It is estimated that only 20% of the nucleic acids
in the human genome are encoded into proteins, while the rest, the
so called non-coding nucleic acids, are considered as the 'dark
matter' of the genome (1).
However, several studies have shown that these non-coding RNAs are
involved in important biological processes, such as cell
proliferation, differentiation, apoptosis, metabolism, senescence,
and especially in post-transcriptional regulation (1-3).
Circular RNAs (circRNAs) were first discovered in the early 1970s
(4,5), however, they have been poorly
studied for nearly three decades due to the limited available
technology. Recent advances in RNA sequencing technologies have
promoted the identification of an increasing number of circRNAs
(3,6-8).
Several studies have shown that circRNAs can compete with mRNAs for
the target binding sites on microRNAs (miRNAs) to affect gene
expression. However, the function of the majority of circRNAs
remains unclear. In 2013, Hansen et al (9) reported the overlapping co-expression
of circRNA ciRS-7 and miR-7 in murine brain tissues. In addition,
it was also demonstrated that the circRNA sex-determining region
Y-9 (SRY-9) could act as a sponge for miR-138, thus providing the
first solid evidence for the biological roles of circRNAs. To date,
>183,000 circRNAs have been identified from human transcripts,
to the best of our knowledge (10). The role of circRNAs in different
tissues and different diseases have also been gradually revealed
(11). The cardiovascular system
is considered as one of the two major vascular systems in the human
body. Cardiovascular diseases (CVDs) remain the leading cause of
death worldwide. A study found that the circRNAs solute carrier
family 8 member A1 (SLC8A1), ataxin 10 (ATXN10), SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin subfamily
A member 5 (SMARCA5), chromodomain Y (CDY) and myoblast
determination protein 1 (MyoD) are associated with myocardial
differentiation (12). In
addition, several circRNAs are involved in the pathological process
of atherosclerosis (AS) (13).
Furthermore, stress stimuli such as hypoxia and high temperature
have been associated with the expression of cardiovascular-related
factors (14-16). Emerging evidence has shown that
circRNAs are involved in the progression of CVD, and are therefore
considered promising targets for diagnosis and treatment. The
present review article aimed to summarize the current knowledge on
the biological aspects of circRNAs and highlight the current
research progress on the mechanisms underlying the function of
circRNAs in the physiology and pathology of the cardiovascular
system.
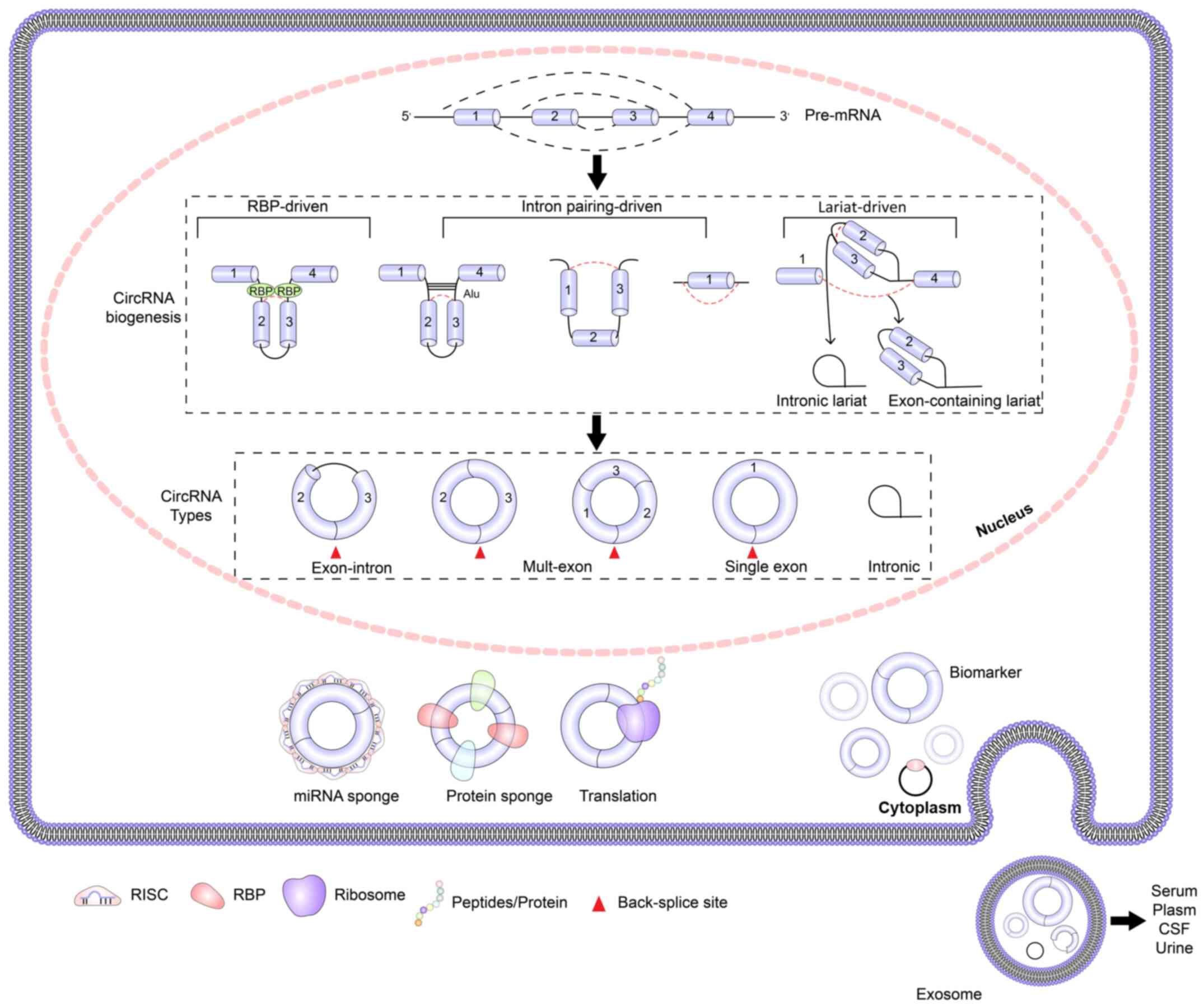
It is considered that circRNAs are derived from the
variable splicing of precursor mRNAs (pre-mRNA), mediated by RNA
polymerase II (17-19). However, to the best of our
knowledge, the mechanism underlying the formation of circRNAs
remains to be elucidated.
CircRNAs may also be formed by complementary pairing
of flanking introns located in dorsal exons, such as short
interspersed nuclear elements or non-repetitive complementary
sequences (23,24). Among them, Alu repetitive elements
are widespread in the expressed RNA and in the non-coding parts
(25,26). Alu repeats serve a key role in the
formation of circRNAs, with >1 million copies in the genome
(24). It is considered that the
probability and intensity of the reverse intron folding, mediated
by the repeats encompassed in the pre-mRNA, may also determine the
frequency of reverse splicing (25,27).
Lariat-driven circularization is another important
process of circRNA formation (28). Exon skipping occurs when the
pre-mRNA is spliced, resulting in a lariat intermediate containing
exons and introns. Subsequently, the internal splicing of the
lariat leads to the removal or retention of introns, thus resulting
in the formation of exon or exon-intron circRNAs (29-32).
