Introduction
Head and neck cancer, including oral and
oropharyngeal cancer, is the 6th most common malignant tumor
worldwide (1). Oral cancer (OC),
cancer in the oral cavity and lip, with 300,000 new cases and
145,000-related deaths per year, is a major public health concern
globally (2,3). Despite the development of various
treatments, the five-year survival rate of patients with OC has not
improved significantly over the past 30 years (4). These daunting statistics emphasize
the importance of enhancing the comprehensive understanding of the
molecular pathogenesis of OC and the need for the development of
effective therapeutic strategies for these patients.
Exosomes are small, lipid-rich, membrane-bound
microvesicles with a diameter of 30-150 nm (5) that are released into the
extracellular environment under normal or pathological conditions
(6,7). As key microvesicles in cell-cell
communications, exosomes contain a large number of biomolecules,
such as microRNAs (miRNAs or miRs), proteins and lipids (8,9).
Most cell types, including cancer cells, fibroblasts, immune cells
and mesenchymal cells have the ability to secrete exosomes.
Furthermore, it has been reported that the functional role of
exosomes is dependent on the cell of origin (10-12). Numerous studies have demonstrated
that tumor cell-derived exosomes (TDEXs) are involved in tumor
metastasis, drug resistance and tumor immunity, and are widely
distributed in the bodily fluids of cancer patients (13-15). TDEXs are a novel component of the
intercellular communication network and play a vital role in the
tumor microenvironment (16).
Given the diversity of components in TDEXs, the mechanisms through
which TDEXs modulate cancer immunity in OC may be complex.
TDEXs reportedly participate in multiple immune
mechanisms by interfering with the maturation of dendritic cells
(DCs), the function of natural killer (NK) cells and the
'education' of macrophages (17-19). NK cells belong to the innate
immune system and can directly kill tumor cells (20). The activity of NK cells is
regulated by an array of activating and inhibitory receptors on the
cell surface (21). In general,
the inhibitory state is dominant during homeostasis (22,23); however, when activating receptors
are stimulated, NK cells exert cytotoxic effects and exhibit high
levels of antitumor activity (20). NK cells in tumor patients have a
reduced level of activating receptors, such as natural cytotoxicity
triggering receptor 3 (NCR3; also known as NKp30, as used herein),
natural cytotoxicity triggering receptor 1 (NCR1; also known as
NKp46, as used herein), natural cytotoxicity triggering receptor 2
(NCR2; also known as NKp44, as used herein) and killer cell lectin
like receptor K1 (KLRK1; also known as NKG2D, as used herein),
which predicts a poor tumor prognosis (24,25). In addition, TDEXs may influence
natural antitumor immunity by affecting the quantity and function
of NK cells (26,27). Indeed, some studies have
demonstrated that TDEXs stimulate the antitumor immunity of NK
cells (18,28,29), whereas others have indicated that
TDEXs mediate the dysfunction of NK cells through different
mechanisms (30,31). Regardless of this, the functional
role of OC cell-derived exosomes (OCEXs) in NK cell-mediated
antitumor immunity and the underlying mechanisms are far from being
fully understood.
In the present study, exosomes were isolated from OC
cells and the effects of TDEXs on NK cell activity were
investigated. Moreover, the effect of OCEXs on the expression of NK
cell receptors were examined and the underlying mechanisms were
determined. The findings of the present study may enhance the
current understanding of the progression of OCEX-mediated NK cell
education.
Materials and methods
Cells and cell culture
The human OC cell lines, WSU-HN4 (donated by the
University of Maryland) and SCC-9 (purchased from ATCC, CRL-1629)
were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) and DMEM/F-12 (1:1) (basal medium,
Gibco; Thermo Fisher Scientific, Inc.), respectively. The human NK
cell line NK92MI (purchased from ATCC, CRL-2408) was cultured in
RPMI-1640 (basal medium, Gibco; Thermo Fisher Scientific, Inc.).
All media were supplemented with 10% fetal bovine serum (FBS;
Bovogen Biologicals Pty Ltd.) and 1% penicillin-streptomycin
(Sigma-Aldrich; Merck KGaA), and all cells were incubated in a
humidified incubator with 5% CO2 at 37°C. In all OCEX
stimulation experiments, OCEXs were co-cultured with NK92MI cells
for 1, 3 and 7 days. In all transforming growth factor (TGF)-β1
stimulation experiments, recombinant human TGF-β1 (PeproTech, Inc.)
was used at a concentration of 10 ng/ml for 1, 3 and 7 days, as
previously described (32-34).
The SCC-9 cells were seeded in a 96-well plate at a
density of 1,000 cells per well. Approximately 24 h later, the
tumor cells were co-cultured with the NK92MI cells in medium
without FBS and penicillin-streptomycin for 4 h, after which the
culture supernatant was removed. The wells were washed gently with
PBS and observed under an inverted-phase microscope (Zeiss AG).
Exosome isolation
The steps of exosome isolation and purification were
carried out as previously described by Wang et al (28). Briefly, OC cells at 70-80%
confluency were washed 3 times with phosphate-buffered saline (PBS)
and cultured in 10 ml of RPMI-1640 without FBS and
penicillin-streptomycin for 48 h at room temperature. Following
centrifugation for 10 min at 3,214 × g at 4°C, the supernatant was
sequentially microfiltered through polyvinylidene fluoride (PVDF)
membrane filters of 0.45 and 0.22 µm (Merck KGaA) pore size.
The flow-through was ultrafiltered through 100-kDa centrifugal
filter devices (Merck KGaA). Finally, exosomes in the supernatant
were isolated and purified by affinity chromatography using an
ExoEasy Maxi kit (Qiagen GmbH). The quality of the obtained OCEX
fractions was evaluated by transmission electron microscopy (TEM),
nanoparticle tracking analysis (NTA) and western blot analysis.
Transmission electron microscopy
observation
The morphology of the OCEXs was assessed by TEM. In
brief, the isolated exosomes were dropped onto a copper grid for 10
min, after which 2% uranyl acetate was deposited onto the copper
grid for 1 min. The grid was dried for 10 min and examined under a
transmission electron microscope (FEI Tecnai G2 Spirit, Thermo
Fisher Scientific, Inc.).
Nanoparticle tracking analysis
The OCEX size distribution was examined by NTA,
which was performed by a Nano Sight NS300 (Malvern Panalytical,
Ltd.) equipped with rapid video capture and NTA analytical
software. The isolated OCEXs were resuspended in PBS, and the
diameter of the particles was detected.
Western blot analysis
OCEXs were lysed using RIPA lysis buffer (Beyotime
Institute of Biotechnology, Inc.), and the protein concentrations
were measured by a BCA Protein assay (Thermo Fisher Scientific,
Inc.). The protein samples were separated on 10% polyacrylamide
gels at 30 µg/well and transferred to PVDF membranes (Merck
KGaA). The membranes were blocked in 5% skim milk at room
temperature for 1 h and incubated with specific primary antibodies
at 4°C overnight. The membranes were probed with the appropriate
horseradish peroxidase (HRP)-conjugated secondary antibodies
(ProteinTech Group, Inc.) at room temperature for 1 h. Protein
detection was performed with Chemiluminescent HRP Substrate (Merck
KGaA), and signals were captured by an Amersham Imager 600 (GE
Healthcare). Antibodies against the following proteins were used:
CD63 (10628D, mouse monoclonal, 0.5 mg/ml, 1:1,000, Thermo Fisher
Scientific, Inc.), CD9 (10626D, mouse monoclonal, 0.5 mg/ml, 1:500,
Thermo Fisher Scientific, Inc.), Rab5 (A1511-50, rabbit polyclonal,
0.2 mg/ml, 1:500, Biovision, Inc.) and ALG2-interacting protein
(Alix; MA1-83977, mouse monoclonal, 1 mg/ml, 1:500, Thermo Fisher
Scientific, Inc.), TGF-β1 (21898-1-AP, rabbit polyclonal, 300
µg/ml, 1:1,000, ProteinTech Group, Inc.), α-tublin (ab7291,
mouse monoclonal, 1 mg/ml, 1:1,000, Abcam) and GAPDH (10494-1-AP,
rabbit polyclonal, 330 µg/ml, 1:5,000, ProteinTech Group,
Inc.). Secondary antibodies were as follows: Anti-mouse IgG,
HRP-linked antibody (SA0000-1, 1:10,000, Proteintech Group, Inc.),
Anti-rabbit IgG, HRP-linked antibody (SA0000-2, 1:10,000,
Proteintech Group, Inc.).
