Introduction
During the process of bone development and
remodeling, there is a dynamic balance between bone formation and
bone resorption (1,2). When this balance is disrupted,
progressive osseous hyperplasia or osteoporosis eventually occur
(3). Several diseases are caused
by imbalance of bone formation and bone resorption. For instance,
excessive bone formation can lead to joint ankylosis in ankylosing
spondylitis and osteophyte in osteoarthritis (4,5).
Moreover, excessive bone resorption in pathological conditions,
such as immobility and corticosteroid overuse, can lead to
osteoporosis (6). Axial
spondyloarthritis (axSpA) is a chronic inflammatory disease
characterized by structural damage which incorporates aspects of
bone destruction and new bone formation (4).
The cellular activities of osteoblasts and
osteoclasts are the main factors that maintain the dynamic bone
balance; osteoblasts are responsible for bone formation, while
osteoclasts are responsible for bone resorption (1,2).
The cellular activities of osteoblasts are controlled by external
signals. For example, bone morphogenetic proteins, a group of
molecules belonging to the TGF-β superfamily, can induce the
differentiation of osteoblasts (6). Another family of growth factors
that are implicated in osteoblast differentiation is the Wnt
protein family (7). Several
transcription factors have also been identified to engage in the
process of osteoblast differentiation, including RUNX family
transcription factor 2, Osterix and activating transcription factor
4 (8). The transcription factor
Kaiso, which is a unique member of the poxviruses and zinc finger
(POZ) family, exhibits dual-specificity DNA binding at methylated
CpG dinucleotides or at a non-methylated sequence, known as the
Kaiso binding site (KBS) (9,10). Kaiso has diverse functions in the
regulation of inflammation, cell proliferation and cell cycle
(11-15). Several studies have revealed that
Kaiso has an effect on Wnt signaling, which is critical in the
regulation of bone homeostasis (16-18). However, to the best of our
knowledge, no study has investigated the role of Kaiso in
osteoblast differentiation and bone formation to date.
In the present study, a HuProt microarray was used
to identify autoantibodies in patients with axSpA. Gain- and
loss-of-function studies in MC3T3-E1 cells demonstrated that Kaiso
served a critical role in osteoblast differentiation in
vitro. The findings were further confirmed in vivo. An
Illumina HiSeq sequencer was used to analyze the mechanism of Kaiso
in regulating osteoblast differentiation and mineralization. Taken
together, the present results suggested that Kaiso regulated the
differentiation of osteoblasts via the Itga10/PI3K/AKT signaling
pathway.
Materials and methods
Biologic samples
The study was approved by the Medical Ethics
Committee of Changhai Hospital (approval no. CHEC2017-163) and
written informed consent was obtained from all patients (age,
28.57±3.95 years; 5 men; 2 women). Serum samples used for the
protein chip were obtained in patients (seven patients with axSpA
vs. seven healthy controls) recruited from the outpatient clinic of
Changhai Hospital between 2018.01 and 2019.01. The HuProt
microarrays used in this study were provided by CDI Laboratories,
Inc. The array contained duplicate spots of ~20,000 individually
purified human proteins with an N-terminal glutathione
S-transferase tag.
Bone marrow mesenchymal stem cells were extracted
from six patients with femoral neck fractures who underwent
surgical treatment recruited from the Changhai Hospital between
2018.01 and 2019.01.
Cell culture
The MC3T3-E1 (mouse pre-osteoblast) cell line was
obtained from The Cell Bank of Type Culture Collection of Chinese
Academy of Science and grown in α-minimal essential medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 0.1 mg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.). To induce
mineralization, MC3T3-E1 cells were treated for different time
points (1, 3, 5, 7, 14 and 21 days) with osteogenic medium (OM),
which was composed of complete medium supplemented with 50
µg/ml ascorbic acid (Sigma-Aldrich; Merck KGaA), 10 mM
β-glycerophosphate (Sigma-Aldrich; Merck KGaA) and 50 ng/ml BMP2
(Novus Biologicals, LLC) in a humidified incubator at 37°C under 5%
CO2. The medium was changed every 3 days. To validate
the role of the PI3K/AKT signaling pathway in osteogenic
differentiation, 15 mM PI3K inhibitor (LY294002; Selleck Chemicals)
was added to the OM (4°C for 3 days) and the medium changed every 3
days.
Preparation and transfection of
lentivirus
For gene overexpression, a lentivirus expression
vector was constructed by inserting full-length Kaiso cDNA into the
pLenti-EF1a- EGFP-F2A-Puro-CMV-MCS vector (Obio Technology Co.,
Ltd.), to generate LV-Kaiso. Empty vector plasmid was used as
control. For gene silencing, Kaiso-targeting short hairpin RNA
(shRNA) or Itga10-targeting shRNA were inserted into the
PLKD-CMV-EGFP-Puro-U6 vector (Obio Technology Co., Ltd.). The
sequences of the shRNAs are summarized in Table SI (supplied by Obio Technology
Co., Ltd.). The reconstructed vectors were then transfected into
293T cells, and the lentiviral particles were collected at 48 h
post-transfection. The construction and production of the
lentivirus were performed by Obio Technology Co., Ltd. MC3T3-E1
cells were infected with viruses (at a multiplicity of infection of
50) in the presence of 5 µg/ml polybrene (37°C for 24 h),
and replaced it with fresh medium after 24 h. After 72 h of
infection, the fluorescence expression was observed by fluorescence
microscope at a magnification of ×100. Then the cells were selected
using puromycin (2 µg/ml) to establish stable cell lines.
Small interfering (si)RNAs were transfected using
Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of the siRNAs are summarized in
Table SI.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and RT (37°C
for 15 min and 85°C for 5 sec, and cooled to 4°C) was performed
with a PrimeScript RT Reagent kit (Takara Bio, Inc.) according to
the manufacturer's protocols. RT-qPCR was performed (95°C
pre-incubation for 5 min, followed by 40 cycles at 95°C for 20 sec,
60°C for 15 sec and 72°C for 20 sec, and cooled to 4°C) on an ABI
Prism 7500 Sequence Detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using SYBR Premix Ex Taq™ (Takara Bio,
Inc.). The relative expression of each target gene was quantified
by calculating the quantification cycle value, and was normalized
to the levels of β-actin. The relative expression levels were
calculated based on the 2−ΔΔCq method (19). Each sample was analyzed in
triplicate. The primer sequences are listed in Table SII.
Western blot analysis
Cells were lysed with RIPA buffer supplemented with
protease inhibitors (Thermo Fisher Scientific, Inc.). The Pierce™
Rapid Gold BCA Protein Assay kit (Thermo Fisher Scientific, Inc.)
was used for protein determination. Equal quantities of proteins
(20 µg) were separated by 6 or 10% SDS-PAGE and transferred
to PVDF membranes (EMD Millipore). After pre-incubation in 5%
non-fat milk for 1 h, the membranes were incubated with primary
antibodies for 12-16 h at 4°C. The membranes were then incubated
with appropriate horseradish peroxidase-conjugated secondary
antibodies (cat. no. 7074; 1:3,000; Cell Signaling Technology,
Inc.) for 1 h at room temperature. The immunoreactive protein bands
were visualized using an enhanced chemiluminescence detection
system (Cytiva). The primary antibodies used in this study were as
follows: Anti-Kaiso (cat. no. 12723; 1:1,000; Abcam), anti-Itga10
(cat. no. PA5-67829; 1:1,000; Merck KGaA), anti-p85 (cat. no. 4292;
1:1,000; Cell Signaling Technology, Inc.), anti-AKT (cat. no. 9272;
1:1,000; Cell Signaling Technology, Inc.), anti-phosphorylated
(p)-AKT (cat. no. 9271; 1:1,000; Cell Signaling Technology, Inc.)
and anti-β-tubulin (cat. no. 2146; 1:1,000; Cell Signaling
Technology, Inc.). The relative semi-quantification of western
blotting was performed using ImageJ software (1.52a; National
Institutes of Health).
