1. Introduction
Low back pain (LBP) is one of the most prevalent
musculoskeletal diseases worldwide. Approximately 70-85% of adults
suffer from LBP in their lifetime, and a great number of them are
disabled by it (1). The cost for
treatment can reach billions of dollars, creating a huge burden for
the families of patients and society (2). However, as a result of the complex
pathology of LBP and the poor performance of current therapeutic
measures, LBP still constitutes a major threat to the health of
people.
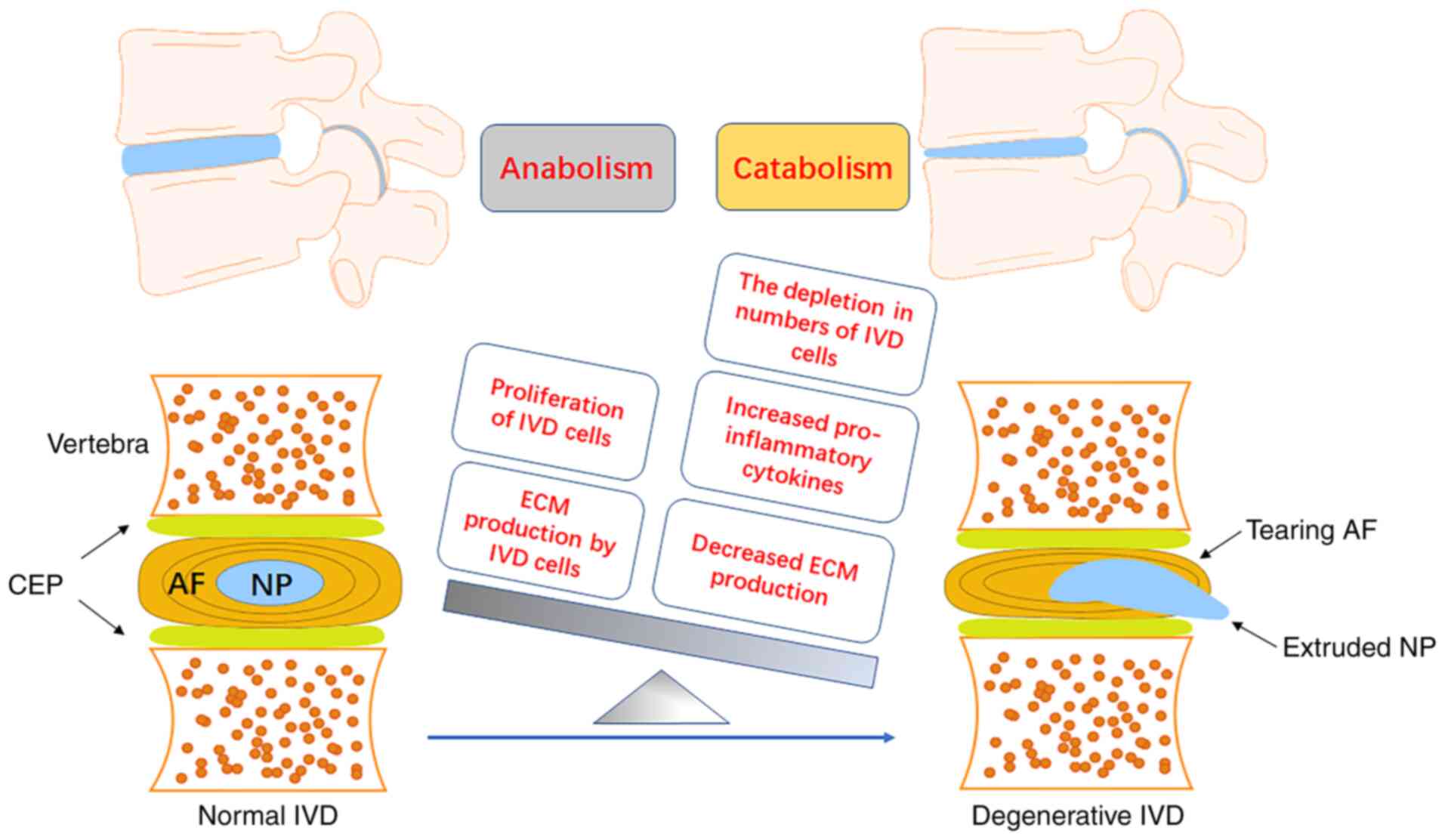
There are numerous pathogenic factors leading to
LBP, with intervertebral disc degeneration (IDD) being the most
common target for diagnosis and intervention (3-5).
As the largest avascular and aneural tissue in the human body, the
normal intervertebral discs (IVDs) are made up of three
morphologically distinct regions, the nucleus pulposus (NP),
annulus fibrosus (AF) and cartilaginous endplates (CEPs) (6-9).
IVDs function through dampening excessive mechanical stresses and
maintaining the stability of the spine (10). It is important to provide thorough
insight into the complicated pathophysiological process of IDD, in
order to develop a strategy for the prevention and treatment of
LBP. IDD is a multifactorial result characterized by an aberrant
cell-mediated response that gradually causes structural failure
(11). Aberrant cell-mediated
responses to the changed microenvironment include an imbalanced
extracellular matrix (ECM) metabolism, an upregulated
proinflammatory phenotype and senescence (12-15). During this process, the
upregulation of proinflammatory and procatabolic phenotypes by NP
cells is the main contributor to the suppression of anabolism and
promotes the catabolism of the ECM (16,17).
Cartilage intermediate layer protein (CILP) is a
monomeric glycoprotein residing in the ECM that is mainly expressed
in the intermediate zone between human IVDs and articular cartilage
(18-20). Previous studies have also revealed
the existence of a tendon ligament (21) and a synovial membrane (22). Notably, CILP expression in disc
tissues has been revealed to be increased as degeneration and aging
progress, contrary to the decreased levels of collagen II and
aggrecans, which are the main components of discs (23). Furthermore, recent studies have
revealed that CILP-overexpression in human NP cells can negatively
regulate matrix synthesis (24).
In the aberrantly expressed genes detected in IDD, CILP is among
the few cartilage matrix proteins whose expression is upregulated
in the early and late stages of cartilage diseases (19,25), and a genetic association has been
revealed between the CILP gene and IDD, suggesting the importance
of CILP beyond that of other structural genes (26). The relationship between CILP and
IDD has received increasing attention in recent years. Herein,
insight was first provided concerning the pathogenesis of IDD, and
the genetic and molecular structure of CILP was described. Next, a
detailed introduction of the function of CILP in IVDs was provided,
and the genetic structure of the association between CILP and
cartilage diseases was described. The regulatory mechanism of CILP
was then summarized. Finally, the discussion focused on the future
perspectives of CILP in biological therapies for IDD.
2. Intervertebral disc degeneration (IDD)
pathogenesis
IDD is a multifactorial result caused by aging,
infection, smoking, mechanical overloading, nutrient deficiency and
genetic predisposition (27-31). Among the numerous factors that
lead to IDD, the destruction of extracellular microenvironmental
homeostasis is considered to be one of the most important factors.
