Introduction
Intervertebral disc degeneration (IDD) is a chronic
and high-incidence musculoskeletal disorder that causes back/neck
and radicular-related pain, with a worldwide socioeconomic impact
(1).
The intervertebral disc is composed of the external
annulus fibrosus (AF) and the inner gel-like nucleus pulposus (NP).
NP cells play a prominent role in both normal intervertebral discs
and the pathogenesis of IDD. Normal NP tissues are responsible for
distributing hydraulic pressure to the AF and endplate, while
reduced numbers of NP cells, loss of proteoglycan and changes in
the biomechanics of the spinal column lead to NP tissue
degeneration and low back pain (2). In addition, mechanical
overloading-induced NP cell apoptosis contributes to the
development of IDD. Therefore, research on NP cells remains a major
challenge in IDD treatment (3-5).
Identifying factors that exert anti-senescent or anti-apoptotic
effects on NP cells appears to be a promising approach. For
example, sirt6 has shown potent anti-senescent and anti-apoptotic
properties in models of age-related degenerative disease (6). Furthermore, overproduction of
reactive oxygen species (ROS) has been reported to be involved in
the senescence and apoptosis of NP cells and may accelerate the
degenerative process (7).
Previous studies also demonstrated that tumor necrosis factor-α
promotes changes in the production of extracellular matrix (ECM)
molecules, tissue degradation (8)
and premature senescence of NP cells (9,10).
However, the molecular mechanisms underlying NP cell growth,
differentiation and senescence are incompletely understood.
Long non-coding RNAs (lncRNAs), which contain
>200 nucleotides (some may contain >100,000 nucleotides),
play an important role in variety of human diseases, including IDD
(11). Several lncRNAs are
dysregulated in human degenerative NP tissues, and some participate
in multiple pathological processes, including inflammatory
responses, ECM degradation, angiogenesis and apoptosis (12). LncRNA PART1 has been shown to be a
potential prognostic biomarker and therapeutic target for tongue
squamous cell carcinoma (13),
hepatocellular carcinoma (14)
and non-small cell lung cancer (15). It was also recently demonstrated
that PART1 regulates apoptosis and proliferation in prostate cancer
cells through toll-like receptor pathways (16). However, the role of lncRNA PART1
in NP cells and IDD has yet to be reported.
The aim of the present study was to investigate the
role of PART1 in NP cells derived from patients with IDD, hoping to
improve our understanding of the mechanisms underlying the
degeneration of NP cells.
Materials and methods
Patients and tissue samples
Human lumbar NP specimens were obtained from 120
patients with spinal cord injury [control (n=30), IDD (n=30),
bulging discs (n=30) and herniated discs (n=30)] from Yanan
University Affiliated Hospital. All the patients underwent surgery
at Yanan University Affiliated Hospital between 2015 and 2017. The
present study was approved by the Ethics Committee of Yanan
University Affiliated Hospital, and written informed consent was
obtained from each participant prior to surgery.
Isolation and culture of human NP
cells
NP tissues from 10 patients with IDD were separated
and washed using Hank's solution (14025126, Gibco; Thermo Fisher
Scientific, Inc.). Subsequently, the samples were cut into
1-mm3 pieces, centrifuged at 500 × g for 5 min at 4°C,
and then treated with type II collagenase (0.2%; 17101015, Gibco;
Thermo Fisher Scientific, Inc.) at 37°C for 4-6 h. Next, the
tissues were centrifuged at 500 × g for 8 min at 4°C, and DMEM/F12
(11320033, Gibco; Thermo Fisher Scientific, Inc.) was added for
cell suspension and centrifugation. When the cell density reached
5×105/ml, the cells were seeded into culture flasks and
cultured at 37°C in DMEM/F12 supplemented with fetal bovine serum
(16140071, Gibco; Thermo Fisher Scientific, Inc.), L-glutamine
(25030081, Gibco; Thermo Fisher Scientific, Inc.), and 1%
streptomycin and penicillin (10378016, Gibco; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2. Cells were
observed under a DMi8 optical microscope (Leica Microsystems GmbH),
at a magnification of ×100 and ×200.
Collagen II immunofluorescence
staining
Collagen II was detected using immunofluorescence
staining. First, NP cells were fixed with 4% paraformaldehyde for
20 min at room temperature, blocked with goat serum (Solarbio) for
30 min, then incubated with collagen II primary antibody (1:200,
ab34712, Abcam) for 1 h, and goat anti-rabbit IgG H&L secondary
antibody (1:500, ab150077, Abcam) for 45 min. Finally, the cells
were stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole
(DAPI; Sigma-Aldrich; Merck KGaA) at room temperature for 10 min,
and observed under a DM13000B fluorescence microscope (Leica
Microsystems GmbH) at a magnification of ×100.
Cell transfection
The small interfering RNA against PART1 (17) (5′-GCA AAU GAA AGC UAC CAA U-3′)
was purchased from GenePharma Co., Ltd. SiPART1
(siG000004313A-1-5), control siRNA (siN0000001-1-5), mimic control
(miR1N0000001-1-5), miR-93 mimic (miR10004976-1-5), inhibitor
control (miR2N0000001-1-5) and miR-93 inhibitor (miR20012845-1-5)
were synthesized by Ribobio Co., Ltd. The full length of matrix
metallopeptidase (MMP)2 was amplified and inserted into the pcDNA
3.1 plasmid (V79020, Invitrogen; Thermo Fisher Scientific, Inc.).
The pcDNA 3.1-MMP2 was then transfected into the cells to achieve
MMP2 overexpression. The vector pcDNA 3.1 plasmid was transfected
into cells as control. Opti-MEM (11058021, Invitrogen; Thermo
Fisher Scientific, Inc.) containing Lipofectamine® 2000
(11668019, Invitrogen; Thermo Fisher Scientific, Inc.) was used to
transfect mimic, control mimic or plasmids into the cells at room
temperature for 10 min. All the transfection procedures were
performed following the manufacturer's instructions. After
incubation for 24 h, the cells were collected for further
experiments.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Tissue specimens were placed in liquid nitrogen and
homogenized using TRIzol reagent (15596018, Invitrogen; Thermo
Fisher Scientific, Inc.) at 4°C. Total RNA was isolated from each
sample at 4°C using the Qiagen RNeasy Plus Mini Kit (74134, Qiagen
GmbH) according to the manufacturer's protocol. The cells were
treated with TRIzol reagent and total RNA was extracted using
chloroform and isopropanol at 4°C. NanoDrop 8000 (ND-8000-GL,
Thermo Fisher Scientific, Inc.) was used to assess the
concentration of RNAs.
For quantification of mRNAs, the PrimeScript™ II 1st
Strand cDNA Synthesis Kit (6210B, Takara Bio, Inc.) was used for
reverse transcription (42°C for 15 min), and SYBR® Green
PCR Master Mix (4312704, Applied Biosystems; Thermo Fisher
Scientific, Inc.) and Bio-Rad CFX 96 Touch Real-Time PCR Detection
System (1855196, Bio-Rad Laboratories, Inc.) were then used for
qPCR. The thermocycling conditions were as follows: 95°C for 5 min,
followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec, and 70°C
for 10 sec. GAPDH served as an internal control and the
2−ΔΔCq method was used to calculate the relative gene
expression (18). All reactions
were performed for three mechanical duplications. All primers for
RT-qPCR are listed in Table
I.
 | Table IPrimers used for reverse
transcription-quantitative PCR analysis. |
Table I
Primers used for reverse
transcription-quantitative PCR analysis.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| PART1 |
AAGGCCGTGTCAGAACTCAA |
GTTTTCCATCTCAGCCTGGA |
| Aggrecan |
ACTCTGGGTTTTCGTGACTCT |
ACACTCAGCGAGTTGTCATGG |
| ADAMTS4 |
GAGGAGGAGATCGTGTTTCCA |
CCAGCTCTAGTAGCAGCGTC |
| Collagen II |
TGGACGATCAGGCGAAACC |
GCTGCGGATGCTCTCAATCT |
| MMP13 |
ACTGAGAGGCTCCGAGAAATG |
GAACCCCGCATCTTGGCTT |
| MMP2 |
ATCCAGACTTCCTCAGGCGG CC |
ATTAGCGCCTCCATCGTAG |
| GAPDH |
TGTGGGCATCAATGGATTTGG |
ACACCATGTATTCCGGGTCAAT |
For miR-93 quantification, cDNA was prepared from
total RNA (2 μg) by using One-Step miRNA RT kit (D1801,
HaiGene). The SYBR Green miRNA RT-qPCR kit for amplifying miR-93
(AP01605, HaiGene) and U6 snRNA (AP02055, HaiGene) were used in
RT-qPCR, and the parameters were set as follows: 95°C for 5 min,
followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec and 70°C
for 10 sec. U6 snRNA served as an internal control. The comparative
quantification cycle method 2−∆∆Cq was used to calculate
the relative gene expression.