Alkaline phosphatase (ALP) activity
assay
After treatment with OM for 1, 3, 5 and 7 days,
MC3T3-E1 cells were rinsed with PBS, solubilized in lysis buffer
(50 mM Tris, 100 mM glycine) for 5 min and centrifuged (2,000 × g;
room temperature; 5 min) to collect the supernatant. The ALP
activity in the supernatant was then determined using ALP Assay
kits (Nanjing Jiancheng Bioengineering Institute) according to the
manufacturer's instructions. The ALP activity was normalized to
total the protein concentration, which was determined using BCA
Protein Assay kits (Thermo Fisher Scientific, Inc.).
ALP staining and Alizarin Red S
staining
MC3T3-E1 cells were cultured in 24-well plates at a
density of 3×104 cells per well. After treatment with OM
(37°C) for a different numbers of days (1, 3, 5, 7, 14 and 21
days), cells were washed with PBS, fixed with 4% formaldehyde for
30 min (room temperature) and rinsed with double-distilled
H2O. For ALP staining, ALP Assay kits (Beyotime
Institute of Biotechnology) were used according to the
manufacturer's protocol. For detection of mineral deposition, the
fixed cells were stained with 1% Alizarin Red S solution
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min. All the
stained cells were rinsed three times with double-distilled
H2O and then imaged using a camera.
Sequencing analysis of mRNA
Total RNA was extracted from stable cell lines
transfected with LV-ctr, LV-Kaiso, Sh-ctr and Sh-Kaiso-1 using
TRIzol reagent. The quantity and purity of total RNA were monitored
using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc.) and an Agilent 2200 Bioanalyzer
(Agilent Technologies, Inc.). Library construction and sequencing
on the Illumina HiSeq 2000 platform (Illumina, Inc.) were performed
by Guangzhou RiboBio Co., Ltd.
Gene Ontology (GO) (http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analyses (https://www.genome.jp/kegg/) were performed to
determine the roles of the differentially expressed genes in
biological pathways. Protein-protein interaction (PPI) network
analysis (https://string-db.org/) was performed to
identify the key genes in the pathways. P<0.05 was considered to
indicate statistically significant enrichment.
Chromatin immunoprecipitation (ChIP)
assay
JASPAR (http://jaspar.genereg.net/) and MatInspector
(https://www.genomatix.de/online_help/help_matinspector/matinspector_help.html)
were used to predict the presence of Kaiso binding site in Itga10
promoter. ChIP assays were performed using an EZ ChIP kit (cat. no.
17-10086; EMD Millipore) according to the manufacturer's protocol.
In brief, cells were fixed with 1% formaldehyde for 10 min (37°C)
to crosslink chromatin complexes before the addition of 125 mM
glycine to stop the reaction. After washing with PBS, the samples
were then sonicated at 4°Cto obtain DNA fragments (400-1,000 bp).
The sonicator Bioruptor was used at mid-range and the cell samples
were sonicated for 10 min, 'ON' for 30 sec, 'OFF' for 30 sec and
centrifuged at 4°C for 10 min at 2,000 × g to remove insoluble
materials. Immunoprecipitation was performed (4°C for 24 h) using a
Kaiso-specific antibody (cat. no. 12723; 1:1,000; Abcam) or an IgG
control antibody. Protein A/G beads (Thermo Fisher Scientific,
Inc.) were used to pull down the immune complexes. The protein-DNA
complexes were eluted with buffer containing 1% SDS and 0.1 M
NaHCO3, and crosslinks were reversed at 65°C overnight.
DNA was purified and used for PCR. The following Itga10 primers
were used for ChIP assay: Forward, 5′-CCG AAT GGA AGA TGA GAG
ACA-3′ and reverse, 5′-TCG GAG GTT AAT ACC GAT GC-3′.
Luciferase reporter assay
To generate the Itga10 luciferase reporter plasmids,
a 1,051-bp DNA fragment upstream of the Itga10 transcription start
site was amplified via PCR from genomic human DNA and cloned into
the pGL4.10-Basic vector (Promega Corporation). The predicted Kaiso
binding sites (5′-AATCCTGCTAC-3′) and the corresponding mutated
Kaiso binding sites (5′-TGG CTC GTG T-3′) were also subcloned into
the vector using standard cloning strategies (20). For luciferase assays, MC3T3-E1
cells were cultured to 70-80% confluence in 24-well plates before
co-transfection with the luciferase reporter vector (0.1 µg)
and Kaiso overexpression vectors or control (0.1 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions
(37°C for 48 h). At 48 h after transfection, the luciferase
activity was measured using the Dual Luciferase Reporter Assay
system (Promega Corporation), and each experiment was repeated in
triplicate. The transfection efficiency was normalized by
determining the activity of Renilla luciferase.
Osteogenesis assay in vivo
NOD/SCID mice (male; age, 8 weeks; 18-22 g; n=21)
purchased from SLRC Laboratory. Mice were bred under specific
pathogen-free conditions at a constant temperature (23±1°C) with
55±15% humidity and a 12-h light-dark cycle (09:00-21:00). All the
mice had free access to food and water. For the in vivo
osteogenic ectopic assay, a total of 2×106 MC3T3-E1 were
seeded on a β-tricalcium phosphate (β-TCP) scaffold (Shanghai
Bio-lu Biomaterials Co., Ltd.). The implants loaded with cells were
implanted into an intramuscular pocket of the right femur of mice
(21). The mice were
anesthetized with an injection of 1% pentobarbital sodium (70
mg/kg) into the abdominal cavity. The mice were randomly divided
into seven groups containing three mice per group: i) β-TCP
(vehicle); ii) β-TCP loading LV-ctr MC3T3-E1 cells (LV-ctr); iii)
β-TCP loading LV-Kaiso MC3T3-E1 cells (LV-Kaiso); iv) β-TCP loading
Sh-ctr MC3T3-E1 cells (Sh-ctr); v) β-TCP loading Sh-Kaiso MC3T3-E1
cells (Sh-Kaiso); vi) β-TCP loading Sh-ctr MC3T3-E1 cells (Sh-ctr);
and vii) β-TCP loading Sh-Itga10 MC3T3-E1 cells (Sh-Itga10). At 8
weeks post-implantation, the mice were sacrificed via carbon
dioxide asphyxia. Briefly, the CO2 flow rate was started
with 8 l/min in a 30-lchamber. The implants were harvested and
fixed in 4% paraformaldehyde (room temperature for 24 h) for
µ-computed tomography (µCT) analysis. The bone
mineral density (BMD) of implants was then generated by skyscan
1176 (22). The implants were
decalcified in 10% EDTA and embedded in paraffin. Then 5-µm
sections were stained with hematoxylin and eosin (cat. no. C0105M;
Beyotime Institute of Biotechnology) for 10 min at room
temperature, and washed with distilled water for 10 min. Sections
were observed and imaged using a light microscope (E200; Nikon
Corporation) at a magnification of ×200. All animal studies were
approved by the Animal Ethics Committee of Changhai Hospital.
Statistical analysis
SPSS software (version 20; IBM Corp.) was used for
statistical analysis. Differences between two groups were observed
using unpaired Student's t-tests, and differences in the results
derived from experiments with >2 groups were analyzed using
one-way ANOVA followed by Bonferroni post-hoc test. P<0.05 was
considered to indicate a statistically significant difference. The
experiment was repeated three times independently.
Results
Autoantibodies in axSpA
The present study used the HuProt microarray to
identify autoantibodies in patients with axSpA and healthy
controls. The results demonstrated that seven candidate
autoantibodies [including microtubule associated protein 9, BAF
chromatin remodelling complex subunit BCL7A, argonaute RISC
component 1, immunoglobulin heavy constant γ1 (G1m marker), myosin
light chain kinase, SGT1 homolog, MIS12 kinetochore complex
assembly cochaperone andKaiso (also known as zinc finger and BTB
domain containing 33)] were present specifically in serum from the
patients with axSpA but not from that of the healthy controls
(Fig. S1).
