1. Introduction
Circular RNAs (circRNAs/circ) are a group of
non-coding RNAs widely found in eukaryotic cells, with a circular
and covalently-bonded structure. The majority of circRNAs consist
of multiple exons and have been found to be conserved in different
species and are specifically expressed during tissue development.
Unlike linear RNAs, circRNAs are continuous, covalent structures
lacking both the 5′ and 3′ ends, which renders circRNAs stable and
resistant to exonuclease-mediated degradation (1). Significant progress has been made
of late in circRNA research, which has highlighted the emerging
role of circRNAs in cancer, neurological diseases and immune
responses (2-4).
The recently acknowledged regulatory role of
circRNAs has led to an increasing awareness of their importance in
viral infections. The present review summarized the role of
circRNAs in frequent viral diseases, with a focus on the possible
use of circRNAs for diagnosis and treatment. The latter is
particularly relevant in light of the recent emergence of the
coronavirus disease 2019 (COVID-19) pandemic caused by the severe
acute respiratory syndrome virus (SARS-CoV-2), which is affecting
tens of millions of individuals worldwide, due to the very high
rate of viral infection (5).
2. Biogenesis and biological functions of
circRNAs
Biogenesis
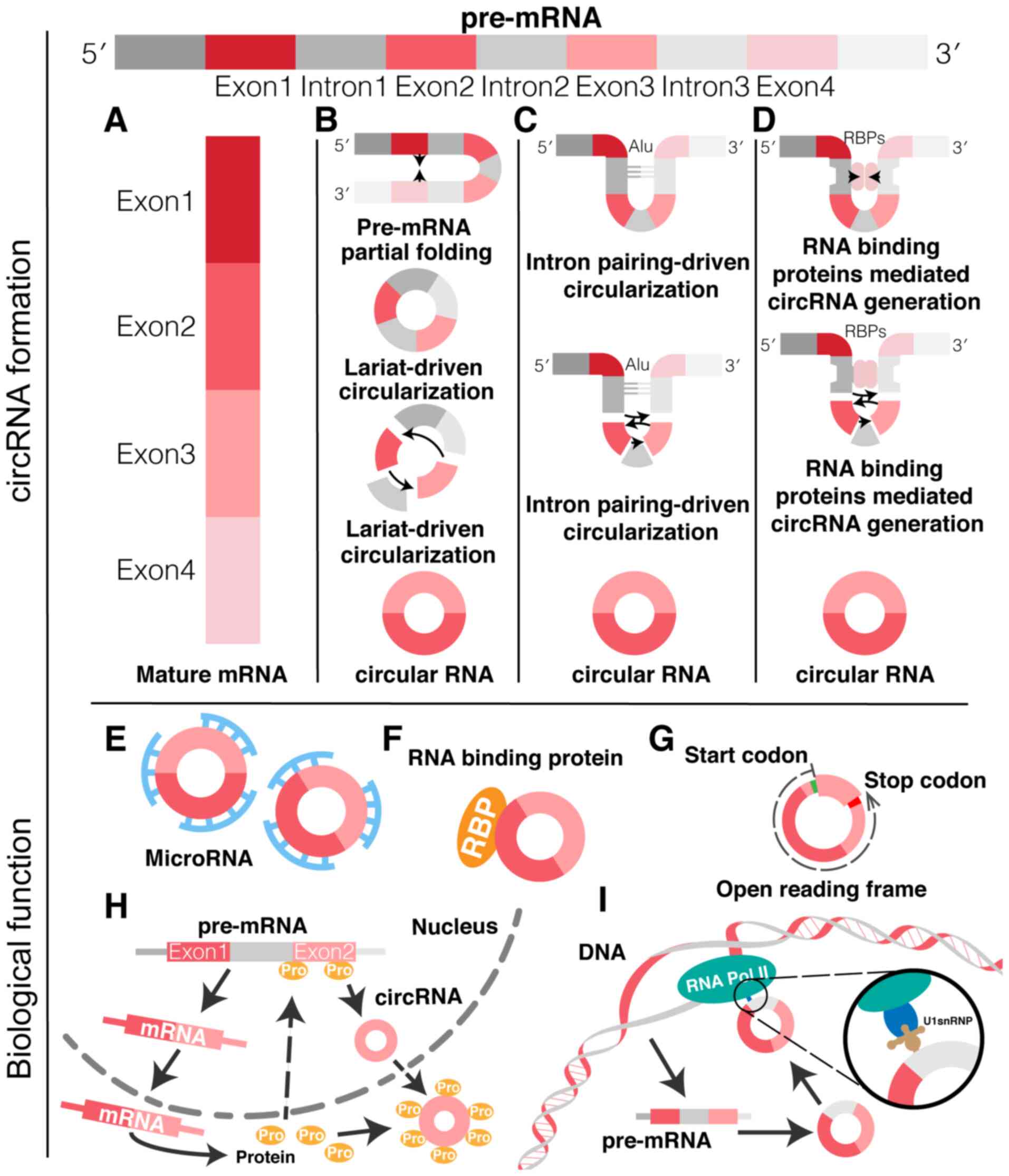
circRNAs are formed by the back-splicing of pre-RNA
catalyzed by RNA polymerase II (RNA Pol II). A downstream 5' splice
donor site is linked to an upstream 3' splice acceptor site to form
a 3′-5′ phosphodiester bond that results in a circularized RNA
(Fig. 1) (6). Circularization is explained by the
'lariat-driven', 'exon-skipping' and 'intron pairing-driven' models
(7). In the lariat model, the
partial folding of the RNA during pre-RNA transcription results in
non-adjacent exons being pulled close to each other (Fig. 1B). This allows exon jumping to
occur, so that the area spanned forms a circRNA intermediate that,
by further back splicing, results in circRNA molecules composed of
exons. In intron pairing-driven circularization (Fig. 1C), the base-pairing of flanking
introns containing complementary sequences (for example, Alu
repeats) occurs. This allows the 5' splice site (SS) of a
downstream exon sequence to approach the 3' SS of an upstream exon
within the intron flanking region, which favors reverse splicing.
In addition, a variety of trans-acting protein factors can actively
participate in the biogenesis of circRNAs (Fig. 1D).
circRNA expression can be regulated at three levels.
First, the transcription of circRNA-producing pre-mRNA occurs via
RNA Pol II. It has been found that the average Pol II transcription
elongation rate (TER), which is an important factor in determining
the outcome of splicing events of nascent circRNA-producing genes,
is higher than that of non-circRNA genes (8). It is possible that the TER of a
parental gene may influence alternative back-splicing selection.
Alternatively, higher processivity might result in less efficient
canonical splicing, and then increase back-splicing, Second, it is
established that cis and trans-regulatory factors
regulate back-splicing during the spliceosome mechanism. The
processing of circRNAs can be facilitated by either the RNA pairing
of reverse complementary sequences across their flanking introns or
protein factors binding to pre-mRNAs to bridge flanking introns
together. When the intronic RNA pairing was disrupted by the
removal of one intronic complementary sequence (ICS) flanking the
circGCN1L1-forming exons by CRISPR/Cas9, circGCN1L1 was completely
eradicated in human cells (8).
Several RNA-binding proteins (RBPs) have also been reported to play
a role in circRNA biogenesis. Muscleblind (MBL) like splicing
regulator 1 (MBNL1) was able to bind to and stabilize RNA pairs
formed between the ICS, leading to the enhanced circRNA biogenesis.
Adenosine deaminase 1 acting on RNA1 (ADAR1) can antagonize circRNA
biogenesis by melting the stem structure (8). A recent study also found that the
depletion of numerous core spliceosomal components, including the
SF3b and SF3a complexes, resulted in preferred circRNA expression,
which suggested a shift from canonical splicing to back-splicing
under spliceosome starving conditions by depleting spliceosomal
factors or chemically inhibiting spliceosomal activities (9). In addition to core spliceosomal
factors, a group of splicing regulators are required for circRNA
expression in human cells (10).
Finally, circRNA turnover may affect circRNA expression. The
expression of certain circRNAs can be detected at the steady-state
level, largely due to the fact that they can accumulate to high
levels post-transcriptionally. A high expression of circRNAs in the
brain, possibly due to discrepant decay rates, was observed between
circRNAs and their linear counterparts (11).
Biological functions
MicroRNA (miRNA/miR) sponges
circRNAs are competing endogenous RNAs that can act
as miRNA sponges. Through their miRNA response elements, circRNAs
can bind to target miRNAs, negatively regulate miRNA expression
levels and influence biological activities, thus regulating the
expression of target genes (Fig.
