Introduction
Diabetes is the leading cause of morbidity and
mortality worldwide, and has affected >415 million of the world
population (1). Type-2 diabetes
mellitus (T2DM) is characterized by an impaired insulin secretion
and insulin resistance in adipose, liver and muscle tissues
(2). Obesity is the principal
causative factor for the development of multiple metabolic
diseases, including cardiovascular disease, neuropathy,
retinopathy, chronic kidney disease and nephropathy, as well as the
increasing healthcare costs (3).
Various genetic and environmental factors may contribute to the
progression of T2DM; however, the mechanisms responsible for the
development of T2DM remain unclear (4).
It has previously been demonstrated that obesity is
one of the most crucial factors that contributes to T2DM (5). As an energy depository, adipose
tissue also produces several bioactive molecules/adipokines to
regulate insulin sensitivity, glucose metabolism and lipid
metabolism-modulating processes, such as adipogenesis,
angiogenesis, hypertension, bone morphogenesis and cell adhesion
(6). The activation of brown
adipose tissue (BAT) can ameliorate metabolic abnormalities in
obesity and can thus resolve the common complications associated
with obesity, including T2DM (7).
The study conducted by Zhao et al revealed that the deletion
of α/β-hydrolase domain 6 promoted the browning of the adipose
tissue and prevented T2DM (8).
Insulin suppresses lipolysis in white adipose tissue (WAT) to
reduce fatty acid flux to the liver, resulting in obesity and T2DM
(9). In spite of these findings
however, the exact mechanisms underlying the association between
obesity and diabetes remain largely unknown.
Long non-coding RNAs (lncRNAs) are a class of
protein incapability transcripts of 200 nucleotides in length, and
have been identified as important regulators of diabetes (10). Carter et al reported that
circulating lncRNA GAS5 expression was associated with the
population of T2DM (11). The
inhibition of lncRNA NONRATT021972 alleviates diabetic neuropathic
pain via the P2X3 receptor in the dorsal root ganglia (12). Leti and DiStefano reported that
lncRNAs function as diagnostic and therapeutic targets in T2DM and
related complications (13).
Recently, the TUG1/TraF5 signaling pathway has been reported to be
involved in podocyte apoptosis in rats with diabetic nephropathy
through the modulation of extracellular matrix accumulation
(14,15). However, the function of TUG1 in
diabetes has rarely been reported, at least to the best of our
knowledge.
Sirtuin 1 (SIRT1) is a member of the mammalian
sirtuins, and is characterized by NAD+-dependent
deacetylases and ADP-ribosyl transferases that function in multiple
cellular process, including DNA repair, gene silencing and
metabolic regulation (16). SIRT1
has been reported to be a therapeutic target for T2DM (17), and has the function of improving
energy efficiency and preventing diabetes in mice (18). miR-204 has been reported to play
key roles in the processes of obesity and diabetes (19). However, the mechanisms of the
miR-204-SIRT1 axis remain unclear as regards the regulation of
diabetes.
In the present study, a diabetic mouse model was
constructed to explore the function of the lncRNA TUG1, SIRT1 and
miR-204 in diabetes. In addition, the regulatory mechanisms of and
associations between TUG1, SIRT1 and miR-204 were confirmed in
3T3-L1 cells, followed by pathway investigation. According to these
investigations, it is hoped the findings of the present study may
provide novel new insight into the pathogenesis and treatment of
diabetes.
Materials and methods
Construction of the diabetic mouse
model
The animal proto-cols used in the present study were
authorized by the Ethics Committee of the Shanghai General Hospital
of Nanjing Medical University. The use of animals was also approved
by the same Ethics Committee. A total of 100 C57BL/6J mice (5
weeks, weighing 24±2 g), were purchased from Slac Laboratory Animal
Co., Ltd., and housed under a 12-h light/dark cycle, SPF
conditions, with free access to food and water. Following 1 week of
adaptation, these mice were randomly divided into 2 groups.
Specifically, mice in the control group (n=20) were fed a normal
diet (3% kcal from fat, Harlan Teklad), and mice in the model group
(n=80) were fed a high-fat diet (45% kcal from fat, D12451,
Research Diet Inc.). After 6 weeks, the mice in the model group
were intraperitoneally injected with streptozotocin (50 mg/kg; EMD
Chemicals, Inc.; Merck KGaA) to establish the diabetic model. Blood
samples (0.1-0.2 ml for each mouse) were then collected via tail
vein blood collection and blood glucose levels were measured
weekly. When the blood glucose of mice was >11.0 mmol/l, the
insulin level was considered to be markedly increased. The mice in
the model group were then further randomly divided into 3 groups as
follows: The model (n=23), TUG1 NC (n=23) and TUG1 (n=24) groups.
The mice in the TUG1 NC group were injected with the TUG1 NC virus
via the tail vein, while the mice in the TUG1 group were injected
with the TUG1 overexpression virus via the tail vein. The
lentivirus of TUG1 or TUG1 NC were purchased from Promega (Beijing
Biotech Co., Ltd.). After 8 weeks, the mice were anesthetized by an
intraperitoneal injection with 10% chloral hydrate (300 mg/kg; no
peritonitis or any signs of discomfort were observed in the mice)
and sacrificed by cervical dislocation, blood samples (1.0-1.2 ml
per mouse) were collected via the tail vein prior to euthanasia by
cervical dislocation. Fatty tissues were also collected for the
following investigations. During the experimental process, mouse
behavior was routinely monitored, the weight of the mice was
measured each day and blood glucose levels were measured weekly. A
total of 8 mice (3 in the model group, 2 in the TUG1 NC group and 3
in the TUG1 group) were excluded due to severe infection. For the
excluded mice, they were anesthetized by an intraperitoneal
injection with 10% chloral hydrate (300 mg/kg) and sacrificed by
cervical dislocation.
Detection of associated indices in mice
with obesity
In the process of the establishment of the model,
body weight and serum glucose levels were detected each week, and
the fasting blood glucose (FBG), triglyceride (TG), glycerol and
non-esterified fatty acid (NEFA) levels were determined at the 8th
week using ELISA kits (Sigma-Aldrich; Merck KGaA), according to the
manufacturer's instructions.
Immunohistochemistry (IHC)
Following collection, part of the tissues was fixed
with 4% paraformaldehyde (PFA) at 4°C overnight. The tissues were
then dehydrated by gradient ethanol, permeabilized by xylene and
embedded in paraffin. Subsequently, these tissues were cut into
4-µm-thick sections. The paraffin tissue slices were then
dewaxed by heating, and rehydrated with gradient ethanol. EDTA
antigen reduction solution (Sigma-Aldrich; Merck KGaA) was then
used to repair the antigens on the sections in a microwave oven at
40°C, followed by natural cooling. After washing twice with
phosphate-buffered saline (PBS, 3 min per time), the sections were
blocked with 10% bovine serum albumin for 30 min, and incubated
with uncoupling protein-1 (UCP-1) antibody (1:200; cat. no.
ab10983, Abcam) at 4°C overnight. After washing 3 times with PBS (3
min per time), the sections were incubated with the secondary
antibody (1:1,000; cat. no. ab205718, Abcam) at room temperature
for 50 min, followed by hematoxylin and eosin (H&E) staining at
room temperature for 10 min. Finally, the sections were washed 3
times with PBS (5 min per time), mounted with neutral resins and
analyzed under a light microscope (Nikon Eclipse 1000, Nikon
Corporation).
