1. Introduction
Respiratory infections are common in the cold
seasons world-wide, and consequently considered trivial and mild.
Affected individuals rarely consult medical professionals, instead
treating themselves with symptomatic medications. Droplet and
aerosol transmission further facilitates rapid dissemination to
numerous individuals at once, which amplifies socioeconomic impact
even with minimal increases to fatality rate, particularly among
patients with comorbidities.
The latest and contemporary outbreak of a
respiratory pathogen, namely severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2) responsible for coronavirus disease
(COVID-19), has brought to attention a hidden threat from an old
enemy. This is the third major coronavirus outbreak over the past
20 years that has had substantial socioeconomic impact, but the
first in the 21st century to affect countries across all continents
except Antarctica. The general panic and insecurity expressed
across all sociopolitical and economic tiers has dramatically
disrupted day-to-day life, international travel and trade.
Notwithstanding severe lifestyle disruptions, depression-associated
disease has been reported due to extreme measures of isolation.
Here we emphasize current knowledge regarding
coronaviruses from a short history to epidemiology, diagnosis,
pathogenesis, clinical manifestation, as well as treatment and
prevention strategies. Forward lessons informing future strategies
to improve surveillance and prevent recurrence are highlighted.
2. A brief overview of coronavirus
infections in human history
SARS-CoV-2 belongs to the Orthocoronavirinae
subfamily, family Coronaviridae, order Nidovirales
(1). It comprises of four
subtypes, α- and β-coronaviruses that can infect humans, and gamma-
and delta-coronaviruses, which are found only in animals. It is a
zoonotic virus that can be transmitted from animals to humans and,
once adapted to do so, between humans via airborne droplets and
aerosols (2). The most involved
carrier animal species for human infections is the bat (3), though animal reservoirs extend to
cattle, pigs, turkeys, camels, mice, dogs, cats, ferrets and mink
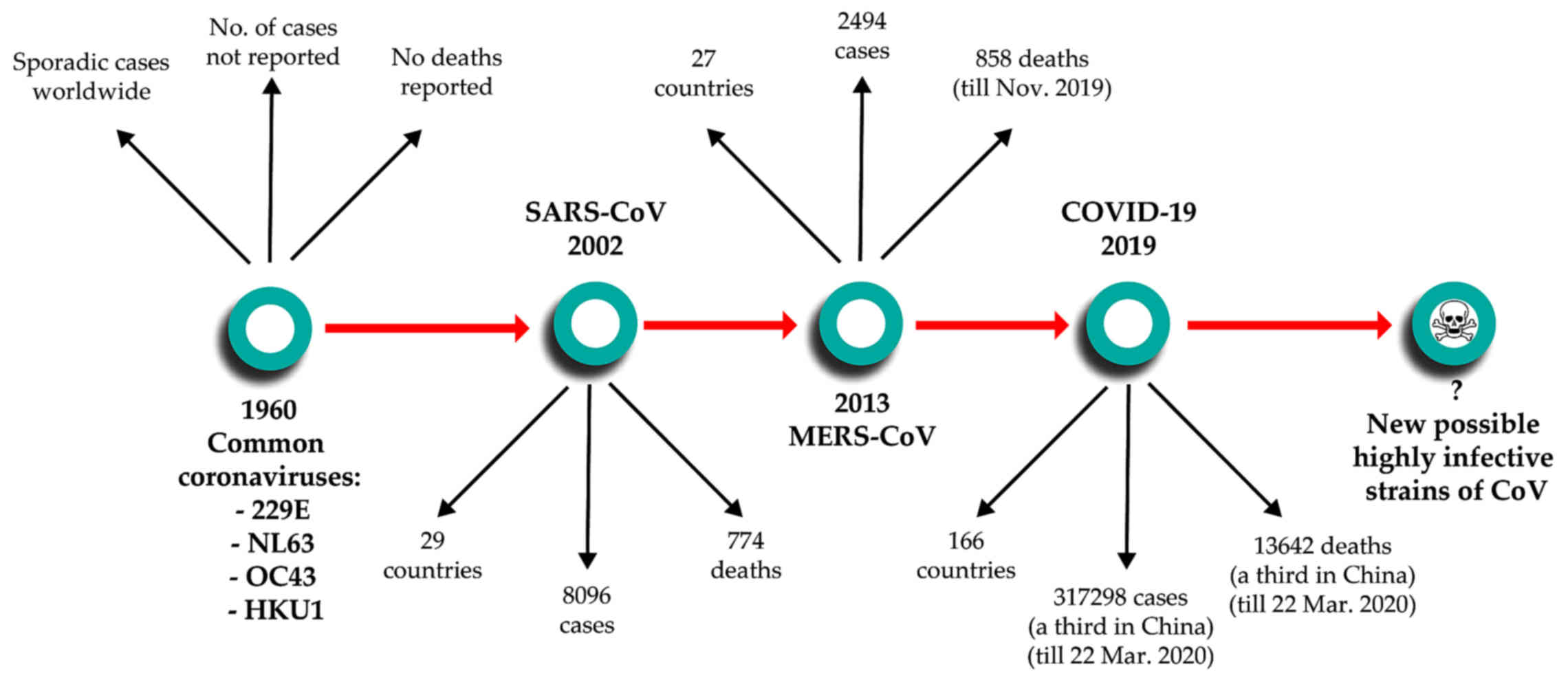
(4,5). The first coronavirus infections were
reported in 1960 as a cause for the common cold. Since then, until
2002, four subtypes of coronaviruses were reported to infect
humans, two α coronaviruses - 229E and NL63 and 2 β coronaviruses -
OC43 and HKU1, which routinely produce noncomplicated infections of
the upper and/or lower respiratory tract (3). Then, 2002 marked a key moment in our
understanding of coronavirus-induced disease with the emergence of
the first lethal severe acute respiratory syndrome (SARS)-causing
coronavirus. Similarly to SARS-CoV-2, the original SARS-CoV emerged
in Guangdong Province in China, spreading through human
transmission chains to infect at least 8,096 individuals in 29
countries and
succumb 774 patients (6). In 2012, a novel β-coronavirus that
had not previously been observed in humans was detected for the
first time in a patient in Saudi Arabia. Since then, the new
coronavirus, which causes Middle Eastern Respiratory Syndrome and
is now known as MERS-CoV, has infected >2,494 individuals across
27 countries and led to the death of at least 858 individuals as of
November 2019, through a series of emergences and re-emergences
from camelid hosts (7). On
December 8, 2019, in Wuhan, Hubei Province, China, the first case
was reported of a new coronavirus that produces pneumonia. Since
then, the new virus first named 2019-nCoV and subsequently renamed
SAS-CoV-2 was identified as a member of the β-coronavirus subtype,
spread rapidly via human-to-human transmission. At the time of
authoring (March 23, 2020), over 317,298 cases have been recorded
worldwide across 166 countries, with over 13,642 deaths attributed
to the virus (8) (Fig. 1).
3. Epidemiology of coronavirus
infections
The emergence of virulence strains of coronavirus
over the past 20 years has increased the interest of scientists
regarding coronavirus strain variability, distribution, and the
ability to be transmitted between animals to humans and further
from humans-to-humans. The strains have slightly different
distributions across sex, age and other demographic attributes. For
example, whilst SARS-CoV-2 affects males more than females with
substantial age-associated mortality increases in frail elderly
subjects, the four societally non-disruptive coronaviruses affect
more females and children between 5 and 14 years (9).
In contrast, SARS-CoV-1 affected more females, and
relatively young people with the median age of SARS infected
patients in Guangzhou province, China at 28.6 years) (10), and mortality rate at ~12% in
people over 65 years (11). In
comparison, the MERS coronavirus was more frequently detected among
males and middle-aged adults aged 50-59 years (12). Emerging data from hospitalized
COVID-19 cases in Wuhan (Hubei province, China) indicate males
(68%) and middle-age adults in the age group of 50-59 years to be
predominantly affected, with a prevalence under 10% in people aged
<39 years. Among these cases, 51% had chronic diseases such as
cardiovascular and cerebrovascular diseases (40%), endocrine system
disease (13%), digestive system disease (11%), respiratory system
disease (1%), malignant tumor (1%), and nervous system disease (1%)
(13). Likewise, data from Italy
as of 15/3/20 indicate a median age of 64 years with middle aged
(51-70 years old) and elderly (75.7%), predominantly male (59.8%)
patients experiencing mild (46.1%) to severe (24.9%) disease and
the burden of mortality (70-79 years old: case fatality ratio
12.5%; 80-89 years old: 19.7%; >90 years old: 22.7%) (14). Whilst the older population
demographics of Italy is believed to drive the higher case fatality
ratio differences (7.9%) vs. China (4.0%) (15), the Italian healthcare system is
experiencing increased requirement for intensive care unit (ICU)
admission (~10% of cases) (16).
Anecdotal reports from frontline clinicians in the principally
affected regions in Italy indicate all ICU beds dedicated to
COVID-19 and patient admission to ICU/mechanic ventilation
restricted to age groups with a higher likelihood of survival. Of
note, these reports indicate 20% of ICU cases to involve
individuals under the age of 65 and as young as 20 years of age.
The reader is strongly advised to consider carefully the
territory-specific diversity of case severity definition as well as
the diagnostic triage/definition criteria before making direct
epidemiological comparisons.Table I
 | Table IThe comparative clinical evolution of
coronavirus infections. |
Table I
The comparative clinical evolution of
coronavirus infections.
| Coronavirus
strain | Incubation
period | Death period | Symptoms |
|---|
| SARS CoV | 4-10 days | 20-25 days | Fever, dry cough,
myalgia, dyspnea, headache, sore throat, sputum production,
rhinorrhea watery diarrhea confusion, poor appetite |
| MERS CoV | 5-6 days | 11-13 days | Myalgia, fever,
chills, malaise associated with confusion, cough, shortness of
breath, dyspnea pneumonia |
| COVID-19 | 3-7 days | 17-24 days | Fever, cough,
dyspnea, muscle ache confusion, headache, sore throat, rhinorrhea,
chest pain, diarrhea, nausea, vomiting, anosmia, dysgeusia |
One additional aspect complicating case
epidemiological oversight pertains to the highly mutagenic nature
of coronaviruses, which is common across RNA viruses, and has led
to inappropriate comparative evolution interpretations and even the
incorrect assertion of mild and sever COVID-19 arising from
separate strains of SARS-CoV-2. By comparing genomes of sequenced
COVID-19 strains, it was shown that synonymous mutations in spike
genes between COVID-19 and rat or bat coronaviruses (RaTG13 and
Bat-SL-CoVZC45) were quite different. The ratio of nucleotide
substitutions to amino acid substitutions was 9.07 in COVID-19
compared with RaTG13, which was significantly higher than the 3.91
ratio from COVID-19 to Bat-SL-CoVZC45 (17). Additionally, there were numerous
concerns that COVID-19 was an engineered bioweapon, hypotheses fed
by speculation that sequences from COVID-19 were identical to those
in HIV. Alignments showed that they are not present in any other
coronavirus strains, but show identity/similarity with sequences in
HIV-1 gp120 and Gag, the former being also a cellular receptor
recognition protein (18).