All the etiologic causes initiate the process of IDD, which is
mediated and characterized by an enhanced proinflammatory phenotype
(32-34). The increased inflammatory
chemokine level, secreted from disc cells, infiltrates immune and
AF cells, greatly destroying the homeostasis of the
microenvironment around disc cells and directly affecting the
metabolism of NP cells, which leads to a disruption of the balance
between the anabolism and degradation of the ECM that directly
accelerate the degradation of the ECM (35-38). As a result, the resident cells are
exposed to excessive mechanical stress, which in turn further
worsens the ECM metabolism of NP cells (24). In addition, the nociceptive nerve
fibers and blood vessels from the dorsal root ganglion intrude into
the herniated disc tissues to cause LBP (Fig. 1) (39,40). Notably, gene susceptibility has
also been revealed to be involved in the initiation and progression
of IDD; CILP is among the susceptible genes that are aberrantly
expressed in IVDs (41,42). Moreover, CILP is restrictively
expressed in few cartilage tissues, including articular cartilage
and disc tissues (18,26), which suggests the importance of
CILP beyond other susceptibility genes, elucidates the function of
CILP and contributes to a better understanding of the pathogenesis
of IDD.
3. Structure and synthesis of CILP
CILP was first identified and isolated by Lorenzo
et al in 1998; this protein was named for its deposition in
the interterritorial matrix without a presence in the superficial
or deepest regions of the articular cartilage (18). CILP is synthesized by cartilage
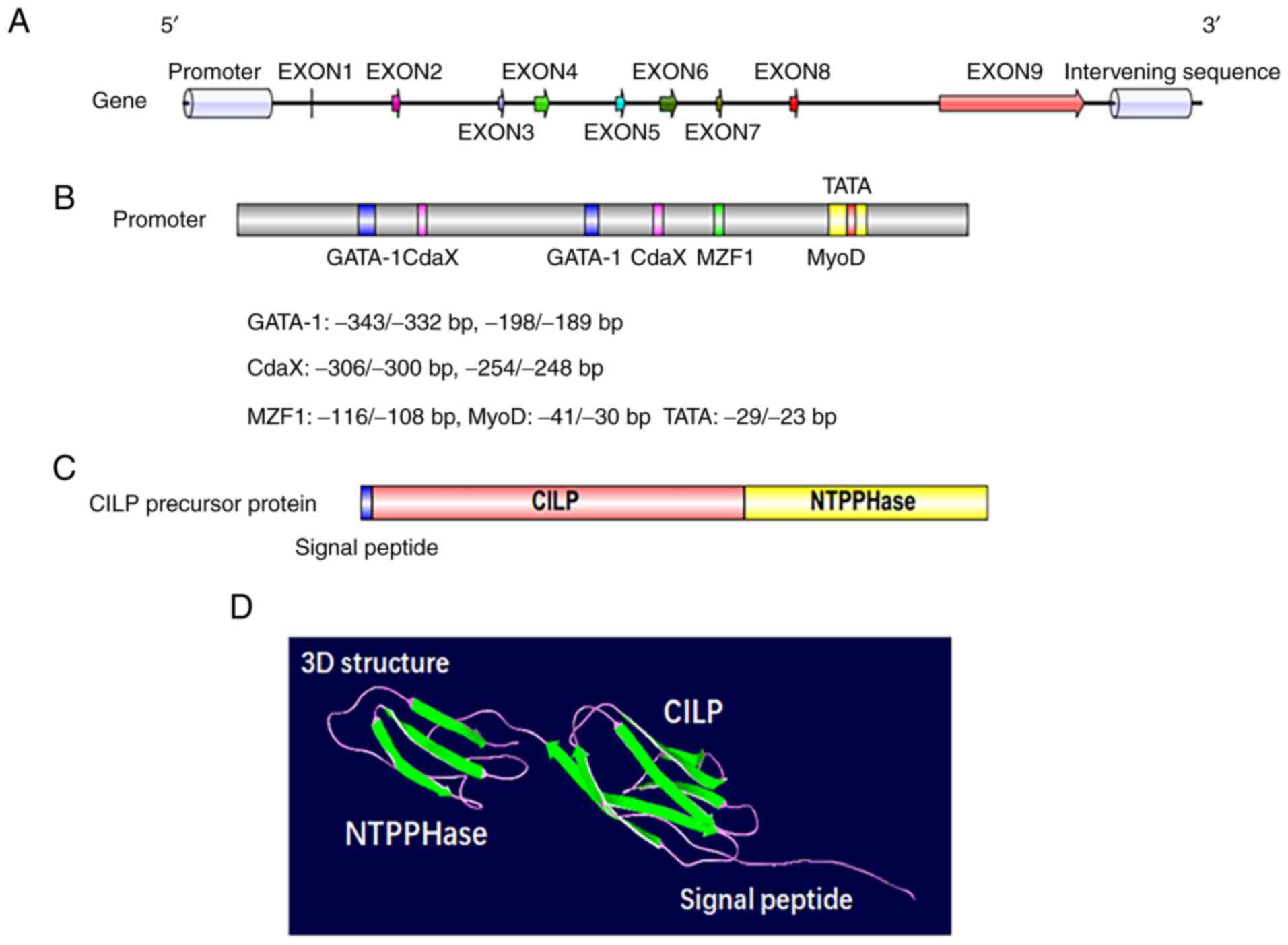
chondrocytes and is a polypeptide of 1,184 amino acids with a
molecular mass of 132.5 kDa. Apart from a putative signal peptide
of 21 amino acids, the protein is comprised of 2 distinct
polypeptides (20). The
N-terminus corresponds to the classical CILP protein, while the
C-terminus corresponds to a homologue of porcine nucleotide
pyrophosphatase phosphodiesterase (NPP) (Fig. 2C and D). The CILP gene, which
evolved from independent ancestral genes spanning 15.3 Kbp of
genomic DNA, resides on chromosome 15q22 (43,44). Human CILP cDNA consists of 9 exons
and 8 introns, of which exons 3-6 are symmetrical, while exons 7
and 8 are asymmetrical (Fig. 2A).
There is a putative promoter region upstream of the encoding start
site at the 5′ flanking region, where regulatory elements such as
GATA-1, MyoD, MZF1 and CdxA have been detected (Fig. 2B) (44). Exon 1 covers 46 bp of the
noncoding region, and the eukaryotic translation of the N-terminal
region corresponding to the CILP protein begins at exon 2 (43,45). Of the exons that are translated,
exons 2 to 8 are 46-154 bp long, and only exon 9 exceeds 2,800 bp
(44). Furthermore, exon 9 not
only participates in the protein translation of CILP but also
encodes the C-terminal protein, a homologue to porcine nucleotide
pyrophosphohydrolase, which has piqued the interest of researchers
due to its possible involvement in calcium pyrophosphate dihydrate
(CPPD) deposition disease (44,46).
By means of transcriptome profiling, a homologue to
CILP1 (the classical CILP) was discovered in mouse cartilage
(47). During the stage before
maturation, CILP2 mainly focuses on the surface of the cartilage.
As maturation progresses, the homologue gradually collocates in the
intermediate zone with CILP1 (47). However, there are also differences
between the two analogs. In surgery-induced osteoarthritis, CILP2
was significantly downregulated, while CILP1 was upregulated.
Ultrastructure analysis suggested that CILP2 may be relevant to
collagen VI, which is a normal component in cartilage tissues, and
that CILP2 may play a role in cartilage by mediating the
interaction among the matrix components; more studies are required
to test this hypothesis (47).