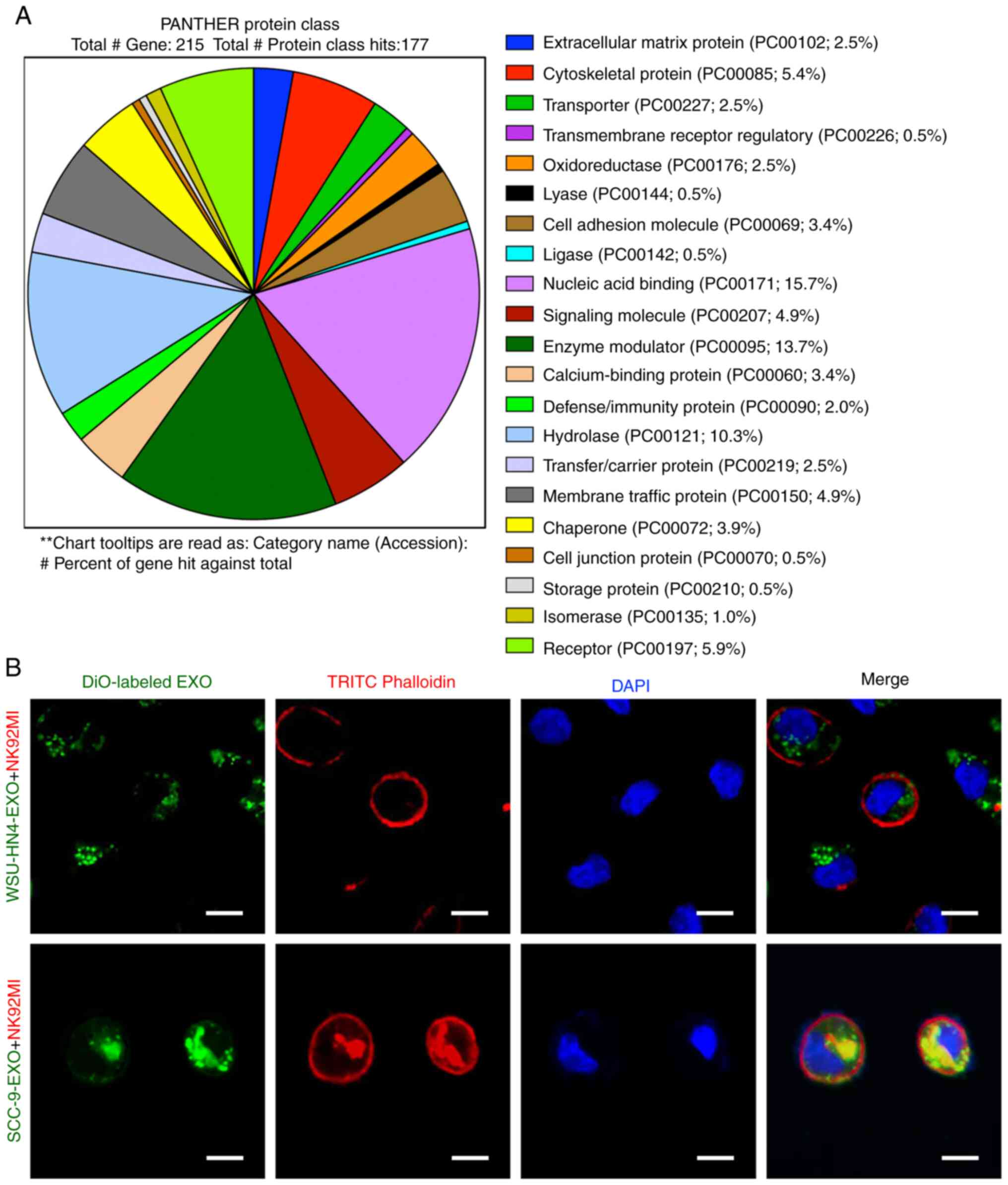
Fluorescence microscopy observation
DiO is a lipophilic fluorescent dye with low
cytotoxicity that can be used to stain cell membranes and other
fat-soluble biological structures. DiO fluorescence is very weak
before entering the cell membrane; however, the intensity is
greatly enhanced when it is combined with the cell membrane and can
be detected with standard FITC filters. Exosomes, as typical
lipid-rich membrane-bound microvesicles, were fluorescently labeled
with DiO. The exosome tracer experiment was implemented with a
green fluorescent DiO cell membrane probe [Yeasen Biotechnology
(Shanghai) Co., Ltd.]. The OCEXs were resuspended and incubated
with 100 µl DiO working solution at 37°C for 20 min.
Following incubation with the OCEXs at 37°C for 24 h, the NK92MI
cells were fixed with 4% paraformaldehyde at room temperature,
stained with TRITC phalloidin [F-actin; Yeasen Biotechnology
(Shanghai) Co., Ltd.] and DAPI [Yeasen Biotechnology (Shanghai)
Co., Ltd.], and observed under a fluorescence microscope (Zeiss
AG).
Cytotoxicity assay
The cytotoxicity of NK92MI cells was determined by a
lactate dehydrogenase (LDH) cytotoxicity assay kit (Beyotime
Institute of Biotechnology, Inc.). Briefly, the WSU-HN4 or SCC-9
cells were seeded in a 96-well plate at a density of 1,000 cells
per well. Approximately 24 h later, the tumor cells were
co-cultured with NK92MI cells in medium without FBS and
penicillin-streptomycin for 4 h. The LDH activity of the samples
was then detected using a multi-well plate reader (Spectra Max i3,
Molecular Devices, LLC) according to the manufacturer's
instructions.
Flow cytometric analysis
The expression of surface markers on NK92MI cells
was analyzed using a BD FACSCalibur flow cytometer (BD
Biosciences). Conjugated fluorescent antibodies were added to an
NK92MI single-cell suspension. Following incubation on ice for 15
min in the dark, the cells were washed twice with PBS and analyzed
by flow cytometry. The following fluorochrome-conjugated monoclonal
antibodies were used according to the manufacturer's instructions:
Anti-NKG2D (cat. no. 558071, clone# 1D11, HU NKG2D APC MAB, BD
Biosciences), anti-NKp30 (cat. no. 558408, clone# p30-15, HU CD337
ALEXA647 MAB, BD Biosciences), anti-NKp44 (cat. no. 558563, clone#
p44-8, HU NKp44 PE MAB, BD Biosciences), anti-NKp46 (cat. no.
557991, clone# 9E2/NKp46, HU NKp46 PE MAB, BD Biosciences) and
anti-NKG2A (cat. no. FAB1059A, clone# 131411, hNKG2A APC MAB,
R&D Systems, Inc.).
Mass spectrometry analysis of OCEXs
OCEXs (50 µg) of were used for mass
spectrometry analyses. The samples were boiled for 10 min and
centrifuged at 14,000 × g at 4°C for 45 min; BCA quantification was
performed on the supernatant, which stored at –80°C. Subsequently,
40 µg per sample was transferred to a new 1.5-ml tube. The
samples were prepared with filter-aided proteome preparation (FASP)
and separated by capillary high-performance liquid chromatography
(HPLC). The samples were transferred from the automatic sampler to
the upper column (EASY column 2 cm × 100 µm × 5 µm
-C18; Thermo Fisher Scientific, Inc.) and separated by the analysis
column (EASY column 75 µm × 100 mm × 3 µm -C18;
Thermo Fisher Scientific, Inc.). Thereafter, the samples were
analyzed by mass spectrometry with a Q-Exactive mass spectrometer
(Thermo Finnigan, LLC). The raw data file was retrieved and
analyzed by MaxQuant_1.3.0.5 software.
Functional enrichment analysis of
exosomal proteins
In the present study, exosomal markers were provided
by ExoCarta databas (http://www.exocarta.org/). An intersection analysis of
exosomal markers and the identified exosomal proteins was carried
out using the Venn diagrams web tool (http://bioin-formatics.psb.ugent.be/webtools/Venn/).
