Introduction
Breast-conservative surgery followed by radiation
therapy (RT) currently is the standard of care for breast cancer
(1). Although
breast-conservative surgery with RT improves overall patient
survival and is universally accepted as the gold standard in breast
cancer treatment. However, improved approaches targeting cancer
cells that develop resistance to RT are still needed (1-3).
Indeed, the appearance of radioresistant cells leads to treatment
failure and local recurrence, thus requiring administration of
higher doses of radiation in these areas of recurrence, which may
cause damage to healthy tissues surrounding the tumor. Therefore,
the identification of new pharmacological approaches that could
overcome radioresistance of cancer cells is crucial (3-5).
It is well established that intrinsic and ionizing
radiation (IR)-induced radioresistance determines cellular
responses to RT. Previous studies have identified numerous
radioresistance-associated genes, including eukaryotic translation
initiation factor 4 γ 1 (eIF4G1) (6), insulin-like growth factor 1
receptor (IGF-1R) (7),
CST telomere replication complex component 1 (CTC1)
(8), and DNA-dependent protein
kinase (DNA-PK), which may represent potential molecular
targets for radiosensitization (9). DNA repair, cancer stemness,
apoptosis (10), cell cycle
arrest (11), autophagy
(12), and hypoxia (13) have been proposed as essential
biological mechanisms leading to radioresistance of cancer cells.
Among the radiosensitizers under active investigation or in use are
agents targeting hypoxic cells, inhibitors of ion channels
(14), regulators of proteins
(such as enzymes) (15-17) or pathways (18) and inhibitors of autophagy
(19). However, several
limitations exist with these compounds, such as lack of
specificity, unclear underlying molecular mechanisms, and adverse
effects (3). Moreover, there is
a paucity of promising radiosensitizers for a variety of cancer
types, including breast cancer.
Drug discovery, including for radiosensitizers, is
both time-consuming and costly. Moreover, the design and synthesis
of new compounds, along with high-throughput screening, remain
challenging (20). Drug
repositioning (also known as drug repurposing or drug reprofiling)
allows the prediction of alternative applications for existing
drugs. This strategy can help reduce the time and cost associated
with new drug development and improve the delivery of new drugs to
patients with severe diseases (21). Thus, drug repositioning may be a
promising strategy for the identification of known compounds, such
as potentially effective radiosensitizers, that could improve the
efficacy and reliability of RT.
Bosutinib is a small BCR-ABL kinase inhibitor
approved by the US Food and Drug Administration (FDA) in 2012 with
improved tolerance and safety, compared with other tyrosine kinase
inhibitors. BCR-ABL kinase plays an important role in the onset and
rapid progression of chronic myelogenous leukemia and is among the
primary targets of bosutinib. Other known targets include LYN, SRC,
CDK2, and MAP2K1 (22,23). It has been reported that
inhibition of SRC could enhance the radiosensitivity of numerous of
cancer cell types (24,25). In the present study, we found
bosutinib and silencing of eIF4G1 following IR had similar
therapeutic effects. Bosutinib was identified as a radiosensitizer
of breast cancer cells for it significantly sensitized cancer cells
to DNA damage induced by RT through the regulation of cell
apoptosis (26) and expression
of several proteins including eIF4G1, ATM, and XRCC4. Additional
experiments were carried out to examine the underlying mechanism
and verify the adjuvant effect of bosutinib in breast cancer cells
exposed to γ-rays. The addition of bosutinib led to an apparent
radiosensitizing effect in breast cancer cells, thus allowing the
use of decreased dosages of both the drug and radiation, with fewer
side effects.
Materials and methods
Library of integrated network-based
cellular signatures (LINCS)
The cellular signatures of all compounds used in
this study were collected and extracted in October 2017 from the
LINCS comprehensive systems biology portal (http://www.lincscloud.org/). It is a large-scale
pharmacogenomics dataset based on HDF5 file format containing
cellular signatures in response to a variety of perturbations
including agents, genetic mutations, micro-environments and
diseases (27). The whole
database of gene expression profiles of treatments and their
corresponding controls in each perturbagen was downloaded from the
portal in homepage of the website. We then selected 55,129 cellular
response signatures of 4,617 chemical reagents at different
time-points and doses and 4,388 signatures of untreated cells as
controls. The signatures were extracted and integrated for
identification and prediction of potential radiosensitizers in this
study.
Gene expression omnibus (GEO)
The raw gene expression profile dataset GSE41627
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41627)
used in this study was extracted in October 2017 from the GEO
(https://www.ncbi.nlm.nih.gov/geo/)
database. The GEO was scanned for gene expression profiles related
to radiotherapy of breast cancer according to 2 criterions for
precise match with LINCS: i) Platform GPL570 or GPL96 was employed.
ii) Cell lines used should be matched accurately. GSE41627 was
selected eventually, comprising expression profiles of several DNA
damage response (DDR) genes in IR in human breast cancer cells.
This dataset by Badura et al (26) reported 12 samples from 4 cell
lines exposed to 10 Gy of γ-ray irradiation, with or without eIF4G1
silencing. In 'Download family' in the website, 'Series Matrix
File(s)' of GSE41627 in '.txt' were downloaded for further
comparison and analysis.
Calculation of enrichment scores
(ES)
Probe IDs in LINCS were converted into official gene
symbols according to annotation files of the platform of GPL96.
However, those in GSE41627 were conducted according to annotation
files of the platform of GPL570. Processing of gene expression
signatures from LINCS and GSE41627 was carried out using the
'contrasts. fit' function in Limma package (v3.24.14) (28) in R software (3.4.4). Cellular
response data of treatment groups and their controls of the two
datasets were inputted and differential expressions of the whole
genome were determined after running of the scripts. The lists of
all differentially expressed genes in LINCS and GSE41627 were
obtained, respectively. ES were used to evaluate the similarity
between the gene expression signatures of cells treated with a
variety of compounds and those of irradiated, eIF4G1-silenced
cells. Gene set enrichment analysis (GSEA) was performed using GSEA
software (v. 3.0) (29). The
gene lists of the two datasets were inputted into the software and
the classic scoring scheme was used to calculate ES for the
silencing and compounds in LINCS. The maximum and average ES in
each case were obtained as a result.
Cell culture and irradiation
The human MX-1, MCF-7, and MDA-MB-231 breast cancer
cell lines were used in this study. MCF-7 (cat. no. CL0206) and
MDA-MB-231 (cat. no. CL0208) cells were purchased from the American
Type Culture Collection (ATCC). MX-1 cells (cat. no. CL0456) were
provided by the Beijing Institute of Transfusion Medicine and
initially purchased from the ATCC.
The cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% FBS (Sigma-Aldrich;
Merck KGaA), 100 U/ml penicillin and 100 µg/l gentamycin and
incubated at 37°C in a humidified atmosphere containing 5%
CO2. The cells were passaged once for 2 days with a
total of three passages. After each resuscitation, 3-5 generations
were used.
Cells were divided into four groups: i) Irradiation
alone; ii) Drug alone; iii) Irradiation and drug; and iv) Control.
Irradiation was carried out using 60Co γ-rays at a dose
rate of 1.98 Gy/min at room temperature in the Beijing Institute of
Radiation Medicine. Then, 2 Gy and 4 Gy of γ-rays were used
according to previous studies and the daily doses in common use in
classical radiotherapy for breast cancer in clinic (30,31).
Drugs and antibodies
The drugs used in the present study included:
Bosutinib (cat. no. E047103; Shanghai EFE Biological Technology
Co., Ltd.), bifonazole (cat. no. B3563; Sigma-Aldrich; Merck KGaA),
and isosorbide (cat. no. 652-67-5; J&K Scientific, Ltd.).
The antibodies used in immunoblotting analyses were
as follows: Anti-eIF4G1 (cat. no. ab2609; Abcam; rabbit;
polyclonal), anti-eIF4G2 (cat. no. 3468; Cell Signaling Technology,
Inc.; rabbit; monoclonal), anti-ATM (cat. no. ab201022; Abcam;
rabbit; monoclonal), anti-γH2AX (Ser139; cat. no. ab11174; Abcam;
rabbit, polyclonal), anti-XRCC4 (cat. no. SC-271087; Santa Cruz
Biotechnology, Inc.; mouse; monoclonal), anti-ATRIP (cat. no.
ab175221; Abcam; rabbit; monoclonal), anti-GADD45A (cat. no.
ab7664; Abcam; rabbit; polyclonal), anti-PARP-1 (cat. no. SC-7150;
Santa Cruz Biotechnology, Inc.; rabbit; polyclonal), anti-Mre11
(cat. no. ab214; Abcam; mouse; monoclonal), anti-pCDK1-Y15 (cat.
no. 9116S; Cell Signaling Technology, Inc.; mouse; monoclonal),
anti-CDK1 (cat. no. ab201008; Abcam; rabbit; monoclonal) and
anti-β-actin (cat. no. TA-09; Beijing Zhongshan Jinqiao
Biotechnology Co. Ltd.; mouse; monoclonal).