Cell Counting Kit (CCK)-8 assay
The cells (1×104 cells/well) were
inoculated onto a 96-well plate containing 100 μl medium,
with each experiment comprising 6 duplicated wells and 5 zero
wells. The cells were cultured with 5% CO2 at 37°C, and
10 μl CCK-8 solution (70-CCK801, MultiSciences) was added
into each well at 24, 48 and 72 h of culture. After incubation for
2 h at 37°C, the absorbance value at 490 nm was detected by the
SpectraMax Plus 384 Microplate Reader (PLUS 384, Molecular Devices,
LLC).
Colony formation assay
The cells (200 cells/well) were seeded in a 12-well
plate and cultured continuously for 14 days. All subsequent clones
were stained using 0.5% crystal violet solution (548-62-9, Aladdin)
for 10 min at room temperature, and counted under a DMi8 optical
microscope (Leica Microsystems GmbH), at a magnification of ×100.
Clusters of >50 cells were considered as colonies, and the
colony formation rate was then calculated in five fields of
view.
Flow cytometry
Apoptosis was evaluated using an Annexin-V kit
(70-AP101-100-AVF, MultiSciences). Briefly, after trypsinization,
1-5×106 cells were pelleted and centrifuged at 450 × g
for 5 min at 4°C. The cells were then resuspended in 300 μl
binding buffer, and 5 μl Annexin-V-FITC solution was added
and incubated with the cells at room temperature for 15 min in the
dark. The nuclei were stained with 5 μl PI for 5 min at room
temperature. Finally, another 200 μl binding buffer was
added, and cell apoptosis was evaluated.
Western blot analysis
Total protein samples were extracted from NP cells
using lysis buffer (89901, Thermo Fisher Scientific, Inc.) with a
protease inhibitor (36978, Thermo Fisher Scientific, Inc.). Equal
amounts of protein (60 μg/lane) were separated on 8%
SDS-polyacrylamide gels and then transferred to a PVDF membrane
(LC2002, Invitrogen; Thermo Fisher Scientific, Inc.). The membrane
was then blocked by 5% non-fat milk (PA201-01, BioMed) for 10 min
at room temperature and then incubated with the primary antibodies:
Anti-Ki-67 (1:5,000, ab92742, Abcam), anti-cleaved (C)-caspase-3
(1:500, ab2302, Abcam), anti-aggrecan (1:100, ab3778, Abcam),
anti-ADAMTS4 (1:1,000, ab185722, Abcam), anti-collagen II (1:2,000,
ab34712, Abcam), anti-MMP13 (1:3,000, ab39012, Abcam) and
anti-GAPDH (1:2,000, ab8245, Abcam) at 4°C overnight. The membrane
was then incubated with HRP-linked anti-rabbit IgG secondary
antibody (1:5,000; 7074, Cell Signaling Technology, Inc.) for
Ki-67, C-caspase-3, ADAMTS4, collagen II, MMP13, or incubated with
HRP-conjugated anti-mouse IgG (1:5,000, 7076, Cell Signaling
Technology, Inc.) for aggrecan and GAPDH at 4°C overnight. Finally,
SignalFire™ ECL reagent (6883, Cell Signaling Technology, Inc.) and
ImageQuant ECL Imager (28-9605-63, GE Healthcare) were used to
detect the signals. Prior to each incubation, 1X TBST (50 mM Tris,
150 mM NaCl and 2% Tween-20, pH 7.5) was used to wash the membrane
three times, 5 min per wash. GADPH served as an internal
control.
Dual-luciferase reporter assay
PART1-wild-type (WT), PART1-mutant (MUT), MMP2-WT
and MMP2-MUT were synthesized by Tsingke Co., Ltd. and separately
cloned into luciferase reporter gene vector (pmirGLO, E1330,
Promega Corporation) to construct luciferase reporter plasmids.
Renilla luciferase in the reporter plasmid served as the internal
control. Briefly, cells cultured in 96-well plates were transfected
with luciferase reporter plasmids (0.05 μg/well), or
co-transfected with luciferase reporter plasmids and miR-93
inhibitor, or miR-93 mimic (15 nM). After transfection for 24 h,
the cells were analyzed for luciferase activity using the
Dual-Glo® Luciferase assay system (E2920, Promega
Corporation) and a Microplate Luminometer (11300010, Berthold
Technologies). The ratio of firefly luciferase activity relative to
that of Renilla luciferase activity was calculated.
Statistical analysis
All statistical analyses were conducted using SPSS
v.19.0 software (IBM Corp.). Graph presentations were created using
GraphPad Prism 5.02 (GraphPad Software, Inc.). The data are
presented as percentage ± standard error of the mean. Data were
analyzed by Student's t-test or ANOVA followed by Tukey's post hoc
test in SPSS. P<0.05 was considered to indicate statistically
significant differences.
Results
Expression of lncRNA PART1 in NP tissues
and the effects of lncRNA PART1 on NP cells
The expression level of lncRNA PART1 was measured in
NP tissues from patients with IDD or controls, and PART1 was found
to be highly expressed in NP tissues from patients with IDD
(P<0.001, Fig. 1A). The
expression level of PART1 in herniated NP tissues was the highest
compared with control and bulging NP tissues (P<0.001, Fig. 1B). Subsequently, NP cells were
isolated from patients with IDD and examined using an optical
microscope. The cultured NP cells were polygonal or spindle-shaped,
displaying short pseudopods. Upon approaching confluence, the cells
exhibited swirling or formation of flame-like cell clusters, and
they were rich in cytoplasm, highly refractive, with large, clear,
oval-shaped nuclei (Fig. 1C).
Collagen II immunofluorescence staining revealed that the cells
emitted green fluorescent light under the fluorescence microscope,
which proved that the expression characteristics of the cultured
cells are consistent with those of NP cells (Fig. 1D). Subsequently, NP cells were
transfected with siPART1, and the expression level of PART1 was
found to be reduced by siPART1 (P<0.001, Fig. 1E). Furthermore, the CCK-8 assay
demonstrated that the proliferation ability of NP cells transfected
with siPART1 was increased (P<0.001, Fig. 1F), and that the colony-forming
ability of NP cells was also increased (P<0.001, Fig. 1G). Furthermore, knockdown of PART1
inhibited the apoptosis of NP cells (P<0.001, Fig. 1H). In addition, the protein levels
of the cell proliferation-associated protein, Ki-67, and the
apoptosis-associated protein, C-caspase-3, were measured in NP
cells after interference with siPART1, and the data revealed that
knockdown of PART1 promoted the expression of Ki-67 and inhibited
the expression of C-caspase-3 (P<0.001, Fig. 1I-J). Aggrecan and collagen II are
ECM synthesis-related genes, while ADAMTS4 and MMP13 are ECM
degradation-related genes; therefore, the levels of aggrecan,
collagen II, ADAMTS4 and MMP13 in NP cells with PART1 knockdown
were detected by western blot and RT-qPCR analyses. The results
demonstrated that siPART1 increased the levels of aggrecan and
collagen II, and decreased the levels of ADAMTS4 and MMP13
(P<0.001, Fig. 1K-M).