Kaiso regulates osteoblast
differentiation in MC3T3-E1 cells
The expression of Kaiso was found to be
downregulated during osteoblast differentiation in MC3T3-E1 cells
(Fig. 1A). To investigate the
role of Kaiso in the regulation of osteoblast differentiation,
MC3T3-E1 cells were infected with a lentivirus expressing Kaiso or
control virus to produce a stable overexpressing cell line
(LV-Kaiso) or control cell line (LV-ctr). Stable overexpression of
Kaiso was confirmed via western blot analyses and qPCR (Fig. 1B and C). To examine the
osteoblast differentiation ability, the stable cell lines were
cultured for the indicated periods in OM, and mRNA expression of
the osteogenic marker genes, including ALP, osteocalcin (OCN) and
bone sialoprotein (BSP), were examined. The mRNA expression levels
of ALP, OCN and BSP were significantly decreased in the LV-Kaiso
group compared with those in LV-ctr group after treatment with OM
for 7 days (Fig. 1D). Moreover,
ALP activity assays and ALP staining demonstrated that Kaiso
overexpression suppressed ALP activity in MC3T3-E1 cells after
induction by OM for different periods (Fig. 1E and F). The effects of Kaiso on
late osteoblast differentiation were examined using Alizarin Red S
staining to assess mineral bone nodule formation. The Alizarin Red
staining results further indicated that Kaiso overexpression
notably suppressed the osteogenic differentiation of MC3T3-E1 cells
(Fig. 1G).
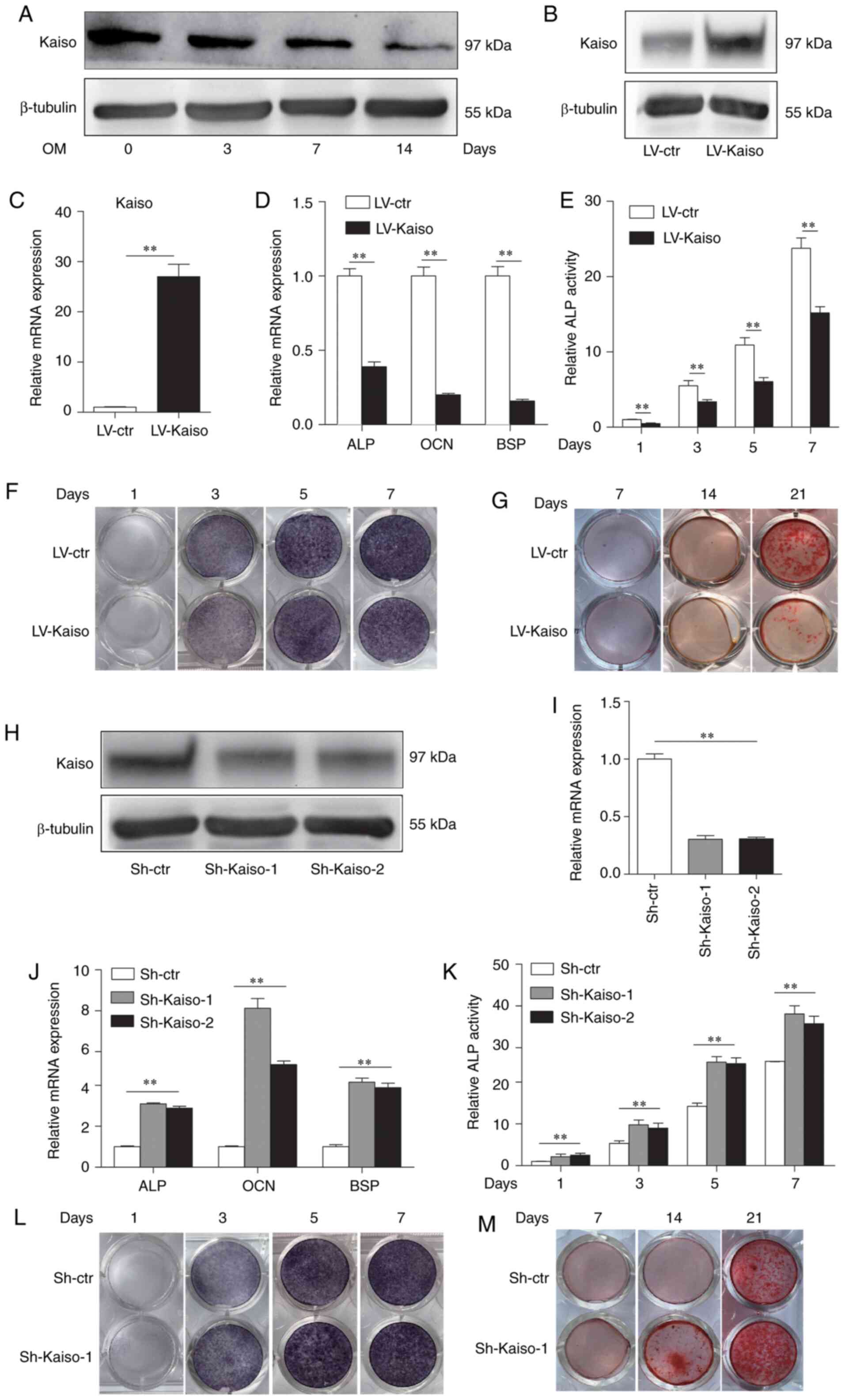 | Figure 1Overexpression or knocking down of
Kaiso can inhibitor promote osteoblast differentiation in MC3T3-E1
cells. (A) Kaiso expression during osteogenic induction was
determined via western blot analysis. (B) Western blot analysis and
(C) RT-qPCR of Kaiso expression in LV-Kaiso and LV-ctr infected
MC3T3-E1 cells. (D) RT-qPCR analysis of ALP, OCN and BSP expression
levels was performed in LV-Kaiso and LV-Ctr infected MC3T3-E1 cells
after 7 days of culture with OM. (E) ALP activity was measured on
days 1, 3, 5 and 7 after culture with OM. The results are presented
as the value relative to that at day 1. (F) ALP staining was
performed on days 1, 3, 5 and 7 in the presence of OM. (G)
Mineralized nodule formation was detected with Alizarin Red S
staining on days 7, 14 and 21. (H) Western blot analysis and (I)
RT-qPCR analysis of Kaiso expression in Sh-Kaiso-1, Sh-Kaiso-2 and
Sh-ctr transfected MC3T3-E1 cells. (J) RT-qPCR analysis of ALP, OCN
and BSP expression levels was performed in Sh-ctr and Sh-Kaiso
transfected MC3T3-E1 cells after 7 days of culture with OM. (K) ALP
activity. (L) ALP staining. (M) Mineralized nodule formation. All
the data were analyzed in triplicate. Data are presented as the
mean ± SD. **P<0.01. P-values of C-E were based on
Student's t-test and P-values of I-K were analyzed using one-way
ANOVA. RT-qPCR, reverse transcription-quantitative PCR; OM,
osteogenic medium; ALP, alkaline phosphatase; OCN, osteocalcin;
BSP, bone sialoprotein; LV, lentivirus; shRNA, short hairpin RNA;
Ctr, control. |
To further clarify the role of Kaiso in osteoblast
differentiation, Kaiso knockdown was conducted in MC3T3-E1 cells
using shRNA (Sh-Kaiso-1 andSh-Kaiso-2). Control cells were also
prepared with Sh-ctr (Fig. 1H and
I). By contrast to those in Sh-ctr cells, the mRNA expression
levels of ALP, OCN and BSP were increased in Sh-Kaiso MC3T3-E1
cells (Fig. 1J). As Sh-Kaiso-1
was more active, this transfectant was used for further
experiments. ALP activity assays, ALP staining and Alizarin Red S
staining at different timepoints demonstrated stronger osteogenic
differentiation of Sh-Kaiso MC3T3-E1 cells compared with that of
Sh-ctr MC3T3-E1 cells (Fig.
1K-M). Similar results were observed in primary human bone
marrow-derived mesenchymal stem cells (Fig. S2). The expression of Kaiso was
also found to be downregulated during osteoblast differentiation in
primary human bone marrow-derived mesenchymal stem cells (BMSCs).
The mRNA expression levels of ALP, OCN and BSP were increased after
knockdown of Kaiso in BMSCs (Fig.
S2). These results suggested that Kaiso negatively regulated
osteoblast differentiation.