Moreover, functional enrichment analyses for Gene Ontology (GO)
terms (biological processes, molecular functions and cellular
components) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses were conducted through the Database for
Annotation, Visualization and Integrated Discovery (DAVID) v6.8
(https://david.ncifcrf.gov/), and the
data were further developed by using the clusterProfiler package in
R (version 3.6.1). In addition, a functional analysis of these
identified exosomal proteins was conducted using the PANTHER
protein class analysis (http://geneontology.org/; http://pantherdb.org/).
Statistical analyses
Differences between 3 groups were assessed by
one-way ANOVA (with Dunnett's multiple comparisons test) and those
between 2 groups were assessed using an unpaired Student's t-test.
All statistical analyses were performed using GraphPad Prism 6
(GraphPad Software, Inc.). Data were considered statistically
significant when the P-value was <0.05.
Results
Characterization of OCEXs
The OCEXs used in the present study were isolated
from cell culture supernatants of OC cells via ultrafiltration, as
previously described (28). The
morphological characteristics and size of the OCEXs were analyzed
by TEM and NTA. The OCEXs exhibited saucer-like particle shapes
with membranous structures, when examined by TEM (Fig. 1B), which is consistent with the
morphology of exosomes described previously (35). For further accurate diameter
detection, the average size of the OCEXs was detected by NTA. The
diameters of the exosomes derived from the WSU-HN4 and SCC-9 cells
were approximately 149 and 113 nm, respectively (Fig. 1A). Moreover, the common exosomal
protein markers, CD63, Rab5, CD9 and Alix, were detected in the
OCEXs using western blot analysis (Fig. 1C). Collectively, the
above-mentioned data confirmed that the particles isolated from the
OC cells were exosomes.
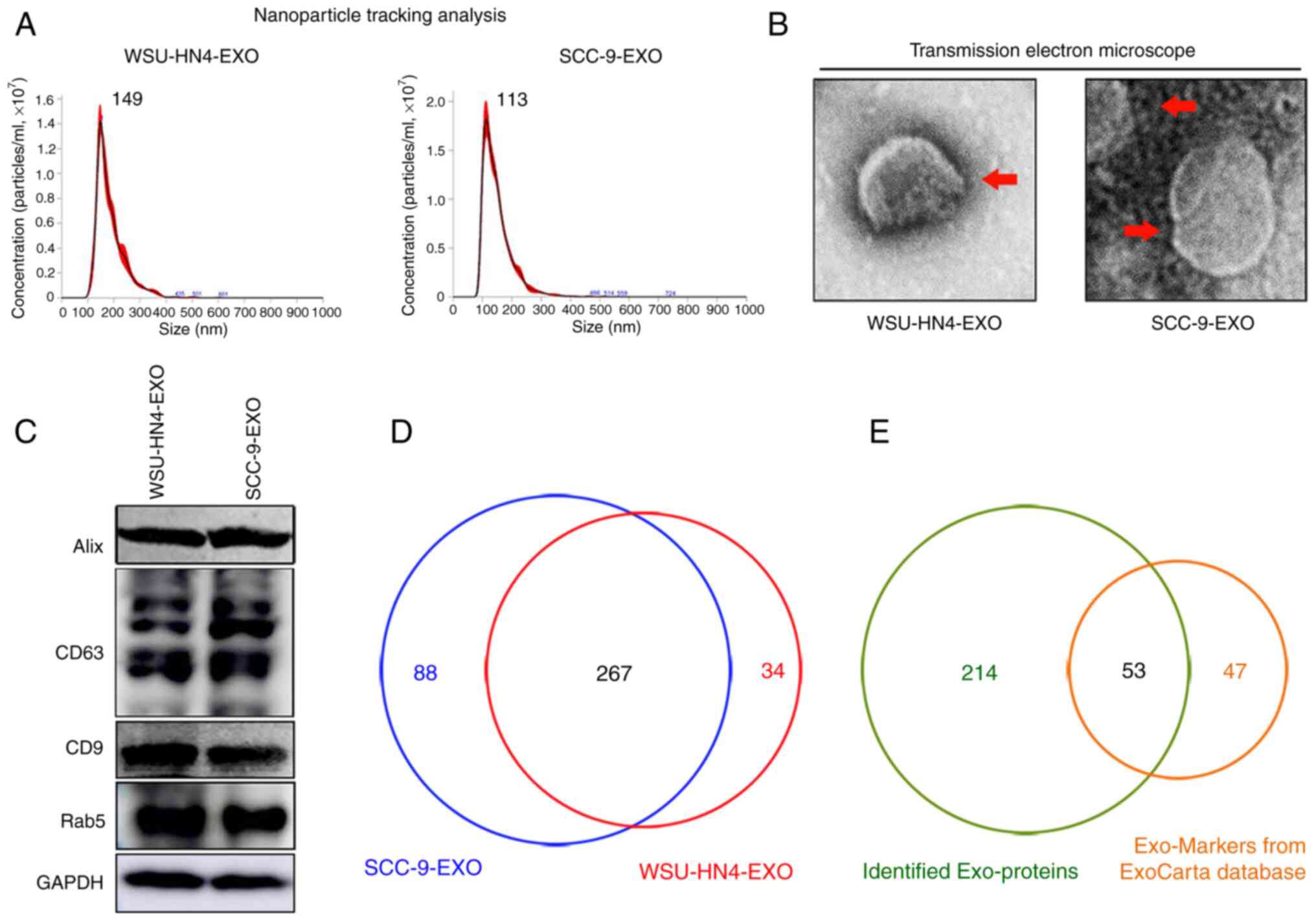 | Figure 1Characterization of OCEXs. (A) The
sizes of the WSU-HN4- and SCC-9-derived exosomes were determined by
NTA. (B) Representative images of exosomes derived from WSU-HN4 and
SCC-9 cells, as detected by TEM. Scale bars, 50 nm. (C) Expression
of the exosomal markers, Alix, CD63, CD9 and Rab5, in WSU-HN4- and
SCC-9-derived exosomes was determined by western blot analysis. (D)
Co-expressed proteins in WSU-HN4- and SCC-9-derived exosomes were
detected by mass spectrometry. (E) Overlap of OCEX proteins and the
top 100 frequently identified exosomal protein markers from the
ExoCarta database. OCEXs, oral cancer-derived exosomes; NTA,
nanoparticle tracking analysis; TEM, transmission electron
microscopy; EXO/Exo, exosomes. |
Comprehensive proteomics analysis of
OCEXs
The proteomics profile of the OCEXs was analyzed by
mass spectrometry. As shown in Fig.
1D, approximately 267 proteins were detected in both the
WSU-HN4- and SCC-9-derived exosomes, with a significant overlap
between the 2 samples. Moreover, 53 of the top 100 most frequently
identified exosomal protein markers according to the ExoCarta
database (www.exocarta.org) were detected in the
OCEXs, indicating the accuracy of the results (Table SI). Cellular component analysis
(https://david.ncifcrf.gov/) was then
performed on these 267 identified proteins, and the majority of the
identified proteins (215/267) were annotated as components of
extracellular exosomes (Fig.
S1A). To further elucidate the functions of these exosomal
proteins, these 215 proteins were subjected to molecular function,
biological process and KEGG pathway analyses (https://david.ncifcrf.gov/). The molecular function
analysis revealed the enrichment of proteins related to protein
binding, protein kinase binding, protein heterodimerization
activity, protein domain specific binding, poly(A) RNA binding,
identical protein binding, GTP binding, DNA binding, calcium ion
binding and cadherin binding and involvement in cell-cell adhesion
(Fig. S1B). According to
biological process analysis, the proteins in the OCEXs were
involved in small GTPase-mediated signal transduction, signal
transduction, protein transport, platelet degranulation, nucleosome
assembly, negative regulation of apoptotic processes, leukocyte
migration, extracellular matrix organization, chromatin silencing
and cell adhesion (Fig. S1C). In
addition, KEGG pathway analysis revealed the identified proteins to
be enriched in alcoholism, systemic lupus erythematosus, pathways
in cancer, viral carcinogenesis, endocytosis and the PI3k-Akt
signaling pathway, among others, which are closely associated with
cancer progression (Fig. S1D).
The results of the PANTHER protein class (http://geneontology.org/) analysis revealed that the
exosomes derived from the WSU-HN4 and SCC-9 cells contained various
types of proteins (Fig. 2A).