1E). For instance, circRNA-7 (ciRS-7) is mainly transcribed
from the antisense strand corresponding to cerebellar
degeneration-related protein 1 (CDR1), and they always combine with
each other as a complex, which is abundant in human and mouse
brains. The miR-7 binding sites present in ciRS-7 can adsorb miR-7.
A low ciRS-7 level can therefore result in an increase in free
miR-7 and the ensuing enhancement of ubiquitin ligase A inhibition
(12,13).
RBPs
RBPs are mainly involved in the regulation of RNA
transcription (14), alternative
splicing (15), transport
(16) and translation (17). circRNAs can combine RBPs and
therefore participate in the regulation of gene expression and
protein translation (Fig. 1F).
For example, it has been shown that exon-intron circRNAs, such as
circ-ETF3 and circ-PAIP2, can bind to U1 small nuclear
ribonucleoprotein (U1snRNP) and regulate gene expression in the
nuclei of in vitro cultured 293 and HeLa cells (18).
Encoding messengers for
transcription
Although circRNAs are non-coding RNAs, the presence
of an internal ribosome entry site (IRES) upstream of the circRNA
start codon can allow the 40S subunit of eukaryotic ribosomes to
bind and initiate translation (Fig.
1G). This was shown by Legnini et al (19), who reported that circ-ZNF609
contains an open reading frame with start and stop codons that can
translate into proteins. In addition, Pamudurti et al
(20) used ribosome imprinting
technology to discover whether circRNA is sufficient to encode
proteins.
Competitor of linear RNA
circRNA is the product of RNA processing, thus
competition between circRNA and its linear RNA is possible
(Fig. 1H). For instance,
Ashwal-Fluss's research reported that circMbl is produced by exon 2
of the muscleblind (MBL) gene. The MBL gene product specifically
binds to the sequences of circRNA (circMbl) and its flanking
introns of the pre-mRNA, and promotes the biosynthesis of circMbl,
while circMbl-MBL binding competitively inhibits classical mRNA
splicing (21).
Regulating the maternal gene
expression
A recent study revealed that circRNAs could regulate
the expression of maternal genes at the transcriptional or
post-transcriptional level (Fig.
1I) (22). Another study
reported a special class of circRNAs, termed Exon-intron circular
RNAs (EIciRNAs), which are associated with RNA Pol II in human
cells and include circ eukaryotic translation initiation factor 3
subunit J (EIF3J) and circ poly(A) binding protein interacting
protein 2. EIciRNAs are mainly localized in the nucleus, using the
5' splicing site of the retained intron to interact with U1 snRNP
and improve EIF3J transcription efficiency (23). Thus, EIciRNA can function in
transcriptional regulation through RNA-RNA interactions and play
different roles from those of circRNAs composed of exonic sequences
previously localized in the cytoplasm.
3. Effect of circRNAs on viral
infections
CircRNAs are involved in the regulation of viral
infections, such as hepatitis B virus (HBV) and human herpesvirus
(HHV) infections (24). In
addition, circRNAs have been proposed as biomarkers to distinguish
viral from non-viral pneumonia (25). The crucial role of circRNAs in
viral infections is associated with controlling miRNA levels and
regulating innate immune responses (Table I).
 | Table IRoles of circRNAs in viral
infections. |
Table I
Roles of circRNAs in viral
infections.
| Virus | Diseases or
infections | circRNA | Role | (Refs.) |
|---|
| HBV | HBV-related
hepatitis |
hsa_circ_0005389 | Associated with the
transcription of viral cccDNA and HBV replication in
hepatocytes. | (29) |
|
hsa_circ_0000038 | Associated with HBV
Enhancer I, HBV enhancer II and core promoter. | (31) |
| Liver fibrosis |
hsa_circ_0000650 | Involved in liver
fibrosis through the hsa_circ_0000650-miR-6873-3p-TGFβ
pathway. | (34) |
| HBV infection |
hsa_circ_0000650 | Interacts with
miR-210, which binds to the pre-S1 antigenic domain of the HBV
envelope protein (HBsAg). | (35) |
| HBV-related
carcinoma |
hsa_circ_00004812 | Associated with the
oncogenesis induced by HBV infection through
circ_00004812-miR-1287-5p-FSTL1 pathway. | (37) |
| hsa_circ_4099 | Associated with HBV
infection and liver fibrosis. | (39) |
|
hsa_circ_0000976 | Differentially
expressed in HBV-related HCC. | (41) |
|
hsa_circ_0007750 | Associated with
chemokine-and cytokine-mediated immune responses. | (42) |
|
hsa_circ_0027089 | Promote HBV
replication. | (43) |
|
hsa_circ_100338 | Correlated with low
survival and metastatic progression. | (45) |
| KSHV | KSHV infection |
hsa_circ_0001400 | Inhibiting the
expression of certain viral genes. | (50) |
| HSV | HSV-1
infection | circRNAs | Implicated in
signaling and apoptosis pathways, the cell cycle and cell
death. | (53) |
| EBV | EBV infection | ebv-circRPMS1 | A novel regulation
agent acting on transcription and/or interfering with splicing as a
miRNA sponge. | (54) |
| EBV-related
tumor | circLMP2, circBHLF1
circBARTs | Associated with
proliferation of tumor cells of EBV-positive patients. | (48) |
| HCMV | HCMV infection |
hsa_circ_0001445
hsa_circ_0001206 | Participated in a
HCMV latency mechanism. | (59) |
| HIV | HIV infection | circRNAs | Associated with HIV
replication and T cell infection. | (60) |
| HPV | HPV-related
cervical cancer | HPV-circE7 | Essential for the
transformation and growth of cervical cancer cells, also promote
virus replication and host cell transformation. | (64) |
| MERS-CoV | MERS-CoV
infection | circFNDC3B
circCNOT1 | Associated with the
regulation of MERS-CoV cellular proliferation. | (67) |
| HTNV | HTNV infection |
circ0002470
circ0006132
circ0000479 | Associated with
HTNV infection. | (71) |
| SARS-CoV-2 | SARS-CoV-2
infection | Ppp1r10
C330019G07Rik | Related with
SARS-CoV-2 infection. | (77) |
| IAV | Influenza virus
infection | circ-GATAD2A | In association with
viral replication. | (79) |
| EBOV | EBOV infection | circRNA_chr19 | May have effect on
EBOV infection through the circRNA_chr19-miR-30b-3p-CLDN18
pathway. | (80) |
| DENV | Dengue fever |
hsa_circ_0006459
hsa_circ_0015962 | Associated with
reductions in white blood cell (WBC) and platelet (PLT)
counts. | (84) |
HBV
HBV is a partially double-stranded DNA virus of
~3,200 base pairs (26).
Infections by HBV are among the most common chronic viral diseases
and can result in the later development of cirrhosis or
hepatocellular carcinoma (HCC). The prevalence of hepatitis B
surface antigen (HBsAg) seropositivity has been reported to be
3.61% worldwide, with a 95% confidence interval (27). The expression levels of host
circRNAs can be affected by HBV infections. Research on the
differential expression of RNAs following HBV infection has
indicated the involvement of four circRNA/miRNA/mRNA pathways. Both
the hsa_circ_0005389-miR-4505/miR-6752-5p/miR-5787-interferon
regulatory factor 7 (IRF7) and hsa_circ_0000038-miR-370/miR-939/HBV
pathways are significantly upregulated, while the
hsa_circ_0000650/miR-6873-3p/transforming growth factor (TGF)β and
hsa_circ_0000650/miR-210-5p/HBV pathways are downregulated
(28). Studies on the downstream
genes of these four pathways have shown that circRNAs are involved
in HBV replication, as well as in the liver inflammation and
fibrosis caused by HBV infection. hsa_circ_0005389 and
hsa_circ_0000038 are associated with HBV replica-tion. IRF7 of the
0005389 pathway is involved in interferon (IFN)-α-mediated
antiviral immunity (29). IFN-α
reduces the acetylation level of histone H4 bound to HBV covalently
closed circular DNA (cccDNA), which inhibits the transcription of
viral cccDNA and HBV replication in hepatocytes (30). miR-370 of the 0000038 pathway
restrains HBV gene expression by targeting the 3′-untranslated
region of the transcription factor nuclear factor IA (NFIA) mRNA,
with NFIA being able to regulate the activity of HBV enhancer I
(31). miR-939, also of the
0000038 pathway, suppresses HBV replication by reducing the
activity of the HBV enhancer II/core promoter (32).
