Zinc (Zn) is an essential metal involved in cell
signalling, proliferation, differentiation, oxidative stress, the
immune response and numerous other important cellular processes
(1-4). The role of Zn in cells is primarily
associated with Zn binding as a cofactor in enzymes, or for
structural and/or regulatory functions of proteins (5). The immune system is highly dependent
on Zn homeostasis for proper and efficient function. Zn is an
integral part of the signalling pathways involved in regulating
both the innate and adaptive immune responses (3). In individuals with Zn deficiency,
these signals are highly perturbed, affecting both T-cell and
B-cell development and function, natural killer cell production and
monocyte cytotoxicity (3). Due to
these perturbations individuals with Zn deficiency are more
susceptible to infection (6). In
this regard, Zn supplements are heralded to boost the immune
system.
The use of Zn against viruses has been studied from
the 1970s to present, where Zn was shown to affect viral
replication, protein synthesis and processing, membrane fusion and
RNA polymerase activity (7-21).
A summary of the influence of Zn on several respiratory viruses is
provided in Table I.
Clinical studies have linked Zn supplementation with
less severe and reduced duration of symptoms along with lower
recurrent infections for viral infections (6-7,22).
Although there is an observed benefit of Zn in antiviral therapy,
this is largely dependent on the type of infection as well as the
concentration, formulation and subsequent redox species of Zn used
(7). For example, the use of Zn
to treat the common cold often caused by rhinoviruses has been
extensively reviewed with large variability in treatment
effectiveness (7,23-27). While there is evidence of the role
of Zn in inhibiting other respiratory viruses such as severe acute
respiratory syndrome (SARS)-coronavirus (CoV), the efficacy of Zn
in clinical trials against these has not been sufficiently studied
with good rigour (7,11).
With the emergence of coronavirus disease 2019
(COVID-19), several studies have explored the therapeutic potential
of compounds previously used against similar coronaviruses, such as
SARS-CoV and Middle East respiratory syndrome (MERS)-CoV (28,29). Two essential proteins in
coronaviruses include: i) RNA-dependent RNA-polymerase (RdRp),
which is necessary for proper viral replication, a core enzyme of
the viruses' multiprotein replication and transcription complex
(30) and ii) 3C-like proteinase
(3CLpro) or main protease, a cysteine protease that has
two domains each containing β-barrel chymotrypsin-like folds
(31). The active site of
3CLpro is located in the cleft between the two domains
and is characterized by a catalytic Cys-His dyad, which is
necessary for polyprotein processing and essential for viral
replication (30,31). For this reason, compounds with the
ability to inhibit these proteins are often used as antivirals
(32,33).
Zn is often delivered as a complex with
N-ethyl-N-phenyl dithiocarbamic acid zinc (EPDTC) or
toluene-3,4-dithiolato zinc (TDT) (13). These Zn ionophores also contribute
to protein binding and inhibit these enzymes (11,12). Zn-ligating compounds are proposed
to aid in coordinating Zn in the catalytic site of
3CLpro, thus inhibiting proteinase activity (12,13). Alternatively, Zn ionophores are
only thought to aid Zn cell entry where Zn2+ ions then
act alone to inhibit RdRp, though how this inhibition occurs has
not been fully elucidated (11,14).
The presents study performed bioinformatics analysis
and modelled Zn binding sites onto RdRp and 3CLpro and
proposed the hypothesis that Zn would modulate COVID-19 replication
and ameliorate the infection and severity of symptoms.
The nucleotide sequence of RdRp for COVID-19
(GenBank accession no. MT042778.1), SARS RdRp (GenBank accession
no. AY340092.1), influenza A PB1 (GenBank accession no.
AJ620348.2), hepatitis C virus (HCV) NS5B (GenBank accession no.
AJ608785.1), calcivirus RdRp (GenBank accession no. Y13703.1) and
T7 Phage RdRp (GenBank accession no. M3830s28.1) was retrieved in
FASTA format from the National Center for Biotechnology Information
(NCBI; http://www.ncbi.nlm.nih.gov).
The amino acid sequence for COVID-19 nsp 12 (GenBank
accession no. YP_009725307.1), SARS rep (UniProtKB accession no.
R1AB_CVHSA), influenza A PB1 (GenBank accession no. AAK18013.1) and
T7 Bacteriophage (T7 Phage) PHA (PDB accession no. 4RNP_C) was
obtained from the following databases: NCBI (https://www.ncbi.nlm.nih.gov/protein/), UniProt
(https://www.uniprot.org/), Protein Data Bank In
Europe (https://www.ebi.ac.uk/pdbe/) and
Worldwide Protein Data Bank (http://www.wwpdb.org/). For all DNA and protein
phylogenetic trees and multiple sequence alignments, ClustalW and
ClustalX were used (http://www.clustal.org/).
The nucleotide sequence of COVID-19 orf1ab (GenBank
accession no. MT049951.1), SARS 3CLpro (GenBank
accession no. AY609081.1), black queen cell virus (BQV)
3CLpro (GenBank accession no. KM232906.1), yellow head
virus (YHV) 3CLpro (GenBank accession no. EU977577.1)
and avian infectious bronchitis virus 3CLpro (GenBank
accession no. Q157446.1) was retrieved in FASTA format from the
same databases used for RdRp.
The previously determined crystal structures of the
RdRp of SARS-CoV (PDB accession no. 6NUR) (34) and COVID-19 (PDB accession no.
6M71) (35) were aligned using
PyMOL Molecular Graphics system (version 1.2r3pre; Schrödinger,
Inc.). The alignment was performed iteratively five times with a
cut-off of 2.0 Å and a resulting root-mean-square deviation (RMSD)
value of 0.588 for 7,027 atoms aligned out of a total 8,040 atoms.