Among these protein types, transmembrane receptor regulatory
proteins (PC00226; 0.5%), cell adhesion molecules (PC00069; 3.4%),
calcium-binding proteins (PC00060; 3.4%), membrane traffic proteins
(PC00150; 4.9%) and receptors (PC00197; 5.9%) have been reported to
be associated with exosome uptake, i.e., internalization by
recipient cells (9,36). To determine whether OCEXs can be
internalized by NK cells, the OCEXs were stained with DiO (green)
and added to the culture medium of NK92MI cells, followed by
incubation for 24 h. Under a fluorescence microscope, it was
observed that the DiO-labeled OCEXs appeared in the cytoplasm of
the NK92MI cells (Fig. 2B). Taken
together, these results indicate that OCEXs are incorporated by NK
cells, suggesting their potential role in regulating the functions
of NK cells.
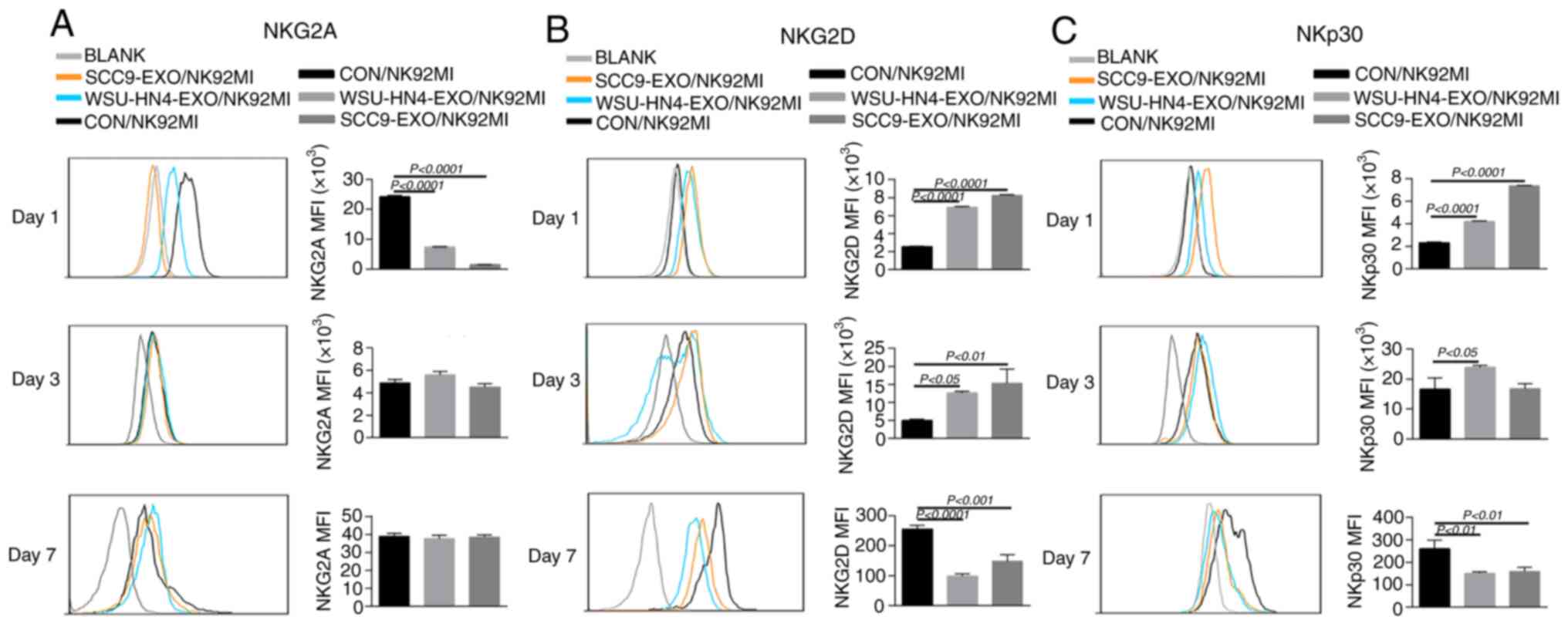
Changes in receptor expression on NK92MI
cells induced by OCEXs
The function of NK cells is tightly regulated by
activating and inhibitory receptors. NKG2D and NKp30, NKp44, NKp46
are important activating receptors on NK cells, and their
expression levels are positively associated with their antitumor
capacity. Additionally, the expression of NKG2A, one of most
important inhibitory receptors of NK cells, is negatively
associated with the antitumor activity of NK cells (37). Hence, the present study
investigated the effects of OCEXs on NKG2D, NKp30, NKp44, NKp46 and
NKG2A expression by treating the NK92MI cells with OCEXs for 1, 3
and 7 days. As a result, the expression of activating receptors
(NKG2D, NKp30, NKp44 and NKp46) on the NK92MI cells was
significantly upregulated after 1 day (Figs. 3B and C and S2A and B), whereas the expression of
NKG2A was evidently decreased following treatment with the OCEXs
for 1 day (Fig. 3A). The
above-mentioned data suggest that short-term treatment with OCEXs