4. Association between cartilage
intermediate layer protein (CILP) and IDD
In a 2005 study, Seki et alfirst revealed
that the CILP is a key regulatory factor in IDD development
(19). At the mammalian model
level, Seki et al used transgenic mice that overexpressed
CILP, and although there was no significant change in the X-ray
analysis results, blood-test values or body weight, MRI analysis
detected an obviously lower intensity in the area where CILP was
deposited (25). In addition,
through the detection and analysis of CILP content in IVDs of
rabbits of different ages, it was revealed that the expression
level of CILP in IVDs in aged rabbits was significantly increased
(23). Previous research has not
reported CILP expression levels in human IVDs. Therefore, we
collected human IVDs with varying degrees of degeneration and
assessed their CILP expression levels. Human IVD tissues were
collected from 18 patients (male:female, 1:1) ranging in age from
20-65 years who were undergoing lumbar spinal surgery for IDD
between October 2019 and May 2020, at the Department of Orthopedics
of The Second Affiliated Hospital (Chongqing, China). The present
study was approved by the Ethics Committee of Xinqiao Hospital
(Chongqing, China) on October 1, 2019 at the implementation of the
study. All subjects provided their informed consent before
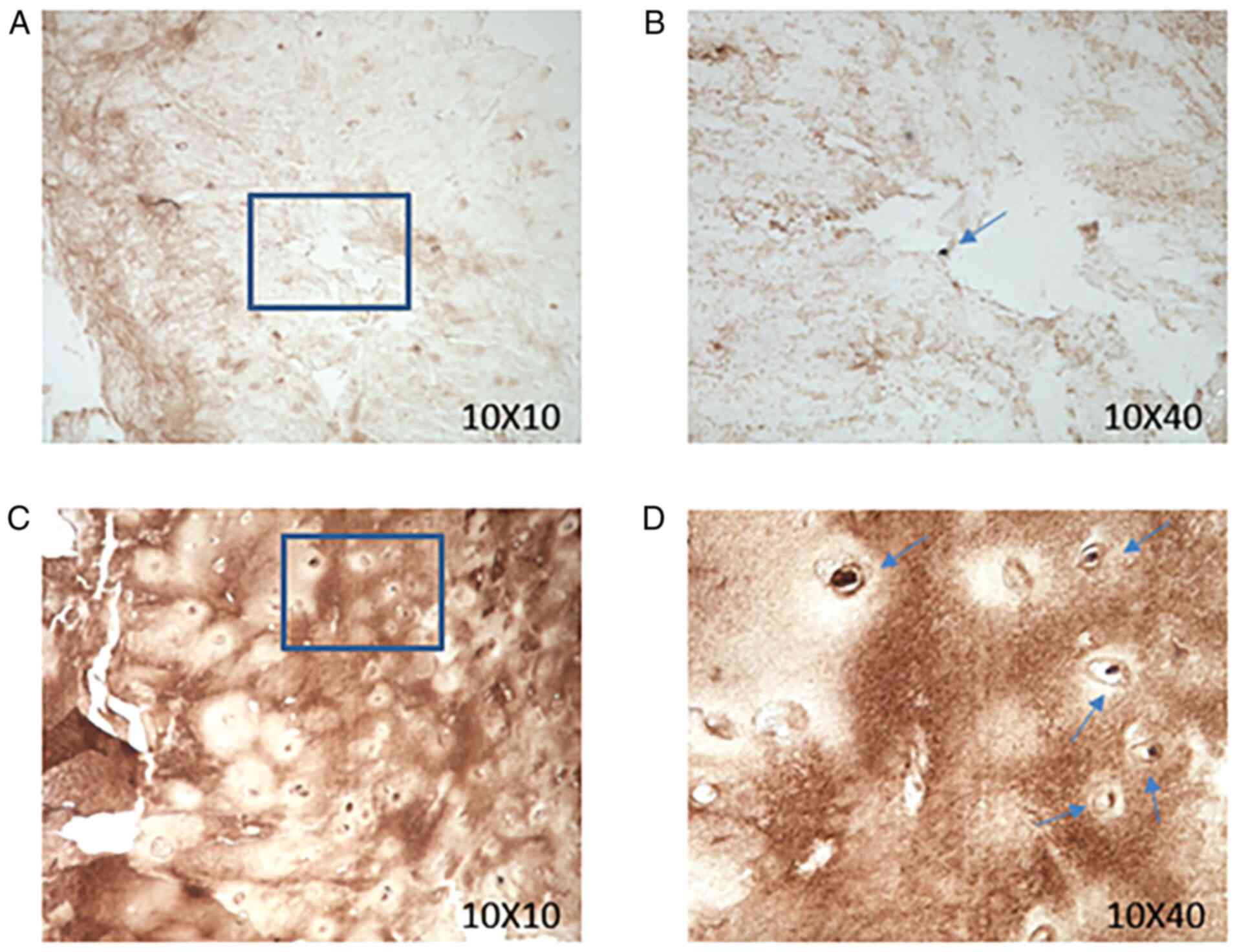
participating in the present study. Immunohistochemical staining of
a CILP antibody was performed in human disc paraffin sections with
different degrees of degeneration in Pfirrmann grades. In brief,
the tissues were fixed in 4% paraformaldehyde at 25°C for 48 h.
Dehydrated tissue was immersed in xylene for 2 h for transparent
treatment and then immersed in section paraffin for 2.5 h. The
tissue sections were 5-µm thick. Then, the sections were
blocked in normal goat serum (cat. no. SL038; Solarbio Life
Sciences) that was diluted 20 times with PBS and added directly to
the slices for 10-30 min at 37°C. Tissue sections were then
incubated with CILP Polyclonal Antibody (rabbit/IgG; dilution,
1:200; cat. no. PA5-51856; Thermo Fisher Scientific, Inc.) at 4°C
for 12 h. Following primary incubation, the sections were incubated
with HRP-conjugated goat anti-rabbit IgG (dilution, 1:100; cat. no.
SA134; Solarbio Life Sciences) at room temperature (25°C) for 1 h.
The sections were observed using a light microscope
(magnifications, 10 X 10 and 10 X 40). The expression of CILP was
revealed to be increased in human disc paraffin sections with
higher degrees of degeneration (Fig.
3). Quantitative proteomic analysis of the IVDs of different
IDD level groups revealed that CILP expression was significantly
increased in the IVDs of people with severe IDD, specifically, in
NP tissue, there was more CILP expression in the degenerate sample
(26). In recent years, genetic
susceptibility analysis of diseases to obtain the degree of genetic
correlation with diseases has been widely used. A study consisting
of 467 Japanese patients and 654 controls revealed that the single
nucleus polymorphism (SNP) rs2073711 at the 1,184 allele (T to C)
was genetically correlated with IDD (19). Furthermore, the substitution from
T to C increased the binding of CILP to transforming growth
factor-β (TGF-β), which enhanced the CILP-mediated suppression of
the pro-anabolic effect mediated by TGF-β; this substitution plays
an important role in the pathogenesis and origin of IDD (19). A meta-analysis of genetic
association studies of IVDs using a total of 1,551 IVD cases and
1,793 controls from the 5 studies which were used in this study,
and comprising four Asian populations and two European populations,
confirmed the positive association between the CILP gene and IVDs
(48). However, the correlation
was absent in a Chinese sample with 691 cases, a Finnish sample
with 502 cases and an Indian sample with 342 cases (49,50). It appears that the genetic
predisposition of CILP for IDD varies with population differences,
which can explain the discrepancy among different ethnicities.
Moreover, two studies further complicated the differential
predisposition of CILP, as they revealed the genetic association
between IDD and the C allele in CILP in Japanese male collegiate
athletes and judokas (42,51).
In another study with a Finnish population, the rs2073711 SNP was
associated with IDD among women (52). It appears reasonable that the
genetic susceptibility of CILP is also gender-dependent, in
addition to the existing race-dependence and the substitution from
the T to the C allele that changes the character of CILP, which
enhances the risk of degeneration. However, given that these
athletes, especially male athletes or male judokas, experience a
higher mechanical loading than that of non-athletes, this
phenomenon can also be explained by mechanical overloading that
leads to the odds ratio discrepancy between males and females
(51). Therefore, more studies
are required to provide a deeper understanding of the association
between CILP and IDD.