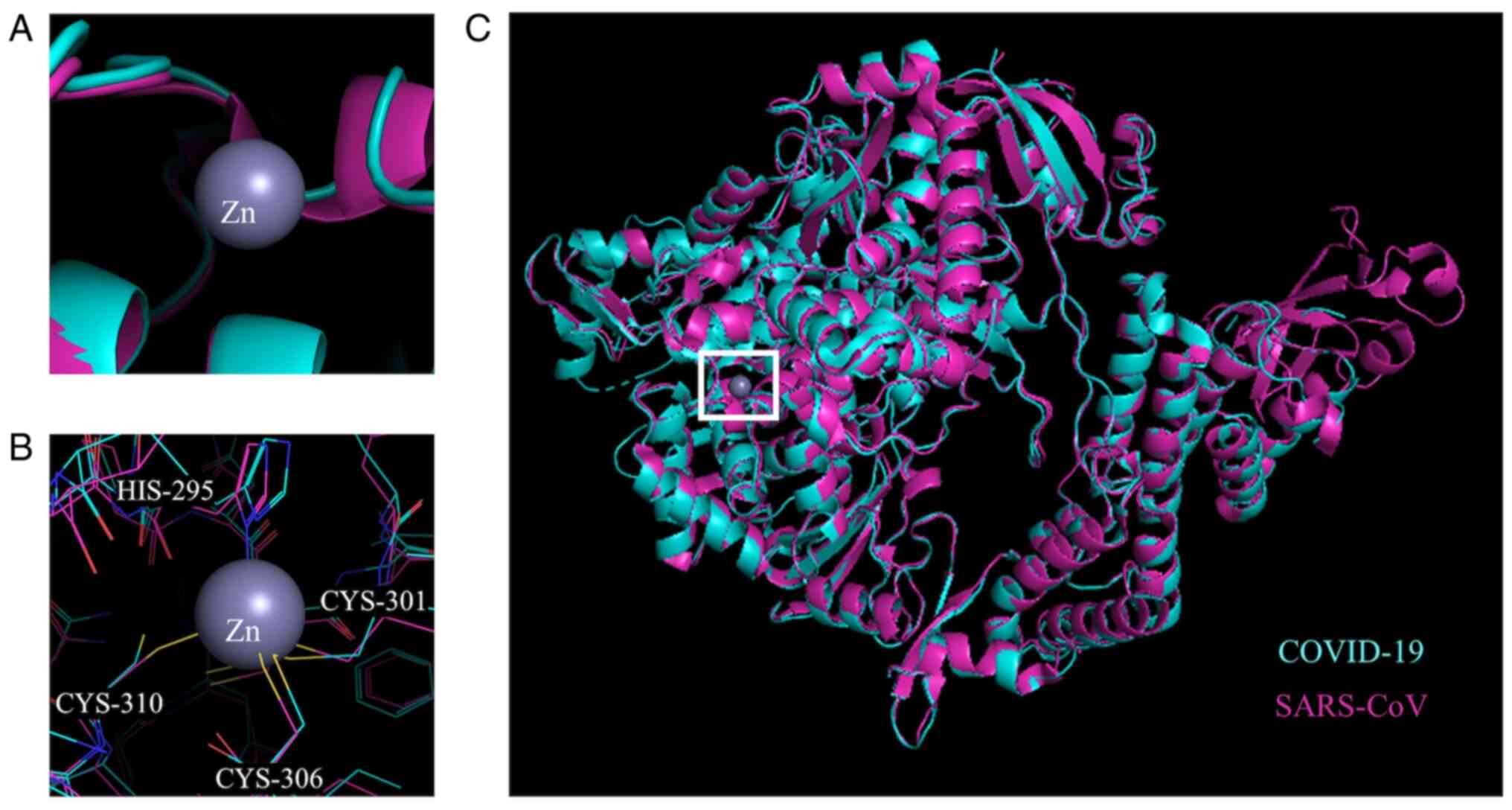
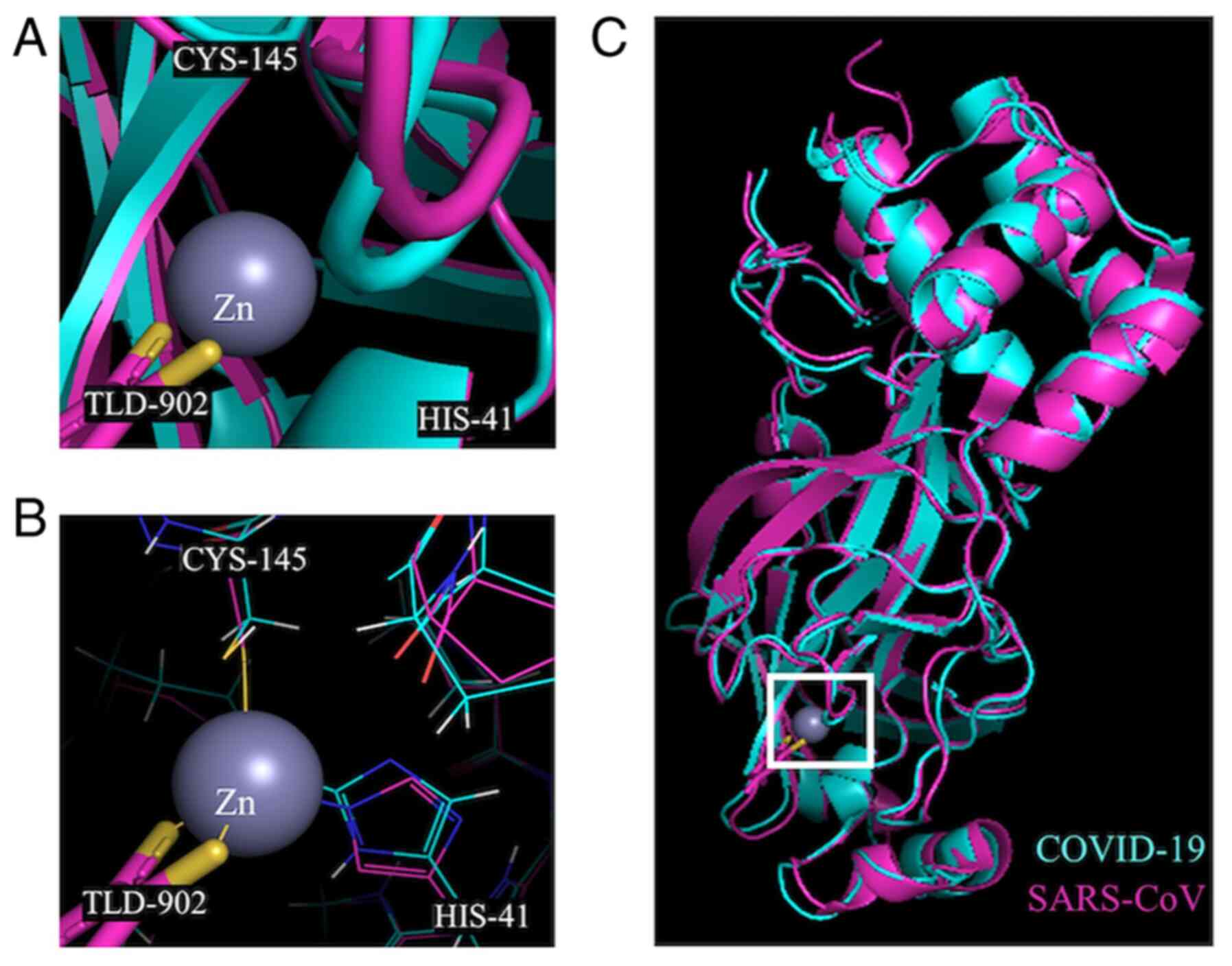
The crystal structures of the 3CL-protease of SARS-CoV bound to a
Zn coordinating compound (TLD902; TDT) (PDB accession no. 2Z94)
(13) and SARS-CoV-2 (PDB
accession no. 6W63) (36) were
also aligned iteratively five times with a cut-off of 2.0 Å and a
resulting RMSD value of 0.621 for 1,985 atoms aligned out of a
total 2,339 atoms in PyMol. The Zn binding sites were illustrated
based on the location of Zn in the crystal structure of these
proteins for SARS-CoV.
