Cells can sense the rigidity of the extracellular
matrix (ECM) through integrin adhesions, which have been reported
to be a significant factor in various processes, including tissue
repair, maintenance and formation (1,2).
The mechanical signals of the extracellular microenvironment
regulate cell growth, proliferation, differentiation and integrin
activity (3). The physical
connection between cells and the ECM is based on multi-component
molecular complexes that are controlled by integrins, known as the
core structure of transmembrane proteins (4). Through integrins, the mechanical
signals of the extracellular microenvironment can be transmitted
intracellularly (4). Integrins
are transmembrane heterodimeric glycoprotein receptors consisting
of α- and β-heterodimers, providing a connection between the ECM
and intracellular actin cytoskeleton through their large
extracellular domains, via adapter proteins, including Talin-1 and
vinculin (5-7). As the main activator of integrin,
Talin-1 has been reported to connect the cytoplasmic domain of
integrin with the cytoskeleton of actin to form focal adhesions
(FAs) (8). FAs are the
congregates of the intracellular proteins that function as
tension-sensing anchoring points, integrating cells with the
extracellular microenvironment (9,10). In addition, FAs have been
reported to promote intracellular reorganization, leading to
dynamic alterations in cell morphology and function (9-12). In recent years, researchers have
discovered that the specific structure of Talin-1 with mechanical
sensitivity plays a defining role in mechanical properties and
interaction networks in cellular mechanotransduction.
Talin-1 is mainly expressed in the liver, kidneys,
stomach, spleen, lungs and vascular smooth muscle, and is a large,
270-kDa protein with 18 domains, containing a 50-kDa globular head,
a long rod comprised of 62 helices forming 13 helical bundle
domains (R1-R13) (13,14) and a dimerization (DD) motif at
the C-terminus (15). A
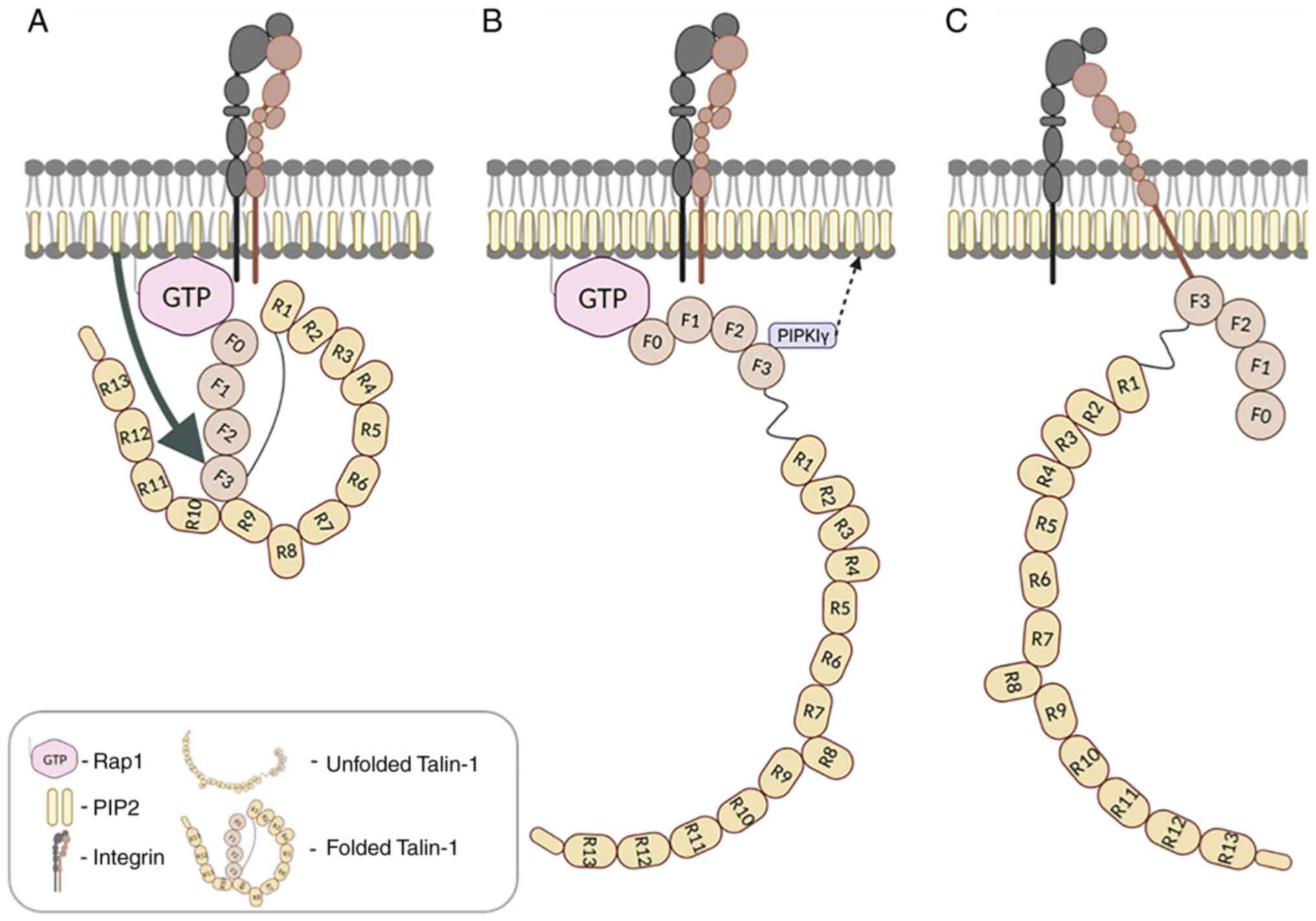
schematic view of Talin-1 structure is illustrated in Fig. 1. The head four-point-one, ezrin,
radixin, moesin (FERM) domain consists of four subdomains (F0-F3),
being a common structural feature of several integrin tail-binding
proteins, while the rod domain includes 13 helical bundles
(R1-R13), forming an extendible and flexible chain with a dimer
domain (DD) at the end of the structure, through the connection
with short linkers. The unique conformation of Talin-1 allows it to
function as a 'spring' and unfold into a 60-100 nm linear rod
(16-18). This conformational change is
responsible for binding to various FA components, including actin
and vinculin (19).
The rod part of Talin-1 is the main structure, with
the ability to sense mechanical forces and regulate the assembly
and maturation of the FA complexes (FACs). R9 and R12 shield the
binding sites of integrin and phosphatidylinositol-4,5-bisphosphate
(PIP2) of the FERM structure (11). R3 is extremely sensitive to
mechanical forces (20). As the
'goalkeeper' of FAC, it is the first to release the folded
four-helix bundle under the action of relatively low-intensity
mechanical force (5 pN), and the compact structure of Talin-1
begins to collapse (19-21). The second messenger, PIP2,
promotes Talin-1 binding to integrin, subsequently stretching
Talin-1 conformation further and binding to actin, while exposing
vinculin binding sites (VBS) (22,23). Moreover, R3 can also directly
bind Rap1-GTP interacting protein (RIAM), which results in VBS
exposure (24). Talin-1 contains
up to 11 VBS, rendering vinculin a crucial regulator for the
performance of signal transformation (25). Vinculin has been identified to
trigger the phosphorylation and nuclear localization of the
transcription factor, Yes-associated protein (YAP) (26).
Subdomains F0 and F1 of Talin-1 have been reported
to bind to Ras-associated protein 1 (Rap1) or RIAM and participate
in integrin activation (27). F3
binds to integrin cytoplasm domains and related proteins, including
FA kinase (FAK), Layilin, T-cell lymphoma invasion and metastasis 1
(TIAM1), phosphatidylinositol phosphokinase Iγ (PIPKIγ) and RIAM,
as well as F1-F3 contain actin-binding site (ABS1), R4-R8 (ABS2)
and R13-DD (ABS3) (19,28). Corresponding to F3, R8 can also
bind to deleted in liver cancer 1 (DLC1), RIAM, vinculin and
paxillin (19,29), being able to form a competitive
interaction network. Among these, DLC1 is the Rho GTPase activation
protein (RhoGAP), being able to inhibit the contraction of actin
and promotes the refolding of Talin-1 (30). The combination of DLC1 and TIAM1
has been reported to provide the capability to balance Rac and Rho
when they are involved in forming tension fibers (30). Subsequently, this combination
plays a counter-regulatory role in the process of FA maturation
driven by actin polymerization (30). Therefore, under the promotion of
multiple factors, Talin-1 functions as a 'spring' at the core of FA
transmitting tension between the ECM and actin (26,29). Based on its unique structure,
Talin-1 is able to sense and respond to mechanical signals from the
matrix and transmit mechanical forces to the surrounding cells
(19). The matrix, in turn, can
convert forces into intracellular biochemical signals, thereby
regulating nuclear transcription (26,31). Furthermore, mechanotransduction
is triggered due to Talin-1 unfolding above the mechanical
threshold (26). Integrins have
been reported to unbind and release forces before Talin-1
unfolding, provided that stiffness remains below the mechanical
threshold; in contrast, when the stiffness supersedes the
threshold, Talin-1 unfolds and binds to vinculin, leading to
adhesion growth and YAP nuclear translocation (26,31). Thus, it has been reported that
the combination of Talin-1 unfolding dynamics with a theoretical
clutch model could quantitatively predict cell response (26,31). Overall, the specific structure of
Talin-1 including the active and inactive forms provides the
possibility for proteins to convert mechanical signals into
chemical signals. In the inhibited state, the rod structure folds
into a closed spherical conformation with a diameter of 15 nm,
based on charge interaction (11). The inhibition of Talin-1 is
crucial to cell function and development, and its disruption has
been reported to greatly contribute to the migration of metastatic
cancer cells (32-34). The active form of Talin-1 has
been well-characterized; however, its inhibited state has not been
extensively studied.
