7.2 years;
range, 44-65 years; 13 men and 16 women) from the Affiliated
Hospital of Guizhou Medical University (Guizhou, China) were
enrolled in the present study between January 2018 and March 2019.
The inclusion criteria were as follows: i) The tissues were
obtained during surgery and diagnosed as CCA by two pathologists;
ii) patients diagnosed and treated for the first time; and iii)
patients consented to participate. The exclusion criteria were as
follows: i) Cases complicated with other malignancies; ii) presence
of other systemic diseases; iii) patients receiving treatment prior
to admission; and iv) patients and/or their families refusing to
participate. All 29 patients provided CCA tissue samples, while
adjacent normal tissues were also obtained from 20 of the patients.
The Human Trial Ethics Committee of Guizhou Medical University
approved the study. Written informed consent was obtained from the
patients who provided the specimens. The present study was
performed in accordance with the principles outlined in the
Declaration of Helsinki. All samples were kept in liquid nitrogen
until used.
Cell culture and lentivirus
infection
The four CCA cell lines (TFK-1, HuCCT1, RBE and
QBC939) and human intrahepatic biliary epithelial cells (HIBEpiC)
were purchased from the Cell Bank of Chinese Academy of Sciences.
All cell lines used in the present study were cultured in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
Human miR-137 overexpression lentivirus (LV-miR-137) and
corresponding negative control (NC) lentivirus were obtained from
Shanghai GeneChem Co., Ltd. The whole infection procedure was
performed based on the protocol provided by the manufacturer
(MOI=10 for both TFK-1 and HuCCT1 cells). The infection efficacy
was evaluated by a fluorescence microscope (magnification, ×200)
and reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
According to the protocol provided by the
manufacturer, total RNA was extracted from cells or tissues using
TRIzol (Yeasen). cDNA was obtained via RT of total RNA using Prime
Script RT Master Mix (Yeasen) and a miRNA First Strand cDNA
Synthesis (Stem-loop Method) kit (Yeasen) for mRNA and miRNA,
respectively. The protocol for RT of mRNA and miRNA was 37°C for 15
min, 85°C for 30 sec and 4°C for 5 min. The expression levels were
measured using SYBR reagent (Yeasan), while GAPDH and U6 were
employed as the reference gene for mRNAs and miRNA, respectively.
Relative expression levels of genes were analyzed using the
2−ΔΔCq method (18).
The primers used were as follows: WNT2B: Forward, 5'-GGG GCA CGA
GTG ATC TGT G-3' and reverse, 5′-GCA TGA TGT CTG GGT AAC GCT-3′;
c-Myc: Forward, 5′-GGC TCC TGG CAA AAG GTC A-3′ and reverse, 5′-CTG
CGT AGT TGT GCT GAT GT-3′; N-cadherin: Forward, 5′-TCA GGC GTC TGT
AGA GGC TT-3′ and reverse, 5′-ATG CAC ATC CTT CGA TAA GAC TG-3′;
vimentin: Forward, 5′-GAC GCC ATC AAC ACC GAG TT-3′ and reverse,
5′-CTT TGT CGT TGG TTA GCT GGT-3′; cyclin D1: Forward, 5′-GCT GCG
AAG TGG AAA CCA TC-3′ and reverse, 5′-CCT CCT TCT GCA CAC ATT TGA
A-3′; cyclin-dependent kinase (CDK)2: Forward, 5′-CCA GGA GTT ACT
TCT ATG CCT GA-3′ and reverse, 5′-TTC ATC CAG GGG AGG TAC AAC-3′;
GAPDH: Forward, 5′-GGA GCG AGA TCC CTC CAA AAT-3′ and reverse,
5′-GGC TGT TGT CAT ACT TCT CAT GG-3′; U6: Forward, 5′-TCG CTT CGG
CAG CAC ATA TAC T-3′ and reverse, 5′-ACG CTT CAC GAA TTT GCG TGT
C-3′. The thermocycling conditions in the present study were: 95°C
for 5 min, 40 cycles of 95°C for 30 sec, annealing at 60°C for 30
sec and a final elongation step at 72°C for 30 sec.
Cell Counting Kit-8 (CCK-8) assay
TFK-1 and HuCCT1 cells at a density of
2×103 cells/well were cultured in 96-well plates (Wuhan
Boster Biological Technology, Ltd.) with 100 ml RPMI-1640 following
infection with LV-miR-137 and NC lentivirus. After culturing for
24, 48, 72 and 96 h, a total of 10 µl CCK-8 reagent (Wuhan
Boster Biological Technology, Ltd.) was added into each well,
according to the manufacturer's protocol. After culturing for 2 h,
cell proliferation ability was measured by a spectrophotometer
(Bio-Rad Laboratories, Inc.) at an optical density (OD) of 450
nm.
Colony formation assay
Cells were seeded onto 6-well plates (Wuhan Boster
Biological Technology, Ltd.) at a density of 2×103 and
cultured in RPMI-1640 medium containing 10% FBS. After 2 weeks of
incubation, the colonies were fixed for 30 min at room temperature
using 4% paraformaldehyde. The number of colonies was counted by
naked eye after staining with 0.1% crystal violet solution for 20
min at room temperature.
Cell cycle distribution analysis
TFK-1 and HuCCT1 cells were resuspended in serum
free RMPI-1640 medium and seeded into 6-well plates at a density of
2×105 cells per well. Then, cells were starved for 24 h
using RPMI-1640 without FBS for cell cycle synchronization. After
cell cycle synchronization, cells were treated with RMPI-1640
medium containing 10% FBS and allowed to proliferate for 48 h.
Then, the cells were harvested and fixed using 75% ethanol
(Servicebio) for 24 h at −20°C. After washing with PBS, the cells
were treated with 5 µl propidium iodide (Invitrogen; Thermo
Fisher Scientific, Inc.) and 1X binding buffer at room temperature
for 30 min. Finally, the cell cycle distribution was examined using
a BD FACScan™ flow cytometer (BD Biosciences; Becton,
Dickinson and Company; version no. 343039) and FlowJo software was
used to analyze the results (version 7.4.1; FlowJo LLC).
Migration and invasion Transwell
assays
TFK-1 and HuCCT1 cells (2×105) were
resuspended in 200 µl RPMI-1640 medium without FBS and added
into the upper Transwell chamber (EMD Millipore) with or without
Matrigel (Invitrogen; Thermo Fisher Scientific, Inc.), while 700
µl medium containing 10% serum was placed into the lower
Transwell chamber. After culturing cells for 24 h, cells not
migrating or invading through the membranes (0.8 µm) were
removed and cells crossing the membranes were fixed using 4%
paraformaldehyde (Servicebio) for 20 min at room temperature and
stained with 1% crystal violet solution (Servicebio) for 30 min at
room temperature. A total of five different fields were
photographed using an inverted optical microscope (Nikon
Corporation; magnification, ×40) and the cells were counted.