Based on their biogenesis, circRNAs are divided into
exon circRNAs, encompassing a single exon or multiple exons,
exon-intron circRNAs, and intron circRNAs (Fig. 1). The majority of circRNAs belong
to the first subtype and mainly exist in the cytoplasm (33). However, a few circRNAs are
produced in the nucleus and can be secreted into the body fluids,
such as the serum, saliva and cerebrospinal fluid (34-36). The exon-intron circRNAs are mainly
located in the nucleus and interact with RNA polymerase II to
promote the transcription of their parental genes (37). Finally, the intron circRNAs exist
in the nucleus (38).
Gradually, circRNAs have been found to exert more
specific biological functions, and their mechanism of action
remains in the focus of attention (39,40). To date, several functions of
circRNAs have been verified (Fig.
1).
Apart from a small number of circRNAs generated by
intron circularization, which are located in the nucleus, the
majority are mainly located in the cytoplasm (41). Several studies have shown that
circRNAs can compete with mRNAs for binding to miRNAs located in
the cytoplasm, thereby regulating the expression of mRNAs (3,9).
ciRS-7/cerebellar degeneration-related protein 1 antisense RNA
(CDR1as) sponging miR-7 and mouse SRY sponging miR-138 are
considered as the most representative circRNAs supporting this
biological function (3). Notably,
only a few circRNAs have been identified to encompass multiple
target binding sites for miRNAs (42). Furthermore, the binding of
circRNAs by miRNAs may initiate circRNA decay (43). It has been also demonstrated that
the CDR1as-miR-7 axis is modulated by Cyrano, a long non-coding RNA
(lncRNA) (44). In the non-coding
regulatory network, miR-671 shares a highly complementary sequence
with CDR1as, which triggers the rare argonaute2-mediated RNA
cleavage targeted by miRNAs in mammals and even vertebrates
(43). The aforementioned
findings indicate that circRNA-miRNA interactions not only mediate
miRNA sequestration, but also exert other functions that are worth
studying.
CircRNAs are formed as a result of the pre-mRNA
atypical splicing, whereas mRNA is the final product of the
pre-mRNA typical linear splicing (45). A study revealed that improving the
efficiency of typical linear splicing could lead to a significant
reduction in the number of generated circRNAs (17). When the length of the intron
flanking the exon is longer, the efficiency of typical linear
splicing is significantly decreased, and cyclization occurs
(46). These findings indicate
that circRNAs can compete with pre-mRNA during transcription.
CircRNAs located in the nucleus can bind RNA
polymerase II or U1 small nuclear ribonucleoprotein to regulate the
transcription of their parental genes (47,48). A study has shown that the
expression of the parental genes is significantly attenuated when
circRNAs are knocked out (47).
Furthermore, back-splicing was observed in RNAs transcribed from
centromeric retrotransposons in maize, while the resulting circular
CRM1 RNAs could bind to maize centromeres through R-loops to
promote the formation of chromatin loops. Previous studies have
demonstrated that QKI attenuated doxorubicin (DOX)-induced
cardiotoxicity via binding to titin (Ttn)-, formin homology 2
domain containing 3 (Fhod3)- and striatin 3 (Strn3)-derived
circRNAs expressed in the heart (49). In addition, circRNAs contain
binding sites for the RBPs of the host and can regulate their
expression. For example, circRNA zinc finger protein 609 (ZNF609)
can regulate the proportion of phosphorylated (p)-Rb/Rb and the
levels of p-protein kinase B (Akt), thus affecting the G1/S phase
progression in cells (50).
Several laboratories and institutions have developed
specialized databases and analytical tools to study circRNAs
(10,54-65) (Table
I). These databases provide the convenience to further
investigate the already identified circRNAs. RNA sequencing
analysis of rRNA-depleted total RNAs and microarrays are currently
applied for the discovery of novel circRNAs (66). Nevertheless, the verification of
the predicted circRNAs by in vitro/in vivo
experiments is always necessary. Several widely used molecular
biology techniques have been applied to detect and verify circRNAs,
including northern blot analysis, which is considered as the gold
standard for circRNAs verification (3,30),
reverse transcription (RT)-PCR, which is the most commonly used and
basic verification technique (17), quantitative PCR which is used to
identify and quantitatively measure differentially expressed
circRNAs, droplet digital PCR, which is performed to quantify the
expression of circRNAs in challenging samples such as plasma
(67,68), and fluorescence in situ
hybridization, which is used to detect circRNA localization and
interactions. Furthermore, overexpression and silencing techniques
are performed to investigate the role of circRNAs in vitro
or in vivo. For example, for the overexpression experiments,
following amplification of a target DNA sequence encompassing the
characteristic flanking Alu sequences, the sequence is digested
with a restriction endonuclease and ligated to the pEGFP-C1 vector.
Subsequently, the overexpression plasmid is transfected into the
corresponding cell sample, thus resulting in the overexpression of
the circRNAs (69,70). Additionally, for silencing
experiments, small interfering RNA (siRNA) sequences complementary
to the back-splice junction are synthesized and the RNA-induced
silencing complex specifically targets circRNA, leading to its
cleavage and degradation, thus silencing its expression (52,71). Notably, the effectiveness of
siRNAs depends on transfection efficiency, and these RNAs only
temporarily inhibit the expression of their corresponding targets
(72). However, short hairpin
RNAs (shRNAs) or Ago shRNAs exert fewer off-target effects, thus
resulting in more stable target-circRNA knockdown (73). The progress on studies concerning
the effects of circRNAs will promote the merging of specific
versions of databases to expand research in the field of
circRNAs.
Several studies have reported the comprehensive
expression profiles of circRNAs in rat, murine and human hearts
(74-76), including the first study on the
development and expression of circRNAs in human induced pluripotent
stem cell-derived cardiomyocytes (76). A study reported for the first time
a catalog of 575 candidate circRNAs in murine hearts. Among them,
several candidate genes were found to be associated with different
diseases (74). Furthermore,
another study extensively compared human, rat and murine circRNA
conservation in a specific tissue (75). Only 10% of these circRNAs were
conserved among the three species. In addition, this study
demonstrated that the overall circRNA expression was significantly
decreased in the heart of adult animals compared with neonatal
rats. The expression of circRNAs in the human heart was
approximately twice the baseline level of adult rats and mice. A
study analyzed the characteristics of circRNA expression in 12
human hearts, 25 mouse hearts and across a 28-day differentiation
time-course from human embryonic stem cell-derived cardiomyocytes
(76). A total of 15,318 and
3,017 circRNAs were identified in human and murine myocardial
tissue, respectively. The expression abundance of these circRNAs
was basically associated with their cognate linear RNA. The
parental genes of circRNAs with the highest content were also
considered as key cardiac genes, such as ryanodine receptor 2
(Ryr2), Ttn and dystrophin. Among them, circRNA SLC8A1-1 was the
most abundantly expressed circRNA in the myocardium, whereas 402
different circRNA isoforms were expressed from the Ttn gene locus.