TGFβ inhibits the anti-fibrotic function of natural
killer cells in liver fibrosis; therefore, hsa_circ_0000650 may be
involved in liver fibrosis through the
hsa_circ_0000650/miR-6873-3p/TGFβ signaling pathway (33). circRNA hsa_circ_0000650 also
plays a role in inflammation through the
hsa_circ_0000650/miR-210-5p/HBV pathway. In fact, inflammation
results in liver hypoxia, which leads to increases in liver and
serum miR-210 levels (34). In
addition, bioinformatics analysis has indicated that miR-210 binds
to the HBV pre-S1 region and reduce HBsAg expression, which leads
to inhibition of hepatitis B virus replication (35). However, no significant
correlation between miR-210 and HBsAg levels has been proven by
experiments (36), casting
doubts on such an association.
Other circRNA/miRNA/mRNA pathways have also been
associated with HBV infectivity. The expression level of
circ_00004812 [circ_00004812/miR-1287-5p/follistatin like 1 (FSTL1)
pathway] has been found to be increased in HBV-infected cells. The
knockdown of circ_00004812 has been shown to lead to an increase in
the mRNA and protein expression levels of IFN-α and IFN-β (37). FSTL1 and IFN-β have been reported
to participate in the oncogenesis process induced by HBV infection
(38). With regards to the
association between HBV infection and liver fibrosis, hsa_circ_4099
enhances H2O2-mediated fibrosis, as well as
exerts a sponge effect on miR-706 (39). miR-706 is known to inhibit
oxidative stress-induced fibrosis in hepatocytes (40). Furthermore, the expression levels
of circRNAs have been reported to be altered in the plasma of
HBV-related patients with cancer, where hsa_circ_0000976 and
hsa_circ_0007750 were also found to be differentially expressed
(41). According to the database
Circular RNA Interactome, hsa_circ_0000976 has a binding site for
hsa-miR-486-3p. The latter is significantly upregulated in infected
hepatocytes and is associated with chemokine-and cytokine-mediated
immune responses (42).
hsa_circ_0027089 is another circRNA upregulated in the plasma of
patients suffering from HBV-related HCC. It has a binding site for
hsa-miR-15b-3p and hsa-miR-141-5p (43). miR-141 down-regulates the
peroxisome proliferator-activated receptor α and inhibits HBV
replication by reducing the HBV promotor activity (44). The upregulation of circRNA_100338
of the circRNA-100338/miR-141-3p pathway is closely correlated with
a low survival and metastatic progression in HBV-related HCC and
has indeed been suggested as a biomarker of disease progression
(45). Not only can HBV induce
changes in host circRNA levels, but it can also be the source of
circRNAs of viral origin that arise during its replication. The
generation of viral circRNAs is under the influence of host
factors, as demonstrated by the observation that knocking down the
host protein DexH-box helicase 9 results in an increase in circRNAs
of viral origin (46).
HHV
A number of circRNAs have been suggested to
participate in herpes virus infections by acting as miRNA sponges,
as well as by encoding proteins. These circRNAs appear to affect
the host's cellular environment by linking the viral behavior to
pathogenic mechanisms (47).
Kaposi's sarcoma-associated herpesvirus
(KSHV)/HHV8
KSHV/HHV8 infection can result in Kaposi's sarcoma,
endothelium-derived angiogenesis-induced tumors, primary exudative
lymphomas (PEL) and multicenter Castleman disease. KSHV-coding
circRNAs are mostly derived from the viral interferon regulatory
factor 4 (vIRF4)/K10 site, the polyadenylated nuclear site and the
K7.3 site (48-50). Abere et al (51) determined the expression
characteristics of circ-vIRF4, circPANs and circK7.3s in PEL cell
lines, tumor tissues and patients' sera. Varying degrees of
expression of KSHV-encoded circRNAs were commonly observed in PEL
cells. These circRNAs are easy to detect because they are packaged
into particles. Some of the circRNAs generated during KSHV
infection possess miRNA binding sites, such as circRNA
hsa_circ_0001400, which can inhibit the expression of certain viral
genes (50). It has also been
reported that host circRNAs can be exported from infected cells in
extracellular vesicles acting as vectors that deliver antiviral
signals to surrounding cells (52). On the whole, it is known that the
production of antiviral circRNAs is activated by KSHV infection,
with their expression being induced during the lytic phase of
infection.
Herpes simplex virus (HSV-1)/HHV1
HSV-1/HHV1 is a member of the herpesvirus and α
herpesvirus subfamilies, which feature double-stranded linear DNA
genomes. HSV-1 infection in human lung fibroblast cell lines
results in a vast number of circRNAs, genes and miRNAs being
involved in responses to the viral infection along the
circRNA-miRNA-gene regulatory axis. Such genes are implicated in
signaling and apoptosis pathways, the cell cycle and cell death
(53).
Epstein-Barr virus (EBV)/HHV4
EBV/HHV4 is a virus from the lymphotropic genus of
the herpesvirus family with linear, double-stranded DNA. It can
cause a variety of human lymphoid and non-lymphoid malignancies and
is also associated with nasopharyngeal and gastric cancer. Huang
et al (54) identified
the EBV-encoded circRNA ebv-circRPMS1 in the cytoplasm and nucleus
of EBV-infected cell lines, which has been regarded as a novel
regulatory agent acting on transcription and/or interfering with
splicing as an miRNA sponge. The following EBV-encoded circRNAs
have been identified to be associated with oncogenes: Circ
proteasome 20S subunit β9 (LMP2), circBHLF1 and circ barttin CLCNK
type accessory subunit β. The latter has been shown to play a role
in the proliferation of tumor cells of EBV-positive patients
(48). Host miRNAs, such as
miR-15b-5p, miR-30c-1-3p, miR-30c-2-3p, miR-424-5p and miR-4286,
have been recognized as targets of EBV circRNAs. These miRNAs,
which are strongly linked to herpes infections, have been found to
also be linked to tumorigenesis and tumor invasion (55-57).
Human cytomegalovirus (HCMV)/HHV5
The HCMV/HHV5 protein TRS1 (pTRS1) has been shown to
enhance the activity of the IRES through its binding to a circRNA
reporter. The expression of pTRS1 also increases the activity of
IRESs, which controls the expression of proteins needed for
efficient HCMV replication within the cell (58). This indicates that circRNAs can
play a relevant role in HCMV infections. Lou et al (59) reported the downregulation of the
circRNAs hsa_circ_0001445 and hsa_circ_0001206 in latently infected
THP-1 monocyte-macrophage cell line and patients infected with HCMV
compared with normal controls. These differentially expressed
circRNAs can bind to miRNAs such as hsa-miR-21, therefore
participating in a HCMV latency mechanism.
Human immunodeficiency virus (HIV)
HIV, widely known as the cause of the acquired
immune deficiency syndrome, is a retrovirus. Three series of
circRNAs have been reported to be involved in three different
regulatory pathways in the early stages of HIV infection:
circRNA-miR-542-3p/miR-101-3p/let-7c-5p-CDKN1A,
circRNA/miR-27b-3p/CCNK and circRNA/miR-548ah-3p/interleukin
(IL)-15 (60). CDKN1A encodes
the cyclin-dependent kinase inhibitor 1 that inhibits HIV
replication (61). Cyclin K,
encoded by CCNK, inhibits HIV gene expression and replication by
using HIV-1 Nef protein to displace Cyclin-T1 from the positive
elongation factor b complex (62). IL-15 facilitates CD4+
T cell infection, therefore creating an HIV reservoir (63).
Human papilloma virus (HPV)
HPV is a small double-stranded DNA virus that
infects stratified epithelial cells. The majority of HPV infections
are asymptomatic or cause benign wart growth, but some high-risk
HPVs can promote the development of cervical and oropharyngeal
cancer. Zhao et al (64)
observed that HPVs can generate circRNAs containing the E7 oncogene
(circE7), which encodes the E7 oncoprotein. HPV16 circE7 is
essential for the transformation and growth of CaSki cervical
cancer (CC) cells. circE7 may also promote virus replication and
host cell transformation, suggesting a link between circE7 and
high-risk HPV, and making circE7 abundance of prognostic value.