Kaiso regulates osteoblast
differentiation by modulating the PI3K/AKT signaling pathway in
MC3T3-E1 cells
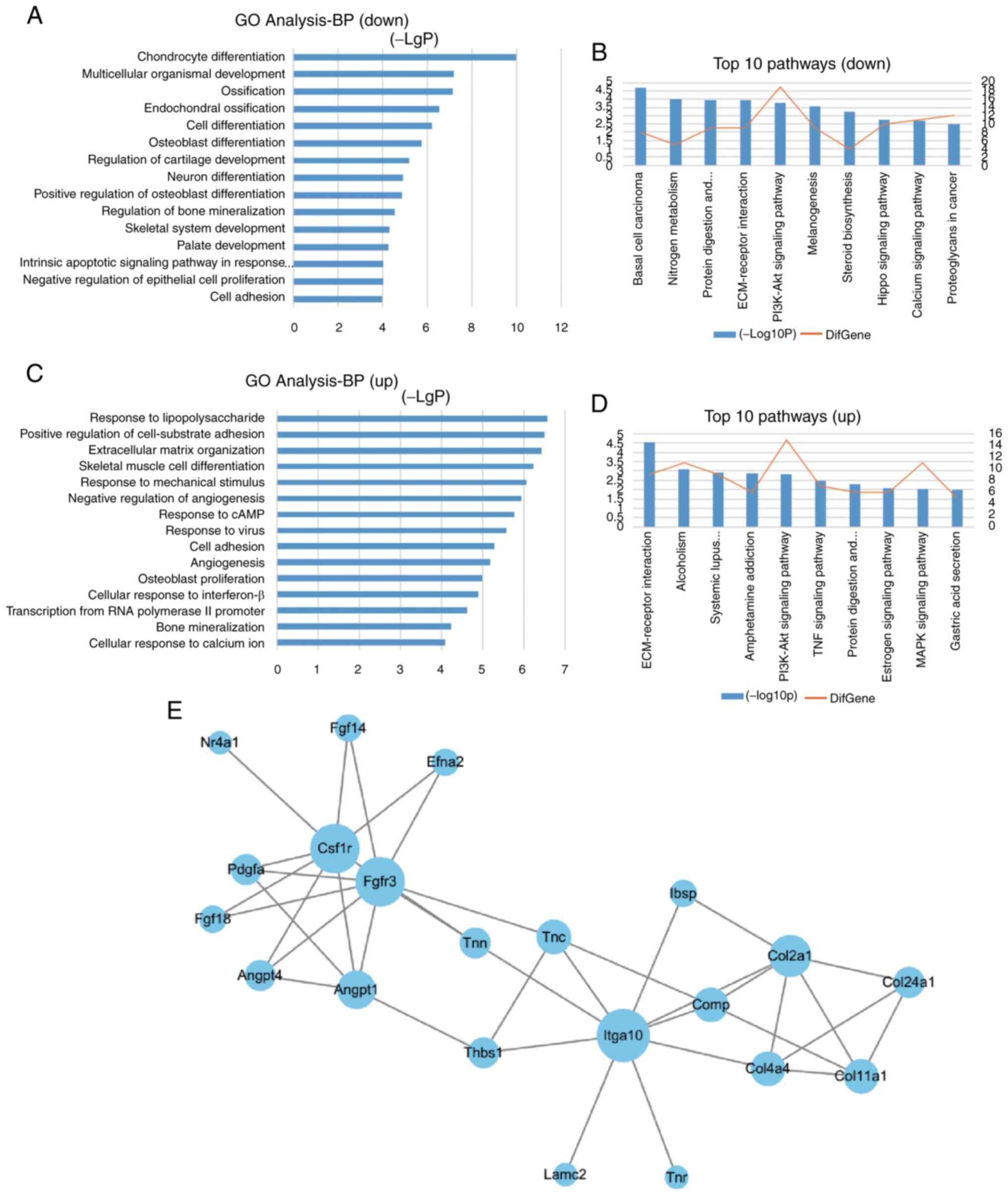
To gain insights into the molecular mechanisms via
which Kaiso regulates osteoblast differentiation, the Illumina
HiSeq sequencer was used to analyze changes in mRNA expression in
Kaiso-overexpressing or Kaiso-knocked down stable cell lines. GO
analysis of biological processes demonstrated that the
downregulated genes in LV-Kaiso cells and the upregulated genes in
Sh-Kaiso cells were significantly enriched in 'osteoblast
differentiation' (Fig. 2A and
C). KEGG pathway analysis of the differentially expressed genes
identified that the 'PI3K/AKT pathway' was inhibited in LV-Kaiso
MC3T3-E1 cells and enhanced in Sh-Kaiso MC3T3-E1 cells (Fig. 2B and D). PPI network analysis of
the differentially expressed genes enriched in the PI3K/AKT pathway
indicated that Itga10, colony stimulating factor 1 receptor (Csf1r)
andfibroblast growth factor receptor 3 (Fgfr3), with higher degrees
of connectivity than other genes, were network-centric proteins
(Fig. 2E).
To confirm the ability of Kaiso in regulating the
PI3K/AKT pathway, the expression levels of p85 and p-AKT were
determined via western blot analysis. The results demonstrated that
the expression levels of p85 and p-AKT were decreased in LV-Kaiso
MC3T3-E1 cells and increased in Sh-Kaiso MC3T3-E1 cells (Fig. 3A). To further evaluate the
involvement of the PI3K/AKT signaling pathway in the enhanced
osteoblast differentiation observed in Sh-Kaiso MC3T3-E1 cells, the
PI3K-specific inhibitor LY294002 was added into the cell culture,
and its effect on osteoblast differentiation was examined. LY294002
markedly blocked the enhancement of p-AKT and p85 protein
expression levels observed in Sh-Kaiso MC3T3-E1 cells (Fig. 3A). The intensity of p85 and p-AKT
was determined via densitometry using ImageJ software and
normalized to the loading control β-tubulin (Fig. S3). LY294002 also had significant
effects on osteoblast differentiation, as demonstrated by the
decreased mRNA expression levels of ALP, OCN and BSP (Sh-ctr vs.
Sh-ctr+LY; Sh-Kaiso-1vs. Sh-Kaiso-1+LY;Sh-ctr+LY vs. Sh-Kaiso-1+LY)
(Fig. 3B-D). The results of ALP
activity assays, ALP staining and Alizarin Red S staining further
demonstrated that LY294002 inhibited the increased ALP activity and
mineral bone nodule formation in Sh-Kaiso MC3T3-E1cells (Sh-ctr vs.
Sh-ctr+LY; Sh-Kaiso-1 vs. Sh-Kaiso-1+LY; Sh-ctr+LY vs.
Sh-Kaiso-1+LY) (Fig. 3E and F).
Thus, these findings indicated that Kaiso regulated the osteoblast
differentiation of MC3T3-E1 cells via a mechanism involving the
PI3K/AKT pathway.