These circRNAs were dynamically and highly expressed during heart
development. In addition, it has been reported that the circRNAs
SLC8A1, circ-calcium voltage-gated channel subunit α1 D, SPHKAP and
α kinase 2 are also specifically expressed in the heart (77). Another study showed that the
circRNAs ATXN10, SMARCA5, CDY, MYOD, SLC8A1, ATXN7 and PHF2 were
involved in cardiomyocyte differentiation (12). Furthermore, a notable study
investigated the changes in the expression of lncRNAs, circRNAs and
protein-coding genes via recording the RNA sequencing data at
sequential stages of cardiomyocyte differentiation (78). The stages of differentiation
studied were the following: Undifferentiated cardiomyocytes,
mesoderm cardiomyocytes, cardiac progenitors and differentiated
cardiomyocytes. This study described the dynamic expression pattern
of non-coding RNAs during cardiogenesis, where the expression of
circRNA SLC8A1 was gradually upregulated during cardiac
differentiation. CircRNAs are derived from exons containing
annotated start codons, which are also termed AUG circRNAs
(79). Jakobi et al
(80) described the AUG circRNAs
in a heart model system for the first time, to the best of our
knowledge. The authors demonstrated that m6A methylation was
enriched in AUG circRNAs. They also reported the potential negative
regulatory effect of AUG circRNAs on the translation of host genes.
A recent study demonstrated that 40 ribosome-associated cardiac
circRNAs could be translated into microproteins with different
biological functions (81). Among
them, the circRNAs CASP8 and FADD like apoptosis regulator, SLC8A1,
myosin binding protein C3 and Ryr2 were reported for the first
time. Therefore, the research on circRNAs will open a new chapter
in heart biology.
It is understood that cardiomyocytes play an
important role in the cardiovascular system (75,82). It has been reported that several
circRNAs, such as hsa_circ_0001879, hsa_circ_0004104 and
hsa_circTCF25, can be isolated from peripheral blood mononuclear
cells, serving as biomarkers of coronary artery disease (CAD)
(83,84). Another study revealed that the
expression of circANRIL subtypes were inversely correlated with the
risk and the severity of AS (85). Furthermore, a recent study
documented for the first time the association between the
expression of circRNAs and atrial fibrillation (86). Therefore, a total of five circRNAs
were identified with notable biological significance, including
chr9:15474007-15490122, chr16:75445723-75448593, hsa_circ_0007256,
chr12:56563313-56563992 and hsa_ circ_0003533. Regarding vascular
endothelial cells, circRNAs may directly or indirectly affect the
occurrence of CVDs through functional changes, such as endothelial
cell apoptosis, endothelial-mesenchymal transition and angiogenesis
(87-90). For example, in vitro
results demonstrated that the expression of circRNA ZNF292 in human
umbilical vein endothelial cells (HUVECs) was significantly
upregulated, suggesting that and circRNA ZNF292 could exert
pro-angiogenic effects (71). Li
et al (91) reported that
the expression of hsa_circ_0003575 and hsa_circ_0003204 were
significantly increased in oxidized low-density lipoprotein
(ox-LDL)-induced HUVECs. Furthermore, hsa_circ_0003575 silencing
significantly inhibited apoptosis and promoted proliferation and
angiogenesis of ox-LDL-induced HUVECs. Yang et al (92) found that the circRNA HECT, C2 and
WW domain containing E3 ubiquitin protein ligase 2 induced the
expression of autophagy-related 5 (ATG5) in methamphetamine- or
lipopolysaccharide (LPS)-induced human brain microvascular
endothelial cells via binding to miR-30d, and ATG5 was shown to
affect the endothelial-mesenchymal transition in an
autophagy-independent manner. Liu et al (93) revealed the mechanism underlying
the microvascular dysfunction mediated by diabetes. This mechanism
also mediates the biological changes in human retinal vascular
endothelial cells following circRNA PWWP domain-containing protein
2A transfer from vascular pericytes to HUVECs through exosomes
(93). Vascular smooth muscle
cells (VSMCs) are another notable type of cardiovascular cells. A
recent study reported that the circRNA low-density lipoprotein
receptor-related protein 6 (Lrp6) can promote the phenotypic
transformation of VSMCs via inhibiting miR-145, while silencing of
circRNA Lrp6, using shRNA technology, attenuated intimal
hyperplasia in a mouse carotid artery injury model (94). Additionally, Zheng et al
(95) demonstrated that
hsa_circ_000595 is upregulated in human aortic aneurysm patients
and hypoxic VSMCs. In addition, hsa_circ_000595 can directly target
cyclooxygenase 2, hypoxia-inducible factor 1α (HIF-1α) and nuclear
factor-κB to promote its biological function in regulating cell
apoptosis via targeting miR-19a. By contrast, hsa-circ-000595
knockdown significantly attenuated apoptosis (95). Sun et al (96) reported that angiotensin II
inhibits apoptosis of mouse aortic smooth muscle cells through
regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Several circRNAs
have been identified to regulate the phenotypic transformation of
VSMCs, including WD repeat domain 77, diaphanous related formin 3
and actin α 2 (97,98). Regarding cardiomyocytes and
cardiac fibroblasts, Gupta et al (49) demonstrated that QKI could inhibit
DOX-induced cardiotoxicity via regulating the expression of the
circRNAs Ttn, Fhod3 and Strn3. Furthermore, QKI could regulate
circRNA expression to protect cardiomyocytes against
adriamycin-induced injury in primary rat, H9C2 and HL-1
cardiomyocytes in vitro. CircRNAs are also considered as
potential targets for treating cardiac fibrosis (99-101). Therefore, a study showed that
the expression of circRNA_000203 of the myosin I9A transcripts was
increased in the myocardium of diabetic mice and in angiotensin
II-induced cardiac fibroblasts (102). It has been also revealed that
the circRNA_000203/miR-26b-5p/collagen type I α 2 and connective
tissue growth factor axes regulate the anti-fibrotic effects of
cardiac fibroblasts (102). Wang
et al (82) demonstrated
that the mitochondrial fission and apoptosis-related circRNA
(MFACR) could mediate cardiomyocyte mitochondrial fission and
apoptosis via regulating the MFACR/miR-652-3p/mitochondrial
trifunctional protein 18 axis. These findings provided novel
knowledge for understanding the molecular events associated with
mitochondrial fission. The circRNAs involved in the cardiovascular
system are listed in Table
II.