Animal experiments
Female BALB/c nude mice (n=12), aged 4 weeks and
weighing ~17 g, were purchased from the Beijing Vital River
Laboratory Animal Technology Co., Ltd. The mice were housed under
specific pathogen-free conditions at 25°C with a 12-h light/dark
cycle and free access to food and water. First, the
miR-137-overexpressing HuCCT1 cell line and normal control were
constructed. Next, a total of 1×107 cells suspended in
PBS were injected into the subcutaneous tissues of the right upper
flank in mice (n=6 per group). The animal health and behavior were
monitored daily, and the volume of tumor tissue was measured weekly
and calculated using the following formula: Volume
(mm3)=(length x width2)/2. When the maximum
length of any tumor reached 15 mm or the volume of any tumor
reached 800 mm3, the experiment was terminated. All mice
(n=12; no mice died during the experiment) were sacrificed using
cervical dislocation and death was verified by the absence of a
heartbeat and the onset of rigor mortis. Subsequently, the tumor
tissues were isolated and weighed. All animal experiments were
approved by the Ethics Committee of Guizhou Medical University.
Immunohistochemistry
After fixation using 4% paraformaldehyde for 24 h at
room temperature, dehydration and embedding in paraffin
(Servicebio), the tissues were cut into 4-µm sections. The
specimens were then deparaffinized using xylene and rehydrated
through graded alcohols. After restoration with sodium citrate and
blocking using H2O2 and bovine serum albumin
(Servicebio), the specimens were incubated with the primary
antibodies (both 1:400), including Ki-67 (cat. no. 27309-1-AP) and
proliferating cell nuclear antigen (PCNA; cat. no. 10205-2-AP;
ProteinTech Group, Inc.) for 12 h at 4°C. Subsequently, the
sections were immunohistochemically stained with horseradish
peroxidase (HRP)-conjugated goat anti-rabbit secondary antibodies
(cat. no. G1213; Servicebio) for 2 h at room temperature. Following
incubation with the Cell and Tissue Staining HRP-DAB kit for 5 min
at room temperature (Servicebio), an orthophotomicroscope (Nikon
Corporation; magnification, ×400) was used to capture images.
miRNA target prediction
Potential targets of miR-137 was predicted using
online website TargetScan (http://www.targetscan.org/; version no. 7.2). Target
genes with an absolute value of cumulative weighted context ++
score >0.2 were considered credible. Credible target genes of
miR-137 were imported to the online tool Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov/; version no. 6.8) to
perform Kyoto Encyclopedia of Genes and Genomes analysis. P<0.05
was set as the cutoff to consider significant enrichment. The
binding site of vital target genes in significantly enriched
pathways was shown and the association between miR-137 and the
vital target gene was further examined.
Dual luciferase reporter assays
In order to verify whether miR-137 binds to WNT2B
and regulates its expression, a dual luciferase reporter assay was
performed. The wild-type (Wt) and mutant (Mut) type 3'-untranslated
regions (UTRs) of WNT2B, which contain predicted binding sites for
miR-137, were synthesized and subcloned into the psiCHECK-2
luciferase reporter vector (Promega Corporation). After culturing
for 24 h, TFK-1 and HuCCT1 cells were co-transfected with Wt/Mut
type WNT2B luciferase reporter vector and miR-137 mimic (sequence:
5'-UUA UUG CUU AAG AAU ACG CGU AG-3'). miR-137 NC (sequence: 5'-UCA
CAA CCU CCU AGA AAG AGU AGA-3'). Finally, the Dual Luciferase
Reporter Assay System (Promega Corporation) was employed to measure
the luciferase activities, and the activities were normalized with
Renilla luciferase activity. Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for transient
transfection, and the duration between transfection and activity
measurement was 24 h.
Western blotting
Cells were lysed using a RIPA buffer (Wuhan Boster
Biological Technology, Ltd.) containing protease inhibitor cocktail
(Boster Biological Technology) and PMSF (Wuhan Boster Biological
Technology, Co., Ltd.). Following centrifugation (8,000 × g/15 min)
at 4°C, proteins were collected from cellular debris and the
bicinchoninic acid method was employed to determine the
concentration. Protein samples (30 µg/well) were separated
using 10% SDS-PAGE under 90 V voltage and transferred onto PVDF
membranes (EMD Millipore). After blocking with TBST (0.1% Tween-20)
containing 5% skimmed milk at room temperature for 2 h, the
membranes were then incubated with primary antibodies (1:1,000)
against WNT2B (cat. no. ab50575; Abcam), β-catenin (cat. no.
51067-2-AP; Proteintech), TCF4 (cat. no. 22337-1-AP; Proteintech),
N-cadherin (cat. no. 22018-1-AP; Proteintech), vimentin (cat. no.
10366-1-AP; Proteintech), cyclin D1 (cat. no. 26939-1-AP;
Proteintech), CDK2 (cat. no. 10122-1-AP; Proteintech), c-Myc (cat.
no. 10828-1-AP; Proteintech) and GAPDH (cat. no. 60004-1-Ig;
Proteintech) for 16 h at 4°C. After washing with TBST to remove
unbound antibodies, the membranes were incubated with the
horseradish peroxidase conjugated secondary antibodies goat
anti-rabbit (1:3,000; cat. no. SA00001-2, Proteintech) and goat
anti-mouse (1:3,000; cat. no. SA00001-1, Proteintech) for 2 h at
room temperature. Finally, the signals were detected by the
Photoshop Image Analysis software CS3 (Adobe Systems).
Statistical analyses
All experiments were repeated three times and the
data are presented as the mean ± standard deviation. All
statistical analyses in the present study were performed using SPSS
21.0 (IBM Corp.). Comparisons between two groups were conducted
using two-tailed Student's t-test, while comparisons among multiple
groups were performed using one-way analysis of variance combined
with LSD-t test. The co-expression relationship between two genes
was analyzed using Pearson relationship analysis. P<0.05 was
considered to indicate statistically significant differences.
