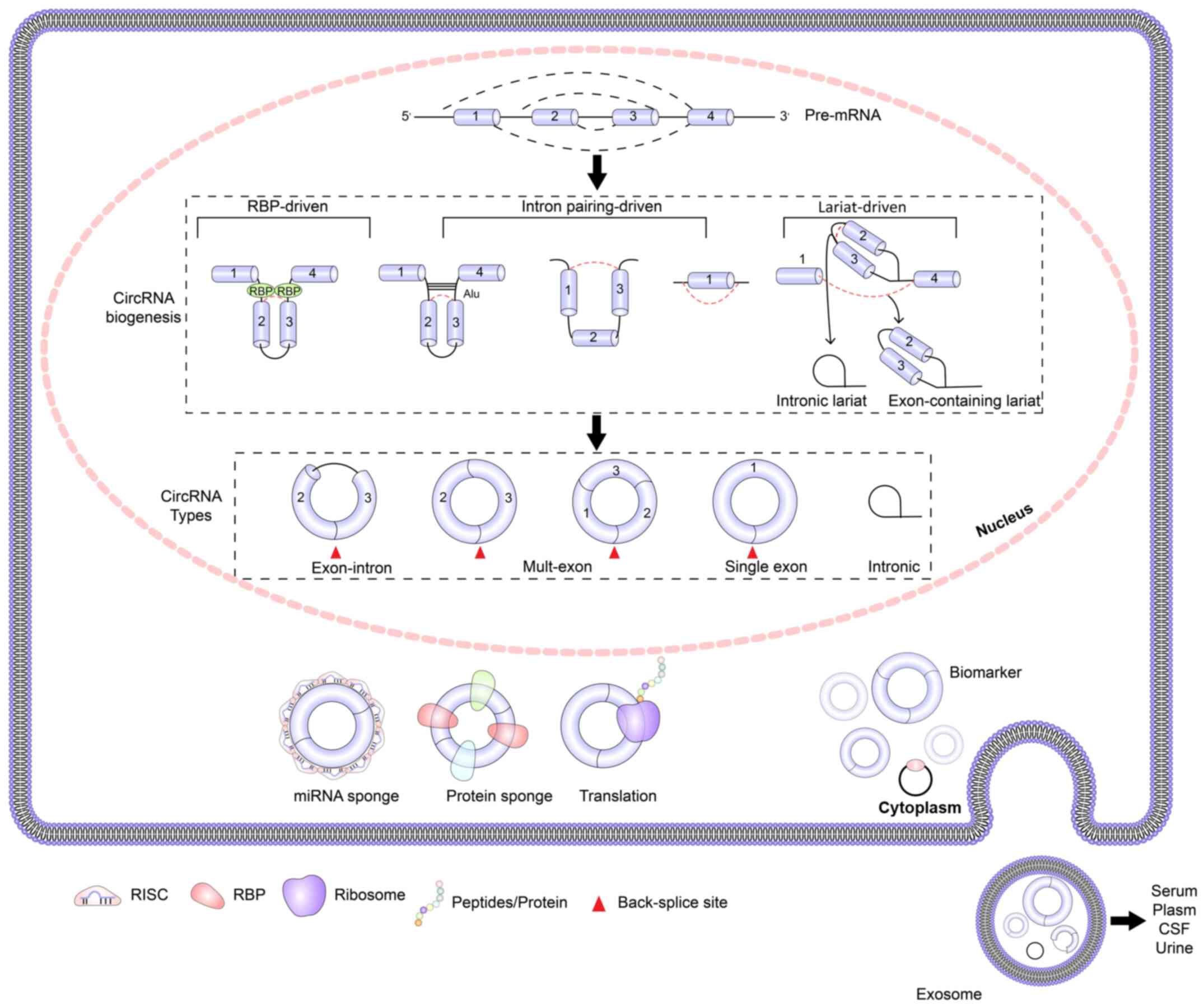
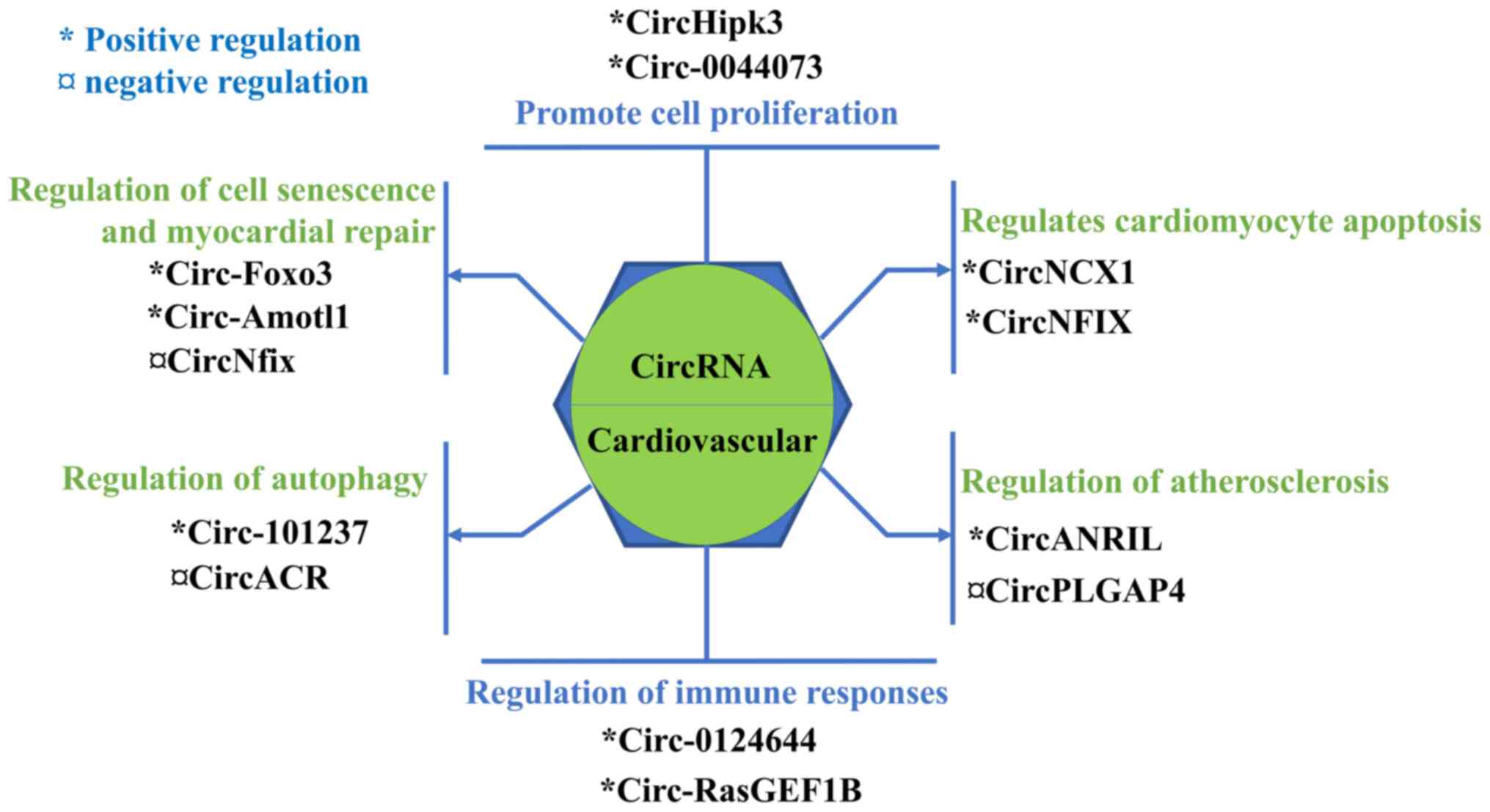
|
1
|
Pennisi E: Shining a light on the genome's
'dark matter'. Science. 330:16142010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15(Suppl 1): R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong R, Ma XK, Li GW and Yang L: CIRCpedia
v2: An updated database for comprehensive circular RNA annotation
and expression comparison. Genomics Proteomics Bioinformatics.