Cell culture and transfection
The 3T3-L1 cells were purchased from the Chinese
Center of Type Culture Collection and maintained in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.), supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 µg/ml of streptomycin
(Sigma-Aldrich; Merck KGaA) and 100 µg/ml of penicillin
(Sigma-Aldrich; Merck KGaA), in a humidity incubator with 5%
CO2 at 37°C. When the cells grew to 70% confluency, they
were transfected with miR-204 mimics, miR-204 inhibitor,
mimics/inhibitor NC, si-SIRT1 and si-SIRT1 NC, as well as the
pshRNA-H1-Luc lentivirus vector (all 10 nM for the above) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to manufacturer's protocol. For transfection, the
mimics/inhibitor, siRNA or vectors were incubated with
Lipofectamine 2000 solution at room temperature for 20 min,
following by 48 h of incubation in a humidity incubator with 5%
CO2 at 37°C. The miR-204 mimic, miR-204 inhibitor, NCs
and plasmids were purchased from KeyGen Biotech. These cells were
used for the downstream investigation at 48 h following
transfection. The miR-204 inhibitor and inhibitor NC were provided
by KeyGen Biotech with no sequence information. The sequences for
miR-204 NC, miR-204 mimics, si-SIRT1 NC and si-SIRT1are listed as
follows: miR-204 mimics NC, 5′-AUC CGG AAU UUU AAG CCU AUC UGA-3′;
miR-204 mimics, 5′-UUC CCU UUG UCA UCC UAU GCC U-3′; si-SIRT1 NC,
5′-UUA UGC UAG CAU CGA UCA AGC-3′; si-SIRT1, 5′-GCA GGT TGC AGG AAT
CCA AAG-3′. To establish the high glucose-induced 3T3-L1 fatty
model, 3T3-L1 cells were exposed to 25 mM glucose [D(+) glucose,
G-5400, Sigma-Aldrich; Merck KGaA] at 48 h post-transfection for an
additional 48 hours. The control group was left untreated.
Dual-luciferase reporter activity
assay
Briefly, the 3T3-L1 cells were seeded in 48-well
plates, and maintained in 200 µl of DMED containing 10% FBS
overnight. For the determination of the regulatory mechanisms
between miR-204 and TUG1, the cells were transfected using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to manufacturer's protocol, and divided into the
following groups: The pGL6-TUG1-WT+miR-204 NC, pGL6-TUG1-WT +
miR-204 mimic, pGL6 -TUG1-MUT + miR-204 NC and pGL6-TUG1-MUT +
miR-204 mimic groups. For the regulatory confirmation between
mR-204 and SIRT1, the cells were respectively transfected according
to the requirement of each assay, and divided into the following
groups: The pGL6-SIRT1-WT + miR-204 NC, pGL6-SIRT1-WT + miR-204
mimic, pGL6-SIRT1-MUT + miR-204 NC and the pGL6-SIRT1-MUT + miR-204
mimic groups. The plasmids and miR-204 mimics were purchased from
Promega (Beijing Biotech Co., Ltd.). At 48 h following
transfection, these cells were washed 3 times with PBS, and the
luciferase activity was measured using the Dual-Luciferase reporter
assay system (GloMax; Promega Corporation), according to
manufacturer's protocol. The Firefly luciferase activity was
normalized to the Renilla luciferase activity.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the tissues or cell
samples using TRIzol reagent (Takara Biotechnology Co., Ltd.),
according to manufacturer's protocols. Subsequently, 2 µg of
total RNA were reverse transcribed into first-strand cDNA using the
5X PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd.) or
the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems),
according to manufacturer's instructions. The amplifications of the
genes or miRNAs were performed using SYBR-Green (Varioskan Flash;
Thermo Fisher Scientific, Inc.) under the following conditions:
95°C for 10 min, 95°C for 3 min, and 40 cycles of 95°C for 10 sec
and 60°C for 30 sec. The housekeeping gene, GAPDH, was set as the
gene internal control, and U6 was used as the miRNA internal
control. The primers used for RT-qPCR are listed in Table I.
 | Table IList of primers used for RT-qPCR. |
Table I
List of primers used for RT-qPCR.
| Gene | Forward primer | Reverse primer |
|---|
| TUG1 |
5′-CTGAAGAAAGGCAATCCATC-3′ |
5′-GTAGGCTACTACAGGTCATTTG-3′ |
| SIRT1 |
5′-TGCTGGCCTAATAGAGTGGCA-3′ |
5′-CTCAGCGCCATGGAAAATGT-3′ |
| miR-204 |
5′-ACACTCCAGCTGGGTTCCCTTTGTCATCCT-3′ |
5′-CTCAACTGGTGTCGTGGAGTCGG-3′ |
| ATGL |
5′-CCCACTTCAACTCCAAGGACGA-3′ |
5′-GCAGGTTGTCTGAAATGCCACC-3′ |
| PPARα |
5′-TCGGCGAACTATTCGGCTG-3′ |
5′-GCACTTGTGAAAACGGCAGT-3′ |
| PGC-α |
5′-AAACTTGCTAGCGGTCCTCA-3′ |
5′-TGGCTGGTGCCAGTAAGAG-3′ |
| UCP-1 |
5′-CAAAAACAGAAGGATTGCCGAAA-3′ |
5′-TCTTGGACTGAGTCGTAGAGG-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
| GAPDH |
5′-ATCACTGCCACCCAGAAGAC-3′ |
5′-TTTCTAGACGGCAGGTCAGG-3′ |
Western blot analysis
Protein was extracted from the tissue and cell
samples using RIPA lysis buffer containing a protease inhibitor
cocktail (Roche Diagnostics), and qualified using the BCA kit
(Pierce; Thermo Fisher Scientific, Inc.). Proteins (50 µg
per lane) were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and
transferred onto polyvinylidene difluoride (PVDF) membranes
(Amersham Biosciences). The membranes were then blocked with TBS
Tween containing 5% non-fat milk (Inner Mongolia Yili Industrial
Group Co., Ltd.) at room temperature for 1 h, followed by
incubation with primary antibodies to SIRT1 (1:800, cat. no.
ab110304, Abcam), adipose triglyceride lipase (1:1,000, ATGL; cat.
no. ab207799, Abcam), MAPK (1:1,000, cat. no. ab31828, Abcam),
p-acetyl-CoA carboxylase (1:800, ACC; cat. no. ab68191, Abcam), ACC
(1:800, cat. no. ab45174, Abcam), peroxisome proliferator-activated
receptor α (1:1,000, PPARα; cat. no. ab215270, Abcam), peroxisome
proliferator-activated receptor gamma coactivator 1-α (1:1,000,
PGC-1α; cat. no. ab54481, Abcam), UCP-1 (1:1,000, cat. no. ab10983,
Abcam) and GAPDH (1:2,000, cat. no. sc-365062, Santa Cruz
Biotechnology, Inc.) at 4°C overnight. The membranes were then
incubated with goat anti-rabbit IgG and goat anti-mouse IgG
(1:2,000, cat. no. ab205718 and cat. no. ab6708, Abcam) at room
temperature for 1 h, and developed using the ECL plus kit
(PerkinElmer, Inc.). Densitometry was performed using the
Alphalmager™ 2000 Imaging System (Alpha Innotech Corporation).
Statistical analysis
All experiments in the present study were performed
in triplicate, and data are presented as the means ± standard
deviation (SD). Statistical analyses in the present study were
performed using GraphPad Prism 6.0. Statistical significance was
assessed by a Student's t-test or one-way analysis of variance
(ANOVA) with Turkey's post hoc tests, and P<0.05 was considered
to indicate a statistically significant difference.