Considering that these inserts appear in hyper-variable regions of
the protein and are as short as 6 residues in length, it is most
probable that they arose naturally: as a consequence the article
describing these similarities was recently withdrawn from
publication. To date, no evidence supports that SARS-CoV-2 is
man-made: COVID-19 closely resembles two other coronaviruses that
have triggered outbreaks in recent decades, SARS-CoV-1 (79%
sequence identity) and MERS-CoV (51.8% identity) (19), and all three viruses are most
likely to have originated in bats (17,20): SARS-CoV-2 has a 96% sequence
identity with a recently sequenced bat coronavirus recovered from
the wild by random sampling (21).
4. The transmission model
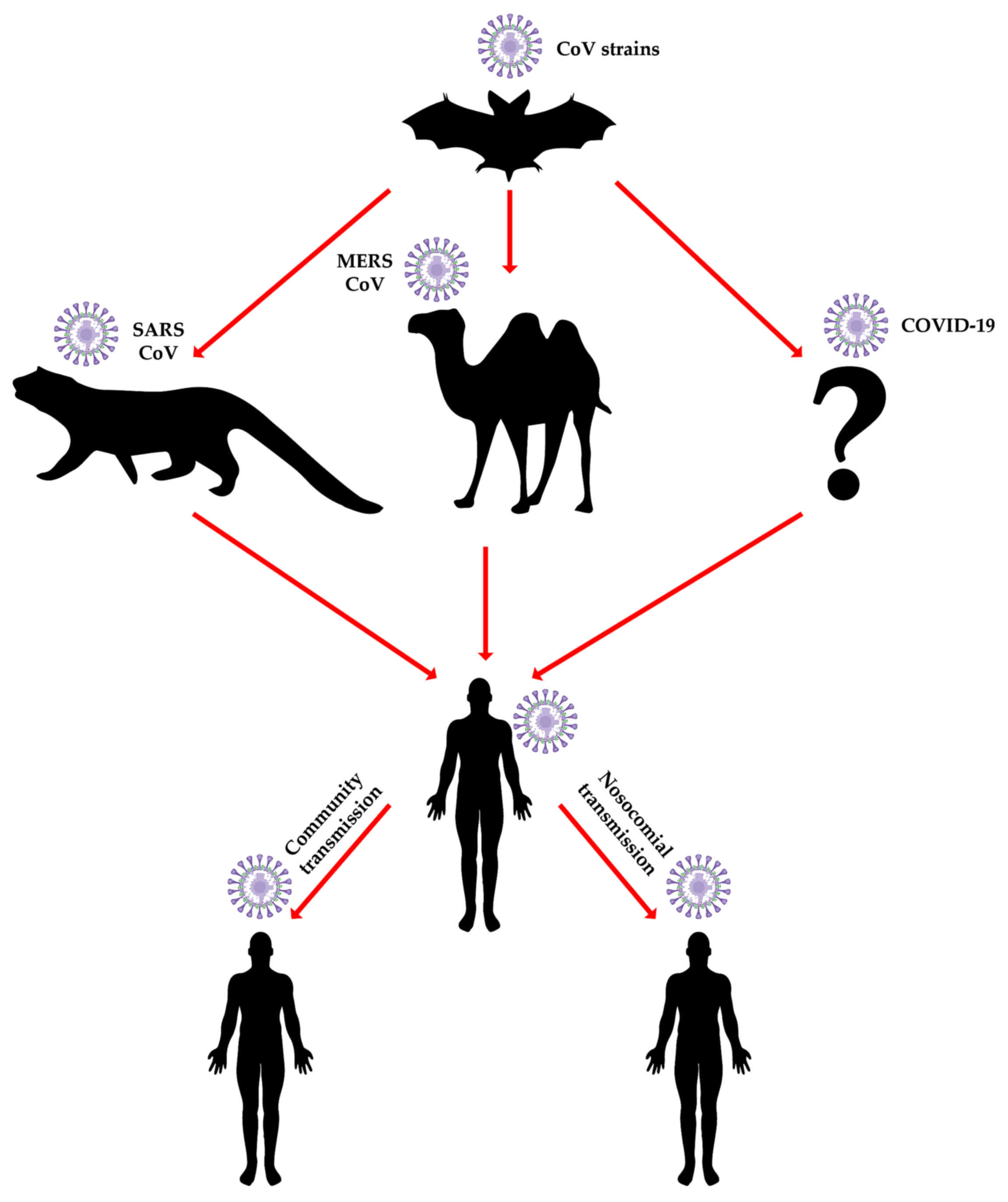
A major question regarding coronavirus epidemiology
is 'Why does the bat of all animals play such a central role in
coronavirus epidemiology?'. Studies in bats have identified viruses
originating in this species, as the potential vector to human
infections. Additionally, as bats live in colonies, they present a
high risk of transmitting the viruses horizontally (intra-species)
which contributes to the vertical (cross-species) spreading
ability. This hypothesis is strongly favored by data on another
high socioeconomic impact bat-derived virus, the Ebola virus, which
was shown to efficiently infect both bat and human cells (22) and to date remains the favored
hypothesis for index case infection in the 2014-16 West African
Ebolavirus disease outbreak.
Additional evidence of bat to human indirect
transmission comes from the COVID-19's Spike protein and ACE2
receptor interactions (23).
Indeed, the two highly pathogenic strains of coronaviruses -
SARS-CoV and MERS-CoV were identified both in bat species and in
animals involved in transmission to humans (20). It is interesting to note that the
first highly pathogenic strain of coronavirus, SARS-CoV-1, has a
low genetic similarity with other known coronaviruses (39% with
bovine coronavirus and 46% with porcine epidemic diarrhea virus)
(24). Recently, three comparison
studies were made with coronavirus strains from pangolins. The
first (February 18, 2020) compared the sequences of COVID-19 with
the coronaviruses in illegally trafficked pangolins to show a
sequence similarity between 85.5 and 92.4% (25), with the subsequent papers
(February 20, 2020), reporting sequence similarities with pangolin
coronaviruses at 90.23% (26) and
91.02% (27), respectively.
However, the α-coronavirus strains that cause human disease were
also originally identified in bats (28); it is therefore not surprising that
COVID-19 shares 96% genetic similarity (whole genome level) with a
coronavirus isolated from Chinese bat species (21). Another important question is 'Why
China?' China is the third-largest country by surface area
worldwide but the most populated country. Its varied land
characteristics and diverse climate range supports huge
biodiversity, which contributes to enabling transmission of viruses
between animal populations. Principally, however, co-habitation of
animals and humans in close proximity, and gastronomic customs
involving the consumption of a variety of exotic animals, including
wild fauna such as bats, increases the chance of vertical
transmission throughout the food supply chain. Officially,
live-bird markets have been closed; however, black-market vendors
run illegal slaughterhouses, that are crowded places with poor
ventilation, where a number of species are hoarded together: These
create ideal conditions for the spreading of the virus through
airborne droplets of blood and other secretions, or shared cages,
trade tools, and utensils, allowing for an 'amplification' of
vertical transmission risk. The prevalence of such culinary and
wild animal handling practices in Southern China have been causally
linked to both the 2002 SARS-CoV-1 outbreak starting from a market
in Shenzen (Guangdong, China) (29) and the entry of SARS-CoV-2 into the
human population. Of the original 41 cases presenting with
pneumonia of unknown origin, two thirds had links to Huanan Seafood
Wholesale Market that also sold live animals (30).
Such a species barrier bridge has been also
documented with filovirus disease outbreaks in Africa, wherein high
risk infectious agents are routinely detected in bushmeat markets
and the associated human population both by PCR and
immunoprecipitation methods (31). As with China, human and animal
close contact and often co-habitation remains common in rural
African areas with practically no barriers to wild environments
(e.g., tropical forests). Such exposure data have fed debates on
the role of so-called 'herd immunity' through natural exposure to
emerging tropical, or perhaps rural, zoonoses. It is noteworthy
that, comparable risks have been documented with viruses not
typically associated with highly biodiverse geographies or the
consumption of wild meat. For example, the 2012 outbreak of
MERS-CoV in the Middle East and the Arab peninsula specifically,
was shown to involve both local and international city dwellers
with no previous exposure to the natural camelid host that
continues to be in close contact with rural populations (Fig. 2) (32,33). The direct impact on individual
health of zoonoses, and indirect impact on health-care systems as
evidenced through the COVID-19 outbreak, underscore how
increasingly urban population organisation even among developing
nations amplifies the risk of onward human transmission, despite
past exposure and indeed immunity in settings with blurred
wild-human habitat barriers.