Interactions between cells and the ECM are
fundamental features of multicellular organisms (35). All living cells are constantly
subjected to various mechanical signals from the surrounding cells
or from the ECM (36).
Mechanotransduction is a process that helps cells to adapt to
changes in the microenvironment by transforming the physical
signals into biological ones (36). In recent years, researchers have
demonstrated a growing interest in mechanical forces as key
regulators of cellular behavior. Mechanical forces can be directed
on the cell externally from the ECM or generated internally from
the active cytoskeleton (37). A
number of intracellular molecules defined as mechanosensors have
been discovered, which can react to mechanical stress, including
Talin-1 and vinculin. Conformational changes of mechanosensors in
response to mechanical stress can alter their binding partners,
thereby revealing new potential binding sites. This switch of
binding partners is the key to converting mechanical signals into
intracellular biochemical signals (38). FAs are the major connection
between ECM and cytoskeleton, with multiple components including
scaffolding molecules, GTPases, and many enzymes, including
kinases, phosphatases, proteases, and lipases (39-42). In FAs, integrins and
proteoglycans mediate adherent junctions from the actin
cytoskeleton to the actin cytoskeleton (43). The integrin family of type I
transmembrane adhesion receptors has been reported to mediate
cell-matrix attachment, as well as cell-cell attachment. Integrins
are involved in cell anchorage to the ECM and its binding to the
cytoskeleton and cytoplasmatic signaling, thus directly affecting
tissue architecture (44).
The signals transmitted by integrins are
bidirectional. Firstly, through 'inside-out' signaling,
intracellular signals may induce the binding of Talin-1 and Kindlin
to the cytoplasmic domains of integrin β subunits, thereby
activating integrin ligand binding function (45-48). Conversely, a second type of
signaling termed 'outside-in' signaling, is mediated by
interactions between integrins and their multiple ligands across
the membrane, enabling cells to sense and respond to the
extracellular environment (49,50), including cell spreading,
retraction, migration, proliferation and survival.
Talin-1 and vinculin are mechanosensitive cellular
proteins that can be folded and unfolded via mechanical forces
(51). This unfolding causes the
disruption of the tertiary structure of the protein, revealing the
cryptic sites for various ligand binding, molecule activation or
domain cleavage by proteases (52). In addition to its role as an
integrin activator, Talin-1 also functions as a mechanosensor
responsible for the transmission of tension between the actomyosin
machinery and the ECM, within the FA (11). To perform its role as a
mechanosensor, Talin-1 requires at least two anchorage points:
ECM-anchored integrins and actin (the cytoskeleton). Talin-1 head
domain binds to the integrin cytosolic tail with its tail binding
to the actin, thus connecting integrins with the cytoskeleton
(53). This bridging position
allows Talin-1 to be folded and unfolded either by mechanical
forces generated from the outside or internally by cytoskeletal
contraction. Talin-1 participates in both 'inside-out' and
'outside-in' signaling, and is capable of transmitting mechanical
forces imposed on cells externally, or generated internally
(45,53-56). Through its binding to the
cytoplasmic tail of integrin β subunits, Talin-1 controls integrin
activation, linking the integrin β subunit to the actin bundles rod
through by the C-terminal rod (14). The bridging position of the
Talin-1 molecule leads to its unfolding and stretching due to the
actomyosin contraction, and these conformation changes affect the
affinity and interaction with its binding partners (57). Talin-1 is required for the
initial weak connection between small clusters of integrins and the
cytoskeleton (58), and for the
reinforcement of integrin linkages to the cytoskeleton when cells
encounter mechanical forces, again suggesting the role of Talin-1
in FA formation (42,59). Thus, similar to the activation
and inactivation of integrins, the activation and inhibition of
Talin-1 has been reported to play a crucial role in the regulation
of FA dynamics and the transduction of mechanical signals.
Rap is a part of the Ras family of small GTPases
with five isoforms reported: Ras-associated protein 1A (Rap1A),
Rap1B, Rap2A, Rap2B and Rap2C, expressed in mammalian cells
(60). Rap2A and Rap2C are
considered as the central membrane-associated small GTPases, which
recruit proteins to the plasma membrane for the modulation of
various cellular responses (61-64), such as exocytosis (65), junction formation (66-68), cell adhesion (69,70), migration and invasion (71), cell proliferation (72), apoptosis (73) and polarity (74,75). Consequently, they are also
important for cardiovascular function (76), carcinogenesis (77,78) and liver physiopathology
regulation (79). The mechanisms
responsible for the recruitment of Talin-1 to the plasma membrane
remain to be clearly elucidated. As suggested in a previous study,
Talin-1 is recruited to the plasma membrane through the Rap1 GTPase
and its effector, RIAM (80).
However, even though RIAM binding to F3 of the Talin-1 head domain
helps the process of Talin-1 activation (81), the function of the
Rap1-RIAM-Talin-1 pathway appears to be leukocyte-specific
(82-84). Nevertheless, RIAM is still an
important factor for cell signaling, which also binds to the folded
R3 of the Talin-1 rod (13).
When exposed to forces, R3 structure is disrupted, leading to the
exposure of cryptic VBS and the disruption of RIAM binding
(21). This exchange of ligands
thus functions as a mechanochemical switch for Rap1 signaling, with
RIAM binding to Talin-1 in the absence of force and to vinculin in
the presence of force (19).
Recent studies have suggested that membrane recruitment can be
controlled via the direct binding of Rap1 to the ubiquitin-like F0
and F1 Talin-1 domains (60,85-88). Even though Talin-1 F0 and F1 both
bind to Rap1 very weakly in solution, Talin-1 F0 and F1 may bind
intracellularly to membrane-anchored Rap1 and lead to strong
binding and efficient recruitment of Talin-1 to the membrane
(60,85-88). The phosphorylation of
membrane-bound Rap1 and its activation promotes Talin-1 binding and
its recruitment to initiate FA formation (Fig. 1A). Mutations interfering with
either Rap1/Talin-1-F0 or Rap1/Talin-1-F1 interaction lead to
impaired FA assembling, decreased integrin activation in CHO cells
and malfunctioning leukocytes and platelets in mouse models
(89). In case both interactions
are disrupted, more severe defects in FA assembly, cell spreading
and adhesion may be observed (60,85-88).
The amount of PIP2 present in membrane phospholipids
is very low (0.1 to 5% of inner plasma membrane bilayer lipids)
(90). However, it has been
reported to play a crucial role in the regulation of various
cellular activities, including vesicle trafficking, actin
polymerization, and integrin signaling complex formation (91-94). PIP2 can recognize multiple
motifs, including epsin N-terminal homology (ENTH), phosphotyrosine
binding (PTB), pleckstrin homology (PH), and can assist the
recruitment of different proteins to the membrane (95). During the cell attachment to the
ECM via integrins, PIP2 functions as a scaffolding molecule,
recruiting various molecules to the activated integrin site and as
a signaling molecule, regulating target molecule activities
(96-100). It has been previously revealed
that PIPKIγ interacts with Talin-1 N-terminal head PTB domain and
is recruited to the FA sites (Fig.
1B) (95,101). When PIP2 binds to Talin-1, it
induces a change in the conformation of the Talin-1 head domain,
further leading to the unmasking of the integrin-binding site
(Fig. 1C) (89). By contrast, the F3 domain of
Talin-1 has been reported to directly bind to the integrin β
cytoplasmic tail through the same PTB domain. Even though PIPKIγ
may appear to impede Talin-1 F3 PTB domain binding to integrin, it
has been reported that PIPKIγ requires only small quantities of
Talin-1, being abundant in cells, to be recruited (89). As a kinase, the concentration of
PIPKIγ has been reported to be usually decreased (102). However, it is capable of
enzymatically producing PIP2, for the activation of the PIPKIγ-free
Talin-1 (89). Thus, the
interaction between PIPKIγ and Talin-1 promotes PIP2 production,
binding and activating in turn Talin-1, leading to the enhanced
Talin-1-integrin binding (Fig.
1C) (89). It has been
reported in a previous study that in migrating cells, integrins
reassembled FAs polarized towards the leading edge, and PIPKIγ,
together with Talin-1 and FAK, regulating endocytosed integrin
activation-induced FA assembly polarization (103).