CILP is associated with other degenerative
conditions, such as osteoarthritis and myocardial fibrosis. CILP
was significantly increased in articular cartilage where
osteoarthritis occurred, and as a key regulatory factor, it has
been revealed to play an important role in the occurrence and
development of osteoarthritis (53). In recent years, CILP has been
regarded as an important indicator protein for myocardial fibrosis,
and its expression level indicates the severity of myocardial
fibrosis and plays a positive role in clinical diagnosis; decreased
levels of CILP are generally considered to indicate severe
myocardial fibrosis (54).
5. Function of CILP in IDD
With the further study on the mechanism of CILP in
the development of IDD, it was revealed that the expression level
of CILP in NP cells has an important effect on the ECM, an
important component of the extracellular microenvironment (24). Aggrecan and collagen II are the
traditional degenerative markers of IDD, which are the main
components of the ECM. CILP siRNA effectively inhibited CILP
expression in NP cells and significantly increased the expression
of aggrecan and collagen II. In addition, treatment of NP cells
with a high concentration of rhCILP resulted in significantly
decreased expression of aggrecan and collagen II (24). The primary function of the
intervertebral disc ECM is to ensure physical and biomechanical
strength (55). The ECM plays
important biological roles in chondrocyte metabolism by regulating
growth factors, including TGF-β (56,57). Studies have revealed that TGF-β
induces the synthesis of proteoglycans and cell proliferation in
IVDs (56,58). Furthermore, an injection of an
adenoviral TGF-β expression vector was revealed to increase
proteoglycan synthesis in human IVDs (59). The TGF-β signaling pathway is
broadly involved in the growth and differentiation of cells and is
responsible for the anabolism of the ECM, which is critical for the
homeostasis of discs (60,61).
The ECM protein decorin binds to TGF-β to form a complex that
controls the accessibility of TGF-β to receptors (62,63); similarly, the binding of CILP to
TGF-β may interfere physically with the binding of TGF-β to its
receptor, or may render TGF-β inaccessible to its receptor by
sequestering TGF-β (19). CILP
has a thrombospondin type 1 repeat domain that contains the WSXW
motif, a well-defined consensus sequence that binds to the active
form of TGF-β; CILP coexists with TGF-β in disc tissues and the
territorial matrices of TGF-β in IVDs (19). This interference results in the
inhibition of phosphorylation of mothers against decapentaplegic
homolog (SMAD)2/3, the key factors of the TGF-β/SMAD signaling
pathway (19). Furthermore, CILP
is capable of suppressing the interaction of TGF-β with its special
receptor by directly binding to the growth factor, a binding that
inhibits the TGF-β signaling pathway in NP cells (25). Moreover, the phosphorylation of
SMAD3, a downstream effector of TGF-β, was revealed to be
suppressed in transgenic mice overexpressing CILP (19,64). Alternatively, the binding of CILP
may hinder the activation mechanism of TGF-β by altering its
interaction with the latency complex, and may hinder the efficacy
of enzymes in releasing the active form of TGF-β (19). In addition, the SNP (rs2073711) in
the CILP gene increases the binding ability of CILP to TGF-β,
consequently enhancing its suppression of the TGF-β signaling
pathway (19). In conclusion, the
overexpression of CILP upsets the balance of the control of TGF-β
in chondrocyte metabolism and intervertebral disc tissue
maintenance, leading to lumbar degenerative disease susceptibility
caused by an inadequate response of intervertebral disc cells to
injury and mechanical stress (25).
Insulin-like growth factor-1 (IGF-1) is a naturally
occur-ring polypeptide protein hormone that plays an important role
in stimulating growth during childhood and helps build and repair
tissues in adults (65). In
particular, IGF-1 is a key player in IVD homeostasis by
upregulating both cell proliferation and the biosynthesis of ECM
components in a dose-dependent manner. Once in the IVDs, IGF-1
binds to IGF-1 cell surface receptors (IGFR1), initiating the
phosphatidylinositol-3 kinase/AKT signaling pathway, stimulating
cell growth and proliferation, and inhibiting programmed cell
death, which leads to an increase in IVD cell population and the
production of new ECM (66,67). CILP is capable of suppressing
ligand-induced IGFR1 autophosphorylation and counteracting
IGF-1-mediated chondrocyte proliferation and proteoglycan synthesis
(68-70), thus interfering with the anabolism
and catabolism of ECM, which leads to the acceleration of IDD.
Other studies have reported that IGF-1 can reduce inorganic
pyrophosphate (PPi), which can be generated via the alkaline
nucleotide phosphodiesterase I activity of the isozymes of the NPP
family (71) and is able to
promote the progression of CPPD crystal deposition in aging
cartilage tissues (46); in
addition, CILP can affect chondrocyte IGF-1 responsiveness via
N-terminal domain-mediated inhibition (68), leading to a PPi increase that can
stimulate cartilage pathological calcification as CPPD crystal
deposition (72). Cartilage
pathological calcification can cause the degeneration of the CEP,
which decreases the availability of nutrients and the exchange of
metabolites, resulting in irreversible and progressive IDD
(73-75).
These studies have revealed that CILP can interfere
with the binding of >1 growth factor to their designated
receptor, and can suppress downstream signal transduction,