5. Pathogenesis of coronavirus
infection
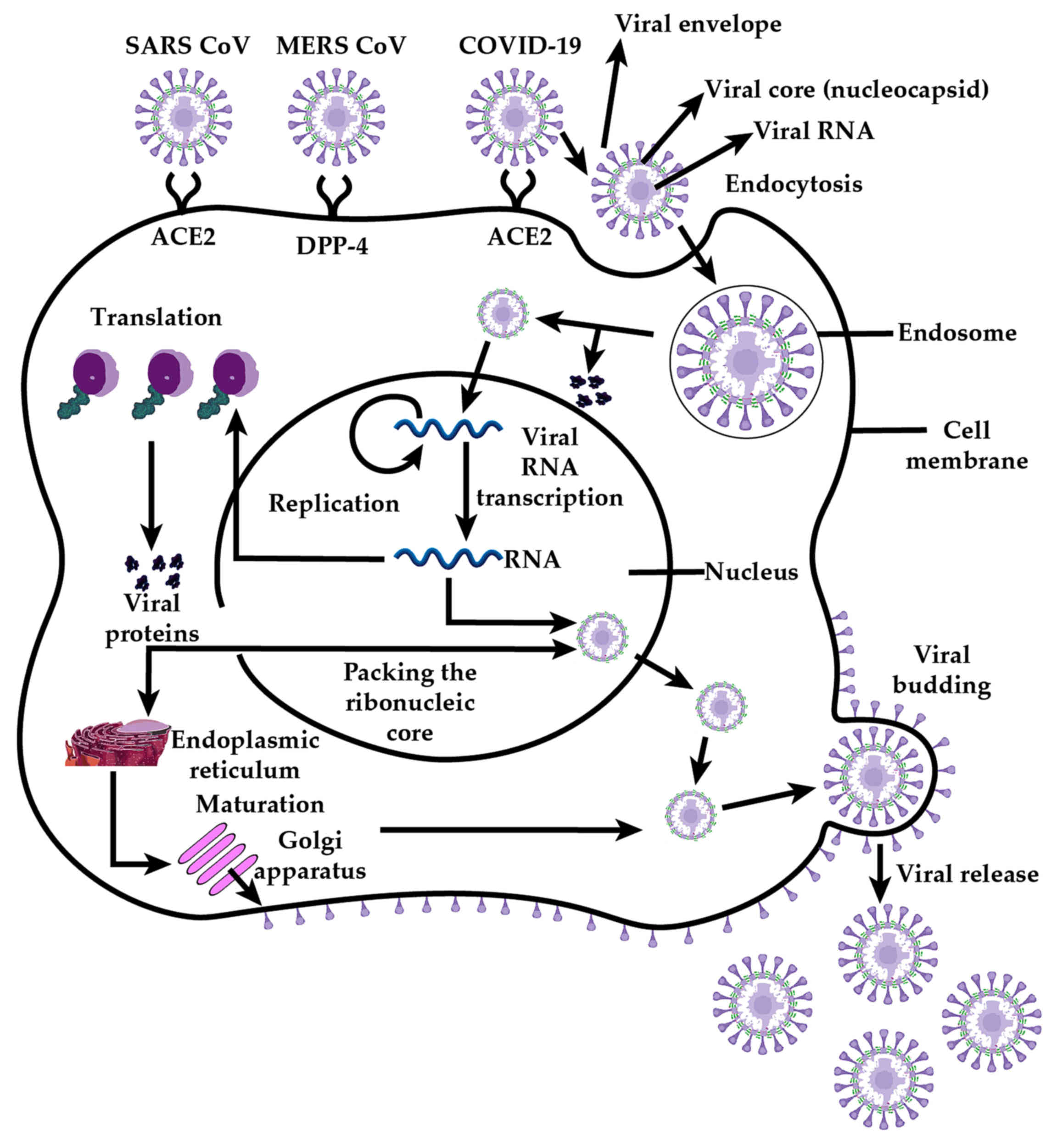
The genome of coronaviruses contains genes for the 4
structural proteins: Envelope (E), membrane (M), nucleocapsid (N)
and spike (S). Coronavirus virions are lipid bilayer-enveloped
particles variable in size (80-160 nm), characterized by multiple
20-nm spike-like extensions on the surface in the form of 'corolla'
or 'flower petals'. At the virion core, a nucleocapsid with
icosahedral symmetry contains an electron-dense layer with a center
that is clear. Its genomic nucleic acid consists of is
single-stranded positive RNA which requires a negative RNA
replication cycle intermediate that generates subgenomic protein
coding RNAs as well as genomic RNA for virion assembly. The core
also features accessory proteins that differ considerably between
various types of coronaviruses.
The lipidic envelope features the
trimerically-organised Type I transmembrane spike glycoproteins
which consist of the ectodomain subunits S1 and S2 protruding
externally, a transmembrane anchor, and a tail extending towards
the viral core (34). Cell
attachment involves the S1 subunit interacting with host cell
surface receptors driving endocytosis and, after membrane fusion
with the involvement of subunit S2 (35), release of the virion core into the
cytosolic milieu. The glycoproteins of coronaviruses mediate
attachment, fusion, and entry in the host cells, but different
parts of those glycoproteins are involved in each of these
processes. These class I viral membrane-fusion proteins undergo
structural rearrangements that produce fusion between the viral and
cellular membranes. Glycoprotein conformational changes and
cathepsin L proteolysis within endosomes are also involved in the
pathogenesis of coronavirus (Fig.
3). Thus, whilst cleavage of S protein is required to expose
the hydrophobic fusion peptide, it seems that receptor interaction
is required for the cleavage to occur (36). Historically, immunotherapies and
many exploratory small molecule treatments have targeted such
host-pathogen interactions to disrupt infection. However,
experience across multiple virus strains indicates such monotherapy
to be inefficient due to mutational escape.
Accessory proteins play a definitive role in
infectivity, however, their functions are not yet fully understood.
It is speculated that some proteins play important roles in
countering the immune response; thus, viruses that lack these have
a lower infectivity. For example, SARS-CoV-1 has two accessory
proteins, 3a and 3b, that play potential roles in the virulence of
this strain (37). On the other
hand, sometimes a complete infectious particle can be assembled
without spike proteins, indicating that accessory proteins can
substitute these (38).
Therefore, there is a potential risk that a vaccine targeting
structural proteins alone, such as the spike protein, might be
inefficient, driving evolutionary escape of the virus, either
through target protein mutation, or by favouring virions utilizing
other accessory proteins for attachment. A some-what more viral
escape-proof approach involves targeting of the receptor-binding
domain (RBD) which is a conserved domain of S protein (39). Notably, other respiratory viruses
that efficiently cross species barriers (e.g., influenza) require
close monitoring and annual adjustment of epitope targeting to
minimize spread and impact.
The primary receptor used by coronaviruses to enter
into target cells is the angiotensin-converting-enzyme II (ACE2)
receptor (21,40), although some strains also use
other alternative receptors, such as CD209L, for which they have a
lower affinity (41). Whilst
there is no evidence in pre- or post-publication literature
currently that SARS-CoV-2 also utilizes CD209L for cell entry this
potential attachment mechanism cannot be excluded until
experimental data thereto are produced. Likewise, other, as yet
unidentified receptors, facilitate coronavirus cellular entry in
the absence of ACE2 in hepatocytes and some enterocytes (42). SARS-CoV-2 in particular, also
appears to use the SARS-CoV receptor ACE2 to enter cells. Although
the spike protein between SARS-CoV-1 and -2 vary in sequence, Xu
et al (43) suggested that
the Spike protein binding affinity for ACE2 is conserved as the 3D
structure of the receptor binding domain is identical with that of
SARS-CoV-1, which would translates to equal infectivity. The
analysis of protein-protein interaction using bioinformatics showed
that SARS nucleocapsidal proteins bind to human cyclophilin A
(hCypA) and this binding was demonstrated by surface plasmon
resonance (SPR) technology. The 3D modelling detected the probable
binding sites and allowed deduction of important interaction
between residue pairs (44). Wan
and colleagues observed that several amino acids in the
receptor-binding motif of SARS-CoV-2 allow binding to human ACE2,
even though with suboptimal strength (45). A recent study demonstrated that a
mutation in the Spike protein (N501T) of SARS-CoV-2 enhanced the
affinity for ACE2 (45).
ACE2 is highly expressed in the respiratory tract,
particularly in epithelial cells of the bronchi, alveoli (both type
I and II cells), trachea and bronchial serous glands, as well as in
macrophages and alveolar monocytes. Notably, the expression in the
lung cells is much higher than in trachea (46). In line with the expression profile
of ACE2, viral genome load has been consistently reported to be
both more elevated in the lower than in the upper respiratory
tract, with lower respiratory specimens being additionally less
prone to false negative results (47,48). ACE2 is also diffusely located on
other cells, such as mucosal cells of the intestines, endothelial
cells of veins and arteries including heart cells, epithelial cells
of the renal tubules, epithelial cells of the kidneys, immune cells
and cerebral neuronal cells, which may also be susceptible to
coronavirus infections. The observation of COVID-19 patient demise
on account of severe heart failure brought about by SARS-CoV-2
infection and the higher risk among patients with previous
cardiovascular and hypertensive disease has driven multiple
hypotheses regarding potential direct mechanisms of viral action on
the circulatory system. Thus, ACE2 expression can be increased as a
result of using drugs such as ACE inhibitors and angiotensin II
type-I receptor blockers (ARBs). Indeed, it was shown that
expression of ACE2 is increased in diabetes patients (another high
risk COVID-19 group) treated with ACE inhibitors and hypertension
patients, treated with ARBs. ACE2 expression can also be increased
by thiazolidinediones and ibuprofen. Therefore, in these categories
of patients, the risk of infection with COVID-19 is proposed to be
higher (49). Yet, whilst the
NL63 coronavirus strain binds to the same ACE2 receptor as
SARS-CoV-2, it produces only upper respiratory tract disease. This
indicates that there are other unknown factors, apart from the
presence of receptors that influence the susceptibility of cells to
coronavirus infection. Insights into the differentiation factors
influencing coronavirus strain-specific outcomes come from recent
data on host co-factors mediating SARS-CoV-2 fusion. Thus,
SARS-CoV-2 uses the serine protease TMPRSS2 for S protein priming;
camostat mesylate, an inhibitor of TMPRSS2, blocked COVID-19
infection of lung cells, but inhibition was more substantial when
the endosomal cysteine protease cathepsin B/L (CatB/L) was also
inhibited e.g., with E-64d (40).
These results point to a complex interplay of host factors in the
endosome with the spike protein in mediating virion fusion. It is
thus speculated that in infected cells furin-mediated precleavage
at the S1/S2 site can promote TMPRSS2-dependent entry, as in the
case of MERS-CoV (40). MERS-CoV
however is an exception to the SARS coronavirus set, as it uses at
hedipeptidyl peptidase 4 (DPP-4) surface antigen as a receptor, and
not ACE2. The relative expression of these endosomal factors and
similarly active proteins across cell types, as well as alternative
receptors that mediate cellular attachment require urgent scrutiny
to understand cell type tropism, and, more crucially,
extra-pulmonary reservoirs and sites of replication. Some of these
extrapulmonary reservoirs have been suggested by CT scans showing
shadows and interstitial changes in tissues separate to the lungs
(50), which require molecular
and cellular studies to confirm direct involvement in virus
replication as opposed to pathological findings secondary to
SARS-CoV-2 infection.