In recent years, there is an increasing interest in
Kindlin, a FERM-domain-containing protein, that may play a crucial
role in integrin activation (104,105). The Kindlin family consists of
three members, Kindlin-1, Kindlin-2 and Kindlin-3. Kindlin-2 is
expressed in the majority of cell types, whereas Kindlin-1 and -3
are mainly expressed in hematopoietic and epithelial cells
(89). The loss of Kindlin-1 may
lead to the blistering and fragility of the skin in humans and
mice, while Kindlin-2 plays an essential role in embryonic
development. The lack of Kindlin-3 has been suggested to cause
various immune problems and bleeding disorders (89). The head domain of Kindlin
consists of four subdomains, ubiquitin-like F0, F2 subdomain with
an inserted PH domain and FERM domain comprised of three subdomains
F1, F2 and F3 (89). β-integrin
tails, Kindlin, and Talin-1 head form a ternary complex in
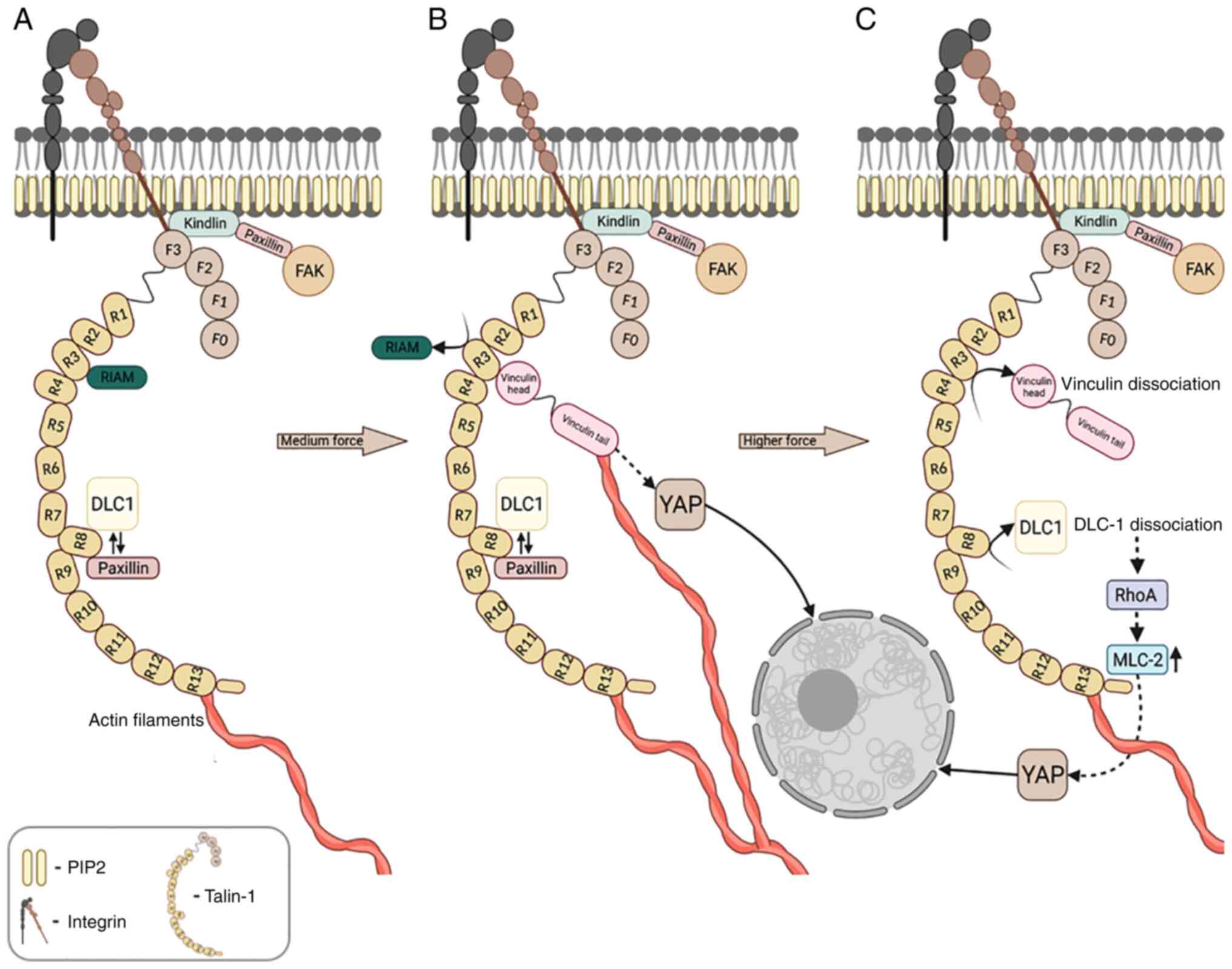
vitro (Fig. 2A) (89,106,107). The direct Kindlin-integrin
interaction is essential for maximal integrin activation (108). Kindlin is recruited to the cell
membrane by binding directly to the anionic membrane phospholipids
(PIP2) utilizing its positively charged residues in F0 domain and
the second NPXY motif in the β-integrin tails using F3 head
subdomain The inhibition of Kindlin binding leads to the disruption
of Talin-1-mediated integrin activation (106,107,109). Nevertheless, a recent study
revealed that Kindlin binding disruption still allows integrins to
be activated via Talin-1; however, the enhancing effect of Kindlin
is obstructed (110). Thus, the
cooperative interaction of Talin-1 and Kindlin leads to the stable
active conformation of integrin and promotes ligands binding and
clustering (89). The binding of
Kindlin with Paxillin, which, in turn, binds to FAK, giving rise to
an interesting 'Kindlin-Paxillin-FAK' pathway that can regulate FAK
activation, and can participate in dynamic cell adhesion and FA
assembly (111-113).
Paxillin is a 68-kDa cytoskeletal protein and its
main function is the recruitment of FAK to the FA site, further
promoting in turn the tyrosine phosphorylation of paxillin and
rendering Paxillin an important docking protein for the integrin
signal transduction (114).
Paxillin is recruited to the FA site mainly through its interaction
with the Kindlin and Talin-1 R8 domain (Fig. 2A) (89) and its head domain, and a recent
study suggested that it may bridge Kindlin and Talin-1 (115). There is evidence to suggest
that the interaction between paxillin and FAK markedly contributes
to cell motility and FA assembly and disassembly (116,117). FAK is 125-kDa non-receptor
tyrosine kinase that is expressed throughout the human body and
participates in signal transduction from the adhesions for the
modulation of diverse biological cellular functions, including cell
migration, survival and cancer cell invasion (118-120). FAK has been also reported to be
involved in binding various proteins and in their recruitment into
larger protein complexes (121), thus functioning as an adaptor
protein utilizing its multiple domains, including the FERM domain
(122). There are multiple
pathways associated with FAK activation, including bioactive
lipids, growth factors and mitogenic neuropeptides (118). Following its recruitment to the
FA site, FAK interacts with the PIP2-rich membrane to relieve the
autoinhibitory interaction between its FERM and kinase domains
(123). This results in the
exposure of the autophosphorylation site, allowing FAK to function
as a molecular scaffold and recruit SRC kinases to further
phosphorylate its main activation loop inside the kinase domain
(123). FAK may also act as a
scaffold for the nuclear transcriptional regulatory complexes
affecting the expression of target genes, including chemokine
encoding genes which are responsible for antitumor immunity and
microenvironment (124,125).
The Talin-1 rod has been reported to respond to
mechanical force, as well as adjusting the contractility of FAC
through the Rho-GTPase signaling pathway (126). This pathway is responsible for
the regulation of FA assembly and contractility, and can be
affected by the tension on Talin-1 (127). Originally, the members of the
Rho branch were considered only as actin cytoskeleton regulators.
However, a wide range of other functions including expression,
morphogenesis, motility and proliferation have been discovered
(128-130). In human cancers, the activity
of Rho-GTPase is often altered and may have an effect on tumor
invasiveness and growth (131,132). In an active state, it has been
suggested that Rho proteins may activate various effector
molecules, including phospholipases, lipid and protein kinases
(128-130). Additionally, Rho family
proteins may function as a molecular switch via a
nucleotide-controlled conformational change (133,134). There are two classes of
regulatory proteins that are controlling the GTP-GDP cycle of Rho:
Guanine nucleotide exchange factors (GEFs) stimulate the binding of
GTP to activate the protein, and GTPase activating proteins (GAPs),
which are responsible for inactivation and GTP hydrolysis (135). It has been suggested that, both
RhoGEFs and RhoGAPs are regulated through interactions with
different molecules. Some of the RhoGEFs can be activated as a
response to the stimulation of receptors on the cell surface, while
other GEFs form various complexes with scaffolding proteins and Rho
effectors, elevating specificity and efficiency of Rho signaling
(136,137). Although the RhoGAP signaling
pathways have not been studied in detail, it has been suggested
that they are linked to normal cell function alterations in diverse
human diseases (138). One of
the RhoGAP family members is DLC1, which has been of increased
interest as a potential tumor suppressor and cytoskeletal
organization and cell proliferation (138). It has been reported that DLC1
mRNA is expressed in the majority of tissues in adults; however, it
is completely absent or downregulated in various human cancers,
including gastric (140),
ovarian (141,142), lung (135,141,143,144), pancreatic (145), renal (141), prostate (146), colon (141), breast tumors (141,147,148) and hepatocellular carcinoma
(HCC) (149-151). DLC1 is a negative regulator of
Ras homolog family member A (RhoA) signaling, binding the
unstretched form of Talin-1 and decreasing the contractility of
myosin-driven machinery (Fig.
2A) (29,30,152), and released in response to
tension (29). Provided that
DLC1 is localized to the FA site through interaction with
Talin-1-R8, it may function as a RhoGTPase regulator and affect
cellular function (152). Since
the tension on large adhesions is often much weaker than tension on
smaller adhesions (54,153), FAs growth promoted by
contractility and tension on Talin-1 results in the redistribution
of force between increased numbers of FA components, consequently
decreasing the level of tension experienced by single Talin-1
molecule. The reduction of mechanical stress may facilitate the
refolding of the Talin-1 domain bound by DLC1, thus decreasing the
contractility by inhibiting RhoA (29). Subsequently, the second important
molecular switch is revealed, thus being able to convert mechanical
signals to the chemical with subsequent effect on downstream
signaling, including myosin light chain 2 (MLC-2), with its
activation being directly proportional to the quantities of active
RhoA (29).
