Introduction
Retinoblastoma (RB) is an eye malignancy that
affects children primarily triggered by mutations in RB genes, the
aberrant expression of intracellular molecules and by the
activation of oncogenic pathways during the development of the
retina (1-3). Approximately 8,000 children are
diagnosed with RB each year (4).
The clinical symptoms of RB include white pupillary reflex,
conjunctival congestion, vitreous opacity, corneal edema,
strabismus and increasing intraocular pressure (5,6).
Currently, focal therapy (laser and cryotherapy), surgery and
chemotherapy are the major treatment strategies for RB (7,8).
RB can be cured over time with a graduated-intensity approach based
on pathology in developed or even developing countries (9-11). However, the therapeutic effect
remains unsatisfactory for children with metastatic and/or advanced
RB (9). Therefore, the
development of novel targets at the molecular level for RB therapy
is crucial.
Long non-coding RNAs (lncRNAs), a type of non-coding
RNA composed of >200 nucleotides, regulate the expression of
genes by regulating transcription, post-transcription and chromatin
modifications (12,13). lncRNAs function as important
regulators in the progression of RB (14-16). For instance, the overexpression
of lncRNA RB-associated transcript-1 can accelerate the
tumorigenesis of RB (14). The
downregulation of lncRNA ILF3 antisense RNA 1 and lncRNA LINC00324
has been shown to suppress the proliferation and invasion of RB
cells, as well as tumor growth in mice (15,16). lncRNA HLA complex P5 (HCP5), a
member of the new pseudogene family P5 (17), is mainly expressed in the immune
system, such as in the spleen, blood and thymus (18). HCP5 plays a role in the
progression of several types of human cancers (19-21). The study by Bai et al
indicated that the downregulation of HCP5 suppressed the viability,
migration and invasion of colorectal cancer (CRC) cells and impeded
the malignant behavior of CRC in vivo (19). Wang et al revealed that
the knockdown of HCP5 exerted an inhibitory effect on the growth
and metastasis of ovarian cancer (OC) in mice (20). Similarly, Yuan et al
suggested that the progression of pancreatic cancer was
significantly suppressed by the silencing of HCP5 (21). Nevertheless, the specific role
and potential mechanisms of HCP5 in RB are relatively unknown.
The participation of microRNAs (miRNAs or miRs) in
the pathogenesis of RB has been uncovered (22,23). Wan et al reported that
miR-25-3p exertd a protective effect against RB tumorigenesis by
dampening the proliferation, migration and invasion of RB (22). Li and You demonstrated that
miR-758 functioned as a suppressor of the metastasis of RB
(23). Recent studies
demonstrated that miR-3619-5p played an anti-tumor role in liver
cancer and RB (24,25). More importantly, the interaction
of miR-3619-5p with lncRNA LINC00202 has been shown to considerably
contribute to the suppression of the progression of RB (25). It remains unclear however, as to
whether the inhibitory effect of miR-3619-5p in RB tumorigenesis is
modulated by HCP5.
Histone deacetylase 9 (HDAC9), a member of class II
HDACs, possesses a conserved domain of HDAC and can interact with
tissue-specific transcription factors and co-repressors (26). In recent years, the
carcinogenesis of HDAC9 in RB has attracted increasing attention
(27-29). Both Zhang et al (27) and Xu et al (29) suggested that the downregulation
of HDAC9 suppressed the proliferation of RB cells, thus,
attenuating RB in vitro (27,29). Jin et al revealed that the
overexpression of HDAC9 reversed the anti-proliferative effect of
miR-101-3p on RB cells (28).
Nevertheless, the interactions between miR-3619-5p and HDAC9, as
well as the HCP5/miR-3619-5p/HDAC9 axis in the progression of RB
remain relatively unknown.
The present study thus focused on investigating the
expression and roles of HCP5 and miR-3619-5p in RB, and on
exploring the association between HCP5, miR-3619-5p and HDAC9 in RB
cells. Collectively, the findings of the present study may provide
a novel target for the treatment of RB.
Materials and methods
Collection of samples
RB tissues and adjacent normal tissues in pairs
(n=71) were obtained from patients who were diagnosed with RB
(unilateral, 51; bilateral, 20) from June, 2017 to July, 2019 at
the Affiliated Hospital of Weifang Medical University. Among the
patients, 41 lymph node metastases and 2 distant metastasis cases
(1 cerebral spinal fluid and 1 brain) were included. The tumor node
metastasis (TNM) stage (30),
intraocular international RB classification (IIRC) stage (31) and Reese Ellsworth (RE) stage
(32) were used to classify RB.
The present study was conducted in accordance with the Declaration
of Helsinki and approved by the Ethics Committee of the Affiliated
Hospital of Weifang Medical University [no. 2020(12)]. Written informed consent was
obtained from all patients, and parental consent was obtained in
cases where the patient was <18 years old.
Cells and cell culture
A human normal retinal pigmented epithelium cell
line (APRE-19) and 4 human RB cell lines (Y79, HXO-RB44, WERI-Rb-1
and SO-RB50) were purchased from the American Type Culture
Collection (ATCC). The ARPE-19 cells were cultured in Dulbecco's
modified Eagle's medium (Invitrogen; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.). The Y79, Weri-RB1, SO-RB50 and HXO-RB44 cells
were cultured in modified Roswell Park Memorial Institute-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS.
All cells were incubated at 37°C in a humidified atmosphere with 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA isolated from the tissues or cells was
analyzed using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The extracted RNA was reverse transcribed into
complementary DNA (cDNA) using a Prime-Script RT reagent kit
(Takara Biotechnology Co., Ltd.). Subsequently, SYBR®
Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) was utilized for
the amplification of cDNA. The reaction conditions were 95°C for 5
min, 40 cycles of 95°C for 15 sec, 60°C for 20 sec, and 70°C for 15
sec. The primer sequences used in the present study are listed in
Table I. The relative expression
of HCP5, miR-3619-5p and HDAC9 was calculated using the
2−ΔΔCq method (33).
The endogenous controls were U6 and GAPDH.
 | Table IPrimer sequences used for RT-qPCR in
the present study. |
Table I
Primer sequences used for RT-qPCR in
the present study.
| Gene | Forward | Reverse |
|---|
| HCP5 |
5′-CCGCTGGTCTCTGGACACATACT-3′ |
5′-CTCACCTGTCGTGGGATTTTGC-3′ |
| miR-3619-5p |
5′-UCAGCAGGCAGGCUGGUGCAGC-3′ |
5′-GCUGCACCAGCCUGCCUGCUGA-3′ |
| HDAC9 |
5′-ATGGTTTCACAGCAACGCATT-3′ |
5′-ACCTTGCCTAAGCGTCTGC-3′ |
| GAPDH |
5′-GGAGCGAGATCCCTCCAAAAT-3′ |
5′-GGCTGTTGTCATACTTCTCATGG-3′ |
| U6 |
5′-TCCGATCGTGAAGCGTTC-3′ |
5′-GTGCAGGGTCCGAGGT-3′ |
Cell transfection
Short hairpin (sh) RNA against HCP5 (sh-HCP5),
sh-negative control (sh-NC), miR-NC and miR-3619-5p mimics, as well
as miR-3619-5p inhibitor, pcDNA-HDAC9, pcDNA-MS2, pBobi-MS2-GFP and
pcDNA-HCP5-MS2 were purchased from Guangzhou RiboBio Co., Ltd.
sh-HCP5 or sh-NC were firstly cloned into a lentiviral vector.
These factors (all, 50 nM) were then transfected into Y79 and/or
HXO-RB44 cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Then, 48 h after transfection, the cells were
harvested for further experiments.