After coronaviruses infect primary cells, the mature
virions can be released and infect other target cells (51). Infective viral particles can be
found in sweat, stool, urine and respiratory secretions from
patients with other coronaviruses, with viable SARS-CoV-2 so far
documented in respiratory droplets, saliva, mechanically generated
aerosols, and faeces. After excretion environmental contamination
can be substantial (fomites) presenting what is currently believed
to be the primary mode of human-to-human transmission for up to 7
days after surface deposition of the virus. The development of
atypical pneumonia with rapid respiratory deterioration and failure
determined by coronavirus infection is associated with increased
levels of activated pro-inflammatory chemokines and cytokines
(52). Thus, it has been proposed
that Vitamin D plays a role in the modulation of the immune
response to infectious agents, based on laboratory findings and
observational studies. However, randomized clinical trials have
returned inconclusive, often controversial results. Therefore, it
has been suggested that cholecalciferol at elevated doses might be
useful for the prevention and therapy of infection with COVID-19
(53,54).
In the pathogeny of coronavirus infection, an
important role is played by the amplitude of host immunity; for
example, canonical interferon levels terminate protein synthesis or
even induce cell death. However, the intensity of the immune
response can vary, depending on other comorbidities of the patient,
explaining the role of these in the evolution of the disease.
Indeed, the majority of deaths have occurred among individuals with
comorbidities (55). In a
nationwide study from China, 2.1% of patients with COVID-19 had
cancer, however no patient had liver transplantation or
inflammatory bowel disease, 7.4% of the patients had diabetes and
15% hypertension (56). The death
rate is expected to be higher in immunocompromised patients,
however this is not established to date. On the other hand, it
seems that the exacerbated immune reaction in COVID-19 infection is
the one that leads to the greatest pulmonary and systemic
damage.
MERS-CoV has a lower infectivity compared to
SARS-CoV-2; thus, an intense and prolonged exposure is necessary in
order for the virus to enter the lungs. MERS-CoV uses DPP-4 as a
cell receptor (57). DPP-4 is
expressed in a wide variety of epithelial cells with localization
in the alveoli, kidneys, liver, small intestine, prostate and
activated leukocytes that determine the higher tropism of MERS-CoV
compared to other coronaviruses (58).
The genome of SARS-CoV-2 was sequenced during the
early stages of the outbreak, enabling the ultrafast development of
point-of-care tests based on RT-PCR for the new COVID-19 infection
within 2 weeks of its discovery (45), in line with past predictions of
rapid response (59) (https://doi.org/10.1039/C7SC03281A). To date,
however, these are available only in kit format for benchtop real
time PCR systems requiring substantial manual processing (5-8 h
minimal data turnaround). Emergency use authorization (EUA) for the
use of these/similar assays in a handful of the numerous
regulator-approved point-of-care integrated
extraction-amplification systems (e.g., BioFire, GeneXpert, etc.)
was obtained only on March 22, 2020, an unacceptable 3 months into
the outbreak. Consequently, despite the tremendous need for mass
screening and isolation of infected cases, sample processing
bottlenecks have forced most health authorities and governments
worldwide to restrict testing only to clinically complex,
hospitalized cases. Conversely, experience from countries with
early outbreak experience such as Korea, Singapore, Hong Kong,
Taiwan, Italy, and indeed China shows that mass testing, case
isolation and contact tracing can reduce human transmission even to
zero. Indeed, presently the risk to these countries appears
restricted to new case introduction from abroad, justifying
measures seeking to halt international travel.
Diagnostic process notwithstanding, a major drawback
lies with the diagnostic sensitivity of preferred as opposed to
available samples. Thus, data from the Chinese Centres for Disease
Control and Prevention, as well as clinical centres at the
epicenter of the outbreak, have reinforced World Health
Organisation technical diagnostic guidelines (60) that require a lower respiratory
sample as well as an upper respiratory sample to rule out
SARS-CoV-19 as the causal agent of viral pneumonia and
COVID-19-like disease. The simplest upper respiratory sample is
presented in the form of an oral and/or nasal swab (or combination
of both, given the handful of reported nasal swab negatives in the
presence of oral swab positives). Lower respiratory specimens are
difficult to obtain as they require either intubated patients
(tracheal aspirates), or anesthesia (bronchoalveolar lavage,
bronchial brushings). Notwithstanding risk of injury to the
patient, these invasive procedures create infectious aerosol
generation risks and are contraindicated. The only suitable
surrogate sample is lung sputum, but only if this is naturally
available, to minimize aerosol generation risk. Unfortunately, the
Chinese experience has been that <30% of COVID-19 suspected
cases generate sputum, thereby restricting testing to oral or nasal
swabs. These, however, have a reported false negative rate of
between 10-37% (47,48), with oral swab false negative rates
on account of low analytical sensitivity reaching up to 68%. These
statistics point underscore the impact of sampling process
variability beyond anatomical differences - one healthcare
professionals' swabbing technique may differ vastly to another- and
emphasize the need for standardizing/automating the sampling
process to increase diagnostic test reliability, even for
respiratory samples. A significant additional complicating factor
is the limited understanding of sample viral load across the
exposure-symptomatic-convalescence continuum, onset of shedding,
and disparity between genome copy detected by RT-PCR vs infectious
virion copy number, which collectively amplify the well-established
issues around operator error in sampling methods. Of interest,
early data suggest viral shed-ding may indeed start as early as one
week ahead of symptoms and transmission may peak up to 3 days
before symptoms (61). The data,
if accepted and validated in separate studies, will compound the
impact of transmission risk attributed to asymptomatic or
subclinical individuals and will emphasize the need for more
aggressive contact tracing. Testing even onto random mass
population screening will be needed to arrest transmission, as
anecdotal evidence suggests from a single town in Italy where the
combination of lockdown, contact tracing and testing eliminated new
cases whilst transmission remained rampant elsewhere in the country
(62).
On the other hand, there is particular importance
attributed by a number of Western governments to serological assays
that monitor the production of antibody responses to pathogens. At
first glance these tests are exceedingly appealing due to their
lower cost (50-100×) and time-to-results (30-48×) as compared to
RT-qPCR in a diagnostic lab, or indeed when using automated point
of care RT-qPCR systems (20× lower cost and 6-12× faster). In
addition, antibody tests can offer a clear answer as to the spread
of exposure that may involve asymptomatic or low-grade respiratory
disease, supporting so called 'herd immunity'. However, evidence
from multiple Asian countries and Italy show transmission
containment was achieved and can be achieved using RT-qPCR which
documents infectious cases: antibody responses do not initiate
until at least a week after the onset of symptoms, meaning that the
deployment of antibody tests for mass screening will have an
enhanced risk of false negatives as compared to oronasal swab
RT-qPCR by missing pre-symptomatic, asymptomatic, and newly
symptomatic cases. Whilst the research and academic value in
understanding asymptomatic spread is undisputed, the magnitude of
the socioeconomic damage by COVID-19 in redirecting resources away
from confirmation of active infection in mildly symptomatic cases
and asymptomatic contacts onto exposure numbers, is likely to
extend transmission long beyond the period of a few weeks
documented in China and Korea into months, extending the impact
onto healthcare systems. Furthermore, such an extension of spread
risks the development of cycles of infection and socioeconomic
damage with the re-introduction of new index cases within country
regions and into countries free of the disease. The increasing risk
of virus evolution, and its establishment as a seasonal, endemic
respiratory disease agent, crucially, does not take into account
how immunity to other coronaviruses appears to be short-lived: we
simply do not know yet if COVID-19 convalescence will protect from
re-infection, and if it does, for how long.
6. Clinical manifestations of
coronavirus-associated diseases
The predominant symptoms of SARS-CoV-1 are
respiratory in nature, associated with fever and diarrhea. Clinical
manifestations among patients are diverse, from mild-moderate to
severe-life threatening symptoms. The virus has a mean incubation
period of 4.6 days (63), however
in the majority of cases, symptoms appear after 10 days (64). In severe cases, the median time
from the appearance of symptoms until artificial ventilation is 11
days, and 23.7 days until the patient's death (65). The clinical manifestations are
similar to flu-like symptoms, such as fever, a dry cough, myalgia,
dyspnea, headache, sore throat, sputum production, rhinorrhea
(66,67). In 40-70% of cases, watery diarrhea
appears within 1 week after the first manifestations (42). In the elderly, the symptoms can
also be associated with confusion, a poor appetite and a decrease
in general well-being. In is noteworthy that the symptomatology in
children below 12 years is mild; no mortality has been registered
among children and teenagers. Asymptomatic cases are reported to be
rare (63,68). Increased levels of C-reactive
protein, lymphopenia, thrombocytopenia, and increased levels of
lactate dehydrogenase have been observed in laboratory tests. The
infection manifests in two different stages, with the first-week
characterized by flu-like symptoms that improve even if the viral
loads persist, and a recurrent period during the second week in
which respiratory failure can appear and more than 20% of patients
may require mechanic ventilation (69). The respiratory system is the main
affected area in the human body where the infection determines the
appearance of diffuse alveolar damage, squamous metaplasia,
giant-cell infiltrates and increased macrophage levels in the
interstitium and the alveoli (46).
Unlike SARS-CoV-1, Middle East Respiratory Syndrome
coronavirus (MERS-CoV) has a wide variety of clinical
manifestations from asymptomatic to severe respiratory symptoms.
The mean incubation period is 5-6 days (70-73). In severe cases, death can occur
within 11-13 days after the onset of the disease (70,73). The clinical manifestations are
similar to flu-like symptoms, such as general myalgia, fever,
chills, malaise associated with confusion, pulmonary symptoms, such
as cough, shortness of breath, dyspnea and pneumonia.
Extra-pulmonary clinical manifestations include abdominal
disorders, nausea, diarrhea, vomiting, acute renal failure and
neurological complications (74).