Adherent cells are anchored to the ECM via FA
proteins, whereas cell-cell contacts connect via FA junction
proteins. A previous study revealed that myosin-driven cell
contractility or externally applied stresses greatly contribute to
the strengthening of these connections (154). The mechanisms through which the
proteins involved in these connections sense, transmit and respond
to mechanical signals have not yet been fully elucidated. Vinculin
is one of the potential candidate molecules for the role of key
cell-matrix and cell-cell adhesion protein, establishing a strong
physical connection for force transmission between ECM and
cytoskeleton (154).
Hippo effectors YAP and TAZ are proteins with a
molecular weight of 65 and 43 kDa, respectively, and have been
reported to act as transcription factors responsive to the changes
in ECM mechanics and composition (168,169). YAP contains a coiled-coil
domain, an Src homology domain-3 binding motif (SH3-BM), an WW
domain and a tea domain transcription factor (TEAD). In its
C-terminus, YAP has a PDZ-binding motif (PDZ-BM) and, in contrast
to TAZ, YAP contains a proline rich region in the N-terminus
(170). Since YAP lacks a
DNA-binding site, it has been reported to bind to the TEAD
DNA-binding transcription factors in order to take effect in
antiapoptotic, cell fate, and growth promoting genes (171). Moreover, YAP/TAZ plays a vital
role in cell shape, polarity and cytoskeletal organization
(171), YAP nuclear expression
is connected to Rho signaling (Fig.
2C) and the presence of vinculin spikes in the FA (169). FAs act as a bridge between
ECM-integrin connection and cytoskeleton and may be crucial for
cellular mechanosensing (172).
FA formation is controlled by cell spreading and RhoA GTPase
activity, in order to stabilize the anchorage of the actin
cytoskeleton to the cell membrane, requiring also YAP
co-transcriptional function (169). Therefore, YAP mechanosensing
activity may be the key determinant of cell mechanics in response
to ECM stimuli.
According to previous reports, the nuclear
localization of Yap is significantly increased in cells on rigid
substrates compared with cells on soft substrates (173). On the rigid ECM, YAP/TAZ
accumulation within the nucleus becomes transcriptionally active,
while on the soft ECM, the accumulation occurs within the cytosol
(173). An increased ECM
stiffness positively affects the amount of cytoskeleton-associated
vinculin and elevates the levels of nuclear-localized
transcriptional coactivator paralogs, YAP and TAZ, and their
activity. On the rigid ECM, vinculin participates in nuclear
translocation and the activity regulation of YAP/TAZ. The process
of YAP/TAZ nuclear translocation has been reported to be partly
regulated by vinculin, through the organization of the actin
cytoskeleton. Talin-1 molecule only unfolds when a certain
stiffness threshold is surpassed. Above this threshold, Talin-1
molecule unfolds and binds to various binding partners, including
vinculin, leading to YAP nuclear translocation and adhesion growth
(169). Even though the actin
organization and intracellular tension appears to take part in the
regulation of this process, the mechanisms through which ECM
stiffness regulates YAP/TAZ remain to be elucidated. The
explanation of the interaction between Talin-1 and Yap will further
broaden the understanding of the cellular mechanotransduction role
of Talin-1.
As the main integrin-activating and mechanosensing
protein, Talin-1 is important for FA assembly, as well as for
various cellular events. Although Talin-1 itself is a protein with
a vast interaction network, its interactions with vinculin, DLC1,
Rap1 and RIAM, paxillin, and FAK subsequently affect multiple
signaling pathways inside the cell. Through interactions with its
binding targets (Table I),
Talin-1 converts mechanical signals into chemical, consequently
affecting a plethora of cellular responses.
Recent discoveries in the field of mechanobiology
have a marked effect on other disciplines. It has previously been
revealed that even the alterations in the physical environment of
the cell, ECM in particular, can cause a malignant phenotype in the
cancer cells (174). The
presence of Talin-1 is insufficient for proper cell function. It
must be subjected to force, either through cell contraction or
through the application of shear stress (34). The function of Talin-1 in
cancerous phenotypes is not limited on the influence on adhesion
and motility. In contrast, it also depends on downstream signaling,
as emphasized by Talin-1 overexpression in nonadherent cells. Until
recently, the role of Talin-1 has been studied in oral squamous
cell (175), colon (176) and prostate carcinoma (177). Studies have revealed that
Talin-1 could be used as a biomarker for HCC infiltration and
metastasis (178,179). Moreover, the co-localization of
Talin-1 and Vinculin has been determined to be in the liver
infected by Ebola virus (180).
In the process of HBV-induced liver fibrosis, the increase in PIP2
levels increases the risk for liver fibrosis (181), and additionally, HBV X protein
(HBx) can mediate the decrease of Talin-1 protein monomer levels
(182). However, no evidence
has yet been reported in relation to the regulatory role of Talin-1
in liver pathophysiological processes, at least to the best of our
knowledge. Although Talin-1 has been found to promote tumor
formation, migration and metastasis in liver cancer and colon
cancer (33,183,184), the role of Talin-1 remains
controversial, particularly in HCC (185). Even though, to the best of our
knowledge, there is no evidence available to date of the direct
regulatory role of Talin-1in pathogenesis, understanding its
interaction network in cellular mechanotransduction is of utmost
significance for disease diagnosis and prevention.
Not applicable.
YZ and CT conceived the research idea. YZ and NL
wrote and reviewed the manuscript. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by a grant from National Natural
Science Foundation of China (no. 31701279).
|
1
|
Edelman GM: Cell adhesion molecules in the
regulation of animal form and tissue pattern. Ann Rev Cell Biol.
2:81–116. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mammoto T and Ingber DE: Mechanical
control of tissue and organ development. Development.
137:1407–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Orr AW, Helmke BP, Blackman BR and
Schwartz MA: Mechanisms of mechanotransduction. Dev Cell. 10:11–20.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schwartz MA: Integrins and extracellular
matrix in mechanotransduction. Cold Spring Harbor Perspectives
Biol. 2:a0050662010. View Article : Google Scholar
|
|
5
|
Horwitz A, Duggan K, Buck C, Beckerle MC
and Burridge K: Interaction of plasma membrane fibronectin receptor
with talin-a transmembrane linkage. Nature. 320:531–533. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burridge K and Mangeat P: An interaction
between vinculin and talin. Nature. 308:744–746. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Humphries JD, Wang P, Streuli C, Geiger B,
Humphries MJ and Ballestrem C: Vinculin controls focal adhesion
formation by direct interactions with talin and actin. J Cell Biol.
179:1043–1057. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Attia Gaballah MS: The combined role of
serum Talin-1 with traditional liver biomarkers in diagnosis of
hepatocellular carcinoma. Adv Microb Res. 3:0082019.
|
|
9
|
Schwartz MA and DeSimone DW: Cell adhesion
receptors in mechanotransduction. Curr Opin Cell Biol. 20:551–556.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rao RK and Wu Z: Chapter 7-Cell Adhesion
and the Extracellular Matrix. Goodman's Medical Cell Biology.
Goodman SR: 4th edition. Academic Press; pp. 203–247. 2021
|
|
11
|
Dedden D, Schumacher S, Kelley CF,
Zacharias M, Biertümpfel C, Fässler R and Mizuno N: The
architecture of Talin1 reveals an autoinhibition mechanism. Cell.
179:120–131.e13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang N: Mechanotransduction in liver
diseases. Seminars in liver disease. Thieme Medical Publishers; New
York, NY: pp. 84–90. 2020
|
|
13
|
Goult BT, Zacharchenko T, Bate N, Tsang R,
Hey F, Gingras AR, Elliott PR, Roberts GCK, Ballestrem C, Critchley
DR and Barsukov IL: RIAM and vinculin binding to talin are mutually
exclusive and regulate adhesion assembly and turnover. J Biol Chem.
288:8238–8249. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calderwood DA, Campbell ID and Critchley
DR: Talins and kindlins: Partners in integrin-mediated adhesion.