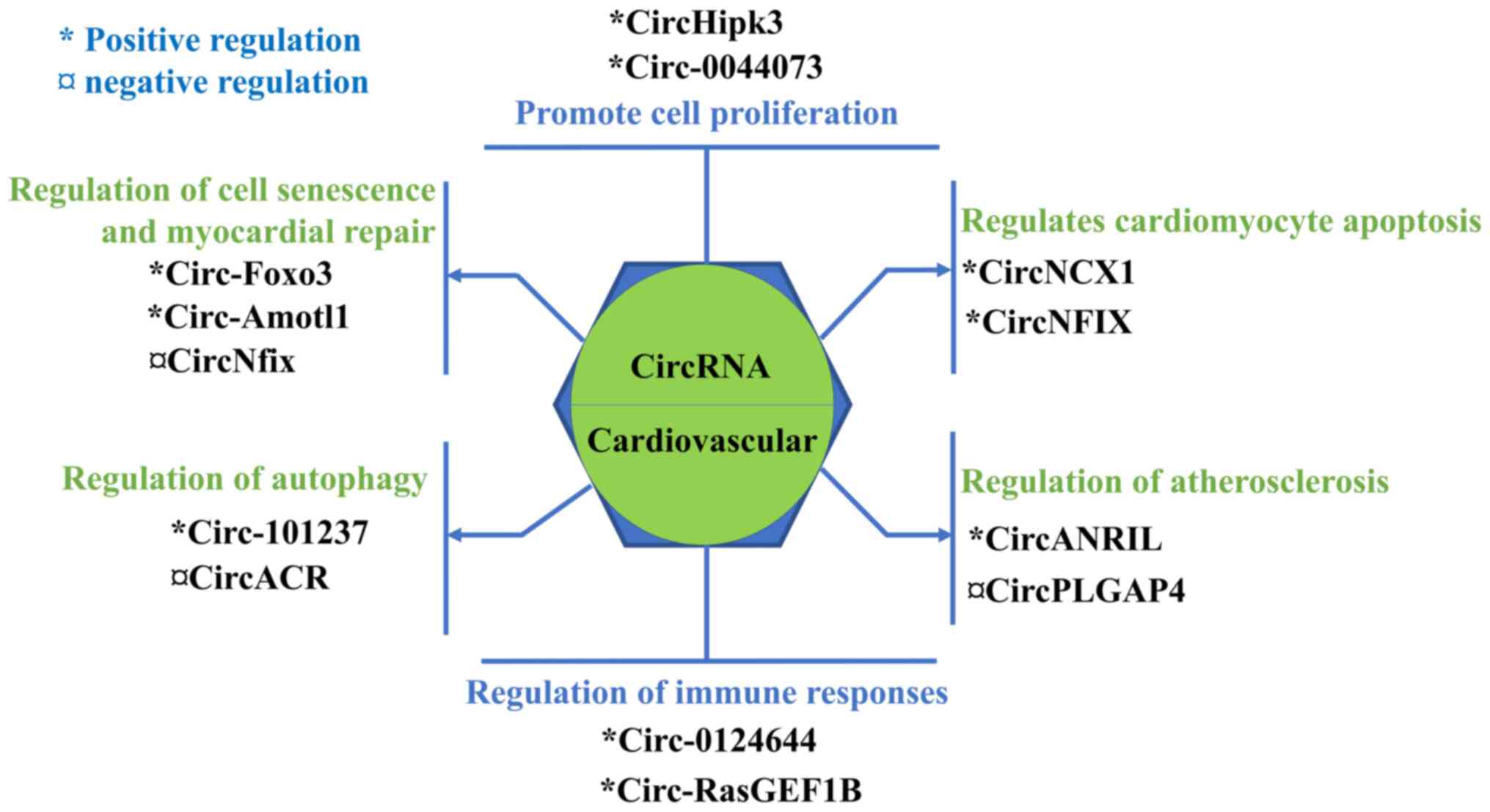
It is understood that circRNAs are involved in
numerous pathological mechanisms underlying CVDs, such as AS,
immune response, apoptosis, cell proliferation, autophagy, cell
senescence and myocardial repair (103-109). Emerging evidence has suggested
that circRNAs play an important role in the occurrence and
development of CVD (110). The
documented regulatory mechanisms of circRNAs on CVDs are listed in
Fig. 2.
The proliferation of cells involved in the
cardiovascular system plays an important role in the development of
CVDs, such as hypertension, AS and restenosis (110). CircRNAs serve an important role
in the development of CVDs (103,111). Shen et al (103) demonstrated that circRNA-0044073
is highly expressed in patients with AS. In addition, it was shown
that circRNA-0044073 promotes the proliferation and invasion of
human umbilical vein smooth muscle cells and HUVECs, and
accelerates the development of AS via targeting miR-107 and
activating the Janus kinase/signal transducer and activator of
transcription. Wang et al (112) found that the circRNA homeodomain
inter-acting protein kinase 3 could act as a sponge to inhibit
miR-29a activity, resulting in increased insulin-like growth factor
1 expression, which in turn could attenuate the dysfunction of
oxidative stress-induced cerebral microvascular endothelial cells
(CMVECs). Additionally, the circRNA HIPK3 promoted the
proliferation and survival of CMVECs under hypoxic conditions.
Another study demonstrated that HIPK3 could sponge miR-29b-3p to
regulate the proliferation, migration and development of cardiac
fibroblasts in angiotensin II-induced myocardial fibrosis (111).
Cardiomyocyte apoptosis is an important factor
affecting the occurrence and development of CVDs (104,113-115). Geng et al (104) investigated the effect of the
co-expression of circ-Cdr1as with miR-7a in mouse myocardial
infarction injury model. Cdr1as over-expression upregulated the
expression of poly (ADP-ribose) polymerase and specific protein 1
to promote cardiomyocyte apoptosis, but was then reversed by miR-7a
overexpression. Li et al (113) suggested that reactive oxygen
species could upregulate circ-NCX1, which in turn could promote
cardiomyocyte apoptosis. Mechanistically, circ-NCX1 could
competitively bind to miR-133a-3p to increase the expression levels
of the pro-apoptotic protein gene cell death-inducing protein 1
(CDIP1). Therefore, circNCX1 knockdown reduced the expression of
CDIP1 in murine cardiomyocytes and heart tissues, thereby improving
the condition of myocardial ischemia-reperfusion injury. Several
other circRNAs have been identified to play a role in promoting
cardiomyocyte apoptosis, including circ-MFACR and circRNA_0010729
(82,114).
As early as 2010, a study revealed that the INK4/ADP
ribosylation factor (ARF) gene locus is adjacent to the chromosome
9p21, which is associated with CAD (116). Circ-ANRIL, the antisense
transcript of the INK4/ARF gene, was shown to be regulated by a
single nucleotide polymorphism in the 9p21 chromosome and is
associated with the risk of AS. Song et al (117) demonstrated that circ-ANRIL
downregulation could prevent coronary AS via attenuating vascular
endothelial cell apoptosis and the expression of inflammatory
factors. However, another study suggested that circ-ANRIL exerts a
protective effect against AS (85). The proportion of circ-ANRIL/lncRNA
ANRIL in patients with coronary heart disease is higher, and
patients with increased expression of circ-ANRIL rarely develop
coronary heart disease. Furthermore, circ-ANRIL could induce
nucleolar stress and p53 activation in VSMCs and macrophages, thus
inducing and inhibiting apoptosis and proliferation, respectively.
Therefore, the role of circ-ANRIL in regulating AS should be
further investigated. In addition, a study revealed that circ-DLG
associated protein 4 (DLGAP4) is negatively associated with the
risk of coronary heart disease (118). Therefore, it was demonstrated
that circ-DLGAP4 was down-regulated in ox-LDL-induced foam cells in
a time-dependent manner. In addition, circ-DLGAP4 could sponge
endogenous miR-143 to reduce the size of the atherosclerotic plaque
and macrophage infiltration in mice (119,120). This finding indicated that
circ-DLGAP4 could be considered as a protective factor against
monocyte-macrophage foaming, and as a potential inhibitory factor
of AS plaque forming.
CircRNAs are involved in autophagy by regulating the
transcription and post-transcriptional modifications of
autophagy-related genes (126).