Results
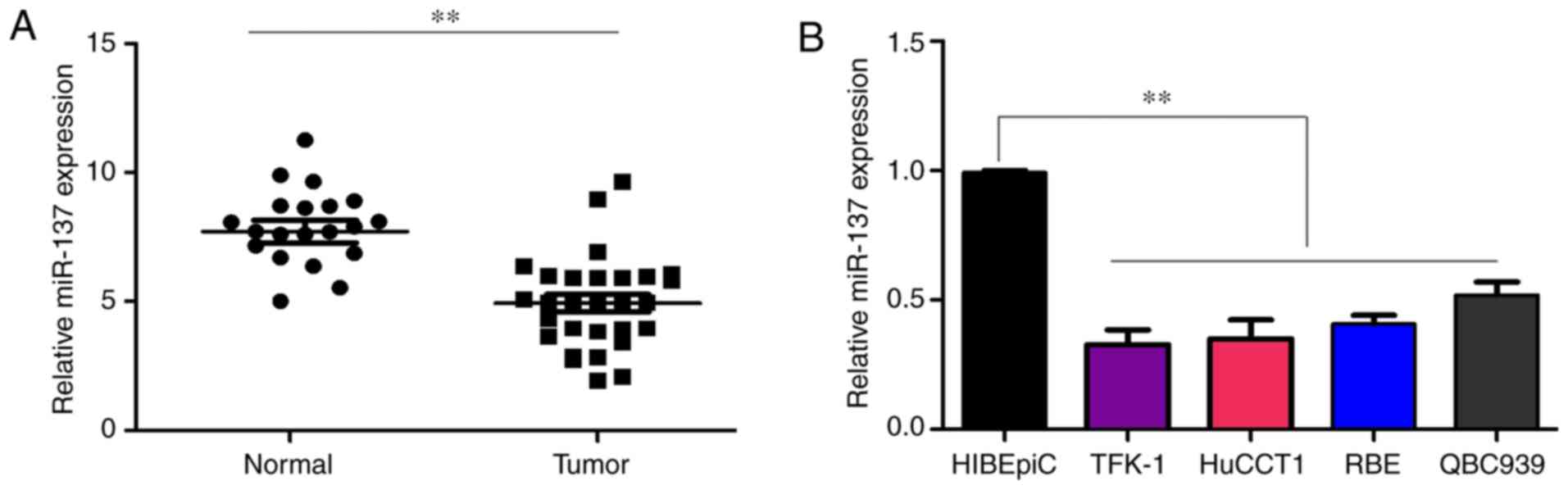
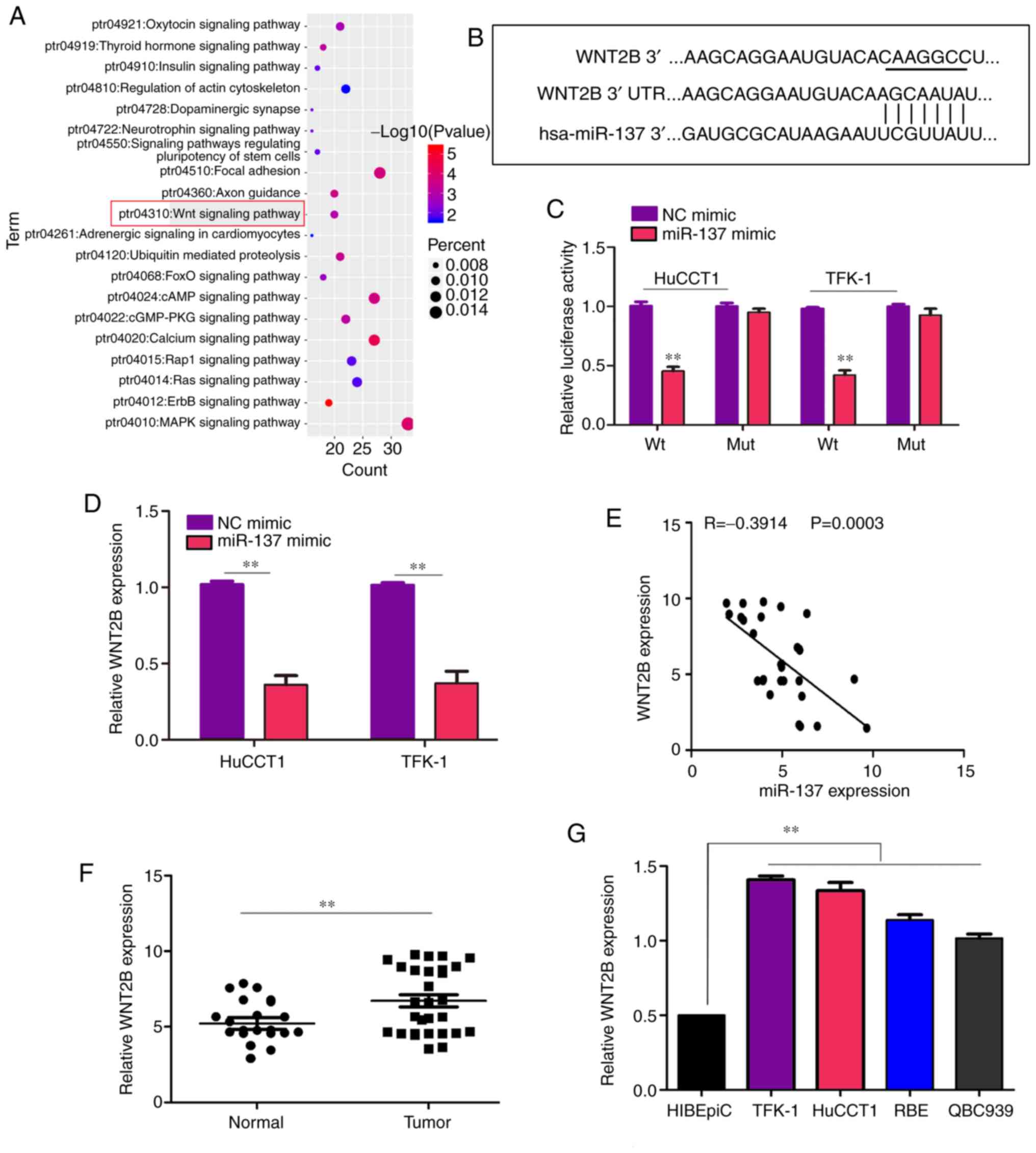
miR-137 is significantly downregulated in
CCA
To determine whether miR-137 was involved in the
progression of CCA, its expression in 29 histologically confirmed
CCA samples and 20 adjacent non-tumor samples was detected using
RT-qPCR. The results demonstrated the expression of miR-137 was
significantly decreased in the CCA tissues compared with that in
adjacent non-tumor tissues (Fig.
1A). Similarly, it was demonstrated that the expression level
of miR-137 was lower in CCA cell lines, including TFK-1, HuCCT1,
RBE and QBC939, compared with that in HIBEpiCs (Fig. 1B). Taken together, these results
indicate that the loss of expression of miR-137 may be involved in
the development of CCA.
