Introduction
Inflammatory bowel diseases (IBDs) are remittent and
progressive inflammatory conditions that may affect the entire
gastrointestinal tract (1).
Multiple factors, such as genes, diet, environment and the
microbiota contribute to complex etiologies (2). Among these factors, research into
the microbiome is a rapidly advancing field in IBD. Accumulating
evidence has indicated that the gut microbiota play a key role in
the process of IBD. The microbiota in the human intestine may be
closely related to the development of IBD via their influence on
the physiological functions of the colorectum (3-5).
However, the inflammatory pathogens which cause IBD remain
incompletely explored.
It was previously demonstrated that the novel
intestinal bacterium, Fusobacterium nucleatum (F.
nucleatum), was associated with an enhanced gut inflammation in
patients with IBD. The proportion of patients with IBD identified
with F. nucleatum infection in the colonic tissues was
higher than the controls. Moreover, F. nucleatum strains
from the inflamed tissues of patients with IBD were more invasive
than strains that were isolated from healthy tissue (6). However, the underlying mechanisms
associated with the inflammatory pathogenesis of this bacterium
remain largely unknown.
Autophagy is a 'self-eating' process that results in
the breakdown of intracellular proteins or organelles for recycling
within the lysosome to maintain cellular homeostasis (7). Excessive or impaired autophagy can
result in pathological processes (8-11).
More importantly, accumulating evidence has indicated an
association between autophagy and IBD. Autophagy plays multiple
roles in the pathogenesis of IBD by altering processes that include
anti-inflammatory cytokine production, antigen presentation and the
endoplasmic reticulum stress response (12). Moreover, autophagy plays an
important role in the epithelial cell-autonomous mechanisms of the
antibacterial defense that protects against the dissemination of
intestinal bacteria (13).
However, the role of autophagy in F. nucleatum-induced
inflammation in intestinal epithelial cells has not yet been
investigated, at least to the best of our knowledge.
It is widely accepted that probiotics exert
beneficial effect against colitis (14-16). Probiotic supplementation appears
to be potentially well tolerated, effective, and safe in patients
with IBD (17). As previously
demonstrated, the probiotic Lactobacillus rhamnosus (L.
rhamnosus) improved the clinical symptoms of patients with mild
to moderately active IBD (18).
However, information on the role of L. rhamnosus and
autophagy in the induction and progression during F.
nucleatum-induced inflammation is limited. Thus, the aim of the
present study was to examine the effects of L. rhamnosus on
F. nucleatum-induced intestinal inflammation, and to
elucidate the underlying mechanisms, with particular focus on
autophagy. The present study wished to provide novel insight into
F. nucleatum-related gut disorders.
Materials and methods
Reagents
Primary antibodies against microtubule-associated
light chain 3B (LC3B), p-AKT and p-mammalian target of rapamycin
(mTOR) were purchased from Cell Signaling Technology, Inc. (cat.
nos. 3868, 4060 and 5536); antibodies against GAPDH, SQSTM1/p62,
Atg16L1, p-p85, p85, AKT and mTOR were obtained from ABclonal (cat.
nos. AC001, A7758, A1871, AP0854, A11526, A11016 and A2445);
HRP-conjugated goat anti rabbit antibody was from Antgene (cat. no.
ANT020); biotin conjugated donkey anti rabbit anti-body was from
Wuhan Boster Biological Technology, Ltd. (cat. no. BA1002);
3-methyladenine (3-MA) and chloroquine (CQ) were from
Sigma-Aldrich; Merck KGaA; Atg16L1 small interfering RNA (siRNA)
and negative control siRNA was from JTS Scientific; dextran sulfate
sodium (DSS) was obtained from MP Biomedicals; the enzyme-linked
immune sorbent assay (ELISA) kits for interleukin (IL)-6, IL-8 and
tumor necrosis factor (TNF)-α were from Neobioscience Technology
Co., Ltd.
Cell and bacterial strain culture
The human colonic adenoma cell line, Caco-2, was
purchased from the American Type Culture Collection (ATCC) and
cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.), with
10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml streptomycin/penicillin (Gibco; Thermo Fisher
Scientific, Inc.) and 1% HEPES (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C under 5% CO2. F. nucleatum (ATCC
25586) and L. rhamnosus (ATCC 11982) were purchased from
ATCC and cultured by the Wuhan Research Institute of First Light
Industry (Wuhan, China). The methods of bacterial pellets and
conditioned medium were as previously described (19). Caco-2 cells were treated with the
supernatant of F. nucleatum and/or L. rhamnosus to
examine the effects of F. nucleatum and L. rhamnosus
(20,21). 3-MA (5 mM) and CQ (10 µM)
treatment for 2 h was used to inhibit the autophagic flux.
Animals
Male C57BL/6 mice (aged 6 weeks, weighing 20-24 g)
were purchased from Beijing Huafukang Biosciences Co., Ltd. and
housed under specific pathogen-free (SPF) conditions with a fixed
12 light/dark cycle at 23°C, with free access to water and food.
The animal experiments in the present study were approved by the
Animal Research Ethics Committee of Tongji Medical College,
Huazhong University of Science and Technology (Approval ID
2016-0057).
To investigate the role of F. nucleatum and
L. rhamnosus in the model of acute colitis, the mice were
randomly divided into 6 groups as follows: i) The control; ii)
L. rhamnosus; iii) F. nucleatum; iv) DSS; v) DSS +
F. nucleatum; and vi) DSS + F. nucleatum + L.
rhamnosus groups. Each group contained 7 mice. The model of
acute colitis was established by the administration of 3% (wt/vol)
DSS in the drinking water for 7 days, with or without the daily
gavage of 109 CFU bacteria (F. nucleatum or/and
L. rhamnosus) solution in phosphate-buffered saline (PBS)
for 7 days prior to DSS administration as previously described
(22,23). In the control and DSS groups, the
same volume of PBS was administered by gavage as the vehicle
control. The DSS solution was refreshed every 2 days and the
leftover DSS solution was measured. The body weight of the mice, as
well as stool consistency and any bleeding were examined each day.
The disease activity index (DAI) was calculated as follows: For
weight loss: 0, no loss; 1, 1-5%; 2, 5-10%; 3, 10-20% and 4,
>20%; for stool consistency: 0, normal; 2, loose stool; 4,
diarrhea; and for stool bleeding: 0, no blood; 2, presence; and 4,
gross blood (24). The mice were
sacrificed on day 8 (mice which exhibited weight loss >20% were
euthanized immediately), and tissues and blood were collected for
subsequent analysis. Mice were anesthetized by an intraperitoneal
injection of 50 mg/kg pentobarbital and 250 mg/kg for euthanasia
(25).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the collected cells or
colonic tissues using TRIzol reagent (Life Technologies; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions and reverse transcribed into complementary DNA using
Prime Script RT Master Mix (Takara Biotechnology, Inc.). qPCR was
performed using the LightCycler® 480 SYBR I Master Mix
(Roche Diagnostics), running on a Roche LightCycle R480 system
(Roche Diagnostics). Gene expression was normalized relative to
GAPDH and calculated using the 2−ΔΔCq method (26). The primer sequences are presented
in Table I.
 | Table ISequences of primers used for
RT-qPCR. |
Table I
Sequences of primers used for
RT-qPCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| Human genes | | |
| IL-6 |
ATGAGGAGACTTGCCTGGTG |
GGCATTTGTGGTTGGGTCAG |
| IL-8 |
CACTGCGCCAACACAGAAAT |
AACTTCTCCACAACCCTCTGC |
| TNF-α |
TACTCCCAGGTCCTCTTCAAGG |
TTGATGGCAGAGAGGAGGTTG |
| GAPDH |
ACCCACTCCTCCACCTTTGA |
AAAGTGGTCGTTGAGGGCAA |
| Mouse genes | | |
| IL-1β |
CTGAACTCAACTGTGAAATGCC C |
TTGTTGATGTGCTGCTGCG |
| IL-6 |
ACAAAGCCAGAGTCCTTCAGAG |
CCACTCCTTCTGTGACTCCA |
| TNF-α |
ACCCTCACACTCACAAACCAC |
TAGCAAATCGGCTGACGGTG |
| IFN-γ |
CAGCAACAGCAAGGCGAAA |
TTGAATGCTTGGCGCTGGAC |
| Beclin1 |
AGGAGCTGGAAGATGTGGAAA |
CACTATACTCCCGCTGGTACTGA |
| Atg7 |
ATGACCGCATGAATGAGCCT |
GGTGAATCCTTCTCGCTCGT |
| Atg16L1 |
GACCTCAGACCACACAGAAGA |
TCCTGGCAGCATCAGAAGAATGA |
| GAPDH |
CATGGCCTTCCGTGTTCCTA |
TACTTGGCAGGTTTCTCCAGG |
Western blot analysis
Proteins were extracted from the cells or tissues
using RIPA lysis buffer (Beyotime Institute of Biotechnology, Inc.)
supplemented with phenylmethyl sulfonyl fluoride (PMSF) protease
inhibitor and phosphatase inhibitor. The total protein content in
the supernatant were measured by bicinchoninic acid (BCA) assay
(Thermo Fisher Scientific, Inc.). A total of 40 µg cellular
proteins and 80 µg tissue proteins per lane were separated
by 12 or 8% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE), and then transferred to a PVDF membrane
(Millipore Corp.). This was followed by blocking with 5% non-fat
milk or 5% bovine serum albumin (BSA) prior to incubation overnight
at 4°C with primary antibodies for GAPDH, LC3B, SQSTM1/p62,
Atg16L1, p-p85, p85, p-AKT, AKT, p-mTOR and mTOR (1:1,000
dilution). The membranes were then washed and incu-bated with goat
anti rabbit secondary antibodies conjugated to HRP (1:2,000
dilution) for 1 h at room temperature the second day. Enhanced
chemiluminescent reagents (Beyotime Institute of Biotechnology,
Inc.) were used to detect the HRP on the immunoblots. Quantitative
analysis was performed using ImageJ1 software (NIH).