Cell proliferation
Cell proliferation was analyzed using a
Cell-Counting Kit-8 (CCK-8) assay kit (Engreen) following the
manufacturer's instructions. Briefly, cells were seeded in 96-well
plates and exposed to increasing concentrations of test drugs
alone. To assess cell proliferation, CCK-8 solution (cat. no. CK04;
Dojindo Molecular Technologies, Inc.) was added 48 and 72 h after
the treatment (32,33). The absorbance was measured at 450
nm using a microplate reader. Half-maximal inhibitory
concentrations (IC50) were calculated using GraphPad
Prism 8.0.1 (244) software (GraphPad Software, Inc.). Each
experiment was performed in triplicate.
Cell viability
Cell viability was assessed using colony formation
assays. Cells were seeded into 60-mm culture plates at a density of
3×102 cells/plate and exposed to various concentrations
of bosutinib for 24 h (34).
Following irradiation, the cells were cultured in normal medium for
10-15 days. Surviving tumor cells were fixed with ethanol and
stained with Giemsa, and the colonies were then counted.
Immunoblotting analyses
The cells were treated with bosutinib for 12 and 24
h (35). Total proteins were
extracted from cultured cells on ice with lysis buffer supplemented
with protease inhibitor cocktail. The BCA method was used for
protein quantification. Cell lysates were separated using SDS-PAGE
with a Bio-Rad Bis-Tris Gel system (percentages of SDS-PAGE were:
8% for ATM, XRCC4 and ATRIP; 10% for eIF4G1 and eIF4G2; 12% for
γH2AX and GADD45A). Separated proteins were then transferred to
PVDF membranes. The membranes were blocked with 5% non-fat dry milk
at room temperature for 1 h. Subsequently, the membranes were
incubated overnight at 4°C with the primary antibodies. The
antibodies used were as follows: Anti-eIF4G1 (cat. no. ab2609),
anti-eIF4G2 (cat. no. 3468), anti-ATM (cat. no. ab201022),
anti-γH2AX (Ser139; cat. no. ab11174), anti-XRCC4 (cat. no.
SC-271087), anti-ATRIP (cat. no. ab175221), anti-GADD45A (cat. no.
ab7664), anti-PARP-1 (cat. no. SC-7150), anti-Mre11 (cat. no.
ab214), anti-pCDK1-Y15 (cat. no. 9116S), anti-CDK1 (cat. no.
ab201008) and anti-β-actin (cat. no. TA-09). The dilutions of
anti-eIF4G1 and anti-γH2AX were 1:3,000 and 1:2,000, respectively.
The dilution of all other antibodies was 1:1,000. The membranes
were then washed three times with TBST and incubated with the
indicated secondary antibodies conjugated with HRP (1:4,000; 1 h)
at room temperature. The antibodies were as follows: anti-mouse
(cat. no. 140193; Jackson ImmunoResearch, Inc.) and anti-rabbit
(cat. no. 138442; Jackson ImmunoResearch, Inc.). The target bands
were visualized using the Image Quant LAS500 system. Densitometry
analysis was conducted by ImageJ software (1.51j8).
Measurement of cell apoptosis
Cells (3×105-6×105) were
treated with bosutinib for 24 h before irradiation with 4 Gy
60Co γ-rays, then subjected to trypsinization with
trypsin/EDTA (HyClone; Cytiva) 8, 12, and 24 h following
irradiation. For apoptosis measurement, the cells were centrifuged
with 179 × g at 4°C for 3 min. The cell pellets were re-suspended
in 100 µl binding buffer and stained with 5 µl
propidium iodide (50 µg/ml) and 5 µl Annexin V-FITC
(cat. no. DA10; Dojindo Molecular Technologies, Inc.). Apoptosis
was then analyzed using a flow cytometer (ACEA Novocyte).
Immunofluorescence (IF)
MDA-MB-231 cells were cultured on coverslips and
treated with 4 Gy of IR. The cells were washed three times with PBS
at 4°C at 1, 4 and 8 h after IR and then incubated with 4%
paraformaldehyde at room temperature for 15 min. The cells were
subsequently permeabilized with PBS containing 0.5% Triton X-100 at
room temperature for 10 min, and blocked with 3% of Albumin Bovine
Fraction V (CAS 9048-46-8; KainuoBio) in PBS at room temperature
for 1 h. Then the cells were incubated with γH2AX antibody (1:200
for IF, Ser139; cat. no. 05-636; Merck Millipore; mouse,
monoclonal) for 1 h at room temperature. Cells were subsequently
washed three times with PBS and then incubated with the secondary
antibody (1:400, Ser139; cat. no. A11029; Invitrogen; mouse) in
phosphatase at room temperature for 1 h. Then DAPI staining was
performed at room temperature for 5 min. Slides were imaged using
the Nikon Application for Inverted Research Microscope ECLIPSE Ti2
Series and oil immersion lens of ×100 was used.
Calculation of combination indices
Combination indices (CI) of bosutinib and radiation
were calculated with the tool CompuSyn (ComboSyn, Inc.) using the
Chou-Talalay method based on the median-effect equation (36). It was derived from the
mass-action law principle describing interactions among multiple
entities and first order and higher order dynamics. Dose-effect
data points of the components and combinations were input,
respectively, and the resulting CI in 'CompuSyn Report' accurately
indicates additive effect (CI=1), synergism (CI <1), and
antagonism (CI >1) for drug combinations. This tool provided an
easy and flexible approach for drug efficacy evaluation in drug
combination studies.
Statistical analysis
Statistical analysis of the GEO dataset was
conducted using an empirical phenotype-based permutation test
procedure (29). The phenotype
labels were permuted randomly, and the ES of the gene set in the
gene list of fold changes for the permuted data was recomputed. The
P-value of the observed ES was then calculated relative to this
distribution. The number of permutations was set to 1,000, and
P<0.05 was considered to indicate a statistically significant
difference.
Experiments were performed at least in triplicate.
Data from three or more independent experiments are presented as
the mean ± SD. The data were analyzed using one-way ANOVA.
Differences between two groups were analyzed using Student's
t-test. Differences among four groups were analyzed using Tukey's
post hoc test. Statistical analyses were performed using SPSS 18.0
software (IBM Corp.). P<0.05 was considered to indicate a
statistically significant difference.