consequently affecting the general homeostasis of cells (Fig. 4).
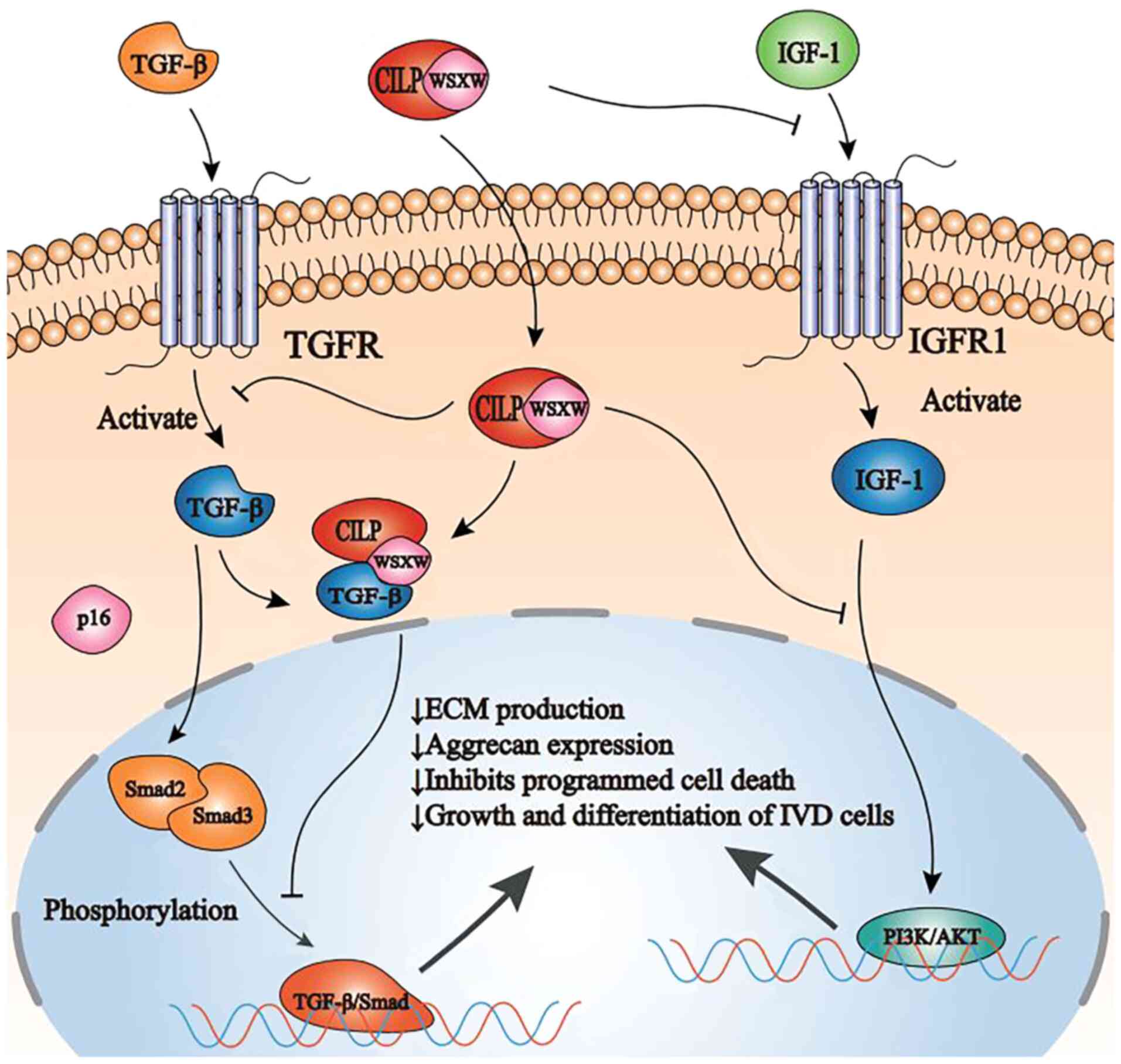 | Figure 4Schematic representation of CILP
function and regulation in NP cells by TGF-β and IGF-1. CILP
inhibits SMAD2/3 phosphorylation, either directly or by interfering
with TGF-β binding to its receptor TGFR, ultimately inhibiting the
TGF-β/SMAD signaling pathway. In addition, CILP can inhibit the
binding of IGF-1 and its receptor IGFR1. Furthermore, CILP can
inhibit the function of the combination of IGF-1 and IGFR1 in NP
cells and interfere with the PI3K/AKT signaling pathway, eventually
leading to a decrease in ECM production, aggrecan expression, and
growth and differentiation of intervertebral disc cells. CILP,
cartilage intermediate layer protein; NP, nucleus pulposus; TGF-β,
transforming growth factor-β; IGF-1, insulin-like growth factor-1;
TGFR, transforming growth factor-β receptors; SMAD, mothers against
decapentaplegic homolog; PI3K, phosphatidylinositol-3 kinase;
IGFR1, IGF-1 cell surface receptors; ECM, extracellular matrix. |
6. Regulation of CILP
As CILP has been revealed to function as a
contributor to IDD, it is imperative to provide insight into the
regulatory mechanism underlying CILP expression. First, CILP
expression is increased as age and degeneration progress (23,26); therefore, aging and degeneration
are among the causes that promote CILP expression. Notably, as a
structural component in the matrix secreted by chondrocytes, CILP
was expressed without the effect of SOX9 (45), which is the core transcription
factor in chondrogenesis (76-78). Instead, TGF-β, another key
regulator of chondrocyte differentiation and proliferation
(60), is able to promote the
secretion of CILP by NP cells through the SMAD and MAPK signaling
pathways (45). With regard to
the CILP-mediated suppression of the TGF-β signaling pathway via
the binding of CILP to TGF-β (19), there appears to be a negative
feedback loop between CILP and TGF-β. Similarly, bone morphogenetic
protein 2 (BMP-2) also significantly increases CILP expression by
increasing CILP promoter activity through the SMAD signaling
pathway, an effect that increases with age (23). However, in contrast to TGF-β and
BMP-2, IGF-1 downregulates CILP expression by binding to the
N-terminal polypeptide domain of CILP (68). These results indicated that the
regulatory effect on CILP by growth factors varies by type, and
that the dysregulated secretion of growth factors may contribute to
the aberrant expression of CILP. In addition, since IDD is
characterized by an enhanced level of inflammatory chemokines,
including IL and TNF (17,79),
which have a significant influence on the secretion of NP cells, it
is possible that a high level of inflammatory chemokines may
regulate CILP expression. However, it was revealed CILP expression
does not undergo a significant change even in a conditioned medium
with a high level of IL-1, markedly increasing the expression of
MMPs and ADAMTs (80). It remains
unknown whether CILP is also unaffected by other inflammatory
factors, and this requires further study for clarification. CILP
expression is also influenced by mechanical factors; in human NP
cells, CILP expression is regulated by mechanical stress, which
affects the synthesis of ECM (24). In conclusion, these results
suggested that, as a structural protein resident in the ECM, CILP
expression is upregulated by aging and degeneration but is
unaffected by IL-1; in addition, various growth factors exert
different, even contrary, regulations on CILP, which in turn affect
the secretion of those growth factors.
7. Conclusions and future directions
As an NP matrix protein, CILP is specifically
expressed in degenerative IVD tissues, which can accelerate the
progression of IDD by altering the balance of intervertebral disc
matrix metabolism.
CILP is an ECM glycoprotein that is highly expressed
in degenerative disc tissues and accelerates the process of disc
degeneration by altering the balance of the intervertebral disc
matrix metabolism (18). Since
the first discovery of CILP, the regulation, expression and
function of CILP have been elucidated in numerous studies (Table I). Disc degeneration is a chronic
metabolic disorder of the extracellular microenvironment. Several
ECM proteins, such as CILP and connective tissue growth factor
(81), are involved in this
process. The end result of these cytokine pathways is an imbalance
of catabolism and anabolism within the disc, leading to disc
degeneration, herniation, and radicular pain. A recent study has
revealed that the expression and function of CILP are regulated by
specific tissue and cell types (82). In intervertebral disc-related
studies, CILP regulates the role of cytokines such as TGF-β and
IGF-1 in IVDs (25,68,82). The TGF-β/CILP mutual regulation is
important for ECM production and the two-way regulation of TGF-β
and CILP (25,82). The TGF-β/SMAD axis is inhibited by
CILP eventually leading to the decrease of ECM production and
aggrecan expression, as well as inhibiting programmed cell death
and growth and differentiation of IVD cells (25). The activation of BMP-2 also
increases CILP expression through the SMAD signaling pathway
(25). In contrast to TGF-β and
BMP-2, the activation of IGF-1 downregulates CILP expression,
thereby inhibiting the progression of disc degeneration. IGF-1
binds to IGFR1, activating the PI3K/AKT signaling pathway,
promoting cell growth and proliferation, and inhibiting programmed
cell death, which leads to an increase in IVD cell population and
the production of new ECM (68).