16:226–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar
|
|
12
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang F, Zhang R, Zhang X, Wu Y, Li X,
Zhang S, Hou W, Ding Y, Tian J, Sun L and Kong X: Comprehensive
analysis of circRNA expression pattern and circRNA-miRNA-mRNA
network in the pathogenesis of atherosclerosis in rabbits. Aging
(Albany NY). 10:2266–2283. 2018. View Article : Google Scholar
|
|
14
|
Santos-Ribeiro D, Godinas L, Pilette C and
Perros F: The inte-grated stress response system in cardiovascular
disease. Drug Discov Today. 23:920–929. 2018. View Article : Google Scholar
|
|
15
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
16
|
Pan T, Sun X, Liu Y, Li H, Deng G, Lin H
and Wang S: Heat stress alters genome-wide profiles of circular
RNAs in Arabidopsis. Plant Mol Biol. 96:217–229. 2018. View Article : Google Scholar
|
|
17
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu XD and Ares M Jr: Context-dependent
control of alternative splicing by RNA-binding proteins. Nat Rev
Genet. 15:689–701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar
|
|
20
|
Kramer MC, Liang D, Tatomer DC, Gold B,
March ZM, Cherry S and Wilusz JE: Combinatorial control of
drosophila circular RNA expression by intronic repeats, hnRNPs, and
SR proteins. Genes Dev. 29:2168–2182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Errichelli L, Dini Modigliani S, Laneve P,
Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R,
Peruzzi G, et al: FUS affects circular RNA expression in murine
embryonic stem cell-derived motor neurons. Nat Commun. 8:147412017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
24
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sela N, Mersch B, Gal-Mark N, Lev-Maor G,
Hotz-Wagenblatt A and Ast G: Comparative analysis of transposed
element insertion within human and mouse genomes reveals Alu's
unique role in shaping the human transcriptome. Genome Biol.
8:R1272007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ruskin B and Green MR: An RNA processing
activity that debranches RNA lariats. Science. 229:135–140. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar :
|
|
30
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar :
|
|
31
|
Talhouarne GJS and Gall JG: Lariat
intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad
Sci USA. 115:E7970–E7977. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Noto JJ, Schmidt CA and Matera AG:
Engineering and expressing circular RNAs via tRNA splicing. RNA
Biol. 14:978–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pasman Z, Been MD and Garcia-Blanco MA:
Exon circularization in mammalian nuclear extracts. RNA. 2:603–610.
1996.PubMed/NCBI
|
|
34
|
Lyu D and Huang S: The emerging role and
clinical implication of human exonic circular RNA. RNA Biol.
14:1000–1006. 2017. View Article : Google Scholar :
|
|
35
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC,
Dong JF, Tian Y, Yuan HJ and Zhang JN: Circular ribonucleic acid
expression alteration in exosomes from the brain extracellular
space after traumatic brain injury in mice. J Neurotrauma.
35:2056–2066. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu Q and Zhou T: EIciRNA-mediated gene
expression: Tunability and bimodality. FEBS Lett. 592:3460–3471.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Panda AC, De S, Grammatikakis I, Munk R,
Yang X, Piao Y, Dudekula DB, Abdelmohsen K and Gorospe M:
High-purity circular RNA isolation method (RPAD) reveals vast
collection of intronic circRNAs. Nucleic Acids Res. 45:e1162017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar
|
|
40
|
Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang
J, Cheng H, Yan J, Zhang S, Yang P and Zhao F: Expanded expression
landscape and prioritization of circular RNAs in mammals. Cell Rep.
26:3444–3460.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
anti-sense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kleaveland B, Shi CY, Stefano J and Bartel
DP: A network of noncoding regulatory RNAs acts in the mammalian
brain. Cell. 174:350–362.e17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The output of protein-coding genes
shifts to circular RNAs when the pre-mRNA processing machinery is
limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gupta SK, Garg A, Bär C, Chatterjee S,
Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J and Thum T:
Quaking inhibits doxorubicin-mediated cardiotoxicity through
regulation of cardiac circular RNA expression. Circ Res.