Results
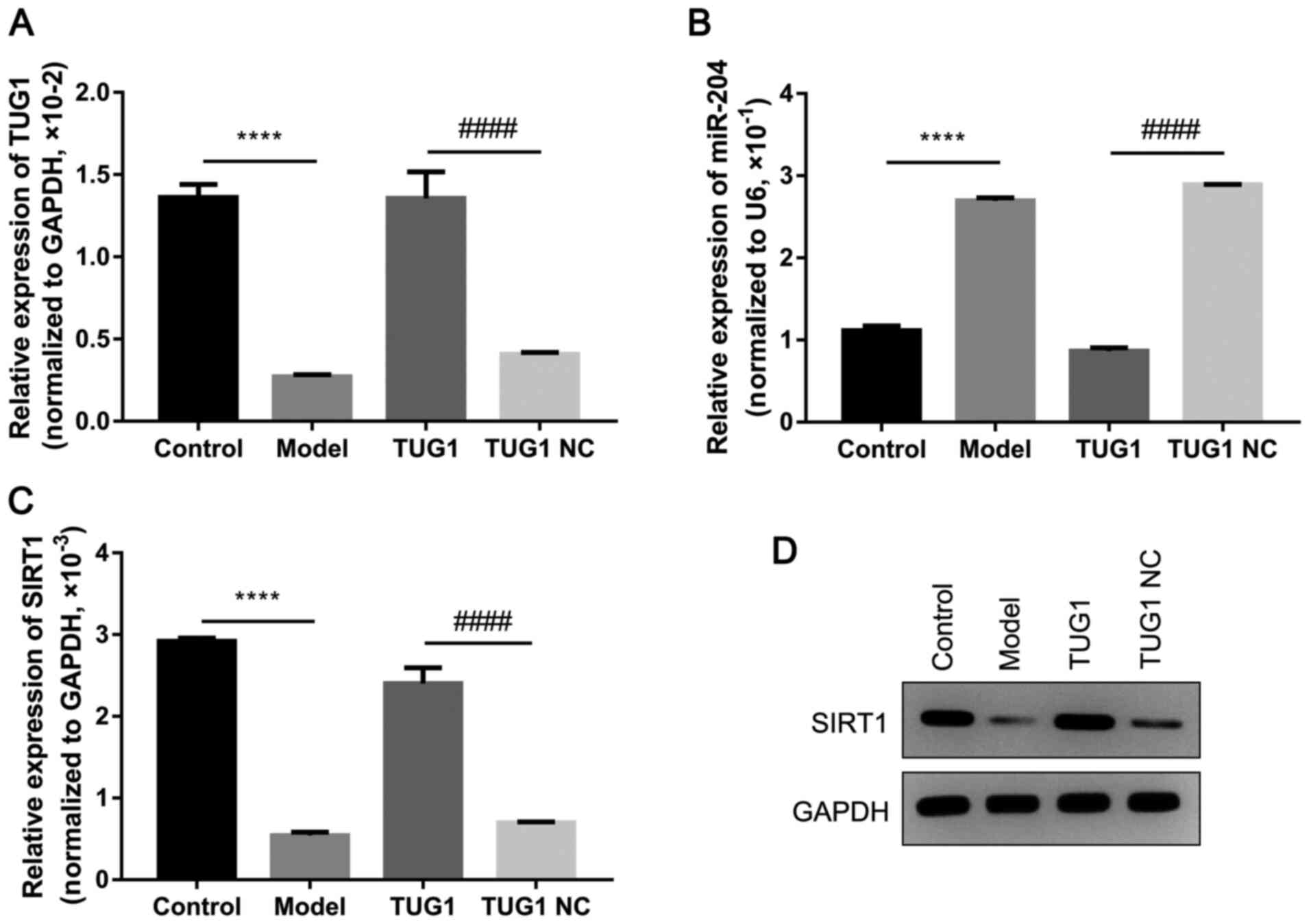
Overexpression of TUG1 decreases miR-204
expression, but increases SURT1 expression
After establishing the diabetic mouse model, the
abdominal subcutaneous adipose tissues of the mice were obtained to
detect the expression of miR-204 and SIRT1. The results of RT-qPCR
revealed that TUG1 was significantly downregulated in the diabetic
mice; however, the injection of TUG1 significantly increased the
expression of TUG1 in the adipose tissues (all, P<0.05; Fig. 1A). Furthermore, the expression of
miR-204 was significantly increased in the adipose tissues of the
diabetic mice, and the overexpression of TUG1 significantly
inhibited the expression of miR-204 in the diabetic mice (all,
P<0.05; Fig. 1B). In addition,
the mRNA and protein levels of SIRT1 were significantly
downregulated in the diabetic mice; however, the upregulation of
TUG1 markedly increased the expression of SIRT1 in the diabetic
mice (all, P<0.05; Fig. 1C and
D).
Overexpression of TUG1 attenuates the
weight gain and abnormal metabolism of diabetic mice
As important indices in diabetes, the variations of
weight and metabolism were also measured in the present mouse
model. The results revealed that the injection of TUG1
significantly attenuated the elevated weight and serum glucose
levels in the diabetic mice (all, P<0.05; Fig. 2A and B). In addition, the TUG1
injection also markedly suppressed the upregulation of the
diabetes-associated metabolic indices, including FBG, TG, glycerol
and NEFA (all, P<0.05; Fig.
2C).
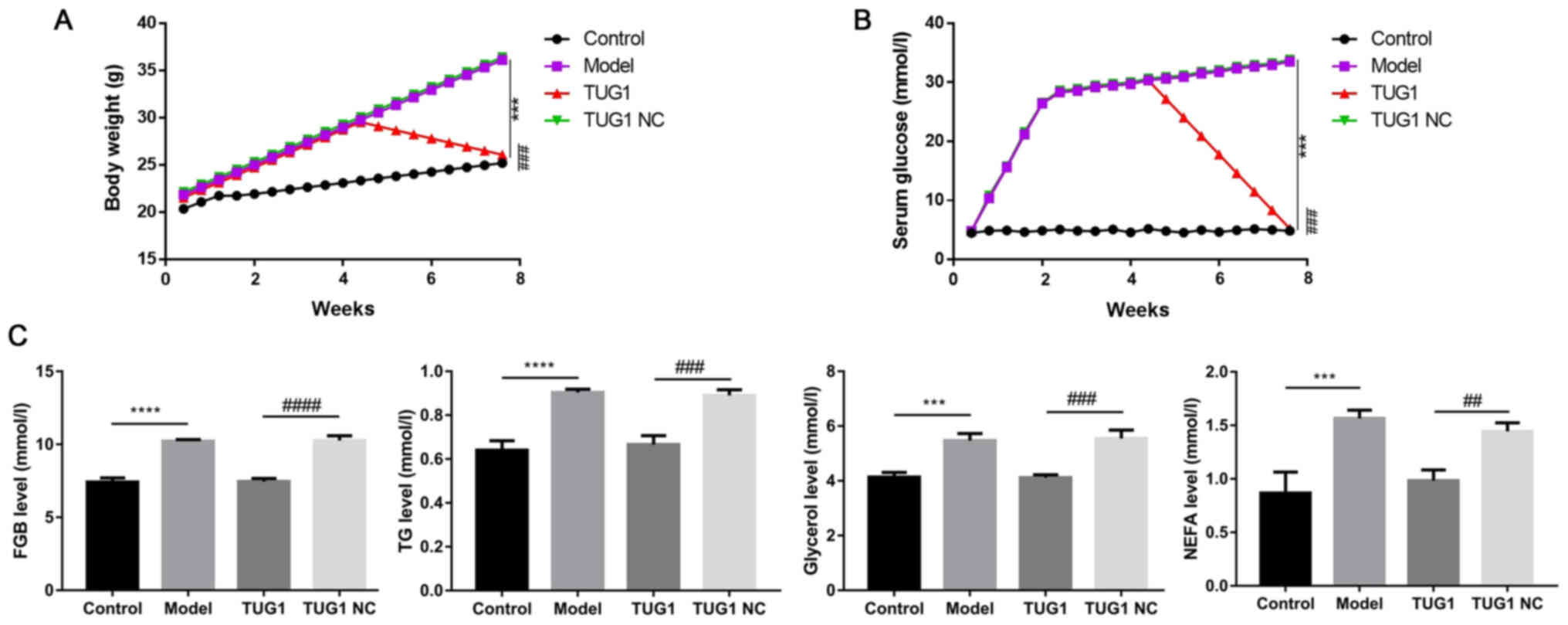 | Figure 2TUG1 improves weight accumulation and
promotes adipose metabolism in diabetic mice. (A) Body weight of
the diabetic mice in the control, model, TUG1 and TUG1 NC groups,
respectively. (B) Serum glucose levels in these 4 groups. (C)
Levels of serum fasting blood glucose (FBG), TG, glycerol and NEFA
were detected. NC, negative control; TG, triglyceride; NEFA,
non-esterified fatty acid. ***P<0.001 and
****P<0.0001, compared with the control group;
##P<0.01, ###P<0.001 and
####P<0.0001, compared with the TUG1 NC group. Each
experiment was repeated at least 3 times. |
Overexpression of TUG1 promotes fat
degradation, fatty acid oxidation and brown fat construction
In order to further reveal the biofunction of TUG1,
the testicular adipose tissues of mice were collected, and it was
found that the injection of TUG1 markedly attenuated the
accumulation of testicular adipose tissue in diabetic mice (all,
P<0.05; Fig. 3A). H&E
staining revealed that the adipocytes were significantly larger in
the diabetic mice when compared to those in the control group, and
that the TUG1 injection markedly inhibited this augmentation, when
compared to the NC group (Fig.