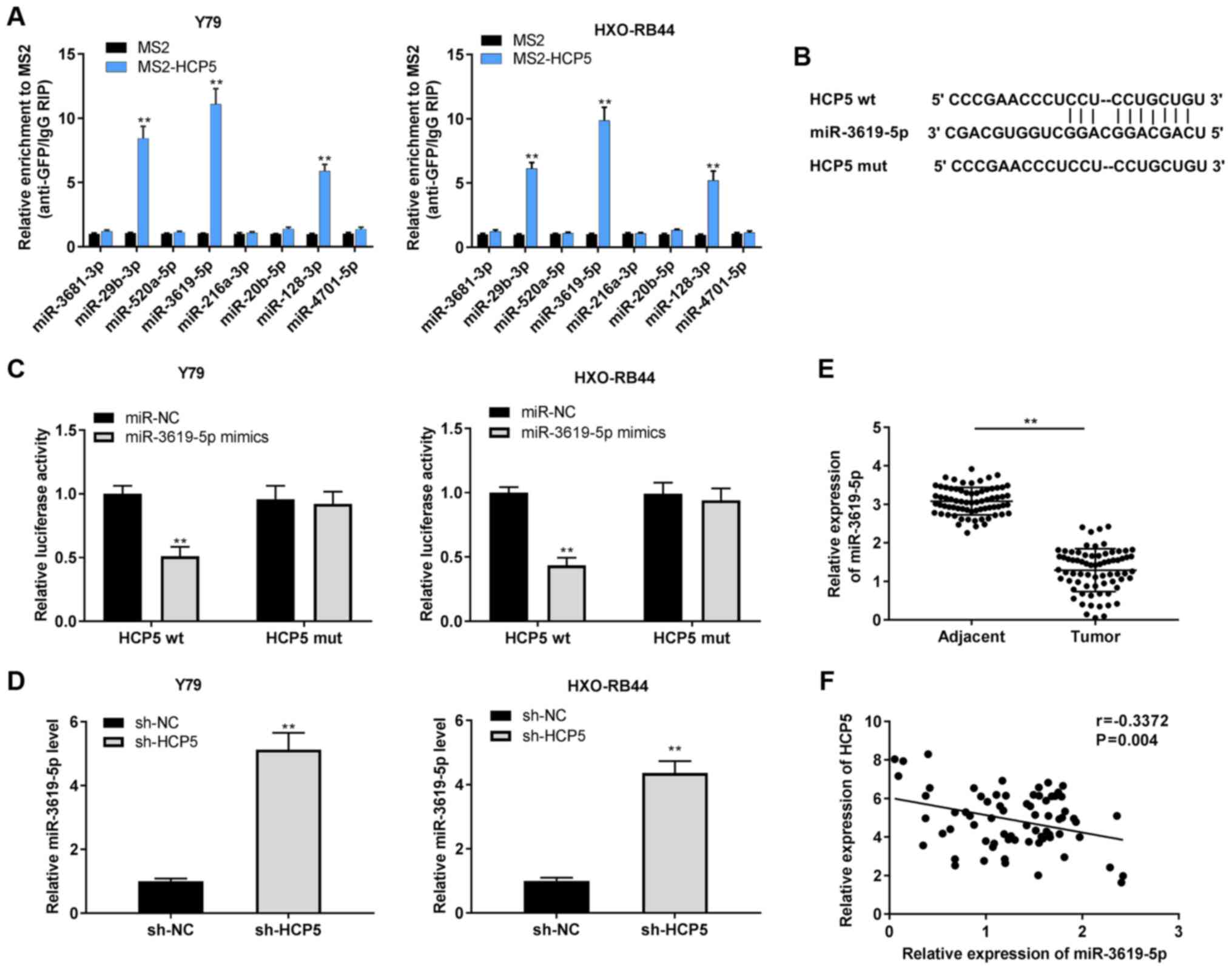
Target prediction
The target miRNAs of HCP5 were predicted using
StarBase v2.0 software (http://starbase.sysu.edu.cn). A total of 8 miRNAs
(miR-3681-3p, miR-29b-3p, miR-520a-5p, miR-3619-5p, miR-216a-3p,
miR-20b-5p, miR-128-3p, and miR-4701-5p) were selected for
verification via RNA immunoprecipitation (RIP) assay. Similarly,
the target genes of miR-3619-5p were also predicted using StarBase
v2.0 software. HDAC9 was selected due to its important role in
RB.
RIP assay
pcDNA-MS2, pBobi-MS2-GFP and pcDNAHCP5-MS2 were
transfected into the Y79 or HXO-RB44 cells. The RIP assay was
performed with anti-GFP antibody (1:5,000; ab6673, Abcam), as well
as a MagnaRIP RNA-Binding Protein Immunoprecipitation kit (EMD
Millipore). The expression of the aforementioned miRNAs was
detected by RT-qPCR.
Dual luciferase reporter (DLR) assay
The 3′-UTR fragment of HCP5, including the predicted
or mutated binding site for miR-3619-5p, was introduced into
psiCHECK2 (Promega Corporation) to establish HCP5 wild-type (wt) or
HCP5 mutant-type (mut). Similarly, the 3′-UTR sequence of HDAC9
containing the predicted or mutated binding site for miR-3619-5p
was inserted into psiCHECK2 (Promega Corporation) to construct
HDAC9 wt or mut. For reporter assays, one of the above-mentioned
vectors (80 ng) along with miR-3619-5p mimics or miR-NC (50 nM)
were co-transfected into the Y79 and HXO-RB44 cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following transfection for 48 h, relative luciferase activity was
examined using a DLR assay System (Promega Corporation). The
activity of Firefly luciferase was normalized to that of
Renilla luciferase.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT) assay
The transfected cells were plated into 96-well
plates at a density of 5,000 cells/well and cultured for 48 h at
37°C. MTT reagent (20 µl; Sigma-Aldrich; Merck KGaA) was
then added to each well at different time points (24, 48, 72 and 96
h) followed by incubation for 4 h at 37°C. After discarding the
medium, 150 µl of dimethyl sulfoxide (Sigma-Aldrich; Merck
KGaA) were added to each well. The optical density was measured at
490 nM using an enzyme immunoassay instrument (Bio-Tek Instruments,
Inc.).
Cell migration and invasion assays
The 24-well Transwell chambers (8 µM pore
size; BD Biosciences) coated with Matrigel (BD Biosciences) were
used to evaluate cell invasion. Cells (1×105) were
resuspended in 200 µl of serum-free medium and then plated
into the upper chambers of each Transwell apparatus. A total of 600
µl of medium containing 10% FBS was added to the lower
chambers followed by incubation at 37°C for 48 h. Subsequently, the
cells in the upper chambers were wiped off using a cotton swab, and
those adhering to the lower chambers were fixed with 4%
paraformaldehyde and stained with 0.5% crystal violet (TCI
chemicals) at 37°C for 30 min. Stained cells were imaged using an
inverted light microscope (Olympus Corporation) and analyzed using
ImageJ software [version 1.46, National Institutes of Health
(NIH)].
For the cell migration assay, the procedure was
similar to the cell invasion assay, except that the 24-well
Transwell chambers were not pre-coated with Matrigel.
Western blot analysis
Total protein was extracted from the transfected Y9
and/or HXO-RB44 cells using RIPA lysis buffer (Beyotime Institute
of Biotechnology). The protein concentration was detected using the
BCA Protein Assay kit (Abcam). Proteins were separated by 10%
sodium dodecyl sulphate-polyacrylamide gel electrophoresis and
transferred onto polyvinylidene fluoride membranes
(MilliporeSigma). The membranes were then blocked with 5% non-fat
milk for 2 h followed by incubation with the primary antibodies
anti-HDAC9 (1:1,000; ab109446, Abcam) and anti-GAPDH (1:1,000;
ab9485, Abcam) overnight at 4°C. Tris-buffered saline Tween-20 was
then used to wash the membranes 3 times. Subsequently,
HRP-conjugated anti-rabbit IgG secondary antibody (1:5,000;
ab205718, Abcam) was added followed by incubation for 1 h at 37°C.
Finally, protein bands were visualized using an enhanced
chemiluminescence system (Thermo Fisher Scientific, Inc.). The
relative protein expression over GAPDH was quantified using Alpha
Innotech imaging software (ProteinSimple).
Xenograft tumor model
Female BALB/c nude mice (4-5 weeks of age, weighing
20-25 g) were obtained from the Shanghai Laboratory Animal Centre
(Shanghai, China). All mice were housed under controlled conditions
(25°C, 50% humidity, 12 h light/dark cycle) and were provided with
free access to food and water. Subsequently, the mice were randomly
divided into the 2 following groups: The sh-NC group (n=5) and the
sh-HCP5 group (n=5). sh-HCP5 or sh-NC were integrated into a
lentiviral vector and transfected into the Y79 cells. Furthermore,
the transfected Y79 cells (2×106 cells/100 µl,
s.c.) were injected into the right flanks of the nude mice. Tumor
volumes were measured every other week using the following formula:
(A × B2)/2, (A, the longest diameter; B, the shortest
diameter). After 4 weeks, the mice were anaesthetized with
pentobarbital sodium (50 mg/kg) and euthanized by cervical
dislocation. The tumor xenografts were separated from the mice and
weighed. The animal experiments were approved by the Ethics
Committee of the Affiliated Hospital of Weifang Medical University
[no. 2020(12)] and were
performed in accordance with the institutional guide for the care
and use of laboratory animals (National Institutes of Health).
Statistical analysis
In vitro experiments were performed in
triplicate, and each experiment was repeated 3 times. In
vivo experiments were performed using 5 mice per group.