In a recent study, it was revealed that mechanical changes in the
disc are another important regulatory factor of CILP (24). Increased CILP expression induced
by mechanical changes in the disc NP cells can promote the process
of disc tissue degeneration. In the intervertebral disc, mechanical
alteration is a physiological niche condition, and in order to
maintain its physiological level, it may be necessary to reduce
unnecessary mechanical alteration. During disc degeneration, the
blood oxygen status of NP is thought to be altered due to vascular
infiltration. Therefore, a reasonable reduction of TGF-β and
increase of IGF-1 expression can reduce the expression level of
CILP in the disc. Further research on the role of CILP in the
degenerative disc and surrounding tissues is required to determine
the ultimate role of CILP in this process.
 | Table IInformation on the relationship
between CILP and IDD. |
Table I
Information on the relationship
between CILP and IDD.
| Authors | Date | Important
events | (Refs.) |
|---|
| Lorenzo et
al | 1998 | CILP is first
identified and isolated | (18,20) |
| Lorenzo et
al | 1999 | Human CILP gene is
isolated and characterized | (44) |
| Hirose et
al | 2002 | Increased CILP mRNA
expression in chondrocytes promotes the formation of calcium
pyrophosphate dihydrate crystals in aged cartilage | (46) |
| Johnson et
al | 2003 | CILP is revealed to
promote osteoarthritis by regulating the IGF-1/PI3K/AKT signaling
pathway | (68) |
| Seki et
al | 2005 | A functional SNP in
CILP, encoding cartilage intermediate layer protein, is associated
with susceptibility to lumbar disc disease | (19) |
| Seki et
al | 2005 | CILP is revealed to
promote IDD by regulating the TGF-β/SMAD signaling pathway | (19) |
| Virtanen et
al | 2007 | Differences of CILP
gene susceptibility are revealed in different populations of IDD
patients | (49) |
| Seki et
al | 2014 | CILP is revealed to
promote lumbar disc degeneration in transgenic mice | (25) |
| Wang et
al | 2016 | Association between
cartilage intermediate layer protein and degeneration of
intervertebral disc: A meta-analysis | (48) |
| He et
al | 2018 | CILP is regulated
by mechanical stress and affects extracellular matrix synthesis to
promote the progression of IDD | (24) |
In vivo and in vitro studies have
clearly revealed that CILP affects the anabolic effects of nucleus
pulposus (CMCS) on stroma production, and that it may be able to
use the properties of this protein as part of a regenerative mix to
treat degenerative discs. However, there have been no clinical
trials of CILP-related therapy. To further investigate CILP-related
therapy, understanding the relationship between CILP and tissue
inflammation will be important for the successful treatment of disc
disease.
Funding
This research was funded by the National Natural
Science Foundation of China (grant nos. 81902255, 81874028 and
81672215).
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
LL and JH were responsible for conceiving the study,
and are the co-first authors. CLiu, MY, JF, ML, YoZ, JY and XA were
responsible for literature collection and summary. YaZ, BH, CLi,
YuZ and CF were responsible for reviewing and editing the
manuscript. YZ and CF were responsible for supervising the study.
All authors have read and approved the published version of the
manuscript.
Ethics approval and consent for
publication
All subjects provided their informed consent before
participating in the study. The study was conducted in accordance
with the Declaration of Helsinki. The study was approved by the
Ethics Committee of Xinqiao Hospital (Chongqing, China) on October
1, 2019 at the implementation of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank the Department of
Orthopedics of Xinqiao Hospital (Chongqing, China) for their
support.
References
|
1
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin BI, Deyo RA, Mirza SK, Turner JA,
Comstock BA, Hollingworth W and Sullivan SD: Expenditures and
health status among adults with back and neck problems. Jama.
299:656–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang W, Yu XH, Wang C, He WS, Zhang SJ,
Yan YG, Zhang J, Xiang YX and Wang WJ: Interleukin-1beta in
intervertebral disk degeneration. Clin Chim Acta. 450:262–272.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luoma K, Riihimäki H, Luukkonen R,
Raininko R, Viikari-Juntura E and Lamminen A: Low back pain in
relation to lumbar disc degeneration. Spine (Phila Pa 1976).
25:487–492. 2000. View Article : Google Scholar
|
|
5
|
Takatalo J, Karppinen J, Niinimäki J,
Taimela S, Näyhä S, Mutanen P, Sequeiros RB, Kyllönen E and
Tervonen O: Does lumbar disc degeneration on magnetic resonance
imaging associate with low back symptom severity in young finnish
adults? Spine (Phila Pa 1976). 36:2180–2189. 2011. View Article : Google Scholar
|
|
6
|
Antoniou J, Goudsouzian NM, Heathfield TF,
Winterbottom N, Steffen T, Poole AR, Aebi M and Alini M: The human
lumbar endplate. Evidence of changes in biosynthesis and
denaturation of the extracellular matrix with growth, maturation,
aging, and degeneration. Spine. 21:1153–1161. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guterl CC, See EY, Blanquer SB, Pandit A,
Ferguson SJ, Benneker LM, Grijpma DW, Sakai D, Eglin D, Alini M, et
al: Challenges and strategies in the repair of ruptured annulus
fibrosus. Eur Cell Mater. 25:1–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang X and X Li: Nucleus pulposus tissue
engineering: A brief review. Eur Spine J. 18:1564–1572. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Edgar MA: The nerve supply of the lumbar
intervertebral disc. J Bone Joint Surg Br. 89:1135–1139. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pattappa G, Li Z, Peroglio M, Wismer N,
Alini M and Grad S: Diversity of intervertebral disc cells:
Phenotype and function. J Anat. 221:480–496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine.
31:2151–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Antoniou J, Steffen T, Nelson F,
Winterbottom N, Hollander AP, Poole RA, Aebi M and Alini M: The
human lumbar intervertebral disc: Evidence for changes in the
biosynthesis and denaturation of the extracellular matrix with
growth, maturation, ageing, and degeneration. J Clin Invest.
98:996–1003. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wuertz K, Vo N, Kletsas D and Boos N:
Inflammatory and catabolic signalling in intervertebral discs: The
roles of NF-kB and MAP kinases. Eur Cell Mater. 23:103–119.
2012.
|
|
14
|
Kepler CK, Markova DZ, Dibra F, Yadla S,
Vaccaro AR, Risbud MV, Albert TJ and Anderson DG: Expression and
relationship of proinflammatory chemokine RANTES/CCL5 and cytokine
IL-1β in painful human intervertebral discs. Spine. 38:873–880.
2013. View Article : Google Scholar
|
|
15
|
Feng C, Liu H, Yang M, Zhang Y, Huang B
and Zhou Y: Disc cell senescence in intervertebral disc
degeneration: Causes and molecular pathways. Cell Cycle.
15:1674–1684. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sivan SS, Hayes AJ, Wachtel E, Caterson B,
Merkher Y, Maroudas A, Brown S and Roberts S: Biochemical
composition and turnover of the extracellular matrix of the normal
and degenerate intervertebral disc. Eur Spine J. 23(Suppl 3):
S344–S353. 2014. View Article : Google Scholar
|
|
17
|
Molinos M, Almeida CR, Caldeira J, Cunha
C, Gonçalves RM and Barbosa MA: Inflammation in intervertebral disc
degeneration and regeneration. J R Soc Interface. 12:201411912015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lorenzo P, Bayliss MT and Heinegard D: A
novel cartilage protein (CILP) present in the mid-zone of human
articular cartilage increases with age. J Biol Chem.
273:23463–23468. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seki S, Kawaguchi Y, Chiba K, Mikami Y,
Kizawa H, Oya T, Mio F, Mori M, Miyamoto Y, Masuda I, et al: A
functional SNP in CILP, encoding cartilage intermediate layer
protein, is associated with susceptibility to lumbar disc disease.