The severity of the disease is associated with an increasing age,
and with individuals with comorbidities, such as diabetes mellitus,
renal and pulmonary diseases. Death can occur after the development
of acute respiratory distress syndrome (ARDS), multiorgan failure
or septic shock (7). The
neurological complications of the infection with MERS-CoV are
associated with the in vitro ability of the virus to invade
the central nervous system (75),
although it was not detected in the CSF. Among the neurological
manifestations, confusion has been reported in 25.7% and seizures
in 8.6% of cases (76). Severe
neurological complications are encephalitis, stroke,
polyneuropathy, acute disseminated encephalomyelitis, Guillain
Barré syndrome and Bickerstaff's encephalitis. Usually, the
neurological complications appear at a delayed time point from the
respiratory symptoms, namely 2-3 weeks from the onset of the
clinical manifestation of the disease and can be underdiagnosed
(74,77-79). Children are rarely affected
(74). In laboratory tests,
thrombocytopenia, lymphopenia, neutrophilia, leucocytosis, reduced
renal functions and increased inflammatory markers have been
observed. Similar to SARS-CoV-1, the pathogenesis has not yet been
fully elucidated. Viral-mediated lung damage is characterized by
diffuse alveolar damage. The virus has been determined in
multinucleated epithelial cells, pneumocytes and bronchial
submucosal glands. Apart from the respiratory system, the virus has
also been identified in the epithelial cells of the proximal renal
tubules (58). Acute tubular
sclerosis and tubulointerstitial nephritis have been observed in
renal biopsies collected from persons infected with MERS-CoV
(80).
The latest high clinical burden coronavirus,
SARS-CoV-2, presents a wide variety of clinical manifestations
which are not yet fully characterized (81,82). The virus has a mean incubation
period of 3-7 days, and no more than 14 days (30) - although anecdotal reports likely
related to poor contact tracing have suggested potentially up to 24
days of incubation. Outside Hubei province, the median incubation
period has been estimated to be 5.1 days (95% CI: 4.5 to 5.8), with
97.% of infected subjects developing symptoms within 11.5 days of
infection (83). In a
retrospective study on 99 patients that have confirmed COVID-19
pneumonia treated in an infec-tious diseases hospital in Wuhan,
China, it was shown that the main clinical manifestations were
flu-like symptoms, such as fever and cough (83 and 81% of the
cases), shortness of breath (31% of cases), muscle ache (11% of
cases), and less frequently confusion (9%), headache (8%), sore
throat, rhinorrhea, and chest pain. In less than 2% of patients,
extra-pulmonary symptoms and diarrhea, nausea and vomiting were
also observed. As complications, ARDS was reported in 17% of cases
followed by acute respiratory injury (8%), septic shock (4%), acute
renal injury (3%), and ventilator-associated pneumonia (1%). Death
occurred in 11% of participants in the study determined by multiple
organ failure. The main finding in laboratory tests was
lymphopenia. The severity of the disease was associated with
co-infections, smoking history, hypertension and age (13). Another retrospective study on 137
patients infected with COVID-19 revealed the same pattern of
clinical manifestations with fever, cough and muscle pain, and less
frequently diarrhea, headache and even heart palpitations in some
cases (50). Lymphopenia was
observed in 72.3% of cases associated with normal or decreased
white blood cell counts (50).
The lung is the main organ affected. In the imaging examination by
computer tomography or X-ray, lesions were identified in multiple
lung lobes presented as patchy/punctate ground-glass opacities
(GGO), patchy consolidation, irregular solid nodules and fibrous
stripes (50,84-86). Studies that have investigated the
pulmonary pattern of COVID-19 pneumonia have demonstrated that the
disease progresses rapidly, and CT re-examination after 3-14 days
has revealed significant changes in lung structure. The single GGO
observed in the early stages can increase significantly in
short-term re-examination. The same has also occurred with fibrous
stripes observed in the early stages that consolidate in short-term
re-examination. In some cases, the irregular solid nodules observed
in the early stages increased and merged in short-term
re-examination. Short-term repeated imaging scans should be carried
out in patients with COVID-19 pneumonia for monitoring the
evolution and for specific management of the patient as the disease
can rapidly progress or indeed resolve (86,87). The peak of lung abnormalities was
reached on day 10 following the onset of symptoms, followed by a
decrease in symptomatology (87).
It was documented that in one third of the severely
ill patients, death occurs because the COVID-19 coronavirus produce
acute myocardial injury and damage to the cardiovas-cular system in
general, that further degrade the state of these already very ill
patients (88).
7. Treatment of coronavirus-associated
diseases
The treatment for all types of coronaviruses is
mainly supportive, as no specific treatment has been discovered to
date. For the treatment of SARS-CoV-1, apart from supportive
measures, several therapeutic schemes have been implemented, with
varying success rates such as antiviral therapy with riba-virin
(89), protease inhibitors -
lopinavir and ritonavir, alone or associated with ribavirin
(67,90), interferon-alfacon-1 (91), systemic corticosteroids (89,92), and convalescent plasma for passive
immunotherapy (93). Apart from
convalescent plasma, none of these treatments have exhibited
significant benefits compared to the side-effects produced.
For MERS-CoV infections, taking as an example the
early SARS-CoV-1 epidemic, supportive care was primarily used, and
in some instances broad-spectrum antibacterials, antivirals such as
ribavirin alone or in combination with interferon-2α2b (94,95) and antifungals have been used to
prevent the co-infection with other opportunistic pathogens
(96,97). Mycophenolic acid is another drug
that exhibited efficacy in monotherapy on a small number of
patients; however further studies are required to confirm these
effects (98,99).
In the case of the new COVID-19-associated
pneumonia, treatment also focuses on supportive care measures and
symptomatic treatment. Following the example of SARS-CoV-1
treatment, the administration of systemic corticosteroids was
attempted, although this did not exhibit any benefits. In the case
of patients with severe evolution, immunoglobulin G was
administered (50). The
application of early respiratory support improved the prognosis and
the recovery of the patients (50). In a 54-year-old Korean man
infected with SARS-CoV-2, the administration of the combination
lopinavir/ritonavir from day 10 of illness determined the decrease
in viral load till no detectable levels (100). The question of whether the
decrease in viral load was associated with the natural course of
the healing process or with antiviral treatment or both, is being
currently investigated in order to confirm the efficacy of
antiviral treatment for COVID-19. The efficacy of remdesivir
(Gilead Sciences), a nucleotide analog, in inhibiting in
vitro and in vivo SARS-CoV-1, MERS-CoV and bat CoV
strains that are capable to replicate in human airway epithelial
cells render it a good choice for testing on newly affected
COVID-19 patients (85,101-103). Taking into consideration that
COVID-19 binds to human ACE2 in order to penetrate human cells and
to induce severe pneumonia, the renin-angiotensin system, ACEI
(angiotensin-converting enzyme inhibitors) and AT1R (angiotensin II
type-I receptor) inhibitors have been suggested to modify
individual susceptibility to COVID-19 by influencing SARS-Cov-2
virulence. However, further studies are needed to confirm the
mechanisms involved (including the role of high ACE2 expression)
and to consider possible changes in administration and dosage of
antihypertensive drugs (46,49,104). Thus far, the Professional
Association for Hypertension Therapy, USA, has warned against
changes in hypertensive drugs following concerns about a possible
increased susceptibility to COVID-19 (https://ish-world.com/news/a/A-statement-from-the-International-Society-of-Hypertension-on-COVID-19/).
Interest has arisen also on the 'old', off-patent antimalarial drug
chloroquine and its less toxic hydroxychloroquine analog, both of
which are quinine derivatives (cinchoca tree bark, South America).
Preliminary support from observations of potential inhibitory
activity against SARS-CoV-2 (105,106), clearly needs to be confirmed
through randomized controlled trials. There are also ongoing trials
using monoclonal antibodies. One of them is tocilizumab, an
anti-IL6 antibody, currently in phase III clinical trials, with
commercial name Actemra produced by Roche (107). Sarilumab, a drug used in
rheumatoid arthritis is about to enter clinical trials for COVID-19
infection, according to Regeneron Pharmaceuticals and Sanofi,
respectively (108).
In order to facilitate COVID-19 virus replication,
certain in vivo conditions, such as the pH value, the
temperature, humidity, and the sufficiency of oxygen supply should
be ensured. Consequently, in order to hinder the reproduction
procedures of the virus, it would be expedient to disturb the
environment of the virus, by interfering with the temperature or
the pH values or both. SARS-CoV-2 has been speculated to be a
seasonal virus, in which case a hindrance of the rapid spread that
characterizes it at ambient temperatures over 25°-30° Celsius would
be observed. Unfortunately, the sup-tropical/arid environments
favored by SARS-CoV-1 and MERS-CoV, as well as the comparatively
higher temperatures in Israel, Egypt, the Arab peninsula and now
sub-Saharan Africa compared to Hubei province indicate that
temperature probably does not affect COVID-19 replication.
Likewise, the ambient humidity levels thought to impact aerosol and
droplet suspension time/travel distance, and by extension virus
transmission, do not seem to affect onward transmission chains
across geographical regions. Alteration of the pH value inside the
human body within tolerable levels however is achievable; such an
intervention would not only suppress replication of the virus, but
also may have - although possibly weak - virucidal effect. A pH
value change with no negative impact on the health of the patient
could be achieved by administering a drug that would lead to urine
acidosis and endocellular alkalosis. This drug could be a simple
small molecule with hydrophobic weak base properties, which would
accumulate in lysosomes after having crossed both plasma and
lysosomal membranes by diffusion. Perhaps such a medicine may
express both a therapeutic and a prophylactic effect by increasing
the environmental barriers to effective virus attachment in early
endosomal pathways, and preventing spike proteolysis required for
capsid release and cell membrane fusion within lysosomes, in line
with cathepsin protease-targeted therapies. Given the success of
combination therapy strategies in other antiviral diseases centered
on virus fusion and/or replication, promoting endocellular
alkalosis during antiviral drug therapy (e.g., replication
inhibitors) could amplify antiviral effect and thereby contribute
to the reduction of the treatment period (109-113).