Cell autophagy is considered as a double-edged sword in health and
disease, while it exerts protective effects on restoring
homeostasis, it has also been reported that excessive autophagy
mediates cardiomyocyte death (127). Zhou et al (128) demonstrated that circacrosin
(ACR) attenuates autophagy and cell death in cardiomyocytes. In
addition, ACR protects against ischemia-reperfusion injury and
reduces myocardial infarct sizes in a myocardial
ischemia/reperfusion injury model. The same study revealed that
circ-ACR could induce the expression of putative kinase 1 (PINK-1)
via directly binding to DNA meth-yltransferase 3B (Dnmt3b), thus
blocking the Dnmt3b-mediated DNA methylation of the PINK-1
promoter. Furthermore, PINK-1 could phosphorylate its downstream
target, Rho family-interacting cell polarization regulator 2 on
serine 46 and inhibit cardiac autophagy and cell death. Another
study on autophagy demonstrated that circRNA_101237 acts as a
sponge of let-7a-5p to regulate insulin like growth factor 2 mRNA
binding protein 3 (IGF2BP3)-dependent autophagy (106). Therefore, circRNA_101237
downregulation decreases the expression of IGF2BP3, which in turn
attenuates the apoptosis of primary cardiomyocytes and inhibits
hypoxia/reoxygenation-induced autophagy. The aforementioned studies
indicated that autophagy could serve an important role in the
regulation of CVDs.
The senescence and repair of tissues or cells is
closely associated with their function (107,129-131). A study showed that the circRNA
forkhead box O3 (FOXO3) is significantly upregulated in heart
samples of elderly patients and mice and is associated with cell
senescence (15). Therefore,
circ-FOXO3 is mainly distributed in the cytoplasm, where it can
interact with the anti-senescence protein inhibitor of DNA
binding-1, the transcription factor E2F1 and the anti-stress
proteins focal adhesion kinase and HIF-1α, thereby promoting
cardiomyocyte senescence. Additional mechanistic studies suggested
that the pro-senescent effect of circ-FOXO3 could be associated
with the regulation of the cell cycle. Therefore, in this study,
the ectopically expressed circ-FOXO3 could form a ternary complex
by binding with cyclin kinase 2 (CDK2) and cycle dependent kinase
inhibitor 1 (p21). The formed circ-FOXO3-p21-CDK2 ternary complex
blocks the function of CDK2, thereby inhibiting the progression of
the cell cycle (129). Zeng
et al (130) investigated
the role of circRNA in myocardial repair by screening the
myocardial tissues of patients of different ages. The results
revealed that circRNA angiomotin Like 1 (Amotl1) is significantly
upregulated in myocardial tissues, thus enhancing the myocardial
function in the neonatal heart. Furthermore, circ-Amotl1 induces
the phosphorylation of Akt and its nuclear transportation via
binding Akt and phosphoinositide dependent kinase 1. This process
promotes the activation of the Akt signaling pathway, resulting in
enhanced cell survival and proliferation. In addition, circ-Amotl1
could play a protective role in the DOX-induced cardiomyopathy
(130). In another study, the
authors investigated the positive and negative regulatory effects
of the signaling pathways Meis1/circ-nuclear factor 1 X-type
(Nfix)/miR-214/Gsk3β/β-catenin and circ-Nfix/Ybx1/cyclin A2 and
cyclin B1 by functional verification experiments (107). The results revealed a close
association between the expression of circ-Nfix and heart
regeneration and repair. The aforementioned studies indicated that
circRNAs could serve an important role in regulating cell
senescence and cell regeneration, and could be used as targets for
treating heart failure and myocardial infarction in the future.
Nowadays, the interest of the scientific community
regarding circRNAs is increasing with their comprehensive
exploration, and the mystery of circRNAs has been slowly revealed.
Although several functions of circRNAs have been identified,
numerous remain to be elucidated. Due to their high stability and
resistance to RNases, circRNAs may accumulate in quiescent or
post-mitotic cells, such as neurons (132). Several studies revealed that the
expression of circRNAs in proliferative cells and tissues,
including cancer cells, is lower compared with terminally
differentiated cells, indicating a negative association between
circRNA abundance and cell proliferation (133). Previous studies have shown that
circRNAs are enriched in extracellular vesicles and are secreted by
cells in the form of vesicles (133-137). Notably, smaller circRNAs are
more prone to vesicle-mediated transport (35). In addition, circRNAs maintain
their circular structure in vesicles and perform their activities
in recipient cells, suggesting that circRNA transport may be
important for cell-to-cell communication (136). In cardiac metabolic diseases,
serum markers in peripheral blood, such as cardiac troponin and
creatine kinase, serve a significant role in the diagnosis of
several diseases. It has been reported that certain linear
non-coding RNAs can be used as biomarkers in the diagnosis of CAD
(137,138). Therefore, circRNAs could be
considered as potential biomarkers for the diagnosis and treatment
of CVDs. However, whether circRNAs selectively enter into exosomes,
and the mechanism and significance of exosomal circRNA formation
should be verified first. Certain circRNAs accumulate in the
nucleus, while the majority are effectively transported to the
cytoplasm (41). The accumulation
of circRNAs in the nucleus is mediated by the ATP-dependent RNA
helicase DexH/D-box, and their nuclear export depends on the length
of the mature circRNA (139).
m6A modification widely occurs on circRNAs (140). In view of the influence of
m6A-binding protein YTH domain-containing 1 (YTHDC1) on the nuclear
export of methylated mRNA (141), the effect of m6A on the export
of circRNAs remains to be elucidated, to the best of our knowledge.
Several circRNA-mediated degradation mechanisms have been reported
(121,142,143). Therefore, the binding of
circRNAs by miRNAs can initiate circRNA decay (121). Furthermore, circRNA can also be
degraded by the cytoplasmic endonuclease RNase L, which is
activated in the presence of pathogenic double-strand RNA during
viral infection (142). In
addition, it has been reported that the YTHDF2-HRSP12-RNase P/MRP
and UPF1-G3BP1 complexes mediate m6A-modified and high-structured
circRNAs, respectively (143).
However, to the best of our knowledge, how the degradation
mechanisms of circRNAs affect the disease course remains to be
investigated. It is understood that circRNAs are conserved among
various species (75), and are
specifically expressed in different tissues and different
developmental stages (12,76).
Nevertheless, the reasons for the conservatism and specificity of
circRNAs and how they affect the functional development of tissues
and organs have not been reported to date. Although it has been
documented that circRNAs exert a folded secondary structure
(144), to the best of our
knowledge, the association between their secondary structure and
their function remains unclear, and it is worthy of further
investigation. There are a number of limited reports regarding the
translational function of circRNAs, including the IRES and m6A
modification pathways (52,53,81,145). Recently, researchers have
combined the stability of circRNAs and their potential for
continuous translation to develop a new expression system that
could produce a protein in a sustainable and stable manner
(146). This finding indicated
that the translational function of circRNAs could be applied in the
future for biological therapy.
The present study was supported by the Key
Scientific and Technological Project of Meizhou People's Hospital
(grant no. MPHKSTP-20180101 to Dr ZZ), the Guangdong Provincial Key
Laboratory of Precision Medicine and Clinical Translation Research
of Hakka Population (grant no. 2018B030322003) and the Science and
Technology Program of Meizhou (grant no. 2019B0202001).