ELISA
The culture supernatants of the Caco-2 cells were
collected following 12 h of treatment with or without the
supernatant of bacteria and stored at -80°C. The levels of IL-8 and
TNF-α in the supernatants were measured using respective ELISA kits
according to the manufacturer's instructions, as indicated above.
The absorbance was obtained at a relative nanometer wavelength
using a microplate reader (Biotek Instruments, Inc.).
GFP-mCherry-LC3 plasmid transfection and
confocal microscopy
Caco2 cells at the logarithmic growth phase were
seeded on coverslips at 50-70% confluence and transfected with
GFP-mCherry-LC3 plasmid DNA (Changsha Yingrun Biotechnology Co.,
Ltd.) using Lipofectamine 3000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions and as previously described (27). The cells were transfected for 24 h
prior to stimulation with the supernatants of F. nucleatum
or L. rhamnosus. The cells were then washed 3 times with PBS
and fixed with 4% paraformaldehyde for 15 min. Images of red, green
and merged yellow dots representing LC3 on autolysosomes and
autophagosomes were captured using a confocal laser scanning
microscope (Olympus Corporation). The average number of punctate
fluorescent structures per cell was determined.
Transmission electron microscopy
(TEM)
Following stimulation with F. nucleatum or
L. rhamnosus, the cells were fixed with 2.5%
phosphate-buffered glutaraldehyde, post-fixed in 1% osmium
tetroxide for 1 h, rinsed with 0.1 M phosphate buffer (pH 7.4), and
then dehydrated with increasingly graded alcohols before being
embedded in Epon 812 (SPI Supplies). Ultrathin sections were cut
using a ultramicrotome and observed using a FEI Tecnai G2 12TEM
(FEI Company). The number of autophagic vacuoles per cell was
calculated.
Atg16L1 siRNA transfection
Caco-2 cells (100,000 cells/well) were seeded in
12-well culture dishes and transfected with 100 nM siRNA against
Atg16L1 or a scrambled siRNA for negative control (NC siRNA) at 50%
confluence using Lipofectamine 3000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Gene silencing efficiency was examined
by western blot analysis following 48 h of transfection.
Histological analysis
For histological analysis, distal colon specimens
were fixed in 4% formaldehyde for 24 h and embedded in paraffin.
The 4-µm-thick sections were stained with hematoxylin and
eosin (H&E) according to appropriate standard procedures
(28), and then analyzed by a
pathologist blinded to the identity of the data. Histological
analysis was performed as previously described (29). Briefly, scores were allocated as
follows: For inflammation severity: 0, none; 1, mild; 2, moderate;
3, severe; for the extent of inflammation: 0, none; 1, mucosa; 2,
mucosa and sub-mucosa; 3, transmural; for crypt damage: 0, none; 1,
basal 1/3 damage; 2, basal 2/3 damage; 3, crypts lost and surface
epithelium present; 4, crypts and surface epithelium loss; for
percentage involvement: 0, 0%; 1, 1-25%; 2, 26-50%; 3, 51-75%; 4,
76-100%. The scores of the 4 items were added to obtain the
histological score.
Immunohistochemistry (IHC)
IHC was performed as previously described (30). In brief, paraffin-embedded tissues
sections (4 µm thickness) were deparaffinized. Sections were
heated in 10 mM citrate buffer for 10 min for antigen-retrieval and
blocked with 10% goat serum for 1 h at room temperature. The
sections were then incubated with LC3B and p62 antibodies (1:200
dilution) overnight at 4°C and then incubated with matched
secondary antibody (1:200 dilution) for 1 h at room temperature the
following day. The DAB kit was used to detect the signal. Images
were analyzed using IHC Profiler plugin in ImageJ1 (NIH).
Statistical analysis
Each experiment was performed at least in triplicate
and data are expressed as the means ± standard error of mean (SEM).
Differences between groups were performed using a two-tailed
Student's t-test and one-way analysis of variance (ANOVA) followed
by post-hoc Tukey's tests. Statistical analyses were performed with
IBM SPSS Statistics 25 and visualized by GraphPad Prism 7.0.
Differences were defined as statistically significant or highly
statistically significant at P<0.05, P<0.01 and
P<0.001.
Results
L. rhamnosus alleviates DSS-induced acute
colitis aggravated by F. nucleatum
For the in vivo experiments, preliminary
experiments were performed to assess whether L. rhamnosus
alone exerts any effect on the mice. As shown in Fig. S1A and B, the mice treated with
L. rhamnosus alone exhibited no differences compared with
the control group as regards colonic tissue destruction or
pro-inflammatory cytokine expression. Therefore, the L.
rhamnosus group was not taken into account in the following
experiments.
Subsequently, the present study determined whether
F. nucleatum contributes to the progression and exacerbation
of IBD. F. nucleatum was administered to the mice in the
model of DSS-induced acute colitis. The mice were treated with or
without F. nucleatum or/and L. rhamnosus via gavage
daily as described in the Materials and methods section for the
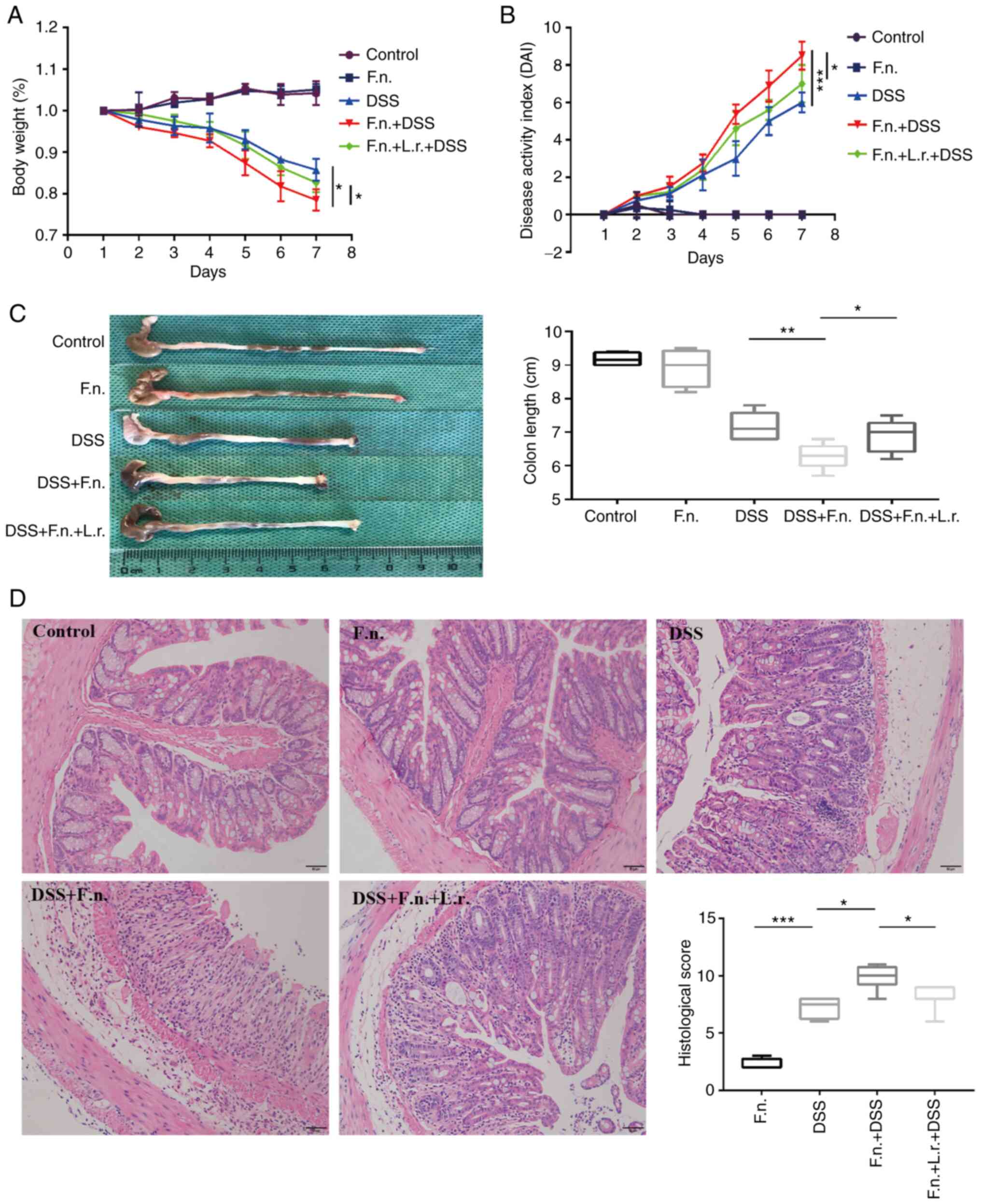
group design at 7 days prior to the DSS administration. As shown in
Fig. 1A and B, following F.
nucleatum treatment, the mice presented severer complications,
including hematochezia, body weight loss (three mice lost 24.2,
21.1, 22.8% wight respectively) and a higher DAI score when
compared with the mice in the DSS group (P<0.05 and P<0.001,
respectively). These more severe phenomena were also observed
evidenced by the shortening of colon length in the DSS + F.
nucleatum group (P<0.01, Fig.