Nat Rev Mol Cell Biol. 14:503–517. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gingras AR, Bate N, Goult BT, Hazelwood L,
Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann
N, et al: The structure of the C-terminal actin-binding domain of
talin. EMBO J. 27:458–469. 2008. View Article : Google Scholar
|
|
16
|
Liu J, Wang Y, Goh WI, Goh H, Baird MA,
Ruehland S, Teo S, Bate N, Critchley DR, Davidson MW and
Kanchanawong P: Talin determines the nanoscale architecture of
focal adhesions. Proc Natl Acad Sci USA. 112:E4864–E4873. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Molony L, McCaslin D, Abernethy J, Paschal
B and Burridge K: Properties of talin from chicken gizzard smooth
muscle. J Biol Chem. 262:7790–7795. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Winkler J, Lünsdorf H and Jockusch BM:
Energy-filtered electron microscopy reveals that talin is a highly
flexible protein composed of a series of globular domains. Eur J
Biochem. 243:430–436. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goult BT, Yan J and Schwartz MA: Talin as
a mechanosensitive signaling hub. J Cell Biol. 217:3776–3784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haining AWM, von Essen M, Attwood SJ,
Hytönen VP and del Río Hernández A: All subdomains of the talin rod
are mechanically vulnerable and may contribute to cellular
mechanosensing. ACS Nano. 10:6648–6658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao M, Goult BT, Klapholz B, Hu X,
Toseland CP, Guo Y, Cong P, Sheetz MP and Yan J: The mechanical
response of talin. Nat Commun. 7:119662016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ye X, McLean MA and Sligar SG:
Phosphatidylinositol 4,5-bisphosphate modulates the affinity of
Talin-1 for phospholipid bilayers and activates its autoinhibited
form. Biochemistry. 55:5038–5048. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Atherton P, Lausecker F, Carisey A,
Gilmore A, Critchley D, Barsukov IL and Ballestrem C:
Force-independent interactions of talin and vinculin govern
integrin-mediated mechanotransduction. bioRxiv. https://doi.org/10.1101/629683.
|
|
24
|
Vigouroux C, Henriot V and Le Clainche C:
Talin dissociates from RIAM and associates to vinculin sequentially
in response to the actomyosin force. Nat Commun. 11:31162020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fillingham I, Gingras AR, Papagrigoriou E,
Patel B, Emsley J, Critchley DR, Roberts GC and Barsukov IL: A
vinculin binding domain from the talin rod unfolds to form a
complex with the vinculin head. Structure. 13:65–74. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elosegui-Artola A, Oria R, Chen Y,
Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X and
Roca-Cusachs P: Mechanical regulation of a molecular clutch defines
force transmission and transduction in response to matrix rigidity.
Nat Cell Biol. 18:540–548. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lagarrigue F, Gingras AR, Paul DS, Valadez
AJ, Cuevas MN, Sun H, Lopez-Ramirez MA, Goult BT, Shattil SJ,
Bergmeier W and Ginsberg MH: Rap1 binding to the talin 1 F0 domain
makes a minimal contribution to murine platelet GPIIb-IIIa
activation. Blood Adv. 2:2358–2368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Watanabe T, Matsuzawa K, Katsumi
A, Kakeno M, Matsui T, Ye F, Sato K, Murase K, Sugiyama I, et al:
Tiam1 interaction with the PAR complex promotes talin-mediated Rac1
activation during polarized cell migration. J Cell Biol.
199:331–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haining AWM, Rahikainen R, Cortes E,
Lachowski D, Rice A, von Essen M, Hytönen VP and Del Río Hernández
A: Mechanotransduction in talin through the interaction of the R8
domain with DLC1. PLoS Biol. 16:e20055992018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zacharchenko T, Qian X, Goult BT, Jethwa
D, Almeida TB, Ballestrem C, Critchley DR, Lowy DR and Barsukov IL:
LD motif recognition by talin: Structure of the talin-DLC1 complex.
Structure. 24:1130–1141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Elosegui-Artola A, Andreu I, Beedle AEM,
Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres
P, Le Roux AL, et al: Force triggers YAP nuclear entry by
regulating transport across nuclear pores. Cell. 171:1397–1410.e14.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Desiniotis A and Kyprianou N: Significance
of talin in cancer progression and metastasis. Int Rev Cell Mol
Biol. 289:117–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang KP, Dai W, Ren YH, Xu YC, Zhang SM
and Qian YB: Both Talin-1 and Talin-2 correlate with malignancy
potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC
Cancer. 16:452016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Haining AW, Lieberthal TJ and Hernández
AdR: Talin: A mechanosensitive molecule in health and disease.
FASEB J. 30:2073–2085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Humph rey JD, Dufresne ER and Schwar tz
MA: Mechanotransduction and extracellular matrix homeostasis. Nat
Rev Mol Cell Biol. 15:802–812. 2014. View Article : Google Scholar
|
|
36
|
Martino F, Perestrelo AR, Vinarský V,
Pagliari S and Forte G: Cellular mechanotransduction: From tension
to function. Front Physiol. 9:8242018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maruthamuthu V, Aratyn-Schaus Y and Gardel
ML: Conserved F-actin dynamics and force transmission at cell
adhesions. Curr Opin Cell Biol. 22:583–588. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Yan J and Goult BT:
Force-dependent binding constants. Biochemistry. 58:4696–4709.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Geiger B, Bershadsky A, Pankov R and
Yamada KM: Transmembrane crosstalk between the extracellular
matrix-cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2:793–805.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geiger B and Bershadsky A: Exploring the
neighborhood: Adhesion-coupled cell mechanosensors. Cell.
110:139–142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
DeMali KA, Wennerberg K and Burridge K:
Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol.
15:572–582. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wozniak MA, Modzelewska K, Kwong L and
Keely PJ: Focal adhesion regulation of cell behavior. Biochim
Biophys Acta. 1692:103–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim SH, Turnbull J and Guimond S:
Extracellular matrix and cell signalling: The dynamic cooperation
of integrin, proteoglycan and growth factor receptor. J Endocrinol.
209:139–151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yue B: Biology of the extracellular
matrix: An overview. J Glaucoma. 23(8 Suppl 1): S20–S23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tadokoro S, Shattil SJ, Eto K, Tai V,
Liddington RC, de Pereda JM, Ginsberg MH and Calderwood DA: Talin
binding to integrin beta tails: A final common step in integrin
activation. Science. 302:103–106. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Moser M, Nieswandt B, Ussar S, Pozgajova M
and Fässler R: Kindlin-3 is essential for integrin activation and
platelet aggregation. Nat Med. 14:325–330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma YQ, Qin J, Wu C and Plow EF: Kindlin-2
(Mig-2): A co-activator of beta3 integrins. J Cell Biol.
181:439–446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim C, Ye F and Ginsberg MH: Regulation of
integrin activation. Ann Rev Cell Dev Biol. 27:321–345. 2011.
View Article : Google Scholar
|
|
49
|
Ginsberg MH, Partridge A and Shattil SJ:
Integrin regulation. Curr Opin Cell Biol. 17:509–516. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Legate KR, Wickström SA and Fässler R:
Genetic and cell biological analysis of integrin outside-in
signaling. Genes Dev. 23:397–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao M, Goult BT, Chen H, Cong P, Sheetz MP
and Yan J: Mechanical activation of vinculin binding to talin locks
talin in an unfolded conformation. Sci Rep. 4:46102014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kerstein PC: Mechanochemical regulation of
growth cone motility and axon guidance. The University of
Wisconsin-Madison; 2015
|
|
53
|
Calderwood DA, Zent R, Grant R, Rees DJG,
Hynes RO and Ginsberg MH: The Talin head domain binds to integrin
beta subunit cytoplasmic tails and regulates integrin activation. J
Biol Chem. 274:28071–28074. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kumar A, Ouyang M, Van den Dries K, McGhee
EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI and
Schwartz MA: Talin tension sensor reveals novel features of focal
adhesion force transmission and mechanosensitivity. J Cell Biol.
213:371–383. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Calderwood DA, Yan B, de Pereda JM,
Alvarez BG, Fujioka Y, Liddington RC and Ginsberg MH: The
phosphotyrosine binding-like domain of talin activates integrins. J
Biol Chem. 277:21749–21758. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Austen K, Ringer P, Mehlich A,
Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M
and Grashoff C: Extracellular rigidity sensing by talin
isoform-specific mechanical linkages. Nat Cell Biol. 17:1597–1606.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Karthik L, Kumar G, Keswani T,
Bhattacharyya A, Chandar SS and Bhaskara Rao K: Protease inhibitors
from marine actinobacteria as a potential source for antimalarial
compound. PLoS One. 9:e909722014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang G, Giannone G, Critchley DR,
Fukumoto E and Sheetz MP: Two-piconewton slip bond between
fibronectin and the cytoskeleton depends on talin. Nature.
424:334–337. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Giannone G, Jiang G, Sutton DH, Critchley
DR and Sheetz MP: Talin1 is critical for force-dependent
reinforcement of initial integrin-cytoskeleton bonds but not
tyrosine kinase activation. J Cell Biol. 163:409–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lagarrigue F, Paul DS, Gingras AR, Valadez
AJ, Sun H, Lin J, Cuevas MN, Ablack JN, Lopez-Ramirez MA, Bergmeier
W and Ginsberg MH: Talin-1 is the principal platelet Rap1 effector
of integrin activation. Blood. 136:1180–1190. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gloerich M and Bos JL: Regulating Rap
small G-proteins in time and space. Trends Cell Biol. 21:615–623.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chrzanowska-Wodnicka M: Rap1 in
endothelial biology. Curr Opin Hematol. 24:2482017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
White GC, Crawford N and Fischer TH:
Cytoskeletal interactions of Raplb in platelets. Adv Exp Med Biol.
344:187–194. 1993. View Article : Google Scholar
|
|
64
|
Hancock JF: Ras proteins: Different
signals from different locations. Nat Rev Mol Cell Biol. 4:373–385.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Branham MT, Bustos MA, De Blas GA, Rehmann
H, Zarelli VE, Treviño CL, Darszon A, Mayorga LS and Tomes CN: Epac
activates the small G proteins Rap1 and Rab3A to achieve
exocytosis. J Biol Chem. 284:24825–24839. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kooistra MR, Dubé N and Bos JL: Rap1: A
key regulator in cell-cell junction formation. J Cell Sci.