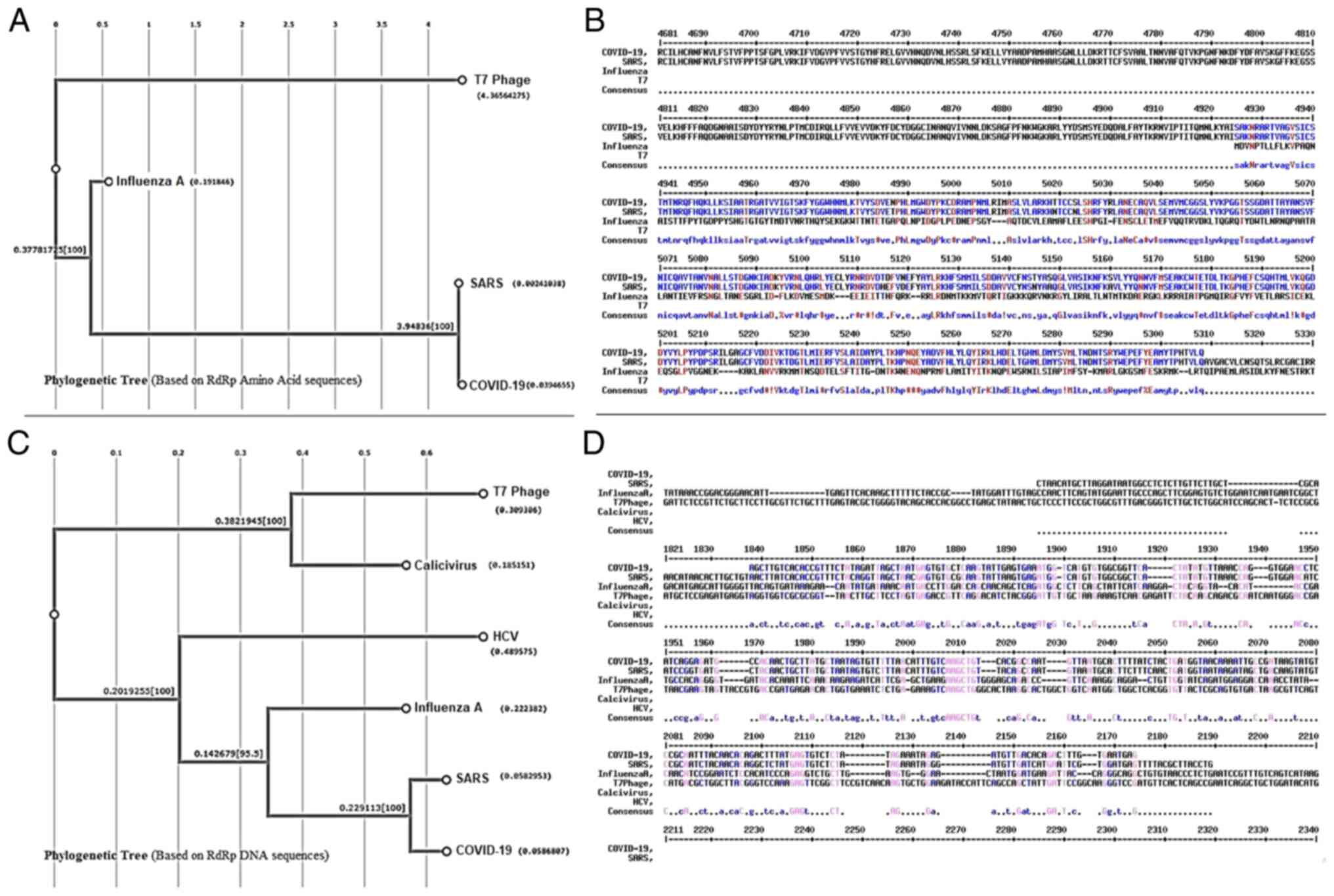
The present analysis revealed a high level of
identity (81.5 for DNA and 96.2 for protein alignment) of COVID-19
RdRp with the enzyme from the SARS virus that belongs in the same
virus family (Coronaviridae). The score, identity and similarity of
RdRp DNA and amino acid sequences are shown in Tables SI and SII. Alignment of the DNA
sequences of COVID-19 RdRp (GenBank accession no. MT042778.1) and
SARS RdRp (GenBank accession no. AY340092.1) showed an 87.7%
aligned score of the two sequences (Fig. 1 and Table SI). Moreover, an amino acid
sequence alignment of COVID-19 nsp 12 (GenBank accession no.
YP_009725307.1) and SARS rep (UniProtKB accession no. R1AB_CVHSA)
showed an aligned score of 96.3% for the two sequences (Fig. 1 and Table SII). The alignment score,
identity and similarity of RdRp DNA and amino acid sequences are
shown in Tables SI and SII.