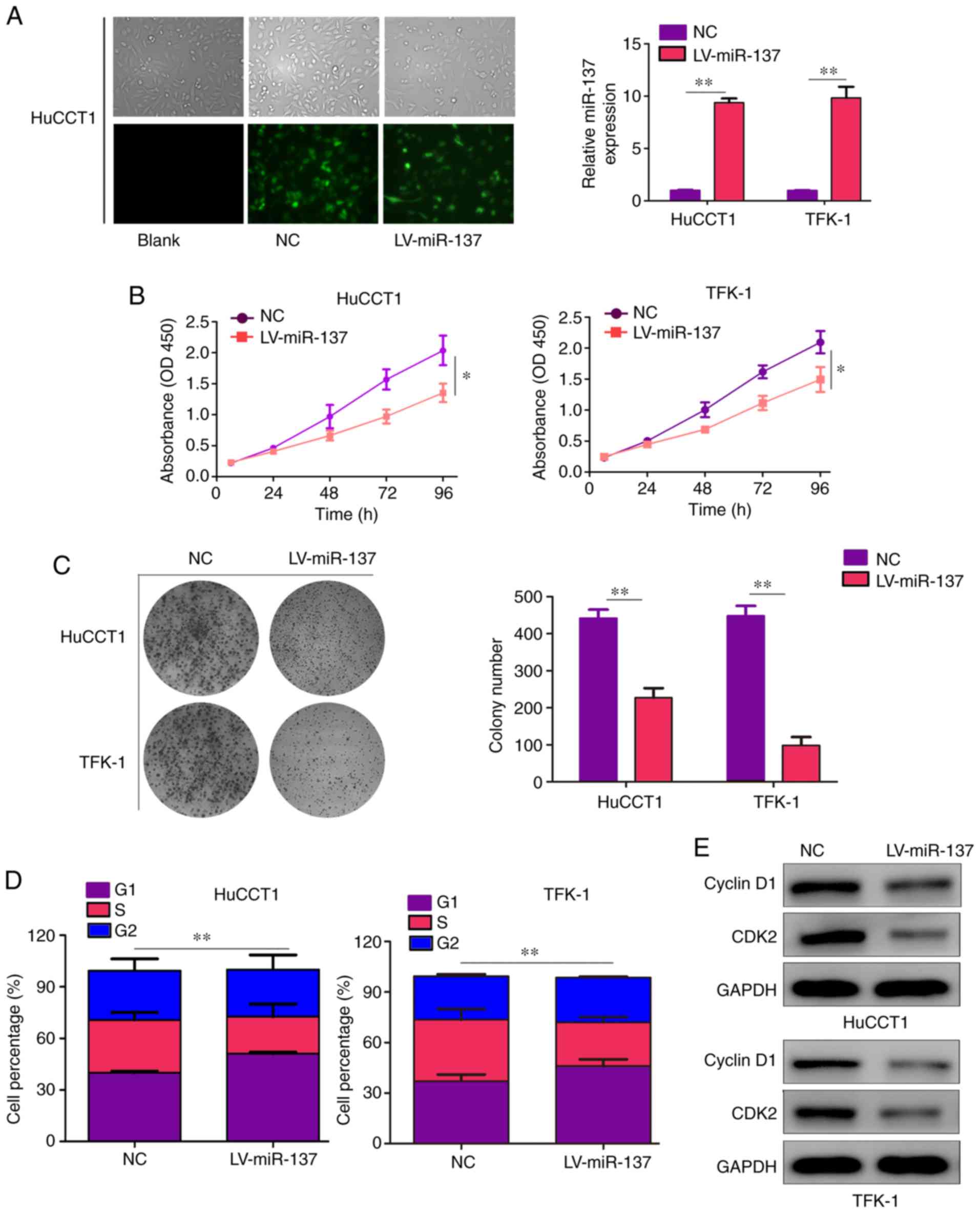
miR-137 represses CCA cell proliferation
in vitro
First, the CCA cell lines HuCCT1 and TFK-1 were
stably infected with miR-137 overexpression (LV-miR-137) and NC
lentiviruses. The results revealed that both LV-miR-137 and NC
lentiviruses exhibited high infection efficiency in HuCCT1 cells
after infection for 24 h (>80%). Similarly, compared with cells
infected with NC lentiviruses, the mRNA levels of miR-137 were
significantly increased in cells infected with LV-miR-137
lentiviruses (Fig. 2A). CCK-8
assays were used to detect the effect of miR-137 on CCA cell
proliferation ability and the results revealed that miR-137
overexpression notably inhibited the proliferation ability of CCA
cells (Fig. 2B). Additionally,
the results of colony formation assays demonstrated that increased
expression of miR-137 inhibited the colony formation ability of CCA
cells (Fig. 2C). Moreover, cell
cycle distribution analysis revealed that the percentage of HuCCT1
and TFK-1 cells in the G1 phase was significantly increased in the
LV-miR-137 group compared with the NC groups, while the number of
cells in the S phase was markedly decreased (Fig. 2D). Furthermore, western blotting
was performed to measure the expression levels of CDK2 and cyclin
D1, which are involved in regulating cell transition from the G1 to
the S phase. The results of western blotting revealed that the
expression of CDK2 and cyclin D1 decreased following infection with
miR-137 overexpression lentiviruses (Fig. 2E).
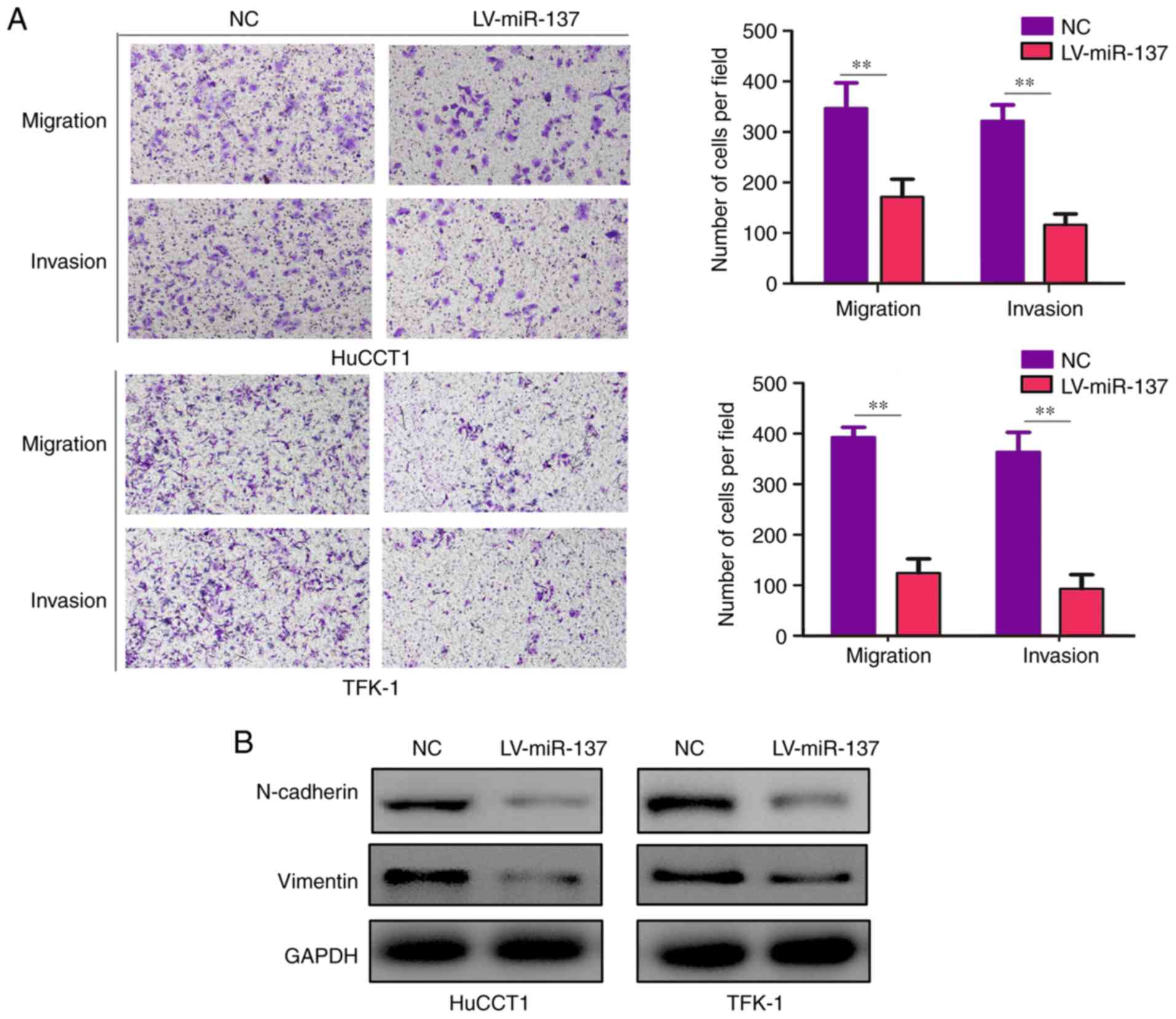
miR-137 represses CCA cell migration and
invasion in vitro
The results of the Transwell assays demonstrated
that both the migration and invasion abilities in the miR-137
overexpression groups were decreased compared with the control
groups (Fig. 3A). Previous
studies demonstrated that epithelial-to-mesenchymal transition
(EMT) is a crucial process for cancer cell mobility (19-21). To determine whether miR-137
inhibits the mobility of HuCCT1 and TFK-1 cells via affecting the
EMT phenotype, western blotting was conducted to detect the protein
levels of two key markers of EMT, N-cadherin and vimentin. It was
demonstrated that the protein levels of both N-cadherin and
vimentin were decreased with miR-137 overexpression (Fig. 3B).