120:17–22. 2007. View Article : Google Scholar
|
|
67
|
Pannekoek WJ, Kooistra MR, Zwartkruis FJ
and Bos JL: Cell-cell junction formation: The role of Rap1 and Rap1
guanine nucleotide exchange factors. Biochim Biophys Acta.
1788:790–796. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hogan C, Serpente N, Cogram P, Hosking CR,
Bialucha CU, Feller SM, Braga VM, Birchmeier W and Fujita Y: Rap1
regulates the formation of E-cadherin-based cell-cell contacts. Mol
Cell Biol. 24:6690–6700. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ohba Y, Ikuta K, Ogura A, Matsuda J,
Mochizuki N, Nagashima K, Kurokawa K, Mayer BJ, Maki K, Miyazaki J
and Matsuda M: Requirement for C3G-dependent Rap1 activation for
cell adhesion and embryogenesis. EMBO J. 20:3333–3341. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Katagiri K, Maeda A, Shimonaka M and
Kinashi T: RAPL, a Rap1-binding molecule that mediates Rap1-induced
adhesion through spatial regulation of LFA-1. Nat Immunol.
4:741–748. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
71
|
Priego N, Arechederra M, Sequera C,
Bragado P, Vázquez-Carballo A, Gutiérrez-Uzquiza Á, Martín-Granado
V, Ventura JJ, Kazanietz MG, Guerrero C and Porras A: C3G
knock-down enhances migration and invasion by increasing
Rap1-mediated p38α activation, while it impairs tumor growth
through p38α-independent mechanisms. Oncotarget. 7:450602016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Altschuler DL and Ribeiro-Neto F:
Mitogenic and oncogenic properties of the small G protein Rap1b.
Proc Natl Acad Sci USA. 95:7475–7479. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Maia V, Sanz M, Gutierrez-Berzal J, de
Luis A, Gutierrez-Uzquiza A, Porras A and Guerrero C: C3G silencing
enhances STI-571-induced apoptosis in CML cells through p38 MAPK
activation, but it antagonizes STI-571 inhibitory effect on
survival. Cell Signal. 21:1229–1235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shimonaka M, Katagiri K, Nakayama T,
Fujita N, Tsuruo T, Yoshie O and Kinashi T: Rap1 translates
chemokine signals to integrin activation, cell polarization, and
motility across vascular endothelium under flow. J Cell Biol.
161:417–427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schwamborn JC and Püschel AW: The
sequential activity of the GTPases Rap1B and Cdc42 determines
neuronal polarity. Nat Neurosci. 7:923–929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jeyaraj SC, Unger NT and Chotani MA: Rap1
GTPases: An emerging role in the cardiovasculature. Life Sci.
88:645–652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen CH, Chuang HC, Huang CC, Fang FM,
Huang HY, Tsai HT, Su LJ, Shiu LY, Leu S and Chien CY:
Overexpression of Rap-1A indicates a poor prognosis for oral cavity
squamous cell carcinoma and promotes tumor cell invasion via
Aurora-A modulation. Am J Pathol. 182:516–528. 2013. View Article : Google Scholar
|
|
78
|
Minato N: Rap G protein signal in normal
and disordered lymphohematopoiesis. Exp Cell Res. 319:2323–2328.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kumari S, Arora M, Singh J, Kadian LK,
Yadav R, Chauhan SS and Chopra A: Molecular associations and
clinical significance of RAPs in hepatocellular carcinoma. Front
Mol Biosci. 8:6779792021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lee HS, Lim CJ, Puzon-McLaughlin W,
Shattil SJ and Ginsberg MH: RIAM activates integrins by linking
talin to ras GTPase membrane-targeting sequences. J Biol Chem.
284:5119–5127. 2009. View Article : Google Scholar :
|
|
81
|
Yang J, Zhu L, Zhang H, Hirbawi J, Fukuda
K, Dwivedi P, Liu J, Byzova T, Plow EF, Wu J and Qin J:
Conformational activation of talin by RIAM triggers
integrin-mediated cell adhesion. Nat Commun. 5:58802014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Stritt S, Wolf K, Lorenz V, Vögtle T,
Gupta S, Bösl MR and Nieswandt B: Rap1-GTP-interacting adaptor
molecule (RIAM) is dispensable for platelet integrin activation and
function in mice. Blood. 125:219–222. 2015. View Article : Google Scholar :
|
|
83
|
Klapproth S, Sperandio M, Pinheiro EM,
Prünster M, Soehnlein O, Gertler FB, Fässler R and Moser M: Loss of
the Rap1 effector RIAM results in leukocyte adhesion deficiency due
to impaired β2 integrin function in mice. Blood. 126:2704–2712.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Su W, Wynne J, Pinheiro EM, Strazza M, Mor
A, Montenont E, Berger J, Paul DS, Bergmeier W, Gertler FB and
Philips MR: Rap1 and its effector RIAM are required for lymphocyte
trafficking. Blood. 126:2695–2703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhu L, Yang J, Bromberger T, Holly A, Lu
F, Liu H, Sun K, Klapproth S, Hirbawi J, Byzova TV, et al:
Structure of Rap1b bound to talin reveals a pathway for triggering
integrin activation. Nat Commun. 8:17442017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Bromberger T, Klapproth S, Rohwedder I,
Zhu L, Mittmann L, Reichel CA, Sperandio M, Qin J and Moser M:
Direct Rap1/Talin1 interaction regulates platelet and neutrophil
integrin activity in mice. Blood. 132:2754–2762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gingras AR, Lagarrigue F, Cuevas MN,
Valadez AJ, Zorovich M, McLaughlin W, Lopez-Ramirez MA, Seban N,
Ley K, Kiosses WB and Ginsberg MH: Rap1 binding and a
lipid-dependent helix in talin F1 domain promote integrin
activation in tandem. J Cell Biol. 218:1799–1809. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Bromberger T, Zhu L, Klapproth S, Qin J
and Moser M: Rap1 and membrane lipids cooperatively recruit talin
to trigger integrin activation. J Cell Sci. 132:jcs2355312019.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhu L, Plow EF and Qin J: Initiation of
focal adhesion assembly by talin and kindlin: A dynamic view.
Protein Sci. 30:531–542. 2021. View Article : Google Scholar :
|
|
90
|
Raucher D, Stauffer T, Chen W, Shen K, Guo
S, York JD, Sheetz MP and Meyer T: Phosphatidylinositol 4,
5-bisphosphate functions as a second messenger that regulates
cytoskeleton-plasma membrane adhesion. Cell. 100:221–228. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Toker A: The synthesis and cellular roles
of phosphatidylinositol 4, 5-bisphosphate. Curr Opin Cell Biol.
10:254–261. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Czech MP: PIP2 and PIP3: Complex roles at
the cell surface. Cell. 100:603–606. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yin HL and Janmey PA: Phosphoinositide
regulation of the actin cytoskeleton. Ann Rev Physiol. 65:761–789.
2003. View Article : Google Scholar
|
|
94
|
Nayal A, Webb DJ and Horwitz AF: Talin: An
emerging focal point of adhesion dynamics. Curr Opin Cell Biol.
16:94–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kong X, Wang X, Misra S and Qin J:
Structural basis for the phosphorylation-regulated focal adhesion
targeting of type Igamma phosphatidylinositol phosphate kinase
(PIPKIgamma) by talin. J Mol Biol. 359:47–54. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
McNamee HP, Ingber DE and Schwartz MA:
Adhesion to fibronectin stimulates inositol lipid synthesis and
enhances PDGF-induced inositol lipid breakdown. J Cell Biol.
121:673–678. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chong LD, Traynor-Kaplan A, Bokoch GM and
Schwartz MA: The small GTP-binding protein Rho regulates a
phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell.
79:507–513. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Gilmore AP and Burridge K: Regulation of
vinculin binding to talin and actin by
phosphatidyl-inositol-4-5-bisphosphate. Nature. 381:531–535. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Han J, Luby-Phelps K, Das B, Shu X, Xia Y,
Mosteller RD, Krishna UM, Falck JR, White MA and Broek D: Role of
substrates and products of PI 3-kinase in regulating activation of
Rac-related guanosine triphosphatases by Vav. Science. 279:558–560.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Martel V, Racaud-Sultan C, Dupe S, Marie
C, Paulhe F, Galmiche A, Block MR and Albiges-Rizo C: Conformation,
localization, and integrin binding of talin depend on its
interaction with phosphoinositides. J Biol Chem. 276:21217–21227.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
de Pereda JM, Wegener KL, Santelli E, Bate
N, Ginsberg MH, Critchley DR, Campbell ID and Liddington RC:
Structural basis for phosphatidylinositol phosphate kinase type
Igamma binding to talin at focal adhesions. J Biol Chem.
280:8381–8386. 2005. View Article : Google Scholar
|
|
102
|
Wu Z, Li X, Sunkara M, Spearman H, Morris
AJ and Huang C: PIPKIγ regulates focal adhesion dynamics and colon
cancer cell invasion. PLoS One. 6:e247752011. View Article : Google Scholar
|
|
103
|
Nader GPF, Ezratty EJ and Gundersen GG:
FAK, talin and PIPKIγ regulate endocytosed integrin activation to
polarize focal adhesion assembly. Nat Cell Biol. 18:491–503. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Karaköse E, Schiller HB and Fässler R: The
kindlins at a glance. J Cell Sci. 123:2353–2356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Plow EF, Qin J and Byzova T: Kindling the
flame of integrin activation and function with kindlins. Curr Opin
Hematol. 16:3232009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bledzka K, Liu J, Xu Z, Perera HD, Yadav
SP, Bialkowska K, Qin J, Ma YQ and Plow EF: Spatial coordination of
kindlin-2 with talin head domain in interaction with integrin β
cytoplasmic tails. J Biol Chem. 287:24585–24594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yates LA, Füzéry AK, Bonet R, Campbell ID
and Gilbert RJ: Biophysical analysis of Kindlin-3 reveals an
elongated conformation and maps integrin binding to the
membrane-distal β-subunit NPXY motif. J Biol Chem. 287:37715–37731.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kahner BN, Kato H, Banno A, Ginsberg MH,
Shattil SJ and Ye F: Kindlins, integrin activation and the
regulation of talin recruitment to αIIbβ3. PLoS One. 7:e340562012.