Results
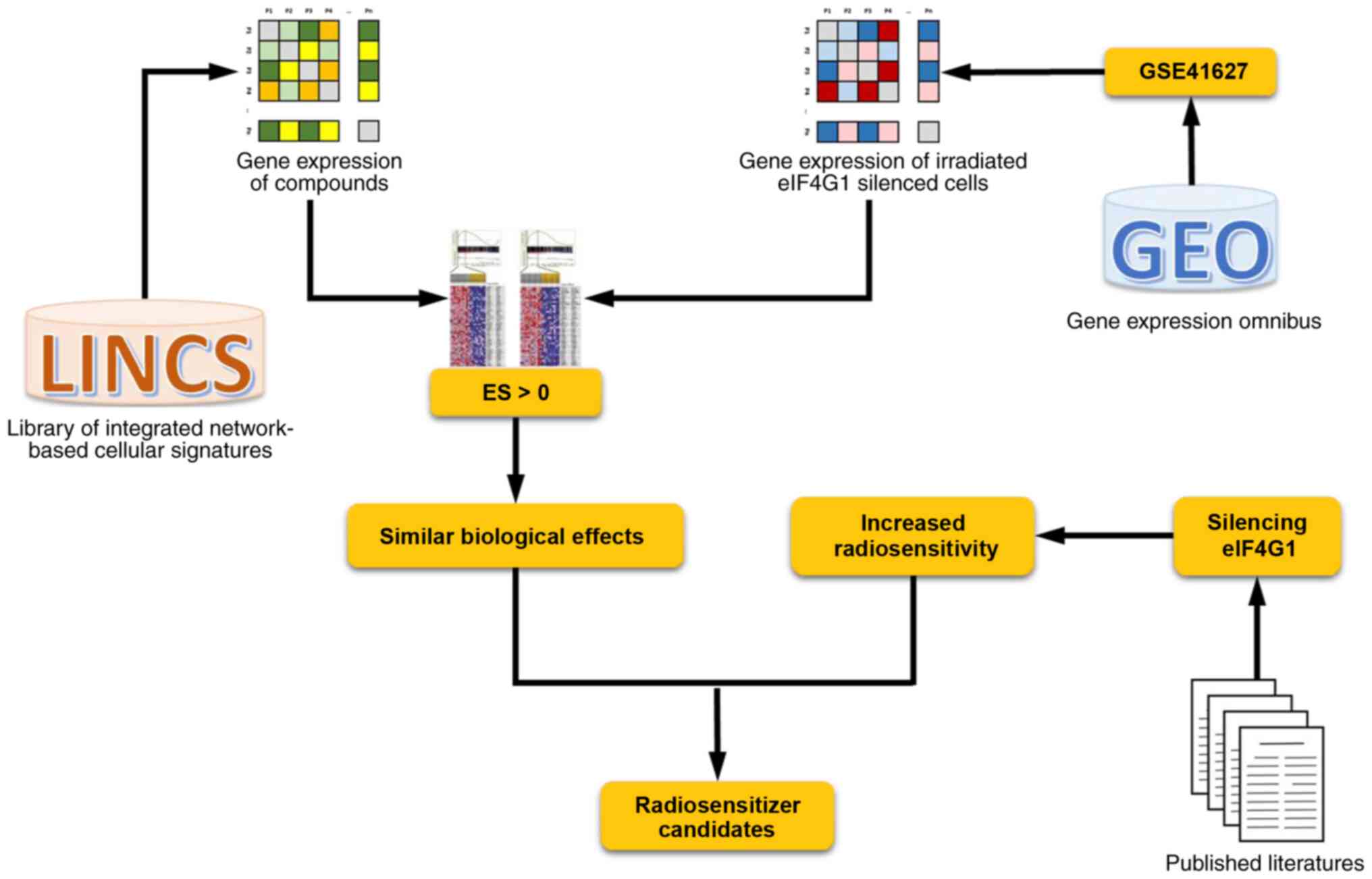
Extraction and integration of gene
expression profiles
Following screening and manual curation, the gene
expression profiles from the GSE41627 dataset were selected for the
identification of potential radiosensitizers. Treatment groups were
divided into four groups, including irradiated cells,
eIF4G1-silenced cells, irradiated eIF4G1-silenced cells, and the
control group. The silencing of eIF4G1 has been shown to
significantly sensitize breast cancer cells to IR. Therefore,
radiosensitizer candidates were identified by comparing the
biological effects of the gene expression profiles from the
GSE41627 dataset and those of breast cancer cells from LINCS in
response to treatment with different chemical compounds. The flow
diagram is shown in Fig. 1.
Identification of potential
radiosensitizers
Gene expression signatures of the GSE41627 dataset,
as well as those generated from breast cancer cells treated with
various compounds, were analyzed using R code. The integrated
genome-wide differential expression signature lists of irradiated
eIF4G1-silenced cells and compounds from LINCS were inputted and ES
between -1 and 1 were outputted indicating their similarity in
biological effects. A positive ES for a known compound indicates
that the drug-induced profile is similar to the effect of silencing
eIF4G1 in response to irradiation, suggesting that the queried
compounds can increase the radiosensitivity of breast cancer
cells.
A total of 2,089 entries from LINCS, including
different concentrations of each drug, were evaluated. These were
obtained at 96 and 144 h following eIF4G1 knockdown. The maximum
and the average ES were calculated for each entry. Thresholds for
screening were selected based on the ES of each compound to ensure
that ≤5% of compounds were screened in each case. In total, 11
compounds were proposed as candidate radiosensitizers for
consideration of further study.
For compounds examined 96 h after eIF4G1 knockdown,
both the maximum and the average ES of 369 drugs were positive
(Table SI). Considering 0.70
and 0.56 as thresholds for the maximum ES and average ES,
respectively, two experimental drugs (SU-11652 and latrunculin-b)
were selected. SU-11652 is a multi-targeting receptor tyrosine
kinase inhibitor currently investigated as an anticancer drug
(37). Latrunculin-b is a
cell-permeable actin polymerization inhibitor against cancer
(38). Considering only the
drugs approved by the FDA, four drugs (floxuridine, palbociclib
isethionate, raltitrexed, and bosutinib) were screened when the
thresholds for maximum and average ES were set at 0.66 and 0.48,
respectively.
At 144 h following eIF4G1 knockdown, 179 drugs
exhibited positive maximum and average ES (Table SI). PD-0325901, an experimental
MEK kinase inhibitor with marked anti-tumor activity (39) was identified with 0.49 and 0.36
as thresholds for the maximum ES and average ES, respectively.
Considering only drugs that were FDA approved were used, no
candidates had maximum ES and average ES above the thresholds
simultaneously. Thus, we changed the thresholds ensuring <10% of
the compounds would be screened. The maximum ES of 10 FDA-approved
drugs was >0.47, including lepirudin, pentoxifylline, and
trametinib. The average ES of 14 approved drugs was >0.30,
including bifonazole, isosorbide mononitrate, and anastrozole.
Selection of promising candidate
radiosensitizers
The comparison of transcriptional signatures
identified several candidate radiosensitizers. The selected
candidates are listed in Table
I. Compounds with both the maximum ES and average ES higher
than thresholds were selected, except in the case of approved drugs
at 144 h following eIF4G1 knockdown, as no compounds met these
criteria. Therefore, the top two compounds based on maximum ES and
average ES in this case were selected.
 | Table ITop-ranked compounds compared with
irradiated eIF4G1-silenced breast cancer cells. |
Table I
Top-ranked compounds compared with
irradiated eIF4G1-silenced breast cancer cells.
| Drugbank ID | Pert name | Drug group | Time-point | Max ES | Ave ES |
|---|
| DB08009 | SU-11652 | aExperimental | 96 h | 0.7094 | 0.6366 |
| DB08080 | Latrunculin-b | Experimental | 96 h | 0.7043 | 0.5687 |
| DB00322 | Floxuridine | bApproved | 96 h | 0.6880 | 0.5784 |
| DB09073 | Palbociclib | Approved | 96 h | 0.6827 | 0.5613 |
| DB00293 | Raltitrexed | Approved; cInvestigational | 96 h | 0.6699 | 0.6010 |
| DB06616 | Bosutinib | Approved | 96 h | 0.6616 | 0.5124 |
| DB07101 | PD-0325901 | Experimental | 144 h | 0.4970 | 0.4006 |
| DB00001 | Lepirudin | Approved | 144 h | 0.5649 | 0.2470 |
| DB00806 | Pentoxifylline | dApproved; Investigational | 144 h | 0.4983 | 0.0669 |
| DB04794 | Bifonazole | Approved | 144 h | 0.4320 | 0.3882 |
| DB01020 | Isosorbide | Approved | 144 h | 0.3874 | 0.3836 |
Bosutinib, bifonazole, and isosorbide were selected
for further validation and additional studies on the role and
mechanism of eIF4G1 in the cellular response to IR-induced DNA
damage. Among them, bosutinib has been used in the treatment of
Philadelphia chromosome-positive (Ph+) chronic
myelogenous leukemia with resistance or intolerance to prior
therapy, according to DrugBank.