Not applicable.
ZY, HW and ZZ contributed to the conception of the
study, performed the literature search and wrote the manuscript.
QH, QZ and HW edited the manuscript, assisted in the literature
search and critically revised the article for important
intellectual content. All the authors have read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Pennisi E: Shining a light on the genome's
'dark matter'. Science. 330:16142010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15(Suppl 1): R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong R, Ma XK, Li GW and Yang L: CIRCpedia
v2: An updated database for comprehensive circular RNA annotation
and expression comparison. Genomics Proteomics Bioinformatics.
16:226–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar
|
|
12
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang F, Zhang R, Zhang X, Wu Y, Li X,
Zhang S, Hou W, Ding Y, Tian J, Sun L and Kong X: Comprehensive
analysis of circRNA expression pattern and circRNA-miRNA-mRNA
network in the pathogenesis of atherosclerosis in rabbits. Aging
(Albany NY). 10:2266–2283. 2018. View Article : Google Scholar
|
|
14
|
Santos-Ribeiro D, Godinas L, Pilette C and
Perros F: The inte-grated stress response system in cardiovascular
disease. Drug Discov Today. 23:920–929. 2018. View Article : Google Scholar
|
|
15
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
16
|
Pan T, Sun X, Liu Y, Li H, Deng G, Lin H
and Wang S: Heat stress alters genome-wide profiles of circular
RNAs in Arabidopsis. Plant Mol Biol. 96:217–229. 2018. View Article : Google Scholar
|
|
17
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu XD and Ares M Jr: Context-dependent
control of alternative splicing by RNA-binding proteins. Nat Rev
Genet. 15:689–701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar
|
|
20
|
Kramer MC, Liang D, Tatomer DC, Gold B,
March ZM, Cherry S and Wilusz JE: Combinatorial control of
drosophila circular RNA expression by intronic repeats, hnRNPs, and
SR proteins. Genes Dev. 29:2168–2182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Errichelli L, Dini Modigliani S, Laneve P,
Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R,
Peruzzi G, et al: FUS affects circular RNA expression in murine
embryonic stem cell-derived motor neurons. Nat Commun. 8:147412017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
24
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sela N, Mersch B, Gal-Mark N, Lev-Maor G,
Hotz-Wagenblatt A and Ast G: Comparative analysis of transposed
element insertion within human and mouse genomes reveals Alu's
unique role in shaping the human transcriptome. Genome Biol.
8:R1272007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ruskin B and Green MR: An RNA processing
activity that debranches RNA lariats. Science. 229:135–140. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar :
|
|
30
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar :
|
|
31
|
Talhouarne GJS and Gall JG: Lariat
intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad
Sci USA. 115:E7970–E7977. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Noto JJ, Schmidt CA and Matera AG:
Engineering and expressing circular RNAs via tRNA splicing. RNA
Biol. 14:978–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pasman Z, Been MD and Garcia-Blanco MA:
Exon circularization in mammalian nuclear extracts. RNA. 2:603–610.
1996.PubMed/NCBI
|
|
34
|
Lyu D and Huang S: The emerging role and
clinical implication of human exonic circular RNA. RNA Biol.
14:1000–1006. 2017. View Article : Google Scholar :
|
|
35
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC,
Dong JF, Tian Y, Yuan HJ and Zhang JN: Circular ribonucleic acid
expression alteration in exosomes from the brain extracellular
space after traumatic brain injury in mice. J Neurotrauma.
35:2056–2066. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu Q and Zhou T: EIciRNA-mediated gene
expression: Tunability and bimodality. FEBS Lett. 592:3460–3471.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Panda AC, De S, Grammatikakis I, Munk R,
Yang X, Piao Y, Dudekula DB, Abdelmohsen K and Gorospe M:
High-purity circular RNA isolation method (RPAD) reveals vast
collection of intronic circRNAs. Nucleic Acids Res. 45:e1162017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar
|
|
40
|
Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang
J, Cheng H, Yan J, Zhang S, Yang P and Zhao F: Expanded expression
landscape and prioritization of circular RNAs in mammals. Cell Rep.
26:3444–3460.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
anti-sense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kleaveland B, Shi CY, Stefano J and Bartel
DP: A network of noncoding regulatory RNAs acts in the mammalian
brain. Cell. 174:350–362.e17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The output of protein-coding genes
shifts to circular RNAs when the pre-mRNA processing machinery is
limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gupta SK, Garg A, Bär C, Chatterjee S,
Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J and Thum T:
Quaking inhibits doxorubicin-mediated cardiotoxicity through
regulation of cardiac circular RNA expression. Circ Res.
122:246–254. 2018. View Article : Google Scholar :
|
|
50
|
Rossi F, Legnini I, Megiorni F, Colantoni
A, Santini T, Morlando M, Di Timoteo G, Dattilo D, Dominici C and
Bozzoni I: Circ-ZNF609 regulates G1-S progression in
rhabdomyosarcoma. Oncogene. 38:3843–3854. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cardona-Monzonís A, García-Giménez JL,
Mena-Mollá S, Pareja-Galeano H, de la Guía-Galipienso F, Lippi G,
Pallardó FV and Sanchis-Gomar F: Non-coding RNAs and coronary
artery disease. Adv Exp Med Biol. 1229:273–285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Glažar P, Papavasileiou P and Rajewsky N:
CircBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar
|
|
55
|
Panda AC, Dudekula DB, Abdelmohsen K and
Gorospe M: Analysis of circular RNAs using the web tool
CircInteractome. Methods Mol Biol. 1724:43–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44(D1): D209–D215. 2016. View Article : Google Scholar :
|
|
57
|
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang
ZL, Zhou H, Yang JH and Qu LH: DeepBase v2.0: Identification,
expression, evolution and function of small RNAs, LncRNAs and
circular RNAs from deep-sequencing data. Nucleic Acids Res. 44(D1):
D196–D202. 2016. View Article : Google Scholar :
|
|
58
|
Liu M, Wang Q, Shen J, Yang BB and Ding X:
Circbank: A comprehensive database for circRNA with standard
nomenclature. RNA Biol. 16:899–905. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46(D1):
D925–D929. 2018. View Article : Google Scholar :
|
|
60
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database Issue): D92–D97. 2014. View Article : Google Scholar
|
|
62
|
Wang D: hppRNA-a snakemake-based handy
parameter-free pipeline for RNA-Seq analysis of numerous samples.
Brief Bioinform. 19:622–626. 2018.
|
|
63
|
Wu W, Ji P and Zhao F: CircAtlas: An
integrated resource of one million highly accurate circular RNAs
from 1070 vertebrate transcriptomes. Genome Biol. 21:1012020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang
T, Shan T and Peng Y: VirusCircBase: A database of virus circular
RNAs. Brief Bioinform. Apr 29–2020.Epub ahead of print. View Article : Google Scholar
|
|
65
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar
|
|
68
|
Chen DF, Zhang LJ, Tan K and Jing Q:
Application of droplet digital PCR in quantitative detection of the
cell-free circulating circRNAs. Biotechnol Biotechnol Equip.