122:246–254. 2018. View Article : Google Scholar :
|
|
50
|
Rossi F, Legnini I, Megiorni F, Colantoni
A, Santini T, Morlando M, Di Timoteo G, Dattilo D, Dominici C and
Bozzoni I: Circ-ZNF609 regulates G1-S progression in
rhabdomyosarcoma. Oncogene. 38:3843–3854. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cardona-Monzonís A, García-Giménez JL,
Mena-Mollá S, Pareja-Galeano H, de la Guía-Galipienso F, Lippi G,
Pallardó FV and Sanchis-Gomar F: Non-coding RNAs and coronary
artery disease. Adv Exp Med Biol. 1229:273–285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Glažar P, Papavasileiou P and Rajewsky N:
CircBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar
|
|
55
|
Panda AC, Dudekula DB, Abdelmohsen K and
Gorospe M: Analysis of circular RNAs using the web tool
CircInteractome. Methods Mol Biol. 1724:43–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44(D1): D209–D215. 2016. View Article : Google Scholar :
|
|
57
|
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang
ZL, Zhou H, Yang JH and Qu LH: DeepBase v2.0: Identification,
expression, evolution and function of small RNAs, LncRNAs and
circular RNAs from deep-sequencing data. Nucleic Acids Res. 44(D1):
D196–D202. 2016. View Article : Google Scholar :
|
|
58
|
Liu M, Wang Q, Shen J, Yang BB and Ding X:
Circbank: A comprehensive database for circRNA with standard
nomenclature. RNA Biol. 16:899–905. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46(D1):
D925–D929. 2018. View Article : Google Scholar :
|
|
60
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database Issue): D92–D97. 2014. View Article : Google Scholar
|
|
62
|
Wang D: hppRNA-a snakemake-based handy
parameter-free pipeline for RNA-Seq analysis of numerous samples.
Brief Bioinform. 19:622–626. 2018.
|
|
63
|
Wu W, Ji P and Zhao F: CircAtlas: An
integrated resource of one million highly accurate circular RNAs
from 1070 vertebrate transcriptomes. Genome Biol. 21:1012020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang
T, Shan T and Peng Y: VirusCircBase: A database of virus circular
RNAs. Brief Bioinform. Apr 29–2020.Epub ahead of print. View Article : Google Scholar
|
|
65
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar
|
|
68
|
Chen DF, Zhang LJ, Tan K and Jing Q:
Application of droplet digital PCR in quantitative detection of the
cell-free circulating circRNAs. Biotechnol Biotechnol Equip.
32:116–123. 2018. View Article : Google Scholar
|
|
69
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kamens J: The addgene repository: An
international nonprofit plasmid and data resource. Nucleic Acids
Res. 43(Database Issue): D1152–D1157. 2015. View Article : Google Scholar :
|
|
71
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bramsen JB, Laursen MB, Nielsen AF, Hansen
TB, Bus C, Langkjaer N, Babu BR, Højland T, Abramov M, Van Aerschot
A, et al: A large-scale chemical modification screen identifies
design rules to generate siRNAs with high activity, high stability
and low toxicity. Nucleic Acids Res. 37:2867–2881. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
de Bruyns A, Geiling B and Dankort D:
Construction of modular lentiviral vectors for effective gene
expression and knockdown. Methods Mol Biol. 1448:3–21. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jakobi T, Czaja-Hasse LF, Reinhardt R and
Dieterich C: Profiling and validation of the circular RNA
repertoire in adult murine hearts. Genomics Proteomics
Bioinformatics. 14:216–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tan WL, Lim BT, Anene-Nzelu CG,
Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD,
et al: A landscape of circular RNA expression in the human heart.
Cardiovasc Res. 113:298–309. 2017.PubMed/NCBI
|
|
77
|
Lei W, Feng T, Fang X, Yu Y, Yang J, Zhao
ZA, Liu J, Shen Z, Deng W and Hu S: Signature of circular RNAs in
human induced pluripotent stem cells and derived cardiomyocytes.
Stem Cell Res Ther. 9:562018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li Y, Zhang J, Huo C, Ding N, Li J, Xiao
J, Lin X, Cai B, Zhang Y and Xu J: Dynamic organization of lncRNA
and circular RNA regulators collectively controlled cardiac
differentiation in humans. EBioMedicine. 24:137–146. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Stagsted LV, Nielsen KM, Daugaard I and
Hansen TB: Noncoding AUG circRNAs constitute an abundant and
conserved subclass of circles. Life Sci Alliance. 2:e2019003982019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Jakobi T, Siede D, Eschenbach J, Heumüller
AW, Busch M, Nietsch R, Meder B, Most P, Dimmeler S, Backs J, et
al: Deep characterization of circular RNAs from human
cardiovascular cell models and cardiac tissue. Cells. 9:16162020.