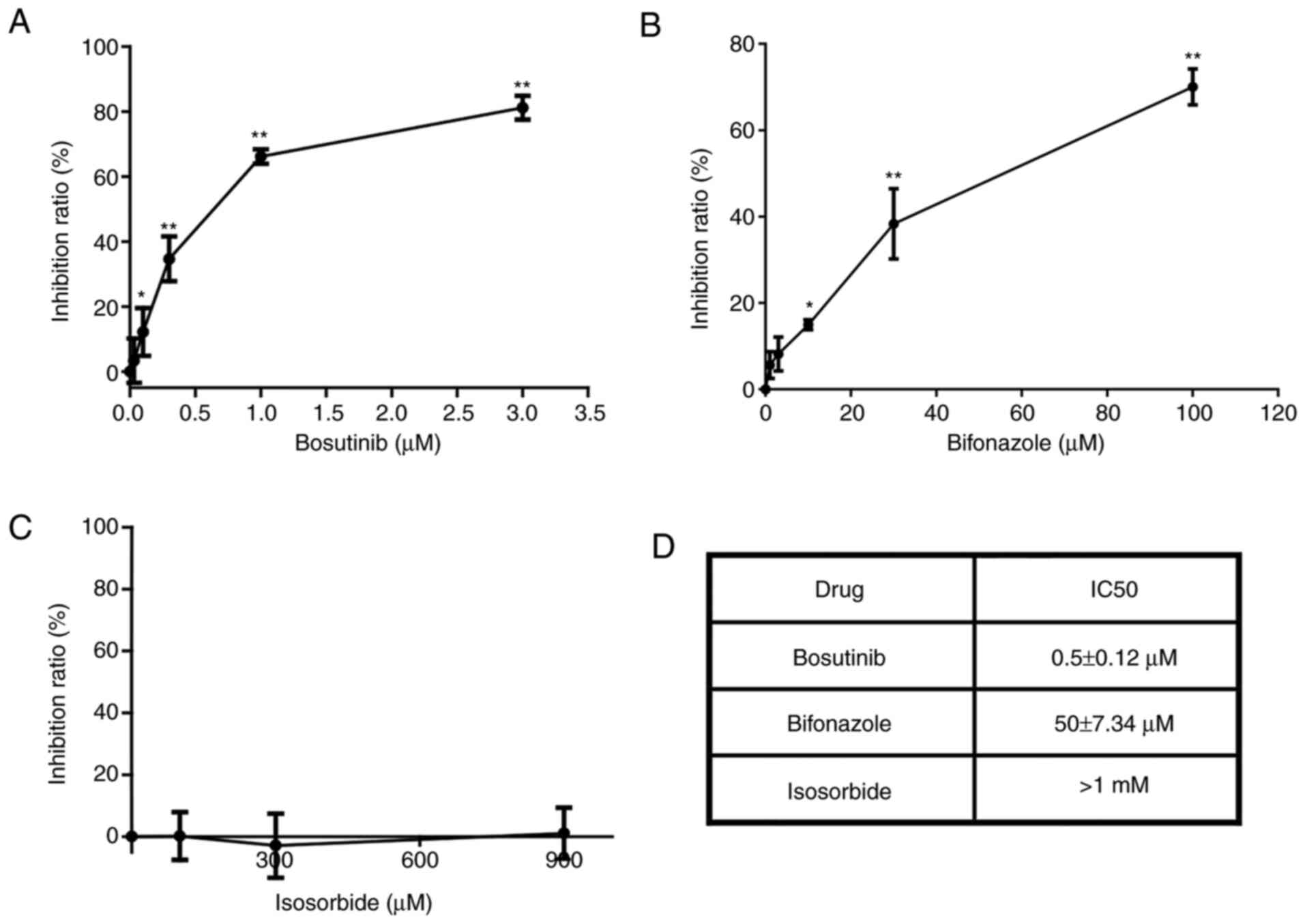
Bosutinib inhibits cell viability
The inhibitory effects of the selected candidate
drugs (bosutinib, bifonazole, and isosorbide) on the viability in
MX-1 cells are presented in Fig.
2. Of these, isosorbide exhibited no evident inhibitory effect
at concentrations <1 mM (Fig. 2C
and D). Bosutinib was more effective (Fig. 2A and B), as its IC50
was only 1% of that of bifonazole (Fig. 2D). Thus, bosutinib was used in
subsequent experiments.
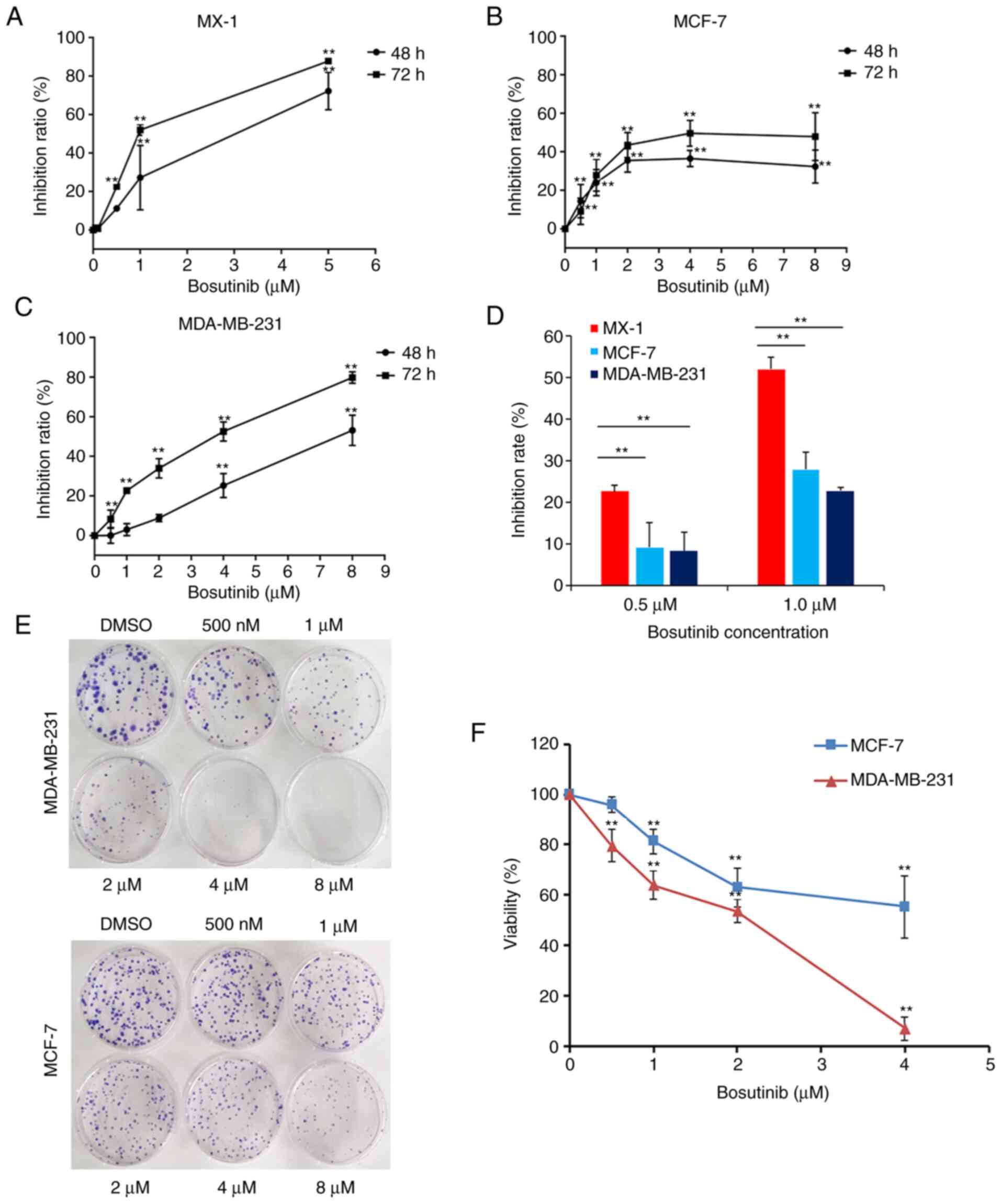
Bosutinib was used to treat MX-1, MCF-7, and
MDA-MB-231 cells for 48 or 72 h. The inhibition rates changed with
the concentration of bosutinib. In MX-1 and MDA-MB-231 cells, the
inhibition rates markedly increased in response to bosutinib
(Fig. 3A and C). However, in
MCF-7 cells, although at the concentration of 2 µM, the
inhibition rates of bosutinib were even higher than that in
MDA-MB-231 cells, this was only the case at concentrations <4
µM of bosutinib, and higher concentrations had no effect
(Fig. 3B). The IC50
of bosutinib in MCF-7 and MDA-MB-231 cells were 5.4±3.63 µM
and 3.2±0.07 µM, respectively. MX-1 cells were much more
sensitive to bosutinib than the other two cell lines at
concentrations of 0.5 and 1.0 µM (Fig. 3D).
Colony formation assays were conducted in MDA-MB-231
and MCF-7 cells. Cell proliferation in the two cell lines was
inhibited by increasing concentrations of bosutinib (Fig. 3E and F). MDA-MB-231 cells were
more sensitive to bosutinib than MCF-7 cells (Fig. 3F).