Middle East respiratory syndrome
coronavirus (MERS-CoV)
MERS-CoV is a coronavirus that affects the human
respiratory system by causing fever, chills and severe dyspnea
(65). The pathogenesis of this
virus is not yet fully understood (66). Recent research has reported that
the knockdown of circ fibronectin type II domain containing 3B
(FNDC3B) and circ CCR4-NOT transcription complex subunit 1 (CNOT1)
with specific small interfering RNAs (siRNAs) reduces the viral
load of infected cells and their target mRNA expression in human
Calu-3 lung adenocarcinoma epithelial cells, resulting in a
reduction in the expression of image result for map3k9 definition
(MAP3K9) (67). MAP3K9 is an
upstream modulator of the MAPK signaling pathway that is involved
in cell proliferation (68), as
well as in MERS-CoV infection (69). Therefore, circRNAs may play an
important role in the regulation of MERS-CoV cellular
proliferation, and could prove useful for the development of novel
antiviral strategies.
In addition, it has been found that the knockdown of
circF-NDC3B and circCNOT1 significantly reduce the MERS-CoV viral
load in Calu-3 and HFL cells, which may be associated with the
downregulation of target genes regulated by circFNDC3B and
circCNOT1 (67). MAP3K9 is an
upstream modulator of the MAPK pathway that can affect many aspects
of cell proliferation, migration and apoptosis (68), while siRNA knockdown of circFND3B
or circCNOT1 leads to a significant decrease in the expression of
MAP3K9. This may provide novel insight into the modulation of the
extracellular signal-regulated kinase (ERK)/MAPK pathway as a
host-targeted antiviral strategy against MERS-CoV infection.
Hantavirus (HTNV)
HTNV is a zoonotic virus that infects humans and
leads to hemorrhagic fever with renal syndrome, hantavirus
pulmonary syndrome or hantavirus cardiopulmonary syndrome (70). In total, 4 circRNA/miRNA/mRNA
regulatory axes have been found to be involved in HTNV infections
by ceRNA analysis: circ0002470/miR-3182/interferon induced protein
44 (IFI44), circ0006132/miR-1304-3p/2'-5'-oligoaenylate synthetase
1 (OAS1), circ0000479/miR-149-5p/retinoic acid-inducible gene I
(RIG-I) and circ0000479/miR-337-3p/caspase 1 (CASP1) (71).
IFI44 is an IFN-inducible protein associated with
viral infections. It can rescue the inhibition of IFN-induced viral
growth. More precisely, it inhibits the promoter activity of HIV-1
LTR, thereby affecting viral transcription (72). Furthermore, IFI44 interacts with
the immunoregulatory protein FK506-binding protein 5, which binds
to cellular kinases that are inhibitors of the nuclear factor κB
kinases (IKK)α, IKKβ and IKKε. Such inhibition results in a
negative modulation of IFN responses (73). The antiviral protein, OAS1, is
induced by viral infection [Sendai or influenza A virus (IAV)], an
induction that requires a type I IFN response (74). RIG-I is a pattern recognition
receptor that, upon recognition of RNA viruses, activates the
expression of type I interferons, hence promoting an antiviral
response (75). CASP1 is a
component of the inflammasome, a complex involved in the activation
of the pro-inflammatory cytokines IL-18 and IL-1β that lead to
inflammatory cell death (76).
In brief, circRNAs may provide new targets for
interventions aimed at controlling HTNV infections.
SARS-CoV-2
In the case of SARS-CoV-2 infection, one miRNA
(miR-124-3p), one lncRNA (Gm26917), one transcription factor
(STAT2), one mRNA (DeXD/H-box helicase 58) and two circRNAs
(protein phosphatase 1 regulatory subunit 10 and C330019G07Rik)
have been found to comprise a quintuple network (77). miR-124-3p has been confirmed to
be upregulated in the case of HIV-1 infection, resulting in the
downregulation of p21 and potassium two pore domain channel
subfamily K member, which leads to increased virion release
(78). However, the main targets
of this network remain unclear, and an in-depth study of the
release of certain inflammatory mediators may provide new insight
into the diagnosis of SARS-CoV-2 infection.
Other viral infections
circRNAs have also been found to be linked to
several other viral infections as major regulatory molecules of
signaling pathways, and even targets for disease diagnosis and
treatment.
IAV
The infection of A549 human alveolar basal
epithelial cells with IAV H1N1 promotes the formation of high
levels of circ-GATA zinc finger domain containing 2A (GATAD2A). In
parallel, circ-GATAD2A knockdown enhances autophagy, as established
by determining LC3-II and p62 levels, while it inhibits H1N1
replication. Conversely, circ-GATAD2A overexpression impairs
autophagy and favors H1N1 replication. The enhancement of viral
replication is thought to be due to the interaction of circ-GATAD2A
with the vacuolar protein sorting 34, since the knockdown of the
latter blocks autophagy and results in an increase of H1N1
replication. In conclusion, circ-GATAD2A may serve as a potential
therapeutic target in connection with influenza virus infections
(79).
Ebola virus
In Ebola virus infection, circRNA_chr19 regulates
the expression of claudin 18 (CLDN18) by targeting miR-30b-3p, thus
reducing the inhibitory effect of miR-30b-3p on CLDN18 through the
sponge effect (80). CLDN18 is
likely to be involved in preserving epithelial integrity (81), with its effect on Ebola virus
infections requiring further research.
Dengue fever (DF) virus (DENV)
DF is an acute infectious disease caused by Aedes
aegypti mosquito-transmitted DENV. The incidence of DF has
increased substantially worldwide, with its clinical manifestations
including acute attacks, sudden high fever, fatigue, aches, loss of
appetite and nausea (82). The
main diagnostic tool of DF is the reduction in white blood cell
(WBC) and platelet (PLT) counts (83). circRNA hsa_circ_0006459 levels
have been found to be negatively associated with WBC, PLT and
lymphocyte counts in the peripheral blood of patients with DF,
while hsa_circ_0015962 levels have been found to be positively
associated with the same parameters. With regards to the mechanisms
of action, hsa_circ_0015962 binds to miR-4683 and hsa_circ_0006459
binds to miR-133b. The levels of these circRNAs may thus be
considered as potential biomarkers (84).
4. circRNAs as tools for the diagnosis and
prognosis of viral infection-related diseases
The rapid development of high-throughput sequencing
and bioinformatics in recent years has allowed for the detection of
circRNAs highly expressed in virus infection-related diseases. The
fact that circRNAs are abundant, ubiquitous, stable, conserved
across species and tissue-cell specific indicates a diagnostic
potential greater than that of linear RNA (85). circRNAs can therefore serve as
novel diagnostic biomarkers of viral infections (Table II).
 | Table IIcircRNAs as tools for the diagnosis
and prognosis of viral infection-related diseases. |
Table II
circRNAs as tools for the diagnosis
and prognosis of viral infection-related diseases.
| Diseases | | Diagnostic
indicators | (Refs.) |
|---|
| HBV-related
hepatocellular carcinoma |
hsa_circ_0000976
hsa_circ_0007750
hsa_circ_0139897 | The plasma circRNA
panel (CircPanel) containing hsa_circ_0000976, hsa_circ_0007750 and
hsa_circ_0139897 is used to detect the changes of circRNA contents
in plasma for clinical diagnosis of HBV-related hepatocellular
carcinoma (HCC). This method performs better than alpha-fetoprotein
(AFP) in the diagnosis of small-HCC and could also identify
AFP-negative HCC and AFP-negative small-HCC. | (41) |
| EBV-related
nasopharyngeal carcinoma | circRPMS1 | CircRPMS1 was found
to be significantly increased in EBV-positive NPC tissues compared
with adjacent tissues, while circRPMS1 was not expressed in
EBV-negative NPC tissues. | (86) |
|
hsa_circRNA_001387 | In EBV infection,
hsa_circRNA_001387 is significantly upregulated. | (87) |
| HPV-positive
cervical cancers | circE7 | circE7 as a
possible sensitive marker for high-risk HPV. | (64) |
Recent research has demonstrated that a plasma
circRNA panel (CircPanel) containing hsa_circ_0000976,
hsa_circ_0007750 and hsa_circ_0139897 could be used as a biomarker
for the clinical diagnosis of HBV-related hepatocellular carcinoma.
This HCC detection CircPanel was constructed based on microarray
screening and reverse transcription-quantitative PCR to identify
plasma circRNAs that are increased in patients with HCC with HBV
compared with the controls. The diagnostic accuracy of the method
was evaluated by constructing a logistic regression model and
analyzing the area under the receiver operating characteristic
(ROC) curve (AUC). This CircPanel performed better than
α-fetoprotein (AFP) in the diagnosis of small HCC and could also
identify AFP-negative HCC and AFP-negative small HCC (41).