View Article : Google Scholar
|
|
81
|
van Heesch S, Witte F, Schneider-Lunitz V,
Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S,
Sandmann CL, et al: The translational landscape of the human heart.
Cell. 178:242–260.e29. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao
JN, Chen C, Yan KW, Ponnusamy M, Zhang YH and Li PF: Circular RNA
mediates cardiomyocyte death via miRNA-dependent upregulation of
MTP18 expression. Cell Death Differ. 24:1111–1120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_ circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zou TY, Li L, Huang JF, Yang B and Wang
LY: Expression and clinical significance of circular RNA circTCF25
in patients with coronary artery disease. Mol Cardiol China.
3:27–31. 2018.
|
|
85
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu X, Chen L, Wu S, Xu K, Jiang W, Qin M,
Zhang Y and Liu X: Integrative analysis reveals key circular RNA in
atrial fibrillation. Front Genet. 10:1082019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ludmer PL, Selwyn AP, Shook TL, Wayne RR,
Mudge GH, Alexander RW and Ganz P: Paradoxical vasoconstriction
induced by acetylcholine in atherosclerotic coronary arteries. N
Engl J Med. 315:1046–1051. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Simionescu M: Implications of early
structural-functional changes in the endothelium for vascular
disease. Arterioscler Thromb Vasc Biol. 27:266–274. 2007.
View Article : Google Scholar
|
|
89
|
Chen PY, Qin L, Baeyens N, Li G, Afolabi
T, Budatha M, Tellides G, Schwartz MA and Simons M:
Endothelial-to-mesenchymal transition drives atherosclerosis
progression. J Clin Invest. 125:4514–4528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu C, Yao MD, Li CP, Shan K, Yang H, Wang
JJ, Liu B, Li XM, Yao J, Jiang Q and Yan B: Silencing Of Circular
RNA-ZNF609 ameliorates vascular endothelial dysfunction.
Theranostics. 7:2863–2877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li CY, Ma L and Yu B: Circular RNA
hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells
proliferation and angiogenesis. Biomed Pharmacother. 95:1514–1519.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu
G and Yao H: Engagement of circular RNA HECW2 in the nonautophagic
role of ATG5 implicated in the endothelial-mesenchymal transition.
Autophagy. 14:404–418. 2018. View Article : Google Scholar :
|
|
93
|
Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen
X, Yao MD, Li XM, Yao J, Zhou RM, et al: Targeting
pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A
inhibition aggravates diabetes-induced microvascular dysfunction.
Proc Natl Acad Sci USA. 116:7455–7464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hall IF, Climent M, Quintavalle M, Farina
FM, Schorn T, Zani S, Carullo P, Kunderfranco P, Civilini E,
Condorelli G and Elia L: Circ_Lrp6, a Circular RNA enriched in
vascular smooth muscle cells, acts as a sponge regulating miRNA-145
function. Circ Res. 124:498–510. 2019. View Article : Google Scholar
|
|
95
|
Zheng C, Niu H, Li M, Zhang H, Yang Z,
Tian L, Wu Z, Li D and Chen X: Cyclic RNA has-circ-000595 regulates
apoptosis of aortic smooth muscle cells. Mol Med Rep. 12:6656–6662.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sun Y, Zhang S, Yue M, Li Y, Bi J and Liu
H: Angiotensin II inhibits apoptosis of mouse aortic smooth muscle
cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell
Death Dis. 10:3622019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen J, Cui L, Yuan J, Zhang Y and Sang H:
Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle
cells proliferation and migration by sponging miR-124. Biochem
Biophys Res Commun. 494:126–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang
ML, Zhao XS, Zhao HY, Suzuki T and Wen JK: A novel regulatory
mechanism of smooth muscle α-actin expression by
NRG-1/circACTA2/miR-548f-5p Axis. Circ Res. 121:628–635. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao
L and Wang L: Upregulation of circular RNA circNFIB attenuates
cardiac fibrosis by sponging miR-433. Front Genet. 10:5642019.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM
and Bie ZD: Circ_LAS1L regulates cardiac fibroblast activation,
growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem
Funct. 38:443–450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar
|
|
103
|
Shen L, Hu Y, Lou J, Yin S, Wang W, Wang
Y, Xia Y and Wu W: CircRNA-0044073 is upregulated in
atherosclerosis and increases the proliferation and invasion of
cells by targeting miR-107. Mol Med Rep. 19:3923–3932.