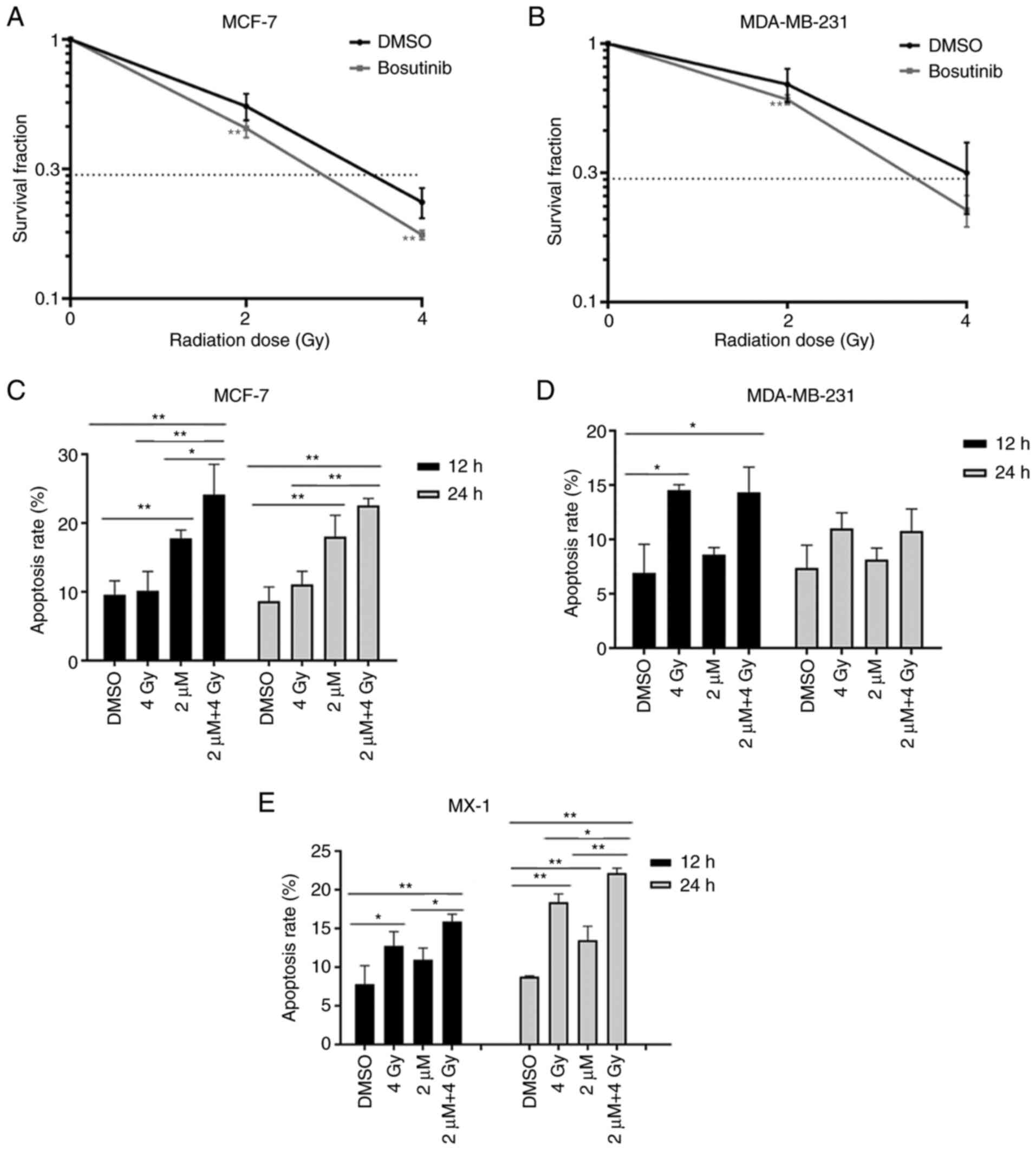
Bosutinib increases the radiosensitivity
of breast cancer cells
Clonogenic assays were used to examine the
radiosensitivity of MCF-7 and MDA-MB-231 cells. The cells were
treated with 2 µM bosutinib for 24 h, then exposed to 2 or 4
Gy of γ-rays. Survival fractions (SF) following 2-Gy irradiation
were 55.18 and 69.64% in MCF-7 and MDA-MB-231 cells, respectively.
SF following 4-Gy irradiation were 23.56 and 31.64% in MCF-7 and
MDA-MB-231 cells, respectively. Exposure to γ-rays alone killed
breast cancer cells, and the viability rates decreased by 44.82 and
30.36% following 2-Gy irradiation in MCF-7 and MDA-MB-231 cells,
respectively. After 4-Gy irradiation, the viability rates were
further reduced by 76.44 and 68.36%, respectively (Fig. 4A and B), suggesting that
MDA-MB-231 cells were more resistant to radiation. Bosutinib alone
decreased the viability rates of the two cell lines by
approximately 40%. When the treatment with irradiation and
bosutinib was applied, SF following 2-Gy irradiation were 27.4 and
36.79% in MCF-7 and MDA-MB-231 cells, respectively. SF following
4-Gy irradiation were 10.63 and 13.65% in MCF-7 and MDA-MB-231
cells, respectively. Compared with irradiation or bosutinib alone,
the combined treatment reduced the viability of MCF-7 and
MDA-MB-231 cells irradiated with 2 Gy from 55.18±6.51% to
27.4±0.74% and 69.64±10.28% to 36.79±2.05%, respectively, and
irradiated with 4 Gy from 23.56±3.14% to 10.63±1.18% and
31.64±9.76% to 13.65±0.79%, respectively (Fig. S1). The combination indices (CI)
in the two cell lines were calculated based on Chou-Talalay method
with the tool CompuSyn. Inhibition rates of MCF-7 and MDA-MB-231
cells treated with bosutinib, radiation alone and their
combinations (Table II) were
used. The CI was 0.62 in MCF-7 cells treated with 2 µM
bosutinib and 2-Gy radiation and 0.63 for 2 µM of bosutinib
with 4-Gy of radiation. In MDA-MB-231 cells, the CI was 0.86 and
0.77 for 2 µM bosutinib with 2-Gy radiation and for 2
µM bosutinib with 4-Gy radiation, respectively. These
findings indicated synergy between bosutinib and radiation,
according to the Chou-Talalay method. Thus, bosutinib may notably
improve the efficiency of radiation in killing cancer cells. These
data suggested that the viability of breast cancer cells was
reduced when bosutinib and irradiation were used in combination,
highlighting bosutinib as a promising candidate radiosensitizer for
its adjuvant effect to irradiation.
 | Table IIInhibition rates of different
concentrations of bosutinib, different doses of radiation and their
combinations in cell lines MCF-7 and MDA-MB-231. |
Table II
Inhibition rates of different
concentrations of bosutinib, different doses of radiation and their
combinations in cell lines MCF-7 and MDA-MB-231.
| Variable | MCF-7 | MDA-MB-231 |
|---|
| DMSO | 0 | 0 |
| Bosu 0.5
µM | 14.37% | 1.71% |
| Bosu 1
µM | 23.93% | 7.24% |
| Bosu 2
µM | 39.77% | 39.36% |
| Bosu 4
µM | 36.53% | 28.74% |
| Radi 2 Gy | 44.82% | 30.38% |
| Radi 4 Gy | 76.44% | 68.36% |
| 2 µM+2
Gy | 72.76% | 63.21% |
| 2 µM+4
Gy | 89.37% | 86.35% |
Apoptosis was detected in all three breast cancer
cells at 12-96 h after irradiation. After 4-Gy irradiation alone,
apoptosis was significantly increased at 12 h in MDA-MB-231 cells
from 6.92 to 14.54% (P<0.01; Fig.
4D). In MX-1 cells, apoptosis increased significantly at 12 h
from 7.83 to 12.77% (P<0.05) and at 24 h from 8.79 to 18.40%
(P<0.01; Fig. 4E). This was
not the case in MCF-7 cells (Fig.
4C). Treatment with bosutinib alone induced apoptosis in MCF-7
and MX-1 cells, but not in MDA-MB-231 cells. Apoptosis induced by
the combined treatment of radiation and bosutinib in MCF-7 cells
was 24.14% at 12 h and 22.56% at 24 h after irradiation. These
apoptotic rates were much higher than those induced by radiation
alone, which were nearly similar to the controls, suggesting an
apparent radiosensitizing effect of bosutinib in MCF-7 cells
(Fig. 4C). In MX-1 cells, the
addition of bosutinib increased the rate of apoptosis from 12.77 to
15.90%, and from 18.40 to 22.15% at 12 and 24 h after irradiation,
respectively, further indicating bosutinib as a potential adjuvant
drug of irradiation (Fig. 4E).
However, bosutinib did not promote apoptosis following irradiation
in MDA-MB-231 cells, both at 12 and 24 h (Fig. 4D). Representative flow cytometry
dot plots for apoptosis in the three cell lines are shown in
Figs. S5-S7.