1C). No apparent changes in the appearance of the colon were
observed between the control and F. nucleatum alone groups
(P=0.436, Fig. 1C). Subsequently,
whether L. rhamnosus modifies the course of DSS-induced
acute colitis induced by F. nucleatum was determined. As was
expected, the DDS + F. nucleatum + L. rhamnosus group
exhibited more evident positive changes than the DDS + F.
nucleatum group as regards body weight loss, DAI score and
colon length (P<0.05, Fig.
1A-C). It was further found that compared to the mice in the
DDS group, the mice in the DDS + F. nucleatum group
exhibited more severe mucosal ulceration, inflammatory cell
infiltration, crypt and gland destruction (Fig. 1D). Finally, it was found that the
expression of pro-inflammatory cytokines in colon tissue, as well
as the mRNA levels of IL-1β, IL-6, TNF-α and interferon (IFN)-γ
were significantly decreased in DDS + F. nucleatum group
following L. rhamnosus treatment (for IL-1β, P<0.01; for
the other level, P<0.05, respectively; Fig. S1C). On the whole, these results
indicate that F. nucleatum aggravates DSS-induced acute
colitis in vivo and that L. rhamnosus may induce
antagonistic inflammatory functions through the downregulation of
pro-inflammatory cytokine expression in mice administered DDS and
F. nucleatum.
L. rhamnosus decreases the production of
pro-inflammatory cytokines induced by F. nucleatum infection in
vitro
To verify the pro-inflammatory effects of F.
nucleatum in vitro, human epithelial colorectal cells Caco2
were treated with the supernatant of F. nucleatum for 12 h
at different multiplicities of infection (MOIs). Higher gene
expression levels of IL-6, IL-8 and TNF-α were observed in a
dose-dependent manner. The results revealed a gradual increase in
the levels of these cytokines along with the increasing MOI
(Fig. 2A). Notably, the mRNA
levels of these cytokines were significantly decreased following
the addition of the supernatant of L. rhamnosus (for IL-8,
P<0.0001; for the other levels, P<0.001; respectively;
Fig. 2B). The HT29 and HCT116
cell lines were used in preliminary experiments and similar results
were obtained (data not shown). However, the Caco2 cells exhibited
the most significant changes in response to the stimulation.
Therefore, the Caco2 cell line was selected for use the following
experiments. The results of ELISA also indicated that the secretion
levels of IL-6, IL-8 and TNF-α in the Caco2 cell culture medium
were greater in the F. nucleatum group than the control
group (P<0.0001 and P<0.001, respectively). The levels of
these cytokines were significantly decreased by L. rhamnosus
(all P<0.05 respectively; Fig.
2C). Collectively, these results demonstrate that L.
rhamnosus reduces the production of pro-inflammatory cytokines
induced by F. nucleatum infection in vitro.
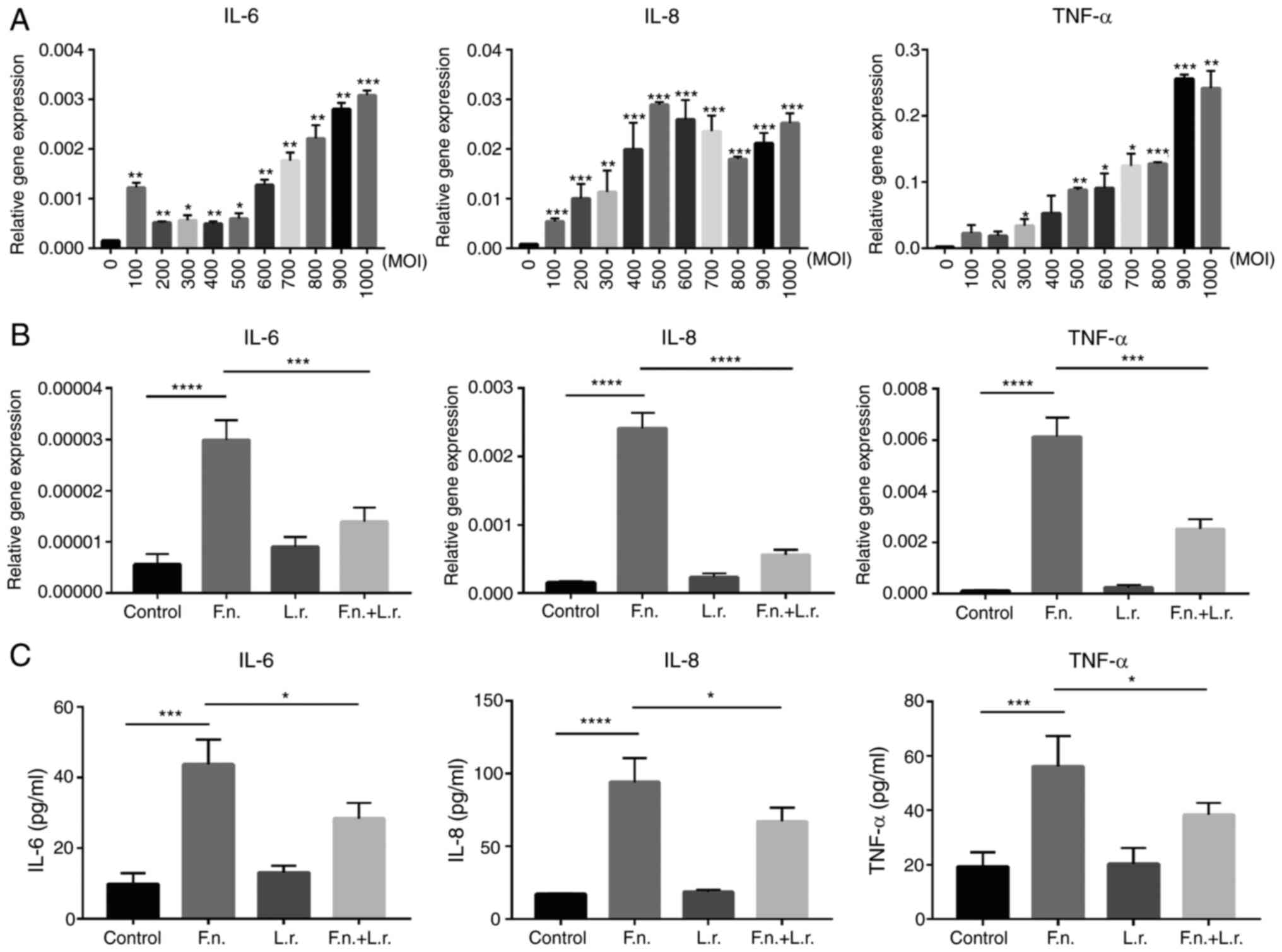 | Figure 2Lactobacillus rhamnosus
decreases pro-inflammatory cytokine production induced by F.
nucleatum infection in vitro. (A) The relative gene
expres-sion of IL-6, IL-8, TNF-α in Caco2 cells treated with F.
nucleatum supernatant at increasing MOIs. (B) The relative gene
expression of IL-6, IL-8, TNF-α in Caco2 cells treated with F.
nucleatum and Lactobacillus rhamnosus supernatant. (C)
The secretion of IL-6, IL-8, TNF-α proteins was detected by ELISA
in cell culture medium. Data are presented as the means ± SEM of at
least 3 repeated experiments. *P≤0.05,
**P≤0.01, ***P<0.001,
****P<0.0001. F. nucleatum/F.n.,
Fusobacterium nucleatum; L.r., Lactobacillus
rhamnosus. |
L. rhamnosus restores the impaired
autophagic flux induced by F. nucleatum in vitro
Subsequently, the present study further explored and
elucidated the mechanisms through which L. rhamnosus
downregulates the production of pro-inflammatory cytokines induced
by F. nucleatum infection. Since it has been previously
indicated that autophagy plays a key role in intestinal
inflammation and the maintenance of intestinal homeostasis
(31), the present study then
detected the autophagic reaction following stimulation with F.