2019.PubMed/NCBI
|
|
104
|
Geng HH, Li R, Su YM, Xiao J, Pan M, Cai
XX and Ji XP: The circular RNA Cdr1as promotes myocardial
infarction by mediating the regulation of miR-7a on its target
genes expression. PLoS One. 11:e01517532016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gan J, Yuan J, Liu Y, Lu Z, Xue Y, Shi L
and Zeng H: Circular RNA_101237 mediates anoxia/reoxygenation
injury by targeting let-7a-5p/IGF2BP3 in cardiomyocytes. Int J Mol
Med. 45:451–460. 2020.PubMed/NCBI
|
|
107
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated circRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL,
Annadoray L, Sian S, Ackers-Johnson MA and Foo RS: Targeting the
highly abundant circular RNA circSlc8a1 in cardiomyocytes
attenuates pressure overload induced hypertrophy. Cardiovasc Res.
115:1998–2007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhang S, Song G, Yuan J, Qiao S, Xu S, Si
Z, Yang Y, Xu X and Wang A: Circular RNA circ_0003204 inhibits
proliferation, migration and tube formation of endothelial cell in
atherosclerosis via miR-370-3p/TGFβR2/phosph-SMAD3 axis. J Biomed
Sci. 27:112020. View Article : Google Scholar
|
|
110
|
Altesha MA, Ni T, Khan A, Liu K and Zheng
X: Circular RNA in cardiovascular disease. J Cell Physiol.
234:5588–5600. 2019. View Article : Google Scholar
|
|
111
|
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y,
Shen G and Wang F: Inhibition of circHIPK3 prevents angiotensin
II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol.
292:188–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wang Y, Zhao R, Liu W, Wang Z, Rong J,
Long X, Liu Z, Ge J and Shi B: Exosomal circHIPK3 released from
hypoxia-pretreated cardiomyocytes regulates oxidative damage in
cardiac micro-vascular endothelial cells via the miR-29a/IGF-1
pathway. Oxid Med Cell Longev. 2019:79546572019. View Article : Google Scholar
|
|
113
|
Li M, Ding W, Tariq MA, Chang W, Zhang X,
Xu W, Hou L, Wang Y and Wang J: A circular transcript of ncx1 gene
mediates ischemic myocardial injury by targeting miR-133a-3p.
Theranostics. 8:5855–5869. 2018. View Article : Google Scholar
|
|
114
|
Jin Q and Chen Y: Silencing circular RNA
circ_0010729 protects human cardiomyocytes from oxygen-glucose
deprivation-induced injury by up-regulating microRNA-145-5p. Mol
Cell Biochem. 462:185–194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y,
Xu J, Chen S and Liu S: Circular RNA Ttc3 regulates cardiac
function after myocardial infarction by sponging miR-15b. J Mol
Cell Cardiol. 130:10–22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Holdt LM, Beutner F, Scholz M, Gielen S,
Gäbel G, Bergert H, Schuler G, Thiery J and Teupser D: ANRIL
expression is associated with atherosclerosis risk at chromosome
9p21. Arterioscler Thromb Vasc Biol. 30:620–627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Song CL, Wang JP, Xue X, Liu N, Zhang XH,
Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y and Yang YB: Effect of
Circular ANRIL on the inflammatory response of vascular endothelial
cells in a rat model of coronary atherosclerosis. Cell Physiol
Biochem. 42:1202–1212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Menglan LI, Siying HE, Jialing R, Bin L,
Xiaokang Z and Fang Z: The possible protective role of circDLGAP4
from peripheral blood in coronary heart disease. Chin J Clin Lab
Sci. 37:109–112. 2019.
|
|
119
|
Parahuleva MS, Euler G, Mardini A, Parviz
B, Schieffer B, Schulz R and Aslam M: Identification of microRNAs
as potential cellular monocytic biomarkers in the early phase of
myocardial infarction: A pilot study. Sci Rep. 7:159742017.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sala F, Aranda JF, Rotllan N, Ramírez CM,
Aryal B, Elia L, Condorelli G, Catapano AL, Fernández-Hernando C
and Norata GD: MiR-143/145 deficiency attenuates the progression of
atherosclerosis in Ldlr-/-mice. Thromb Haemost. 112:796–802. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo
SK, Xue W, Cui Y, Dong K, Ding H, et al: Structure and degradation
of circular RNAs regulate PKR activation in innate immunity. Cell.