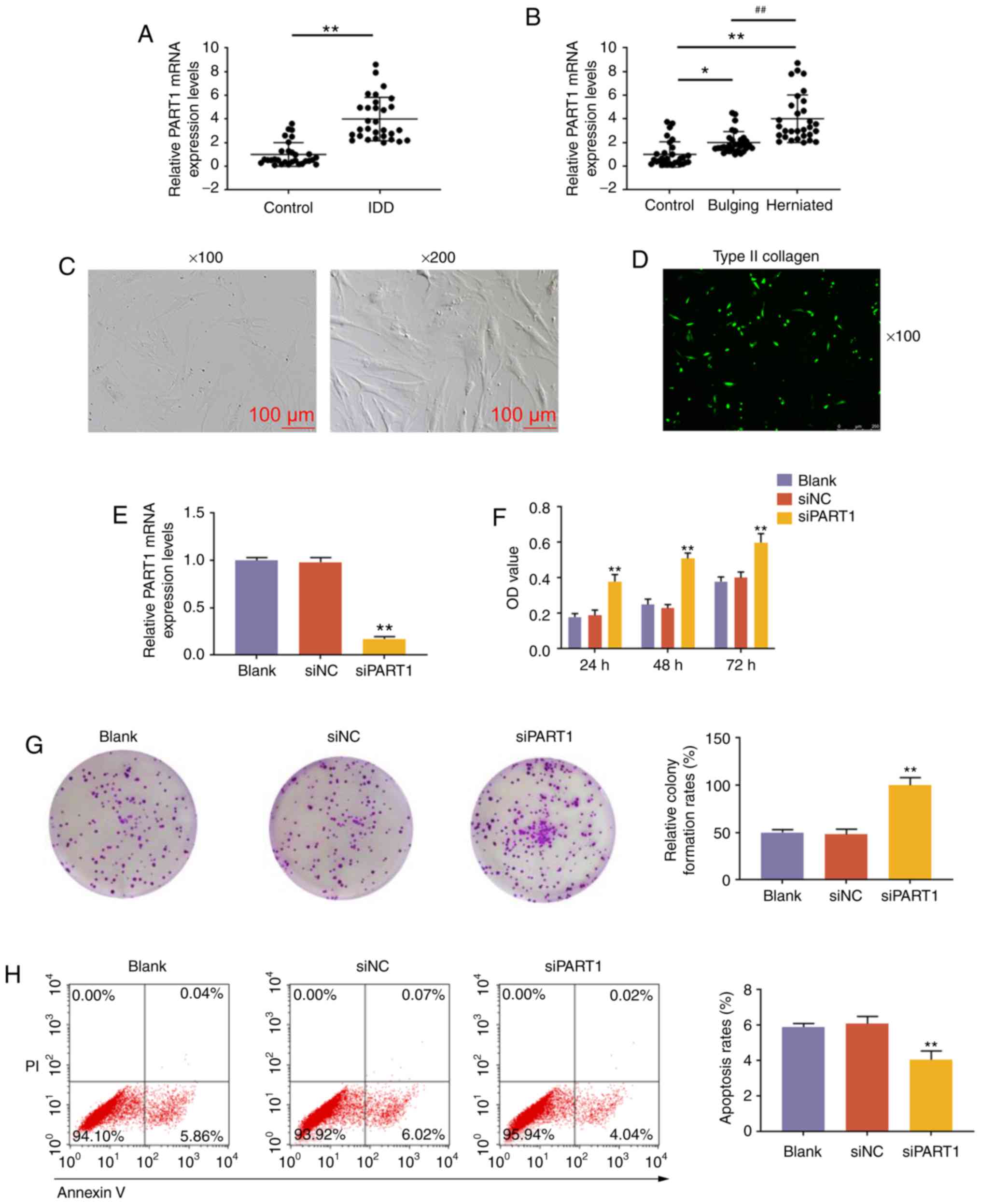 | Figure 1Expression pattern and effects of
lncRNA PART1 on NP tissues. (A) Expression level of PART1 in NP
tissues from IDD patients and controls. **P<0.001 vs.
control. (B) The expression level of PART1 from control, bulging
and herniated NP tissues was measured by quantitative PCR analysis.
*P<0.05 and **P<0.001 vs.
control;##P<0.001 vs. bulging. (C) NP cells were
observed under an optical microscope, at a magnification of ×100
and ×200. (D) Immunofluorescence staining of type II collagen at a
magnification of ×100. (E) Expression level of PART1 in NP cells
isolated from patients with IDD. (F) The proliferative capacity of
NP cells was measured by Cell Counting Kit-8 assays after
non-interference, or interference with siNC or siPART1 at 24, 48
and 72 h. (G) Images of NP cell colonies. The colony formation
rates are shown on the right. (H) The apoptosis of NP cells was
measured by flow cytometry. PI+/Annexin V+
cells were considered to be apoptotic. The apoptosis rates of NP
cells are shown on the right. (I and J) The expression levels of
Ki67 and C-caspase-3 in NP cells were measured by western blotting.
(K-M) The expression levels of aggrecan, ADAMTS4, collagen II and
MMP13 in NP cells were measured by (K and L) western blotting and
(M) reverse transcription-quantitative PCR. **P<0.001
vs. siNC. Blank, non-transfection; transfection with siNC, siRNA
for negative control; siPART1, siRNA for PART1; NP, nucleus
pulposus; IDD, intervertebral disc degeneration. |
LncRNA PART1 regulates the expression of
miR-93-5p via sponging miR-93-5p in NP cells
DIANA-LncBase V2 (http://carolina.imis.athena-innovation.gr/, accessed
January 2018) and StarBase (http://starbase.sysu.edu.c, accessed January 2018)
predicted that PART1 could sponge miR-93 (Fig. 2A). StarBase predicted that PART1
could bind 35 miRNAs, and DIANA-LncBase V2 predicted that 34 miRNAs
could be combined. However, only miR-93 was reported in the
literature and jointly predicted by the two websites. Other miRNAs
may also play a targeting role, which may be of value for future
research. The prediction was confirmed by dual-luciferase reporter
assay, as the results demonstrated that miR-93 inhibitor caused an
increase in the luciferase activity of NP cells transfected with
PART1-WT plasmid (P<0.001), while the luciferase activity of NP
cells transfected with PART1-MUT plasmid was not affected
(P>0.05, Fig. 2B). We then
measured the expression of miR-93 in NP cells from patients with
IDD or controls, and the RT-qPCR results revealed that miR-93 was
downregulated in NP tissues from patients with IDD (P<0.001,
Fig. 2C). Further experiments
demonstrated that knockdown of PART1 could increase the level of
miR-93 (P<0.001, Fig. 2D). The
rescue assay revealed that miR-93 mimic increased the level of
miR-93, and knockdown of PART1 rescued the decrease in the miR-93
level caused by miR-93 inhibitor (P<0.001, Fig. 2E).