Statistical analyses were performed using SPSS Statistics 22.0. The
Student's t-test was used for comparisons between 2 groups (paired
t-test for the data in Figs. 1A,
4E, and 6C; unpaired t-test for the data in
Figs. 1B, C, D, 2C, D, 3A, C, 4D, 5C,
D and 6F). One-way ANOVA was
used to assess differences among multiple groups, followed by
Tukey's post hoc test. Pearson's correlation analysis was used to
determine the correlation between HCP5 and miR-3619-5p expression,
as well as that between miR-3619-5p and HDAC9, and HDAC9 and HCP5
expression in RB tissues. Data are presented as the means ±
standard deviation (SD). The Chi-squared test was used to analyze
the categorical data presented in Table II. Statistically significant
differences were regarded as those with values of P<0.05.
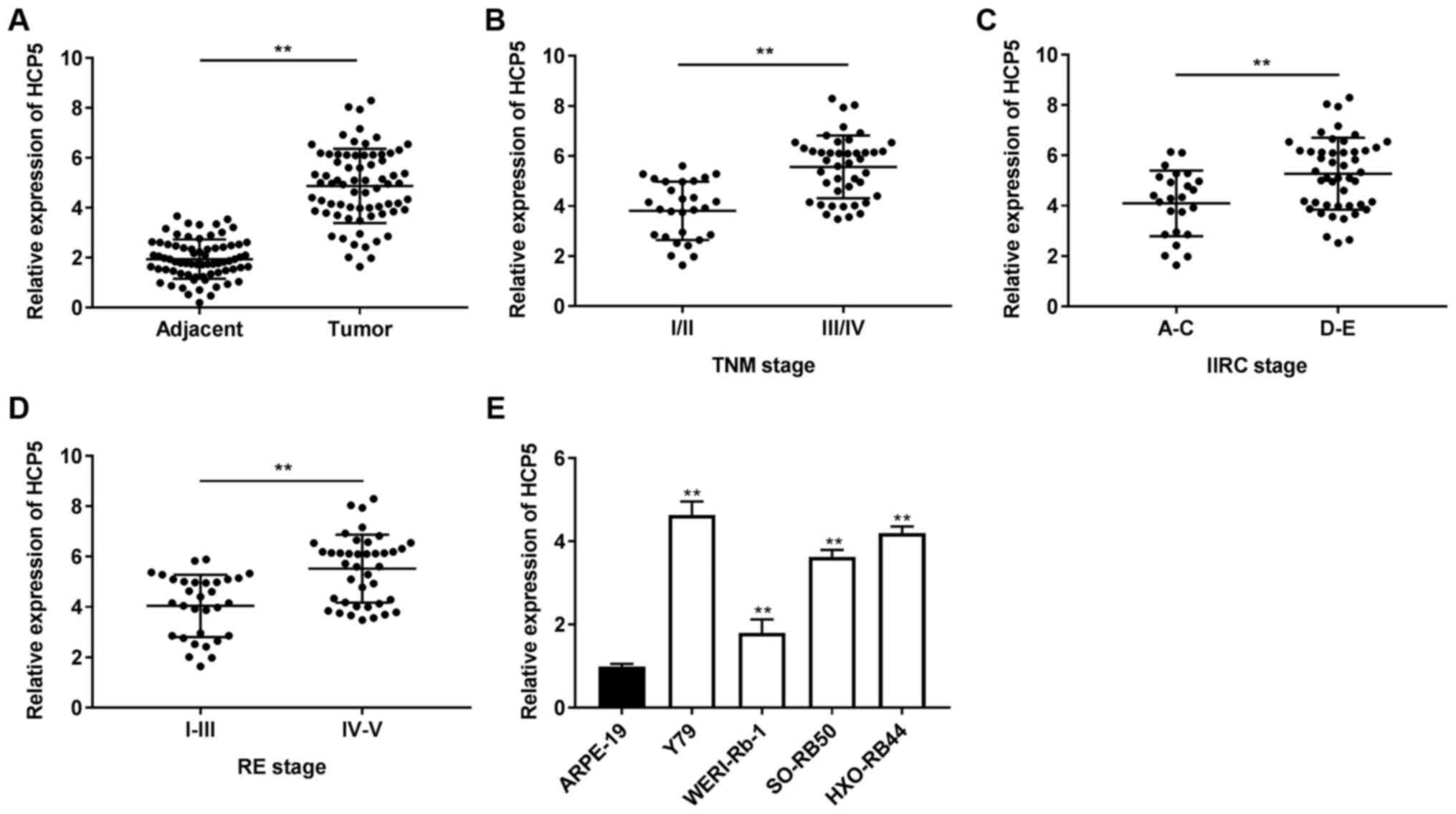 | Figure 1lncRNA HCP5 is highly expressed in
retinoblastoma tissues and cells. (A) Relative expression of HCP5
in tumor tissues and adjacent normal tissues was determined by
RT-qPCR. **P<0.01, vs. adjacent normal tissues. (B)
Relative expression of HCP5 in tumors at TNM stage I/II and III/IV
was determined by RT-qPCR. **P<0.01, vs. TNM stage
I/II. (C) Relative expression of HCP5 in IIRC stage A/B/C and IIRC
stage D/E was determined by RT-qPCR. **P<0.01, vs.
IIRC stage A/B/C. (D) Relative expression of HCP5 in RE stage
I/II/III and RE stage IV/V was determined by RT-qPCR.
**P<0.01, vs. RE stage I/II/III. (E) Relative
expression of HCP5 in ARPE-19, Y79, HXO-RB44, WERI-Rb-1 and SO-RB50
cells was determined by RT-qPCR. **P<0.01, vs.
ARPE-19 cells. The experiments were performed in triplicate and
repeated 3 times. Data are presented as the means ± standard
deviation (SD). TNM, tumor node metastasis; IIRC, intraocular
international retinoblastoma classification; RE, Reese
Ellsworth. |
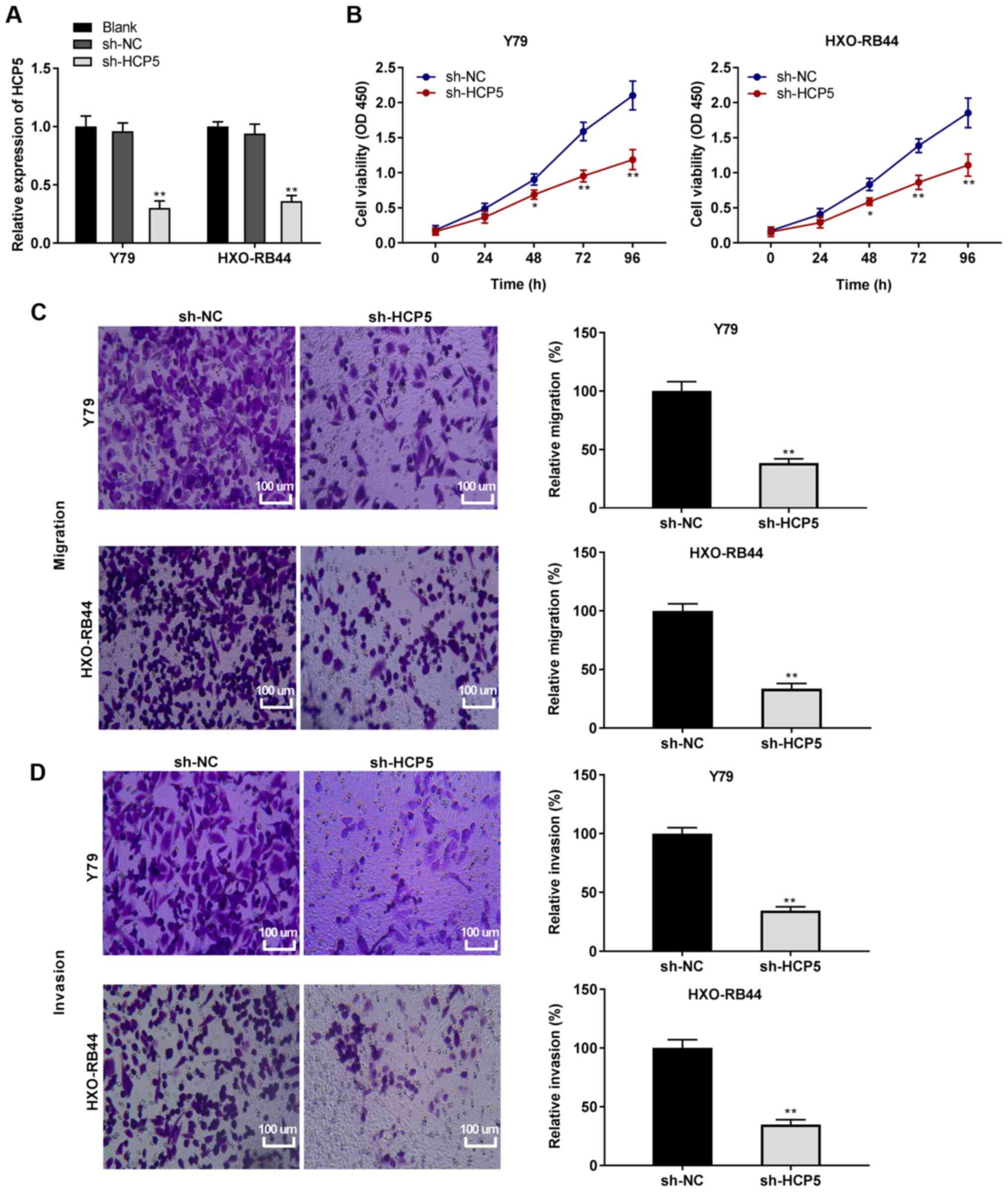 | Figure 2Silencing of lncRNA HCP5 suppresses
the viability, migration and invasion of retinoblastoma cells. (A)
Following transfection with short hairpin (sh)-HCP5 and sh-negative
control (sh-NC), the relative expression of HCP5 in Y79 and
HXO-RB44 cells was detected by RT-qPCR. **P<0.01, vs.
sh-NC. (B) Following transfection with sh-HCP5 and sh-NC, the
viability of Y79 and HXO-RB44 cells was detected MTT assay.