increases the cytotoxicity of NK cells. Of note, it was found that
the expression of activating receptors on NK cells decreased
gradually over time, with the expression of the activating
receptors, NKG2D, NKp30 and NKp44, diminishing following co-culture
with the OCEXs for 7 days (Figs. 3B
and C and S2A and B).
Conversely, no significant changes in NKG2A expression were
observed at 7 days (Fig. 3A).
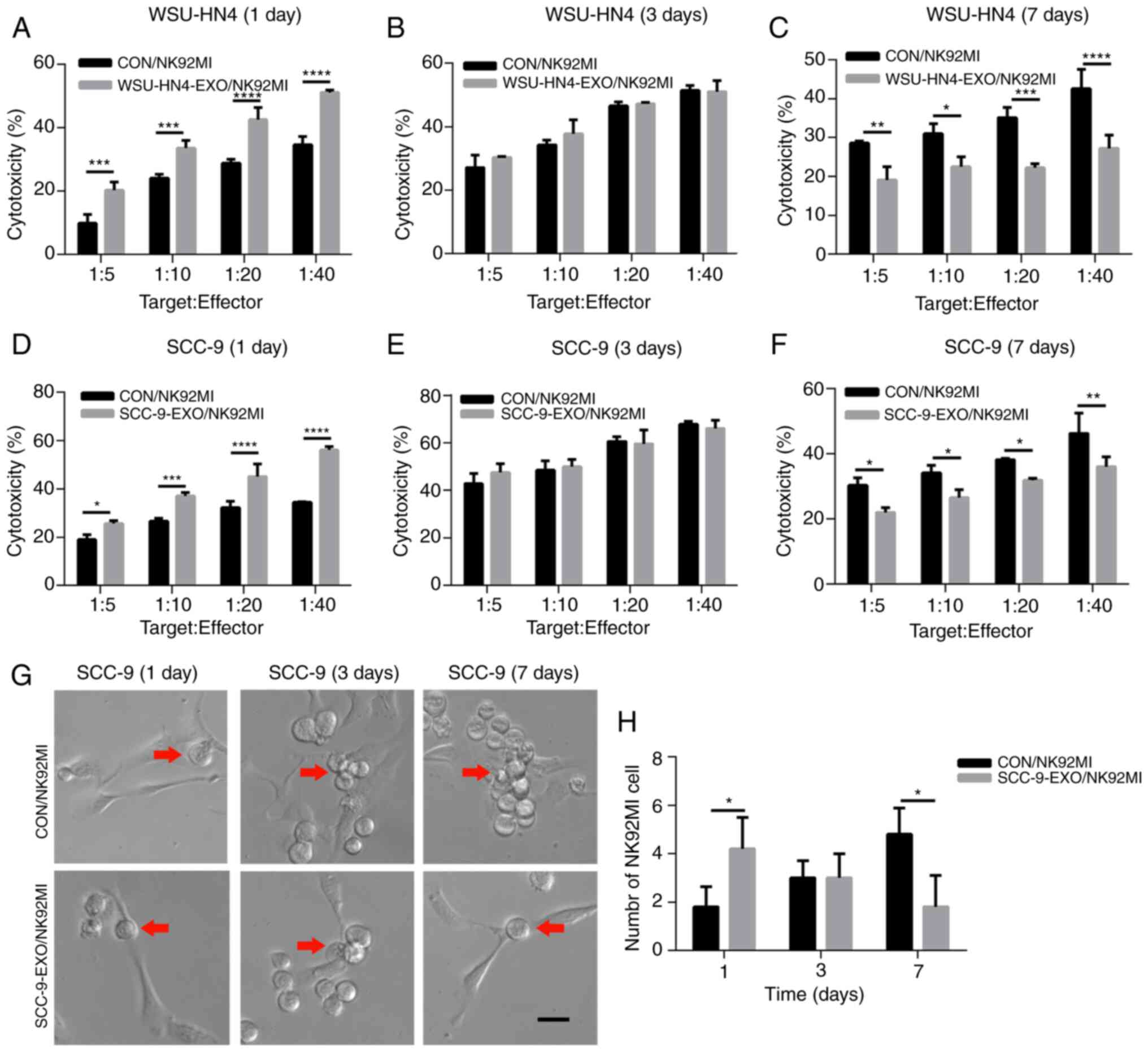
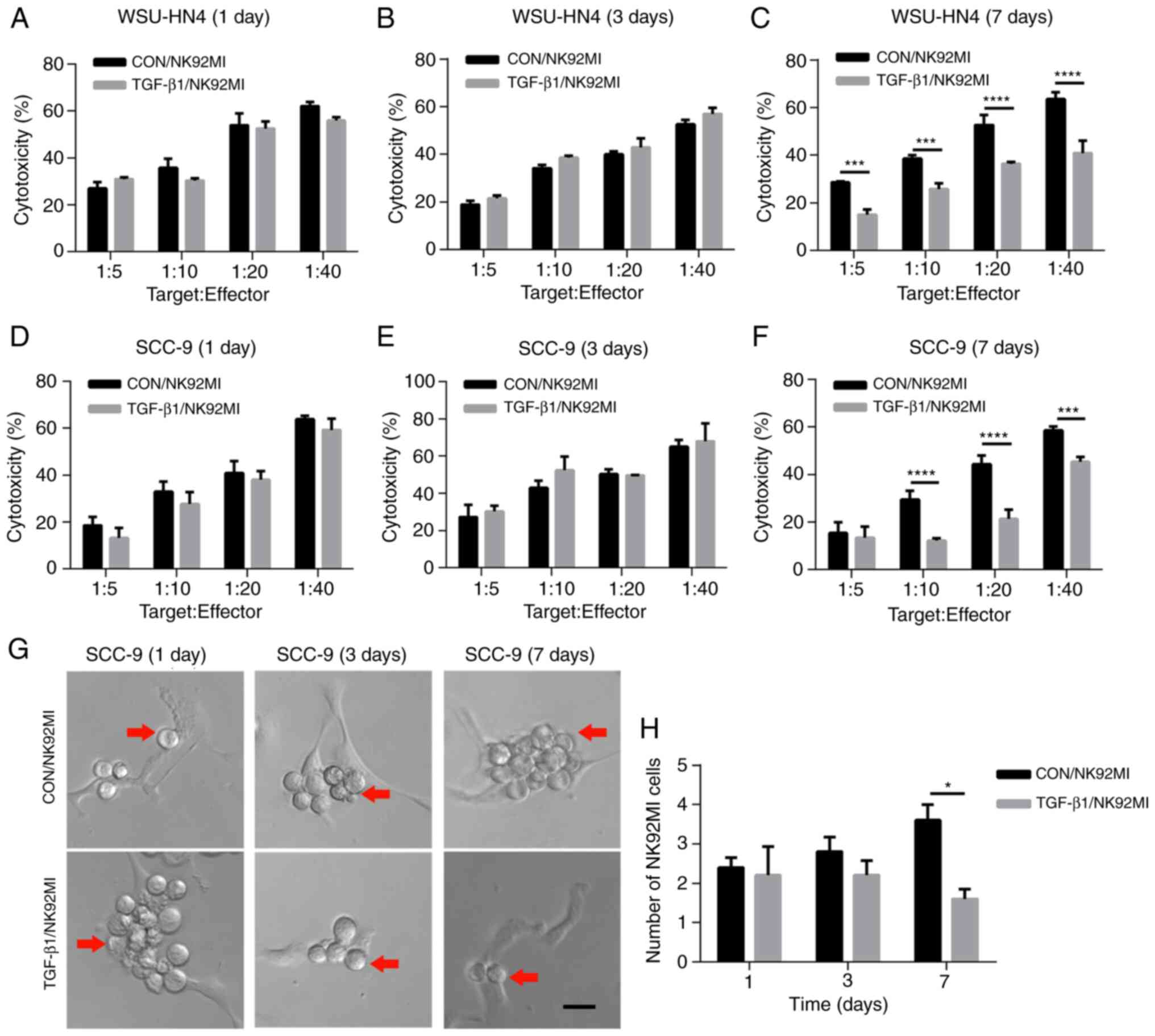
Changes in the cytotoxicity of NK92MI cells induced by OCEXs
To investigate whether the trend of NK cell
cytotoxicity was consistent with that of NK cell receptor
expression, the NK92MI cells were treated with the OCEXs for 1, 3
and 7 days, and NK92MI cell cytotoxicity was detected by LDH assay.
Functionally, the OC cell-killing effect of the NK92MI cells was
increased following incubation with the OCEXs for 1 day (Fig. 4A and D). In addition, increased
numbers of NK92MI cells attached to the SCC-9 cells were observed
at 1 day (Fig. 4G and H). As the
OCEX incubation time increased, the cytotoxicity of the NK92MI
cells compared to that of the control cells was decreased at 7 days
(Fig. 4C and F), and the number
of NK92MI cells adhering to the SCC-9 cells was reduced at the same
time point (Fig. 4G and H).
However, there were no clear differences in cytotoxicity between
the OCEX-treated and control cells at 3 days (Fig. 4B and E). Hence, the
above-mentioned data demonstrated that the OCEXs stimulated NK
cytotoxicity at 1 day and this was then inhibited with time up to 7
days.
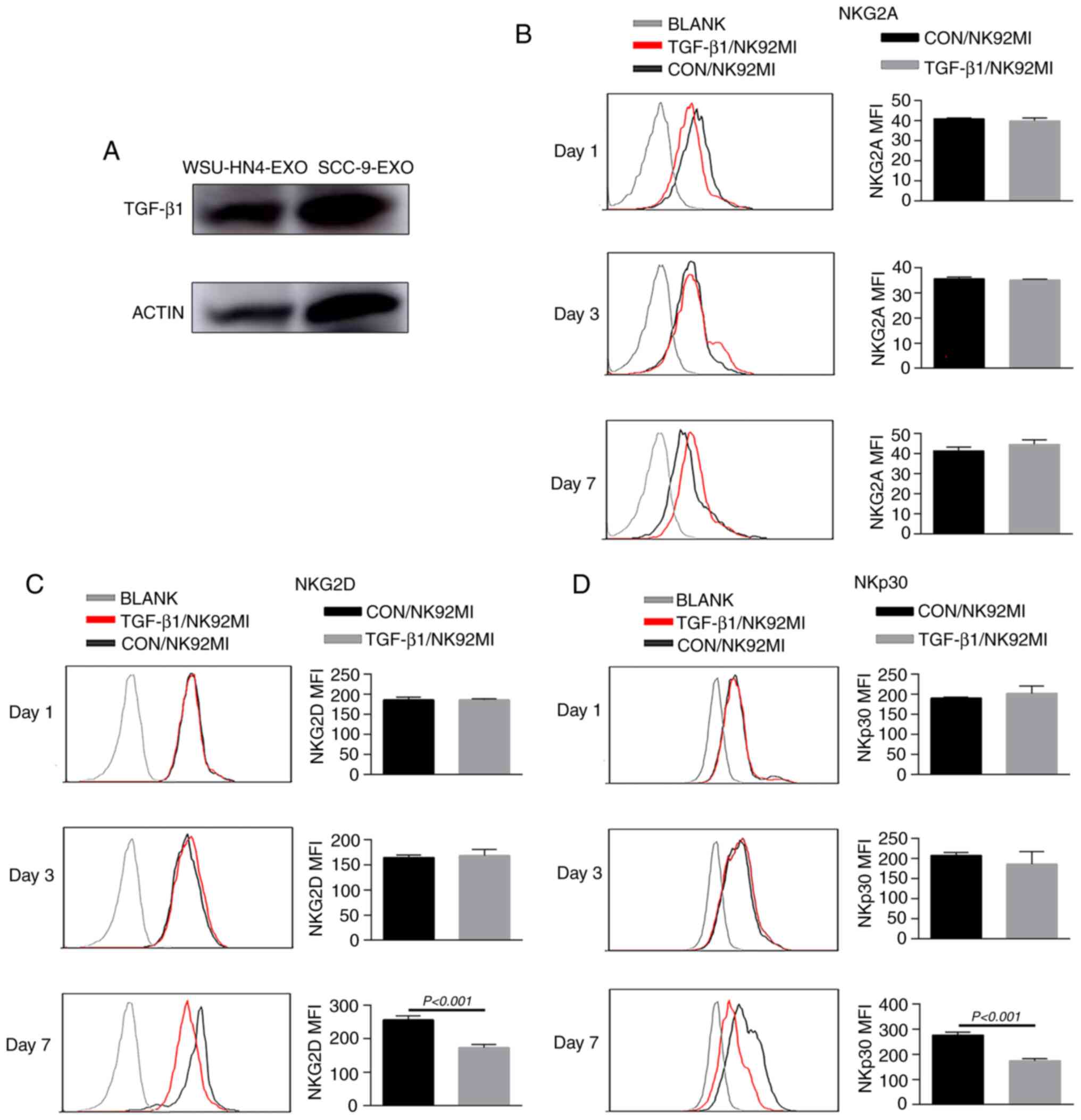
OCEX-derived TGF-β1 inhibits the
cytotoxicity of NK cells
TGF-β1, a cytokine of the bone morphogenetic protein
(BMP)-activin family, participates in a wide range of immune system
regulatory processes (38-41).