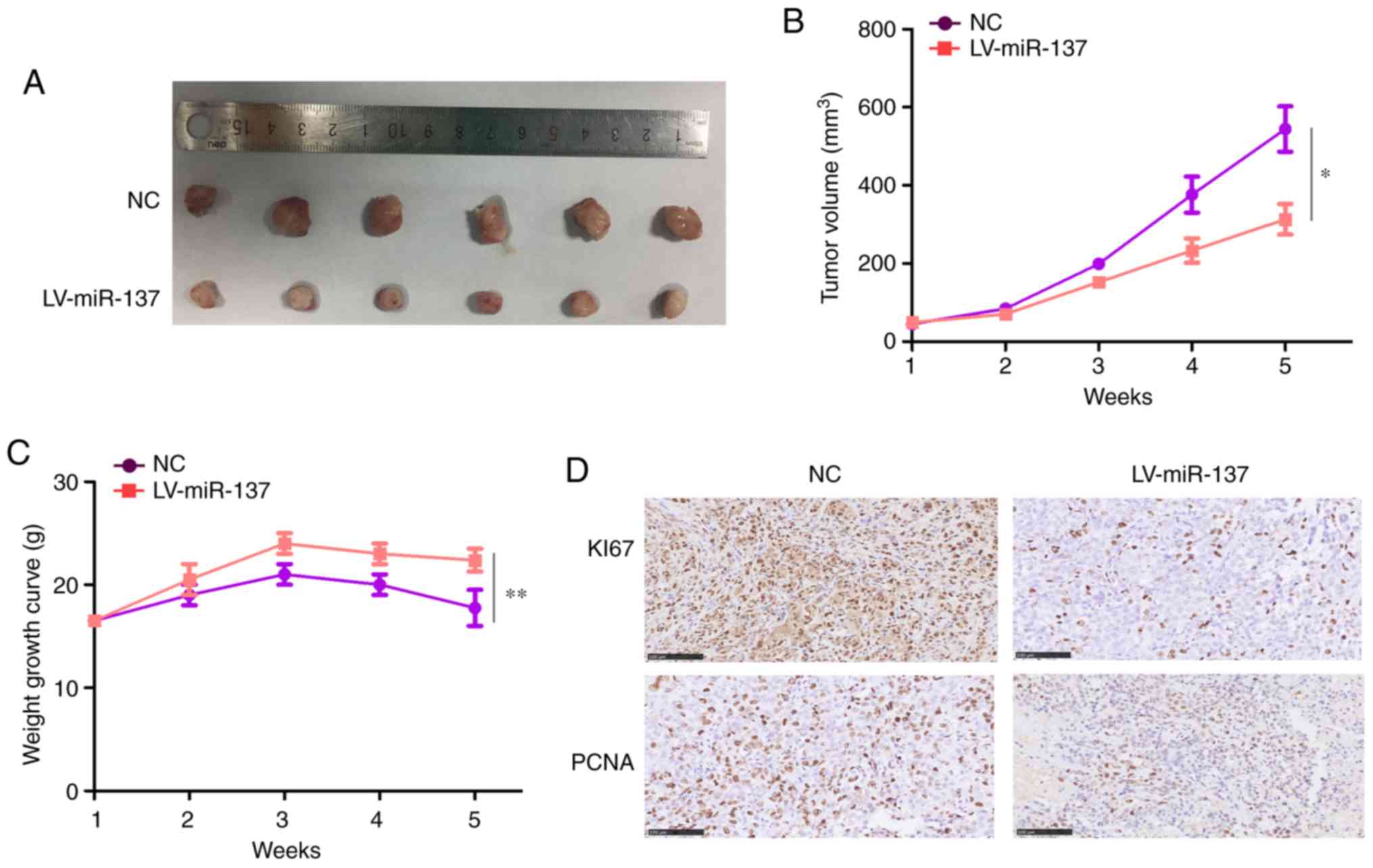
miR-137 significantly inhibits tumor
growth in vivo
Having uncovered the vital impact of miR-137 in the
biology of CCA cells in vitro, the effect of miR-137 on
tumor growth in vivo was next investigated. For this
purpose, HuCCT1 cells stably expressing miR-137 or miR-NC were
injected into the subcutaneous tissues of nude mice and tumor
growth was monitored. The results revealed that the growth rate of
tumors derived from miR-137-overexpressing HuCCT1 cells was
significantly slower and the formed tumors were significantly
smaller compared with those originating from miR-NC cells (Fig. 4A and B). In addition, the weight
of the mice decreased more slowly in the miR-137 overexpression
group (Fig. 4C). Furthermore,
miR-137-overexpressing tumors excised after 5 weeks exhibited
markedly decreased levels of the proliferation marker Ki-67 and
PCNA proteins compared with miR-NC tumors, as determined by
immunohistochemical examination (Fig.
4D).
WNT2B is a key target of miR-137 in
CCA
To uncover the molecular mechanism underlying the
role of miR-137 in regulating the function of CCA cells, the online
bioinformatics tool TargetScan was employed to identify mRNAs
containing 3'UTR sequences complementary to miR-137. As the results
demonstrated, one of the key pathways in which the credible target
genes of miR-137 were enriched was the Wnt signaling pathway
(Fig. 5A). In addition, the 3'UTR
of WNT2B, which plays a key role in the Wnt signaling pathway,
contained a putative miR-137-binding site (Fig. 5B). Therefore, WNT2B may be an
important target of miR-137. To validate the prediction, the 3'UTR
of WNT2B, either Wt or Mut, in the putative binding site of miR-137
was cloned into a luciferase reporter vector, which was transfected
into TFK-1 and HuCCT1 cells together with miR-137 or miR-NC. The
results indicated that co-transfection with miR-137 decreased
luciferase activity driven by WNT2B-Wt, but not by WNT2B-Mut
(Fig. 5C). Similarly, increased
expression of miR-137 decreased the mRNA level of WNT2B in both
TFK-1 and HuCCT1 cells (Fig. 5D).
Subsequently, correlation analysis proved that the mRNA levels of
WNT2B were negatively associated with miR-137 in the 29 human CCA
samples (Fig. 5E). Furthermore,
the mRNA level of WNT2B was higher in CCA samples and cell lines
compared with normal samples (Fig. 5F
and G).
miR-137 regulates the Wnt signaling
pathway via WNT2B
Based on the fact that WNT2B is a key factor in the
Wnt signaling pathway (22,23), whether miR-137 may regulate Wnt
signaling pathway via WNT2B was investigated. The results of
western blotting demonstrated that miR-137 overexpression decreased
the protein level of WNT2B, β-catenin and TCF4, while ectopic
expression of WNT2B was able to reverse this effect induced by
miR-137 (Fig. 6A and B).