*P<0.05, **P<0.01, vs. sh-NC. (C)
Following transfection with sh-HCP5 and sh-NC, the relative
migration of Y79 and HXO-RB44 cells was detected by Transwell
assay; magnification, ×400; scale bar, 100 µm;
**P<0.01, vs. sh-NC. (D) Following transfection with
sh-HCP5 and sh-NC, the relative invasion of Y79 and HXO-RB44 cells
was detected by Transwell assay; magnification, ×400; scale bar,
100 µm; **P<0.01, vs. sh-NC. The in
vitro experiments were performed in triplicate and repeated 3
times. Data are presented as the means ± standard deviation (SD).
HCP5, HLA complex P5. |
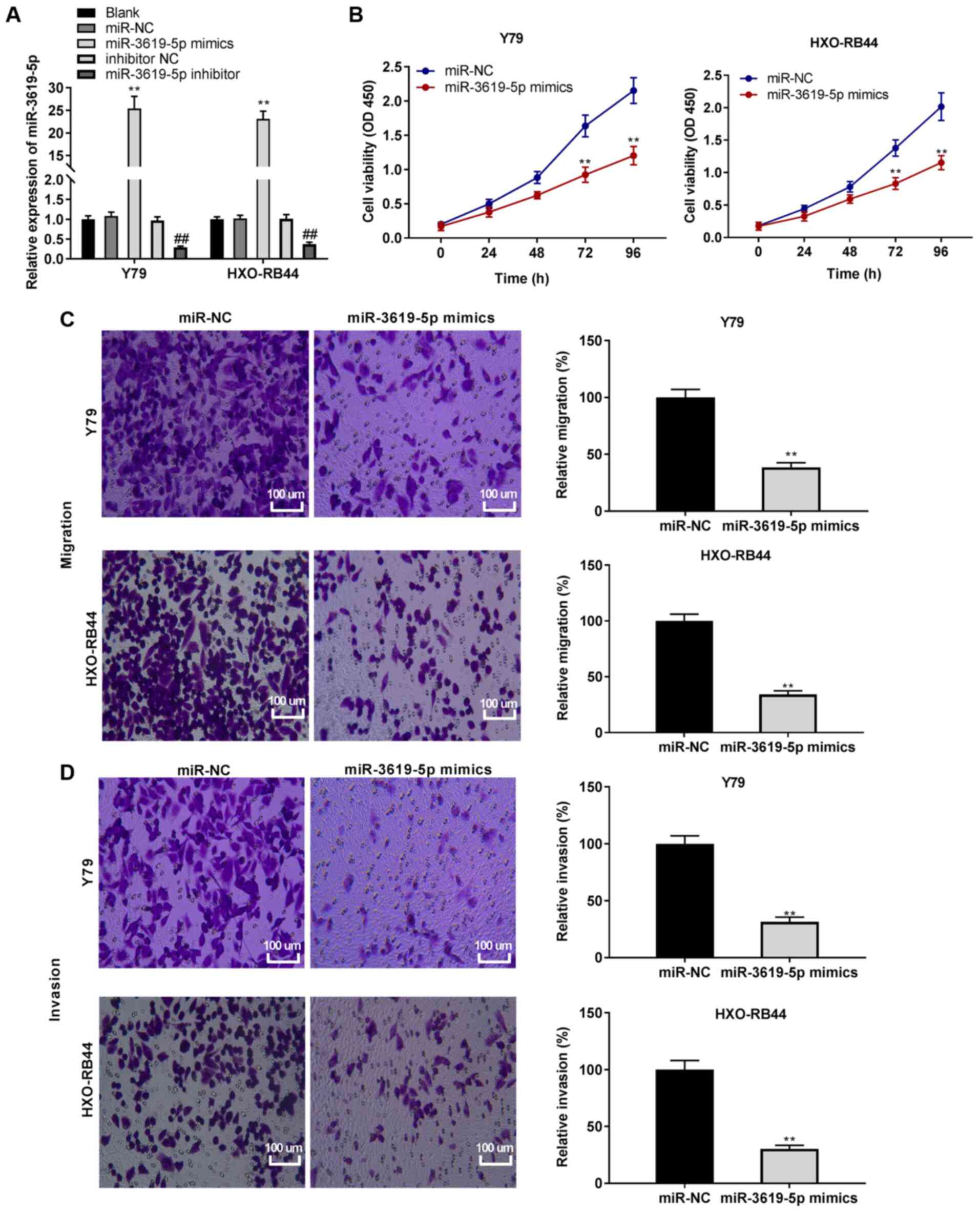 | Figure 5Overexpression of miR-3619-5p
inhibits the viability, migration and invasion in retinoblastoma
cells. (A) Following transfection with miR-3619-5p
mimics/miR-negative control (NC) or miR-3619-5p inhibitor/inhibitor
NC, the relative expression of miR-3619-5p was detected by RT-qPCR.
**P<0.01, vs. miR-NC. (B) Following transfection with
miR-3619-5p mimics and miR-NC, cell viability was determined by MTT
assay in Y79 and HXO-RB44 cells. **P<0.01, vs.
miR-NC. (C) Following transfection with miR-3619-5p mimics and
miR-NC, the relative migration of Y79 and HXO-RB44 cells was
determined by Transwell assay; magnification, ×400; scale bar, 100
µm; **P<0.01, vs. miR-NC. (D) Following
transfection with miR-3619-5p mimics and miR-NC, the relative
invasion of Y79 and HXO-RB44 cells was detected by Transwell assay;
magnification, ×400; scale bar, 100 µm;
**P<0.01, vs. miR-NC. The experiments were performed
in triplicate and repeated 3 times. Data are presented as the means
± standard deviation (SD). |
 | Table IIClinical parameters of the patients
with retinoblastoma included in the present study. |
Table II
Clinical parameters of the patients
with retinoblastoma included in the present study.
| Variable | Total | HCP5 expression
| P-value |
|---|
| Low (n=35) | High (n=36) |
|---|
| Age | | | | 0.540 |
| <3 years | 40 | 21 | 19 | |
| ≥3 years | 31 | 14 | 17 | |
| Sex | | | | 0.124 |
| Male | 32 | 19 | 13 | |
| Female | 39 | 16 | 23 | |
| Tumor size
(mm) | | | | 0.546 |
| <10 | 38 | 20 | 18 | |
| ≥10 | 33 | 15 | 18 | |
| TNM stage | | | | 0.003b |
| I/II | 28 | 20 | 8 | |
| III/IV | 43 | 15 | 28 | |
| Lymph node
metastasis | | | | 0.043a |
| No | 30 | 19 | 11 | |
| Yes | 41 | 16 | 25 | |
| Distant
metastasis | | | | |
| No | 69 | 35 | 34 | 0.157 |
| Yes | 2 | 0 | 2 | |
| Laterality | | | | 0.131 |
| Unilateral | 51 | 28 | 23 | |
| Bilateral | 20 | 7 | 13 | |
| IIRC stage | | | | 0.009b |
| A/B/C | 24 | 17 | 7 | |
| D/E | 47 | 18 | 29 | |
| RE stage | | | | 0.024a |
| I/II/III | 31 | 20 | 11 | |
| IV/V | 40 | 15 | 25 | |
Results
lncRNA HCP5 is highly expressed in RB
tissues and cells
First, the expression of HCP5 was determined by
RT-qPCR and it was found that the expression of HCP5 in the tumor
tissues was distinctly increased compared to that in adjacent
tissues (P<0.01; Fig. 1A).