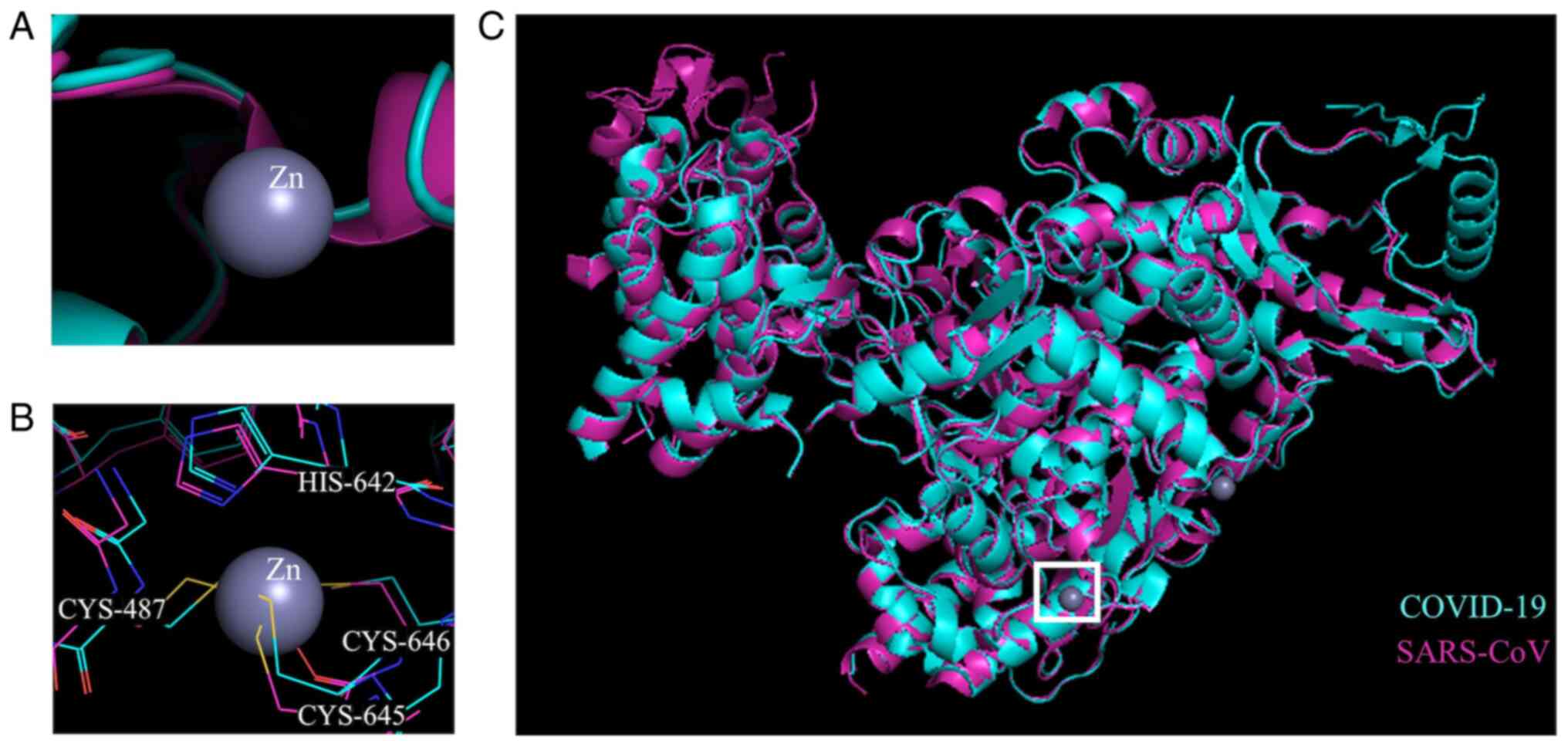
Based on bioinformatic similarities, structural
analyses were performed to evaluate the structural similarity
between the RdRp of SARS-CoV and COVID-19 (Figs. 3 and 4). A structural alignment was performed
on previously determined crystal structures for RdRp of SARS-CoV
(PDB accession no. 6NUR) (34)
and COVID-19 (PDB accession no. 6M71) (35). The alignment produced an RMSD
value of 0.588 for 7,027 atoms aligned out of a total 8,040 atoms.
The Zn binding sites, based on the crystal structure of the
SARS-CoV RdRp, were conserved in the COVID-19 RdRp.
The antiviral activity of Zn was reported by several
studies and shown to effect viral replication, protein synthesis
and processing, membrane fusion and RNA polymerase activity
(7-21). A previous study by Kirchdoerfer
and Ward (34) indicated that
RdRp-targeted drugs for SARS have the potential for COVID-19
treatment, and the present analysis suggested that there is similar
potential of Zn-targeting RdRp enzymes from this group of viruses
(37). Likewise, a phylogenetic
tree based on COVID-19 and SARS RdRp DNA and amino acid sequences
also supported in hypothesis. Two Zn binding sites were previously
identified in the structure of SARS-CoV RdRp, which the present
study has shown to be conserved in the COVID-19 RdRp. These sites
were hypothesized by the authors of the structure to be important
for proper folding of RdRp based on their location in the protein
(34). However, it is possible
that binding of Zn may also be allosterically regulatory and lead
to catalytic inhibition of RdRp in SARS-CoV (11). More enzymology studies would be
required to confirm the importance of these sites for either
inhibition or folding by Zn atom binding. Previous structural
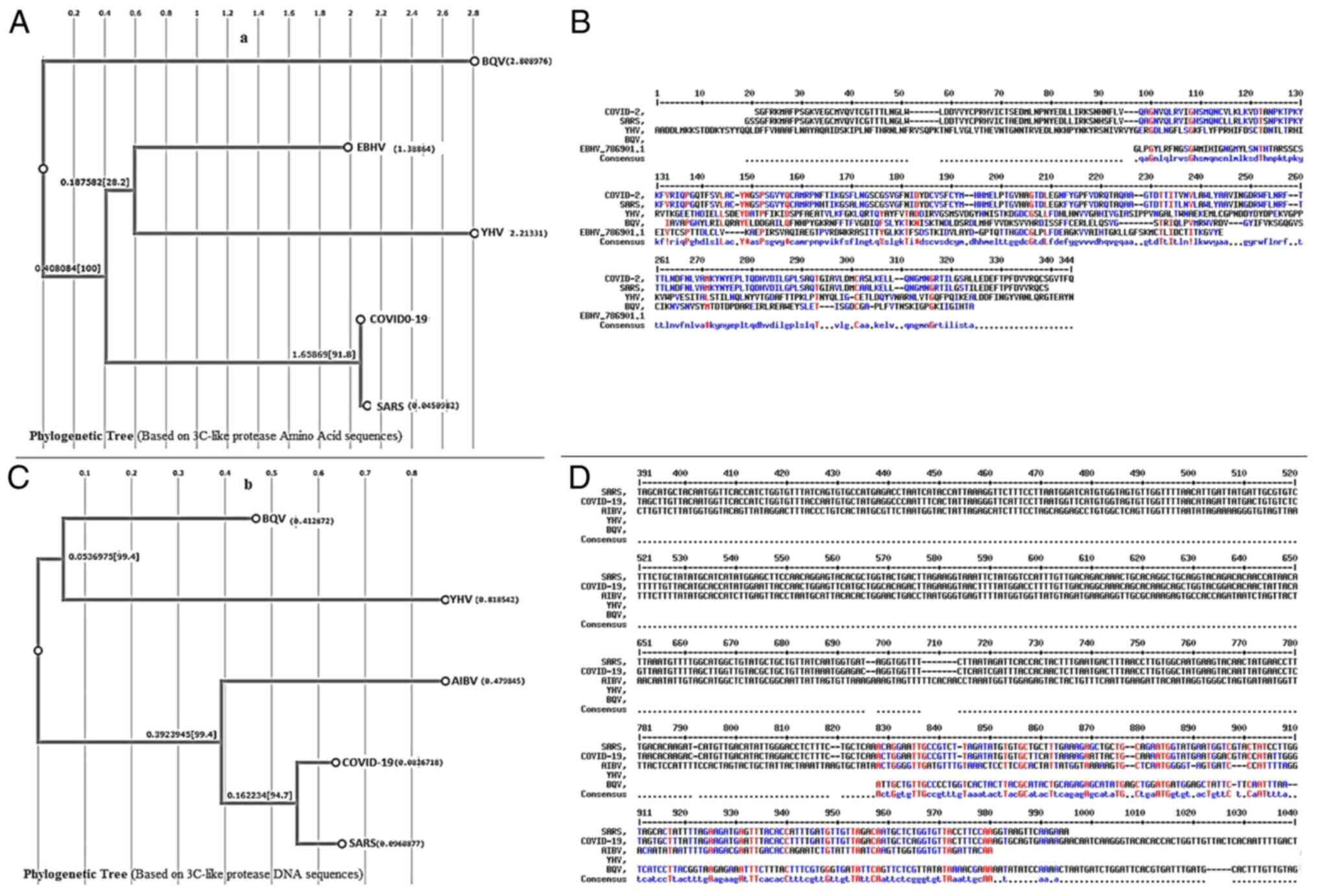
studies with Zn-coordinating compounds and 3CLpro of
SARS-CoV revealed that Zn bound to the catalytic dyad present in
3CLpro with the help of TDT (38). These residues are also found in
the aligned structure of COVID-19 3CLpro at the same
position, indicating that Zn would also bind to the COVID-19 enzyme
catalytic residues. Both Zn alone and the Zn coordinating compounds
were effective inhibitors of 3CLpro of SARS-CoV activity
with a Ki of 1.1, 1.4 and 1.0 µM for Zn alone,
TDT and EPDTC, respectively (12). Therefore, considering the current
COVID-19 pandemic and the present data, the present study
hypothesized that Zn supplementation would be applicable in
clinical practice to modulate symptoms and replication of the
virus.
With the emergent threat of the COVID-19 virus,
several studies have explored the therapeutic potential of
compounds previously used against similar coronaviruses (SARS-CoV
and MERS-CoV) (28,29). Recent meta-analyses by our
research group showed the similarities of COVID-19 with other
respiratory viral infections such as SARS, MERS and influenzas
(39,40). Clinical studies have linked Zn
supplementation with less severe and reduced duration of symptoms
along with lower recurrent infections for viral infections
(6,7,22).
Several studies have identified lungs as one of the
earlier organs to fail due to inflammation in COVID-19 cases
(41-44). Lung failure is one of the most
important leading causes of severe outcomes, including death, in
these cases (41-46). Our recent meta-analysis on 52,251
confirmed cases of COVID-19 indicated an increase to
pre-inflammatory factors such as IL-6 present in 52% of cases