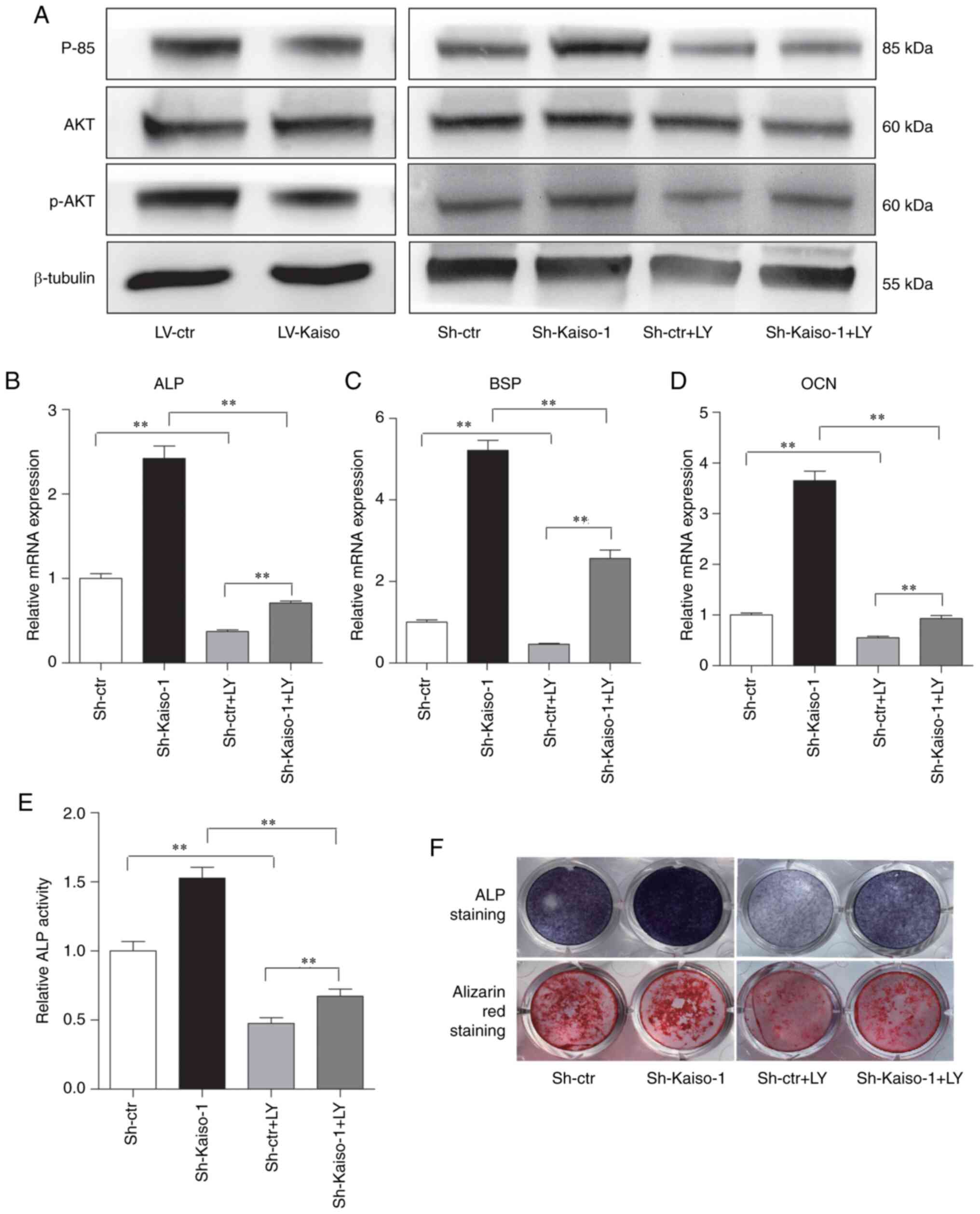 | Figure 3Kaiso regulates osteoblast
differentiation by modulating the PI3K/AKT pathway in MC3T3-E1
cells. (A) Western blot analysis was used to detect the protein
expression levels of p85, AKT and p-AKT in MC3T3-E1 cells with
Kaiso overexpression or Kaiso knockdown. Sh-Kaiso-1 and Sh-ctrl
cells were treated with LY, and the expression levels of p85, AKT
and p-AKT were examined via western blot analysis. RT-qPCR analysis
of (B) ALP, (C) BSP and (D) OCN expression levels in Sh-ctr and
Sh-Kaiso transfected MC3T3-E1 cells after 7 days of culture with
osteogenic medium in the presence of LY. (E) ALP activity was
measured on day 7. (F) ALP staining was performed on day 7, and
Alizarin Red S staining was performed on day 21. Data are presented
as the mean ± SD (n=3). **P<0.01. LY, LY294002;
RT-qPCR, reverse transcription-quantitative PCR; ALP, alkaline
phosphatase; OCN, osteocalcin; BSP, bone sialoprotein; LV,
lentivirus; shRNA, short hairpin RNA; Ctr, control; p-,
phosphorylated. |
Kaiso regulates the PI3K/AKT pathway via
Itga10
To further investigate the mechanism via which Kaiso
regulates the PI3K/AKT pathway, the present study focused on
Itga10, Csf1r and Fgfr3, which demonstrated a high degree of
connectivity in the PPI network analysis. The results of the
western blot analysis indicated that Itga10 was downregulated in
LV-Kaiso MC3T3-E1 cells and upregulated in Sh-Kaiso MC3T3-E1 cells
(Fig. 4A and B). The existence
of the KBS (5′-TCCTGCNA-3′) in the promoter region of Itga10
(5′-AATCCTGCTAC-3′) was predicted using JASPAR and MatInspector
databases. For further verification, ChIP assays were performed on
LV-Kaiso and control LV-ctr cell lines using an anti-Kaiso
monoclonal antibody. qPCR of the ChIP DNA products using an Itga10
promoter primer set demonstrated the binding of Kaiso to the Itga10
promoter (Fig. 4C). To further
assess these findings, the promoter activity of wild-type (WT) and
KBS-mutated (MUT) Itga10 was detected in MC3T3-E1 cells. It was
identified that Kaiso overexpression significantly inhibited the
promotor luciferase reporter activity of Itga10. Moreover, the MUT
of Itag10 could partially restore the suppressed Itga10 promoter
activity (relative luciferase activity, 0.39±0.04 in Itga10-WT vs.
0.50±0.04 in Itga10-MUT) (Fig. 4D
and E).
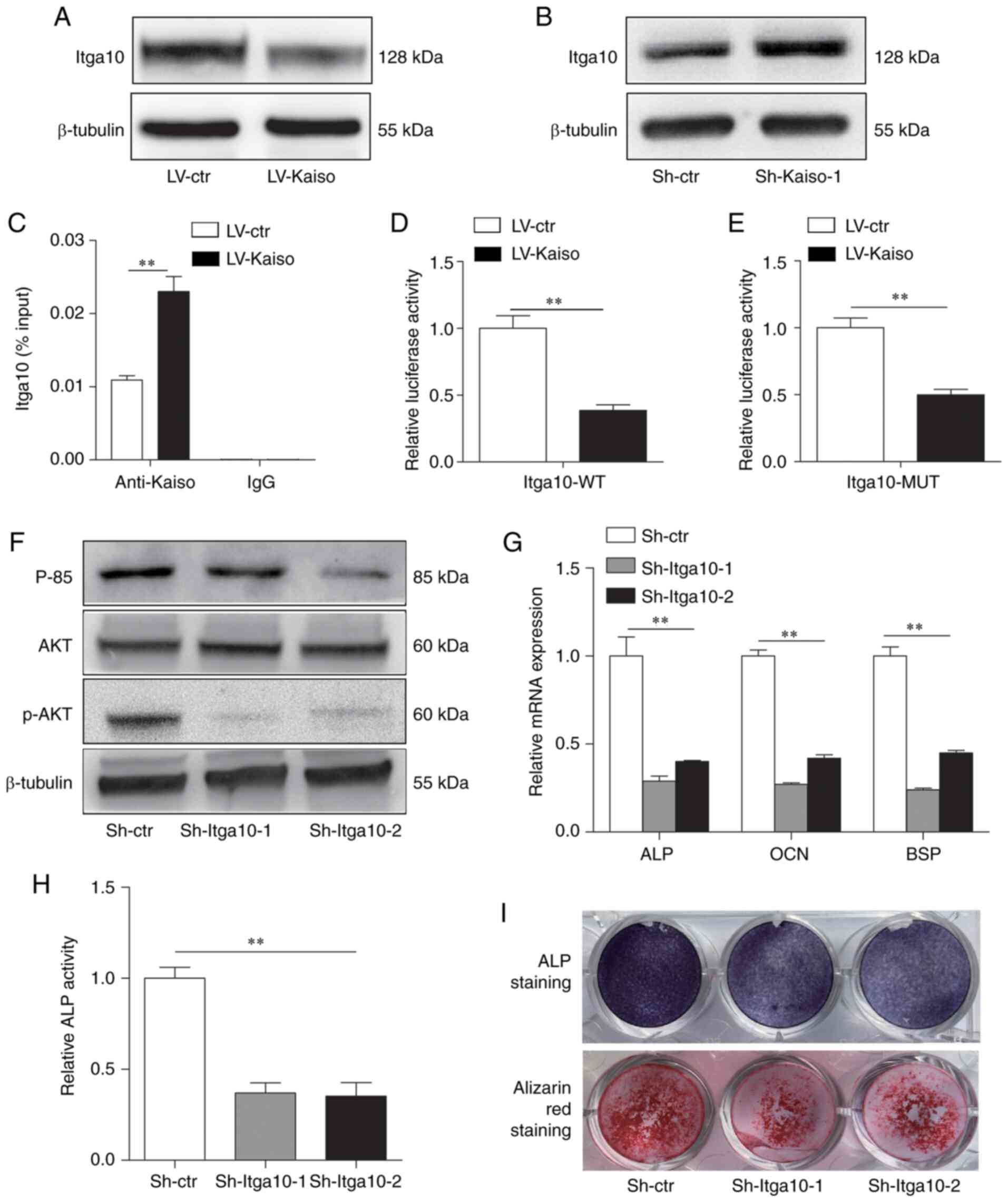 | Figure 4Kaiso regulates the PI3K/AKT pathway
via Itga10. Western blot analysis of Itga10 expression in (A)
Kaiso-overexpressing or (B) Kaiso-knockdown MC3T3-E1 cells. (C)
Chromatin immunoprecipitation analysis of the binding of Kaiso to
the promoter of Itga10 in LV-ctr and LV-Kaiso MC3T3-E1 cells. Input
DNA and DNA immunoprecipitated with IgG were included as positive
and negative controls, respectively. The results are expressed as
percentages of the input level. (D) WT and (E) Kaiso binding
site-MUT Itga10 promoter-luciferase reporter vectors were
co-transfected with Kaiso-overexpression vectors or control vector
in MC3T3-E1 cells. Luciferase activity was normalized to
Renilla luciferase activity and presented as the fold change
to LV-ctr. (F) Expression levels of p85, AKT and p-AKT were
determined via western blot analysis in Sh-Itga10-1, Sh-Itga10-2
and Sh-ctr transfectedMC3T3-E1 cells. (G) RT-qPCR analysis of ALP,
OCN and BSP expression levels in Sh-ctr and Sh-Kaiso transfected
MC3T3-E1 cells after 7 days culture with osteogenic medium. (H) ALP
activity was measured on day 7. (I) ALP staining was performed on
day 7, and Alizarin Red S staining was performed on day 21. All
quantitative data are presented as the mean ± SD (n=3).