Nat Genet. 37:607–612. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lorenzo P, Neame P, Sommarin Y and
Heinegård D: Cloning and deduced amino acid sequence of a novel
cartilage protein (CILP) identifies a proform including a
nucleotide pyrophospho-hydrolase. J Biol Chem. 273:23469–23475.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cardenal A, Masuda I, Haas AL, Ono W and
McCarty DJ: Identification of a nucleotide pyrophosphohydrolase
from articular tissues in human serum. Arthritis Rheum. 39:252–256.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Masuda I, Cardenal A, Ono W, Hamada J,
Haas AL and McCarty DJ: Nucleotide pyrophosphohydrolase in human
synovial fluid. J Rheumatol. 24:1588–1594. 1997.PubMed/NCBI
|
|
23
|
Wang Z, Kim JH, Higashino K, Kim SS, Wang
S, Seki S, Hutton WC and Yoon ST: Cartilage intermediate layer
protein (CILP) regulation in intervertebral discs. The effect of
age, degeneration, and bone morphogenetic protein-2. Spine (Phila
Pa 1976). 37:E203–E208. 2012. View Article : Google Scholar
|
|
24
|
He J, Feng C, Sun J, Lu K, Chu T, Zhou Y
and Pan Y: Cartilage intermediate layer protein is regulated by
mechanical stress and affects extracellular matrix synthesis. Mol
Med Rep. 17:6130–6137. 2018.PubMed/NCBI
|
|
25
|
Seki S, Tsumaki N, Motomura H, Nogami M,
Kawaguchi Y, Hori T, Suzuki K, Yahara Y, Higashimoto M, Oya T, et
al: Cartilage intermediate layer protein promotes lumbar disc
degeneration. Biochem Biophys Res Commun. 446:876–881. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yee A, Lam MP, Tam V, Chan WC, Chu IK,
Cheah KS, Cheung KM and Chan D: Fibrotic-like changes in degenerate
human intervertebral discs revealed by quantitative proteomic
analysis. Osteoarthritis Cartilage. 24:503–513. 2016. View Article : Google Scholar
|
|
27
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88(Suppl 2): S10–S14. 2006.
|
|
28
|
Kanayama M, Togawa D, Takahashi C, Terai T
and Hashimoto T: Cross-sectional magnetic resonance imaging study
of lumbar disc degeneration in 200 healthy individuals. J Neurosurg
Spine. 11:501–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Adams MA, Freeman BJ, Morrison HP, Nelson
IW and Dolan P: Mechanical initiation of intervertebral disc
degeneration. Spine (Phila Pa 1976). 25:1625–1636. 2000. View Article : Google Scholar
|
|
30
|
Battie MC, Videman T, Kaprio J, Gibbons
LE, Gill K, Manninen H, Saarela J and Peltonen L: The Twin Spine
Study: Contributions to a changing view of disc degeneration. Spine
J. 9:47–59. 2009. View Article : Google Scholar
|
|
31
|
Wang D, Nasto LA, Roughley P, Leme AS,
Houghton AM, Usas A, Sowa G, Lee J, Niedernhofer L, Shapiro S, et
al: Spine degeneration in a murine model of chronic human tobacco
smokers. Osteoarthritis Cartilage. 20:896–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Le Maitre CL, Freemont AJ and Hoyland JA:
The role of interleukin-1 in the pathogenesis of human
intervertebral disc degeneration. Arthritis Res Ther. 7:R732–R745.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Le Maitre CL, Hoyland JA and Freemont AJ:
Catabolic cytokine expression in degenerate and herniated human
intervertebral discs: IL-1beta and TNFalpha expression profile.
Arthritis Res Ther. 9:R772007. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Karli P, Martlé V, Bossens K, Summerfield
A, Doherr MG, Turner P, Vandevelde M, Forterre F and Henke D:
Dominance of chemokine ligand 2 and matrix metalloproteinase-2 and
-9 and suppression of pro-inflammatory cytokines in the epidural
compartment after intervertebral disc extrusion in a canine model.
Spine J. 14:2976–2984. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamamoto J, Maeno K, Takada T, Kakutani K,
Yurube T, Zhang Z, Hirata H, Kurakawa T, Sakai D, Mochida J, et al:
Fas ligand plays an important role for the production of
pro-inflammatory cytokines in intervertebral disc nucleus pulposus
cells. J Orthop Res. 31:608–615. 2013. View Article : Google Scholar
|
|
36
|
Kepler CK, Markova DZ, Hilibrand AS,
Vaccaro AR, Risbud MV, Albert TJ and Anderson DG: Substance P
stimulates production of inflammatory cytokines in human disc
cells. Spine (Phila Pa 1976). 38:E1291–E1299. 2013. View Article : Google Scholar
|
|
37
|
Singh K, Masuda K, Thonar EJ, An HS and
Cs-Szabo G: Age-related changes in the extracellular matrix of
nucleus pulposus and anulus fibrosus of human intervertebral disc.
Spine (Phila Pa 1976). 34:10–16. 2009. View Article : Google Scholar
|
|
38
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Freemont AJ, Watkins A, Le Maitre C, Baird
P, Jeziorska M, Knight MT, Ross ER, O'Brien JP and Hoyland JA:
Nerve growth factor expression and innervation of the painful
intervertebral disc. J Pathol. 197:286–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Melrose J, Roberts S, Smith S, Menage J
and Ghosh P: Increased nerve and blood vessel ingrowth associated
with proteoglycan depletion in an ovine anular lesion model of
experimental disc degeneration. Spine (Phila Pa 1976).
27:1278–1285. 2002. View Article : Google Scholar
|
|
41
|
Harshitha SM, Sibin MK, Chetan GK and
Dhananjaya I Bhat: Association of CILP, COL9A2 and MMP3 gene
polymorphisms with lumbar disc degeneration in an Indian
population. J Mol Neurosci. 66:378–382. 2018. View Article : Google Scholar
|
|
42
|
Min SK, Nakazato K, Yamamoto Y, Gushiken
K, Fujimoto H, Fujishiro H, Kobayakawa Y and Hiranuma K: Cartilage
intermediate layer protein gene is associated with lumbar disc
degeneration in male, but not female, collegiate athletes. Am J
Sports Med. 38:2552–2557. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nakamura I, Okawa A, Ikegawa S, Takaoka K
and Nakamura Y: Genomic organization, mapping, and polymorphisms of
the gene encoding human cartilage intermediate layer protein
(CILP). J Hum Genet. 44:203–205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lorenzo P, Aman P, Sommarin Y and
Heinegård D: The human CILP gene: Exon/intron organization and
chromosomal mapping. Matrix Biol. 18:445–454. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mori M, Nakajima M, Mikami Y, Seki S,
Takigawa M, Kubo T and Ikegawa S: Transcriptional regulation of the
cartilage intermediate layer protein (CILP) gene. Biochem Biophys
Res Commun. 341:121–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hirose J, Ryan LM and Masuda I:
Up-regulated expression of cartilage intermediate-layer protein and
ANK in articular hyaline cartilage from patients with calcium
pyrophosphate dihydrate crystal deposition disease. Arthritis
Rheum. 46:3218–3129. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bernardo BC, Belluoccio D, Rowley L,
Little CB, Hansen U and Bateman JF: Cartilage intermediate layer
protein 2 (CILP-2) is expressed in articular and meniscal cartilage
and down-regulated in experimental osteoarthritis. J Biol Chem.
286:37758–33767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang W, Hao J, Zheng S, Xiao X, Wen Y, He
A, Guo X and Zhang F: Association between cartilage intermediate
layer protein and degeneration of intervertebral disc: A
meta-analysis. Spine (Phila Pa 1976). 41:E1244–E1248. 2016.
View Article : Google Scholar
|
|
49
|
Virtanen IM, Song YQ, Cheung KM, Ala-Kokko
L, Karppinen J, Ho DW, Luk KD, Yip SP, Leong JC, Cheah KS, et al:
Phenotypic and population differences in the association between
CILP and lumbar disc disease. J Med Genet. 44:285–288. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rajasekaran S, Kanna RM, Senthil N,
Raveendran M, Cheung KM, Chan D, Subramaniam S and Shetty AP:
Phenotype variations affect genetic association studies of
degenerative disc disease: Conclusions of analysis of genetic
association of 58 single nucleotide polymorphisms with highly
specific phenotypes for disc degeneration in 332 subjects. Spine J.