Nasopharyngeal carcinoma (NPC), mainly induced by
EBV, is one of the most difficult cancers to detect. Thus, specific
biomarkers of NPC are very helpful for a timely diagnosis of NPC. A
recent study reported that circRPMS1 was found to be significantly
increased in EBV-positive NPC tissues compared with adjacent
tissues, and was not expressed in EBV-negative NPC tissues,
suggesting that circRPMS1 could indeed be a useful NPC diagnostic
biomarker (86). In addition, a
marked upregulation of hsa_circRNA_001387, which could be used as a
novel NPC biomarker, has been reported. The ROC curve for the
determination of the diagnostic value of hsa_circRNA_001387 showed
a high accuracy, indicating that this circRNA could be a specific
and sensitive NPC marker (87).
circE7 is present in RNA-Seq data from The Cancer
Genome Atlas concerning HPV-positive cancers and cell lines with
only episomal HPVs. Given the stability of circRNA and the
importance of the E7 oncoprotein in the genesis of CC, it has been
considered appropriate to examine circE7 as a possible, sensitive
marker of high-risk HPVs (64).
Zhou et al (88) examined
the diagnostic potential of ciRS-7 regarding CC by assessing ROC
curves, and concluded that it is indeed an indicator of the
activity of CC cells.
The altered expression of circRNAs is expected to
become an important molecular tool for the early diagnosis of viral
infection-related diseases, which can result in an increased
patient long-term survival, reducing the mortality rate and
improving prognosis.
5. circRNAs and the treatment of viral
infections
circRNAs can also be used as therapeutic targets in
the management of viral infections (Table III). circRNA-10156 is
abundantly expressed in HBV-related liver cancer and can regulate
the proliferation of malignant cells through the
circRNA_10156/miR-149-3p/AKT1 signaling pathway (89). The depletion of circRNA_10156 can
increase the content of miR-149-3p and decrease AKT1 levels. AKT1
mediates the epithelial-mesenchymal conversion of HCC cells through
the AKT/glycogen synthase kinase-3β/β-catenin signaling pathway and
inhibits HBV replication by suppressing gene transcription
(90,91). In addition, the increased
expression of hsa_circ_0006942 (circ-ATP5H) and tumor necrosis
factor α-induced protein 3 (TNFAIP3) were observed in HCC cells
associated with HBV infection, while the expression of miR-138-5p
was downregulated. Further experiments proved that circ-ATP
synthase peripheral stalk subunit d (ATP5H) acts as an miR-138-5p
sponge and regulates the expression of TNFAIP3, thus circ-ATP5H may
be a potential therapeutic target for HBV-related HCC (92). The circ_0004812/miR-1287-5p/FSTL1
pathway regulates HBV-induced immune system suppression (37). circ_0004812 inhibits miR-1287-5p,
which promotes FSTL1 expression leading to the expansion of HCC
cells, an increase in cellular proliferation and the inhibition of
cellular apoptosis. EBV(+) NPC tissues express high levels of
circRPMS1. circRPMS1 knockdown can reduce the invasiveness of NPC
cells. Cell invasion is mediated by miR-203, miR-31 and miR-451, as
shown by the fact that inhibitors of these miRNAs can reverse the
circRPMS1 knockdown effects. circRPMS1 can sponge the
aforementioned miRNAs, therefore mediating NPC oncogenesis, which
occurs by promoting epithelial-mesenchymal transition. As a
consequence, circRPMS1 can likely become a convenient therapeutic
target for the treatment of EBV-associated NPC (86,93,94).
 | Table IIIcircRNAs and the treatment of viral
infections. |
Table III
circRNAs and the treatment of viral
infections.
| circRNA | Role in viral
infection | Therapeutic
potential | (Refs.) |
|---|
| circ-10156 | Acting as a
molecular sponge for miR-149-3p, regulating the proliferation of
HBV-related liver cancer cells through the miR-149-3p/Akt1
pathway. | It was observed
that circ-10156 was up-regulated in liver cancer tissues, and
inhibition of circ-10156 expression in liver cancer tissues could
inhibit the proliferation of liver cancer cells. | (89-91) |
| circ-ATP5H | Acts as miR-138-5p
sponge and regulates the expression of TNFAIP3. | Inhibit the
expression of circ-ATP5H, and enhance the anticancer effect of
miR-138-5p. | (92) |
| circ-0004812 | By inhibiting
miR-1287-5p, it promotes the expression of FSTL1, thereby
regulating the immune suppression induced by HBV. | Releasing the
inhibitory effect of circ-0004812, thereby inhibiting the
expression of FSTL1, and inhibiting its HCC cell proliferation
promotion and apoptosis inhibitory effect. | (37) |
| circ-RPMS1 | Sponge multiple
miRNAs (miR-203, miR-31 and miR-451) and promote
epithelial-mesenchymal transition to promote the occurrence of
nasopharyngeal carcinoma. | Reduce or eliminate
the sponge effect of circRPMS1 and inhibit the
epithelial-mesenchymal transition of nasopharyngeal carcinoma. | (86,93,94) |
| circ-LMP2A (from
EBV) | Attenuate the tumor
suppressor effect of miR-3908/TRIM59/p53 axis to promote
EBV-related gastric cancer cell metastasis and tumor
progression. | Inhibit circ-LMP2A
derived from EBV and strengthen the tumor suppressive effect of
miR-3908/TRIM59/p53 axis. | (96) |
| circ-FNDC3B and
circ-CNOT1 | Associated with the
MERS-CoV load in Mers-CoV-infected lung adenocarcinoma and its
target mRNA expression, and significantly affect mitogen-activated
protein kinase (MAPK) and ubiquitination pathway. | Knockout of
circFNDC3B and circCNOT1 will result in decreased cell virus
content. | (67) |
| Artificial circRNA
sponge | Artificially
synthesized circRNA | Isolate miR-122,
thereby inhibiting the production of viral proteins in the HCV cell
culture system. | (97) |
circRNAs have been found to be involved in
EBV-associated gastric cancer. In fact, circRPMS1_E4_E3a and
circRPMS1_ E4_E2 were detected in EBV-positive stomach cancer
patient specimens (95). In a
recent study, the expression of the EBV-encoded ebv-circLMP2A was
found to be associated with the induction of stem cell-like
phenotypes in a tumor cell line through targeting the
miR-3908/tripartite motif containing 59 (TRIM59)/p53 axis. Of note,
a high expression of ebv-circLMP2A has been found to be
significantly associated with metastasis and a poor prognosis of
patients with gastric cancer, indicating the potential of this
EBV-encoded circRNA as a therapeutic target (96).
The expression of hsa_circ_0067985/circFNDC3B and
hsa_circ_0006275/circCNOT1 has been found to be upregulated in
MERS-CoV-infected human lung adenocarcinoma epithelial cells. The
knockdown of circFNDC3B and circCNOT1 results in a reduction of
both cellular virus content and the expression of the target mRNAs,
which modulate MAPK and ubiquitination pathways. These results
provided insights into potential antiviral strategies for MERS-CoV
infections (67).
circRNAs can be engineered as miRNA sponges. In a
model system, advantage was taken of the fact that the life cycle
of HCV relies on host miR-122 (97). Artificial circRNA sponges were
devised to sequester miR-122, which resulted in the inhibition of
HCV production in cell culture systems. These findings uncovered
the potential application of circRNAs to tackle HCV infection
(98).
6. Conclusions
circRNAs are specific non-coding RNAs involved in
the regulation of miRNAs, and thus affect gene expression. These
RNAs titrate miRNAs and regulate transcription and/or interfere
with splicing. The regulatory role of circRNAs in the immunity to
viral infections has received increasing attention. The analysis of
the specific mechanisms through which circRNAs can influence viral
infections would facilitate a deeper understanding of disease
etiologies. In addition, such studies will likely unveil novel
therapeutic targets and possible treatments for infectious diseases
that involve changes in circRNA levels (Fig. 2), including COVID-19. These new
approaches are expected to lead to improved prognoses of viral
diseases.
Availability of data and materials
Not applicable.
Authors' contributions
LT conceived and designed the study. YM, YL, AL, HZ
and LT discussed, wrote and edited the manuscript. YL edited the
figures. LT revised the manuscript and acquired the financial
support for the project leading to this publication. LT, YM and YL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Rodolfo Garcia
(Biomedicine Research Honorary Consultant, London, United Kingdom)
for commenting on and revising the manuscript.