(39). Therefore, researchers are
focusing on anti-inflammatory drugs such as anti-IL-6 for treatment
of patients with COVID-19 (42,47-49). Previous studies revealed a key
role for Zn in the regulation of inflammation, especially for
lungs; Gammoh and Rink (50)
reported that Zn is critical in the prevention of host-tissue
damage by inflammation, controlling oxidative stress and regulating
inflammatory cytokines. Zn is involved in modulating inflammation
by decreasing IL-6 and pro-inflammatory responses via reducing
NF-κB, the master regulator of pro-inflammatory responses (50). NF-κB can regulate inflammatory
responses by targeting genes, such as TNF-α and IL-1β, as well as
increasing the expression of A20 and peroxisome
proliferator-activated receptors-α genes (50,51). Moreover, a study by Knoell et
al (52) showed that
insufficient Zn can actually enhance lung inflammation.
Additionally, the overuse and abuse of antibiotics
is of increasing concern, particularly during treatment of
respiratory infections (53-56). Although co-infection of viruses
and bacteria can occur, identifying these cases can be challenging
(57,58). Lack of appropriate antimicrobial
stewardship programs and overprescription and use of antibiotics in
viral infections such as the novel COVID-19 can lead to antibiotic
side effects and antimicrobial resistance (AMR) (59,60). Both suspected and confirmed cases
of COVID-19 have received broad-spectrum antibiotics as there is
currently no rapid method that distinguishes between cases which
need antibiotic treatment and those that do not (61,62). Although it is lifesaving in some
patients, in others this treatment may be excessive and lead to
antibiotic side effects such as septic shock, killing normal
microbiota and contribute to AMR (62-64).
Our research group has been investigating
metal-based antimicrobials in response to the AMR era (65-69). A number of different metal
elements being reintroduced into regular infection control
applications have been observed (68). Studies have now established the
antibacterial potency of Zn, as either a metal salt or metal oxide
nanoparticle, against common pathogenic strains (70) and clinical isolates (71-73). Therefore, the use of Zn can be
considered for use in both viral and bacterial disease states.
Zn is recommended by the National Institutes of
Health (NIH) for inducing the immune system and preventing viral
infections; however, the amount of Zn people requires each day
depends on age (74). While Zn
supplementation is necessary to correct any deficiency, an
overabundance of Zn can also lead to a variety of physiological
dysfunctions. Excess Zn can lead to copper deficiency, alter
lymphocyte response and inhibit T-cell function (75). Therefore, the use of Zn for
therapeutic purposes should still be monitored based on food intake
and use of supplements. Although Zn is relatively non-toxic to
humans with an median lethal dose of 3 g/kg weight, extreme excess
Zn (>100-300 mg/day) should be avoided; the NIH considers 40 mg
of zinc a day for adults and 4 mg of zinc a day for infants under 6
months to be the upper limit dose (75).
The aforementioned points support the potential use
of Zn in the clinical treatment of COVID-19 patients. However, the
main obstacle for the current study is limited supportive clinical
data for prevention and treatment potency of Zn in patients with
COVID-19.