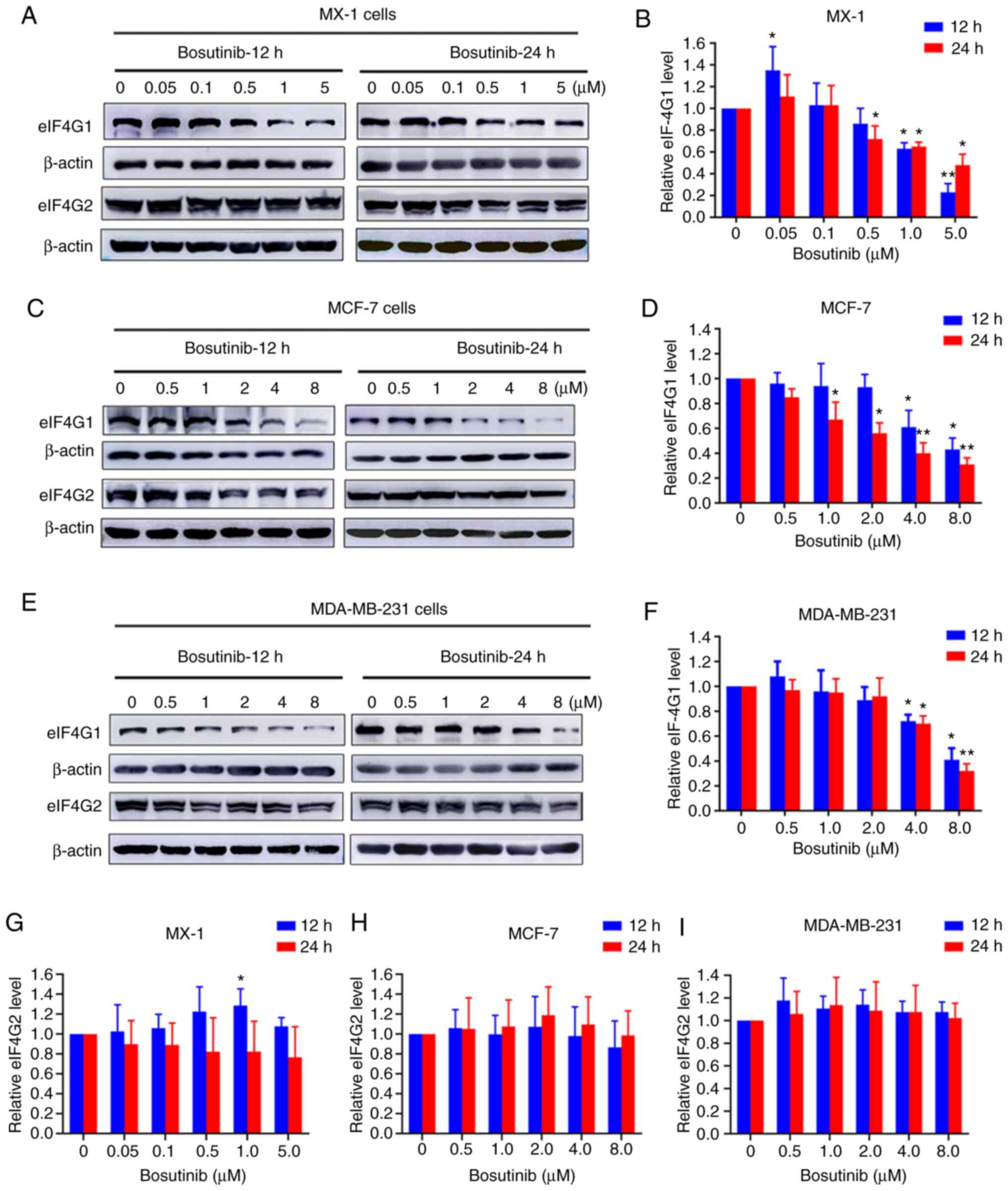
Bosutinib inhibits expression of
eIF4G1
Western blot assays were conducted to detect the
protein expression levels of eIF4G1 and its homolog eIF4G2 in all
three breast cancer cell lines. The expression of eIF4G1 in
MDA-MB-231 was lower than that in the other two cell lines
(Fig. S2A and F). In MX-1
cells, eIF4G1 expression levels decreased in a dose-dependent
manner following treatment with bosutinib (Fig. 5A and B). This decrease was
apparent at the 12-h time-point at bosutinib concentrations of 1
and 5 µM, and at 24 h with concentrations ≥0.5 µM
(Fig. 5B). However, eIF4G2
expression levels did not change markedly (Fig. 5G). In MCF-7 cells, eIF4G1
expression significantly decreased at 12 h at concentrations of ≥4
µM, and at 24 h at concentrations >1 µM (Fig. 5C and D). In MDA-MB-231 cells,
eIF4G1 expression decreased significantly at >4 µM of
bosutinib at both time-points (Fig.
5E and F). However, bosutinib did not affect the expression of
eIF4G2 in MCF-7 and MDA-MB-231 cells (Fig. 5H and I).
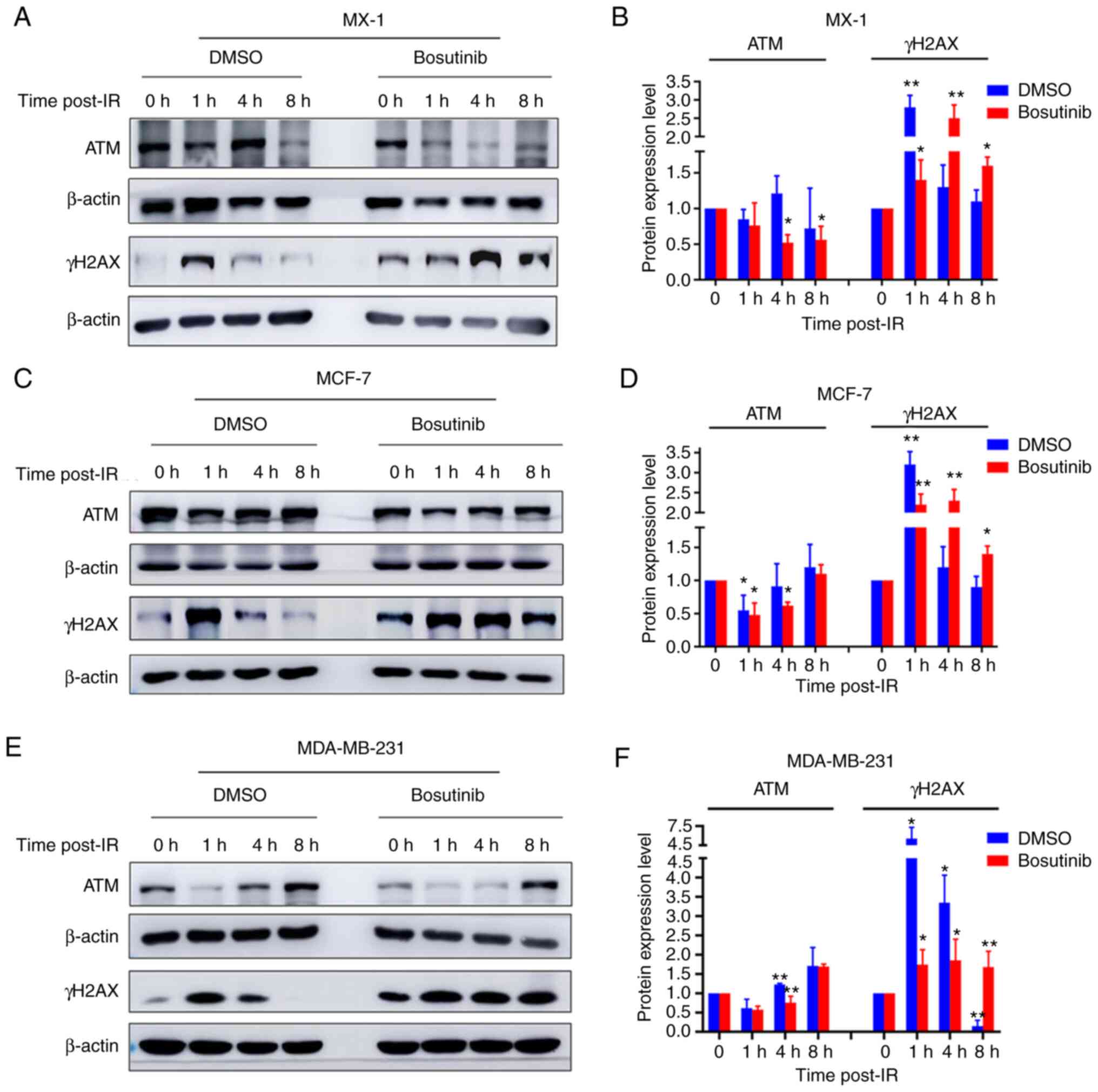
Effect of bosutinib on the expression of
DDR proteins
The effect of bosutinib on the expression of several
DDR proteins was then evaluated. Bosutinib increased the levels of
γH2AX expression, a biomarker of DNA double-strand breaks, or
prolonged its vanishing effect in all three cell lines (Fig. 6), indicating increased DNA damage
or reduced DNA repair efficiency (Fig. S4). The expression levels of
other IR-induced DDR proteins were also measured. The expression
levels of the ATM DNA repair protein were reduced after treatment
with bosutinib, compared with before treatment, in MX-1 (Fig. 6A and B), MCF-7 (Fig. 6C and D) and MDA-MB-231 cells
(Fig. 6E and F). In addition,
the expression levels of DDR proteins XRCC4, ATRIP and GADD45A were
also reduced in MDA-MB-231 and MCF-7 cells after bosutinib
treatment with or without irradiation (Fig. S2B-E). Bosutinib did not alter
the expression levels of PARP-1, Mre11, CDK1 and pCDK1 (Fig. S3).