3B). Furthermore, IHC assay revealed that the expression of
UCP-1 significantly decreased in the diabetic mice, and that the
TUG1 injection significantly reversed these alterations in the
diabetic mice (Fig. 3C). In
addition, the underlying signaling pathway involved in this process
was also investigated in these tissues. The results revealed that
the expression of ATGL, PPARα, PGC-1α and UCP-1 significantly
decreased in the diabetic mice, and that the TUG1 injection
significantly reversed these alterations in the diabetic mice
(P<0.001, Fig. 3D). The
results of western blot analysis revealed that the expression of
ATGL, PPARα, PGC-1α and UCP-1, and the phosphorylation levels of
AMPK and ACC were significantly downregulated in the diabetic mice,
while the overexpression of TUG1 markedly attenuated the effects
induced by diabetes (Fig.
3E).
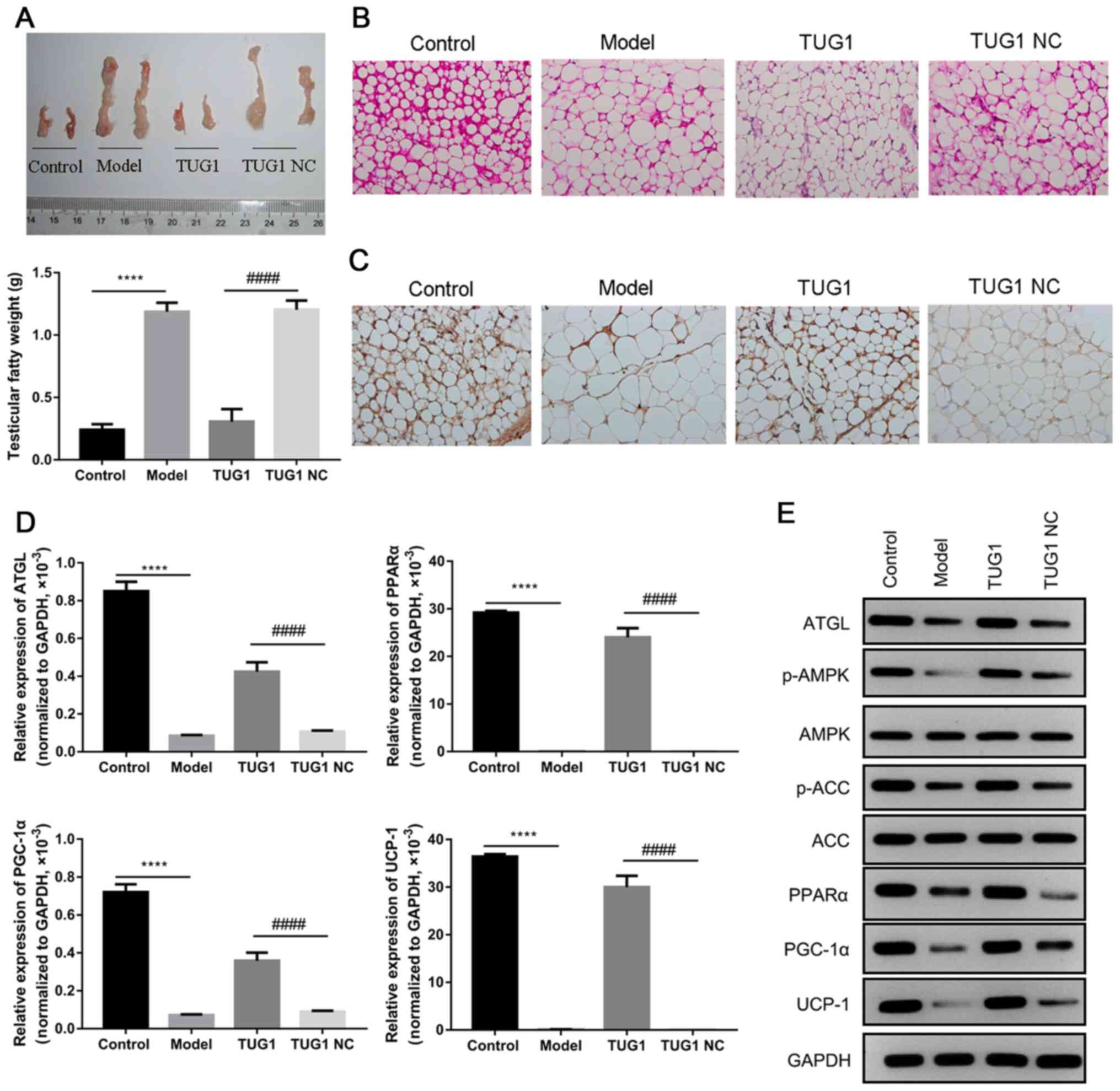 | Figure 3The overexpression of TUG1 reduces
testicular adipose accumulation and promotes fat degradation, fatty
acid oxidation and brown fat construction. (A) The weight of
testicular fatty tissues was measured using ruler, and
statistically analyzed in columns. (B) Hematoxylin and eosin
(H&E) staining was applied to examine the testicular fatty
tissues. (C) The expression of UCP-1 in testicular fatty tissues
was analyzed by immunohistochemistry (IHC). (D) The expression of
ATGL, PGC-1α, PPARα and UCP-1 was statistically evaluated by
RT-qPCR. (E) The protein expression levels of AMPK, p-AMPK, ACC,
p-ACC, ATGL, PGC-1α, PPARα and UCP-1 were detected by western blot
analysis. ATGL, adipose triglyceride lipase; PGC-1α, peroxisome
proliferator-activated receptor gamma coactivator 1-α; PPARα,
peroxisome proliferator-activated receptor α; UCP-1, uncoupling
protein-1; AMPK, AMP-activated protein kinase; ACC, acetyl-CoA
carboxylase; NC, negative control. ****P<0.0001,
compared with the control group; ####P<0.0001,
compared with the TUG1 NC group. Each assay, apart from western
blot analysis, was performed in triplicate. |
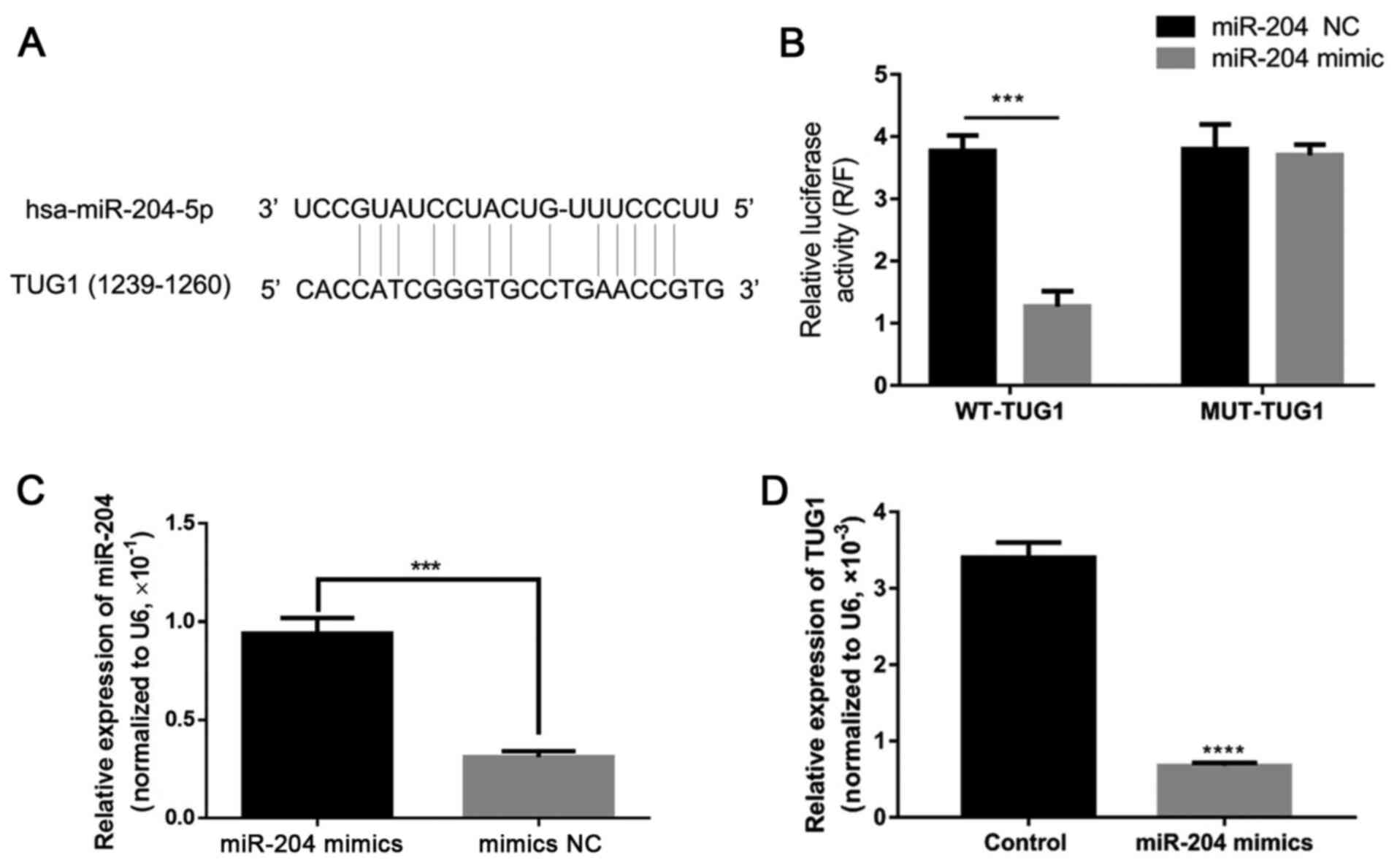
TUG1 can interact with miR-204 and
negatively regulate its expression
Considering the association between the expression
of TUG1 and miR-204, the regulatory relation association ship
