Mitochondria are semi-autonomous organelles found in
most eukaryotic cells with a bilayered structure consisting of an
outer membrane, an intermembrane space and an inner membrane. They
serve key roles in a variety of cellular processes, including cell
metabolism, signal transduction and the regulation of cell death.
Mitochondria have numerous biological functions, including the
production of ATP for cellular energy, regulation of the dynamic
balance of intracellular Ca2+, production of reactive
oxygen species (ROS), the release of cytochrome c and regulation of
intracellular environmental homeostasis. As an important signaling
hub in cells, the mitochondrion serves a key role in diseases such
as aging and obesity. Mitochondrial biogenesis and mitochondrial
homeostasis require the expression of nuclear genes and
mitochondria-nuclear signaling pathways to be regulated (1). On the one hand, it depends on the
regulatory pathways of nuclear gene transcription and anterograde
signaling. Mitochondria, on the other hand, pass intracellular
signaling molecules, such as Ca2+, mitochondrial DNA
(mtDNA), reactive oxygen species (ROS), adenosine triphosphate
(ATP), coenzyme Q (CoQ) and nicotinamide adenine dinucleotide (NAD)
and then present mitochondrial abnormalities and cellular metabolic
change signals to the nucleus (retrograde signaling). This triggers
the nucleus to activate important signaling pathways by mobilizing
a series of nuclear transcription factors (2-5),
mitochondrial transcription and mitochondrial biosynthesis. Among
them, the activation of signaling pathways is closely related to
inflammation and tumorigenesis (6). During cellular stress and virus
infection, mtDNA and ROS are released from abnormal mitochondria
and retrogradely presented to the nucleus as danger signals. The
nucleus can promote the expression of PTEN-induced kinase 1 (PINK1)
and then upregulate mitophagy to clear abnormal mitochondria and
maintain a stable intracellular environment. When too many abnormal
mitochondria cannot be completely removed, mtDNA can activate
Toll-like receptor 9 (TLR9) and its downstream inflammatory
pathways and lead to inflammation. Excessive ROS can cause DNA
damage by oxidizing nucleic acid bases, which is closely related to
tumorigenesis. Abnormalities in mitochondrial structure and
function can lead to a variety of intracellular signaling cascades,
oxidative stress and the initiation of programmed cell death,
thereby contributing to the development and progression of nearly
all diseases. Therefore, the detection of mitochondrial
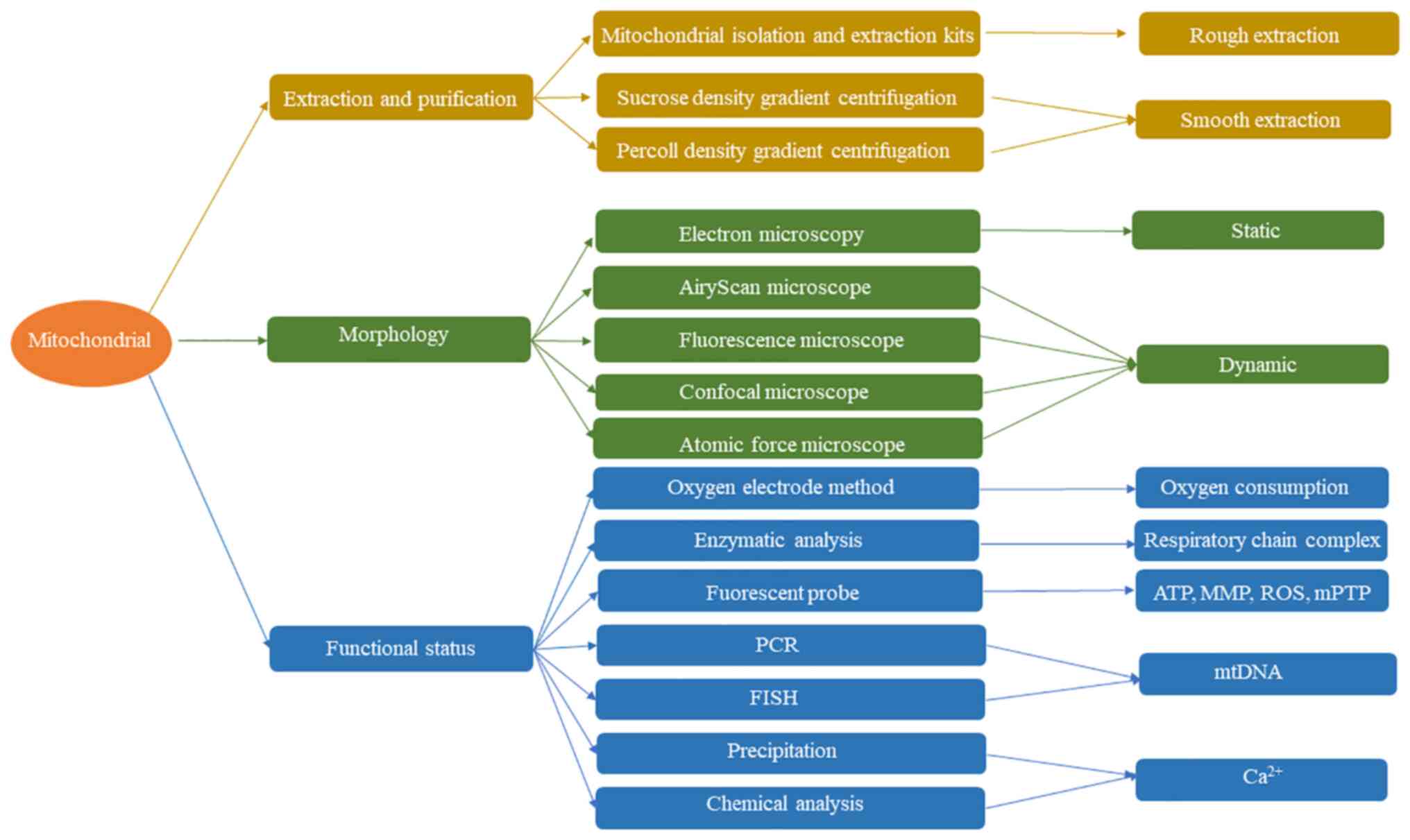
abnormalities is crucial and various mitochondrial assays (Fig. 1) developed in the last century
have contributed substantially to the differential diagnosis of
mitochondrial diseases. The present study reviewed common
experimental methods (Table I) in
mitochondrial research. In particular, it discussed a wide range of