nucleatum. The transformation of LC3-I to LC3-II is necessary
for the formation of autophagosomes and is a widely accepted
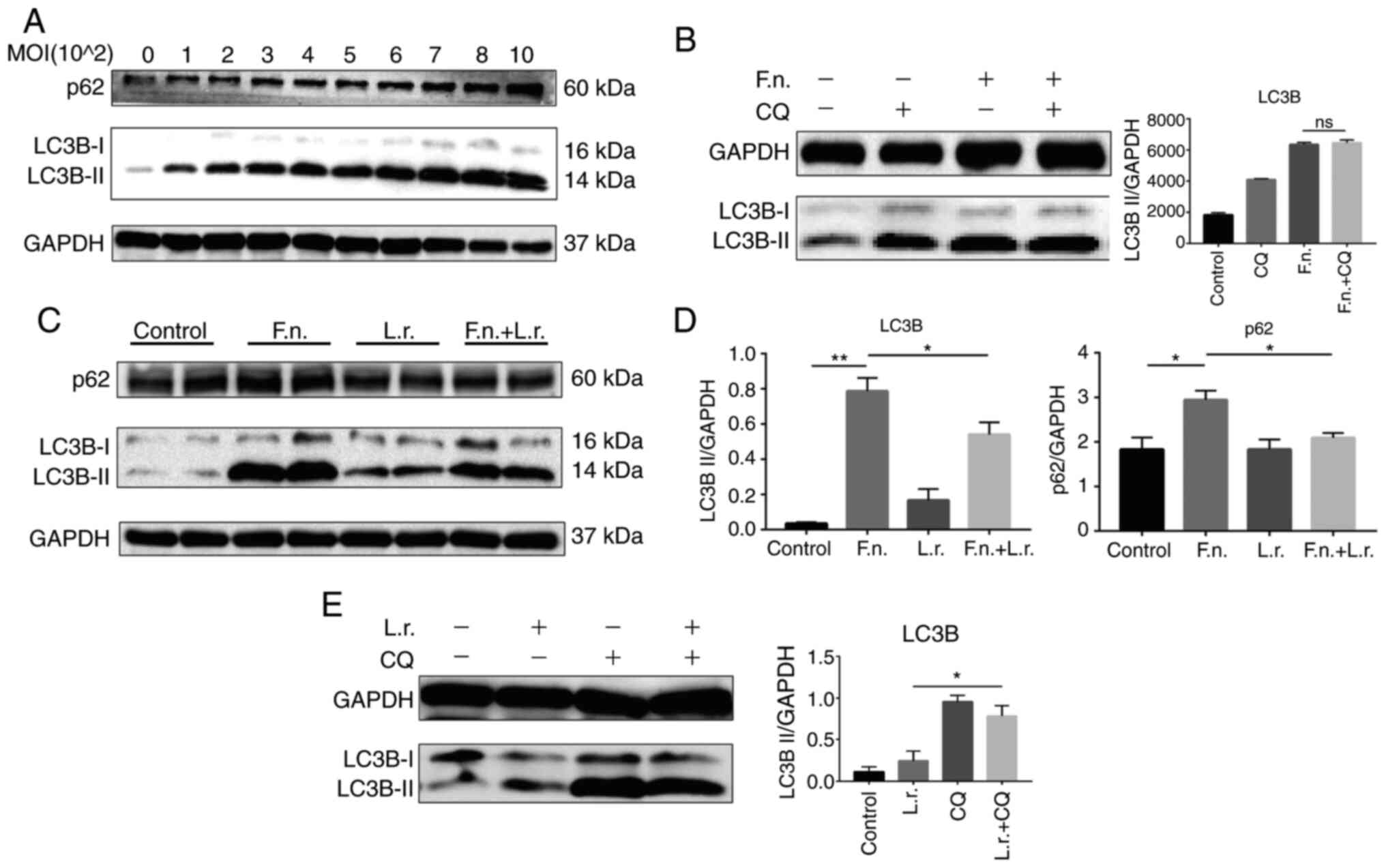
molecular marker for autophagy activation. Western blot analysis
revealed that following exposure to F. nucleatum at
different MOIs, a significant upregulation of LC3-II protein
expression and SQSTM1/p62 expression was observed in a
MOI-dependent manner. The protein levels of p62 and LC3-I also
increased correspondingly, which indicated that the autophagic flux
may be blocked by F. nucleatum (Fig. 3A).
In order to describe the detailed condition of the
autophagic flux, CQ, a key autophagy inhibitor which can block
autophagy by inhibiting the function of lysosome and then inhibit
the degradation of LC3-II, was used to examine the condition of the
autophagic flux. As shown in Fig.
3B, the protein level of LC3-II in the F. nucleatum + CQ
group did not exhibit an obvious increase following pre-treatment
with 10 µM CQ for 2 h compared to treatment with F.
nucleatum supernatant only (P=0.59), indicating that F.
nucleatum impairs the autophagic flux process.
Subsequently, the effects of L. rhamnosus
were evaluated and it was found that the ratio of LC3-II/GAPDH and
p62/GAPDH was lower (P<0.05) following the addition of L.
rhamnosus (Fig. 3C and D).
Furthermore, L. rhamnosus treatment not only increased the
expression of LC3B-II, but also the protein level of LC3-II in the
L. rhamnosus + CQ group exhibited an obvious increase
compared to the L. rhamnosus only group, indicating that
L. rhamnosus promoted the autophagic process (Fig. 3E).
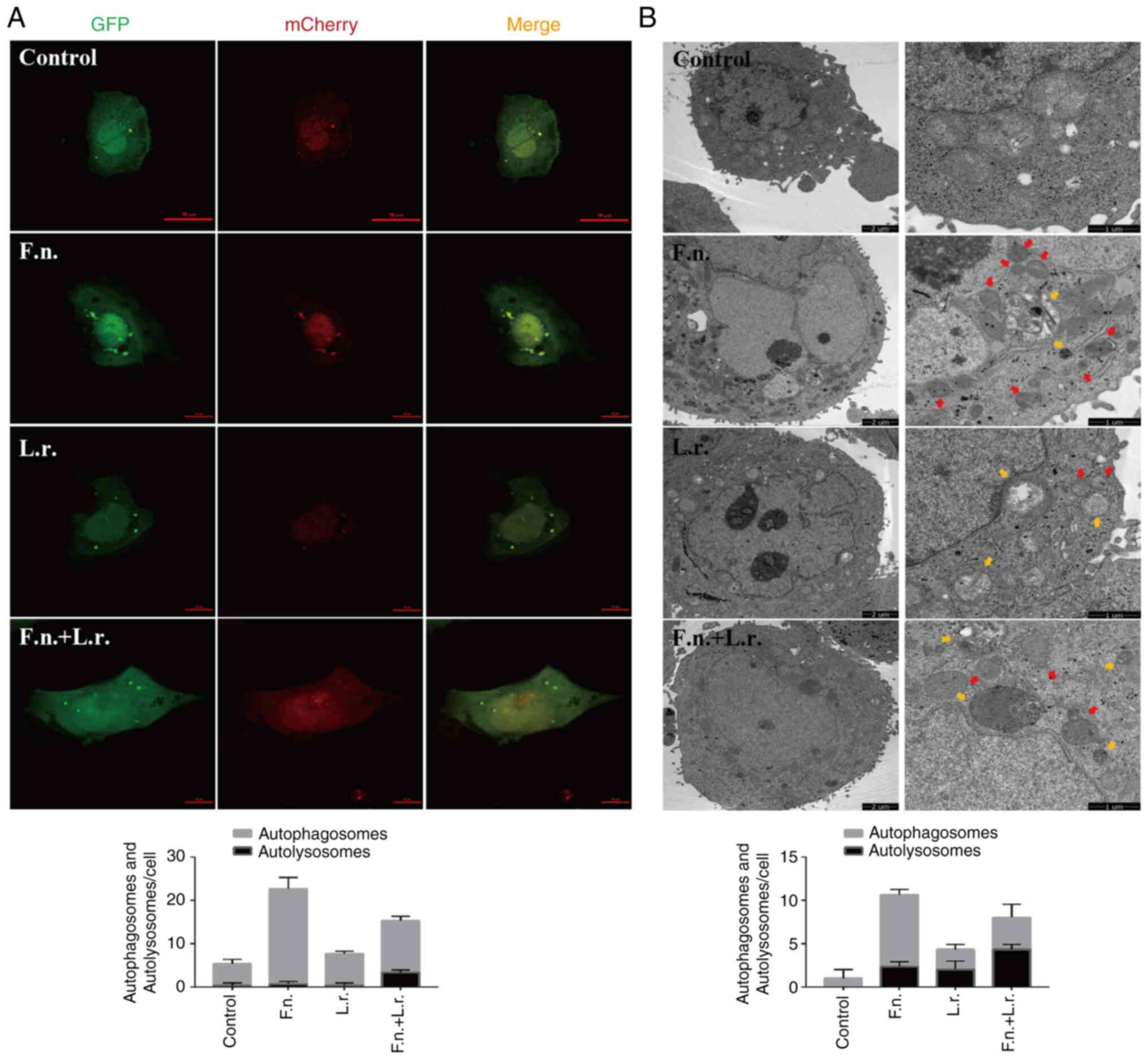
To further confirm the autophagic flux status,
GFP-mCherry-LC3 plasmid was transfected into Caco2 cells. Following
transfection for 24 h and treatment with the indicated
interventions, confocal microscopic analysis revealed that F.
nucleatum stimulation resulted in the accumulation of red
puncta with the significant augmentation of of green puncta,
suggesting that the fusion of autophagosomes and lysosomes was
blocked (Fig. 4A). From electron
microscopic analysis at the ultrastructural level in the F.
nucleatum-treated cells, autophagosome structures were also
more prevalent (Fig. 4B). When
the transfected cells were treated with L. rhamnosus culture
supernatants, the number of red fluorescence signals markedly
increased (Fig. 4A). More
autolysosomes were also observed in the F. nucleatum group
when exposed to the L. rhamnosus supernatants (Fig. 4B). Collectively, these results
demonstrate that in the Caco2 cells, L. rhamnosus probiotics
can regulate and restore the impaired autophagic flux associated
with F. nucleatum stimulation.