Most people obtain their daily required Zn through a
healthy diet. However, the dietary oral intake supplements of 15-25
mg Zn tablets per day is recommended to help aid immune response in
the short term (4). Currently,
there is no consensus that Zn is helpful for the prevention and
treatment of COVID-19 infection. However, the present
bioinformatics and molecular modeling analysis supported the
hypothesis that Zn would bind and regulate the enzymatic activities
of 3CLpro and RdRp of SARS-CoV-2 and thus inhibit viral
replication. Further studies would be necessary to identify the
exact mechanism by which this could occur in the COVID-19
viral-cell cycle processes. More studies are necessary to
understand the molecular mechanisms, effective concentration and
delivery formulations. Zn may be considered a candidate for the
prevention and treatment of COVID-19 infection.
This study was supported by the National Sciences
Engineering Research Council of Canada to RJT.
All data generated or analyzed during this study are
included in this published article.
AP and RJT conceived and designed the study; NKM and
AP performed comprehensive research; AP and NKM analyzed the data;
AP, NKM and RJT wrote and revised the paper; AP, NKM and RJT
participated in data analysis and manuscript editing. All authors
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Beyersmann D and Haase H: Functions of
zinc in signaling, proliferation and differentiation of mammalian
cells. Biometals. 14:331–341. 2001. View Article : Google Scholar
|
|
2
|
Marreiro D do N, Cruz KJ, Morais JB,
Beserra JB, Severo JS and Soares de Oliveira AR: Zinc and oxidative
stress: Current mechanisms. Antioxidants (Basel). 6:242017.
View Article : Google Scholar
|
|
3
|
Maywald M, Wessels I and Rink L: Zinc
signals and immunity. Int J Mol Sci. 18:22222017. View Article : Google Scholar :
|
|
4
|
Miller BD and Welch RM: Food system
strategies for preventing micronutrient malnutrition. Food Policy.
Wolters Kluwer-Medknow Publications; pp. 115–128. 2013, View Article : Google Scholar
|
|
5
|
Kochańczyk T, Drozd A and Krężel A:
Relationship between the architecture of zinc coordination and zinc
binding affinity in proteins-Insights into zinc regulation.
Metallomics. 7:244–257. 2015. View Article : Google Scholar
|
|
6
|
Prasad AS: Impact of the discovery of
human zinc deficiency on health. J Am Coll Nutr. 28:257–265. 2009.
View Article : Google Scholar
|
|
7
|
Read SA, Obeid S, Ahlenstiel C and
Ahlenstiel G: The role of zinc in antiviral immunity. Adv Nutr.
10:696–710. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Korant BD and Butterworth BE: Inhibition
by zinc of rhinovirus protein cleavage: Interaction of zinc with
capsid polypeptides. J Virol. 18:298–306. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaushik N, Subramani C, Anang S,
Muthumohan R, Shalimar, Nayak B, Ranjith-Kumar CT and Surjit M:
Zinc salts block hepatitis E virus replication by inhibiting the
activity of viral RNA-dependent RNA polymerase. J Virol.
91:e00754–e00717. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Korant BD, Kauer JC and Butterworth BE:
Zinc ions inhibit replication of rhinoviruses. Nature. 248:588–590.
1974. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
te Velthuis AJ, van den Worml SH, Sims AC,
Baric RS, Snijder EJ and van Hemert MJ: Zn2+ inhibits
coronavirus and arterivirus RNA polymerase activity in vitro and
zinc ionophores block the replication of these viruses in cell
culture. PLoS Pathog. 6:e10011762010. View Article : Google Scholar
|
|
12
|
Hsu JTA, Kuo CJ, Hsieh HP, Wang YC, Huang
KK, Lin CPC, Huang PF, Chen X and Liang PH: Evaluation of
metal-conjugated compounds as inhibitors of 3CL protease of
SARS-CoV. FEBS Lett. 574:116–120. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee CC, Kuo CJ, Hsu MF, Liang PH, Fang JM,
Shie JJ and Wang AH: Structural basis of mercury- and
zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
FEBS Lett. 581:5454–5458. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krenn BM, Gaudernak E, Holzer B, Lanke K,
Van Kuppeveld FJ and Seipelt J: Antiviral activity of the zinc
ionophores pyrithione and hinokitiol against picornavirus
infections. J Virol. 83:58–64. 2009. View Article : Google Scholar :
|
|
15
|
Lanke K, Krenn BM, Melchers WJ, Seipelt J
and van Kuppeveld FJ: PDTC inhibits picornavirus polyprotein
processing and RNA replication by transporting zinc ions into
cells. J Gen Virol. 88:1206–1217. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Geist FC, Bateman JA and Hayden FG: In
vitro activity of zinc salts against human rhinoviruses. Antimicrob
Agents Chemother. 31:622–624. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hung M, Gibbs CS and Tsiang M: Biochemical
characterization of rhinovirus RNA-dependent RNA polymerase.
Antiviral Res. 56:99–114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krenn BM, Holzer B, Gaudernak E, Triendl
A, van Kuppeveld FJ and Seipelt J: Inhibition of polyprotein
processing and RNA replication of human rhinovirus by pyrrolidine
dithiocarbamate involves metal ions. J Virol. 79:13892–13899. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suara RO and Crowe JE: Effect of zinc
salts on respiratory syncytial virus replication. Antimicrob Agents
Chemother. 48:783–790. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Srivastava V, Rawall S, Vijayan VK and
Khanna M: Influenza a virus induced apoptosis: Inhibition of DNA
laddering & caspase-3 activity by zinc supplementation in
cultured HeLa cells. Indian J Med Res. 129:579–586. 2009.PubMed/NCBI
|
|
21
|
Ghaffari H, Tavakoli A, Moradi A,
Tabarraei A, Bokharaei-Salim F, Zahmatkeshan M, Farahmand M,
Javanmard D, Kiani SJ, Esghaei M, et al: Inhibition of H1N1
influenza virus infection by zinc oxide nanoparticles: Another
emerging application of nanomedicine. J Biomed Sci. 26:702019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shankar AH and Prasad AS: Zinc and immune
function: The biological basis of altered resistance to infection.
Am J Clin Nutr. 68(Suppl 2): 447S–463S. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hulisz D: Efficacy of zinc against common
cold viruses: An overview. J Am Pharm Assoc 2003. 44:594–603. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hemilä H, Fitzgerald JT, Petrus EJ and
Prasad A: Zinc acetate lozenges may improve the recovery rate of
common cold patients: An individual patient data meta-analysis.