Subsequently, the association between HCP5 expression and the
pathological features of RB cases was analyzed. It was found that a
high and low expression of HCP5 exhibited significant differences
in TNM stage (P=0.003), lymph node metastasis (P=0.043), IIRC stage
(P=0.009) and RE stage (P=0.024) (Table II). As illustrated in Fig. 1B-D, an increased expression of
HCP5 was observed in TNM stage III/IV, IIRC stage D/E and RE stage
IV/V (P<0.01), compared to TNM stage I/II, IIRC stage A/C and RE
stage I/II/III, respectively. The expression of HCP5 was also
determined in RB cells and an elevated expression of HCP5 was found
in the Rb cell lines (Y79, HXO-RB44, WERI-Rb-1 and SO-RB50)
(P<0.01; Fig. 1E). In
addition, a relatively higher expression of HCP5 was found in the
HOX-RB44 and Y79 cell lines among the 4 RB cell lines. Therefore,
the HOX-RB44 and Y79 cells were used in the following
experiments.
Silencing of lncRNA HCP5 suppresses the
viability, migration and invasion of RB cells
To explore the function of HCP5 in RB cells, the
transfection efficiency of sh-HCP5 in both the HXO-RB44 and Y79
cell lines was initially detected. Transfection with sh-HCP5
markedly reduced the expression of HCP5 (all P<0.01; Fig. 2A). Subsequently, functional
experiments were conducted on HCP5. MTT assay revealed that
transfection with sh-HCP5 led to a notable decrease in the
viability of the HXO-RB44 and Y79 cells (P<0.05; Fig. 2B). Additionally, the relative
migration and invasion of the Y79 and HXO-RB44 cells were reduced
by the knockdown of HCP5 (all P<0.01; Fig. 2C and D).
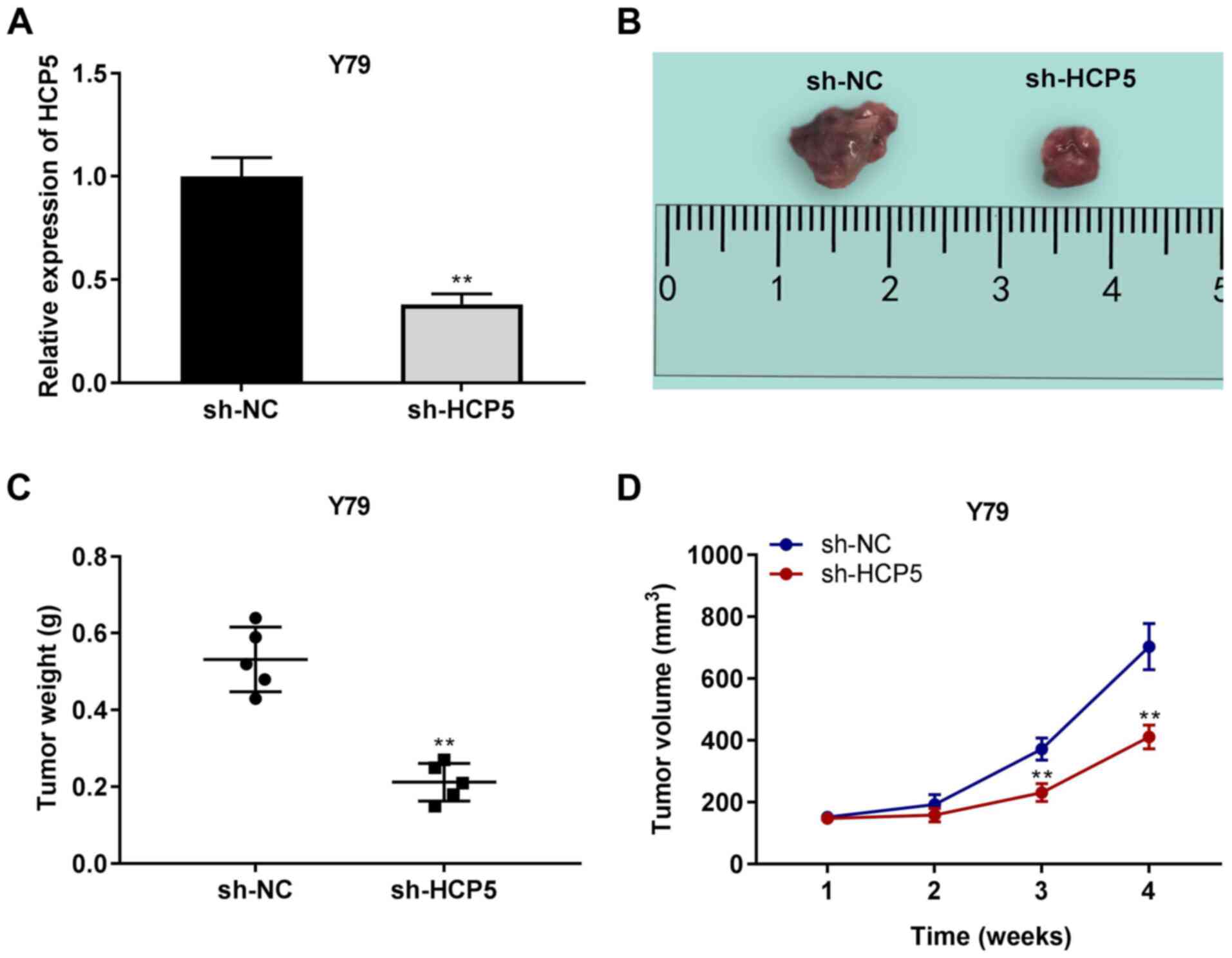
Silencing of lncRNA HCP5 suppresses the
growth of tumor xenografts in vivo
The expression of HCP5 in mouse tissues was also
determined. The results of RT-qPCR demonstrated that HCP5
expression was decreased by the injection of sh-HCP5 (P<0.01,
Fig. 3A). The effect of sh-HCP5
on tumor xenografts was then examined. As illustrated in Fig. 3B-D, it was found that tumor
weight in the sh-HCP5 group was lower than that in the sh-NC group,
and the tumor volume was markedly decreased by sh-HCP5 on the 28th
day after the injection in mice (P<0.01).
HCP5 serves as a competing endogenous RNA
for miR-3619-5p
To explore the regulatory mechanisms of HCP5 in RB,
the target miRNAs of HCP5 were predicated using StarBase v2.0
software. RIP assay was performed to verify 8 selected miRNAs. As
shown in Fig. 4A, the Y79 and
HXO-RB44 cells in the MS2-HCP5 group were enriched with miR-29b-3p,
miR-3619-5p and miR-128-3p compared with the MS2 group. Among these
3 miRNAs, miR-3619-5p was selected due to its relatively high
enrichment with HCP5. The predicted binding site of HCP5 and
miR-3619-5p is shown in Fig. 4B.
DLR assay further revealed that the relative luciferase activity of
the Y79 and HXO-RB44 cells co-transfected with HCP5 wt and
miR-3619-5p mimics was conspicuously lower than that of the Y79 and
HXO-RB44 cells co-transfected with HCP5 wt and miR-NC (all
P<0.01; Fig. 4C).
Subsequently, the effect of HCP5 on miR-3619-5p expression was
examined and it was observed that the expression of miR-3619-5p
increased following the downregulation of HCP5 in the Y79 and
HXO-RB44 cells (all P<0.01; Fig.
4D). Moreover, miR-3619-5p expression in tumor tissues was
diminished in comparison with adjacent normal tissues (P<0.01;
Fig. 4E). A negative correlation
was validated between HCP5 and miR-3619-5p in RB tissues using
Pearson's correlation analysis (P=0.004, r=-0.3372; Fig. 4F).
Overexpression of miR-3619-5p inhibits
the viability, migration and invasion of RB cells
Accordingly, the role of miR-3619-5p in RB cells was
explored; miR-3619-5p mimics and inhibitors were transfected into
RB cells. As was expected, miR-3619-5p expression increased by the
addition of miR-3619-5p mimics, whereas it was decreased by
transfection with miR-3619-5p inhibitor in the HXO-RB44 and Y79
cells (all P<0.01; Fig. 5A).
Importantly, it was discovered that the overexpression of
miR-3619-5p caused a decrease in the viability of the Y79 and
HXO-RB44 cells (all P<0.01; Fig.
5B). Moreover, the overexpression of miR-3619-5p suppressed the
migration and invasion of the Y79 and HXO-RB44 cells (all
P<0.01; Fig. 5C and D).