View Article : Google Scholar
|
|
109
|
Perera HD, Ma YQ, Yang J, Hirbawi J, Plow
EF and Qin J: Membrane binding of the N-terminal ubiquitin-like
domain of kindlin-2 is crucial for its regulation of integrin
activation. Structure. 19:1664–1671. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ye F, Hu G, Taylor D, Ratnikov B, Bobkov
AA, McLean MA, Sligar SG, Taylor KA and Ginsberg MH: Recreation of
the terminal events in physiological integrin activation. J Cell
Biol. 188:157–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Theodosiou M, Widmaier M, Böttcher RT,
Rognoni E, Veelders M, Bharadwaj M, Lambacher A, Austen K, Müller
DJ, Zent R and Fässler R: Kindlin-2 cooperates with talin to
activate integrins and induces cell spreading by directly binding
paxillin. Elife. 5:e101302016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhu L, Liu H, Lu F, Yang J, Byzova TV and
Qin J: Structural basis of paxillin recruitment by kindlin-2 in
regulating cell adhesion. Structure. 27:1686–1697.e5. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Böttcher RT, Veelders M, Rombaut P, Faix
J, Theodosiou M, Stradal TE, Rottner K, Zent R, Herzog F and
Fässler R: Kindlin-2 recruits paxillin and Arp2/3 to promote
membrane protrusions during initial cell spreading. J Cell Biol.
216:3785–3798. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Fujita H, Kamiguchi K, Cho D, Shibanuma M,
Morimoto C and Tachibana K: Interaction of Hic-5, a
senescence-related protein, with focal adhesion kinase. J Biol
Chem. 273:26516–26521. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gao J, Huang M, Lai J, Mao K, Sun P, Cao
Z, Hu Y, Zhang Y, Schulte ML, Jin C, et al: Kindlin supports
platelet integrin αIIbβ3 activation by interacting with paxillin. J
Cell Sci. 130:3764–3775. 2017.PubMed/NCBI
|
|
116
|
Hanks SK, Ryzhova L, Shin NY and Brábek J:
Focal adhesion kinase signaling activities and their implications
in the control of cell survival and motility. Front Biosci.
8:d982–d996. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
117
|
Webb DJ, Donais K, Whitmore LA, Thomas SM,
Turner CE, Parsons JT and Horwitz AF: FAK-Src signalling through
paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell
Biol. 6:154–161. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Lipinski CA and Loftus JC: Targeting Pyk2
for therapeutic intervention. Expert Opin Ther Targets. 14:95–108.
2010. View Article : Google Scholar :
|
|
119
|
Lee BY, Timpson P, Horvath LG and Daly RJ:
FAK signaling in human cancer as a target for therapeutics.
Pharmacol Ther. 146:132–149. 2015. View Article : Google Scholar
|
|
120
|
Sulzmaier FJ, Jean C and Schlaepfer DD:
FAK in cancer: Mechanistic findings and clinical applications. Nat
Rev Cancer. 14:598–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Dawson JC, Serrels A, Stupack DG,
Schlaepfer DD and Frame MC: Targeting FAK in anticancer combination
therapies. Nat Rev Cancer. 21:313–324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Frame MC, Patel H, Serrels B, Lietha D and
Eck MJ: The FERM domain: Organizing the structure and function of
FAK. Nat Rev Mol Cell Biol. 11:802–814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Acebró I, Righetto RD, Schoenherr C, de
Buhr S, Redondo P, Culley J, Rodríguez CF, Daday C, Biyani N,
Llorca O, et al: Structural basis of Focal Adhesion Kinase
activation on lipid membranes. EMBO J. 39:e1047432020.
|
|
124
|
Serrels A, Lund T, Serrels B, Byron A,
McPherson RC, von Kriegsheim A, Gómez-Cuadrado L, Canel M, Muir M,
Ring JE, et al: Nuclear FAK controls chemokine transcription,
Tregs, and evasion of anti-tumor immunity. Cell. 163:160–173. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Canel M, Taggart D, Sims AH, Lonergan DW,
Waizenegger IC and Serrels A: T-cell co-stimulation in combination
with targeting FAK drives enhanced anti-tumor immunity. Elife.
9:e480922020. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Osiak AE, Zenner G and Linder S:
Subconfluent endothelial cells form podosomes downstream of
cytokine and RhoGTPase signaling. Exp Cell Res. 307:342–353. 2005.
View Article : Google Scholar
|
|
127
|
Chen CS, Alonso JL, Ostuni E, Whitesides
GM and Ingber DE: Cell shape provides global control of focal
adhesion assembly. Biochem Biophys Res Commun. 307:355–361. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Van Aelst L and D'Souza-Schorey C: Rho
GTPases and signaling networks. Genes Dev. 11:2295–2322. 1997.
View Article : Google Scholar
|
|
129
|
Ridley AJ: Rho family proteins:
Coordinating cell responses. Trends Cell Biol. 11:471–477. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Jaffe AB and Hall A: Rho GTPases:
Biochemistry and biology. Annu Rev Cell Dev Biol. 21:247–269. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar
|
|
132
|
Del Pulgar TG, Benitah SA, Valerón PF,
Espina C and Lacal JC: Rho GTPase expression in tumourigenesis:
Evidence for a significant link. Bioessays. 27:602–613. 2005.
View Article : Google Scholar
|
|
133
|
Hodge RG and Ridley AJ: Regulating Rho
GTPases and their regulators. Nat Rev Mol Cell Biol. 17:496–510.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Berken A, Thomas C and Wittinghofer A: A
new family of RhoGEFs activates the Rop molecular switch in plants.
Nature. 436:1176–1180. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Bos JL, Rehmann H and Wittinghofer A: GEFs
and GAPs: Critical elements in the control of small G proteins.
Cell. 129:865–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Marinissen MJ and Gutkind JS: Scaffold
proteins dictate Rho GTPase-signaling specificity. Trends
Biochemical Sci. 30:423–426. 2005. View Article : Google Scholar
|
|
137
|
García-Mata R and Burridge K: Catching a
GEF by its tail. Trends Cell Biol. 17:36–43. 2007. View Article : Google Scholar
|
|
138
|
Tcherkezian J and Lamarche-Vane N: Current
knowledge of the large RhoGAP family of proteins. Biol Cell.
99:67–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Durkin ME, Yuan BZ, Zhou X, Zimonjic DB,
Lowy DR, Thorgeirsson SS and Popescu NC: DLC-1: A Rho
GTPase-activating protein and tumour suppressor. J Cell Mol Med.
11:1185–1207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Kim TY, Jong HS, Song SH, Dimtchev A,
Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M and Bang YJ:
Transcriptional silencing of the DLC-1 tumor suppressor gene by
epigenetic mechanism in gastric cancer cells. Oncogene.
22:3943–3951. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Ullmannova V and Popescu NC: Expression
profile of the tumor suppressor genes DLC-1 and DLC-2 in solid
tumors. Int J Oncol. 29:1127–1132. 2006.PubMed/NCBI
|
|
142
|
Zhang X, Feng J, Cheng Y, Yao Y, Ye X, Fu
T and Cheng H: Characterization of differentially expressed genes
in ovarian cancer by cDNA microarrays. Int J Gynecol Cancer.
15:50–57. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Yuan BZ, Jefferson AM, Baldwin KT,
Thorgeirsson SS, Popescu NC and Reynolds SH: DLC-1 operates as a
tumor suppressor gene in human non-small cell lung carcinomas.
Oncogene. 23:1405–1411. 2004. View Article : Google Scholar
|
|
144
|
Healy KD, Kim TY, Shutes AT, Bang YJ,
Juliano RL and Der CJ: RhoGAP DLC-1 tumor suppression and aberrant
Rho GTPase activation in lung cancer. Proc Am Assoc Cancer Res.
47:9702006.
|
|
145
|
Nakamura T, Furukawa Y, Nakagawa H,
Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N,
Miyamoto M, et al: Genome-wide cDNA microarray analysis of gene
expression profiles in pancreatic cancers using populations of
tumor cells and normal ductal epithelial cells selected for purity
by laser microdissection. Oncogene. 23:2385–2400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Guan M, Zhou X, Soulitzis N, Spandidos DA
and Popescu NC: Aberrant methylation and deacetylation of deleted
in liver cancer-1 gene in prostate cancer: Potential clinical
applications. Clin Cancer Res. 12:1412–1419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Plaumann M, Seitz S, Frege R,
Estevez-Schwarz L and Scherneck S: Analysis of DLC-1 expression in
human breast cancer. J Cancer Res Clin Oncol. 129:349–354. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Gatalica Z, Velagaleti G, Kuivaniemi H,
Tromp G, Palazzo J, Graves KM, Guigneaux M, Wood T, Sinha M and
Luxon B: Gene expression profile of an adenomyoepithelioma of the
breast with a reciprocal translocation involving chromosomes 8-16.