32:116–123. 2018. View Article : Google Scholar
|
|
69
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kamens J: The addgene repository: An
international nonprofit plasmid and data resource. Nucleic Acids
Res. 43(Database Issue): D1152–D1157. 2015. View Article : Google Scholar :
|
|
71
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bramsen JB, Laursen MB, Nielsen AF, Hansen
TB, Bus C, Langkjaer N, Babu BR, Højland T, Abramov M, Van Aerschot
A, et al: A large-scale chemical modification screen identifies
design rules to generate siRNAs with high activity, high stability
and low toxicity. Nucleic Acids Res. 37:2867–2881. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
de Bruyns A, Geiling B and Dankort D:
Construction of modular lentiviral vectors for effective gene
expression and knockdown. Methods Mol Biol. 1448:3–21. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jakobi T, Czaja-Hasse LF, Reinhardt R and
Dieterich C: Profiling and validation of the circular RNA
repertoire in adult murine hearts. Genomics Proteomics
Bioinformatics. 14:216–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tan WL, Lim BT, Anene-Nzelu CG,
Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD,
et al: A landscape of circular RNA expression in the human heart.
Cardiovasc Res. 113:298–309. 2017.PubMed/NCBI
|
|
77
|
Lei W, Feng T, Fang X, Yu Y, Yang J, Zhao
ZA, Liu J, Shen Z, Deng W and Hu S: Signature of circular RNAs in
human induced pluripotent stem cells and derived cardiomyocytes.
Stem Cell Res Ther. 9:562018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li Y, Zhang J, Huo C, Ding N, Li J, Xiao
J, Lin X, Cai B, Zhang Y and Xu J: Dynamic organization of lncRNA
and circular RNA regulators collectively controlled cardiac
differentiation in humans. EBioMedicine. 24:137–146. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Stagsted LV, Nielsen KM, Daugaard I and
Hansen TB: Noncoding AUG circRNAs constitute an abundant and
conserved subclass of circles. Life Sci Alliance. 2:e2019003982019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Jakobi T, Siede D, Eschenbach J, Heumüller
AW, Busch M, Nietsch R, Meder B, Most P, Dimmeler S, Backs J, et
al: Deep characterization of circular RNAs from human
cardiovascular cell models and cardiac tissue. Cells. 9:16162020.
View Article : Google Scholar
|
|
81
|
van Heesch S, Witte F, Schneider-Lunitz V,
Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S,
Sandmann CL, et al: The translational landscape of the human heart.
Cell. 178:242–260.e29. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao
JN, Chen C, Yan KW, Ponnusamy M, Zhang YH and Li PF: Circular RNA
mediates cardiomyocyte death via miRNA-dependent upregulation of
MTP18 expression. Cell Death Differ. 24:1111–1120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_ circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zou TY, Li L, Huang JF, Yang B and Wang
LY: Expression and clinical significance of circular RNA circTCF25
in patients with coronary artery disease. Mol Cardiol China.
3:27–31. 2018.
|
|
85
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu X, Chen L, Wu S, Xu K, Jiang W, Qin M,
Zhang Y and Liu X: Integrative analysis reveals key circular RNA in
atrial fibrillation. Front Genet. 10:1082019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ludmer PL, Selwyn AP, Shook TL, Wayne RR,
Mudge GH, Alexander RW and Ganz P: Paradoxical vasoconstriction
induced by acetylcholine in atherosclerotic coronary arteries. N
Engl J Med. 315:1046–1051. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Simionescu M: Implications of early
structural-functional changes in the endothelium for vascular
disease. Arterioscler Thromb Vasc Biol. 27:266–274. 2007.
View Article : Google Scholar
|
|
89
|
Chen PY, Qin L, Baeyens N, Li G, Afolabi
T, Budatha M, Tellides G, Schwartz MA and Simons M:
Endothelial-to-mesenchymal transition drives atherosclerosis
progression. J Clin Invest. 125:4514–4528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu C, Yao MD, Li CP, Shan K, Yang H, Wang
JJ, Liu B, Li XM, Yao J, Jiang Q and Yan B: Silencing Of Circular
RNA-ZNF609 ameliorates vascular endothelial dysfunction.
Theranostics. 7:2863–2877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li CY, Ma L and Yu B: Circular RNA
hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells
proliferation and angiogenesis. Biomed Pharmacother. 95:1514–1519.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu
G and Yao H: Engagement of circular RNA HECW2 in the nonautophagic
role of ATG5 implicated in the endothelial-mesenchymal transition.
Autophagy. 14:404–418. 2018. View Article : Google Scholar :
|
|
93
|
Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen
X, Yao MD, Li XM, Yao J, Zhou RM, et al: Targeting
pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A
inhibition aggravates diabetes-induced microvascular dysfunction.
Proc Natl Acad Sci USA. 116:7455–7464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hall IF, Climent M, Quintavalle M, Farina
FM, Schorn T, Zani S, Carullo P, Kunderfranco P, Civilini E,
Condorelli G and Elia L: Circ_Lrp6, a Circular RNA enriched in
vascular smooth muscle cells, acts as a sponge regulating miRNA-145
function. Circ Res. 124:498–510. 2019. View Article : Google Scholar
|
|
95
|
Zheng C, Niu H, Li M, Zhang H, Yang Z,
Tian L, Wu Z, Li D and Chen X: Cyclic RNA has-circ-000595 regulates
apoptosis of aortic smooth muscle cells. Mol Med Rep. 12:6656–6662.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sun Y, Zhang S, Yue M, Li Y, Bi J and Liu
H: Angiotensin II inhibits apoptosis of mouse aortic smooth muscle
cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell
Death Dis. 10:3622019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen J, Cui L, Yuan J, Zhang Y and Sang H:
Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle
cells proliferation and migration by sponging miR-124. Biochem
Biophys Res Commun. 494:126–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang
ML, Zhao XS, Zhao HY, Suzuki T and Wen JK: A novel regulatory
mechanism of smooth muscle α-actin expression by
NRG-1/circACTA2/miR-548f-5p Axis. Circ Res. 121:628–635. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao
L and Wang L: Upregulation of circular RNA circNFIB attenuates
cardiac fibrosis by sponging miR-433. Front Genet. 10:5642019.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM
and Bie ZD: Circ_LAS1L regulates cardiac fibroblast activation,
growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem
Funct. 38:443–450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar
|
|
103
|
Shen L, Hu Y, Lou J, Yin S, Wang W, Wang
Y, Xia Y and Wu W: CircRNA-0044073 is upregulated in
atherosclerosis and increases the proliferation and invasion of
cells by targeting miR-107. Mol Med Rep. 19:3923–3932.
2019.PubMed/NCBI
|
|
104
|
Geng HH, Li R, Su YM, Xiao J, Pan M, Cai
XX and Ji XP: The circular RNA Cdr1as promotes myocardial
infarction by mediating the regulation of miR-7a on its target
genes expression. PLoS One. 11:e01517532016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gan J, Yuan J, Liu Y, Lu Z, Xue Y, Shi L
and Zeng H: Circular RNA_101237 mediates anoxia/reoxygenation
injury by targeting let-7a-5p/IGF2BP3 in cardiomyocytes. Int J Mol
Med. 45:451–460. 2020.PubMed/NCBI
|
|
107
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated circRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL,
Annadoray L, Sian S, Ackers-Johnson MA and Foo RS: Targeting the
highly abundant circular RNA circSlc8a1 in cardiomyocytes
attenuates pressure overload induced hypertrophy. Cardiovasc Res.