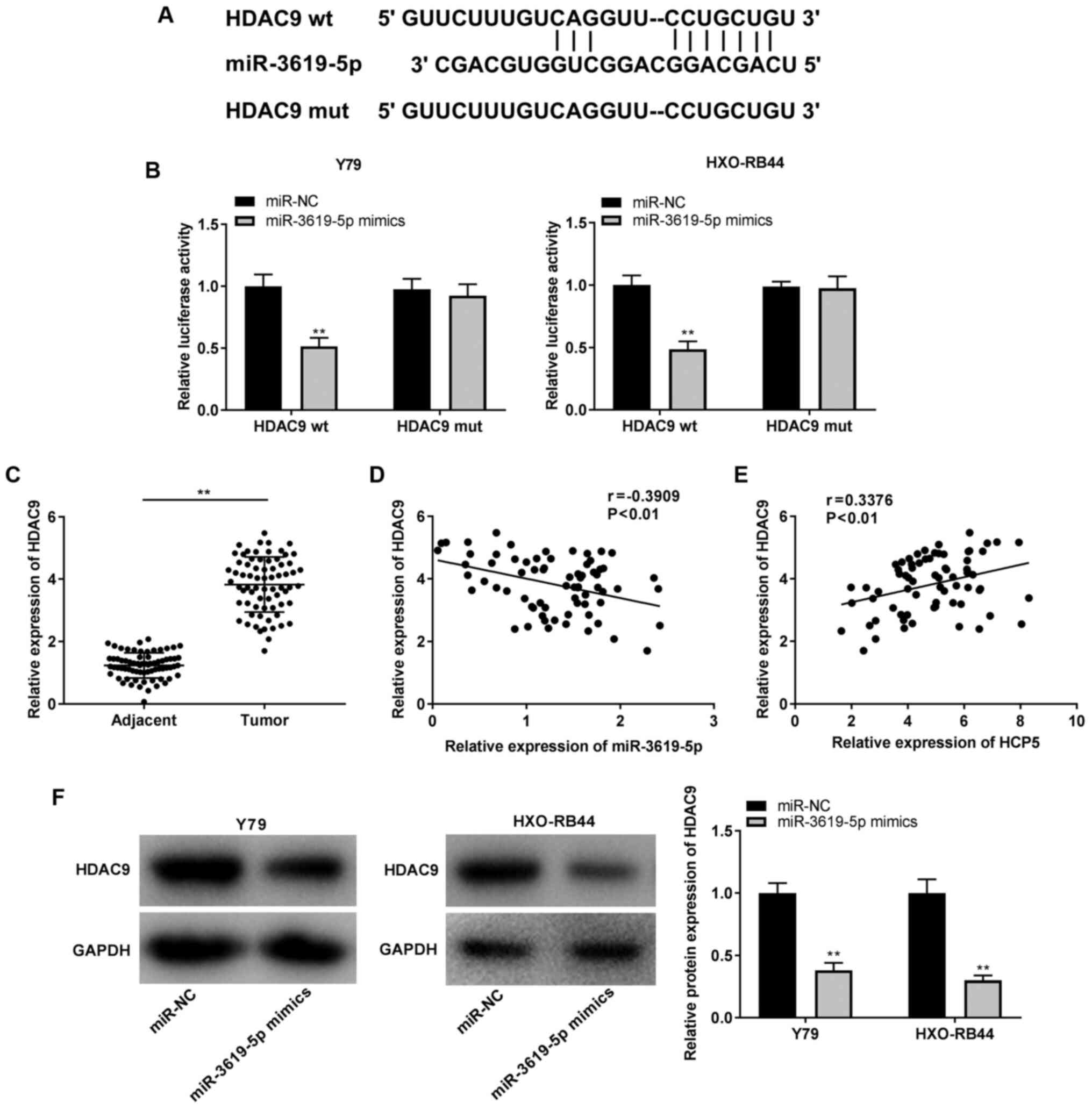
Identification of HDAC9 as a target gene
of miR-3619-5p
To elucidate the mechanisms of miR-3619-5p in the
progression of RB, the target gene of miR-3619-5p was predicted
using StarBase v2.0 software and it was found that miR-3619-5p
shared a complementary binding sequence at the 3-UTR with HDAC9
(Fig. 6A). Furthermore, the DLR
assay verified that transfection with miR-3619-5p mimics markedly
reduced the luciferase activity of the HDAC9 wt vector compared
with that of miR-NC, whereas it did not exert any significant
effect on the luciferase activity of the HDAC9 mut vector in the
Y79 and HXO-RB44 cells (all P<0.01; Fig. 6B). Simultaneously, it was
discovered that HDAC9 expression was markedly increased in tumor
tissues in contrast to adjacent normal tissues (P<0.01; Fig. 6C). Pearson's correlation analysis
demonstrated that HDAC9 expression negatively correlated with
miR-3619-5p expression (r=-0.3909), and HDAC9 expression positively
correlated with HCP5 expression in RB tissues (r=0.3376; all
P<0.01; Fig. 6D and E). The
regulatory association between miR-3619-5p and HDAC9 was then
examined, and it was observed that the relative protein expression
of HDAC9 was reduced by the upregulation of miR-3619-5p in the Y79
and HXO-RB44 cells (all P<0.01; Fig. 6F).
lncRNA HCP5 suppresses RB cell viability,
migration and invasion by sponging miR-3619-5p to target HDAC9
Subsequently, Y79 cells with a relatively high
expression of HCP5 were used to perform rescue experiments. To
investigate whether HCP5 affects RB cells by regulating the
miR-3619-5p/HDAC9 axis, HDAC9 was first overexpressed in Y79 cells.
As shown in Fig. 7A, HDAC9
expression was markedly upregulated by transfection with
pcDNA-HDAC9 in the Y79 cells (P<0.01). Furthermore, rescue
assays were performed. MTT assay revealed that cell viability was
reduced by transfection with sh-HCP5, while the addition of
miR-3619-5p inhibitor or pcDNA-HDAC9 reversed the sh-HCP5-mediated
decrease in the viability of Y79 cells (all P<0.01; Fig. 7B). Transwell assays indicated
that sh-HCP5 suppressed the migration and invasion of Y79 cells,
whereas the suppressive effects of sh-HCP5 on the migratory and
invasive abilities were partially eliminated by miR-3619-5p
inhibitor or pcDNA-HDAC9 in the Y79 cells (all P<0.01; Fig. 7C and D).
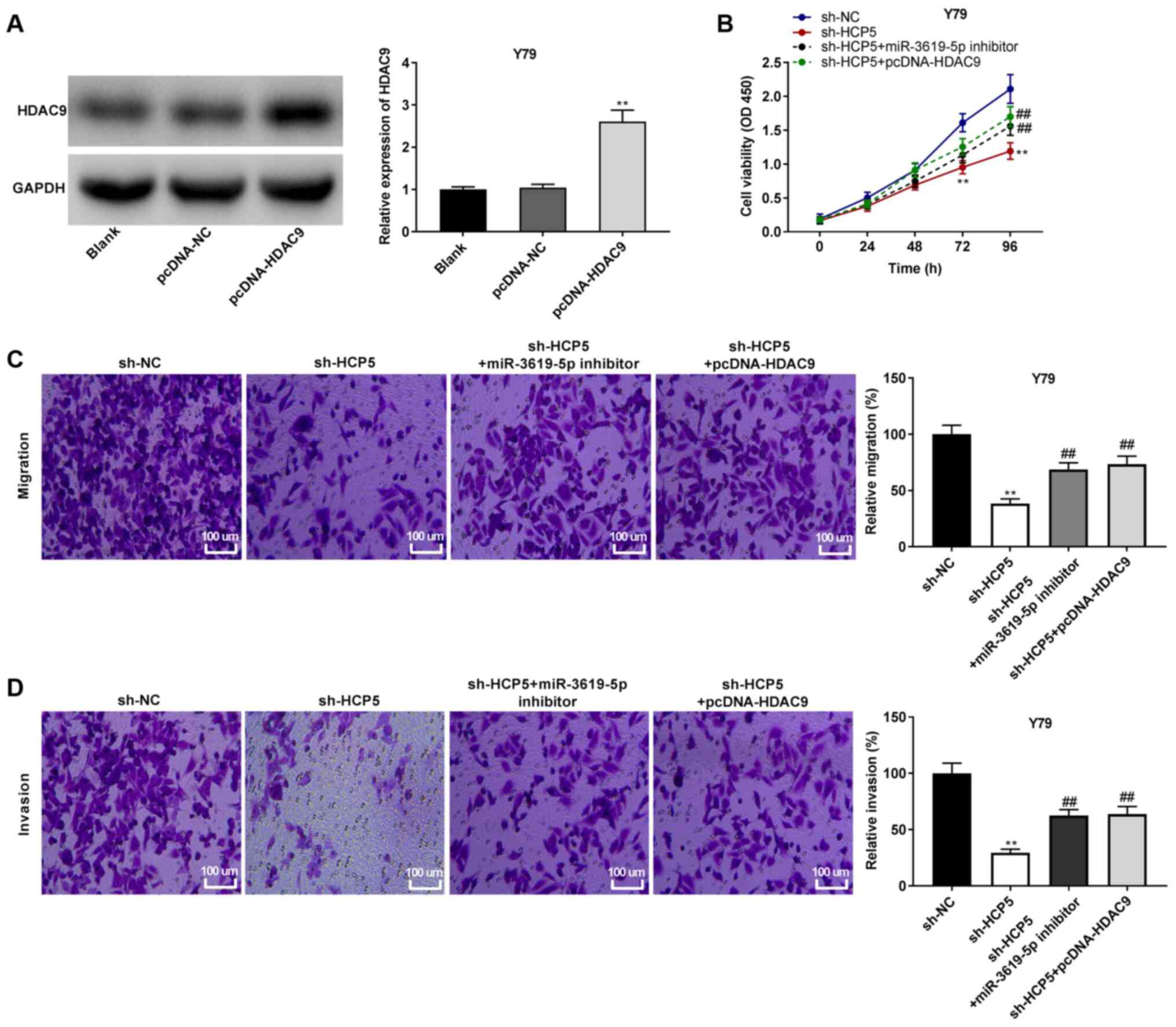 | Figure 7lncRNA HCP5 suppresses cell
viability, migration and invasion by sponging miR-3619-5p to target
HDAC9. (A) The protein expression of HDAC9 in Y79 cells was
detected by western blot analysis. **P<0.01, vs.