L. rhamnosus increases autophagy
inhibited by F. nucleatum infection in vivo
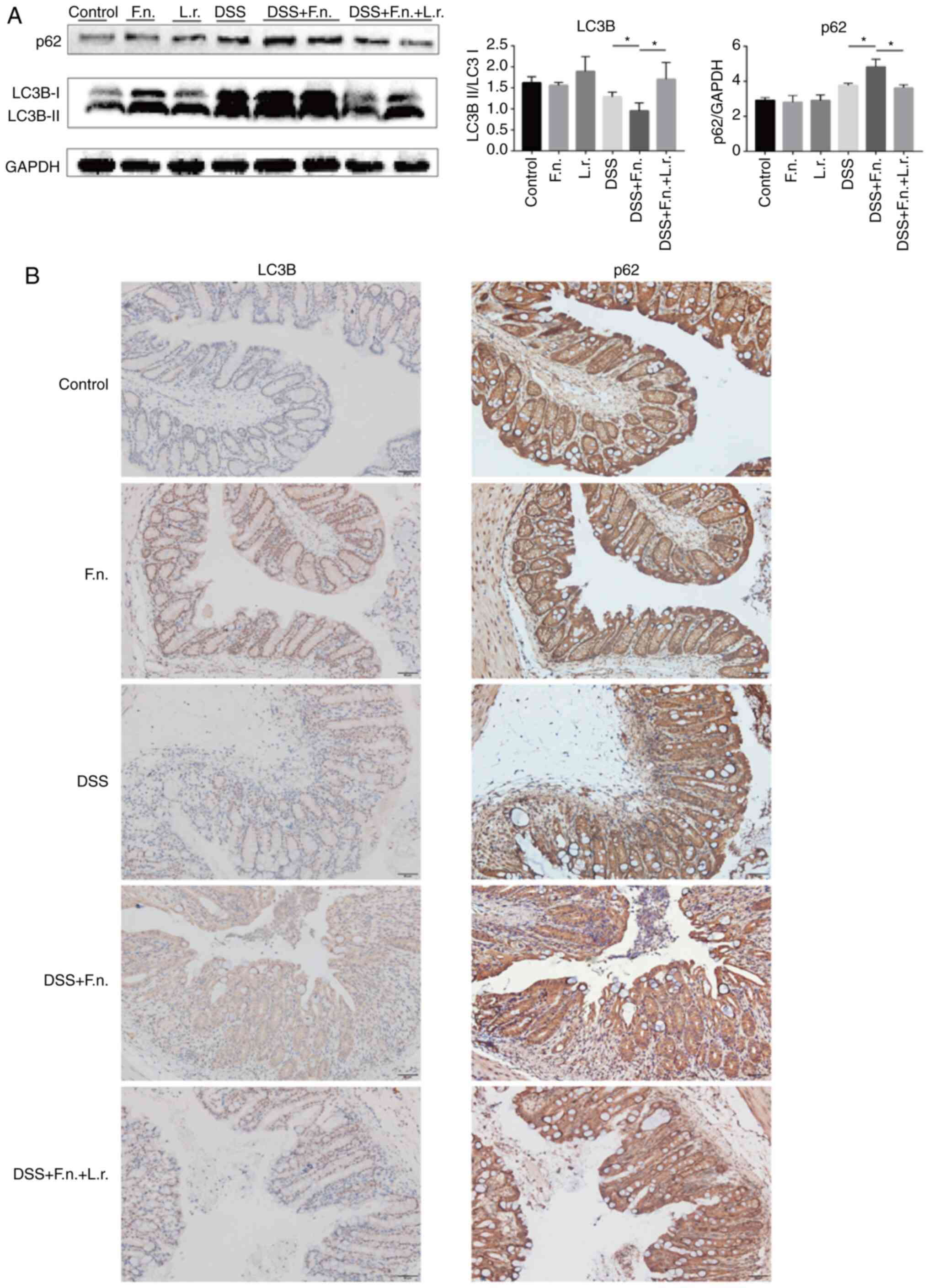
To assess whether the administration of F.
nucleatum alters the course of autophagy in the progression of
colitis, experiments were conducted with the mice from the model of
DSS-induced colitis. Western blot analysis was performed to examine
the ratio of LC3-II/LC3-I and p62/GAPDH. As shown in Fig. 5A, pre-treatment of the mice with
F. nucleatum was associated with a decreased ratio of
LC3-II/LC3-I (although LC3-II expression increased, the increase in
LC3-I expression was relatively more evident compared to the other
groups) and an increased p62/GAPDH ratio in response to DSS
administration (all P<0.05). The ratio was reversed when the
mice in the F. nucleatum + DSS group were treated with L.
rhamnosus (P<0.01 and P<0.05, respectively). Consistent
with the above-mentioned data, the IHC analysis of LC3B and p62
expression in the colon tissues further confirmed the antagonistic
effects of F. nucleatum and L. rhamnosus (Figs. 5B and S2A). Following L. rhamnosus
intervention, F. nucleatum-DSS impaired autophagy was
significantly restored.
In addition, the mRNA levels of Beclin1, Atg7 and
Atg16L1 were also measured, which are key genes associated with
autophagy. The levels of Beclin1, Atg7 and Atg16L1 were
significantly lower in the mice that were administered F.
nucleatum and DSS compared to those administered F.
nucleatum or DSS alone (P<0.05, <0.01 and <0.01,
respectively; Fig. S2B) and were
higher in response to L. rhamnosus treatment (all
P<0.05). Collectively, L. rhamnosus significantly
attenuated the F. nucleatum-induced intestinal blocking of
autophagy in vivo.
L. rhamnosus restores the impaired
autophagic flux via the AKT/mTOR pathway
The AKT/mTOR pathway has been shown to be a key
regulator of the autophagic process. The mTOR complex 1 (mTORC1) is
a major negative regulator of autophagy, while AKT is a major
upstream modulator of mTORC1 (32). The present study therefore
assessed AKT and mTOR expression in the current experimental system
in order to confirm whether F. nucleatum and L.
rhamnosus regulate autophagy through this pathway. It was found
that the protein levels of p-mTOR and p-AKT gradually increased to
a peak in a concentration-depend manner following exposure to F.
nucletum and then gradually decreased (Fig. 6A and B).