**P<0.01. WT, wild-type; MUT, mutant; LV, lentivirus;
shRNA, short hairpin RNA; Ctr, control; p-, phosphorylated; ALP,
alkaline phosphatase; OCN, osteocalcin; BSP, bone sialoprotein;
Itga10, integrin subunit α10; RT-qPCR, reverse
transcription-quantitative PCR. |
To examine the direct role of Itga10 in osteogenesis
of MC3T3-E1cells, two shRNAs were used to knock down Itga10
(Sh-Itga10-1 andSh-Itga10-2). RT-qPCR and western blot analyses
identified that both shRNAs led to efficient knockdown of Itga10 in
MC3T3-E1 cells (Fig. S4A and
B). The results demonstrated that knocking down Itga10
decreased the protein expression levels of p85 and p-AKT (Fig. 4F), as well as the mRNA expression
levels of ALP, OCN and BSP (Fig.
4G). Furthermore, the stable Sh-Itga10-1 and Sh-Itga10-2 cell
lines had suppressed osteoblast differentiation, as demonstrated by
declined ALP activity, ALP staining and mineralized bone nodule
formation (Fig. 4H and I).
To examine whether Itga10 serves as a downstream
target of Kaiso, cells were transfected with lentivirus expressing
Itga10. The efficiency of lentivirus expressing Itga10 in MC3T3-E1
cells was confirmed via western blot analyses and qPCR (Fig. S4C and D). LV-Kaiso cells were
infected with a lentivirus expressing Itga10 to produce a stable
overexpressing cell line (LV-Kaiso+Itga10). The results
demonstrated that overexpression of Itga10 could significantly
alleviate the osteoblast inhibitory effects observed in
LV-Kaiso-stably transfected cells (LV-ctr vs. LV-Kaiso+Itga10;
LV-Kaisovs. LV-Kaiso+Itga10) (Fig.
S5). Taken together, these results suggested that Itga10 serves
an important role in the Kaiso-mediated regulation of the PI3K/AKT
pathway in MC3T3-E1 cells.
Osteogenesis assay in vivo
To investigate the osteogenic potential of LV-Kaiso,
Sh-Kaiso and Sh-Itga10 MC3T3-E1 cells in vivo, β-TCP
scaffolds loaded with cells were implanted into the intramuscular
pocket of nude mice. Experimental mice were sacrificed
usingCO2 at the end of the study. After 8 weeks of
recovery, newly formed bones were analyzed via µCT and
H&E staining. µCT images demonstrated more ectopic bone
formation in the Sh-Kaiso group compared with that in the Sh-ctr
group by comparison of BMD generated by skyscan 1176. The Kaiso
overexpression and Itga10 knockdown groups had decreased
regenerated bone volume compared with that in the control group
(Fig. 5A-D). These results were
confirmed via measurement of BMD (Fig. 5E). The results of H&E
staining provided further evidence for the differences in bone
formation in the β-TCP scaffolds from different groups (Fig. 5F-I). In the H&E staining, the
lightly dyed continuous structure marked by '#' in the figure
represents newly formed mature bone; the small reddish tissue
(indicated by arrowheads) is newly formed osteoid; and the deeply
stained nuclei (indicated by arrows), which can be seen in the
osteoid, are osteoblasts. The Sh-Kaiso group had more newly formed
mature bone compared with the other groups.
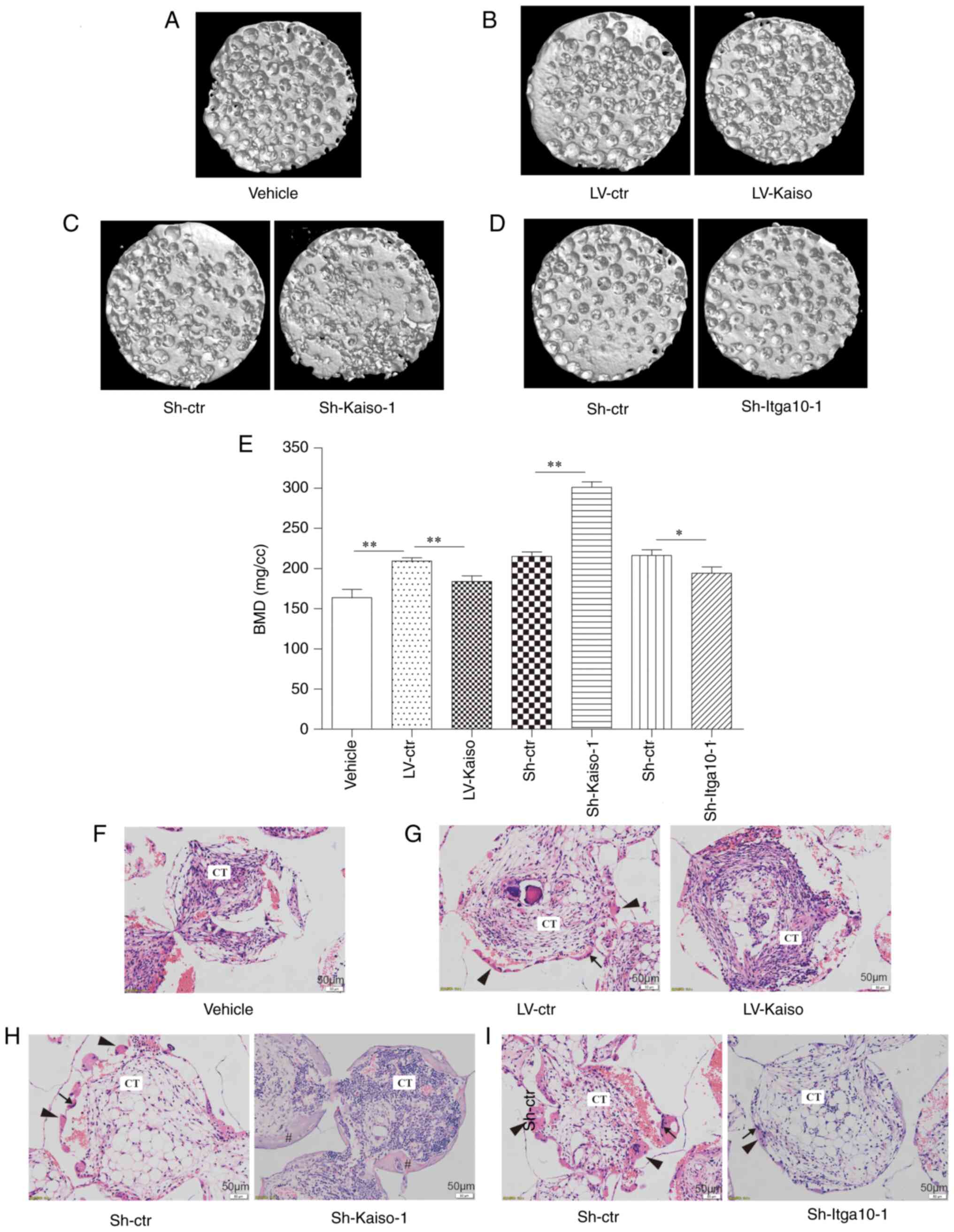 | Figure 5In vivo osteogenesis of
LV-Kaiso, Sh-Kaiso and Sh-Itga10 infected MC3T3-E1 cells.