115:1998–2007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhang S, Song G, Yuan J, Qiao S, Xu S, Si
Z, Yang Y, Xu X and Wang A: Circular RNA circ_0003204 inhibits
proliferation, migration and tube formation of endothelial cell in
atherosclerosis via miR-370-3p/TGFβR2/phosph-SMAD3 axis. J Biomed
Sci. 27:112020. View Article : Google Scholar
|
|
110
|
Altesha MA, Ni T, Khan A, Liu K and Zheng
X: Circular RNA in cardiovascular disease. J Cell Physiol.
234:5588–5600. 2019. View Article : Google Scholar
|
|
111
|
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y,
Shen G and Wang F: Inhibition of circHIPK3 prevents angiotensin
II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol.
292:188–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wang Y, Zhao R, Liu W, Wang Z, Rong J,
Long X, Liu Z, Ge J and Shi B: Exosomal circHIPK3 released from
hypoxia-pretreated cardiomyocytes regulates oxidative damage in
cardiac micro-vascular endothelial cells via the miR-29a/IGF-1
pathway. Oxid Med Cell Longev. 2019:79546572019. View Article : Google Scholar
|
|
113
|
Li M, Ding W, Tariq MA, Chang W, Zhang X,
Xu W, Hou L, Wang Y and Wang J: A circular transcript of ncx1 gene
mediates ischemic myocardial injury by targeting miR-133a-3p.
Theranostics. 8:5855–5869. 2018. View Article : Google Scholar
|
|
114
|
Jin Q and Chen Y: Silencing circular RNA
circ_0010729 protects human cardiomyocytes from oxygen-glucose
deprivation-induced injury by up-regulating microRNA-145-5p. Mol
Cell Biochem. 462:185–194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y,
Xu J, Chen S and Liu S: Circular RNA Ttc3 regulates cardiac
function after myocardial infarction by sponging miR-15b. J Mol
Cell Cardiol. 130:10–22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Holdt LM, Beutner F, Scholz M, Gielen S,
Gäbel G, Bergert H, Schuler G, Thiery J and Teupser D: ANRIL
expression is associated with atherosclerosis risk at chromosome
9p21. Arterioscler Thromb Vasc Biol. 30:620–627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Song CL, Wang JP, Xue X, Liu N, Zhang XH,
Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y and Yang YB: Effect of
Circular ANRIL on the inflammatory response of vascular endothelial
cells in a rat model of coronary atherosclerosis. Cell Physiol
Biochem. 42:1202–1212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Menglan LI, Siying HE, Jialing R, Bin L,
Xiaokang Z and Fang Z: The possible protective role of circDLGAP4
from peripheral blood in coronary heart disease. Chin J Clin Lab
Sci. 37:109–112. 2019.
|
|
119
|
Parahuleva MS, Euler G, Mardini A, Parviz
B, Schieffer B, Schulz R and Aslam M: Identification of microRNAs
as potential cellular monocytic biomarkers in the early phase of
myocardial infarction: A pilot study. Sci Rep. 7:159742017.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sala F, Aranda JF, Rotllan N, Ramírez CM,
Aryal B, Elia L, Condorelli G, Catapano AL, Fernández-Hernando C
and Norata GD: MiR-143/145 deficiency attenuates the progression of
atherosclerosis in Ldlr-/-mice. Thromb Haemost. 112:796–802. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo
SK, Xue W, Cui Y, Dong K, Ding H, et al: Structure and degradation
of circular RNAs regulate PKR activation in innate immunity. Cell.
177:865–880.e21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhang Y, Zhang Y, Li X, Zhang M and Lv K:
Microarray analysis of circular RNA expression patterns in
polarized macrophages. Int J Mol Med. 39:373–379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY
and Ea CK: Inducible RasGEF1B circular RNA is a positive regulator
of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 13:861–871. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen YG, Kim MV, Chen X, Batista PJ,
Aoyama S, Wilusz JE, Iwasaki A and Chang HY: Sensing self and
foreign circular RNAs by intron identity. Mol Cell. 67:228–238.e5.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Valentim L, Laurence KM, Townsend PA,
Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS and
Stephanou A: Urocortin inhibits Beclin1-mediated autophagic cell
death in cardiac myocytes exposed to ischaemia/reperfusion injury.
J Mol Cell Cardiol. 40:846–852. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang
YH, Zhang RC, Liu CY, Dong YH, Wang M, et al: The circular RNA ACR
attenuates myocardial ischemia/reperfusion injury by suppressing
autophagy via modulation of the Pink1/FAM65B pathway. Cell Death
Differ. 26:1299–1315. 2019. View Article : Google Scholar
|
|
129
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A circular RNA binds to
and activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Rep. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar
|
|
134
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extra-cellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar
|
|
135
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Preußer C, Hung LH, Schneider T, Schreiner
S, Hardt M, Moebus A, Santoso S and Bindereif A: Selective release
of circRNAs in platelet-derived extracellular vesicles. J Extracell
Vesicles. 7:14244732018. View Article : Google Scholar
|
|
137
|
Viereck J and Thum T: Circulating
noncoding RNAs as biomarkers of cardiovascular disease and injury.
Circ Res. 120:381–399. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
142
|
Han Y, Donovan J, Rath S, Whitney G,
Chitrakar A and Korennykh A: Structure of human RNase L reveals the
basis for regulated RNA decay in the IFN response. Science.
343:1244–1248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic cleavage of
m6A-containing RNAs by RNase P/MRP complex. Mol Cell.
74:494–507.e8. 2019. View Article : Google Scholar
|
|
144
|
Tapsin S, Sun M, Shen Y, Zhang H, Lim XN,
Susanto TT, Yang SL, Zeng GS, Lee J, Lezhava A, et al: Genome-wide
identification of natural RNA aptamers in prokaryotes and
eukaryotes. Nat Commun. 9:12892018. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar :
|
|
146
|
Costello A, Lao NT, Barron N and Clynes M:
Continuous translation of circularized mRNA improves recombinant
protein titer. Metab Eng. 52:284–292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Xie YZ, Yang F, Tan W, Li X, Jiao C, Huang
R and Yang BB: The anti-cancer components of Ganoderma lucidum
possesses cardiovascular protective effect by regulating circular
RNA expression. Oncoscience. 3:203–207. 2016. View Article : Google Scholar :
|
|
148
|
Dang RY, Liu FL and Li Y: Circular RNA
hsa_circ_0010729 regulates vascular endothelial cell proliferation
and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys
Res Commun. 490:104–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhang Y, Yu J, Kahkoska AR and Gu Z:
Photoacoustic drug delivery. Sensors (Basel). 17. pp. 14002017,
View Article : Google Scholar
|
|
151
|
Lavenniah A, Luu TDA, Li YP, Lim TB, Jiang
J, Ackers-Johnson M and Foo RS: Engineered circular RNA sponges act
as miRNA inhibitors to attenuate pressure overload-induced cardiac
hyper-trophy. Mol Ther. 28:1506–1517. 2020. View Article : Google Scholar : PubMed/NCBI
|