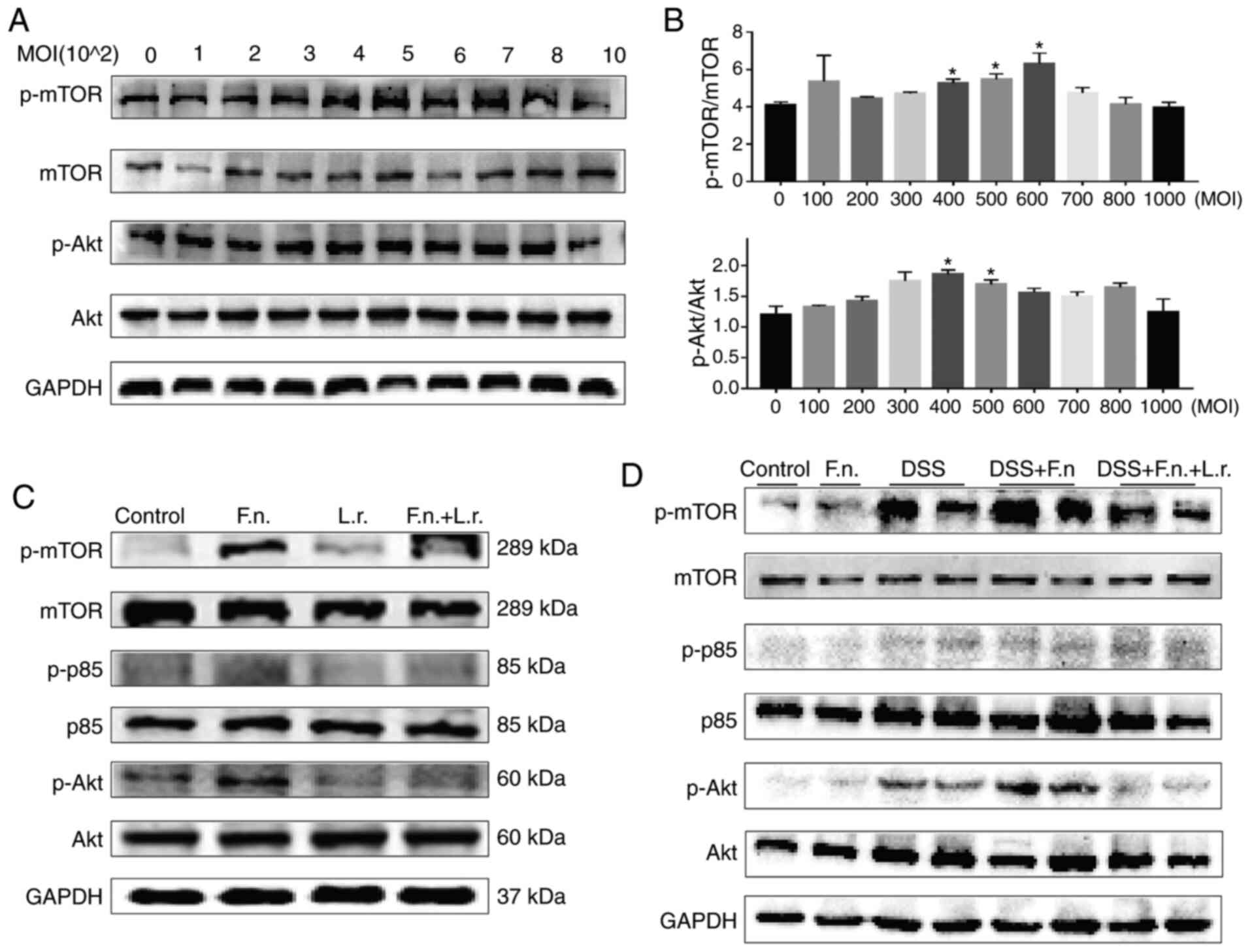 | Figure 6Lactobacillus rhamnosus
restores the impaired autophagic flux via AKT/mTOR pathway. (A and
B) Effects of increasing MOI F. nucleatum on p-mTOR, mTOR,
p-AKT and AKT protein levels in Caco2 cells are indicated by
representative western blots. (C) Western blot analysis of p-mTOR,
mTOR, p-p85, p85, p-AKT and AKT levels in Caco2 cells,
respectively. (D) Western blot analysis of p-mTOR, mTOR, p-p85,
p85, p-AKT and AKT levels in Caco2 cells and colon tissues,
respectively. *P≤0.05. F. nucleatum/F.n.,
Fusobacterium nucleatum; L.r., Lactobacillus
rhamnosus; DSS, dextran sulfate sodium. |
As was expected, the F. nucleatum group
exhibited higher p-mTOR, p-p85 and p-AKT protein expression levels
compared with the control or L. rhamnosus only groups. L.
rhamnosus treatment induced a decrease in these protein levels
and enhanced autophagy (for p-p85, P<0.01; for the other levels,
P<0.05; Figs. 6C and S3A). Consistent with these findings,
the results obtained from the colonic tissues were similar to those
obtained with the cell experiments (Figs. 6D and S3B). The mice in the F.
nucleatum + DSS group exhibited a decreased p-mTOR and p-AKT
protein level following L. rhamnosus intervention, although
the p-p85 levels were not significantly altered. All these results
demonstrated that the AKT/mTOR pathway was involved in the
regulation of the autophagy process and that L. rhamnosus
attenuated intestinal colitis aggravated by F. nucleatum
relative.
Blocking the autophagic flux reinforces
the production of pro-inflammatory cytokines induced by F.
nucleatum and partly inhibits the effects of Lactobacillus
rhamnosus
As illustrated above, F. nucleatum
intensified colonic inflammation in vitro and in
vivo; the mechanisms involved the blocking of the autophagic
flux. To further investigate the phenomenon of the autophagic flux,
2 autophagy chemical inhibitors of autophagy, 3-MA and CQ, were
used prior to the F. nucleatum or L. rhamnosus
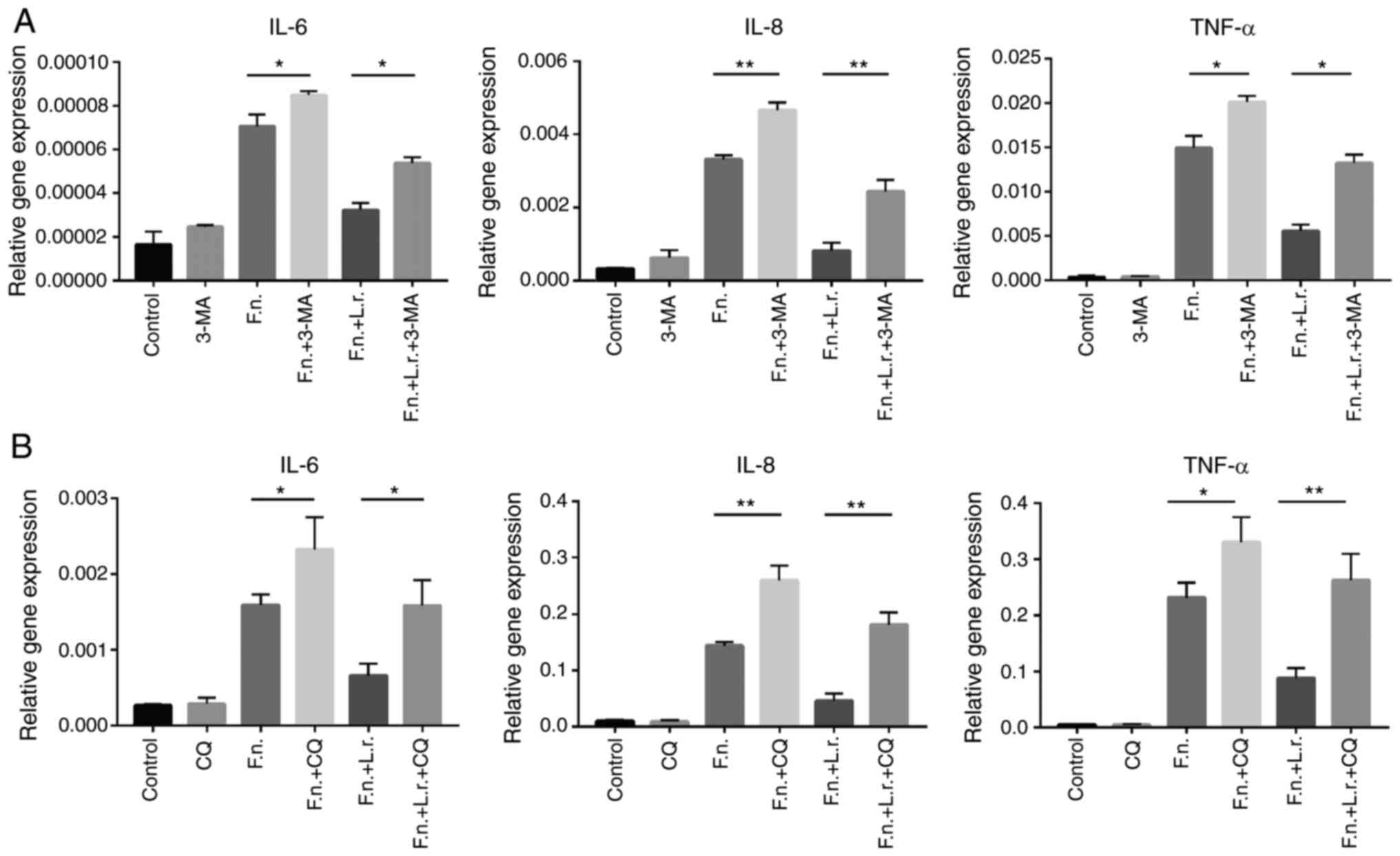
treatment of the Caco2 cells. As shown in Fig. 7, the autophagy inhibitors
significantly enhanced the gene expression levels of IL-6, IL-8 and
TNF-α (for IL-8, P<0.01; for the other levels, P<0.05,
respectively). As was expected, L. rhamnosus did not
markedly reduce the mRNA levels of these pro-inflammatory cytokines
following pre-treatment with the autophagy inhibitors (for IL-8 and
TNF-α in CQ treatment, P<0.01; for the other levels, P<0.05,
respectively).