Open Forum Infect Dis. 4:ofx0592017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Science M, Johnstone J, Roth DE, Guyatt G
and Loeb M: Zinc for the treatment of the common cold: A systematic
review and meta-analysis of randomized controlled trials. CMAJ.
184:E551–E561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
D'Cruze H, Arroll B and Kenealy T: Is
intranasal zinc effective and safe for the common cold? A
systematic review and meta-analysis. J Prim Health Care. 1:134–139.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Caruso TJ, Prober CG and Gwaltney JM Jr:
Treatment of naturally acquired common colds with zinc: A
structured review. Clin Infect Dis. 45:569–574. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y, Hou Y, Shen J, Huang Y, Martin W
and Cheng F: Network-based drug repurposing for novel coronavirus
2019-nCoV/SARS-CoV-2. Cell Discov. 6:142020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lung J, Lin YS, Yang YH, Chou YL, Shu LH,
Cheng YC, Liu HT and Wu CY: The potential chemical structure of
anti-SARS-CoV-2 RNA-dependent RNA polymerase. J Med Virol.
92:693–697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prentice E, McAuliffe J, Lu X, Subbarao K
and Denison MR: Identification and characterization of severe acute
respiratory syndrome coronavirus replicase proteins. J Virol.
78:9977–9986. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan K, Wei P, Feng Q, Chen S, Huang C, Ma
L, Lai B, Pei J, Liu Y, Chen J and Lai L: Biosynthesis,
purification, and substrate specificity of severe acute respiratory
syndrome coronavirus 3C-like proteinase. J Biol Chem.
279:1637–1642. 2004. View Article : Google Scholar
|
|
32
|
Subissi L, Imbert I, Ferron F, Collet A,
Coutard B, Decroly E and Canard B: SARS-CoV ORF1b-encoded
nonstructural proteins 12-16: Replicative enzymes as antiviral
targets. Antiviral Res. 101:122–130. 2014. View Article : Google Scholar
|
|
33
|
Wu YS, Lin WH, Hsu JT and Hsieh HP:
Antiviral drug discovery against SARS-CoV. Curr Med Chem.
13:2003–2020. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kirchdoerfer RN and Ward AB: Structure of
the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors.
Nat Commun. 10:23422019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao
L, Wang T, Sun Q, Ming Z, Zhang L, et al: Structure of the
RNA-dependent RNA polymerase from COVID-19 virus. Science.
368:779–782. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mesecar AD; Center for Structural Genomics
of Infectious Diseases (CSGID): RCSB PDB-6W63: Structure of
COVID-19 main protease bound to potent broad-spectrum non-covalent
inhibitor X77. National Institutes of Health/National Institute of
Allergy and Infectious Diseases (NIH/NIAID); 2020
|
|
37
|
Nitulescu GM, Paunescu H, Moschos SA,
Petrakis D, Nitulescu G, Ion GND, Spandidos DA, Nikolouzakis TK,
Drakoulis N and Tsatsakis A: Comprehensive analysis of drugs to
treat SARS-CoV-2 infection: Mechanistic insights into current
COVID-19 therapies (Review). Int J Mol Med. 46:467–488. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC,
Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, et al: Structural basis
of inhibition specificities of 3C and 3C-like proteases by
zinc-coordinating and peptidomimetic compounds. J Biol Chem.
284:7646–7655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pormohammad A, Ghorbani S, Khatami A,
Farzi R, Baradaran B, Turner DL, Turner RJ, Bahr NC and Idrovo JP:
Comparison of confirmed COVID-19 with SARS and MERS cases-clinical
characteristics, laboratory findings, radiographic signs and
outcomes: A systematic review and meta-analysis. Rev Med Virol.
30:e21122020. View Article : Google Scholar
|
|
40
|
Pormohammad A, Ghorbani S, Khatami A,
Razizadeh MH, Alborzi E, Zarei M, Idrovo JP and Turner RJ:
Comparison of influenza type A and B with COVID-19: A global
systematic review and meta-analysis on clinical, laboratory and
radio-graphic findings. Rev Med Virol. Oct 9–2020.Epub ahead of
print. View Article : Google Scholar
|
|
41
|
Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi
Y, Sun R, Tian Z, Xu X and Wei H: Pathogenic T cells and
inflammatory monocytes incite inflammatory storm in severe COVID-19
patients. Natl Sci Rev. Mar 13–2020.Epub ahead of print. View Article : Google Scholar
|
|
42
|
Conti P, Ronconi G, Caraffa A, Gallenga C,
Ross R, Frydas I and Kritas S: Induction of pro-inflammatory
cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19
(COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J Biol Regul
Homeost Agents. 34:327–331. 2020.PubMed/NCBI
|
|
43
|
Shi Y, Wang Y, Shao C, Huang J, Gan J,
Huang X, Bucci E, Piacentini M, Ippolito G and Melino G: COVID-19
infection: The perspectives on immune responses. Cell Death Differ.
27:14512020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mehta P, McAuley DF, Brown M, Sanchez E,
Tattersall RS and Manson JJ; HLH Across Speciality Collaboration,
UK: COVID-19: Consider cytokine storm syndromes and
immuno-suppression. Lancet. 395:1033–1034. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Skalny AV, Rink L, Ajsuvakova OP, Aschner
M, Gritsenko VA, Alekseenko SI, Svistunov AA, Petrakis D, Spandidos
DA, Aaseth J, et al: Zinc and respiratory tract infections:
Perspectives for CoviD'19 (Review). Int J Mol Med. 46:17–26.
2020.PubMed/NCBI
|
|
46
|
Wessels I, Rolles B and Rink L: The
potential impact of zinc supplementation on COVID-19 pathogenesis.
Front Immunol. 11:17122020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Stebbing J, Phelan A, Griffin I, Tucker C,
Oechsle O, Smith D and Richardson P: COVID-19: Combining antiviral
and anti-inflammatory treatments. Lancet Infect Dis. 20:400–402.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Favalli EG, Ingegnoli F, De Lucia O,
Cincinelli G, Cimaz R and Caporali R: COVID-19 infection and
rheumatoid arthritis: Faraway, so close! Autoimmun Rev.