8. Prevention strategies
Although since 2004, the World Health Organization
issued guidelines for the prevention of re-emergence of new
coronavirus virulent strains (114), implementation has evidently been
inadequate. Key recommendations include: the availability of the
early detection of infected individuals to prevent the spread of
emergent and re-emergent infection, particularly to prevent
international spread; the development of contingency plans by each
country for the management and detection of coronavirus,
individualized by the risks that exist locally; preparing global
and assisting national risk assessments; updating new discoveries;
assisting countries in their efforts to improve protection against
infections; assessing the availability of all necessary resources
in the case of an outbreak; and, supporting the international
scientific collaboration for SARS study. Unfortunately, all of
these strategies could not prevent the current outbreak, and after
17 years, the medical, scientific and governmental structures have
demonstrated that they are not adequately prepared to effectively
contain highly pathogenic viruses. The main measures that should be
taken for preventing the dissemination of pathogens in the general
population are associated with infection control measures both in
healthcare facilities and in the community. Contact tracing and
quarantine or the isolation of those suspected of infection along
with the education of the population has proven essential in
preventing community transmission of the viruses both in developing
nations (West African and Democratic Republic of Congo Ebola virus
outbreaks), developed nations (e.g., Hong Kong, South Korea SARS
and MERS outbreaks), and now practically all G20 nations. The lack
of local implementation of what are demonstrably effective
measures, as learned since the SARS-CoV-1 outbreak due to
scientific chauvinism over healthcare standards between East and
West, population anthropology between Africa and Europe in terms of
communication strategy and behaviour, or indeed local vs
international experience between e.g., the UK and Italy with
emerging threats is lamentable. Going forward, the identification
of natural reservoirs and intermediate hosts for existing and
future risks as identifiable through random sampling and monitoring
(21) will become essential in
preventing future zoonotic virus outbreaks. Unfortunately, these
alarms were raised repeatedly over the past two decades with
minimal heed: perhaps the damage brought by COVID-19 onto global
economy and healthcare systems will transform the field of tropical
emergent disease.
A good example of positive action are the actions of
currently more than 40 teams developing vaccines against SARS-CoV-2
infection (115); interest is
largely spurred by the unprecedented success and rapid development
of Ebolavirus vaccines during the West African outbreak. COVID-19
vaccines can be based on the viral RNA or derivatives: Examples
include Innovio and Moderna in USA, as well as CureVac (Germany),
and another team at Imperial College in London. RNA and DNA
vaccines have the advantage of being quick to develop and high
likelihood of safety, though no RNA vaccine or therapeutic
currently enjoys regulatory approval. The Institute Pasteur in
France is working on a traditional COVID-19 vaccine based on a SARS
vaccine. CanSino Biologics developed a recombinant coronavirus
vaccine incorporating adenovirus type 5 vector (Ad5). To date
clinical trials have been initiated by Moderna Therapeutics and
Cansino Biologics (115) with
further trials shortly anticipated in the UK and other European
countries.
A sobering note, however, is that despite the ample
amounts of research performed on coronaviruses over the past 20
years, particularly following the SARS-CoV-1 outbreak, no vaccine
is unfortunately yet available for either SARS-CoV-1 or MERS,
although some potential vaccines have reached as far as phase I
clinical trials (116-118). Some of the strategies used for
vaccines are eliciting neutralizing-antibody for S proteins and
T-cell responses (119) which
seem to be jointly required for convalescence, rather than simply
neutralizing antibody titers. The quest for protective immunity to
coronavirus is but nascent, and virology offers a plethora of high
socioeconomic impact, high prevalence RNA viruses for whom decades
of vaccine research have yet to deliver success.
9. Conclusions
The Guangdong region of China was considered a
high-risk area for a coronavirus outbreak after the SARS-CoV-1
epidemic in 2002. Nevertheless, although a large amount of research
and efforts have been made worldwide to prevent further outbreaks,
a coronavirus outbreak occurred again in 2019. Unfortunately,
healthcare systems locally, nationally, and now internationally
have again been overcome, as with the other two major coronavirus
outbreaks. The evidence from COVID-19 is clear that highly
aggressive measures are necessary to break the transmission chain
in the regions at risk - practically most countries globally at the
time of authoring. Unfortunately, there is no specific prevention
strategy against the new coronavirus, and its pandemic status
restrict the global community to two options: effective measures
for the immediate prevention of respiratory infections with
predictable and measurable consequences, or ambitions of 'herd' or
vaccine-derived immunity at a non-specific time in the future. We
can only hope that after this third significant outbreak,
sufficient experience has been gained; a joint effort of the
worldwide health authorities will be needed in extinguishing the
current epidemic. It can only be trusted that this experience will
re-emphasize the need for prevention and readiness and for further
studies to limit the transmission of animal viruses to humans and
mitigation should it arise again in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
Conceptualization, AOD, DC, DT, MV, MA, JMD, MG,
VAT, GGO and DAS; validation, research, resources, data reviewing,
and writing AOD, DA, OZ, OC, DT, ND, SAM, MG, VAT, GGO; review and
editing, AOD, AT, DAS, MV, SAM, JMD, MA, ND and DC. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
References
|
1
|
Groot RJ, Baker SC, Baric R, et al: Family
- Coronaviridae. Virus Taxonomy Ninth Report of the International
Committee on Taxonomy of Viruses King AMQ. Lefkowitz EJ, Adams MJ
and Carstens EB: Elsevier; pp. 806–828. 2011
|
|
2
|
van Doremalen N, Bushmaker T, Morris DH,
et al: Aerosol and Surface Stability of SARS-CoV-2 as Compared with
SARS-CoV-1. N Engl J Med. Mar 17–2020.Epub ahead of print.
View Article : Google Scholar :
|
|
3
|
Geller C, Varbanov M and Duval RE: Human
coronaviruses: Insights into environmental resistance and its
influence on the development of new antiseptic strategies. Viruses.
4:3044–3068. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anthony SJ, Johnson CK, Greig DJ, Kramer
S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, et al:
PREDICT Consortium: Global patterns in coronavirus diversity. Virus
Evol. 3:vex0122017. View Article : Google Scholar
|
|
5
|
Goumenou M, Spandidos DA and Tsatsakis A:
[Editorial] Possibility of transmission through dogs being a
contributing factor to the extreme Covid 19 outbreak in North
Italy. Mol Med Rep. In Press.
|
|
6
|
World Health Organization: Summary of
probable SARS cases with onset of illness from 1 November 2002 to
31 July 2003. https://www.who.int/csr/sars/country/table2004_04_21/en/.
Accesed July 24, 2015.
|
|
7
|
World Health Organization: Middle East
respiratory syndrome coronavirus (MERS-CoV). MERS Monthly Summary.
2013, https://www.who.int/emergencies/mers-cov/en/.
Accessed July 9, 2013.
|
|
8
|
World Health Organization: Coronavir us
disease (COVID-2019) situation reports. https://www.who.int/emer-gencies/diseases/novel-coronavirus-2019/situation-reports.
|
|
9
|
Movert E, Wu Y, Lambeau G, Kahn F, Touqui
L and Areschoug T: Using Patient Pathways to Accelerate the Drive
to Ending Tuberculosis. J Infect Dis. 208:2025–2035. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chan-Yeung M and Xu RH: SARS:
Epidemiology. Respirology. 8(Suppl): S9–S14. 2003. View Article : Google Scholar
|
|
11
|
Xu RH, He JF, Evans MR, Peng GW, Field HE,
Yu DW, Lee CK, Luo HM, Lin WS, Lin P, et al: Epidemiologic clues to
SARS origin in China. Emerg Infect Dis. 10:1030–1037. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mobaraki K and Ahmadzadeh J: Current
epidemiological status of Middle East respiratory syndrome
coronavirus in the world from 1.1.2017 to 17.1.2018: A
cross-sectional study. BMC Infect Dis. 19:3512019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen N, Zhou M, Dong X, et al:
Epidemiological and clinical characteristics of 99 cases of 2019
novel coronavirus pneumonia in Wuhan, China: a descriptive study.
Lancet. 395:507–513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livingston E and Bucher K: Coronavirus
Disease 2019 (COVID-19) in Italy. JAMA. Mar 17–2020.Epub ahead of
print. View Article : Google Scholar
|
|
15
|
Novel coronavirus (COVID-19) situation.
https://experience.arcgis.com/experience/685d0ace521648f8a5beeeee1b9125cd.
|
|
16
|
Remuzzi A and Remuzzi G: COVID-19 and
Italy: what next? Lancet. Mar 13–2020.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv L, Li G, Chen J, Liang X and Li Y:
Comparative genomic analysis revealed specific mutation pattern
between human coronavirus SARS-CoV-2 and Bat-SARSr-CoV RaTG13.
bioRxiv. In Press.
|
|
18
|
Pradhan P, Pandey AK, Mishra A, et al:
Uncanny similarity of unique inserts in the 2019-nCoV spike protein
to HIV-1 gp120 and Gag. bioRxiv. In Press.
|
|
19
|
Ren LL, Wang YM, Wu ZQ, et al:
Identification of a novel coro-navirus causing severe pneumonia in
human: a descriptive study. Chin Med J. Feb 11–2020.Epub ahead of
print. View Article : Google Scholar
|
|
20
|
Fan Y, Zhao K, Shi ZL and Zhou P: Bat
Coronaviruses in China. Viruses. 11:2102019. View Article : Google Scholar :
|
|
21
|
Zhou P, Yang XL, Wang XG, Hu B, Zhang L,
Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al: A pneumonia outbreak
associated with a new coronavirus of probable bat origin. Nature.
579:270–273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Leroy EM, Rouquet P, Formenty P, et al:
Multiple Ebola Virus Transmission Events and Rapid Decline of
Central African Wildlife. Science. 303:387–390. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li W, Wong SK, Li F, Kuhn JH, Huang IC,
Choe H and Farzan M: Animal origins of the severe acute respiratory
syndrome coronavirus: Insight from ACE2-S-protein interactions. J
Virol. 80:4211–4219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Drosten C, Günther S, Preiser W, et al:
Identification of a novel coronavirus in patients with severe acute
respiratory syndrome. N Engl J Med. 348:1967–1976. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lam TTY, Shum MHH, Zhu HC, et al:
Identification of 2019-nCoV related coronaviruses in Malayan
pangolins in southern China. bioRxiv. In Press.
|
|
26
|
Liu P, Jiang JZ, Hua Y, et al: Are
pangolins the intermediate host of the 2019 novel coronavirus
(2019-nCoV)? bioRxiv. In Press.
|
|
27
|
Zhang T, Wu Q and Zhang Z: Pangolin
homology associated with 2019-nCoV. bioRxiv. In Press.
|
|
28
|
Wang LF and Cowled C: Bats and Viruses: A
New Frontier of Emerging Infectious Diseases. John Wiley & Sons
Inc; 2015, https://doi.org/10.1002/9781118818824.