between TUG1 and miR-204 was predicted, and an obvious miR-204
binding site was identified on the TUG1 mRNA (Fig. 4A). Dual luciferase reporter
activity assay revealed that the overexpression of miR-204 by
transfection with miR-204 mimic significantly reduced the reporter
activity of TUG1, when compared to the NC group (P<0.05), while
the overexpression of miR-204 had no marked effect on the reporter
activity of MUT-TUG1, when compared to the NC group (P>0.05,
Fig. 4B). In addition,
transfection with miR-204 mimic significantly increased miR-204
expression, and the overexpression of TUG1 significantly inhibited
the expression of miR-204 in the 3T3-L1 cells (P<0.05, Fig. 4C and D). These findings suggest
that TUG1 can directly and negatively affect the expression of
miR-204.
TUG1 promotes fatty oxidation in
adipocytes by negatively regulating miR-204
Following these investigations, the levels of fatty
tissue oxidation and degradation-associated proteins were
determined in the high glucose-induced 3T3-L1 fatty model. The
results revealed that the expression levels of SIRT1, ATGL, PPARα,
PGC-1α and UCP-1, and the phosphorylation levels of AMPK and ACC,
but not AMPK and ACC, significantly decreased in the model groups
when compared to the control group. However, the overexpression of
TUG1 significantly reversed the alteration in protein expression
and phosphorylation in the 3T3-L1 model cells (Fig. 5). These findings suggest that TUG1
promotes fatty degradation and oxidation in adipocytes. However,
the overexpression of miR-204 markedly attenuated the regulatory
effects of TUG1 on fatty oxidation and degradation, namely, the
decrease in the expression of SIRT1, ATGL, PPARα, PGC-1α and UCP-1,
and the phosphorylation levels of AMPK and ACC, without affecting
the expression of AMPK and ACC in the 3T3-L1 cells (Fig. 5).
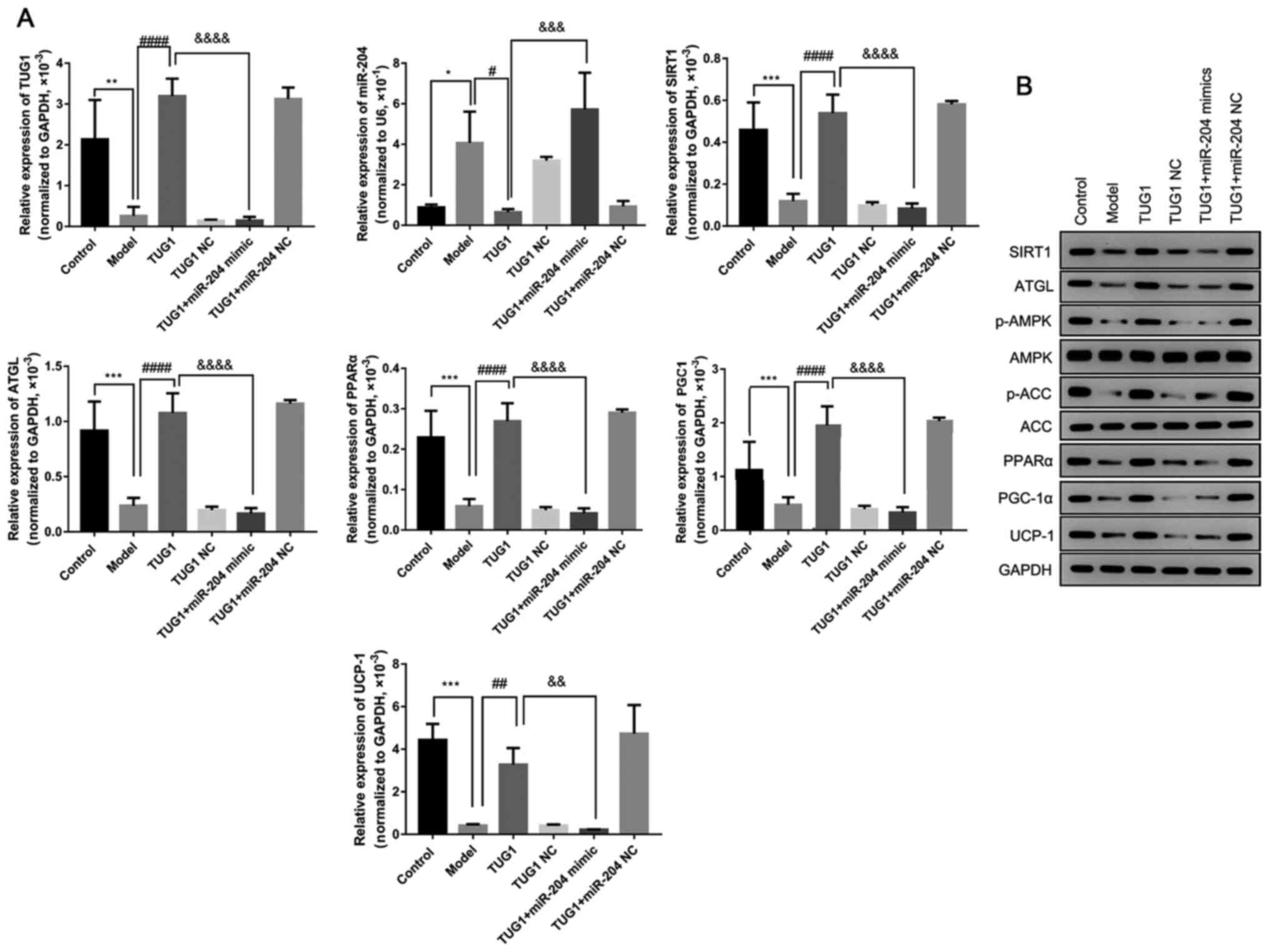 | Figure 5TUG1 promotes fatty oxidation in
adipocytes by negatively regulating miR-204. (A) Relative
expression of TUG1, miR-204, SIRT1, ATGL, PPARα, PGC-α and UCP1 was
measured by RT-qPCR in 3T3-L1 cells after altering the expression
of TUG1 and miR-204. (B) The protein expression of SIRT1, ATGL,
p-AMPK, AMPK, p-ACC, ACC, PPARα, PGC-1α and UCP-1 was evaluated by