13:1309–1320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Min SK, Nakazato K, Ishigami H and
Hiranuma K: Cartilage intermediate layer protein and asporin
polymorphisms are independent risk factors of lumbar disc
degeneration in male collegiate athletes. Cartilage. 5:37–42. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kelempisioti A, Eskola PJ, Okuloff A,
Karjalainen U, Takatalo J, Daavittila I, Niinimäki J, Sequeiros RB,
Tervonen O, Solovieva S, et al: Genetic susceptibility of
intervertebral disc degeneration among young Finnish adults. BMC
Med Genet. 12:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Taipale M, Solovieva S, Leino-Arjas P and
Männikkö M: Functional polymorphisms in asporin and CILP together
with joint loading predispose to hand osteoarthritis. BMC Genetics.
18:1082017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Park S, Ranjbarvaziri S, Zhao P and
Ardehali R: Cardiac fibrosis is associated with decreased
circulating levels of full-length CILP in heart failure. JACC Basic
Transl Sci. 5:432–443. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hayes AJ, Benjamin M and Ralphs JR:
Extracellular matrix in development of the intervertebral disc.
Matrix Biol. 20:107–121. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pattison ST, Melrose J, Ghosh P and Taylor
TK: Regulation of gelatinase-A (MMP-2) production by ovine
intervertebral disc nucleus pulposus cells grown in alginate bead
culture by Transforming Growth Factor-beta(1)and insulin like
growth factor-I. Cell Biol Int. 25:679–689. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen J, Yan W and Setton LA: Static
compression induces zonal-specific changes in gene expression for
extracellular matrix and cytoskeletal proteins in intervertebral
disc cells in vitro. Matrix Biol. 22:573–583. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang H, Kroeber M, Hanke M, Ries R, Schmid
C, Poller W and Richter W: Release of active and depot GDF-5 after
adenovirus-mediated overexpression stimulates rabbit and human
intervertebral disc cells. J Mol Med (Berl). 82:126–134. 2004.
View Article : Google Scholar
|
|
59
|
Nishida K, Kang JD, Gilbertson LG, Moon
SH, Suh JK, Vogt MT, Robbins PD and Evans CH: Modulation of the
biologic activity of the rabbit intervertebral disc by gene
therapy: An in vivo study of adenovirus-mediated transfer of the
human transforming growth factor beta 1 encoding gene. Spine (Phila
Pa 1976). 24:2419–2425. 1999. View Article : Google Scholar
|
|
60
|
Grimaud E, Heymann D and Redini F: Recent
advances in TGF-beta effects on chondrocyte metabolism. Potential
therapeutic roles of TGF-beta in cartilage disorders. Cytokine
Growth Factor Rev. 13:241–257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
de Caestecker M: The transforming growth
factor-beta super-family of receptors. Cytokine Growth Factor Rev.
15:1–11. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Border WA, Noble NA, Yamamoto T, Harper
JR, Yamaguchi YU, Pierschbacher MD and Ruoslahti E: Natural
inhibitor of transforming growth factor-beta protects against
scarring in experimental kidney disease. Nature. 360:361–364. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Isaka Y, Brees DK, Ikegaya K, Kaneda Y,
Imai E, Noble NA and Border WA: Gene therapy by skeletal muscle
expression of decorin prevents fibrotic disease in rat kidney. Nat
Med. 2:418–423. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Laron Z: Insulin-like growth factor 1
(IGF-1): A growth hormone. Mol Pathol. 54:311–316. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Foulstone E, Prince S, Zaccheo O, Burns
JL, Harper J, Jacobs C, Church D and Hassan AB: Insulin-like growth
factor ligands, receptors, and binding proteins in cancer. J
Pathol. 205:145–153. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu ZQ, Zhao S and Fu WQ: Insulin-like
growth factor 1 antagonizes lumbar disc degeneration through
enhanced autophagy. Am J Transl Res. 8:4346–4353. 2016.PubMed/NCBI
|
|
68
|
Johnson K, Farley D, Hu SI and Terkeltaub
R: One of two chondrocyte-expressed isoforms of cartilage
intermediate-layer protein functions as an insulin-like growth
factor 1 antagonist. Arthritis Rheum. 48:1302–1314. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Olmez U, Ryan LM, Kurup IV and Rosenthal
AK: Insulin-like growth factor-1 suppresses pyrophosphate
elaboration by transforming growth factor beta1-stimulated
chondrocytes and cartilage. Osteoarthritis Cartilage. 2:149–154.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Abu Shehab M, Iosef C, Wildgruber R,
Sardana G and Gupta MB: Phosphorylation of IGFBP-1 at discrete
sites elicits variable effects on IGF-I receptor
autophosphorylation. Endocrinology. 154:1130–1143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Russell RG, Bisaz S, Fleisch H, Currey HL,
Rubinstein HM, Dietz AA, Boussina I, Micheli A and Fallet G:
Inorganic pyro-phosphate in plasma, urine, and synovial fluid of
patients with pyrophosphate arthropathy (chondrocalcinosis or
pseudogout). Lancet. 2:899–902. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Johnson K and Terkeltaub R: Inorganic
pyrophosphate (PPI) in pathologic calcification of articular
cartilage. Front Biosci. 10:988–997. 2005. View Article : Google Scholar
|
|
73
|
Liu MH, Sun C, Yao Y, Fan X, Liu H, Cui
YH, Bian XW, Huang B and Zhou Y: Matrix stiffness promotes
cartilage endplate chon-drocyte calcification in disc degeneration
via miR-20a targeting ANKH expression. Sci Rep. 6:254012016.
View Article : Google Scholar
|
|
74
|
Roberts S, Urban JP, Evans H and
Eisenstein SM: Transport properties of the human cartilage endplate
in relation to its composition and calcification. Spine (Phila Pa
1976). 21:415–420. 1996. View Article : Google Scholar
|
|
75
|
Moore RJ: The vertebral endplate: Disc
degeneration, disc regeneration. Eur Spine J. 15(Suppl 3 (Suppl
3)): S333–S337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lefebvre V, Li P and De Crombrugghe B: A
new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in
chondrogenesis and cooperatively activate the type II collagen
gene. Embo J. 17:5718–5733. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
de Crombrugghe B, Lefebvre V, Behringer
RR, Bi W, Murakami S and Huang W: Transcriptional mechanisms of
chondrocyte differentiation. Matrix Biol. 19:389–394. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ikeda T, Kamekura S, Mabuchi A, Kou I,
Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S and Chung UI:
The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides
signals sufficient for induction of permanent cartilage. Arthritis
Rheum. 50:3561–3573. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang C, Yu X, Yan Y, Yang W, Zhang S,
Xiang Y, Zhang J and Wang W: Tumor necrosis factor-α: A key
contributor to intervertebral disc degeneration. Acta Biochim
Biophys Sin (Shanghai). 49:1–13. 2017. View Article : Google Scholar
|
|
80
|
Clutterbuck AL, Smith JR, Allaway D,
Harris P, Liddell S and Mobasheri A: High throughput proteomic
analysis of the secretome in an explant model of articular
cartilage inflammation. J Proteomics. 74:704–715. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tran CM, Shapiro IM and Risbud MV:
Molecular regulation of CCN2 in the intervertebral disc: Lessons
learned from other connective tissues. Matrix Biol. 32:298–306.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang CL, Zhao Q, Liang H, Qiao X, Wang
JY, Wu D, Wu LL and Li L: Cartilage intermediate layer protein-1
alleviates pressure overload-induced cardiac fibrosis via
interfering TGF-β1 signaling. J Mol Cell Cardiol. 116:135–144.
2018. View Article : Google Scholar : PubMed/NCBI
|