TGF-β1 is an acknowledged inhibitory factor of immune system
components, including NK cells. In particular, TGF-β1 has been
reported to reduce the surface expression of crucial activating NK
receptors (NKp30 and NKG2D) on NK cells (42). Of note, in the present study, the
immunoregulatory factor, TGF-β1, was enriched in OCEXs, as
determined by mass spectrometric analysis (Table SII), and the expression of TGF-β1
in the OCEXs was verified by western blot analysis (Fig. 5A). To further assess the effects
of TGF-β1 on NK cell activity, NK92MI cells were treated with or
without TGF-β1 for different periods of time, and expression of NK
cell receptors and cytotoxicity were evaluated. There were no
obvious changes in receptor expression or cytotoxicity to the
NK92MI cells treated with TGF-β1 for 1 and 3 days. However, the
expression levels of activating receptors (NKp30 and NKG2D) and the
cytotoxicity of the TGF-β1-treated cells decreased significantly
compared to those of the control cells at 7 days (Figs. 5 and 6). However, other receptors (NKp44,
NKp46 and NKG2A) exhibited no significant changes in expression
(Figs. 5B and S2C and D). Moreover, the number of
NK92MI cells attached to the SCC-9 cells was reduced (Fig. 6G and H). These findings suggest
that OCEXs deliver TGF-β1 to NK cells, ultimately resulting in the
dysfunction of NK cells.
Discussion
OC is one of the most lethal malignancies worldwide
(2), and the long-term survival
rate and prognosis of patients with advanced OC are poor (43). Therefore, an enhanced
understanding of the pathogenesis and mechanisms of OC is
warranted. Despite tremendous research advances, the underlying
cellular and molecular events involved in OC have yet to be
determined. Over the past few decades, exosomes have attracted
increasing attention due to their roles in the occurrence and
development of tumors. In the present study, OCEXs were isolated
from 2 human oral cancer cell lines, WSU-HN4 and SCC-9, by
ultra-filtration. Comprehensive proteomics analyses are expected to
reveal the potential function of OCEXs. Exosomes are known to carry
a variety of cargo, including diverse RNAs, proteins including
cytokines, growth factors and lipids (43-45). In the present study, the protein
components carried in OCEXs were mainly analyzed by mass
spectrometry. The results of PANTHER protein class analysis
revealed the significant enrichment of proteins related to
localization and biological adhesion, which may promote the ability
of OCEXs to adhere to the surfaces of recipient cells, fuse with
their membranes, and transfer exosomal components to modulate the
biological functions of the target cells.
Exosomes are important cellular crosstalk
nanovesicles that participate in a number of biological processes
(46). In the tumor
microenvironment, exosomal contents can be conveyed not only to
donor cells but also to different types of immune cells, including
NK cells, thus affecting the antitumor immune response (47,48). For example, the antitumor immune
responses of NK cells are mediated by the direct killing of cancer
cells and cytokine production. NK cell activation is tightly
regulated by an interplay between excitatory and inhibitory
signals, the latter mainly occurring through NKG2A. NKG2D and
natural cytotoxicity receptors, such as NKp30, are potent
activating receptors on the surface of NK cells for tumor killing
(37,49). It has been reported that heat
shock protein (HSP)-bearing exosomes secreted by hepatocellular
carcinoma cells enhance the function of NK cells. In this process,
the expression of the activating receptors, CD69, NKG2D and NKp44,
is upregulated, whereas that of the inhibitory receptor CD94 is
downregulated (50). The authors
have previously demonstrated that the cytotoxicity of NK cells is
temporarily enhanced by OCEXs (28). The results of the present study
verified that the short-term incubation of OCEXs with NK cells
indeed promoted the function of NK cells. Consequently, when the NK
cells were co-cultured with the OCEXs for 1 day, the upregulated
expression of the activating receptors, NKp30 and NKG2D, and the
downregulated expression of the inhibitory receptor, NKG2A, were
detected. Simultaneously, the cytotoxicity of NK cells was
enhanced, and the number of NK cells adhering to cancer cells was
increased at 1 day.
Nevertheless, previous studies have demonstrated
that TDEXs exert an inhibitory effect on the immune system via NK
cells (30,51), and an impaired NK cell function in
head and neck, pharyngeal and other solid cancers has been
reported. The effects of TDEXs on NK cells may be affected by
several factors, such as the cancer cell type, cancer progression
stage and the microenvironment (52,53). The results of the present study
demonstrated that activating receptor expression decreased with the
prolonged incubation of NK cells with OCEXs; the killing function
of NK cells also decreased accordingly. These data reveal a change
in NK receptor expression, as well as in NK cytotoxicity with
increasing OCEX and NK cell incubation times. Thus, the interaction
between OCEXs and NK cells increased the tumor-suppressive effects
of NK cells in the short-term, whereas it gradually decreased or
even reversed the effects, such as cytotoxicity, over time.
The activation of NK cells is dependent on a shift
in the signaling balance between inhibitory and activating
receptors (37). The factors
controlling the capacity of balance between the activating and
inhibitory signals have not been fully determined. In the present
study, the function of NK cells was ultimately suppressed by
prolonged stimulation with OCEXs. To examine the in-depth
mechanisms, the levels of proteins in the OCEXs were detected, and
TGF-β1 was found to be enriched. It has been suggested that
patients with head and neck squamous cell carcinoma who exhibit
persistently elevated TGF-β1 levels following chemoradiotherapy
have a poor cancer prognoses (54). In addition, numerous studies have
demonstrated that TGF-β1 exerts an obvious inhibitory effect on NK
cells. When exposed to TGF-β1, NK cells may lose the ability to
kill cancer cells in humans or mice (55). Studies have also indicated that
the level of serum TGF-β1 is associated with cancer metastasis and
a poor prognosis (56).