Funding
The present study was supported by the Changsha Municipal
Natural Science Foundation (grant no. kq2007065), Teacher Research
Fund of Central South University (grant no. 2014JSJJ034) and
Innovation Project of Central South University (grant no.
xcx201990539).
Abbreviations:
|
COVID-19
|
coronavirus disease 2019
|
|
SARS-CoV-2
|
severe acute respiratory syndrome
virus
|
|
RNA Pol II
|
RNA polymerase II
|
|
SS
|
splice site
|
|
TER
|
transcription elongation rate
|
|
ICS
|
intronic complementary sequence
|
|
MBNL1
|
muscleblind like splicing regulator
1
|
|
ADAR1
|
adenosine deaminase 1 acting on
RNA1
|
|
CDR1
|
cerebellar degeneration-related
protein 1
|
|
U1snRNP
|
U1 small nuclear
ribonucleoproteins
|
|
IRES
|
internal ribosome entry site
|
|
MBL
|
muscleblind
|
|
EIciRNAs
|
exon-intron circular RNAs
|
|
EIF3J
|
eukaryotic translation initiation
factor 3 subunit J
|
|
HBV
|
hepatitis B virus
|
|
HHV
|
human herpesvirus
|
|
HCC
|
hepatocellular carcinoma
|
|
IRF7
|
interferon regulatory factor 7
|
|
cccDNA
|
hepatitis B virus covalently closed
circular DNA
|
|
KSHV
|
Kaposi's sarcoma-associated
herpesvirus
|
|
PEL
|
primary exudative lymphoma
|
|
vIRF4
|
viral interferon regulatory factor
4
|
|
HSV
|
herpes simplex virus
|
|
EBV
|
Epstein-Barr virus
|
|
HCMV
|
human cytomegalovirus
|
|
pTRS1
|
protein TRS1
|
|
HIV
|
human immunodeficiency virus
|
|
HPV
|
human papilloma virus
|
|
circE7
|
circRNAs containing the E7
oncogene
|
|
MERS-CoV
|
Middle East respiratory syndrome
coronavirus
|
|
MAPK
|
mitogen-activated protein kinase
|
|
HTNV
|
hantavirus
|
|
IKK
|
inhibitors of the nuclear factor κB
kinases
|
|
DF
|
dengue fever
|
|
DENV
|
dengue fever virus
|
|
CircPanel
|
circRNA panel
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the receiver operating
characteristic curve
|
|
AFP
|
α-fetoprotein
|
|
NPC
|
nasopharyngeal carcinoma
|
|
CC
|
cervical cancer
|
|
Akt
|
protein kinase B
|
|
TNFAIP3
|
tumor necrosis factor α-induced
protein 3
|
|
WBC
|
white blood cell
|
|
PLT
|
platelet
|
References
|
1
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen M, Xu Y, Hong N, Yang Y, Lei W, Du L,
Zhao J, Lei X, Xiong L, Cai L, et al: Epidemiology of fungal
infections in China. Front Med. 12:58–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan KE and Lim YY: Viruses join the
circular RNA world. FEBS J. Nov 25–2020.Epub ahead of print.
View Article : Google Scholar :
|
|
4
|
Xie H, Sun H, Mu R, Li S, Li Y, Yang C, Xu
M, Duan X and Chen L: The role of circular RNAs in viral infection
and related diseases. Virus Res. 291:1982052021. View Article : Google Scholar
|
|
5
|
Garassino MC, Whisenant JG, Huang LC,
Trama A, Torri V, Agustoni F, Baena J, Banna G, Berardi R, Bettini
AC, et al: COVID-19 in patients with thoracic malignancies
(TERAVOLT): First results of an international, registry-based,
cohort study. Lancet Oncol. 21:914–922. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun X, Wang L, Ding J, Wang Y, Wang J,
Zhang X, Che Y, Liu Z, Zhang X, Ye J, et al: Integrative analysis
of Arabidopsis thaliana transcriptomics reveals intuitive splicing
mechanism for circular RNA. FEBS Lett. 590:3510–3516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
8
|
Zhang Y, Xue W, Li X, Zhang J, Chen S,
Zhang JL, Yang L and Chen LL: The biogenesis of nascent circular
RNAs. Cell Rep. 15:611–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The output of protein-coding genes
shifts to circular RNAs when the Pre-mRNA processing machinery is
limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou Y, Zheng S, Deng X, Yang A and Xie X,
Tang H and Xie X: The role of circular RNA CDR1as/ciRS-7 in
regulating tumor microenvironment: A pan-cancer analysis.
Biomolecules. 9:4292019. View Article : Google Scholar :
|
|
13
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao R, Chen JY, Liang Z, Luo D, Chen G,
Lu ZJ, Chen Y, Zhou B, Li H, Du X, et al: Pervasive chromatin-RNA
binding protein interactions enable RNA-Based regulation of
transcription. Cell. 178:107–121.e18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ule J and Blencowe BJ: Alternative
splicing regulatory networks: Functions, mechanisms, and evolution.
Mol Cell. 76:329–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Williams T, Ngo LH and Wickramasinghe VO:
Nuclear export of RNA: Different sizes, shapes and functions. Semin
Cell Dev Biol. 75:70–77. 2018. View Article : Google Scholar
|
|
17
|
Martinez-Salas E, Lozano G,
Fernandez-Chamorro J, Francisco-Velilla R, Galan A and Diaz R:
RNA-binding proteins impacting on internal initiation of
translation. Int J Mol Sci. 14:21705–21726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu KS, Pan F, Mao XD, Liu C and Chen YJ:
Biological functions of circular RNAs and their roles in occurrence
of reproduction and gynecological diseases. Am J Transl Res.
11:1–15. 2019.PubMed/NCBI
|
|
19
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: CircRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Yang L and Chen LL: Life without
A tail: New formats of long noncoding RNAs. Int J Biochem Cell
Biol. 54:338–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Awan FM, Yang BB, Naz A, Hanif A, Ikram A,
Obaid A, Malik A, Janjua HA, Ali A and Sharif S: The emerging role
and significance of circular RNAs in viral infections and antiviral
immune responses: Possible implication as theranostic agents. RNA
Biol. 18:1–15. 2021. View Article : Google Scholar
|
|
25
|
Zhao T, Zheng Y, Hao D, Jin X, Luo Q, Guo
Y, Li D, Xi W, Xu Y, Chen Y, et al: Blood circRNAs as biomarkers
for the diagnosis of community-acquired pneumonia. J Cell Biochem.
120:16483–16494. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Trepo C, Chan HL and Lok A: Hepatitis B
virus infection. Lancet. 384:2053–2063. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schweitzer A, Horn J, Mikolajczyk RT,
Krause G and Ott JJ: Estimations of worldwide prevalence of chronic
hepatitis B virus infection: A systematic review of data published
between 1965 and 2013. Lancet. 386:1546–1555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou TC, Li X, Chen LJ, Fan JH, Lai X,
Tang Y, Zhang L and Wei J: Differential expression profile of
hepatic circular RNAs in chronic hepatitis B. J Viral Hepat.
25:1341–1351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Zhao J, Ren J, Hall KH, Moorman
JP, Yao ZQ and Ning S: Protein phosphatase 1 abrogates
IRF7-mediated type I IFN response in antiviral immunity. Eur J
Immunol. 46:2409–2419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Belloni L, Allweiss L, Guerrieri F,
Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M
and Levrero M: IFN-α inhibits HBV transcription and replication in
cell culture and in humanized mice by targeting the epigenetic
regulation of the nuclear cccDNA minichromosome. J Clin Invest.
122:529–537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan H, Lv P, Lv J, Zhao X, Liu M, Zhang G
and Tang H: MiR-370 suppresses HBV gene expression and replication
by targeting nuclear factor IA. J Med Virol. 89:834–844. 2017.
View Article : Google Scholar
|
|
32
|
Chen C, Wu M, Zhang W, Lu W, Zhang M,
Zhang Z, Zhang X and Yuan Z: MicroRNA-939 restricts hepatitis B
virus by targeting Jmjd3-mediated and C/EBPα-coordinated chromatin
remodeling. Sci Rep. 6:359742016. View Article : Google Scholar
|
|
33
|
Shi J, Zhao J, Zhang X, Cheng Y, Hu J, Li
Y, Zhao X, Shang Q, Sun Y, Tu B, et al: Activated hepatic stellate
cells impair NK cell anti-fibrosis capacity through a
TGF-β-dependent emperipolesis in HBV cirrhotic patients. Sci Rep.