Furthermore, it was observed that both the transcription and
translation levels of targeted genes of the Wnt signaling pathway,
including c-Myc, N-cadherin, vimentin, CDK2 and cyclin D1, were
decreased in miR-137-overexpressing cells, whereas restoring the
expression of WNT2B in miR-137-overexpressing cells reversed this
effect (Fig. 6C and D). Taken
together, these findings indicate that miR-137 may inhibit
Wnt-related signaling pathways via WNT2B.
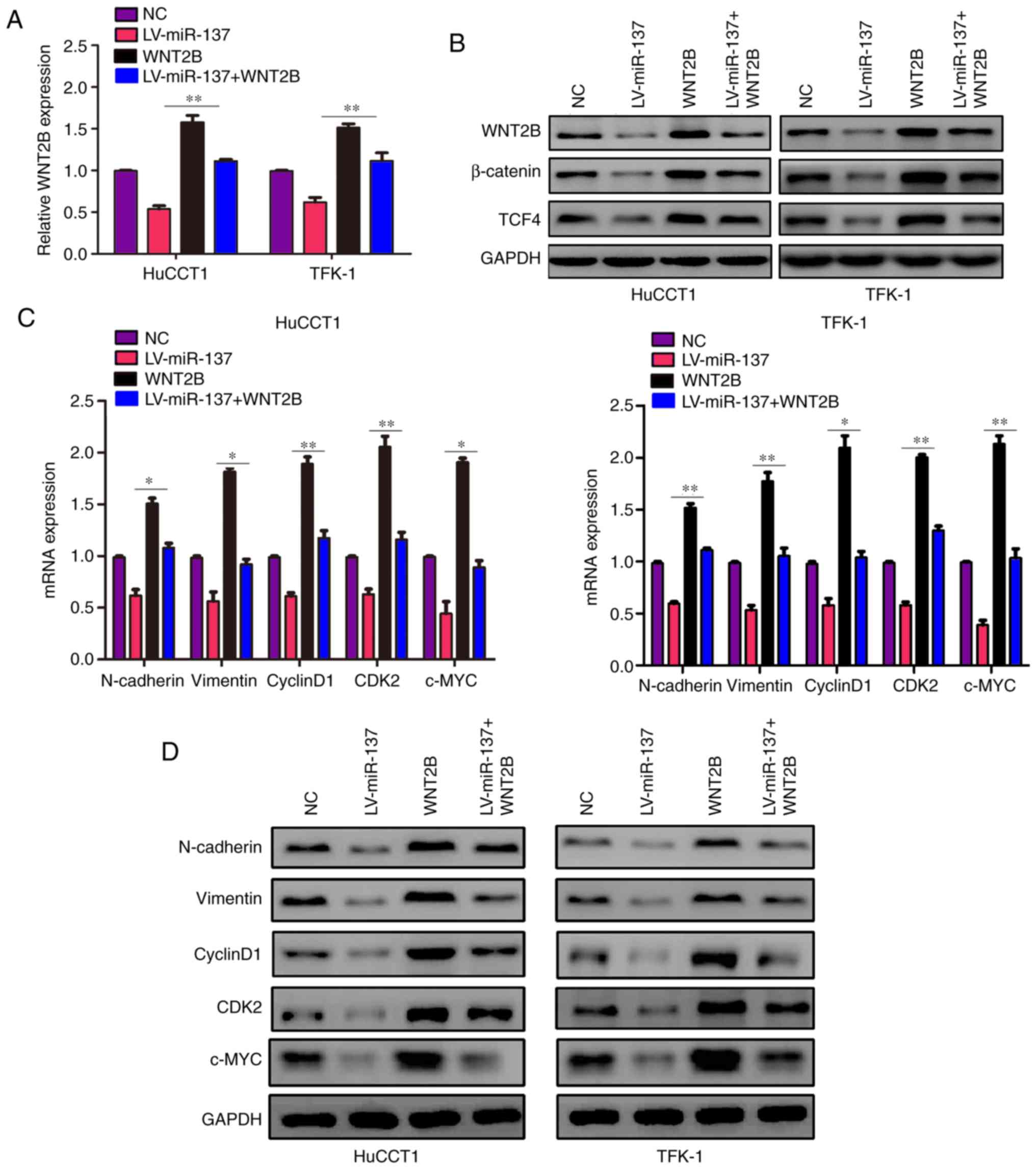 | Figure 6miR-137 regulates the Wnt signaling
pathway via WNT2B. Cells were divided into four groups and
subjected to different treatments: NC lentiviruses; transduction of
miR-137 lentiviruses (LV-miR-137) alone; treatment with WNT2B
plasmid (WNT2B) alone; transduction of miR-137 lentiviruses and
treatment with WNT2B plasmid (LV-miR-137 + WNT2B). (A) The mRNA
level of WNT2B in each group was detected using reverse
transcription-quantitative PCR. (B) The protein expression levels
of WNT2B, β-catenin and TCF4 in each group were detected using
western blotting. (C) The mRNA levels of N-cadherin, vimentin,
cyclin D1, CDK2 and c-Myc in each group was detected using reverse
transcription-quantitative PCR. (D) The protein levels of
N-cadherin, vimentin, cyclin D1, CDK2 and c-Myc in each group were
detected using western blotting. *P<0.05,
**P<0.01. WNT2B, Wnt family member 2B; CDK,
cyclin-dependent kinase; NC, negative control; LV, lentivirus; miR,
microRNA; N, neural. |
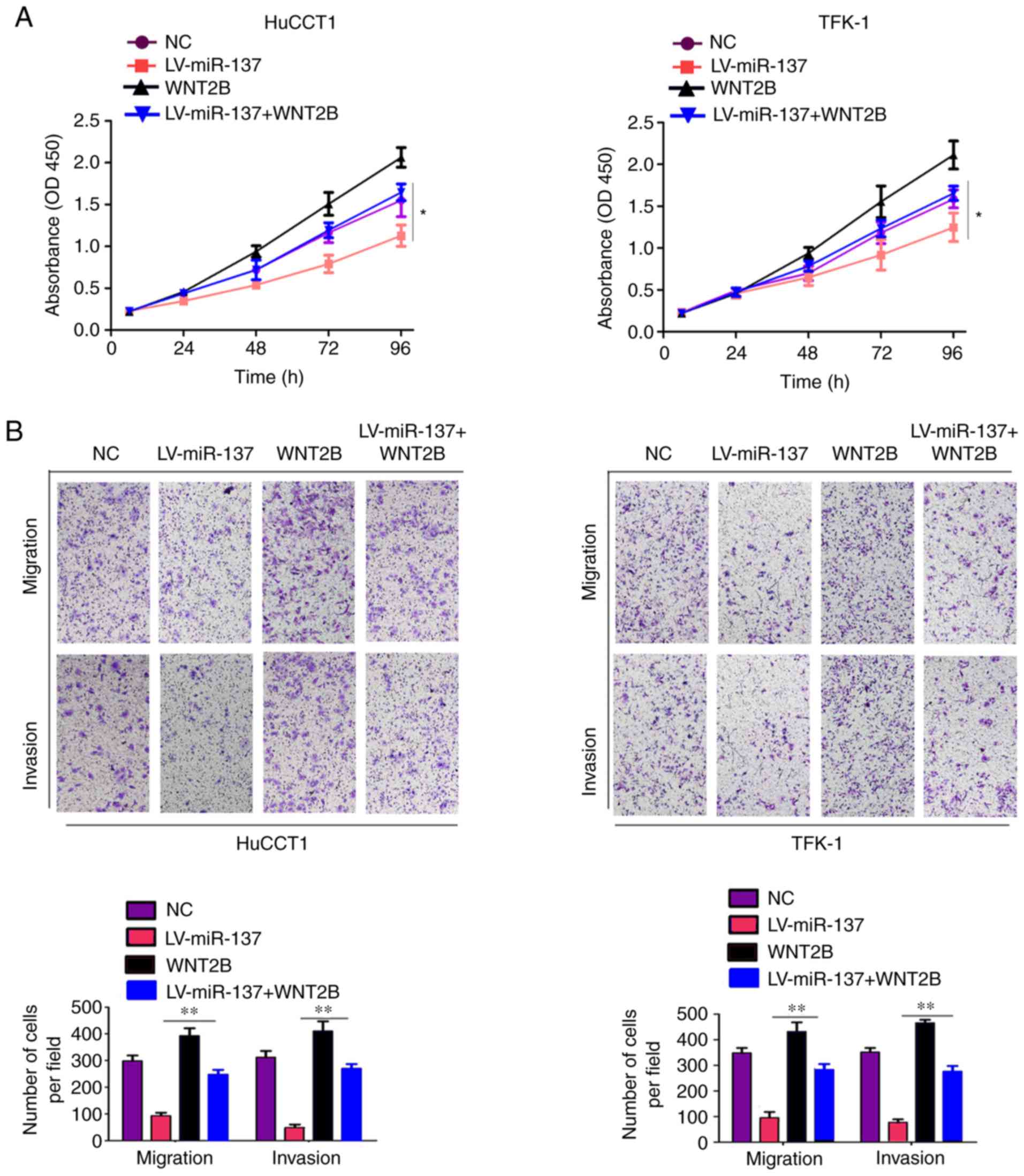
Restoration of WNT2B expression reverses
the inhibitory effects of miR-137
The present study investigated whether WNT2B
overexpression was able to reverse the inhibitory effects of
miR-137 on CCA cells. First, the results of the CCK-8 assays
indicated that restoration of WNT2B expression in cells stably
expressing miR-137 reversed the inhibitory effect of miR-137 on
cell proliferation (Fig. 7A).
Similarly, Transwell assays revealed that restoring the expression
level of WNT2B in miR-137-overexpressing cells relieved the
inhibitory effect of miR-137 on cell mobility (Fig. 7B). Overall, these results
demonstrated that restoration of WNT2B expression can reverse the
inhibitory effects of miR-137 on CCA cells.
Discussion
Various studies have demonstrated that the miRNA
class of non-coding RNAs participate in tumor progression through
regulating a series of target genes involved in tumor proliferation
and metastasis (24).
Dysregulated expression or function of miRNAs has been demonstrated
in a series of malignancies, including CCA (25). For example, miR-142-5p was shown
to decrease the expression of phosphatase and tensin homolog and
promote the progression of CCA (26). It was demonstrated that miR-329
can suppress the expression of pituitary tumor-transforming 1,
inactivates the mitogen-activated protein kinase/extracellular
signal-regulated kinase signaling pathway and has the potential to
suppress CCA cell proliferation (27). Even though numerous miRNAs may be
involved in the development of CCA, the present study only focused
on miR-137, exploring its role and its target gene, WNT2B, in CCA
biology.
The present study verified that the expression of
miR-137 was decreased in CCA tissues compared with that in adjacent
non-tumor tissues. It was also indicated that the expression of
miR-137 was markedly lower in CCA cell lines compared with that in
HIBEpiCs and miR-137 overexpression inhibited CCA cell