Therefore, it was hypothesized that the inhibition of NK cell
function may be associated with TGF-β1 enrichment in OCEXs. In the
present study, the inhibitory function of TGF-β1 on NK92MI cells at
7 days was verified, and it was found that NK cell function was not
significantly altered following stimulation with TGF-β1 for 1 and 3
days. Due to the diverse inclusions of exosomes, the effects of
OCEXs on NK cells may be complex (57). Our previous study also
demonstrated that OCEX-derived NAP1 enhanced cytotoxicity of NK
cells (28). In previous studies,
NK cell receptor expression has been shown to be altered following
co-culture with TGF-β1 for 3 days, and NKG2D and NKp30 have been
shown to be inhibited (39,58). It is possible that the stimulating
effects of TGF-β1-treated OCEXs on NK cells may not be evident at
first, and gradually become prominent following the extension of
the co-culture time. Therefore, it was preliminarily hypothesized
that the OCEX-induced NK cell inhibition during 7 days of
co-incubation may be related to the enrichment of TGF-β1 in OCEXs;
nonetheless, the internal mechanisms need to be further explored
and investigated. The phenotype of NK cells can be modified by
TGF-β1. Recently, it was reported that a high TGF-β1 expression was
associated with a low expression of activating receptors (such as
NKG2D and NKp30) on NK cells in the peripheral blood of human
cancer patients, which is positively related to disease progression
(33). In the present study, the
expression of the activating receptors, NKG2D and NKp30, was
downregulated in impaired NK cells treated with either OCEXs or
TGF-β1, suggesting that TGF-β1 contained in OCEXs is related to NK
cell dysfunction.
In conclusion, the present study found that exosomes
derived from OC cells were internalized by NK cells and caused
changes in receptor expression on the NK cell surface. Short-term
incubation with OCEXs enhanced the function of NK cells, whereas
long-term incubation consistently resulted in decreased NK cell
cytotoxicity. Further analysis revealed that TGF-β1 in OCEXs
gradually inhibited NK cell function, deepening the understanding
of the mechanisms through which tumor-derived exosomes modulate the
functional role of NK cells in the tumor microenvironment. However,
it should also be noted that the results of the present study
require further confirmation in vivo and further studies are
required to identify the specific mechanisms. In addition, whether
there are other factors involved in NK cell dysfunction needs to be
further explored. Taken together, the findings of the present study
provide a different perspective on the important role of exosomes
in the immune system, particularly in OCs.
Supplementary Data
Acknowledgments
The authors would like to thank Professor Li Mao
(Maryland University, USA) for providing the oral cancer cell line,
WSU-HN4.
Funding
The present study was supported by grants from the
National Key Research and Development Program of China (no.
2016YFC0902700), the National Natural Science Foundation of China
(nos. 81874126 and 91229103 and 81672 745), and the Shanghai
Science and Technology Commission (no. 18DZ2291500).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
WCh and JZ made substantial contributions to the
conception and design of the study and gave the final approval of
the version to be published. XZ and XQ collated the data, performed
the data analyses, produced the initial draft of the manuscript and
revised it. JZ, WCa, YW and XW participated in collating and
analyzing the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lala M, Chirovsky D, Cheng JD and Mayawala
K: Clinical outcomes with therapies for previously treated
recurrent/metastatic head-and-neck squamous cell carcinoma (R/M
HNSCC): A systematic literature review. Oral Oncol. 84:108–120.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinomaan update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sasahira T and Kirita T: Hallmarks of
cancer-related newly prognostic factors of oral squamous cell
carcinoma. Int J Mol Sci. 19:24132018. View Article : Google Scholar :
|
|
4
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar
|
|
5
|
Brinton LT, Sloane HS, Kester M and Kelly
KA: Formation and role of exosomes in cancer. Cell Mol Life Sci.
72:659–671. 2015. View Article : Google Scholar
|
|
6
|
Waldenstrom A and Ronquist G: Role of
exosomes in myocardial remodeling. Circ Res. 114:315–324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Syn N, Wang L, Sethi G, Thiery JP and Goh
BC: Exosome-mediated metastasis: From epithelial-mesenchymal
transition to escape from immunosurveillance. Trends Pharmacol Sci.
37:606–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allenson K, Castillo J, San Lucas FA,
Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ,
et al: High prevalence of mutant KRAS in circulating
exosome-derived DNA from early-stage pancreatic cancer patients.
Ann Oncol. 28:741–747. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mathieu M, Martin-Jaular L, Lavieu G and
Thery C: Specificities of secretion and uptake of exosomes and
other extracellular vesicles for cell-to-cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, An
T, Yao S and Zheng L: Single-exosome-counting immunoassays for
cancer diagnostics. Nano Lett. 18:4226–4232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Chopp M, Meng Y, Katakowski M,
Xin H, Mahmood A and Xiong Y: Effect of exosomes derived from
multipluripotent mesenchymal stromal cells on functional recovery
and neurovascular plasticity in rats after traumatic brain injury.
J Neurosurg. 122:856–867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van den Boorn JG, Dassler J, Coch C,
Schlee M and Hartmann G: Exosomes as nucleic acid nanocarriers. Adv
Drug Deliv Rev. 65:331–335. 2013. View Article : Google Scholar
|
|
13
|
Fang T, Lv H, Lv G, Li T, Wang C, Han Q,
Yu L, Su B, Guo L, Huang S, et al: Tumor-derived exosomal
miR-1247-3p induces cancer-associated fibroblast activation to
foster lung metastasis of liver cancer. Nat Commun. 9:1912018.
View Article : Google Scholar :
|
|
14
|
Shedden K, Xie XT, Chandaroy P, Chang YT
and Rosania GR: Expulsion of small molecules in vesicles shed by
cancer cells: Association with gene expression and chemosensitivity
profiles. Cancer Res. 63:4331–4337. 2003.PubMed/NCBI
|
|
15
|
Whiteside TL: Exosomes and tumor-mediated
immune suppression. J Clin Invest. 126:1216–1223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar B, Garcia M, Weng L, Jung X,
Murakami JL, Hu X, McDonald T, Lin A, Kumar AR, DiGiusto DL, et al:
Acute myeloid leukemia transforms the bone marrow niche into a
leukemia-permissive microenvironment through exosome secretion.
Leukemia. 32:575–587. 2018. View Article : Google Scholar :
|
|
17
|
Whiteside TL: The effect of tumor-derived
exosomes on immune regulation and cancer immunotherapy. Future
Oncol. 13:2583–2592. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Q, Huang Q, Huyan T, Wang Y, Huang Q
and Shi J: Bifacial effects of engineering tumour cell-derived
exosomes on human natural killer cells. Exp Cell Res. 363:141–150.
2018. View Article : Google Scholar
|
|
19
|
Cai J, Qiao B, Gao N, Lin N and He W: Oral
squamous cell carcinoma-derived exosomes promote M2 subtype
macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am
J Physiol Cell Physiol. 316:C731–C740. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Childs RW and Carlsten M: Therapeutic
approaches to enhance natural killer cell cytotoxicity against
cancer: The force awakens. Nat Rev Drug Discov. 14:487–498. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guillerey C, Huntington ND and Smyth MJ:
Targeting natural killer cells in cancer immunotherapy. Nat
Immunol. 17:1025–1036. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gasser S and Raulet DH: Activation and
self-tolerance of natural killer cells. Immunol Rev. 214:130–142.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raulet DH and Vance RE: Self-tolerance of
natural killer cells. Nat Rev Immunol. 6:520–531. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mamessier E, Sylvain A, Thibult ML,
Houvenaeghel G, Jacquemier J, Castellano R, Goncalves A, Andre P,
Romagne F, Thibault G, et al: Human breast cancer cells enhance
self tolerance by promoting evasion from NK cell antitumor
immunity. J Clin Invest. 121:3609–3622. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pietra G, Manzini C, Rivara S, Vitale M,
Cantoni C, Petretto A, Balsamo M, Conte R, Benelli R, Minghelli S,
et al: Melanoma cells inhibit natural killer cell function by
modulating the expression of activating receptors and cytolytic
activity. Cancer Res. 72:1407–1415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu MC, Xiong P, Li GL and Zhu M: Could
lung cancer exosomes induce apoptosis of natural killer cells
through the p75NTR-proNGF-sortilin axis? Med Hypotheses.
108:151–153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mincheva-Nilsson L and Baranov V: Cancer
exosomes and NKG2D receptor-ligand interactions: Impairing
NKG2D-mediated cytotoxicity and anti-tumour immune surveillance.
Semin Cancer Biol. 28:24–30. 2014. View Article : Google Scholar
|
|
28
|
Wang Y, Qin X, Zhu X and Chen W, Zhang J
and Chen W: Oral cancer-derived exosomal NAP1 enhances cytotoxicity
of natural killer cells via the IRF-3 pathway. Oral Oncol.