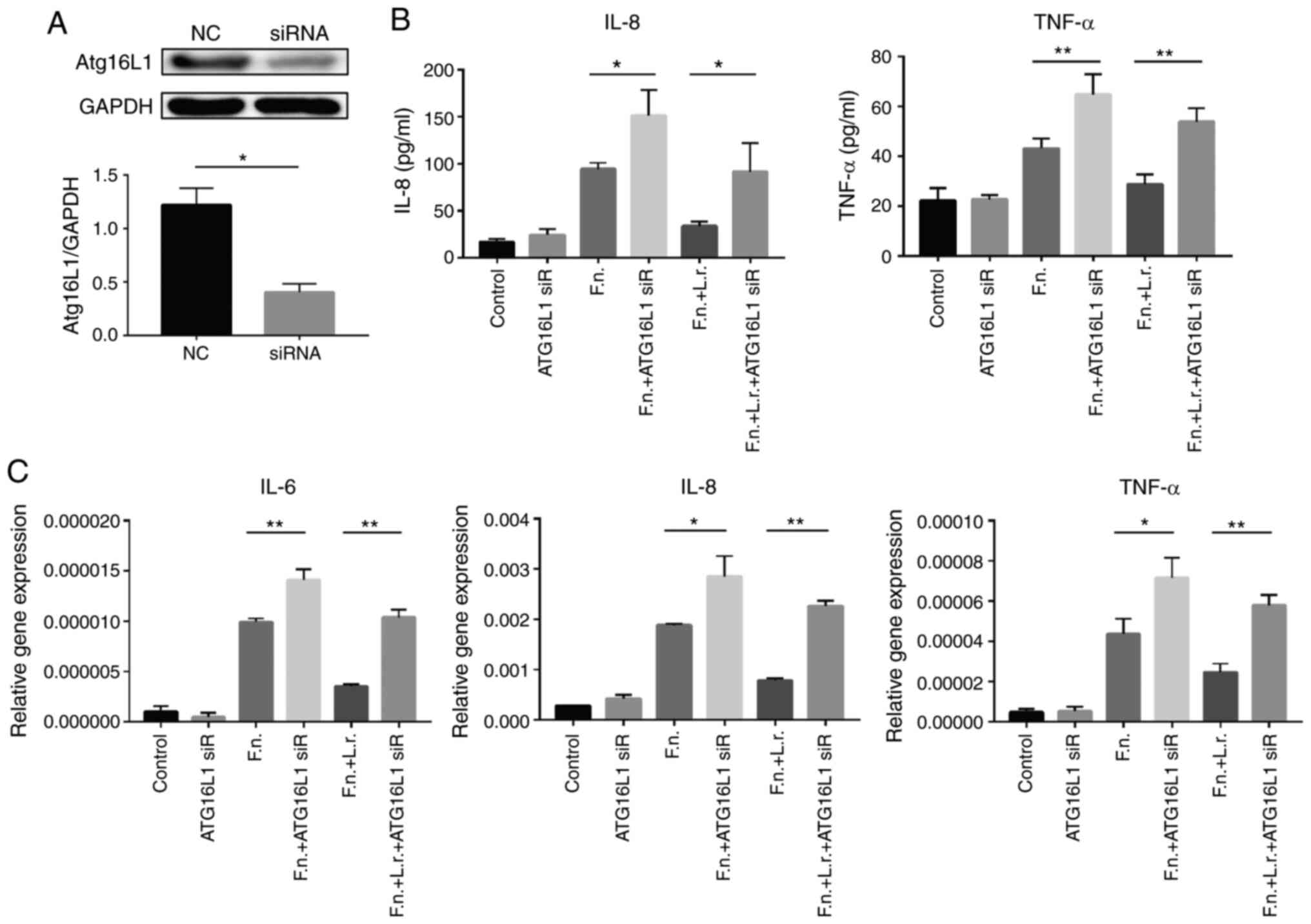
Subsequently, the present study examined the L.
rhamnosus-mediated activation of the F.
nucleatum-induced autophagic flux damage using gene
interference. Caco2 cells were transfected with siRNA designed to
inhibit Atg16L1 expression and this was expected to
attenuate the mediating effects of L. rhamnosus on the F.
nucleatum-induced blocking of autophagy. The knockdown of
Atg16L1 expression by siRNA effectively decreased its
expression by 66.8% compared to the untransfected cells (Fig. 8A). The cytokine production of the
siRNA-transfected cells was subsequently examined in response to
F. nucleatum and L. rhamnosus treatment using ELISA
and RT-qPCR. Consistent with the above-mentioned results, IL-8 and
TNF-a production induced by F. nucleatum treatment increased
in the presence of Atg16L1 siRNA and the effects of L.
rhamnosus were suppressed (Fig.
8B). The results of RT-qPCR revealed a similar variation
tendency for the mRNA levels (Fig.
8C). Taken together, the mediation of autophagy by the
probiotics L. rhamnosus was involved in the protective
effects against F. nucleatum-related intestinal
inflammation.
Discussion
There is increasing evidence of the connection
between intestinal bacterium and defective autophagy in the
pathogenesis of IBD. The present study demonstrated that L.
rhamnosus attenuated F. nucleatum-related intestinal
inflammation by mediating the autophagy of intestinal epithelial
cells. First, establishing an in vitro and in vivo
model revealed that L. rhamnosus effectively alleviated
colitis and attenuated the production of pro-inflammation cytokines
aggravated by F. nucleatum. Subsequently, the status of
autophagy was detected and it was found that L. rhamnosus
restored the autophagic flux impaired by F. nucleatum in
vitro and in vivo. Moreover, the PI3K/AKT/mTOR pathway
was investigated and it was demonstrated that it was involved in
the autophagic process. Finally, autophagy was interfered with
using autophagy inhibitors and siRNAs in Caco2 cells. As a result,
the pro-inflammatory effects of F. nucleatum were enhanced
and the anti-inflammatory of effects L. rhamnosus were
impaired.
Previous studies have demonstrated the association
of F. nucleatum with intestinal inflammation, as it can
generate a pro-inflammatory microenvironment (33). Moreover, it is easier to isolate
Fusobacterium spp. from patients with intestinal
inflammatory disease compared with healthy controls, more than a
half of which are F. nucleatum and these strains are
significantly more invasive than those from healthy controls
(6). Another study found that
F. nucleatum regulated M1 macro-phage polarization and the
secretion of IFN-γ by Th1-related cytokines, thus promoting the
progression of ulcerative colitis (22). Consistent with these findings, the
present study found that F. nucleatum intensified the
severity of experimental colitis. However, the underlying
mechanisms remain unknown.
It is known that probiotics can improve the
intestinal mucosal barrier by promoting the secretion of
anti-inflammatory factors and inhibiting the growth of harmful
bacteria in the intestine (34).
L. rhamnosus is a widely used probiotic with the ability to
restrain pathogenic bacteria. It has been reported L.
rhamnosus can produce factors capable of suppressing
Clostridium difficile-induced inflamed production (35) and attenuate enterohemorrhagic
Escherichia coli-induced changes in paracellular
permeability in epithelial cell monolayers, thus protecting
epithelial barrier function (36). However, to the best of our
knowledge, no study to date has reported the effects of L.
rhamnosus on F. nucleatum. In the present study, it was
found that L. rhamnosus attenuated DSS-induced colitis
aggravated by F. nucleatum, as shown by an improvement in
pathological features, DAI scores, colon length and
pro-inflammatory cytokine expression. Similarly, L.
rhamnosus decreased F. nucleatum-induced
pro-inflammatory cytokine production in Caco2 cells. However, there
are some limitations to the present study. A group administered DSS
+ L. rhamnosus mice may be useful to further support the
findings, since it could clarify the repairing effect of L.
rhamnosus on DSS-induced colitis without F. nucleatum.
However, it may still be difficult to clarify whether L.
rhamnosus purely acts on DSS or on F. nucleatum as well.
Therefore, cell experiments were also performed to further clarify
whether L. rhamnosus can protect intestinal epithelial cells
from F. nucleatum infection without other influencing
factors.
The present study then focused on the underlying
mechanisms. Autophagy plays a vital role in host defenses against
microbial infection and protects the intestine from injury in the
process of IBD (37). With the
conditional knockdown of crucial autophagy-related genes in mice or
colonic epithelial cells, the susceptibility to DSS-induced colitis
is enhanced and an abnormal microflora is formed (12). Crohn's disease-associated adherent
invasive Escherichia coli (AIEC) can reduce the levels of
Atg5 and Atg16L1 and inhibit autophagy, thus increasing the
inflammatory response (38). On
the other hand, probiotics can also activate autophagy to eliminate
intracellular bacteria. Thus, it was hypothesized that autophagy
may be involved in the process of F. nucleatum invasion and
L. rhamnosus protection. In the present study, it was
demonstrated that autophagy was deregulated following the
administration of F. nucleatum in vitro and in vivo,
while L. rhamnosus restored the autophagic flux. Autophagy
interference experiments revealed that the inhibition of autophagy
increased the secretion of pro-inflammatory cytokines induced by
F. nucleatum and suppressed the 'anti-F. nucleatum'
effects of L. rhamnosus. Therefore, it is reasonable to
point out that autophagy is involved in the said circumstances.
However, although autophagy inhibitors or siRNA weakened the
protective effects of L. rhamnosus on F. nucleatum,
these effects were not completely inhibited. This may indicate that
there are other inhibitory and mediating effects of L.
rhamnosus on the actions of F. nucleatum, apart from the
restoration of autophagy. Thus, further studies are warranted to
fully investigate this matter. Furthermore, the present study
examined the changes in the PI3K/AKT/mTOR pathway, as it serves as
one of the main regulatory pathways of autophagy (32). It was found that the changes in
this pathway conformed to the changes in autophagy.
In conclusion, the present study demonstrates that
L. rhamnosus plays a protective role in the pathogenesis of
F. nucleatum-related colitis and that the mediation of
autophagy is involved in this process. The findings of the present
study may provide new insight into the pathogenesis and therapy of
IBD.