7:445442017. View Article : Google Scholar
|
|
34
|
Song G, Jia H, Xu H, Liu W, Zhu H, Li S,
Shi J, Li Z, He J and Chen Z: Studying the association of
microRNA-210 level with chronic hepatitis B progression. J Viral
Hepat. 21:272–280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang GL, Li YX, Zheng SQ, Liu M, Li X and
Tang H: Suppression of hepatitis B virus replication by
microRNA-199a-3p and microRNA-210. Antiviral Res. 88:169–175. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Zhang X, Chen L, Zhang Z, Zhang J,
Wang W, Wu M, Shi B, Zhang X, Kozlowski M, et al: Circulating
miR-210 and miR-22 combined with ALT predict the virological
response to interferon-alpha therapy of CHB patients. Sci Rep.
7:156582017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L and Wang Z: Circular RNA
hsa_circ_0004812 impairs IFN-induced immune response by sponging
miR-1287-5p to regulate FSTL1 in chronic hepatitis B. Virol J.
17:402020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang MH, Chen M, Mo HH, Tsai WC, Chang YC,
Chang CC, Chen KC, Wu HY, Yuan CH, Lee CH, et al: Utilizing
experimental mouse model to identify effectors of hepatocellular
carcinoma induced by HBx antigen. Cancers (Basel). 12:4092020.
View Article : Google Scholar
|
|
39
|
Li Y, Gao X, Wang Z, Liu W, Xu F, Hu Y, Li
Y and Shi L: Circular RNA 4099 aggravates hydrogen peroxide-induced
injury by down-regulating microRNA-706 in L02 cells. Life Sci.
241:1168262020. View Article : Google Scholar
|
|
40
|
Yin R, Guo D, Zhang S and Zhang X: miR-706
inhibits the oxidative stress-induced activation of PKCα/TAOK1 in
liver fibrogenesis. Sci Rep. 6:375092016. View Article : Google Scholar
|
|
41
|
Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu
QG, Yang Y, Sun SH, Liu JF, Qin LX, et al: Plasma circular RNA
panel to diagnose hepatitis B virus-related hepatocellular
carcinoma: A large-scale, multicenter study. Int J Cancer.
146:1754–1763. 2020. View Article : Google Scholar
|
|
42
|
Kohno T, Tsuge M, Murakami E, Hiraga N,
Abe H, Miki D, Imamura M, Ochi H, Hayes CN and Chayama K: Human
microRNA hsa-miR-1231 suppresses hepatitis B virus replication by
targeting core mRNA. J Viral Hepat. 21:e89–e97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia
H, Dai Z, Tang Z, Fan J and Zhou J: Plasma hsa_circ_0027089 is a
diagnostic biomarker for hepatitis B virus-related hepatocellular
carcinoma. Carcinogenesis. 41:296–302. 2020. View Article : Google Scholar :
|
|
44
|
Hu W, Wang X, Ding X, Li Y, Zhang X, Xie
P, Yang J and Wang S: MicroRNA-141 represses HBV replication by
targeting PPARA. PLoS One. 7:e341652012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen
Z, Song W, Zhou J, Tang ZY and Huang XY: Comprehensive circular RNA
profiling reveals the regulatory role of the
circRNA-100338/miR-141-3p pathway in hepatitis B-related
hepatocellular carcinoma. Sci Rep. 7:54282017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sekiba K, Otsuka M, Ohno M, Kishikawa T,
Yamagami M, Suzuki T, Ishibashi R, Seimiya T, Tanaka E and Koike K:
DHX9 regulates production of hepatitis B virus-derived circular RNA
and viral protein levels. Oncotarget. 9:20953–20964. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ungerleider NA, Tibbetts SA, Renne R and
Flemington EK: Gammaherpesvirus RNAs come full circle. mBio.
10:e00071–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Toptan T, Abere B, Nalesnik MA, Swerdlow
SH, Ranganathan S, Lee N, Shair KH, Moore PS and Chang Y: Circular
DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA.
115:E8737–E8745. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ungerleider NA, Jain V, Wang Y, Maness NJ,
Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C, et al:
Comparative analysis of gammaherpesvirus circular RNA repertoires:
Conserved and unique viral circular RNAs. J Virol. 93:e01952–18.
2019.
|
|
50
|
Tagawa T, Gao S, Koparde VN, Gonzalez M,
Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan
R and Ziegelbauer JM: Discovery of Kaposi's sarcoma
herpes-virus-encoded circular RNAs and a human antiviral circular
RNA. Proc Natl Acad Sci USA. 115:12805–12810. 2018. View Article : Google Scholar
|
|
51
|
Abere B, Li J, Zhou H, Toptan T, Moore PS
and Chang Y: Kaposi's sarcoma-associated herpesvirus-encoded
circRNAs are expressed in infected tumor tissues and are
incorporated into virions. mBio. 11:e03027–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Preusser C, Hung LH, Schneider T,
Schreiner S, Hardt M, Moebus A, Santoso S and Bindereif A:
Selective release of circRNAs in platelet-derived extracellular
vesicles. J Extracell Vesicles. 7:14244732018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shi J, Hu N, Mo L, Zeng Z, Sun J and Hu Y:
Deep RNA sequencing reveals a repertoire of human fibroblast
circular RNAs associated with cellular responses to herpes simplex
virus 1 infection. Cell Physiol Biochem. 47:2031–2045. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang JT, Chen JN, Gong LP, Bi YH, Liang
J, Zhou L, He D and Shao CK: Identification of virus-encoded
circular RNA. Virology. 529:144–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun G, Shi L, Yan S, Wan Z, Jiang N, Fu L,
Li M and Guo J: MiR-15b targets cyclin D1 to regulate proliferation
and apoptosis in glioma cells. Biomed Res Int. 2014:6878262014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lovat F, Fassan M, Gasparini P, Rizzotto
L, Cascione L, Pizzi M, Vicentini C, Balatti V, Palmieri D,
Costinean S and Croce CM: miR-15b/16-2 deletion promotes B-cell
malignancies. Proc Natl Acad Sci USA. 112:11636–11641. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Qiao Y, Zhao X, Liu J and Yang W:
Epstein-Barr virus circRNAome as host miRNA sponge regulates virus
infection, cell cycle, and oncogenesis. Bioengineered. 10:593–603.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vincent HA, Ziehr B, Lenarcic EM and
Moorman NJ: Human cytomegalovirus pTRS1 stimulates cap-independent
translation. Virology. 537:246–253. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lou YY, Wang QD, Lu YT, Tu MY, Xu X, Xia
Y, Peng Y, Lai MM and Zheng XQ: Differential circRNA expression
profiles in latent human cytomegalovirus infection and validation
using clinical samples. Physiol Genomics. 51:51–58. 2019.
View Article : Google Scholar
|
|
60
|
Zhang Y, Zhang H, An M, Zhao B, Ding H,
Zhang Z, He Y, Shang H and Han X: Crosstalk in competing endogenous
RNA networks reveals new circular RNAs involved in the pathogenesis
of early HIV infection. J Transl Med. 16:3322018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shi B, Sharifi HJ, DiGrigoli S, Kinnetz M,
Mellon K, Hu W and de Noronha CMC: Inhibition of HIV early
replication by the p53 and its downstream gene p21. Virol J.
15:532018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Khan SZ and Mitra D: Cyclin K inhibits
HIV-1 gene expression and replication by interfering with
cyclin-dependent kinase 9 (CDK9)-cyclin T1 interaction in
Nef-dependent manner. J Biol Chem. 286:22943–22954. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manganaro L, Hong P, Hernandez MM, Argyle
D, Mulder LCF, Potla U, Diaz-Griffero F, Lee B, Fernandez-Sesma A
and Simon V: IL-15 regulates susceptibility of CD4+ T
cells to HIV infection. Proc Natl Acad Sci USA. 115:E9659–E9667.