Cancer Genet Cytogenet. 156:14–22. 2005. View Article : Google Scholar
|
|
149
|
Ng IO, Liang ZD, Cao L and Lee TK: DLC-1
is deleted in primary hepatocellular carcinoma and exerts
inhibitory effects on the proliferation of hepatoma cell lines with
deleted DLC-1. Cancer Res. 60:6581–6584. 2000.PubMed/NCBI
|
|
150
|
Wong CM, Lee JMF, Ching YP, Jin DY and Ng
IO: Genetic and epigenetic alterations of DLC-1 gene in
hepatocellular carcinoma. Cancer Res. 63:7646–7651. 2003.PubMed/NCBI
|
|
151
|
Yuan BZ, Miller MJ, Keck CL, Zimonjic DB,
Thorgeirsson SS and Popescu NC: Cloning, characterization, and
chromosomal localization of a gene frequently deleted in human
liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res.
58:2196–2199. 1998.PubMed/NCBI
|
|
152
|
Li G, Du X, Vass WC, Papageorge AG, Lowy
DR and Qian X: Full activity of the deleted in liver cancer 1
(DLC1) tumor suppressor depends on an LD-like motif that binds
talin and focal adhesion kinase (FAK). Proc Natl Acad Sci USA.
108:17129–17134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Beningo KA, Dembo M, Kaverina I, Small JV
and Wang YL: Nascent focal adhesions are responsible for the
generation of strong propulsive forces in migrating fibroblasts. J
Cell Biol. 153:881–888. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Goldmann WH: Role of vinculin in cellular
mechanotransduction. Cell Biol Int. 40:241–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wang DY, Melero C, Albaraky A, Atherton P,
Jansen KA, Dimitracopoulos A, Dajas-Bailador F, Reid A, Franze K
and Ballestrem C: Vinculin is required for neuronal mechanosensing
but not for axon outgrowth. Exp Cell Res. 407:1128052021.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Margadant F, Chew LL, Hu X, Yu H, Bate N,
Zhang X and Sheetz M: Mechanotransduction in vivo by repeated talin
stretch-relaxation events depends upon vinculin. PLoS Biol.
9:e10012232011. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Bakolitsa C, Cohen DM, Bankston LA, Bobkov
AA, Cadwell GW, Jennings L, Critchley DR, Craig SW and Liddington
RC: Structural basis for vinculin activation at sites of cell
adhesion. Nature. 430:583–586. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Borgon RA, Vonrhein C, Bricogne G, Bois PR
and Izard T: Crystal structure of human vinculin. Structure.
12:1189–1197. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
DeMali KA, Barlow CA and Burridge K:
Recruitment of the Arp2/3 complex to vinculin: Coupling membrane
protrusion to matrix adhesion. J Cell Biol. 159:881–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Brindle NP, Holt MR, Davies JE, Price CJ
and Critchley DR: The focal-adhesion vasodilator-stimulated
phosphoprotein (VASP) binds to the proline-rich domain in vinculin.
Biochemical J. 318:753–757. 1996. View Article : Google Scholar
|
|
161
|
Del Rio A, Perez-Jimenez R, Liu R,
Roca-Cusachs P, Fernandez JM and Sheetz MP: Stretching single talin
rod molecules activates vinculin binding. Science. 323:638–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Goldmann W, Niggli V, Kaufmann S and
Isenberg G: Probing actin and liposome interaction of talin and
talin-vinculin complexes: A kinetic, thermodynamic and lipid
labeling study. Biochemistry. 31:7665–7671. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ling K, Schill NJ, Wagoner MP, Sun Y and
Anderson RA: Movin'on up: The role of PtdIns(4,5)P(2) in cell
migration. Trends Cell Biol. 16:276–284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Kelley CF, Litschel T, Schumacher S,
Dedden D, Schwille P and Mizuno N: Phosphoinositides regulate
force-independent interactions between talin, vinculin, and actin.
Elife. 9:e561102020. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Wen KK, Rubenstein PA and DeMali KA:
Vinculin nucleates actin polymerization and modifies actin filament
structure. J Biol Chem. 284:30463–30473. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Atherton P, Lausecker F, Carisey A,
Gilmore A, Critchley D, Barsukov I and Ballestrem C: Relief of
talin autoinhibition triggers a force-independent association with
vinculin. J Cell Biol. 219. pp. e2019031342020, View Article : Google Scholar
|
|
167
|
Bays JL and DeMali KA: Vinculin in
cell-cell and cell-matrix adhesions. Cell Mol Life Sci.
74:2999–3009. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Chen L, Chan SW, Zhang X, Walsh M, Lim CJ,
Hong W and Song H: Structural basis of YAP recognition by TEAD4 in
the hippo pathway. Genes Dev. 24:290–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Nardone G, Oliver-De La Cruz J, Vrbsky J,
Martini C, Pribyl J, Skládal P, Pešl M, Caluori G, Pagliari S,
Martino F, et al: YAP regulates cell mechanics by controlling focal
adhesion assembly. Nat Commun. 8:153212017. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Chen YA, Lu CY, Cheng TY, Pan SH, Chen HF
and Chang NS: WW domain-containing proteins YAP and TAZ in the
hippo pathway as key regulators in stemness maintenance, tissue
homeostasis, and tumorigenesis. Front Oncol. 9:602019. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Dupont S: Role of YAP/TAZ in cell-matrix
adhesion-mediated signalling and mechanotransduction. Exp Cell Res.
343:42–53. 2016. View Article : Google Scholar
|
|
172
|
Seetharaman S and Etienne-Manneville S:
Integrin diversity brings specificity in mechanotransduction. Biol
Cell. 110:49–64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Kurotsu S, Sadahiro T, Fujita R, Tani H,
Yamakawa H, Tamura F, Isomi M, Kojima H, Yamada Y, Abe Y, et al:
Soft matrix promotes cardiac reprogramming via inhibition of
YAP/TAZ and suppression of fibroblast signatures. Stem Cell Rep.
15:612–628. 2020. View Article : Google Scholar
|
|
174
|
Paszek MJ, Zahir N, Johnson KR, Lakins JN,
Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M,
Boettiger D, et al: Tensional homeostasis and the malignant
phenotype. Cancer Cell. 8:241–254. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Lai MT, Hua CH, Tsai MH, Wan L, Lin YJ,
Chen CM, Chiu IW, Chan C, Tsai FJ and Jinn-Chyuan Sheu J: Talin-1
overexpression defines high risk for aggressive oral squamous cell
carcinoma and promotes cancer metastasis. J Pathol. 224:367–376.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Bostanci O, Kemik O, Kemik A, Battal M,
Demir U, Purisa S and Mihmanli M: A novel screening test for colon
cancer: Talin-1. Eur Rev Med Pharmacol Sci. 18:2533–2537.
2014.PubMed/NCBI
|
|
177
|
Sakamoto S, McCann RO, Dhir R and
Kyprianou N: Talin1 promotes tumor invasion and metastasis via
focal adhesion signaling and anoikis resistance. Cancer Res.
70:1885–1895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Zhang JL, Qian YB, Zhu LX and Xiong QR:
Talin1, a valuable marker for diagnosis and prognostic assessment
of human hepatocelluar carcinomas. Asian Pac J Cancer Prev.
12:3265–3269. 2011.PubMed/NCBI
|
|
179
|
Fang KP, Zhang JL, Ren YH and Qian YB:
Talin-1 correlates with reduced invasion and migration in human
hepatocellular carcinoma cells. Asian Pac J Cancer Prev.
15:2655–2661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Liu Y, Xiao J, Zhang B, Shelite TR, Su Z,
Chang Q, Judy B, Li X, Drelich A, Bei J, et al: Increased
talin-vinculin spatial proximities in livers in response to spotted
fever group rickettsial and Ebola virus infections. Lab Invest.
100:1030–1041. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Murata K, Tsukuda S, Suizu F, Kimura A,
Sugiyama M, Watashi K, Noguchi M and Mizokami M: Immunomodulatory
mechanism of acyclic nucleoside phosphates in treatment of
hepatitis B virus infection. Hepatology. 71:1533–1545. 2020.
View Article : Google Scholar
|
|
182
|
Van de Klundert MA, Van den Biggelaar M,
Kootstra NA and Zaaijer HL: Hepatitis B virus protein X induces
degradation of Talin-1. Viruses. 8:2812016. View Article : Google Scholar :
|
|
183
|
Ji L, Jiang F, Cui X and Qin C: Talin1
knockdown prohibits the proliferation and migration of colorectal
cancer cells via the EMT signaling pathway. Oncol Lett.
18:5408–5416. 2019.PubMed/NCBI
|
|
184
|
Azizi L, Cowell AR, Mykuliak VV, Goult BT,
Turkki P and Hytönen VP: Cancer associated talin point mutations
disorganise cell adhesion and migration. Sci Rep. 11:3472021.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Aboelfotoh AO, Foda EM, Elghandour AM,
Teama NM, Abouzein RA and Mohamed GA: Talin-1; other than a
potential marker for hepatocellular carcinoma diagnosis. Arab J
Gastroenterol. 21:80–84. 2020. View Article : Google Scholar : PubMed/NCBI
|