Representative µCT images (magnification, ×1) from each
group: (A) Vehicle, (B) LV-ctr and LV-Kaiso, (C) Sh-ctr and
Sh-Kaiso-1, (D) Sh-ctr and Sh-Itga10-1. (E) BMD of the implants was
measured based on µCT images. Representative hematoxylin and
eosin staining images from each group: (F) Vehicle, (G) LV-ctr and
LV-Kaiso, (H) Sh-ctr and Sh-Kaiso-1, (I) Sh-ctr and Sh-Itga10-1.
Scale bar, 50 µm. Thin arrows indicate osteoblast;
arrowheads indicate newly formed osteoid; # indicates newly formed
mature bone. Data are presented as the mean ± SD (n=3).
*P<0.05 and **P<0.01. BMD, Bone mineral
density; CT, connective tissue; Itga10, integrin subunit α10; LV,
lentivirus; shRNA, short hairpin RNA; Ctr, control. |
Discussion
The present study identified the essential role of
Kaiso during osteoblast differentiation in MC3T3-E1 cells, and it
was identified that Kaiso regulated this process via the PI3K/AKT
pathway by binding to the promoter of Itga10. To the best of our
knowledge, this was the first study to demonstrate the potential
involvement of Kaiso in osteoblast differentiation and bone
formation.
Previous studies have reported that the
transcription factor Kaiso functions in various biological
processes. For instance, Kaiso regulates the cell cycle viacyclins
D1 and E1 (12). Moreover, Kaiso
acts as an essential transcriptional factor in the proliferation
and migration of cancer cells (13,14), as well as potentiates intestinal
tumorigenesis in the murine intestine (11,15). Kaiso also serves a role in
vascularization, which is critical in bone formation (23). The zinc finger and BTB domain
containing 16 gene, which is also a member of the POZ-zinc finger
family, has been reported to regulate osteoblast differentiation
(24,25). Furthermore, Kaiso is upregulated
in subchondral bone of an early experimental osteoarthritis model
based on transcription factors analysis (26). These findings indicate that Kaiso
may serve important roles in the pathogenesis, including cartilage
destruction or osteophyte formation, of early experimental
osteoarthritis. The present study identified the role of Kaiso in
osteoblast differentiation. The expression of Kaiso was found to be
downregulated during osteoblast differentiation in MC3T3-E1 cells.
Gain- and loss-of-function in vitro studies demonstrated
that overexpression of Kaiso was sufficient to suppress osteoblast
differentiation, whereas gene-specific silencing promoted this
process. In accordance with these findings, the present results
indicated that Kaiso overexpression decreased bone formation, while
Kaiso knockdown increased bone formation in vivo. These
findings suggested that Kaiso negatively regulated osteoblast
differentiation.
Next, the current study investigated the mechanisms
via which Kaiso regulated osteoblast differentiation. The effect of
Kaiso on Wnt has been widely studied; however, the present results
indicated that Kaiso regulated osteoblast differentiation in a Wnt
pathway-independent manner. Previous studies have revealed that the
PI3K/AKT pathway is critical during osteoblast differentiation
(27,28). In the present study, the enhanced
osteoblast differentiation observed in Kaiso-knockdown cells was
accompanied by increased p85 and p-AKT expression levels, and these
effects were almost completely blocked by the PI3K/AKT pathway
inhibitor LY294002. Thus, these findings suggested that the
PI3K/AKT pathway may be the target of Kaiso during osteoblast
differentiation.
Kaiso is a zinc finger transcriptional factor that
contains a BTB/POZ domain for PPI at the N-terminus (12). Previous studies have reported
that Kaiso regulates pathological processes by directly binding to
the promoters of target genes via the KBS or methylated CpG
dinucleotides (12,29,30). To evaluate the potential target
via which Kaiso regulated the PI3K/AKT pathway, the present study
focused on Itga10. Integrins are cell surface receptors that
interact with several signaling molecules and activate
intracellular signaling pathways, such as the PI3K/AKT pathway
(31). The roles of integrins in
osteoblast differentiation, as well as their influence on the
physiology and pathophysiology of cartilage and bone have been
widely studied (32-35). Itga10 has been reported to
modulate chondrocyte differentiation, and can regulate AKT activity
(36-38). The present results suggested that
Kaiso binds to the KBS region of the Itga10 promoter and inhibits
the expression of Itga10. Moreover, mutation of the KBS in the
promoter of the Itga10 gene partially alleviated the inhibition of
luciferase reporter activity observed in Kaiso-overexpressing
MC3T3-E1 cells. This may be attributed to the existence of other
methylated CpG dinucleotide binding sites. Knocking down Itga10
decreased the activation of the PI3K/AKT pathway and suppressed
osteoblast differentiation, while overexpression of Itga10 could
significantly alleviate the osteoblast inhibitory effects observed
in Kaiso-stably transfected cells. These findings indicated that
Kaiso regulated the PI3K/AKT pathway by binding to the Itga10
promoter.
In conclusion, the present study demonstrated that
Kaiso regulated osteoblast differentiation in MC3T3-E1 cells via
the Itga10/PI3K/AKT signaling pathway. Kaiso and the
Itga10/PI3K/AKT signaling pathway may serve an essential role in
bone destruction and bone formation, thus representing a novel
therapeutic target for related diseases.
Supplementary Data
Funding
This research was supported by the National Nature
Science Foundation of China (grant nos. 81601412, 81901654 and
81672126) and the Youth Startup Foundation of Changhai Hospital
(grant no. 2018QNA017).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
Study design: WX and CH. Acquisition of data: WT, XF
and JL. Analysis and interpretation of data: JL, CW and YX.
Drafting the article or revising: WT, JL and XF. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All animal studies were approved by the Animal
Ethics Committee of Changhai Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Harada S and Rodan GA: Control of
osteoblast function and regulation of bone mass. Nature.
423:349–355. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Teitelbaum SL and Ross FP: Genetic
regulation of osteoclast development and function. Nat Rev Genet.
4:638–649. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Loi F, Córdova LA, Pajarinen J, Lin TH,
Yao Z and Goodman SB: Inflammation, fracture and bone repair. Bone.
86:119–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sieper J and Poddubnyy D: Axial
spondyloarthritis. Lancet. 390:73–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang TL, Shen H, Liu A, Dong SS, Zhang L,
Deng FY, Zhao Q and Deng HW: A road map for understanding molecular
and genetic determinants of osteoporosis. Nat Rev Endocrinol.
16:91–103. 2020. View Article : Google Scholar :
|
|
7
|
Day TF, Guo X, Garrett-Beal L and Yang Y:
Wnt/beta-catenin signaling in mesenchymal progenitors controls
osteoblast and chondrocyte differentiation during vertebrate
skeletogenesis. Dev Cell. 8:739–750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Franceschi RT, Ge C, Xiao G, Roca H and
Jiang D: Transcriptional regulation of osteoblasts. Ann NY Acad
Sci. 1116:196–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Daniel JM, Spring CM, Crawford HB,
Reynolds AB and Baig A: The p120(ctn)-binding partner Kaiso is a
bi-modal DNA-binding protein that recognizes both a
sequence-specific consensus and methylated CpG dinucleotides.