2018. View Article : Google Scholar
|
|
64
|
Zhao J, Lee EE, Kim J, Yang R, Chamseddin
B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al: Transforming
activity of an oncoprotein-encoding circular RNA from human
papilloma-virus. Nat Commun. 10:23002019. View Article : Google Scholar
|
|
65
|
Chan JF, Lau SK, To KK, Cheng VC, Woo PC
and Yuen KY: Middle East respiratory syndrome coronavirus: Another
zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol
Rev. 28:465–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zumla A, Chan JF, Azhar EI, Hui DS and
Yuen KY: Coronaviruses-drug discovery and therapeutic options. Nat
Rev Drug Discov. 15:327–347. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang X, Chu H, Wen L, Shuai H, Yang D,
Wang Y, Hou Y, Zhu Z, Yuan S, Yin F, et al: Competing endogenous
RNA network profiling reveals novel host dependency factors
required for MERS-CoV propagation. Emerg Microbes Infect.
9:733–746. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Morrison DK: MAP kinase pathways. Cold
Spring Harb Perspect Biol. 4:a0112542012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kindrachuk J, Ork B, Hart BJ, Mazur S,
Holbrook MR, Frieman MB, Traynor D, Johnson RF, Dyall J, Kuhn JH,
et al: Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling
modulation for Middle East respiratory syndrome coronavirus
infection as identified by temporal kinome analysis. Antimicrob
Agents Chemother. 59:1088–1099. 2015. View Article : Google Scholar :
|
|
70
|
Echterdiek F, Kitterer D, Alscher MD,
Schwenger V, Ruckenbrod B, Bald M and Latus J: Clinical course of
hanta-virus-induced nephropathia epidemica in children compared to
adults in Germany-analysis of 317 patients. Pediatr Nephrol.
34:1247–1252. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lu S, Zhu N, Guo W, Wang X, Li K, Yan J,
Jiang C, Han S, Xiang H, Wu X, et al: RNA-Seq revealed a circular
RNA-microRNA-mRNA regulatory network in hantaan virus infection.
Front Cell Infect Microbiol. 10:972020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Power D, Santoso N, Dieringer M, Yu J,
Huang H, Simpson S, Seth I, Miao H and Zhu J: IFI44 suppresses
HIV-1 LTR promoter activity and facilitates its latency. Virology.
481:142–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
DeDiego ML, Nogales A, Martinez-Sobrido L
and Topham DJ: Interferon-induced protein 44 interacts with
cellular FK506-binding protein 5, negatively regulates host
antiviral responses, and supports virus replication. mBio.
10:e01839–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Melchjorsen J, Kristiansen H, Christiansen
R, Rintahaka J, Matikainen S, Paludan SR and Hartmann R:
Differential regulation of the OASL and OAS1 genes in response to
viral infections. J Interferon Cytokine Res. 29:199–207. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Leung DW: Mechanisms of non-segmented
negative sense RNA viral antagonism of host RIG-I-Like receptors. J
Mol Biol. 431:4281–4289. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lupfer C, Malik A and Kanneganti TD:
Inflammasome control of viral infection. Curr Opin Virol. 12:38–46.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Arora S, Singh P, Dohare R, Jha R and Ali
Syed M: Unravelling host-pathogen interactions: ceRNA network in
SARS-CoV-2 infection (COVID-19). Gene. 762:1450572020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Farberov L, Herzig E, Modai S, Isakov O,
Hizi A and Shomron N: MicroRNA-mediated regulation of p21 and TASK1
cellular restriction factors enhances HIV-1 infection. J Cell Sci.
128:1607–1616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yu T, Ding Y, Zhang Y, Liu Y, Li Y, Lei J,
Zhou J, Song S and Hu B: Circular RNA GATAD2A promotes H1N1
replication through inhibiting autophagy. Vet Microbiol.
231:238–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang ZY, Guo ZD, Li JM, Zhao ZZ, Fu YY,
Zhang CM, Zhang Y and Liu LN, Qian J and Liu LN: Genome-wide search
for competing endogenous RNAs responsible for the effects induced
by ebola virus replication and transcription using a trVLP system.
Front Cell Infect Microbiol. 7:4792017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yao F, Kausalya JP, Sia YY, Teo AS, Lee
WH, Ong AG, Zhang Z, Tan JH, Li G, Bertrand D, et al: Recurrent
fusion genes in gastric cancer: CLDN18-ARHGAP26 induces loss of
epithelial integrity. Cell Rep. 12:272–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Barba-Spaeth G, Dejnirattisai W, Rouvinski
A, Vaney MC, Medits I, Sharma A, Simon-Lorière E, Sakuntabhai A,
Cao-Lormeau VM, Haouz A, et al: Structural basis of potent
Zika-dengue virus antibody cross-neutralization. Nature. 536:48–53.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Swanstrom JA, Plante JA, Plante KS, Young
EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM and
Baric RS: Dengue virus envelope dimer epitope monoclonal antibodies
isolated from dengue patients are protective against Zika virus.
mBio. 7:e01123–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
He J, Ming Y, MinLi Y, Han Z, Jiang J,
Zhou J, Dai B, Lv Y, He ML, Fang M and Li Y: hsa_circ_0006459 and
hsa_circ_0015962 affect prognosis of dengue fever. Sci Rep.
9:194252019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J,
Chen L, He Z, Cai X, Chen M, et al: Hsa_Circ_0001275: A potential
novel diagnostic biomarker for postmenopausal osteoporosis. Cell
Physiol Biochem. 46:2508–2516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liu Q, Shuai M and Xia Y: Knockdown of
EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma
cell proliferation and metastasis through sponging multiple miRNAs.
Cancer Manag Res. 11:8023–8031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shuai M and Huang L: High expression of
hsa_circRNA_001387 in nasopharyngeal carcinoma and the effect on
efficacy of radiotherapy. Onco Targets Ther. 13:3965–3973. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhou Y, Shen L, Wang YZ and Zhou CC: The
potential of ciRS-7 for predicting onset and prognosis of cervical
cancer. Neoplasma. 67:312–322. 2020. View Article : Google Scholar
|
|
89
|
Wang M, Gu B, Yao G, Li P and Wang K:
Circular RNA expression profiles and the pro-tumorigenic function
of CircRNA_10156 in Hepatitis B virus-related liver cancer. Int J
Med Sci. 17:1351–1365. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen J, Chan AW, To KF, Chen W, Zhang Z,
Ren J, Song C, Cheung YS, Lai PB, Cheng SH, et al: SIRT2
overexpression in hepatocellular carcinoma mediates epithelial to
mesenchymal transition by protein kinase B/glycogen synthase
kinase-3β/β-catenin signaling. Hepatology. 57:2287–2298. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Guo H, Zhou T, Jiang D, Cuconati A, Xiao
GH, Block TM and Guo JT: Regulation of hepatitis B virus
replication by the phosphatidylinositol 3-kinase-akt signal
transduction pathway. J Virol. 81:10072–10080. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Jiang W, Wang L, Zhang Y and Li H:
Circ-ATP5H induces hepatitis B virus replication and expression by
regulating miR-138-5p/TNFAIP3 axis. Cancer Manag Res.
12:11031–11040. 2020. View Article : Google Scholar :
|
|
93
|
Cheung CC, Chung GT, Lun SW, To KF, Choy
KW, Lau KM, Siu SP, Guan XY, Ngan RK, Yip TT, et al: miR-31 is
consistently inactivated in EBV-associated nasopharyngeal carcinoma
and contributes to its tumorigenesis. Mol Cancer. 13:1842014.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu N, Jiang N, Guo R, Jiang W, He QM, Xu
YF, Li YQ, Tang LL, Mao YP, Sun Y and Ma J: MiR-451 inhibits cell
growth and invasion by targeting MIF and is associated with
survival in nasopharyngeal carcinoma. Mol Cancer. 12:1232013.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ungerleider N, Concha M, Lin Z, Roberts C,
Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, et al: The
Epstein Barr virus circRNAome. PLoS Pathog. 14:e10072062018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gong LP, Chen JN, Dong M, Xiao ZD, Feng
ZY, Pan YH, Zhang Y, Du Y, Zhang JY, Bi YH, et al: Epstein-Barr
virus-derived circular RNA LMP2A induces stemness in EBV-associated
gastric cancer. EMBO Rep. 21:e496892020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li Y, Masaki T, Yamane D, McGivern DR and
Lemon SM: Competing and noncompeting activities of miR-122 and the
5' exonuclease Xrn1 in regulation of hepatitis C virus replication.
Proc Natl Acad Sci USA. 110:1881–1886. 2013. View Article : Google Scholar
|
|
98
|
Jost I, Shalamova LA, Gerresheim GK,
Niepmann M, Bindereif A and Rossbach O: Functional sequestration of
microRNA-122 from hepatitis C virus by circular RNA sponges. RNA
Biol. 15:1032–1039. 2018.PubMed/NCBI
|