western blot analysis in 3T3-L1 cells after altering the expression
of TUG1 and miR-204. ATGL, adipose triglyceride lipase; PGC-1α,
peroxisome proliferator-activated receptor gamma coactivator 1-α;
PPARα, peroxisome proliferator-activated receptor α; UCP-1,
uncoupling protein-1; AMPK, AMP-activated protein kinase; ACC,
acetyl-CoA carboxylase; NC, negative control. *P<0.05,
**P<0.01 and ***P<0.001, compared with
the control group; #P<0.05, ##P<0.01
and ####P<0.0001, compared with the model group;
&&P<0.01,
&&&P<0.001 and
&&&&P<0.0001, compared with the TUG1
group. Each assay, apart from western blot analysis, was repeated
at least 3 times, and GAPDH or U6 was set as the internal
control. |
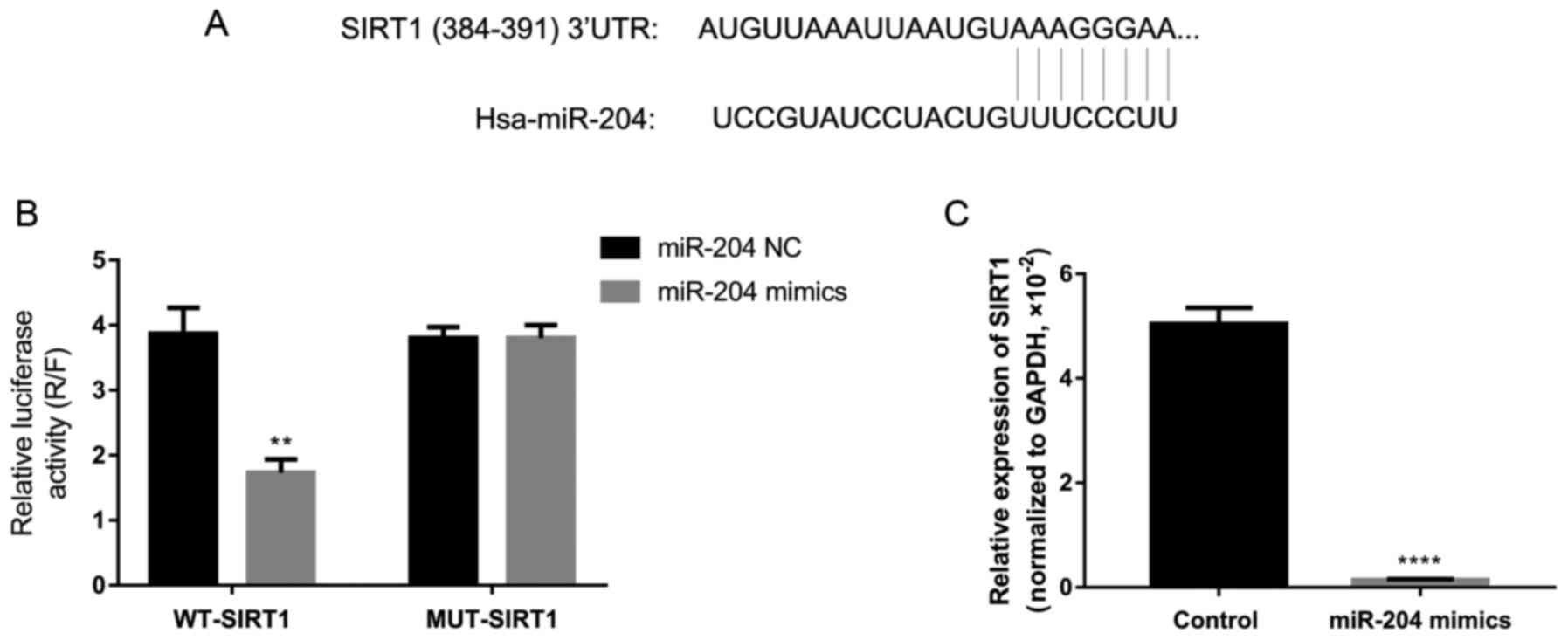
miR-204 directly inhibits the expression
of SIRT1
According to bioinformatics prediction using
TargetScan, it was predicted that SIRT1 and miR-204 shared some
potential binding sites (Fig.
6A). Thus, dual luciferase reporter activity assay was used to
confirm the regulatory association between SIRT1 and miR-204 in
3T3-L1 cells. As a result, the overexpression of miR-204
significantly inhibited the reporter activity of WT-SIRT1, without
affecting the activity of MUT-SIRT1 (Fig. 6B). In addition, the overexpression
of miR-204 by transfection with miR-204 mimic significantly
decreased the expression of SIRT1 in the 3T3-L1 cells (Fig. 6C). These findings indicate that
SIRT1 is a direct target of miR-204.
Inhibiting miR-204 promotes fatty
oxidation in adipocytes by upregulating SIRT1
The present study then determined the effects of
miR-204 on fatty oxidation. The results revealed that transfection
with miR-204 inhibitor significantly elevated the expression of
SIRT1, ATGL, PPARα, PGC-1α and UCP-1, and the phosphorylation
levels of AMPK and ACC in the 3T3-L1 cells, which were decreased by
high glucose. However, the silencing of SIRT1 markedly attenuated
the effects of miR-204 inhibitor, suppressing the expression of
SIRT1, ATGL, PPARα, PGC-1α and UCP-1, and the phosphorylation
levels of AMPK and ACC in the 3T3-L1 cells (Fig. 7). These findings suggest that the
inhibition of miR-204 promotes the high glucose-induced fatty
oxidation in 3T3-L1 cells by upregulating SIRT1.