Discussion
Advances in new physical and biological techniques
in RT, including radiosensitization, are aimed at improving the
clinical effect against cancer invasion and metastasis at a
relatively low dose, with fewer adverse effects (40). Identifying effective
radiosensitizers with a more favorable toxicity profile and
understanding the underlying mechanism of radioresistance and
radiosensitization is needed.
eIF4G1, the most abundant member of the eIF4G
family, is a large scaffolding protein in the eIF4F complex, upon
which ribosomes and eIFs assemble. Increased expression of eIF4G1
is associated with progression and metastasis of several cancer
types (41). Thus, eIF4G1 has
been proposed as a potential anticancer target.
Using computational methods, the present study
investigated the repositioning opportunities of bosutinib, an oral,
first-line, second-generation therapeutic drug approved for the
treatment of newly diagnosed chronic myelogenous leukemia (42), as a new sensitizer for RT. The
gene expression signature of bosutinib was compared with that of
eIF4G1-silenced breast cancer cells in order to determine whether
this drug could improve the sensitivity of the breast cancer cells
to irradiation. The regulatory role of bosutinib in
radiosensitivity and the mechanism underlying the effect of eIF4G1
in DNA double-strand break repair following IR were also
examined.
The results indicated that the rates of apoptosis
following RT in MCF-7 cells was lower compared with the two other
cell lines. The order of sensitivity to higher concentrations of
bosutinib among the three breast cancer cell lines was: MX-1 >
MDA-MB-231 > MCF-7 cells. Apoptosis was not evidently observed
in bosutinib-treated MDA-MB-231 cells, with or without irradiation;
thus, cell killing may be attributed to other mechanisms of cell
death, such as autophagy (41-43), lysosomal membrane
permeabilization (46) and cell
cycle arrest (47). However,
radiation and bosutinib combined significantly enhanced cell
killing. Administration of bosutinib efficiently functioned at
relatively low doses of both radiation and the drug, which may
result in lower toxicity in normal tissues in the clinical
setting.
Furthermore, the present findings also indicated
that bosutinib inhibited eIF4G1 expression in all three cell lines
in a dose-dependent manner. Importantly, bosutinib also inhibited
the expression of several DDR-associated proteins by suppressing
eIF4G1. This could represent a major cause of radiosensitization
through eIF4G1 targeting (26).
Consequently, the efficiency of DNA damage repair was reduced, as
evidenced by the prolonged quenching effect of γH2AX. The mechanism
of radiosensitization mediated by bosutinib could primarily be
associated with eIF4G1. Thus, the present study highlights the
effectiveness of the LINCS gene expression signature strategy in
identifying novel applications of old drugs.
Recent studies have suggested that cancer patients
who have undergone RT may be at increased risk of non-cancer
diseases induced by late effects of radiation, such as
cardiovascular diseases (48)
and stroke (49). A
comprehensive analysis demonstrated that, compared with other
new-generation tyrosine kinase inhibitors, the incidence of
vascular and cardiac treatment-associated adverse events in
patients receiving bosutinib were markedly lower, even after
long-term treatment (50).
Therefore, the addition of bosutinib would not increase the risk of
cardiotoxicity.
Overall, the present study described a powerful
approach to the identification of radiosensitizer candidates, based
on rational computational drug repositioning. The findings on the
mechanism of action of bosutinib in DDR may provide insight into
gene regulation in IR-induced cellular damage. However, further
in vivo experiments are needed to better understand the
mechanisms that could overcome radioresistance in breast cancer
cells. Advances in the development of effective radiosensitizers
and further studies into the mechanism underlying the regulatory
roles of genes involved in IR-induced damage may lead to improved
therapeutic applications that could address radioresistance in
cancer therapy.
In conclusion, using computational methods, the
present study suggested that bosutinib, an FDA-approved drug, may
be a potential radiosensitizer for breast cancer therapy.
Experimental evidence also validated these results and suggested
that eIF4G1 silencing resulted in the downregulation of the DDR
proteins, thereby enhancing radiosensitivity in breast cancer
cells. Thus, bosutinib is a potential candidate radiosensitizer for
breast cancer therapy.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PZ and HG conceived and designed the study. SH, DX,
PZ, XL, XY, BH and HG contributed to the concept, design and
definition of the intellectual content of this study. DX and XY
contributed to the data acquisition, calculation and statistical
analyses. SH, XL and BH contributed to the experimental studies and
statistical analyses. PZ and DX contributed to preparation and
review of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present was supported by the National Key Basic Research
Program (973 Program) of MOST, China (grant no. 2015CB910601), the
National Natural Science Foundation of China (grant nos. 11705283,
81530085 and 31870847), and the Young Elite Scientists Sponsorship
Program of CAST (grant no. CSTQT2017003).
References
|
1
|
Bernier J: Precision medicine for early
breast cancer radiotherapy: Opening up new horizons? Crit Rev Oncol
Hematol. 113:79–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bennett MH, Feldmeier J, Smee R and
Milross C: Hyperbaric oxygenation for tumour sensitisation to
radiotherapy. Cochrane Database Syst Rev. 11:CD0050072012.
|
|
3
|
Schütze A, Vogeley C, Gorges T, Twarock S,
Butschan J, Babayan A, Klein D, Knauer SK, Metzen E, Müller V, et
al: RHAMM splice variants confer radiosensitivity in human breast
cancer cell lines. Oncotarget. 7:21428–21440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang RX and Zhou PK: DNA damage response
signaling pathways and targets for radiotherapy sensitization in
cancer. Signal Transduct Target Ther. 5:602020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai Y, Yu X, Lin X and He S: Inhibition of
mTOR sensitizes breast cancer stem cells to radiation-induced
repression of self-renewal through the regulation of MnSOD and Akt.
Int J Mol Med. 37:369–377. 2016. View Article : Google Scholar :
|
|
6
|
Silvera D, Formenti SC and Schneider RJ:
Translational control in cancer. Nat Rev Cancer. 10:254–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peretz S, Jensen R, Baserga R and Glazer
PM: ATM-Dependent expression of the insulin-like growth factor-I
receptor in a pathway regulating radiation response. Proc Natl Acad
Sci USA. 98:1676–1681. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo YM, Xia NX, Yang L, Li Z, Yang H, Yu
HJ, Liu Y, Lei H, Zhou FX, Xie CH, et al: CTC1 increases the
radioresistance of human melanoma cells by inhibiting telomere
shortening and apoptosis. Int J Mol Med. 33:1484–1490. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dolman MEM, van der Ploeg I, Koster J,
Bate-Eya LT, Versteeg R, Caron HN and Molenaar JJ: DNA-Dependent
protein kinase as molecular target for radiosensitization of
neuroblastoma cells. PLoS One. 10:e01457442015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rachmadi L, Siregar NC, Kanoko M,
Andrijono A, Bardosono S, Suryandari DA, Sekarutami SM and Hernowo
BS: Role of cancer stem cell, apoptotic factor, DNA repair, and
telomerase toward radiation therapy response in stage IIIB cervical
cancer. Oman Med J. 34:224–230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu CC, Huang H, Hung SK, Liao HF, Lee CC,
Lin HY, Li SC, Ho HC, Hung CL and Su YC: AZD2014 radiosensitizes
oral squamous cell carcinoma by inhibiting AKT/mTOR axis and
inducing G1/G2/M cell cycle arrest. PLoS One. 11:e01519422016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gewirtz DA: The four faces of autophagy:
Implications for cancer therapy. Cancer Res. 74:647–651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khoshinani HM, Afshar S and Najafi R:
Hypoxia: A double-edged sword in cancer therapy. Cancer Invest.
34:536–545. 2016. View Article : Google Scholar
|
|
14
|
Saenko YV, Mastilenko AV, Glushchenko ES,
Antonova AV and Svekolkin VP: Inhibition of mitochondrial
voltage-dependent anion channels increases radiosensitivity of K562
leukemic cells. Bull Exp Biol Med. 161:104–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baro M, Sambrooks CL, Quijano A, Saltzman
WM and Contessa J: Oligosaccharyltransferase inhibition reduces
receptor tyrosine kinase activation and enhances glioma
radiosensitivity. Clin Cancer Res. 25:784–795. 2019. View Article : Google Scholar :
|
|
16
|
Segawa T, Fujii Y, Tanaka A, Bando SI,
Okayasu R, Ohnishi K and Kubota N: Radiosensitization of human lung
cancer cells by the novel purine-scaffold Hsp90 inhibitor, PU-H71.