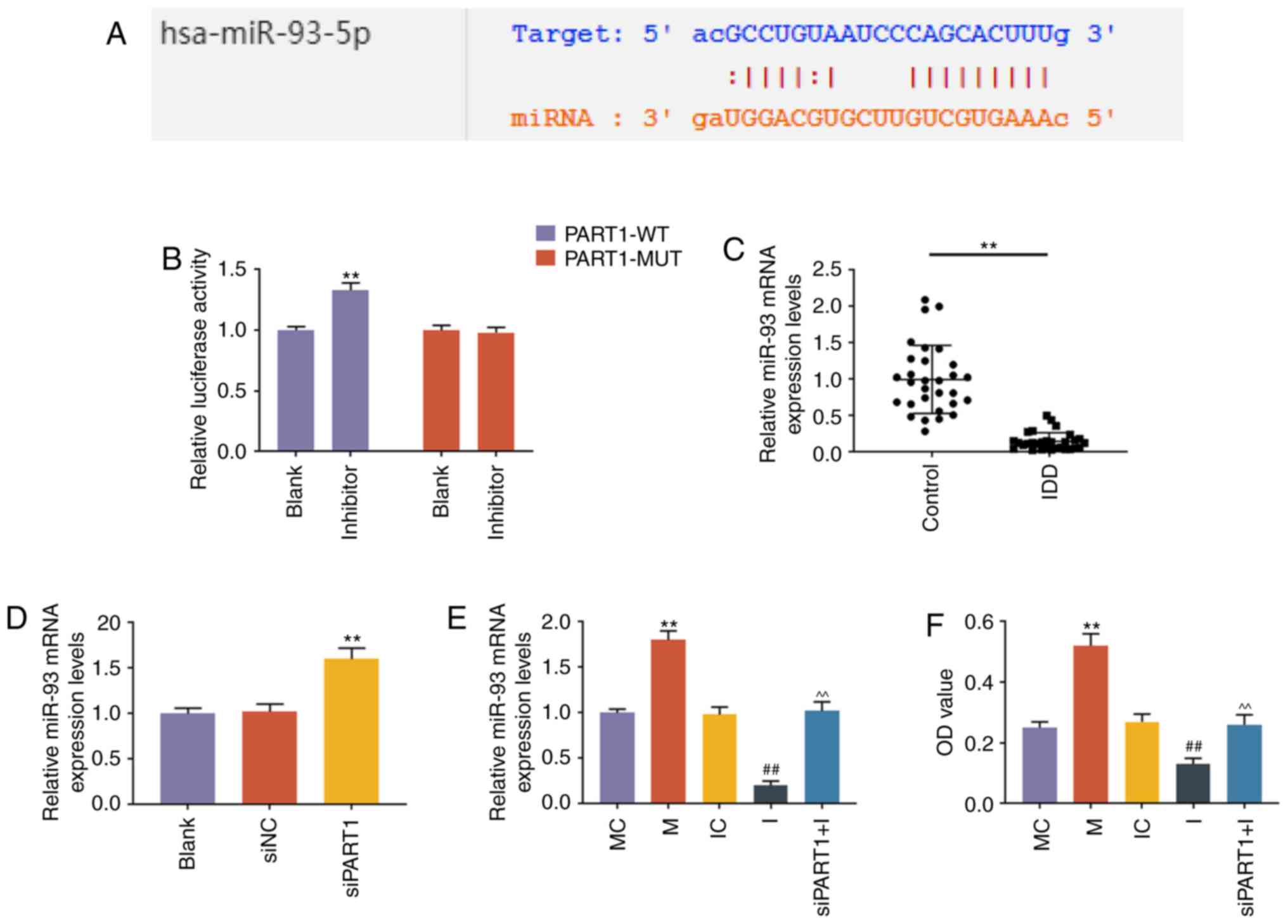 | Figure 2LncRNA PART1 regulated the expression
level of miR-93-5p via sponging miR-93-5p in NP cells. (A) A
potential target site for miR-93-5p in lncRNA PART1 was predicted
by DIANA-LncBase V2 and starBase. (B) Dual-luciferase reporter
assay was used to validate lncRNA PART1 sponging miR-93-5p in NP
cells with PART1-WT reporter plasmid or PART1-MUT reporter plasmid.
**P<0.001 vs. blank. (C) The expression levels of
miR-93-5p in the NP tissues from IDD patients and controls were
detected by RT-qPCR. **P<0.001 vs. control. (D) The
expression level of miR-93-5p in the NP cells was measured by
RT-qPCR. **P<0.001 vs. siNC. Blank, non-transfection;
siNC, siRNA for negative control; siPART1, siRNA for PART1. (E) The
expression level of miR-93-5p in NP cells was measured by RT-qPCR.
**P<0.001 vs. MC; ##P<0.001 vs. IC; and
^^P<0.001 vs. I. MC, mimic for control; M, miR-93-5p
mimics; IC, inhibitor for control; I, inhibitor for miR-93-5p;
siPART1 + I, co-transfection with siRNA for PART1 and miR-93-5p
inhibitor. (F) The proliferative capacity of NP cells was measured
by Cell Counting Kit-8 assays. **P<0.001 vs. MC;
##P<0.001 vs. IC; and ^^P<0.001 vs. I.
MC, mimic for control; M, miR-93-5p mimics; IC, inhibitor for
control; I, inhibitor for miR-93-5p; siPART1 + I, co-transfection
with siRNA for PART1 and miR-93-5p inhibitor. NP, nucleus pulposus;
WT, wild-type; MUT, mutant; IDD, intervertebral disc degeneration;
RT-qPCR, reverse transcription-quantitative PCR. |
LncRNA PART1 regulates cell growth,
apoptosis and ECM-related gene expression via sponging miR-93-5p in
NP cells
CCK-8 and colony formation assays demonstrated that
miR-93 mimic promoted and miR-93 inhibitor suppressed cell growth,
which was abrogated by siPART1 (all P<0.001, Figs. 2F and 3A). Cell apoptosis was also evaluated by
flow cytometry, and the results revealed that the apoptosis rate of
NP cells was reduced by miR-93 mimics and increased by miR-93
inhibitor; moreover, the increase in the apoptosis rate was rescued
by knocking down PART1 (all P<0.001, Fig. 3B). Furthermore, the cell
proliferation-related protein Ki-67 was upregulated by miR-93
mimics and downregulated by miR-93 inhibitor; however, these
effects were abolished by siPART1 (all P<0.001, Fig. 3C and D). In contrast to the change
of the Ki-67 level in NP cells, the level of C-caspase-3 was
reduced by miR-93 mimics, increased by miR-93 inhibitor, and could
be compensated by siPART1 in combination with miR-93 inhibitor
treatment (all P<0.001, Fig. 3C
and D). Moreover, aggrecan and collagen II were upregulated,
whereas ADAMTS4 and MMP13 were downregulated in NP cells
transfected with miR-93 mimic, and these effects were opposite to
those of the miR-93 inhibitor. In addition, the effects of the
miR-93 inhibitor on ECM-related genes in NP cells were abolished by
siPART1 (P<0.001, Fig.
3E-G).