76:34–41. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shoae-Hassani A, Hamidieh AA, Behfar M,
Mohseni R, Mortazavi-Tabatabaei SA and Asgharzadeh S: NK
cell-derived exosomes from NK cells previously exposed to
neuroblastoma cells augment the antitumor activity of
cytokine-activated NK cells. J Immunother. 40:265–276. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berchem G, Noman MZ, Bosseler M, Paggetti
J, Baconnais S, Le Cam E, Nanbakhsh A, Moussay E, Mami-Chouaib F,
Janji B and Chouaib S: Hypoxic tumor-derived microvesicles
negatively regulate NK cell function by a mechanism involving TGF-β
and miR23a transfer. Oncoimmunology. 5:e10629682015. View Article : Google Scholar
|
|
31
|
Clayton A, Mitchell JP, Court J, Linnane
S, Mason MD and Tabi Z: Human tumor-derived exosomes down-modulate
NKG2D expression. J Immunol. 180:7249–7258. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Donatelli SS, Zhou JM, Gilvary DL,
Eksioglu EA, Chen X, Cress WD, Haura EB, Schabath MB, Coppola D,
Wei S and Djeu JY: TGF-β-inducible microRNA-183 silences
tumor-associated natural killer cells. Proc Natl Acad Sci USA.
111:4203–4208. 2014. View Article : Google Scholar
|
|
33
|
Han B, Mao FY, Zhao YL, Lv YP, Teng YS,
Duan M, Chen W, Cheng P, Wang TT, Liang ZY, et al: Altered NKp30,
NKp46, NKG2D, and DNAM-1 expression on circulating NK cells is
associated with tumor progression in human gastric cancer. J
Immunol Res. 2018:62485902018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Viel S, Marçais A, Guimaraes FS, Loftus R,
Rabilloud J, Grau M, Degouve S, Djebali S, Sanlaville A, Charrier
E, et al: TGF-β inhibits the activation and functions of NK cells
by repressing the mTOR pathway. Sci Signal. 9:ra192016. View Article : Google Scholar
|
|
35
|
Contreras-Naranjo JC, Wu HJ and Ugaz VM:
Microfluidics for exosome isolation and analysis: Enabling liquid
biopsy for personalized medicine. Lab Chip. 17:3558–3577. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gonda A, Kabagwira J, Senthil GN and Wall
NR: Internalization of exosomes through receptor-mediated
endocytosis. Mol Cancer Res. 17:337–347. 2019. View Article : Google Scholar
|
|
37
|
Chiossone L, Dumas PY, Vienne M and Vivier
E: Natural killer cells and other innate lymphoid cells in cancer.
Nat Rev Immunol. 18:671–688. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Casu B, Dondero A, Regis S, Caliendo F,
Petretto A, Bartolucci M, Bellora F, Bottino C and Castriconi R:
Novel immunoregulatory functions of IL-18, an accomplice of TGF-β1.
Cancers (Basel). 11:752019. View Article : Google Scholar
|
|
39
|
Rouce RH, Shaim H, Sekine T, Weber G,
Ballard B, Ku S, Barese C, Murali V, Wu MF, Liu H, et al: The
TGF-β/SMAD pathway is an important mechanism for NK cell immune
evasion in childhood B-acute lymphoblastic leukemia. Leukemia.
30:800–811. 2016. View Article : Google Scholar
|
|
40
|
Liénart S, Merceron R, Vanderaa C, Lambert
F, Colau D, Stockis J, van der Woning B, De Haard H, Saunders M,
Coulie PG, et al: Structural basis of latent TGF-β1 presentation
and activation by GARP on human regulatory T cells. Science.
362:952–956. 2018. View Article : Google Scholar
|
|
41
|
Dedobbeleer O, Stockis J, van der Woning
B, Coulie PG and Lucas S: Cutting Edge: Active TGF-β1 released from
GARP/TGF-β1 complexes on the surface of stimulated human B
lymphocytes increases class-switch recombination and production of
IgA. J Immunol. 199:391–396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fujii R, Jochems C, Tritsch SR, Wong HC,
Schlom J and Hodge JW: An IL-15 superagonist/IL-15Rα fusion complex
protects and rescues NK cell-cytotoxic function from
TGF-β1-mediated immunosuppression. Cancer Immunol Immunother.
67:675–689. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carnielli CM, Macedo CCS, De Rossi T,
Granato DC, Rivera C, Domingues RR, Pauletti BA, Yokoo S, Heberle
H, Busso-Lopes AF, et al: Combining discovery and targeted
proteomics reveals a prognostic signature in oral cancer. Nat
Commun. 9:35982018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kourembanas S: Exosomes: Vehicles of
intercellular signaling, biomarkers, and vectors of cell therapy.
Annu Rev Physiol. 77:13–27. 2015. View Article : Google Scholar
|
|
45
|
Filipazzi P, Burdek M, Villa A, Rivoltini
L and Huber V: Recent advances on the role of tumor exosomes in
immunosuppression and disease progression. Semin Cancer Biol.
22:342–349. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Whiteside TL: Exosome and mesenchymal stem
cell cross-talk in the tumor microenvironment. Semin Immunol.
35:69–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li I and Nabet BY: Exosomes in the tumor
microenvironment as mediators of cancer therapy resistance. Mol
Cancer. 18:322019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kruse PH, Matta J, Ugolini S and Vivier E:
Natural cytotoxicity receptors and their ligands. Immunol Cell
Biol. 92:221–229. 2014. View Article : Google Scholar
|
|
50
|
Lv LH, Wan YL, Lin Y, Zhang W, Yang M, Li
GL, Lin HM, Shang CZ, Chen YJ and Min J: Anticancer drugs cause
release of exosomes with heat shock proteins from human
hepatocellular carcinoma cells that elicit effective natural killer
cell antitumor responses in vitro. J Biol Chem. 287:15874–15885.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Reiners KS, Topolar D, Henke A, Simhadri
VR, Kessler J, Sauer M, Bessler M, Hansen HP, Tawadros S, Herling
M, et al: Soluble ligands for NK cell receptors promote evasion of
chronic lymphocytic leukemia cells from NK cell anti-tumor
activity. Blood. 121:3658–3665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reiners KS, Dassler J, Coch C and Pogge
von Strandmann E: Role of exosomes released by dendritic cells
and/or by tumor targets: Regulation of NK cell plasticity. Front
Immunol. 5:912014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Habif G, Crinier A, Andre P, Vivier E and
Narni-Mancinelli E: Targeting natural killer cells in solid tumors.
Cell Mol Immunol. 16:415–422. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen JL, Chang CC, Huang YS, Kuo HY, Chen
TY, Wang CW, Kuo SH and Lin YL: Persistently elevated soluble MHC
class I polypeptide-related sequence A and transforming growth
factor-β1 levels are poor prognostic factors in head and neck
squamous cell carcinoma after definitive chemoradiotherapy. PLoS
One. 13:e02022242018. View Article : Google Scholar
|
|
55
|
Jun E, Song AY, Choi JW, Lee HH, Kim MY,
Ko DH, Kang HJ, Kim SW, Bryceson Y, Kim SC and Kim HS: Progressive
impairment of NK cell cytotoxic degranulation is associated with
TGF-β1 deregulation and disease progression in pancreatic cancer.
Front Immunol. 10:13542019. View Article : Google Scholar
|
|
56
|
Astrom P, Juurikka K, Hadler-Olsen ES,
Svineng G, Cervigne NK, Coletta RD, Risteli J, Kauppila JH, Skarp
S, Kuttner S, et al: The interplay of matrix metalloproteinase-8,
transforming growth factor-β1 and vascular endothelial growth
factor-C cooperatively contributes to the aggressiveness of oral
tongue squamous cell carcinoma. Br J Cancer. 117:1007–1016. 2017.
View Article : Google Scholar
|
|
57
|
Vulpis E, Soriani A, Cerboni C, Santoni A
and Zingoni A: Cancer exosomes as conveyors of stress-induced
molecules: New players in the modulation of NK cell response. Int J
Mol Sci. 20:6112019. View Article : Google Scholar :
|
|
58
|
Klöß S, Chambron N, Gardlowski T, Arseniev
L, Koch J, Esser R, Glienke W, Seitz O and Köhl U: Increased sMICA
and TGFβ1 levels in HNSCC patients impair
NKG2D-dependent functionality of activated NK cells.
Oncoimmunology. 4:e10559932015. View Article : Google Scholar
|