Supplementary Data
Funding
The present study was financially supported by the
National Natural Science Foundation of China (grant nos. 81800467,
81330014, 81720108006 and 81974062).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CD performed the experiments, analyzed the data and
drafted the manuscript. XT, WW and WQ were involved in the
evaluation of the data. XF and XD were involved in the culture of
the bacteria. SZ assisted in the animal experiments. CH and XH
designed the study, revised the manuscript, provided funding and
obtained grants. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The animal experiments in the present study were
approved by the Animal Research Ethics Committee of Tongji Medical
College, Huazhong University of Science and Technology (approval ID
2016-0057).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Kaser A, Zeissig S and Blumberg RS:
Inflammatory bowel disease. Annu Rev Immunol. 28:573–621. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Olivera P, Danese S, Jay N, Natoli G and
Peyrin-Biroulet L: Big data in IBD: A look into the future. Nat Rev
Gastroenterol Hepatol. 16:312–321. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mazmanian SK, Round JL and Kasper DL: A
microbial symbiosis factor prevents intestinal inflammatory
disease. Nature. 453:620–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garrett WS, Gallini CA, Yatsunenko T,
Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L,
Glickman JN, et al: Enterobacteriaceae act in concert with the gut
microbiota to induce spontaneous and maternally transmitted
colitis. Cell Host Microbe. 8:292–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chu H, Khosravi A, Kusumawardhani IP, Kwon
AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, et
al: Gene-microbiota interactions contribute to the pathogenesis of
inflammatory bowel disease. Science. 352:1116–1120. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Strauss J, Kaplan GG, Beck PL, Rioux K,
Panaccione R, Devinney R, Lynch T and Allen-Vercoe E: Invasive
potential of gut mucosa-derived Fusobacterium nucleatum positively
correlates with IBD status of the host. Inflamm Bowel Dis.
17:1971–1978. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lévy J, Cacheux W, Bara MA, L'Hermitte A,
Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain
AM, et al: Intestinal inhibition of Atg7 prevents tumour initiation
through a microbiome-influenced immune response and suppresses
tumour growth. Nat Cell Biol. 17:1062–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Narendra D, Tanaka A, Suen DF and Youle
RJ: Parkin is recruited selectively to impaired mitochondria and
promotes their autophagy. J Cell Biol. 183:795–803. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mialet-Perez J and Vindis C: Autophagy in
health and disease: Focus on the cardiovascular system. Essays
Biochem. 61:721–732. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chai P, Ni H, Zhang H and Fan X: The
evolving functions of autophagy in Ocular health: A double-edged
sword. Int J Biol Sci. 12:1332–1340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuboi K, Nishitani M, Takakura A, Imai Y,
Komatsu M and Kawashima H: Autophagy protects against colitis by
the maintenance of normal gut microflora and secretion of mucus. J
Biol Chem. 290:20511–20526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benjamin JL, Sumpter R Jr, Levine B and
Hooper LV: Intestinal epithelial autophagy is essential for host
defense against invasive bacteria. Cell Host Microbe. 13:723–734.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sanders ME, Guarner F, Guerrant R, Holt
PR, Quigley EM, Sartor RB, Sherman PM and Mayer EA: An update on
the use and investigation of probiotics in health and disease. Gut.
62:787–796. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gionchetti P, Lammers KM, Rizzello F and
Campieri M: Probiotics and barrier function in colitis. Gut.
54:898–900. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ng SC, Plamondon S, Kamm MA, Hart AL,
Al-Hassi HO, Guenther T, Stagg AJ and Knight SC: Immunosuppressive
effects via human intestinal dendritic cells of probiotic bacteria
and steroids in the treatment of acute ulcerative colitis. Inflamm
Bowel Dis. 16:1286–1298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Claes IJ, Lebeer S, Shen C, Verhoeven TL,
Dilissen E, De Hertogh G, Bullens DM, Ceuppens JL, Van Assche G,
Vermeire S, et al: Impact of lipoteichoic acid modification on the
performance of the probiotic Lactobacillus rhamnosus GG in
experimental colitis. Clin Exp Immunol. 162:306–314. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Plaza-Díaz J, Ruiz-Ojeda FJ,
Vilchez-Padial LM and Gil A: Evidence of the anti-inflammatory
effects of probiotics and synbiotics in intestinal chronic
diseases. Nutrients. 9:5552017. View Article : Google Scholar :
|
|
19
|
Neish AS: Microbes in gastrointestinal
health and disease. Gastroenterology. 136:65–80. 2009. View Article : Google Scholar
|
|
20
|
Hussain QA, McKay IJ, Gonzales-Marin C and
Allaker RP: Regulation of adrenomedullin and nitric oxide
production by periodontal bacteria. J Periodontal Res. 50:650–657.
2015. View Article : Google Scholar
|
|
21
|
Sadeghi-Aliabadi H, Mohammadi F, Fazeli H
and Mirlohi M: Effects of Lactobacillus plantarum A7 with probiotic
potential on colon cancer and normal cells proliferation in
comparison with a commercial strain. Iran J Basic Med Sci.
17:815–819. 2014.
|
|
22
|
Liu L, Liang L, Liang H, Wang M, Lu B, Xue
M, Deng J and Chen Y: Fusobacterium nucleatum aggravates the
progression of colitis by regulating M1 macrophage polarization via
AKT2 pathway. Front Immunol. 10:13242019. View Article : Google Scholar :
|
|
23
|
Liu H, Hong X, Sun T, Huang X, Wang J and
Xiong H: Fusobacterium nucleatum exacerbates colitis by damaging
epithelial barrier and inducing aberrant inflammation. J Dig Dis.
May 22–2020.Epub ahead of print. View Article : Google Scholar
|
|
24
|
Cooper HS, Murthy SN, Shah RS and
Sedergran DJ: Clinicopathologic study of dextran sulfate sodium
experimental murine colitis. Lab Invest. 69:238–249.
1993.PubMed/NCBI
|
|
25
|
Dutton JW III, Artwohl JE, Huang X and
Fortman JD: Assessment of pain associated with the injection of
sodium pentobarbital in laboratory mice (Mus musculus). J Am Assoc
Lab Anim Sci. 58:373–379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Zhang Q, Gao M, Zhang Y, Song Y, Cheng H
and Zhou R: The germline-enriched Ppp1r36 promotes autophagy. Sci
Rep. 6:246092016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cardiff RD, Miller CH and Munn RJ: Manual
hematoxylin and eosin staining of mouse tissue sections. Cold
Spring Harbor protocols. 2014:655–658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Horino J, Fujimoto M, Terabe F, Serada S,
Takahashi T, Soma Y, Tanaka K, Chinen T, Yoshimura A, Nomura S, et
al: Suppressor of cytokine signaling-1 ameliorates dextran sulfate
sodium-induced colitis in mice. Int Immunol. 20:753–762. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Misra RM, Bajaj MS and Kale VP:
Vasculogenic mimicry of HT1080 tumour cells in vivo: Critical role
of HIF-1α-neuropilin-1 axis. PLoS One. 7:e501532012. View Article : Google Scholar
|
|
31
|
Baxt LA and Xavier RJ: Role of Autophagy
in the Maintenance of Intestinal Homeostasis. Gastroenterology.
149:553–562. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heras-Sandoval D, Pérez-Rojas JM,
Hernández-Damián J and Pedraza-Chaverri J: The role of
PI3K/AKT/mTOR pathway in the modulation of autophagy and the
clearance of protein aggregates in neurodegeneration. Cell Signal.
26:2694–2701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kostic AD, Chun E, Robertson L, Glickman
JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold
GL, et al: Fusobacterium nucleatum potentiates intestinal
tumorigenesis and modulates the tumor-immune microenvironment. Cell
Host Microbe. 14:207–215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen ZH, Zhu CX, Quan YS, Yang ZY, Wu S,
Luo WW, Tan B and Wang XY: Relationship between intestinal
microbiota and ulcerative colitis: Mechanisms and clinical
application of probiotics and fecal microbiota transplantation.
World J Gastroenterol. 24:5–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boonma P, Spinler JK, Venable SF,
Versalovic J and Tumwasorn S: Lactobacillus rhamnosus L34 and
Lactobacillus casei L39 suppress Clostridium difficile-induced IL-8
production by colonic epithelial cells. BMC Microbiol. 14:1772014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Johnson-Henry KC, Donato KA, Shen-Tu G,
Gordanpour M and Sherman PM: Lactobacillus rhamnosus strain GG
prevents enterohemorrhagic Escherichia coli O157:H7-induced changes
in epithelial barrier function. Infect Immun. 76:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Macias-Ceja DC, Cosin-Roger J, Ortiz-Masiá
D, Salvador P, Hernández C, Esplugues JV, Calatayud S and
Barrachina MD: Stimulation of autophagy prevents intestinal mucosal
inflammation and ameliorates murine colitis. Br J Pharmacol.
174:2501–2511. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nguyen HT, Dalmasso G, Müller S, Carrière
J, Seibold F and Darfeuille-Michaud A: Crohn's disease-associated
adherent invasive Escherichia coli modulate levels of microRNAs in
intes-tinal epithelial cells to reduce autophagy. Gastroenterology.
146:508–519. 2014. View Article : Google Scholar
|