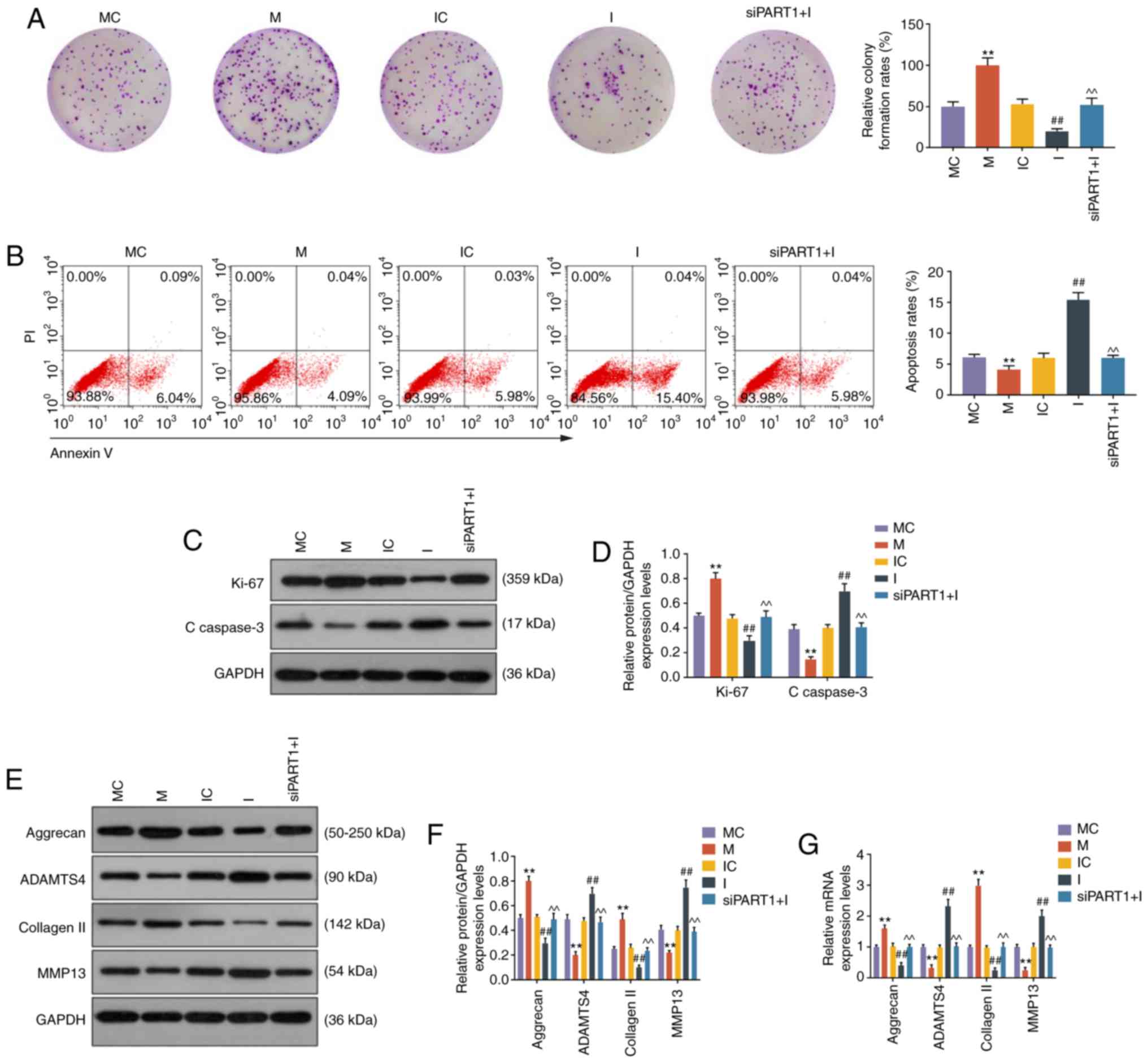 | Figure 3LncRNA PART1 regulated cell growth,
apoptosis and ECM-related genes through targeting miR-93-5p. (A)
Images of NP cell colonies. The colony formation rates are shown on
the right. (B) The apoptosis of NP cells was measured by flow
cytometry. The apoptosis rates of NP cells are shown on the right.
(C and D) The expression levels of Ki67 and C-caspase-3 in NP cells
were measured by western blotting. (E-G) The expression levels of
aggrecan, ADAMTS4, collagen II and MMP13 in NP cells were measured
by (E and F) western blotting and (G) reverse
transcription-quantitative PCR analysis. **P<0.001
vs. MC; ##P<0.001 vs. IC; and ^^P<0.001
vs. I. Blank, non-transfection; siNC, siRNA for negative control;
siPART1, siRNA for PART1; MC, mimic for control; M, miR-93-5p
mimics; IC, inhibitor for control; I, inhibitor for miR-93-5p;
siPART1 + I, co-transfection with siRNA for PART1 and miR-93-5p
inhibitor. ECM, extracellular matrix; NP, nucleus pulposus; MMP,
matrix metallopeptidase. |
miR-93 regulates the expression of MMP2
through targeting the 3′ untranslated region (UTR) of MMP2 in NP
cells
The role of miR-93 in NP cells is unclear; thus, we
sought to identify the target gene of miR-93. TargetScan predicted
that the target genes of miR-93-5p were from a total of 1,647
conserved sites and 914 poorly conserved sites. MMP2 was also
reported to be targeted, and was therefore investigated. A
potential binding site at the 3′UTR of MMP2 mRNA (515-521 nt) was
predicted by TargetScan, and dual-luciferase reporter assay
demonstrated that miR-93 mimic could suppress the luciferase
activity in NP cells transfected with MMP2-WT reporter plasmid
(P<0.001, Fig. 4B), while the
luciferase activity in NP cells transfected with MMP2-MUT reporter
plasmid was not altered (Fig.
4B). The expression level of MMP2 in NP tissues was evaluated
by RT-qPCR, and the results demonstrated that MMP2 was highly
expressed in NP tissues from patients with IDD (P<0.001,
Fig. 4C). A functional rescue
assay revealed that the proliferation ability of NP cells was
increased by miR-93 mimic, but was abrogated by MMP2 overexpression
(P<0.001, Fig. 4D). The
expression level of MMP2 in NP cells was increased by MMP2
overexpression plasmid (P<0.001, Fig. 4E and F).
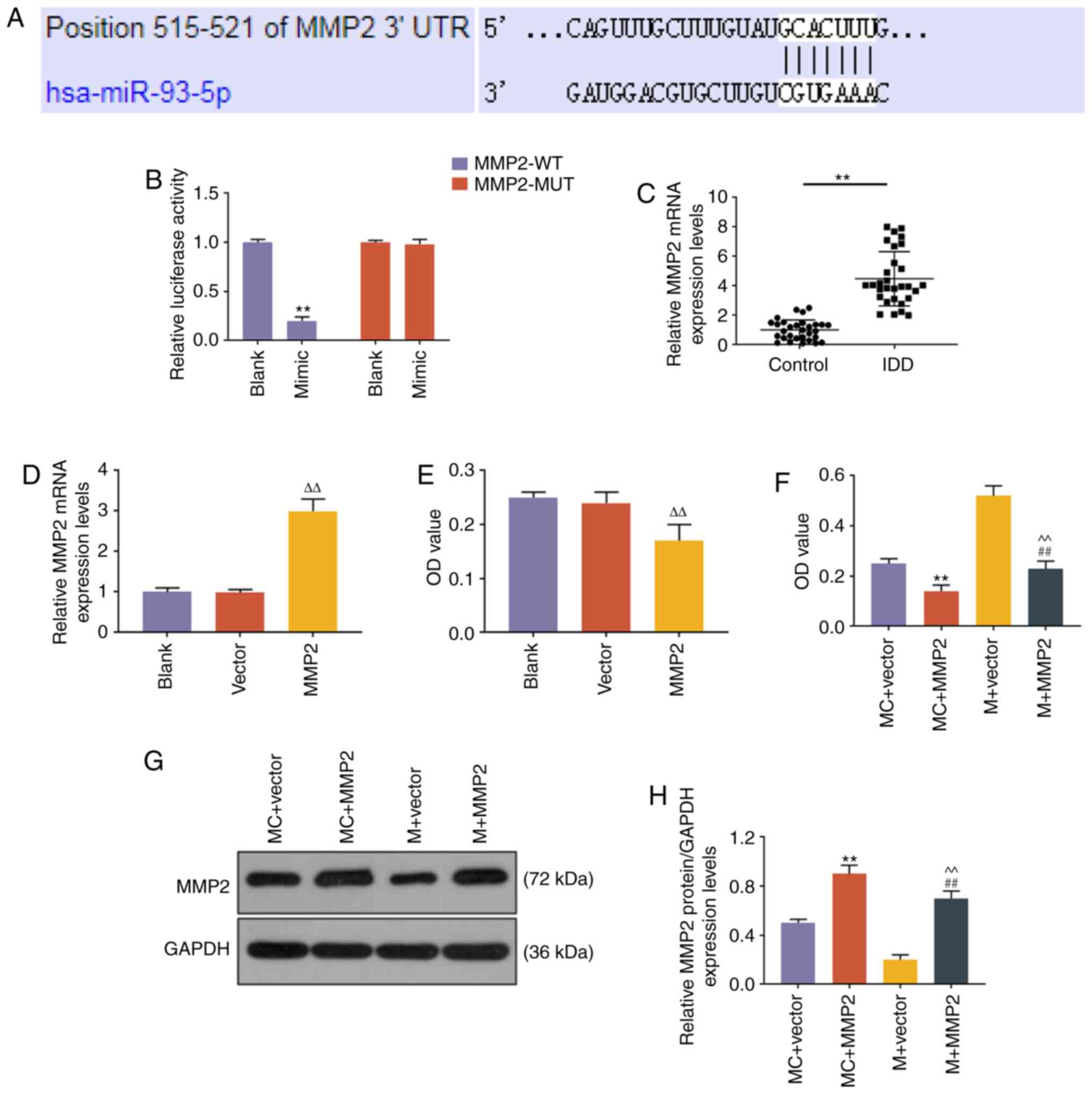 | Figure 4miR-93-5p regulated the expression of
MMP2 through targeting the 3′UTR of MMP2. (A) The miR-93-5p
targeting position 515-521 of MMP2 3′UTR was predicted by
TargetScan. (B) Dual-luciferase reporter assay was used to validate
miR-93-5p targeting MMP2 in NP cells. **P<0.001 vs.