pcDNA-negative control (NC). (B) Cell viability was determined by
MTT assay in Y79 cells. **P<0.01, vs. short hairpin
(sh)-NC; ##P<0.01, vs. sh-HCP5. (C) Relative
migration of Y79 cells was determined by Transwell assay.
Magnification, ×400; scale bar, 100 µm;
**P<0.01, vs. sh-NC; ##P<0.01, vs.
sh-HCP5. (D) Relative invasion of Y79 cells was detected by
Transwell assay. Magnification, ×400; scale bar, 100 µm;
**P<0.01, vs. sh-NC; ##P<0.01, vs.
sh-HCP5. The experiments were performed in triplicate and repeated
3 times. Data are presented as the means ± standard deviation (SD).
HCP5, HLA complex P5; HDAC9, histone deacetylase 9. |
Discussion
RB is an eye malignant tumor that mainly affects
children (8). The abnormal
expression of lncRNA HCP5 plays a clinically predictive role in
tumorigenesis in a number of types of cancer (34-36). For example, HCP5 expression is
higher in cervical cancer tissues than in paracancerous tissues,
which is negatively associated with the survival rate of patients
with cervical cancer (34). The
upregulation of HCP5 has been observed in prostate cancer,
eventually resulting in tumor metastasis (35). Increased HCP5 levels have also
been found in oral squamous cell carcinoma, and it has been shown
to be significantly associated with lymph node metastasis and an
advanced TNM stage (36). In the
present study, it was discovered that HCP5 was upregulated in RB
tissues and cells compared to the controls. Concurrently, it was
found that a high expression of HCP5 was closely associated with
TNM stage and lymph node metastasis. In addition, strong
associations were identified between enhanced HCP5 and IIRC
stage/RE stage, suggesting that HCP5 may be an onco-lncRNA in
RB.
HCP5 plays a vital role in the cellular processes of
several human cancers (19,20,37). Wang et al revealed that
the downregulation of HCP5 suppressed the proliferation of
triple-negative breast cancer (TNBC) cells in vitro and the
tumor growth of TNBC in vivo (37). Bai et al indicated that
HCP5 silencing suppressed the progression of CRC in vivo
(19). Wang et al
demonstrated that the knockdown of HCP5 greatly contributed to the
inhibition of the proliferation, migration and invasion of OC cells
(20). In the present study, the
proliferation, migration and invasion of RB cells, as well as the
growth of tumor xenografts were suppressed by HCP5 downregulation.
Based on these results, it can be conjectured that the silencing of
HCP5 acts as a suppressor in the progression of Rb.
miR-3619-5p plays an anti-tumor role in the
malignancy of several cancers (38,39). For instance, miR-3619-5p is
minimally expressed in non-small cell lung cancer (NSCLC) tissues,
whereas the overexpression of miR-3619-5p suppresses the growth and
invasion of NSCLC cells in vitro (38). A high expression of miR-3619-5p
suppresses the proliferation, migration and invasion of breast
cancer cells (39). In the
present study, a decreased miR-3619-5p expression was identified in
RB tissues, and the upregulation of miR-3619-5p exerted suppressive
effects on the proliferation and metastasis of RB cells. A recent
study by Yan et al revealed that the proliferation,
migration and invasion of RB cells were impeded by transfection
with miR-3619-5p mimics (25).
These results suggest that miR-3619-5p may also be function as an
anti-tumor miRNA in RB. In addition, miR-3619-5p interacts with
lncRNAs and is involved in cancer progression, such as
LINC00202-miR-3619-5p in RB (25), DGCR5-miR-3619-5p in gallbladder
cancer (40) and
PVT1-miR-3619-5p in gastric cancer (41). We hypothesised that the
anti-tumour function of miR-3619-5p in RB may also be regulated by
HCP5. Herein, miR-3619-5p was initially identified as a target of
and was negatively modulated by HCP5. The results of the present
study demonstrated that the reduction of the proliferation and
metastasis of Y79 cells transfected with sh-HCP5 was reversed by
transfection with miR-3619-5p inhibitor. These results suggest that
the silencing of HCP5 attenuates RB carcinogenicity by interacting
with miR-3619-5p.
In recent years, increasing attention has been paid
to the involvement of HDAC9 in the pathological process of RB
(27,29). Xu et al suggested that
HDAC9 expression was increased in RB tissues, and its
downregulation inhibited the proliferation, migration and invasion
of RB cells (29). In addition,
Zhang et al confirmed that the silencing of HDAC9 suppressed
the proliferation of RB cells in vitro (27). Consistent with these studies, an
increased expression of HDAC9 was identified in the present study,
indicating the oncogenicity of HDAC9 in RB. Simultaneously, it was
found that HDAC9 was a target gene of and was negatively regulated
by miR-3619-5p, and a positive correlation was observed between
HDAC9 and HCP5 expression in RB tissues. Therefore, it was
demonstrated miR-3619-5p is involved in the tumorigenesis of Rb
through regulation by HCP5. It was further hypothesized that HDAC9
may also be modulated by HCP5 to play a role in the progression of
Rb. Feedback verification experiments revealed that the inhibitory
effects of HCP5 knockdown on the proliferation, migration and
invasion of Y79 cells were reversed by HDAC9 overexpression, which
confirmed this hypothesis. It was concluded that the downregulation
of HCP5 suppressed the progression of Rb by sponging miR-3619-5p
and regulating HDAC9. However, there may be a limitation in to the
presents tudy. Only one Rb cell line (Y79) was used to perform the
feedback verification experiments, and further experiments using
other cell lines are required to validate this conclusion.
In conclusion, the present study demonstrates that
HCP5 acts as an endogenous sponge of miR-3619-5p to regulate HDAC9.
The knockdown of HCP5 mitigates the progression of RB by regulating
the miR-3619-5p/HDAC9 axis. Overall, the present study provides
perspectives for novel therapeutic targets for RB.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ made substantial contributions to the conception
and design of the study, data acquisition, the drafting of the
article, and gave final approval of the version to be published. FH
made substantial contributions to the conception and design of the
study, data analysis and interpretation, and in the drafting of the
article. Both authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Hospital of Weifang Medical University [No.
2020(12)]. Written informed
consent was obtained from all the patients. The study was conducted
in accordance with the Declaration of Helsinki. The animal
experiments were approved by the Ethics Committee of Affiliated
Hospital of Weifang Medical University, and were performed in
accordance with the institutional guide for the care and use of
laboratory animals (National Institutes of Health).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was funded by the Shandong Medical and Health
Science and Technology Development Project (Project name: Study on
the mechanism of mirNA-21 on retinal microvascular endothelial
cells of RF/6A in diabetic rats; no. 2019WS599).