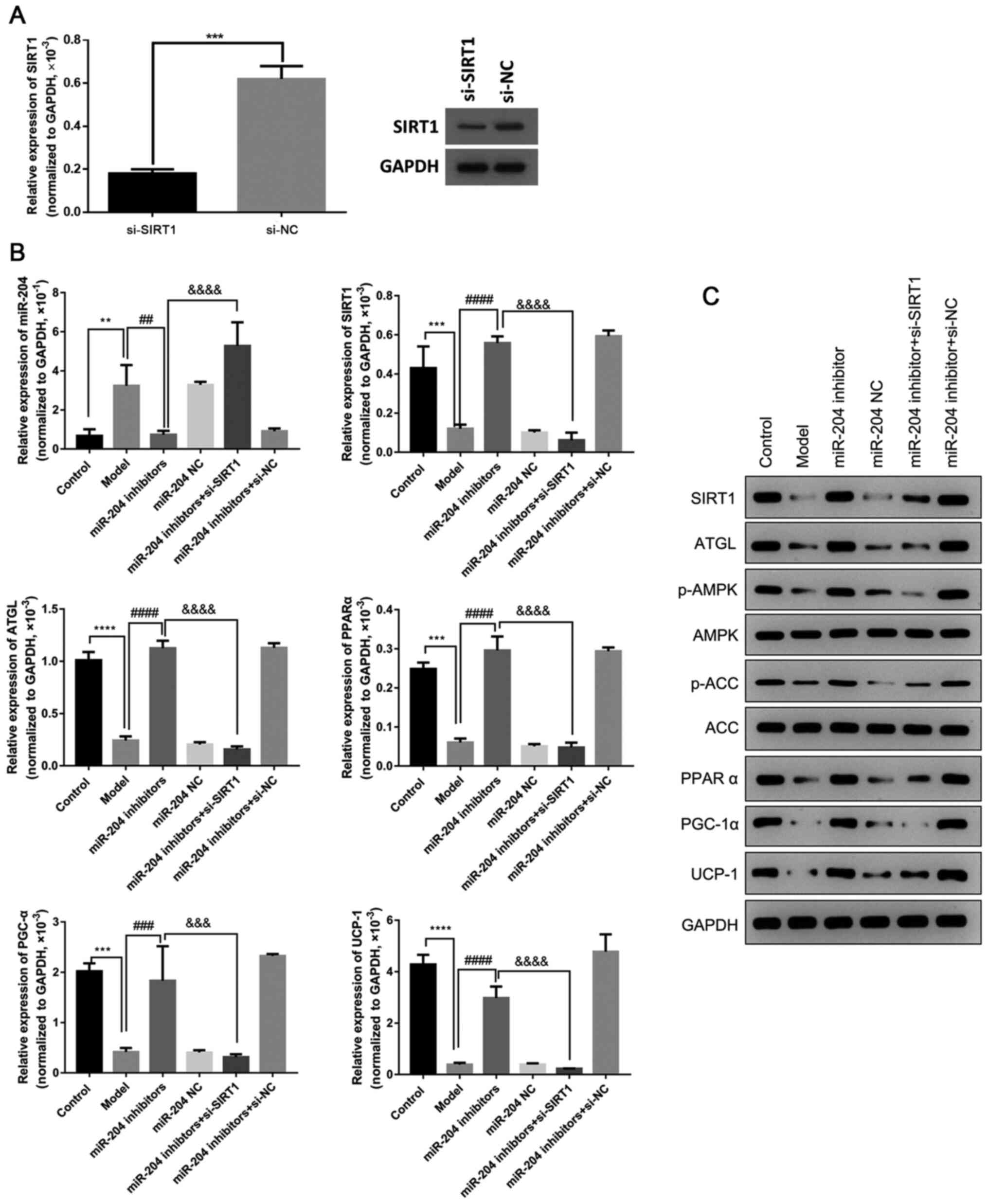 | Figure 7miR-204 inhibitor promotes fatty
oxidation in adipocytes by upregulating SIRT1. (A) Expression of
SIRT1 in model cells transfected with si-SIRT1 or si-NC. (B)
Relative expression of miR-204, SIRT1, ATGL, PPARα, PGC-α and UCP-1
in 3T3-L1 cells was evaluated by RT-qPCR after altering the
expression of SIRT1 and miR-204. (C) The protein expression of
SIRT1, ATGL, p-AMPK, AMPK, p-ACC, ACC, PPARα, PGC-1α and UCP-1 was
evaluated by western blot analysis in 3T3-L1 cells after altering
the expression of SIRT1 and miR-204. ATGL, adipose triglyceride
lipase; PGC-1α, peroxisome proliferator-acti-vated receptor gamma
coactivator 1-α; PPARα, peroxisome proliferator-activated receptor
α; UCP-1, uncoupling protein-1; AMPK, AMP-activated protein kinase;
ACC, acetyl-CoA carboxylase; NC, negative control.
**P<0.01, ***P<0.001 and
****P<0.0001, compared with the control group;
##P<0.01, ###P<0.001 and
####P<0.0001, compared with the model group;
&&&P<0.001 and
&&&&P<0.0001, compared with the
miR-204 inhibitors group. Each RT-qPCR assay was performed at least
in triplicate, and GAPDH or U6 was set as the internal control. |
Discussion
Despite the development of management strategies,
diabetes is becoming one of the most common health concerns
worldwide, and the number of diagnosed cases is significantly
increasing, particularly in developing countries (20). Considering this, multiple efforts
have been made to explore the mechanism of diabetes, in order to
providing new insight into the understanding of the pathogenesis of
diabetes. In the present study, TUG1 and SIRT1 were significantly
downregulated in diabetes, while the expression of miR-204 was
significantly upregulated in diabetes. Therefore, the mechanisms of
TUG1, miR-204 and SIRT1 were investigated in the present study, as
well as the regulatory associations between these factors.
Recently, studies have documented that inhibiting
the expression of TUG1 modulates the insulin secretion in mouse
pancreatic β cells (21,22), suggesting that TUG1 may regulate
the pathogenesis of diabetes. In the present study, TUG1 was
identified to be significantly downregulated in diabetic mice, and
the injection of the TUG1 lentivirus significantly attenuated the
increase in body weight and adipose accumulation, and decreased the
serum insulin, FGB, TG, glycerol and NEFA levels in diabetic mice.
In addition, further investigations revealed that the
overexpression of TUG1 markedly reversed the downregulation of
ATGL, PGC-1α, PPARα and UCP-1 expression, and AMPK and ACC
phosphorylation in diabetic mice. As previously demonstrated, the
silencing of TUG1 increases the apoptosis of pancreatic β cells,
but decreases serum insulin levels in vivo (21). Long et al reported that the
overexpression of TUG1 rescued the expression of PGC-1α and its
transcriptional targets, in order to improve the mitochondrial
biogenetics in diabetic nephropathy (23). Enhancing AMPK phosphorylation
protects the diabetic heart against ischemia/reperfusion injury
(24). Chronic and long-term
exercise improves glucose and lipid metabolism by activating the
AMPK-ACC signaling pathway (25).
These findings suggest that lncRNA decreases adipose accumulation,
in order to ameliorate the development of diabetes via the
activation of the AMPK/ACC signaling pathway.
He et al reported that miR-204 promoted the
audiogenic differentiation of human adipose-derived mesenchymal
stem cells by regulating DVL3 in the Wnt/β-catenin signaling
pathway (26). In the present
study, TUG1 was found to be a target of miR-204, and the
overexpression of miR-204 significantly decreased the expression of
TUG1. Furthermore, the overexpression of TUG1 suppressed miR-204,
and increased ATGL, PGC-1α, PPARα and UCP-1 expression, and AMPK
and ACC phosphorylation in diabetic mice. Yu et al reported
that TUG1 could sponge miR-204 to promote the differentiation of
osteoblasts by elevating the expression of Runx2 in aortic valve
calcification, which is a common complication in the obesity
population (27). Taken together,
these findings suggest that miR-204 promotes the development of
diabetes by attenuating the expression of TUG1, and the activation
of the AMPK/ACC signaling pathway.
The silencing of SIRT1 contributes to hepatic
steatosis in obesity (28). In
the present study, SIRT1 was found to be a of miR-204, and the
overexpression of TUG1 significantly upregulated the expression of
SIRT1. In addition, transfection with miR-204 inhibitor
significantly increased the expression of SIRT1, ATGL, PGC-1α,
PPARα and UCP-1, and AMPK and ACC phosphorylation, whereas the
silencing of SIRT1 could significantly reversed these effects. It
has been reported that α-lipoic acid modulates lipid metabolism by
inducing SIRT1, and activating AMPK (29). In addition, α-lipoic acid improves
hepatic steatosis induced by a high-fat diet by modulating FoxO1
and Nrf2 in the SIRT1/LKB1/AMPK signaling pathway (30). These findings indicate that SIRT1
may inhibit the development of diabetes by reversing the effects of
miR-204 through the SIRT1/AMPK/ACC signaling pathway.
In conclusion, the present study demonstrates that
the abnormal upregulation of miR-204 promotes the development of
diabetes, while the overexpression of TUG1 significantly promotes
adipose oxidation and attenuates the procedure of diabetes via the
upregulation of the SIRT1/AMPK/ACC signaling pathway. The present
study also has some limitations. The time- and dose-dependent
effects of miR-204 on the expression of SIRT1 and TUG1 were not
investigated. Thus, the underlying molecular mechanisms of the role
of miR-204 in diabetes and obesity warrant further in depth
investigations.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
YZ, YM, MG and YP conceived and planned the
experiments. YZ and YM carried out the experiments. YZ and MG
performed the data analysis. YZ and YP wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal protocols used in the present study were
authorized by the Ethics Committee of the Shanghai General Hospital
of Nanjing Medical University. The use of animals was also approved
by the same Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hoch E, Rusu V, Schreiber SL, Florez JC,
Jacobs SB and Lander ES: Type 2 diabetes-associated variants
disrupt function of SLC16A11, a proton-coupled monocarboxylate
transporter, through two distinct mechanisms. FASEB J.
31:S950.22017.
|
|
2
|
Samuel VT and Shulman GI: Mechanisms for
insulin resistance: Common threads and missing links. Cell.