Blank. (C) The expression level of MMP2 in NP tissues from IDD
patients and controls was detected by RT-qPCR analysis.
**P<0.001 vs. control. (D) RT-qPCR was applied to
measure the mRNA expression levels of MMP2 after MMP2
overexpression plasmid transfection. (E) Proliferative capacity of
cells transfected with MMP2. (F) The proliferative capacity of NP
cells was measured by Cell Counting Kit-8 assays.
**P<0.001 vs. MC + vector; ##P<0.001
vs. M + vector; ^^P<0.001 vs. MC + MMP2; and
∆∆P<0.001 vs. vector. (G and H) The expression level
of MMP2 in NP cells was measured by western blotting.
**P<0.001 vs. MC + vector; ##P<0.001
vs. M + vector; ^^P<0.001 vs. MC + MMP2. Vector,
transfection with empty vector for overexpression; MC + vector,
co-transfection with mimic control and empty vector for
overexpression; MC + MMP2, co-transfection with mimic control and
MMP2 overexpression vector; M + vector, co-transfection with
miR-93-5p mimic and empty vector for overexpression; M + MMP2,
co-transfection with miR-93-5p mimic and MMP2 overexpression
vector; MMP, matrix metallopeptidase; UTR, untranslated region; NP,
nucleus pulposus; RT-qPCR, reverse transcription-quantitative PCR;
IDD, intervertebral disc degeneration; WT, wild-type; MUT,
mutant. |
miR-93-5p regulates cell growth,
apoptosis and ECM-related gene expression through targeting MMP2 in
NP cells
Further experiments were performed to confirm the
role of miR-93-5p in targeting MMP2 in NP cells. Colony formation
assay demonstrated that overexpression of MMP2 inhibited cell
growth, miR-93 mimic promoted cell growth, and the effect of miR-93
mimic on cell growth could be rescued by MMP2 overexpression (all
P<0.001, Fig. 5A). Moreover,
flow cytometry analysis demonstrated that the reduction in cell
apoptosis by miR-93 mimic was reversed by MMP2 overexpression (all
P<0.001, Fig. 5B). The
expression of ECM-related genes was also evaluated, and results
revealed that miR-93 mimic upregulated the levels of aggrecan and
collagen II, and reduced the levels of ADAMTS4 and MMP13
(P<0.001, Fig. 5C-E). In
addition, the effects of miR-93 mimic on ECM-related gene
expression were blocked by MMP2 overexpression (P<0.001,
Fig. 5C-E).
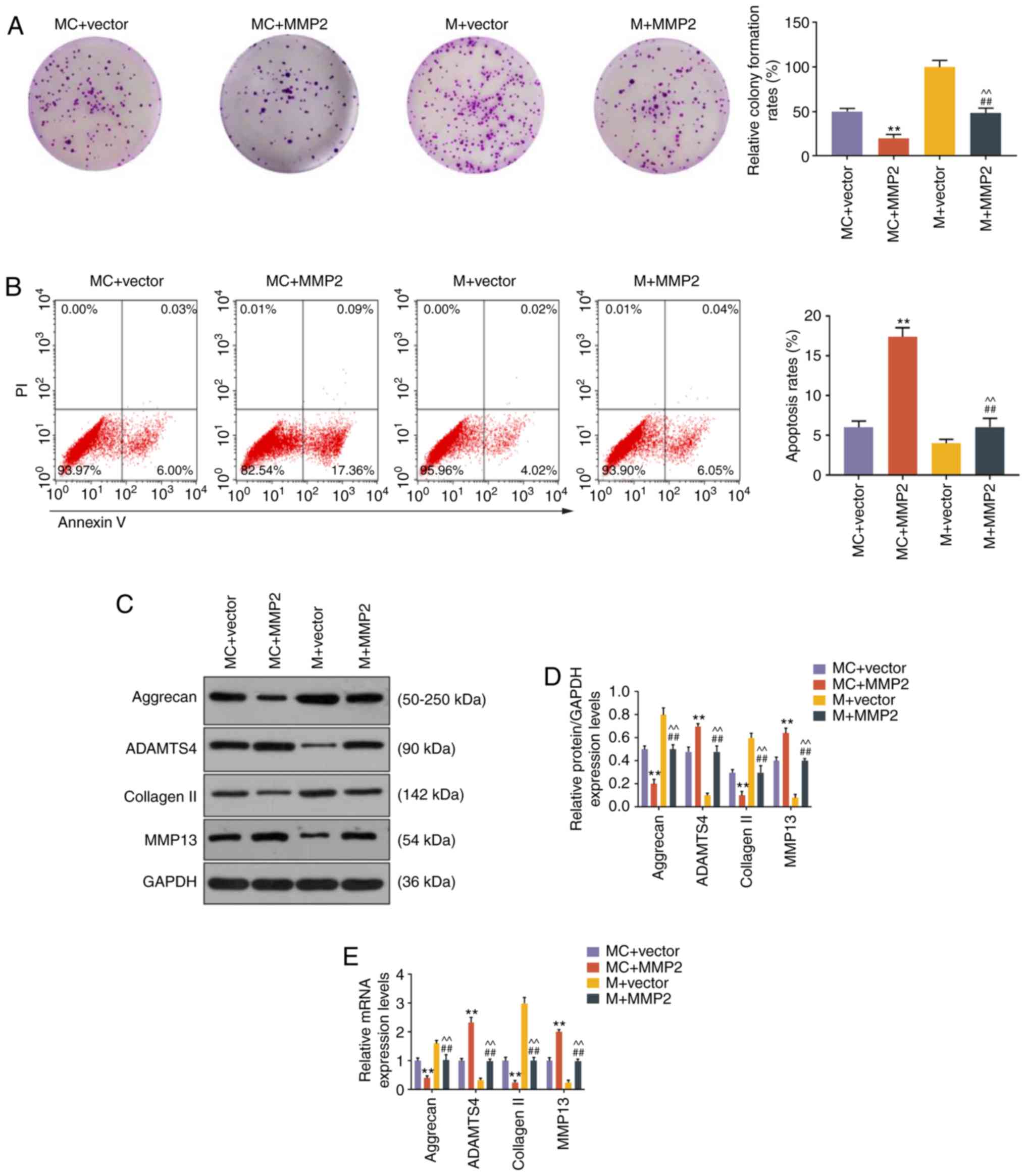 | Figure 5miR-93-5p regulated cell growth,
apoptosis and ECM-related genes through targeting MMP2. (A) Images
of NP cell colonies. The colony formation rates are shown on the
right. (B) The apoptosis of NP cells was measured by flow
cytometry. The apoptosis rates of NP cells are shown on the right.
(C-E) The expression levels of aggrecan, ADAMTS4, collagen II and
MMP13 in NP cells were measured by (C and D) western blotting and
(E) reverse transcription-quantitative PCR analysis.