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
RB
|
retinoblastoma
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
DLR
|
dual-luciferase reporter
|
References
|
1
|
Villegas VM, Hess DJ, Wildner A, Gold AS
and Murray TG: Retinoblastoma. Curr Opin Ophthalmol. 24:581–588.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cimino PJ, Robirds DH, Tripp SR, Pfeifer
JD, Abel HJ and Duncavage EJ: Retinoblastoma gene mutations
detected by whole exome sequencing of merkel cell carcinoma. Mod
Pathol. 27:1073–1087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venkatesan N, Deepa PR, Khetan V and
Krishnakumar S: Computational and in vitro investigation of
miRNA-Gene regulations in retinoblastoma pathogenesis: miRNA mimics
strategy. Bioinform Biol Insights. 9:89–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dimaras H, Corson TW, Cobrinik D, White A,
Zhao J, Munier FL, Abramson DH, Shields CL, Chantada GL, Njuguna F
and Gallie BL: Retinoblastoma. Nat Rev Dis Primers. 1:150212015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okimoto S and Nomura K: Clinical
manifestations and treatment of retinoblastoma in Kobe children's
hospital for 16 years. J Pediatr Ophthalmol Strabismus. 51:222–229.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abu-Ain MS, Shatnawi R, Yousef YA and
Watts P: Heterochromia irides and mistaken identity of
retinoblastoma. BMJ Case Rep. 12:e2310912019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jenkinson H: Retinoblastoma: Diagnosis and
management-the UK perspective. Arch Dis Child. 100:1070–1075. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rao R and Honavar SG: Retinoblastoma.
Indian J Pediatr. 84:937–944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sullivan EM, Wilson MW, Billups CA, Wu J,
Merchant TE, Brennan RC, Haik BG, Shulkin B, Free TM, Given V, et
al: Pathologic risk-based adjuvant chemotherapy for unilateral
retinoblastoma following enucleation. J Pediatr Hematol Oncol.
36:e335–e340. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qaddoumi I, Nawaiseh I, Mehyar M, Razzouk
B, Haik BG, Kharma S, Jaradat I, Rodriguez-Galindo C and Wilson MW:
Team management, twinning, and telemedicine in retinoblastoma: A
3-tier approach implemented in the first eye salvage program in
Jordan. Pediatr Blood Cancer. 51:241–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chantada GL, Dunkel IJ, Schaiquevich PS,
Grynszpancholc EL, Francis J, Ceciliano A, Zubizarreta PA, Fandiño
AC and Abramson DH: Twenty-Year collaboration between north
American and South American retinoblastoma programs. J Glob Oncol.
2:347–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
14
|
He X, Chai P, Li F, Zhang L, Zhou C, Yuan
X, Li Y, Yang J, Luo Y, Ge S, et al: A novel LncRNA transcript,
RBAT1, accelerates tumorigenesis through interacting with HNRNPL
and cis-activating E2F3. Mol Cancer. 19:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han S, Song L, Chen Y, Hou M, Wei X and
Fan D: The long non-coding RNA ILF3-AS1 increases the proliferation
and invasion of retinoblastoma through the miR-132-3p/SMAD2 axis.
Exp Cell Res. 393:1120872020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong Y, Wan G, Yan P, Qian C, Li F and
Peng G: Long noncoding RNA LINC00324 promotes retinoblastoma
progression by acting as a competing endogenous RNA for
microRNA-769-5p, thereby increasing STAT3 expression. Aging (Albany
NY). 12:7729–7746. 2020. View Article : Google Scholar
|
|
17
|
Vernet C, Ribouchon MT, Chimini G,
Jouanolle AM, Sidibe I and Pontarotti P: A novel coding sequence
belonging to a new multicopy gene family mapping within the human
MHC class I region. Immunogenetics. 38:47–53. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Y, Helms C, Liao W, Zaba LC, Duan S,
Gardner J, Wise C, Miner A, Malloy MJ, Pullinger CR, et al: A
genome-wide association study of psoriasis and psoriatic arthritis
identifies new disease loci. PLoS Genet. 4:e10000412008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bai N, Ma Y, Zhao J and Li B: Knockdown of
lncRNA HCP5 suppresses the progression of colorectal cancer by
miR-299-3p/PFN1/AKT axis. Cancer Manag Res. 12:4747–4758. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, He M, Fu L and Jin Y: Role of
lncRNAHCP5/microRNA-525-5p/PRC1 crosstalk in the malignant
behaviors of ovarian cancer cells. Exp Cell Res. 394:1121292020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan B, Guan Q, Yan T, Zhang X, Xu W and
Li J: LncRNA HCP5 regulates pancreatic cancer progression by
miR-140-5p/CDK8 axis. Cancer Biother Radiopharm. 35:711–719. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wan W, Long Y, Li Q, Jin X, Wan G, Zhang
F, Lv Y, Zheng G, Li Z and Zhu Y: MiR-25-3p promotes malignant
phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed
Pharmacother. 118:1091112019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J and You X: MicroRNA758 inhibits
malignant progression of retinoblastoma by directly targeting PAX6.
Oncol Rep. 40:1777–1786. 2018.PubMed/NCBI
|
|
24
|
Hao P, Yue F, Xian X, Ren Q, Cui H and
Wang Y: Inhibiting effect of MicroRNA-3619-5p/PSMD10 axis on liver
cancer cell growth in vivo and in vitro. Life Sci. 254:1176322020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan G, Su Y, Ma Z, Yu L and Chen N: Long
noncoding RNA LINC00202 promotes tumor progression by sponging
miR-3619-5p in retinoblastoma. Cell Struct Funct. 44:51–60. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parra M and Verdin E: Regulatory signal
transduction pathways for class IIa histone deacetylases. Curr Opin
Pharmacol. 10:454–460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H
and Huang Z: Downregulation of HDAC9 inhibits cell proliferation
and tumor formation by inducing cell cycle arrest in
retinoblastoma. Biochem Biophys Res Commun. 473:600–606. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin Q, He W, Chen L, Yang Y, Shi K and You
Z: MicroRNA-101-3p inhibits proliferation in retinoblastoma cells
by targeting EZH2 and HDAC9. Exp Ther Med. 16:1663–1670.
2018.PubMed/NCBI
|
|
29
|
Xu L, Li W, Shi Q, Wang M, Li H, Yang X
and Zhang J: MicroRNA936 inhibits the malignant phenotype of
retinoblastoma by directly targeting HDAC9 and deactivating the
PI3K/AKT pathway. Oncol Rep. 43:635–645. 2020.PubMed/NCBI
|
|
30
|
TNM8: The updated TNM classification for
retinoblastoma. Community Eye Health. 31:342018.
|
|
31
|
Shields CL and Shields JA: Basic
understanding of current classification and management of
retinoblastoma. Curr Opin Ophthalmol. 17:228–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yousef YA, Al-Hussaini M, Mehyar M, Sultan
I, Jaradat I, AlRawashdeh K, Khurma S, Deebajah R and Nawaiseh I:
Predictive value of tnm classification, international
classification, and reese-ellsworth staging of retinoblastoma for
the likelihood of high-risk pathologic features. Retina.
35:1883–1889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Yu Y, Shen HM, Fang DM, Meng QJ and Xin
YH: LncRNA HCP5 promotes the development of cervical cancer by
regulating MACC1 via suppression of microRNA-15a. Eur Rev Med
Pharmacol Sci. 22:4812–4819. 2018.PubMed/NCBI
|
|
35
|
Hu R and Lu Z: Long noncoding RNA HCP5
promotes prostate cancer cell proliferation by acting as the sponge
of miR4656 to modulate CEMIP expression. Oncol Rep. 43:328–336.
2020.
|
|
36
|
Zhao J, Bai X, Feng C, Shang X and Xi Y:
Long non-coding RNA HCP5 facilitates cell invasion and
epithelial-mesenchymal transition in oral squamous cell carcinoma
by miR-140-5p/SOX4 axis. Cancer Manag Res. 11:10455–10462. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L, Luan T, Zhou S, Lin J, Yang Y, Liu
W, Tong X and Jiang W: LncRNA HCP5 promotes triple negative breast
cancer progression as a ceRNA to regulate BIRC3 by sponging
miR-219a-5p. Cancer Med. 8:4389–4403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Niu X, Liu S, Jia L and Chen J: Role of
MiR-3619-5p in β-catenin-mediated non-small cell lung cancer growth
and invasion. Cell Physiol Biochem. 37:1527–1536. 2015. View Article : Google Scholar
|
|
39
|
Lv MH, Mao QX, Li JT, Qiao J, Chen X and
Luo S: Knockdown of LINC00665 inhibits proliferation and invasion
of breast cancer via competitive binding of miR-3619-5p and
inhibition of catenin beta 1. Cell Mol Biol Lett. 25:432020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu S, Chu B, Cai C, Wu X, Yao W, Wu Z,
Yang Z, Li F, Liu Y, Dong P and Gong W: DGCR5 promotes gallbladder
cancer by sponging MiR-3619-5p via MEK/ERK1/2 and JNK/p38 MAPK
pathways. J Cancer. 11:5466–5477. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu C, Hu Y, Ning Y, Zhao A, Zhang G and
Yan L: Long noncoding RNA plasmacytoma variant translocation 1
regulates cisplatin resistance via miR-3619-5p/TBL1XR1 axis in
gastric cancer. Cancer Biother Radiopharm. 35:741–752. 2020.
View Article : Google Scholar : PubMed/NCBI
|