View Article : Google Scholar
|
|
29
|
Guan Y, Zheng BJ, He YQ, et al: Isolation
and characterization of viruses related to the SARS coronavirus
from animals in Southern China. Science. 302:276–278. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang C, Wang Y, Li X, et al: Clinical
features of patients infected with 2019 novel coronavirus in Wuhan,
China. Lancet. 395:497–506. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smiley Evans T, Tutaryebwa L, Gilardi KV,
Barry PA, Marzi A, Eberhardt M, Ssebide B, Cranfield MR, Mugisha O,
Mugisha E, et al: Suspected Exposure to Filoviruses Among People
Contacting Wildlife in Southwestern Uganda. J Infect Dis. 218(Suppl
5): S277–S286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Azhar EI, El-Kafrawy SA, Farraj SA, Hassan
AM, Al-Saeed MS, Hashem AM and Madani TA: Evidence for
camel-to-human transmission of MERS coronavirus. N Engl J Med.
370:2499–2505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Haagmans BL, Al Dhahiry SH, Reusken CB, et
al: Middle East respiratory syndrome coronavirus in dromedary
camels: an outbreak investigation. Lancet Infect Dis. 14:140–145.
2014. View Article : Google Scholar
|
|
34
|
Du L, He Y, Zhou Y, Liu S, Zheng BJ and
Jiang S: The spike protein of SARS-CoV - a target for vaccine and
therapeutic development. Nat Rev Microbiol. 7:226–236. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li F: Evidence for a common evolutionary
origin of coronavirus spike protein receptor-binding subunits. J
Virol. 86:2856–2858. 2012. View Article : Google Scholar :
|
|
36
|
Simmons G, Gosalia DN, Rennekamp AJ,
Reeves JD, Diamond SL and Bates P: Inhibitors of cathepsin L
prevent severe acute respiratory syndrome coronavirus entry. Proc
Natl Acad Sci U S A. 102:11876–11881. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Narayanan K, Huang C and Makino S: SARS
coronavirus accessory proteins. Virus Res. 133:113–121. 2008.
View Article : Google Scholar
|
|
38
|
Schoeman D and Fielding BC: Coronavirus
envelope protein: Current knowledge. Virol J. 16:692019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu C, Zhou Q, Li Y, et al: Research and
Development on Therapeutic Agents and Vaccines for COVID-19 and
Related Human Coronavirus Diseases. ACS Cent Sci. 6:315–331. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hoffmann M, Kleine-Weber H, Schroeder S,
et al: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is
Blocked by a Clinically Proven Protease Inhibitor Article
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by
a Clinically Proven Protease Inhibitor. Cell. 181:1–10. 2020.
View Article : Google Scholar
|
|
41
|
Jeffers SA, Tusell SM, Gillim-Ross L, et
al: CD209L (L-SIGN) is a receptor for severe acute respiratory
syndrome coronavirus. Proc Natl Acad Sci USA. 101:15748–15753.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Leung WK, To KF, Chan PK, et al: Enteric
involvement of severe acute respiratory syndrome-associated
coronavirus infection. Gastroenterology. 125:1011–1017. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu X, Chen P, Wang J, Feng J, Zhou H, Li
X, Zhong W and Hao P: Evolution of the novel coronavirus from the
ongoing Wuhan outbreak and modeling of its spike protein for risk
of human transmission. Sci China Life Sci. 63:457–460. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo C, Luo H, Zheng S, Gui C, Yue L, Yu C,
Sun T, He P, Chen J, Shen J, et al: Nucleocapsid protein of SARS
coronavirus tightly binds to human cyclophilin A. Biochem Biophys
Res Commun. 321:557–565. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang C, Horby PW, Hayden FG and Gao GF: A
novel coronavirus outbreak of global health concern. Lancet.
395:470–473. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kuba K, Imai Y, Rao S, et al: A crucial
role of angiotensin converting enzyme 2 (ACE2) in SARS
coronavirus-induced lung injury. Nat Med. 11:875–879. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fred Hutchinson Cancer Research Center:
INTERIM guidelines for COVID-19 management in hematopoietic cell
transplant and cellular therapy patients, Version 1, 2020.
https://www.fredhutch.org/content/dam/www/coronavirus/COVID-19_Interim_Patient_Guidelines_3_9_20.pdf.
Accessed March 8 2020.
|
|
48
|
Wang W, Xu Y, Gao R, Lu R, Han K, Wu G and
Tan W: Detection of SARS-CoV-2 in Different Types of Clinical
Specimens. JAMA. Mar 11–2020.Epub ahead of print. View Article : Google Scholar
|
|
49
|
Fang L, Karakiulakis G and Roth M: Are
patients with hyper-tension and diabetes mellitus at increased risk
for COVID-19 infection? Lancet Respir Med. 2600:301162020.
|
|
50
|
Kui L, Fang YY, Deng Y, et al: Clinical
characteristics of novel coronavirus cases in tertiary hospitals in
Hubei Province. Chin Med J (Engl). Feb 7–2020.Epub ahead of
print.
|
|
51
|
Qinfen Z, Jinming C, Xiaojun H, et al: The
life cycle of SARS coronavirus in Vero E6 cells. J Med Virol.
73:332-7. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ding Y, He L, Zhang Q, et al: Organ
distribution of severe acute respiratory syndrome (SARS) associated
coronavirus (SARS-CoV) in SARS patients: implications for
pathogenesis and virus transmission pathways. J Pathol.
203:622–630. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Watkins J: Preventing a covid-19 pandemic.
BMJ. 368:m8102020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gruber-Bzura BM: Vitamin D and
Influenza-Prevention or Therapy? Int J Mol Sci. 19:24192018.
View Article : Google Scholar
|
|
55
|
Lau EH, Hsiung CA, Cowling BJ, Chen CH, Ho
LM, Tsang T, Chang CW, Donnelly CA and Leung GM: A comparative
epidemiologic analysis of SARS in Hong Kong, Beijing and Taiwan.
BMC Infect Dis. 10:502010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guan W, Ni Z, Hu Y, et al: Clinical
Characteristics of Coronavirus Disease 2019 in China. N Engl J Med.
Feb 28–2020.Epub ahead of print. View Article : Google Scholar
|
|
57
|
Ng DL, Al Hosani F, Keating MK, et al:
Clinicopathologic, Immunohistochemical, and Ultrastructural
Findings of a Fatal Case of Middle East Respiratory Syndrome
Coronavirus Infection in the United Arab Emirates. Am J Pathol.
186:652–658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Alsaad KO, Hajeer AH, Al Balwi M, et al:
Histopathology of Middle East respiratory syndrome coronovirus
(MERS-CoV) infection - clinicopathological and ultrastructural
study. Histopathology. 72:516–524. 2018. View Article : Google Scholar
|
|
59
|
Shah K, Bentley E, Tyler A, Richards KSR,
Wright E, Easterbrook L, Lee D, Cleaver C, Usher L, Burton JE, et
al: Field-deployable, quantitative, rapid identification of active
Ebola virus infection in unprocessed blood. Chem Sci (Camb).
8:7780–7797. 2017. View Article : Google Scholar
|
|
60
|
World Health Organization: Country &
Technical Guidance - Coronavirus disease (COVID-19). https://www.who.int/emer-gencies/diseases/novel-coronavirus-2019/technical-guidance.
|
|
61
|
Du Z, Xu X, Wu Y, Wang L, Cowling BJ and
Meyers LA: Serial Interval of COVID-19 Among Publicly Reported
Confirmed Cases. Emerg Infect Dis. Mar 19–2020.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Crisanti A and Cassone A: In one Italian
town, we showed mass testing could eradicate the coronavirus
Opinion. The Guardian; 2020, https://www.theguardian.com/comment-isfree/2020/mar/20/eradicated-coronavirus-mass-testing-covid-19-italy-vo.
Accessed March 20 2020.
|
|
63
|
Chiu WK, Cheung PC, Ng KL, et al: Severe
acute respiratory syndrome in children: experience in a regional
hospital in Hong Kong. Pediatr Crit Care Med. 4:279–283. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Donnelly CA, Ghani AC, Leung GM, et al:
Epidemiological determinants of spread of causal agent of severe
acute respiratory syndrome in Hong Kong. Lancet. 361:1761–1766.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Leung GM, Hedley AJ, Ho LM, et al: The
epidemiology of severe acute respiratory syndrome in the 2003 Hong
Kong epidemic: an analysis of all 1755 patients. Ann Intern Med.
141:662–673. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Booth CM, Matukas LM, Tomlinson GA, et al:
Clinical features and short-term outcomes of 144 patients with SARS
in the greater Toronto area. JAMA. 289:2801–2809. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chan JW, Ng CK, Chan YH, et al: Short term
outcome and risk factors for adverse clinical outcomes in adults
with severe acute respiratory syndrome (SARS). Thorax. 58:686–689.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hon KL, Leung CW, Cheng WT, et al:
Clinical presentations and outcome of severe acute respiratory
syndrome in children. Lancet. 361:1701–1703. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hui DS and Sung JJ: Severe acute
respiratory syndrome. Chest. 124:12–15. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Assiri A, McGeer A, Perl TM, et al: KSA
MERS-CoV Investigation Team, 2013b. Hospital outbreak of Middle
East respiratory syndrome coronavirus. N Engl J Med. 369:407–416.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et
al: Epidemiological, demographic, and clinical characteristics of
47 cases of Middle East respiratory syndrome coronavirus disease
from Saudi Arabia: a descriptive study. Lancet Infect Dis.
13:752–761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Memish ZA, Zumla AI, Al-Hakeem RF,
Al-Rabeeah AA and Stephens GM: Family cluster of Middle East
respiratory syndrome coronavirus infections. N Engl J Med.
368:2487–2494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ki M: 2015 MERS Outbreak in Korea:
Hospital-To-Hospital Transmission. Epidemiol Heal. 37:e20150332015.