Nucleic Acids Res. 30:2911–2919. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buck-Koehntop BA, Stanfield RL, Ekiert DC,
Martinez-Yamout MA, Dyson HJ, Wilson IA and Wright PE: Molecular
basis for recognition of methylated and specific DNA sequences by
the zinc finger protein Kaiso. Proc Natl Acad Sci USA.
109:15229–15234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choi SH, Koh DI, Cho SY, Kim MK, Kim KS
and Hur MW: Temporal and differential regulation of
KAISO-controlled transcription by phosphorylated and acetylated p53
high-lights a crucial regulatory role of apoptosis. J Biol Chem.
294:12957–12974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pozner A, Terooatea T and Buck-Koehntop B:
Cell-specific Kaiso (ZBTB33) regulation of cell cycle through
cyclin D1 and cyclin E1. J Biol Chem. 291:24538–24550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng J: Upregulation of MicroRNA-4262
targets Kaiso (ZBTB33) to inhibit the proliferation and EMT of
cervical cancer cells. Oncol Res. 26:1215–1225. 2018. View Article : Google Scholar
|
|
14
|
Pierre CC, Hercules SM, Yates C and Daniel
JM: Dancing from bottoms up-roles of the POZ-ZF transcription
factor Kaiso in cancer. Biochim Biophys Acta Rev Cancer.
1871:64–74. 2019. View Article : Google Scholar
|
|
15
|
Short SP, Barrett CW, Stengel KR, Revetta
FL, Choksi YA, Coburn LA, Lintel MK, McDonough EM, Washington MK,
Wilson KT, et al: Kaiso is required for MTG16-dependent effects on
colitis-associated carcinoma. Oncogene. 38:5091–5106. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim SW, Park JI, Spring CM, Sater AK, Ji
H, Otchere AA, Daniel JM and McCrea PD: Non-canonical Wnt signals
are modulated by the Kaiso transcriptional repressor and
p120-catenin. Nat Cell Biol. 6:1212–1220. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Dong QZ, Wang S, Xu HT, Miao Y,
Wang L and Wang EH: Kaiso interacts with p120-catenin to regulate
β-catenin expression at the transcriptional level. PLoS One.
9:e875372014. View Article : Google Scholar
|
|
18
|
Baron R and Kneissel M: WNT signaling in
bone homeostasis and disease: From human mutations to treatments.
Nat Med. 19:179–192. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Metwally M, Bayoumi A, Romero-Gomez M,
Thabet K, John M, Adams LA, Huo X, Aller R, García-Monzón C, Teresa
Arias-Loste M, et al: A polymorphism in the Irisin-encoding gene
(FNDC5) associates with hepatic steatosis by differential miRNA
binding to the 3′UTR. J Hepatol. 70:494–500. 2019. View Article : Google Scholar
|
|
21
|
Guan J, Zhang J, Zhu Z, Niu X, Guo S, Wang
Y and Zhang C: Bone morphogenetic protein 2 gene transduction
enhances the osteogenic potential of human urine-derived stem
cells. Stem Cell Res Ther. 6:52015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Collings AJ and Richards CT: Digital
dissection of the pelvis and hindlimb of the red-legged running
frog, phlyctimantis maculatus, using diffusible iodine contrast
enhanced computed microtomography (DICE µ CT). PeerJ. 7:e70032019.
View Article : Google Scholar
|
|
23
|
Delgado-Bellido D, Fernández-Cortés M,
Rodríguez MI, Serrano-Sáenz S, Carracedo A, Garcia-Diaz A and
Oliver FJ: VE-cadherin promotes vasculogenic mimicry by modulating
kaiso-dependent gene expression. Cell Death Differ. 26:348–361.
2019. View Article : Google Scholar :
|
|
24
|
Marofi F, Vahedi G, Solali S, Alivand M,
Salarinasab S, Zadi Heydarabad M and Farshdousti Hagh M: Gene
expression of TWIST1 and ZBTB16 is regulated by methylation
modifications during the osteoblastic differentiation of
mesenchymal stem cells. J Cell Physiol. 234:6230–6243. 2019.
View Article : Google Scholar
|
|
25
|
Onizuka S, Iwata T, Park SJ, Nakai K,
Yamato M, Okano T and Izumi Y: ZBTB16 as a downstream target gene
of osterix regulates osteoblastogenesis of human multipotent
mesenchymal stromal cells. J Cell Biochem. 117:2423–2434. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang RK, Li GW, Jiang D, Zhang DW, Yu B
and Yang LK: Transcription factors analysis of subchondral bone in
early experimental osteoarthritis based on gene expression
profiles. Zhongguo Gu Shang. 31:165–169. 2018.In Chinese.
PubMed/NCBI
|
|
27
|
Shen WC, Lai YC, Li LH, Liao K, Lai HC,
Kao SY, Wang J, Chuong CM and Hung SC: Methylation and PTEN
activation in dental pulp mesenchymal stem cells promotes
osteogenesis and reduces oncogenesis. Nat Commun. 10:22262019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye C, Zhang W, Hang K, Chen M, Hou W, Chen
J, Chen X, Chen E, Tang L, Lu J, et al: Extracellular IL-37
promotes osteogenic differentiation of human bone marrow
mesenchymal stem cells via activation of the PI3K/AKT signaling
pathway. Cell Death Dis. 10:7532019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Robinson SC, Klobucar K, Pierre CC, Ansari
A, Zhenilo S, Prokhortchouk E and Daniel JM: Kaiso differentially
regulates components of the Notch signaling pathway in intestinal
cells. Cell Commun Signal. 15:242017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bassey-Archibong BI, Kwiecien JM,
Milosavljevic SB, Hallett RM, Rayner LG, Erb MJ, Crawford-Brown CJ,
Stephenson KB, Bédard PA, Hassell JA and Daniel JM: Kaiso depletion
attenuates transforming growth factor-β signaling and metastatic
activity of triple-negative breast cancer cells. Oncogenesis.
5:e2082016. View Article : Google Scholar
|
|
31
|
Hynes RO: Integrins: Bidirectional,
allosteric signaling machines. Cell. 110:673–687. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Docheva D, Popov C, Alberton P and Aszodi
A: Integrin signaling in skeletal development and function. Birth
Defects Res C Embryo Today. 102:13–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hamidouche Z, Fromigué O, Ringe J, Häupl
T, Vaudin P, Pagès JC, Srouji S, Livne E and Marie PJ: Priming
integrin alpha5 promotes human mesenchymal stromal cell osteoblast
differentiation and osteogenesis. Proc Natl Acad Sci USA.
106:18587–18591. 2009. View Article : Google Scholar
|
|
34
|
Shen B, Vardy K, Hughes P, Tasdogan A,
Zhao Z, Yue R, Crane GM and Morrison SJ: Integrin alpha11 is an
osteolectin receptor and is required for the maintenance of adult
skeletal bone mass. Elife. 8:e422742019. View Article : Google Scholar :
|
|
35
|
Raines AL, Berger MB, Schwartz Z and Boyan
BD: Osteoblasts grown on microroughened titanium surfaces regulate
angiogenic growth factor production through specific integrin
receptors. Acta Biomater. 97:578–586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Varas L, Ohlsson LB, Honeth G, Olsson A,
Bengtsson T, Wiberg C, Bockermann R, Järnum S, Richter J,
Pennington D, et al: Alpha10 integrin expression is up-regulated on
fibroblast growth factor-2-treated mesenchymal stem cells with
improved chondrogenic differentiation potential. Stem Cells Dev.
16:965–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bengtsson T, Aszodi A, Nicolae C, Hunziker
EB, Lundgren-Akerlund E and Fässler R: Loss of alpha10beta1
integrin expression leads to moderate dysfunction of growth plate
chondrocytes. J Cell Sci. 118:929–936. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Okada T, Lee AY, Qin LX, Agaram N, Mimae
T, Shen Y, O'Connor R, López-Lago MA, Craig A, Miller ML, et al:
Integrin-α10 dependency identifies RAC and RICTOR as therapeutic
targets in high-grade myxofibrosarcoma. Cancer Discov. 6:1148–1165.
2016. View Article : Google Scholar : PubMed/NCBI
|