proliferation, decreased the ability of colony formation, induced
cell cycle arrest, and inhibited cell migration and invasion. This
constitutes evidence that miR-137 may act as a tumor suppressor in
CCA.
WNT2B, also referred to as WNT13, is the thirteenth
Wnt gene to be cloned. WNT2B interacts with frizzled receptor and
activates the Wnt signaling pathway, which leads to the increased
expression of β-catenin in the cytoplasm (23). Subsequently, the β-catenin protein
translocates from the cytoplasm into the nucleus and promotes the
transcriptional effect of TCF4 (28,29). Previous studies have reported that
WNT2B acts as an oncogene in a number of tumors and is positively
associated with tumor progression. WNT2B increases cell
proliferation ability and induces EMT via activating
β-catenin-related pathways in non-small cell lung cancer (22). WNT2B inhibition was found to
increase the chemosensitivity of head and neck squamous cell
carcinoma (30). The expression
of WNT2B was also found to be higher in pancreatic cancer tissues
and high mRNA levels of WNT2B were positively associated with
unfavorable prognosis in patients suffering from pancreatic cancer
(31). To the best of our
knowledge, the present study is the first to demonstrate that
miR-137 targets WNT2B in CCA. The expression levels of WNT2B were
negatively associated with miR-137 in CCA tissues. miR-137
decreased the expression of WNT2B and its downstream pathway
molecules, while restoration of WNT2B expression alleviated the
inhibitory effect of miR-137. Overall, the present study provided
evidence that miR-137 has the potential to suppress CCA cell
proliferation and mobility via decreasing WNT2B translation.
Therefore, miR-137 and WNT2B may prove to be useful
diagnostic biomarkers as well as effective targets for the clinical
treatment of CCA. Furthermore, the findings of the present study
may improve understanding of the molecular mechanism underlying the
development of CCA.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81060176) and the
Major Projects of Applied and Basic Research Program of Guizhou
Province [grant no. J-(2015)2003].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL ZZ, SP and JZ performed the experiments. YX, YS,
JL, SX, DM and BG were responsible for analysis and interpretation
of the results. TC designed the experiments and wrote the
manuscript. All the authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The Human Trial Ethics Committee of Guizhou Medical
University approved the study. Written informed consent was
obtained from the patients who provided the specimens. The present
study was performed in accordance with the principles outlined in
the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ma WJ, Wu ZR, Shrestha A, Yang Q, Hu HJ,
Wang JK, Liu F, Zhou RX, Li QS and Li FY: Effectiveness of
additional resection of the invasive cancer-positive proximal bile
duct margin in cases of hilar cholangiocarcinoma. Hepatobiliary
Surg Nutr. 7:251–269. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang W, Xu L, Yang P, Zhang L, Wan D,
Huang Q, Niu T and Chen F: Novel nomogram for preoperative
prediction of early recurrence in intrahepatic cholangiocarcinoma.
Front Oncol. 8:3602018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Beal EW, Tumin D, Moris D, Zhang XF,
Chakedis J, Dilhoff M, Schmidt CM and Pawlik TM: Cohort
contributions to trends in the incidence and mortality of
intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr.