177:865–880.e21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhang Y, Zhang Y, Li X, Zhang M and Lv K:
Microarray analysis of circular RNA expression patterns in
polarized macrophages. Int J Mol Med. 39:373–379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY
and Ea CK: Inducible RasGEF1B circular RNA is a positive regulator
of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 13:861–871. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen YG, Kim MV, Chen X, Batista PJ,
Aoyama S, Wilusz JE, Iwasaki A and Chang HY: Sensing self and
foreign circular RNAs by intron identity. Mol Cell. 67:228–238.e5.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Valentim L, Laurence KM, Townsend PA,
Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS and
Stephanou A: Urocortin inhibits Beclin1-mediated autophagic cell
death in cardiac myocytes exposed to ischaemia/reperfusion injury.
J Mol Cell Cardiol. 40:846–852. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang
YH, Zhang RC, Liu CY, Dong YH, Wang M, et al: The circular RNA ACR
attenuates myocardial ischemia/reperfusion injury by suppressing
autophagy via modulation of the Pink1/FAM65B pathway. Cell Death
Differ. 26:1299–1315. 2019. View Article : Google Scholar
|
|
129
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A circular RNA binds to
and activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Rep. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar
|
|
134
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extra-cellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar
|
|
135
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Preußer C, Hung LH, Schneider T, Schreiner
S, Hardt M, Moebus A, Santoso S and Bindereif A: Selective release
of circRNAs in platelet-derived extracellular vesicles. J Extracell
Vesicles. 7:14244732018. View Article : Google Scholar
|
|
137
|
Viereck J and Thum T: Circulating
noncoding RNAs as biomarkers of cardiovascular disease and injury.
Circ Res. 120:381–399. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
142
|
Han Y, Donovan J, Rath S, Whitney G,
Chitrakar A and Korennykh A: Structure of human RNase L reveals the
basis for regulated RNA decay in the IFN response. Science.
343:1244–1248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic cleavage of
m6A-containing RNAs by RNase P/MRP complex. Mol Cell.
74:494–507.e8. 2019. View Article : Google Scholar
|
|
144
|
Tapsin S, Sun M, Shen Y, Zhang H, Lim XN,
Susanto TT, Yang SL, Zeng GS, Lee J, Lezhava A, et al: Genome-wide
identification of natural RNA aptamers in prokaryotes and
eukaryotes. Nat Commun. 9:12892018. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar :
|
|
146
|
Costello A, Lao NT, Barron N and Clynes M:
Continuous translation of circularized mRNA improves recombinant
protein titer. Metab Eng. 52:284–292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Xie YZ, Yang F, Tan W, Li X, Jiao C, Huang
R and Yang BB: The anti-cancer components of Ganoderma lucidum
possesses cardiovascular protective effect by regulating circular
RNA expression. Oncoscience. 3:203–207. 2016. View Article : Google Scholar :
|
|
148
|
Dang RY, Liu FL and Li Y: Circular RNA
hsa_circ_0010729 regulates vascular endothelial cell proliferation
and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys
Res Commun. 490:104–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhang Y, Yu J, Kahkoska AR and Gu Z:
Photoacoustic drug delivery. Sensors (Basel). 17. pp. 14002017,
View Article : Google Scholar
|
|
151
|
Lavenniah A, Luu TDA, Li YP, Lim TB, Jiang
J, Ackers-Johnson M and Foo RS: Engineered circular RNA sponges act
as miRNA inhibitors to attenuate pressure overload-induced cardiac
hyper-trophy. Mol Ther. 28:1506–1517. 2020. View Article : Google Scholar : PubMed/NCBI
|