19:1025232020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang W, Zhao Y, Zhang F, Wang Q, Li T,
Liu Z, Wang J, Qin Y, Zhang X, Yan X, et al: The use of
anti-inflammatory drugs in the treatment of people with severe
coronavirus disease 2019 (COVID-19): The experience of clinical
immunologists from China. Clin Immunol. 214:1083932020. View Article : Google Scholar
|
|
50
|
Gammoh NZ and Rink L: Zinc in infection
and inflammation. Nutrients. 9:6242017. View Article : Google Scholar :
|
|
51
|
Jarosz M, Olbert M, Wyszogrodzka G,
Młyniec K and Librowski T: Antioxidant and anti-inflammatory
effects of zinc. Zinc-dependent NF-κB signaling.
Inflammopharmacology. 25:11–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Knoell DL, Smith DA, Sapkota M, Heires AJ,
Hanson CK, Smith LM, Poole JA, Wyatt TA and Romberger DJ:
Insufficient zinc intake enhances lung inflammation in response to
agricultural organic dust exposure. J Nutr Biochem. 70:56–64. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fischer KJ, Yajjala VK, Bansal S, Bauer C,
Chen R and Sun K: Monocytes represent one source of bacterial
shielding from antibiotics following influenza virus infection. J
Immunol. 202:2027–2034. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang L, Forst CV, Gordon A, Gussin G,
Geber AB, Fernandez PJ, Ding T, Lashua L, Wang M, Balmaseda A, et
al: Characterization of antibiotic resistance and host-microbiome
interactions in the human upper respiratory tract during influenza
infection. Microbiome. 8:392020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Llor C and Bjerrum L: Antimicrobial
resistance: Risk associated with antibiotic overuse and initiatives
to reduce the problem. Ther Adv Drug Saf. 5:229–241. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kostoff RN, Briggs MB, Porter AL,
Hernández AF, Abdollahi M, Aschner M and Tsatsakis A: The
under-reported role of toxic substance exposures in the COVID-19
pandemic. Food Chem Toxicol. 145:1116872020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Çaǧlayan Serin D, Pullukçu H, Çiçek C,
Sipahi OR, Taşbakan S, Atalay S and Pneumonia Study Group:
Bacterial and viral etiology in hospitalized community acquired
pneumonia with molecular methods and clinical evaluation. J Infect
Dev Ctries. 8:510–518. 2014. View Article : Google Scholar
|
|
58
|
Matson MJ, Stock F, Shupert WL, Bushmaker
T, Feldmann F, Bishop WB, Frank KM, Dekker JP, Chertow DS and
Munster VJ: Compatibility of maximum-containment virus-inactivation
protocols with identification of bacterial coinfections by
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. J Infect Dis. 218(Suppl 5): S297–S300. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Stevens MP, Patel PK and Nori P: Involving
antimicrobial stewardship programs in COVID-19 response efforts:
All hands on deck. Infect Control Hosp Epidemiol. 41:744–745. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Essack S, Bell J, Burgoyne DS, Duerden M
and Shephard A: Topical (local) antibiotics for respiratory
infections with sore throat: An antibiotic stewardship perspective.
J Clin Pharm Ther. 44:829–837. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sohrabi C, Alsafi Z, O'Neill N, Khan M,
Kerwan A, Al-Jabir A, Iosifidis C and Agha R: World Health
Organization declares global emergency: A review of the 2019 novel
coronavirus (COVID-19). Int J Surg. 76:71–76. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Denny KJ, De Wale J, Laupland KB, Harris
PNA and Lipman J: When not to start antibiotics: Avoiding
antibiotic overuse in the intensive care unit. Clin Microbiol
Infect. 26:35–40. 2020. View Article : Google Scholar
|
|
63
|
Song Z, Hu Y, Zheng S, Yang L and Zhao R:
Hospital pharmacists' pharmaceutical care for hospitalized patients
with COVID-19: Recommendations and guidance from clinical
experience. Res Social Adm Pharm. Apr 3–2020.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gupta S, Sakhuja A, Kumar G, McGrath E,
Nanchal RS and Kashani KB: Culture-negative severe sepsis:
Nationwide trends and outcomes. Chest. 150:1251–1259. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lemire JA, Harrison JJ and Turner RJ:
Antimicrobial activity of metals: Mechanisms, molecular targets and
applications. Nat Rev Microbiol. 11:371–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Turner RJ, Gugala N and Lemire J: Can
metals replace traditional antibiotics? Adjac Gov. November;46–47.
2016.
|
|
67
|
Lemire JA and Turner RJ: Mechanisms
underlying the anti-microbial capacity of metals. Stress and
Environmental Regulation of Gene Expression and Adaptation in
Bacteria. John Wiley & Sons, Inc; Hoboken, NJ: pp. 215–224.
2016, View Article : Google Scholar
|
|
68
|
Turner RJ: Metal-based antimicrobial
strategies. Microb Biotechnol. 10:1062–1065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Monych NK, Gugala N and Turner RJ: Chapter
9. Metal-based Antimicrobials. Antimicrobial Materials for
Biomedical Applications. Thomas Graham House; Cambridge: pp.
252–276. 2019, View Article : Google Scholar
|
|
70
|
Gugala N, Lemire JA and Turner RJ: The
efficacy of different anti-microbial metals at preventing the
formation of, and eradicating bacterial biofilms of pathogenic
indicator strains. J Antibiot (Tokyo). 70:775–780. 2017. View Article : Google Scholar
|
|
71
|
Jesline A, John NP, Narayanan PM, Vani C
and Murugan S: Antimicrobial activity of zinc and titanium dioxide
nanoparticles against biofilm-producing methicillin-resistant
Staphylococcus aureus. Appl Nanosci. 5:157–162. 2015. View Article : Google Scholar
|
|
72
|
Wang X, Du Y and Liu H: Preparation,
characterization and anti-microbial activity of chitosan-Zn
complex. Carbohydr Polym. 56:21–26. 2004. View Article : Google Scholar
|
|
73
|
Gugala N, Vu D, Parkins MD and Turner RJ:
Specificity in the susceptibilities of escherichia coli,
pseudomonas aeruginosa and Staphylococcus aureus clinical isolates
to six metal antimicrobials. Antibiotics (Basel). 8:512019.
View Article : Google Scholar
|
|
74
|
National Institutes of Health: Vitamin K -
Fact Sheet for Health Professionals. https://ods.od.nih.gov/factsheets/vita-minK-HealthProfessional/urisimplehttps://ods.od.nih.gov/factsheets/vita-minK-HealthProfessional/
Accessed June 3, 2020.
|
|
75
|
Plum LM, Rink L and Hajo H: The essential
toxin: Impact of zinc on human health. Int J Environ Res Public
Health. 7:1342–1365. 2010. View Article : Google Scholar : PubMed/NCBI
|