7:270–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma-evolving concepts and therapeutic
strategies. Nat Rev Clin Oncol. 15:95–111. 2018. View Article : Google Scholar
|
|
5
|
Burroughs AM and Ando Y: Identifying and
characterizing functional 3' nucleotide addition in the miRNA
pathway. Methods. 152:23–30. 2019. View Article : Google Scholar
|
|
6
|
Lee D and Shin C: MicroRNA-target
interactions: New insights from genome-wide approaches. Ann N Y
Acad Sci. 1271:118–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Puik JR, Meijer LL, Le Large TY, Prado MM,
Frampton AE, Kazemier G and Giovannetti E: miRNA profiling for
diagnosis, prognosis and stratification of cancer treatment in
cholangiocarcinoma. Pharmacogenomics. 18:1343–1358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wan P, Chi X, Du Q, Luo J, Cui X, Dong K,
Bing Y, Heres C and Geller DA: miR-383 promotes cholangiocarcinoma
cell proliferation, migration, and invasion through targeting IRF1.
J Cell Biochem. 119:9720–9729. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu Z, Liu G, Zhang M, Zhang Z, Jia Y, Peng
L, Zhu Y, Hu J, Huang R and Sun X: miR-122-5p inhibits the
proliferation, invasion and growth of bile duct carcinoma cells by
targeting ALDOA. Cell Physiol Biochem. 48:2596–2606. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma J, Weng L, Wang Z, Jia Y, Liu B, Wu S,
Cao Y, Sun X, Yin X, Shang M and Mao A: MiR-124 induces
autophagy-related cell death in cholangiocarcinoma cells through
direct targeting of the EZH2-STAT3 signaling axis. Exp Cell Res.
366:103–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Z, Guo X, Tian S, Zhu C, Chen S, Yu C,
Jiang J and Sun C: MicroRNA-137 reduces stemness features of
pancreatic cancer cells by targeting KLF12. J Exp Clin Cancer Res.
38:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao J, Peng F, Yu C, Wang M, Li X, Li Z,
Jiang J and Sun C: microRNA-137 modulates pancreatic cancer cells
tumor growth, invasion and sensitivity to chemotherapy. Int J Clin
Exp Pathol. 7:7442–7450. 2014.
|
|
13
|
Min L, Wang F, Hu S, Chen Y, Yang J, Liang
S and Xu X: Aberrant microRNA-137 promoter methylation is
associated with lymph node metastasis and poor clinical outcomes in
non-small cell lung cancer. Oncol Lett. 15:7744–7750.
2018.PubMed/NCBI
|
|
14
|
Wu QQ, Zheng B, Weng GB, Yang HM, Ren Y,
Weng XJ, Zhang SW and Zhu WZ: Downregulated NOX4 underlies a novel
inhibitory role of microRNA-137 in prostate cancer. J Cell Biochem.
120:10215–10227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X, Lu C, Chu W, Zhang B, Zhen Q, Wang
R, Zhang Y, Li Z, Lv B, Li H and Liu J: MicroRNA-124 suppresses
proliferation and glycolysis in non-small cell lung cancer cells by
targeting AKT-GLUT1/HKII. Tumour Biol. 39:10104283177062152017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakaguchi M, Hisamori S, Oshima N, Sato F,
Shimono Y and Sakai Y: miR-137 regulates the tumorigenicity of
colon cancer stem cells through the inhibition of DCLK1. Mol Cancer
Res. 14:354–362. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bi WP, Xia M and Wang XJ: miR-137
suppresses proliferation, migration and invasion of colon cancer
cell lines by targeting TCF4. Oncol Lett. 15:8744–8748.
2018.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Zhang XY, Gao PT, Yang X, Cai JB, Ding GY,
Zhu XD, Ji Y, Shi GM, Shen YH, Zhou J, et al: Reduced
selenium-binding protein 1 correlates with a poor prognosis in
intrahepatic cholangiocarcinoma and promotes the cell
epithelial-mesenchymal transition. Am J Transl Res. 10:3567–3578.
2018.
|
|
20
|
Zou Z, Zheng B, Li J, Lv X, Zhang H, Yu F,
Kong L, Li Y, Yu M, Fang L and Liang B: TPX2 level correlates with
cholangiocarcinoma cell proliferation, apoptosis, and EMT. Biomed
Pharmacother. 107:1286–1293. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peng R, Zhang PF, Zhang C, Huang XY, Ding
YB, Deng B, Bai DS and Xu YP: Elevated TRIM44 promotes intrahepatic
cholangiocarcinoma progression by inducing cell EMT via MAPK
signaling. Cancer Med. 7:796–808. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang B, Sun L, Li J and Jiang R: miR-577
suppresses cell proliferation and epithelial-mesenchymal transition
by regulating the WNT2B mediated Wnt/β-catenin pathway in non-small
cell lung cancer. Mol Med Rep. 18:2753–2761. 2018.PubMed/NCBI
|
|
23
|
Jiang Z, Jiang C and Fang J: Up-regulated
lnc-SNHG1 contributes to osteosarcoma progression through
sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin
pathway. Biochem Biophys Res Commun. 495:238–245. 2018. View Article : Google Scholar
|
|
24
|
Plieskatt JL, Rinaldi G, Feng Y, Peng J,
Yonglitthipagon P, Easley S, Laha T, Pairojkul C, Bhudhisawasdi V,
Sripa B, et al: Distinct miRNA signatures associate with subtypes
of cholangiocarcinoma from infection with the tumourigenic liver
fluk Opisthorchis viverrini. J Hepatol. 61:850–858. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Liu D, Liu P, Chen Y, Yu H and
Zhang Q: Identification of biomarkers of intrahepatic
cholangiocarcinoma via integrated analysis of mRNA and miRNA
microarray data. Mol Med Rep. 15:1051–1056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei G, Yuan Y, He X, Jin L and Jin D:
Enhanced plasma miR-142-5p promotes the progression of intrahepatic
cholangiocarcinoma via targeting PTEN. Exp Ther Med. 17:4190–4196.
2019.PubMed/NCBI
|
|
27
|
Liang HQ, Wang RJ, Diao CF, Li JW, Su JL
and Zhang S: The PTTG1-targeting miRNAs miR-329, miR-300, miR-381,
and miR-655 inhibit pituitary tumor cell tumorigenesis and are
involved in a p53/PTTG1 regulation feedback loop. Oncotarget.
6:29413–29427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu X, Yuan C, Tian C, Li C, Nie F, Song
X, Zeng R, Wu D, Hao X and Li L: The plant sesquiterpene lactone
parthenolide inhibits Wnt/beta-catenin signaling by blocking
synthesis of the transcriptional regulators TCF4/LEF1. J Biol Chem.
293:5335–5344. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Lin P, Wang Q, Zheng M and Pang L:
Wnt3a-regulated TCF4/β-catenin complex directly activates the key
Hedgehog signalling genes Smo and Gli1. Exp Ther Med. 16:2101–2107.
2018.PubMed/NCBI
|
|
30
|
Li SJ, Yang XN and Qian HY: Antitumor
effects of WNT2B silencing in GLUT1 overexpressing cisplatin
resistant head and neck squamous cell carcinoma. Am J Cancer Res.
5:300–308. 2014.
|
|
31
|
Jiang H, Li F, He C, Wang X, Li Q and Gao
H: Expression of Gli1 and Wnt2B correlates with progression and
clinical outcome of pancreatic cancer. Int J Clin Exp Pathol.
7:4531–4538. 2014.PubMed/NCBI
|