**P<0.001 vs. MC + vector; ##P<0.001
vs. MC + MMP2; ^^P<0.001 vs. M + vector. MC + vector,
co-transfection with mimic control and empty vector for
overexpression; MC + MMP2, co-transfection with mimic control and
MMP2 overexpression vector; M + vector, co-transfection with
miR-93-5p mimic and empty vector for overexpression; M + MMP2,
co-transfection with miR-93-5p mimic and MMP2 overexpression
vector; ECM, extracellular matrix; NP, nucleus pulposus; MMP,
matrix metallopeptidase. |
Discussion
Delaying or reversing the degeneration of NP tissue
is a promising strategy for the treatment of IDD. However, the
molecular mechanisms underlying NP cell degeneration are
incompletely understood. In the present study, it was observed
that, in IDD patients, upregulation of the lncRNA PART1 in NP
tissues and cells regulated cell growth, apoptosis, ECM production
and degradation through targeting miR-93, and resulted in an
increase of the MMP2 level.
Emerging studies indicate that lncRNAs are important
regulators of NP cells. For example, lncRNA FAM83H-AS1 promotes the
growth of NP cells through the Notch signaling pathway (19); LINC00969 promotes the development
of IDD by targeting miR-335-3p and regulating NLRP3 inflammasome
activation (20). In the present
study, it was observed that PART1 was upregulated in degenerative
NP tissues from patients with IDD, bulging and herniated discs,
suggesting a potential role for PART1 in the degeneration of NP
cells. The degeneration of NP tissues is directly associated with
reduced numbers of NP cells, excessive apoptosis and ECM imbalance
(21). Thus, the effects of PART1
knockdown on cell growth, apoptosis, and the levels of genes
implicated in ECM synthesis and degradation, were investigated. NP
is mainly composed of collagen type II and aggrecan (22). ADAMTS4 and MMP13 are matrix
proteases that are considered to promote disc degenerative changes
during aging (23). Therefore,
these factors were assessed to evaluate the levels of ECM synthesis
and degradation, and the results revealed that siPART1 promoted
cell growth, reduced cell apoptosis and promoted ECM synthesis,
which were similar to the findings of a previous study on prostate
cancer (16).
Preclinical development of a microRNA-based therapy
is a promising approach that is widely applied in the treatment of
IDD. It has been reported that miR-141 promotes the progression of
IDD and, thus, silencing of miR-141 in vivo may represent a
potential therapeutic strategy (24); PART1 acts as a competitive
endogenous RNA for promoting tumor progression through targeting
miR-143 in colorectal cancer (25); and PART1 enhances gefitinib
resistance by binding to miR-129 to facilitate Bcl-2 expression in
esophageal squamous cell carcinoma cells (17). The present study demonstrated that
PART1 targeted and regulated the expression of miR-93 in NP cells.
Conversely, the expression level of miR-93 was reduced in NP
tissues from IDD patients. Previous studies indicated that miR-93
can be sponged by various lncRNAs, such as lncRNA BGL3 (26), lncRNA MEG3 (27), lncRNA MIAT (28), LINC01567 (29) and lnc-NTF3-5 (30), which play important roles in
normal as well as cancerous tissues. In the present study, miR-93
was predicted to be a downstream target of lncRNA PART1 in NP
cells, and it was shown to regulate cell growth, apoptosis and the
expression of ECM-related genes. To the best of our knowledge,
these findings have not been reported previously. The biological
function of miR-93 has been extensively investigated, and miR-93
was found to be a gastric tumor-related miRNA that targets and
regulates E2F1 expression, and forms a negative feedback loop
(31). miR-93 was also found to
be involved in chromatin reorganization and progression of diabetic
nephropathy by modulating Msk2 and its substrate, H3S10 (32). Furthermore, miR-93 induces
repression of cGAS during hypoxia through regulating NCOA3
(33); it also affects the
proliferation of fibroblasts and deposition of ECM through
regulating c-Ski (34). A
previous study also reported that ECM accumulation in uterine
leiomyoma tissues is affected by the expression of certain miRNAs,
including miR-93 (35). It should
be noted that the effects of PART1 on NP cells via miR-93 must be
further verified in vivo.
In the present study, miR-93 was also shown to
target and regulate the expression of MMP2 in NP cells. The
expression level of MMP2 in NP tissues from patients with IDD was
upregulated, which was opposite to the expression of miR-93.
Functional rescue assays also confirmed that miR-93 regulated cell
growth, apoptosis and expression of ECM-related genes through
targeting MMP2. MMP2, which is a member of the MMP gene family, is
an ECM degradation enzyme and is involved in signal transduction.
Integrative bioinformatics analysis and functional experiments
indicated that the MMP2 signaling pathway is closely associated
with the initiation and development of IDD (36,37). MMP2 was also found to be
implicated in the proliferation of pancreatic and breast cancer
cells (38). MMP2 can be targeted
by several other microRNAs, such as miR-29b (39) and miR-34a (40), and it was herein verified that
miR-93 directly targets MMP2 and plays an important role in NP
cells.
In conclusion, lncRNA PART1 promotes degeneration of
NP cells, and it regulates cell proliferation, apoptosis and the
expression of ECM-related genes through targeting miR-93, resulting
in increased MMP2 levels. Taken together, the findings of the
present study indicate the presence of a novel signaling axis in NP
cells that may be further explored for IDD treatment.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DG and LH made substantial contributions to
conception and design; data acquisition, analysis and
interpretation were performed by DG, LH and ZZ; DG, LH and ZZ
drafted the article and critically revised it for important
intellectual content. All authors have read and approved the final
version of the manuscript for publication. All authors agree to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All procedures involving human participants were in
accordance with the ethical standards of the institutional and/or
national research committee and with the 1964 Helsinki Declaration
and its later amendments or comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
NP
|
nucleus pulposus
|
|
IDD
|
intervertebral disc degeneration
|
|
MSC
|
mesenchymal stem cell
|
References
|
1
|
van Uden S, Silva-Correia J, Oliveira JM
and Reis RL: Current strategies for treatment of intervertebral
disc degeneration: Substitution and regeneration possibilities.
Biomater Res. 21:222017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Priyadarshani P, Li Y and Yao L: Advances
in biological therapy for nucleus pulposus regeneration.
Osteoarthritis Cartilage. 24:206–212. 2016. View Article : Google Scholar
|
|
3
|
He F and Pei M: Rejuvenation of nucleus
pulposus cells using extracellular matrix deposited by
synovium-derived stem cells. Spine (Phila Pa 1976). 37:459–469.
2012. View Article : Google Scholar
|
|
4
|
Li P, Liang Z, Hou G, Song L, Zhang R, Gan
Y, Zhang C, Xu Y and Zhou Q: N-Cadherin-mediated activation of
PI3K/Akt-GSK-3β signaling attenuates nucleus pulposus cell
apoptosis under high-magnitude compression. Cell Physiol Biochem.
44:229–239. 2017. View Article : Google Scholar
|
|
5
|
Clarke LE, McConnell JC and Sherratt MJ:
Derby B, Richardson SM and Hoyland JA: Growth differentiation
factor 6 and transforming growth factor-beta differentially mediate
mesenchymal stem cell differentiation, composition, and
micromechanical properties of nucleus pulposus constructs.
Arthritis Res Ther. 16:R672014. View
Article : Google Scholar
|
|
6
|
Chen J, Xie JJ, Jin MY, Gu YT, Wu CC, Guo
WJ, Yan YZ, Zhang ZJ, Wang JL, Zhang XL, et al: Sirt6
overexpression suppresses senescence and apoptosis of nucleus
pulposus cells by inducing autophagy in a model of intervertebral
disc degeneration. Cell Death Dis. 9:562018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W, Li P, Xu J, Wu X, Guo Z, Fan L,
Song R, Wang J, Wei L and Teng H: Resveratrol attenuates high
glucose-induced nucleus pulposus cell apoptosis and senescence
through activating the ROS-mediated PI3K/Akt pathway. Biosci Rep.
38:BSR201714542018. View Article : Google Scholar :
|
|
8
|
Séguin CA, Pilliar RM, Roughley PJ and
Kandel RA: Tumor necrosis factor-alpha modulates matrix production
and catabolism in nucleus pulposus tissue. Spine (Phila Pa 1976).