Int J Mol Med. 33:559–564. 2014. View Article : Google Scholar
|
|
17
|
Zhang L, Yoo S, Dritschilo A, Belyaev I
and Soldatenkov V: Targeting ku protein for sensitizing of breast
cancer cells to DNA-damage. Int J Mol Med. 14:153–159.
2004.PubMed/NCBI
|
|
18
|
Zhang F, Fan B and Mao L: Radiosensitizing
effects of cyclocarya paliurus polysaccharide on hypoxic A549 and
H520 human non-small cell lung carcinoma cells. Int J Mol Med.
44:1233–1242. 2019.PubMed/NCBI
|
|
19
|
Chang L, Graham PH, Hao J, Ni J, Bucci J,
Cozzi PJ, Kearsley JH and Li Y: PI3K/Akt/mTOR pathway inhibitors
enhance radiosensitivity in radioresistant prostate cancer cells
through inducing apoptosis, reducing autophagy, suppressing NHEJ
and HR repair pathways. Cell Death Dis. 5:e14372014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boguski MS, Mandl KD and Sukhatme VP: Drug
discovery. Repurposing with a difference Science. 324:1394–1395.
2009.
|
|
21
|
Rasheed S, Sánchez SS, Yousuf S, Honoré SM
and Choudhary MI: Drug repurposing: In-vitro anti-glycation
properties of 18 common drugs. PLoS One. 13:e01905092018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amsberg GK and Schafhausen P: Bosutinib in
the management of chronic myelogenous leukemia. Biologics.
7:115–122. 2013.PubMed/NCBI
|
|
23
|
Amsberg GKV and Brümmendorf TH: Novel
aspects of therapy with the dual src and abl kinase inhibitor
bosutinib in chronic myeloid leukemia. Expert Rev Anticancer Ther.
12:1121–1127. 2012. View Article : Google Scholar
|
|
24
|
Lee JH, Choi SI, Kim RK, Cho EW and Kim
IG: Tescalcin/c-Src/IGF1Rβ-mediated STAT3 activation enhances
cancer stemness and radioresistant properties through ALDH1. Sci
Rep. 8:107112018. View Article : Google Scholar
|
|
25
|
Cammarata FP, Torrisi F, Forte GI, Minafra
L, Bravatà V, Pisciotta P, Savoca G, Calvaruso M, Petringa G,
Cirrone GA, et al: Proton therapy and src family kinase inhibitor
combined treatments on U87 human glioblastoma multiforme cell line.
Int J Mol Sci. 20:47452019. View Article : Google Scholar :
|
|
26
|
Badura M, Braunstein S, Zavadil J and
Schneider RJ: DNA damage and eIF4G1 in breast cancer cells
reprogram translation for survival and DNA repair mRNAs. Proc Natl
Acad Sci USA. 109:18767–18772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Keenan AB, Jenkins SL, Jagodnik KM, Koplev
S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A,
et al: The library of integrated network-based cellular signatures
NIH program: System-Level cataloging of human cells response to
perturbations. Cell Syst. 6:13–24. 2018. View Article : Google Scholar
|
|
28
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pfeffer RM: Radiotherapy for breast
cancer: Curing the cancer while protecting the heart. Isr Med Assoc
J. 20:582–583. 2018.PubMed/NCBI
|
|
31
|
Taylor CW and Kirby AM: Cardiac
side-effects from breast cancer radiotherapy. Clin Oncol (R Coll
Radiol). 27:621–629. 2015. View Article : Google Scholar
|
|
32
|
Tarpley M, Abdissa TT, Johnson GL and
Scott JE: Bosutinib reduces the efficacy of dasatinib in
triple-negative breast cancer cell lines. Anticancer Res.
34:1629–1635. 2014.PubMed/NCBI
|
|
33
|
Tan DSW, Haaland B, Gan JM, Tham SC, Sinha
I, Tan EH, Lim KH, Takano A, Krisna SS, Thu MM, et al: Bosutinib
inhibits migration and invasion via ACK1 in KRAS mutant non-small
cell lung cancer. Mol Cancer. 13:132014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wie SM, Wellberg E, Karam SD and Reyland
ME: Tyrosine kinase inhibitors protect the salivary gland from
radiation damage by inhibiting activation of protein kinase C-δ.
Mol Cancer Ther. 16:1989–1998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wenzel T, Büch T, Urban N, Weirauch U,
Schierle K, Aigner A, Schaefer M and Kalwa H: Restoration of MARCKS
enhances chemosensitivity in cancer. J Cancer Res Clin Oncol.
146:843–858. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chou TC: Drug combination studies and
their synergy quantification using the chou-talalay method. Cancer
Res. 70:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ellegaard AM, Groth-Pedersen L, Oorschot
V, Klumperman J, Kirkegaard T, Nylandsted J and Jäättelä M:
Sunitinib and SU11652 inhibit acid sphingomyelinase, destabilize
lysosomes, and inhibit multidrug resistance. Mol Cancer Ther.
12:2018–2030. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Evelyn CR, Lisabeth EM, Wade SM, Haak AJ,
Johnson CN, Lawlor ER and Neubig RR: Small-Molecule inhibition of
Rho/MKL/SRF transcription in prostate cancer cells: Modulation of
cell cycle, er stress, and metastasis gene networks. Microarrays
(Basel). 5:132016. View Article : Google Scholar
|
|
39
|
Wainberg ZA, Alsina M, Soares HP, Braña I,
Britten CD, Conte GD, Ezeh P, Houk B, Kern KA, Leong S, et al: A
multi-arm phase I study of the PI3K/mTOR inhibitors PF-04691502 and
gedatolisib (PF-05212384) plus irinotecan or the MEK inhibitor
PD-0325901 in advanced cancer. Target Oncol. 12:775–785. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hamilton DG, Bale R, Jones C, Fitzgerald
E, Khor R, Knight K and Wasiak J: Impact of tumour bed boost
integration on acute and late toxicity in patients with breast
cancer: A systematic review. Breast. 27:126–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jaiswal PK, Koul S, Shanmugam PST and Koul
HK: Eukaryotic translation initiation factor 4 gamma 1 (eIF4G1) is
upregulated during prostate cancer progression and modulates cell
growth and metastasis. Sci Rep. 8:74592018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gourd E: Bosutinib more effective than
imatinib in CML. Lancet Oncol. 18:e7162017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu MB, Zhang JW, Gao JB, Qi YW, Gao Y, Xu
L, Ma Y and Wei ZZ: Atorvastatin induces autophagy in MDA-MB-231
breast cancer cells. Ultrastruct Pathol. 42:409–415. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chang CH, Bijian K, Wernic D, Su J, da
Silva SD, Yu H, Qiu D, Asslan M and Alaoui-Jamali MA: A novel
orally available seleno-purine molecule suppresses triple-negative
breast cancer cell proliferation and progression to metastasis by
inducing cytostatic autophagy. Autophagy. 15:1376–1390. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin SC, Chu PY, Liao WT, Wu MY, Tsui KH,
Lin LT, Huang CH, Chen LL and Li CJ: Glycyrrhizic acid induces
human MDA-MB-231 breast cancer cell death and autophagy via the
ROS-mitochondrial pathway. Oncol Rep. 39:703–710. 2018.
|
|
46
|
Noguchi S, Shibutani S, Fukushima K, Mori
T, Igase M and Mizuno T: Bosutinib, an SRC inhibitor, induces
caspase-independent cell death associated with permeabilization of
lysosomal membranes in melanoma cells. Vet Comp Oncol. 16:69–76.
2018. View Article : Google Scholar
|
|
47
|
Nam AR, Kim JW, Park JE, Bang JH, Jin MH,
Lee KH, Kim TY, Han SW, Im SA, Kim TY, et al: Src as a therapeutic
target in biliary tract cancer. Mol Cancer Ther. 15:1515–1524.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weberpals J, Jansen L, Müller OJ and
Brenner H: Long-Term heart-specific mortality among 347 476 breast
cancer patients treated with radiotherapy or chemotherapy: A
registry-based cohort study. Eur Heart J. 39:3896–3903. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang R, Zhou Y, Hu S, Ren G, Cui F and
Zhou PK: Radiotherapy exposure in cancer patients and subsequent
risk of stroke: A systematic review and meta-analysis. Front
Neurol. 10:2332019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cortes JE, Khoury HJ, Kantarjian H,
Brümmendorf TH, Mauro MJ, Matczak E, Pavlov D, Aguiar JM, Fly KD,
Dimitrov S, et al: Long-Term evaluation of cardiac and vascular
toxicity in patients with philadelphia chromosome-positive
leukemias treated with bosutinib. Am J Hematol. 91:606–616. 2016.
View Article : Google Scholar : PubMed/NCBI
|