148:852–871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nunes MK, Silva AS, de Queiroga
Evangelista IW, Filho JM, Gomes CN, Nascimento RA, Luna RC, de
Carvalho Costa MJ, de Oliveira NF and Persuhn DC: Hypermethylation
in the promoter of the MTHFR gene is associated with diabetic
complications and biochemical indicators. Diabetol Metab Syndr.
9:842017. View Article : Google Scholar
|
|
4
|
American Diabetes Association: 2.
Classification and diagnosis of diabetes: Standards of medical care
in diabetes-2018. Diabetes Care. 41(Suppl 1): S13–S27. 2018.
View Article : Google Scholar
|
|
5
|
Rathwa N, Patel R, Palit SP, Ramachandran
AV and Begum R: Genetic variants of resistin and its plasma levels:
Association with obesity and dyslipidemia related to type 2
diabetes susceptibility. Genomics. 111:980–985. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pramanik S, Rathwa N, Patel R,
Ramachandran AV and Begum R: Treatment avenues for type 2 diabetes
and current perspectives on adipokines. Curr Diabetes Rev.
14:201–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peirce V and Vidal-Puig A: Regulation of
glucose homoeostasis by brown adipose tissue. Lancet Diabetes
Endocrinol. 1:353–360. 2013. View Article : Google Scholar
|
|
8
|
Zhao S, Mugabo Y, Ballentine G, Attane C,
Iglesias J, Poursharifi P, Zhang D, Nguyen TA, Erb H, Prentki R, et
al: α/β-Hydrolase domain 6 deletion induces adipose browning and
prevents obesity and type 2 diabetes. Cell Rep. 14:2872–2888. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Perry RJ, Camporez JP, Kursawe R,
Titchenell PM, Zhang D, Perry CJ, Jurczak MJ, Abudukadier A, Han
MS, Zhang XM, et al: Hepatic acetyl CoA links adipose tissue
inflammation to hepatic insulin resistance and type 2 diabetes.
Cell. 160:745–758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Timothy JP and Guy RA: Could lncRNAs
contribute to β-cell identity and its loss in type 2 diabetes?
Biochem Soc Trans. 41:797–801. 2013. View Article : Google Scholar
|
|
11
|
Carter G, Miladinovic B, Patel AA, Deland
L, Mastorides S and Patel NA: Circulating long noncoding RNA GAS5
levels are correlated to prevalence of type 2 diabetes mellitus.
BBA Clin. 4:102–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng H, Zou L, Xie J, Wu H, Wu B, Zhu G,
Lv Q, Zhang X, Liu S, Li G, et al: lncRNA NONRATT021972 siRNA
decreases diabetic neuropathic pain mediated by the P2X 3 receptor
in dorsal root ganglia. Mol Neurobiol. 54:511–523. 2017. View Article : Google Scholar
|
|
13
|
Leti F and DiStefano JK: Long noncoding
RNAs as diagnostic and therapeutic targets in type 2 diabetes and
related complications. Genes. 8:2072017. View Article : Google Scholar :
|
|
14
|
Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ and
Yu DM: Long noncoding RNA TUG1 alleviates extracellular matrix
accumulation via mediating microRNA-377 targeting of PPARγ in
diabetic nephropathy. Biochem Biophys Res Commun. 484:598–604.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lei X, Zhang L, Li Z and Ren J:
Astragaloside IV/lncrna-TUg1/TraF5 signaling pathway participates
in podocyte apoptosis of diabetic nephropathy rats. Drug Des Dev
Ther. 12:2785–2793. 2018. View Article : Google Scholar
|
|
16
|
Price NL, Gomes AP, Ling AJ, Duarte FV,
Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro
JS, et al: SIRT1 Is required for AMPK activation and the beneficial
effects of resveratrol on mitochondrial function. Cell Metab.
15:675–690. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Milne JC, Lambert PD, Schenk S, Carney DP,
Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al: Small
molecule activators of SIRT1 as therapeutics for the treatment of
type 2 diabetes. Nature. 450:712–716. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Banks AS, Kon N, Knight C, Matsumoto M,
Gutiérrez-Juárez R, Rossetti L, Gu W and Accili D: SirT1 gain of
function increases energy efficiency and prevents diabetes in mice.
Cell Metab. 8:333–341. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu SY, Dong B, Fang ZF, Hu XQ, Tang L and
Zhou SH: Knockdown of lnc RNA AK 139328 alleviates myocardial
ischaemia/reperfusion injury in diabetic mice via modulating
miR-204-3p and inhibiting autophagy. J Cell Mol Med. 22:4886–4898.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gnavi R, Migliardi A, Maggini M and Costa
G: Prevalence of and secular trends in diagnosed diabetes in Italy:
1980-2013. Nutr Metab Cardiovasc Dis. 28:219–225. 2017. View Article : Google Scholar
|
|
21
|
Yin DD, Zhang EB, You LH, Wang N, Wang LT,
Jin FY, Zhu YN, Cao LH, Yuan QX, De W and Tang W: Downregulation of
lncRNA TUG1 affects apoptosis and insulin secretion in mouse
pancreatic β cells. Cell Physiol Biochem. 35:1892–1904. 2015.
View Article : Google Scholar
|
|
22
|
Cao LH, Yin DD, Xia CC, Wang N and De W:
Function of lncRNA TUG1 in insulin secretion from pancreatic beta
cells. Xian Dai Sheng Wu Yi Xue Jin Zhan. 25:4847–4851. 2017.In
Chinese.
|
|
23
|
Long J, Badal SS, Ye Z, Wang Y, Ayanga BA,
Galvan DL, Green NH, Chang BH, Overbeek PA and Danesh FR: Long
noncoding RNA Tug1 regulates mitochondrial bioenergetics in
diabetic nephropathy. J Clin Invest. 126:4205–4218. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paiva MA, Rutter-Locher Z, Gonçalves LM,
Providência LA, Davidson SM, Yellon DM and Mocanu MM: Enhancing
AMPK activation during ischemia protects the diabetic heart against
reperfusion injury. Am J Physiol Heart Circ Physiol.
300:H2123–H2134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yi X, Cao S, Chang B, Zhao D, Gao H, Wan
Y, Shi J, Wei W and Guan Y: Effects of acute exercise and chronic
exercise on the liver leptin-AMPK-ACC signaling pathway in rats
with type 2 diabetes. J Diabetes Res. 2013:9464322013. View Article : Google Scholar
|
|
26
|
He H, Chen K, Wang F, Zhao L, Wan X, Wang
L and Mo Z: MiR-204-5p promotes the adipogenic differentiation of
human adipose-derived mesenchymal stem cells by modulating DVL3
expression and suppressing wnt/β-catenin signaling. Int J Mol Med.
35:1587–1595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu C, Li L, Xie F, Guo S, Liu F, Dong N
and Wang Y: LncRNA TUG1 sponges miR-204-5p to promote osteoblast
differen-tiation through upregulating Runx2 in aortic valve
calcification. Cardiovas Res. 114:168–179. 2017. View Article : Google Scholar
|
|
28
|
Zhanguo G, Zhang J, Kheterpal I, Kennedy
N, Davis RJ and Ye J: Sirtuin 1 (SIRT1) protein degradation in
response to persistent c-jun n-terminal kinase 1 (JNK1) activation
contributes to hepatic steatosis in obesity. J Biol Chem.
286:22227–22234. 2011. View Article : Google Scholar
|
|
29
|
Chen WL, Kang CH, Wang SG and Lee HM:
α-Lipoic acid regulates lipid metabolism through induction of
sirtuin 1 (SIRT1) and activation of AMP-activated protein kinase.
Diabetologia. 55:1824–1835. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Li W, Liu Y, Sun Y, Li Y, Yao Q,
Li J, Zhang Q, Gao Y, Gao L and Zhao J: Alpha-Lipoic acid improves
high-fat diet-induced hepatic steatosis by modulating the
transcription factors SREBP-1, FoxO1 and Nrf2 via the
SIRT1/LKB1/AMPK pathway. J Nutr Biochem. 25:1207–1217. 2014.
View Article : Google Scholar : PubMed/NCBI
|