30:1940–1948. 2005. View Article : Google Scholar
|
|
9
|
Li P, Gan Y, Xu Y, Song L, Wang L, Ouyang
B, Zhang C and Zhou Q: The inflammatory cytokine TNF-α promotes the
premature senescence of rat nucleus pulposus cells via the PI3K/Akt
signaling pathway. Sci Rep. 7:429382017. View Article : Google Scholar
|
|
10
|
Li P, Gan Y, Xu Y, Wang L, Ouyang B, Zhang
C, Luo L, Zhao C and Zhou Q: 17beta-estradiol attenuates
TNF-α-induced premature senescence of nucleus pulposus cells
through regulating the ROS/NF-κB pathway. Int J Biol Sci.
13:145–156. 2017. View Article : Google Scholar :
|
|
11
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif. 50:2017.
View Article : Google Scholar
|
|
13
|
Zhang S, Cao R, Li Q, Yao M, Chen Y and
Zhou H: Comprehensive analysis of lncRNA-associated competing
endogenous RNA network in tongue squamous cell carcinoma. PeerJ.
7:e63972019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lv Y, Wei W, Huang Z, Chen Z, Fang Y, Pan
L, Han X and Xu Z: Long non-coding RNA expression profile can
predict early recurrence in hepatocellular carcinoma after curative
resection. Hepatol Res. 48:1140–1148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li M, Zhang W, Zhang S, Wang C and Lin Y:
PART1 expression is associated with poor prognosis and tumor
recurrence in stage I-III non-small cell lung cancer. J Cancer.
8:1795–1800. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun M, Geng D, Li S, Chen Z and Zhao W:
LncRNA PART1 modulates toll-like receptor pathways to influence
cell proliferation and apoptosis in prostate cancer cells. Biol
Chem. 399:387–395. 2018. View Article : Google Scholar
|
|
17
|
Kang M, Ren M, Li Y, Fu Y, Deng M and Li
C: Exosome-mediated transfer of lncRNA PART1 induces gefitinib
resistance in esophageal squamous cell carcinoma via functioning as
a competing endogenous RNA. J Exp Clin Cancer Res. 37:1712018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei R, Chen Y, Zhao Z, Gu Q and Wu J:
LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via
targeting the Notch signaling pathway. J Cell Physiol.
234:22163–22171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu L, Hao Y, Xu C, Zhu G and Cai Y:
LINC00969 promotes the degeneration of intervertebral disk by
sponging miR-335-3p and regulating NLRP3 inflammasome activation.
IUBMB Life. 71:611–618. 2019. View
Article : Google Scholar
|
|
20
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mern DS, Fontana J, Beierfuß A, Thomé C
and Hegewald AA: A combinatorial relative mass value evaluation of
endogenous bioactive proteins in three-dimensional cultured nucleus
pulposus cells of herniated intervertebral discs: Identification of
potential target proteins for gene therapeutic approaches. PLoS
One. 8:e814672013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patil P, Dong Q, Wang D, Chang J, Wiley C,
Demaria M, Lee J, Kang J, Niedernhofer LJ, Robbins PD, et al:
Systemic clearance of p16INK4a -positive senescent cells
mitigates age-associated intervertebral disc degeneration. Aging
Cell. 18:e129272019. View Article : Google Scholar
|
|
23
|
Ji ML, Jiang H, Zhang XJ, Shi PL, Li C, Wu
H, Wu XT, Wang YT, Wang C and Lu J: Preclinical development of a
microRNA-based therapy for intervertebral disc degeneration. Nat
Commun. 9:50512018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Y, Ma Z, He Y, Liu W, Su Y and Tang Z:
PART-1 functions as a competitive endogenous RNA for promoting
tumor progression by sponging miR-143 in colorectal cancer. Biochem
Biophys Res Commun. 490:317–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen
Y, Ouyang J, Zhang L, Tan H, Chen R, et al: A long noncoding RNA
critically regulates Bcr-Abl-mediated cellular transformation by
acting as a competitive endogenous RNA. Oncogene. 34:1768–1779.
2015. View Article : Google Scholar
|
|
26
|
Zhang L, Liang X and Li Y: Long non-coding
RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and
inactivating PI3K/AKT pathway. Oncol Rep. 38:2408–2416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Wang J, Sun L and Zhu S: LncRNA
myocardial infarction-associated transcript (MIAT) contributed to
cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol.
818:508–517. 2018. View Article : Google Scholar
|
|
28
|
Yu X, Mi L, Dong J and Zou J: Long
intergenic non-protein-coding RNA 1567 (LINC01567) acts as a
'sponge' against microRNA-93 in regulating the proliferation and
tumorigenesis of human colon cancer stem cells. BMC Cancer.
17:7162017. View Article : Google Scholar
|
|
29
|
Peng W, Zhu SX, Wang J, Chen LL, Weng JQ
and Chen SL: Lnc-NTF3-5 promotes osteogenic differentiation of
maxillary sinus membrane stem cells via sponging miR-93-3p. Clin
Implant Dent Relat Res. 20:110–121. 2018. View Article : Google Scholar
|
|
30
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-regulated microRNAs impair TGFbeta-dependent
cell-cycle arrest and apoptosis in gastric cancer. Cancer cell.
13:272–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Badal SS, Wang Y, Long J, Corcoran DL,
Chang BH, Truong LD, Kanwar YS, Overbeek PA and Danesh FR: miR-93
regulates Msk2-mediated chromatin remodelling in diabetic
nephropathy. Nat Commun. 7:120762016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu MZ, Cheng WC, Chen SF, Nieh S, O'Connor
C, Liu CL, Tsai WW, Wu CJ, Martin L, Lin YS, et al: miR-25/93
mediates hypoxia-induced immunosuppression by repressing cGAS. Nat
Cell Biol. 19:1286–1296. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang C, Zhang Y, Zhu H, Hu J and Xie Z:
MiR-34a/miR-93 target c-Ski to modulate the proliferaton of rat
cardiac fibroblasts and extracellular matrix deposition in vivo and
in vitro. Cell Signal. 46:145–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Islam MS, Ciavattini A, Petraglia F,
Castellucci M and Ciarmela P: Extracellular matrix in uterine
leiomyoma pathogenesis: A potential target for future therapeutics.
Hum Reprod Update. 24:59–85. 2018. View Article : Google Scholar
|
|
35
|
Xunlu Y, Minshan F, Liguo Z, Jiawen Z, Xu
W, Jie Y, Shangquan W, He Y, Long L, Tao H and Xuepeng L:
Integrative bioinformatics analysis reveals potential gene
biomarkers and analysis of function in human degenerative disc
annulus fibrosus cells. Biomed Res Int. 2019:98902792019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ,
Chang Q, Chen H and Wang C: Dysregulated miR-98 contributes to
extracellular matrix degradation by targeting IL-6/STAT3 signaling
pathway in human intervertebral disc degeneration. J Bone Miner
Res. 31:900–909. 2016. View Article : Google Scholar
|
|
37
|
Han Y, Ma L, Zhao L, Feng W and Zheng X:
Rosmarinic inhibits cell proliferation, invasion and migration via
up-regulating miR-506 and suppressing MMP2/16 expression in
pancreatic cancer. Biomed Pharmacother. 115:1088782019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Su X and Pan H: Inhibition of lncRNA
PANDAR reduces cell proliferation, cell invasion and suppresses EMT
pathway in breast cancer. Cancer Biomark. 25:185–192. 2019.
View Article : Google Scholar
|
|
39
|
Wang T, Hou J, Jian S, Luo Q, Wei J, Li Z,
Wang X, Bai P, Duan B, Xing J and Cai J: miR-29b negatively
regulates MMP2 to impact gastric cancer development by suppress
gastric cancer cell migration and tumor growth. J Cancer.
9:3776–3786. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang L, Song X, Zhu J, Li M, Ji Y, Wu F,
Chen Y, Cui X, Hu J, Wang L, et al: Tumor suppressor microRNA-34a
inhibits cell migration and invasion by targeting
MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J
Oncol. 51:378–388. 2017. View Article : Google Scholar : PubMed/NCBI
|



