View Article : Google Scholar
|
|
74
|
Arabi YM, Balkhy HH, Hayden FG, et al:
Middle East Respiratory Syndrome. N Engl J Med. 376:584–594. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Desforges M, Le Coupanec A, Stodola JK,
Meessen-Pinard M and Talbot PJ: Human coronaviruses: viral and
cellular factors involved in neuroinvasiveness and
neuropathogenesis. Virus Res. 194:145–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Saad M, Omrani AS, Baig K, et al: Clinical
aspects and outcomes of 70 patients with Middle East respiratory
syndrome coronavirus infection: a single-center experience in Saudi
Arabia. Int J Infect Dis. 29:301–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Al-Hameed FM: Spontaneous intracranial
hemorrhage in a patient with Middle East respiratory syndrome
corona virus. Saudi Med J. 38:196–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Algahtani H, Subahi A and Shirah B:
Neurological complications of Middle East respiratory syndrome
coronavirus: A report of two cases and review of the literature.
Case Rep Neurol Med. 2016:35026832016.PubMed/NCBI
|
|
79
|
Kim JE, Heo JH, Kim HO, et al:
Neurological Complications during Treatment of Middle East
Respiratory Syndrome. J Clin Neurol. 13:227–233. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cha RH, Yang SH, Moon KC, et al: A Case
Report of a Middle East Respiratory Syndrome Survivor with Kidney
Biopsy Results. J Korean Med Sci. 31:635–640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lu H, Stratton CW and Tang YW: Outbreak of
pneumonia of unknown etiology in Wuhan China: the mystery and the
miracle. J Med Virol. 92:401–402. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hui DS, Azhar I, Madani E, et al: The
continuing 2019-nCoV epidemic threat of novel coronaviruses to
global health - The latest 2019 novel coronavirus outbreak in
Wuhan, China. Int J Infect Dis. 91:264–266. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng
Q, Meredith HR, Azman AS, Reich NG and Lessler J: The Incubation
Period of Coronavirus Disease 2019 (COVID-19) From Publicly
Reported Confirmed Cases: Estimation and Application. Ann Intern
Med. Mar 10–2020.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lin X, Gong Z, Xiao Z, Xiong J, Fan B and
Liu J: Novel Coronavirus Pneumonia Outbreak in 2019: Computed
Tomographic Findings in Two Cases. Korean J Radiol. 21:365–368.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Warren TK, Wells J, Panchal RG, et al:
Protection against filovirus diseases by a novel broad-spectrum
nucleoside analogue BCX4430. Nature. 508:402–405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pan Y, Guan H, Zhou S, et al: Initial CT
findings and temporal changes in patients with the novel
coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan,
China. Eur Radiol. Feb 13–2020.Epub ahead of print. View Article : Google Scholar
|
|
87
|
Pan F, Ye T, Sun P, Gui S, Liang B, Li L,
Zheng D, Wang J, Hesketh RL, Yang L, et al: Time Course of Lung
Changes On Chest CT During Recovery From 2019 Novel Coronavirus
(COVID-19) Pneumonia. Radiology. Feb 13–2020.Epub ahead of print.
View Article : Google Scholar
|
|
88
|
Zheng YY, Ma YT, Zhang JY and Xie X:
COVID-19 and the cardiovascular system. Nat Rev Cardiol. Mar
5–2020.Epub ahead of print. View Article : Google Scholar
|
|
89
|
Sung JJ, Wu A, Joynt GM, et al: Severe
acute respiratory syndrome: report of treatment and outcome after a
major outbreak. Thorax. 59:414–420. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chu CM, Cheng VC, Hung IF, et al: Role of
lopinavir/ritonavir in the treatment of SARS: initial virological
and clinical findings. Thorax. 59:252–256. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Loutfy MR, Blatt LM, Siminovitch KA, et
al: Interferon alfacon-1 plus corticosteroids in severe acute
respiratory syndrome: a preliminary study. JAMA. 290:3222–3228.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tsui PT, Kwok ML, Yuen H and Lai ST:
Severe acute respiratory syndrome: clinical outcome and prognostic
correlates. Emerg Infect Dis. 9:1064–1069. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Cheng Y, Wong R, Soo YO, et al: Use of
convalescent plasma therapy in SARS patients in Hong Kong. Eur J
Clin Microbiol Infect Dis. 24:44–46. 2005. View Article : Google Scholar
|
|
94
|
Omrani AS, Saad MM, Baig K, et al:
Ribavirin and interferon alfa-2a for severe Middle East respiratory
syndrome coronavirus infection: a retrospective cohort study.
Lancet Infect Dis. 14:1090–1095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Widagdo W, Okba NMA, Stalin Raj V and
Haagmans BL: MERS-coronavirus: From discovery to intervention. One
Heal. 3:11–16. 2016. View Article : Google Scholar
|
|
96
|
Calina D, Rosu L, Rosu AF, et al:
Etiological diagnosis and pharmacotherapeutic management of
parapneumonic pleuresy. Farmacia. 64:946-52. 2016.
|
|
97
|
Cowling BJ, Lam TTY, Yen HL, Poon LLM and
Peiris M: Evidence-Based Options for Controlling Respiratory Virus
Transmission. Emerg Infect Dis. 23:1712312017. View Article : Google Scholar
|
|
98
|
Zhao Z, Zhang F, Xu M, et al: Description
and clinical treatment of an early outbreak of severe acute
respiratory syndrome (SARS) in Guangzhou, PR China. J Med
Microbiol. 52:715–720. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Al-Qahtani AA, Lyroni K, Aznaourova M, et
al: Middle east respiratory syndrome corona virus spike
glycoprotein suppresses macrophage responses via DPP4-mediated
induction of IRAK-M and PPARγ. Oncotarget. 8:9053–9066. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lim J, Jeon S, Shin HY, Kim MJ, Seong YM,
Lee WJ, Choe KW, Kang YM, Lee B and Park SJ: Case of the Index
Patient Who Caused Tertiary Transmission of COVID-19 Infection in
Korea: The Application of Lopinavir/Ritonavir for the Treatment of
COVID-19 Infected Pneumonia Monitored by Quantitative RT-PCR. J
Korean Med Sci. 35:e792020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Agostini ML, Andres EL, Sims AC, et al:
Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is
Mediated by the Viral Polymerase and the Proofreading
Exoribonuclease. mBio. 9:e00221–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cho A, Saunders OL, Butler T, et al:
Synthesis and antiviral activity of a series of 1′-substituted
4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett.
22:2705-7. 2012. View Article : Google Scholar
|
|
103
|
Sheahan TP, Sims AC, Graham RL, Menachery
VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I,
et al: Broad-spectrum antiviral GS-5734 inhibits both epidemic and
zoonotic coronaviruses. Sci Transl Med. 9:36532017. View Article : Google Scholar
|
|
104
|
Sun ML, Yang JM, Sun YP and Su GH:
Inhibitors of RAS Might Be a Good Choice for the Therapy of
COVID-19 Pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 43:142020.
|
|
105
|
Colson P, Rolain JM, Lagier JC, Brouqui P
and Raoult D: Chloroquine and hydroxychloroquine as available
weapons to fight COVID-19. Int J Antimicrob Agents. Mar 4–2020.Epub
ahead of print. View Article : Google Scholar
|
|
106
|
Gao J, Tian Z and Yang X: Breakthrough:
Chloroquine phosphate has shown apparent efficacy in treatment of
COVID-19 asso-ciated pneumonia in clinical studies. Biosci Trends.
14:72–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Clinical Trials Arena: ROCHE to start
Phase III trial of Actemra in Covid-19 patients. https://www.clinicaltrialsarena.com/news/roche-actemra-covid-19-trial/.
Accessed March 19 2020.
|
|
108
|
Healio: Sarilumab enters clinical trial
for COVID-19, spotlighting 'key role' for IL-6. https://www.healio.com/rheu-matology/rheumatoid-arthritis/news/online/%7B1957db6e-f7a2-4e5d-939e-d4b5964b2dd3%7D/sarilumab-enters-clinical-trial-for-covid-19-spotlighting-key-role-for-il-6.
Accessed March 19, 2020.
|
|
109
|
Zhitomirsky B and Assaraf YG: Lysosomes as
mediators of drug resistance in cancer. Drug Resist Updat.
24:23–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kazmi F, Hensley T, Pope C, Funk RS,
Loewen GJ, Buckley DB and Parkinson A: Lysosomal sequestration
(trapping) of lipophilic amine (cationic amphiphilic) drugs in
immortalized human hepatocytes (Fa2N-4 cells). Drug Metab Dispos.
41:897–905. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Adar Y, Stark M, Bram EE, Nowak-Sliwinska
P, van den Bergh H, Szewczyk G, Sarna T, Skladanowski A, Griffioen
AW and Assaraf YG: Imidazoacridinone-dependent lysosomal
photodestruction: A pharmacological Trojan horse approach to
eradicate multidrug-resistant cancers. Cell Death Dis. 3:e293.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ashfaq UA, Javed T, Rehman S, Nawaz Z and
Riazuddin S: Lysosomotropic agents as HCV entry inhibitors. Virol
J. 8:1632011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kaufmann AM and Krise JP: Lysosomal
sequestration of amine-containing drugs: Analysis and therapeutic
implications. J Pharm Sci. 96:729–746. 2007. View Article : Google Scholar
|
|
114
|
WHO: SARS risk assessment and preparedness
framework. 2004, https://www.who.int/csr/resources/publications/CDS_CSR_ARO_2004_2.pdf.
|
|
115
|
Craven J: COVID-19 Vaccine Tracker.
Regulatory Affairs Professionals Society (RAPS); 2020, https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker.
Accessed March 21, 2020.
|
|
116
|
Martin JE, Louder MK, Holman LA, et al:
VRC 301 Study Team. Vaccine. 26:6338–6343. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Lin JT, Zhang JS, Su N, et al: Safety and
immunogenicity from a phase I trial of inactivated severe acute
respiratory syndrome coronavirus vaccine. Antivir Ther.
12:1107–1113. 2007.PubMed/NCBI
|
|
118
|
Beigel JH, Voell J, Kumar P, et al: Safety
and tolerability of a novel, polyclonal human anti-MERS coronavirus
antibody produced from transchromosomic cattle: a phase 1
randomised, double-blind, single-dose-escalation study. Lancet
Infect Dis. 18:410–418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Janice Oh HL, Ken-En Gan S, Bertoletti A
and Tan YJ: Understanding the T cell immune response in SARS
coronavirus infection. Emerg Microbes Infect. 1:e232012. View Article : Google Scholar : PubMed/NCBI
|