imaging and detection techniques for i) extraction and
purification, ii) analyses of morphology and structure and iii)
analyses of function, with a focus on the clinical implications for
disease detection and treatment.
A suitable method is needed to extract purified
mitochondria from various tissues and cells (7). The basic extraction method mainly
relies on differential centrifugation, while purification mainly
depends on density gradient centrifugation. The specificity of
tissue cells determines the details of the method (8-10).
When extracting mitochondria, because the
homogenization process can heat the sample locally, resulting in
protein denaturation and aggregation, the equipment must be
pre-cooled and the temperature kept low throughout the process
(11). Tissue or cell
homogenization is followed by continuous differential
centrifugation. Unlysed cells, cell debris and nuclei are first
removed by low-speed centrifugation (600 × g or 1,000 × g)
(12-15). As mitochondria can remain in flaky
precipitates generated by low-speed centrifugation, resuspending
the pellet and centrifuging it again at low speed increases
mitochondrial yield. The supernatant obtained by two low-speed
centrifugation steps is collected for high-speed centrifugation
(3,500 × g or 10,000 × g) (12-15), resulting in a coarse-lifted
mitochondria precipitate (16).
The purity of these crudely extracted mitochondria can meet some
applications, including the analysis of the activity of known
mitochondrial proteins, the detection of mitochondrial morphology
and mitochondrial apoptosis; however, they often contain a certain
amount of peroxisomes, endoplasmic reticulum and microsomes.
Mitochondrial purity is low; thus, mitochondrial purification and
reduction of membrane fouling are required when analyzing proteins
present in multiple cells or determining the localization of a
protein (17). Furthermore,
although the mitochondrial extraction method is suitable for most
tissues and cells, the extraction efficiency and quantity of
mitochondria in different tissues and cells are significantly
different. This is determined by the number of mitochondria in the
tissue or cell and the energy consumption of muscles and liver;
larger tissues contain more mitochondria, so these tissues and
cells have higher mitochondrial extraction efficiency than other
tissues, such as the lungs (18-20).
Purified mitochondria are the prerequisites for
mitochondrial proteomics research. Density gradient centrifugation
emerged in the 1950s and has become a common method for separating
extracts owing to its ease of operation and low cost (21). For example, sucrose density
gradient centrifugation suspends the cell or a homogeneous tissue
slurry in a uniform suspension medium according to the density of
each cell component and is separated by differential centrifugation
(22-24). The buffered sucrose solution, the
most commonly used suspension medium, is relatively close to the
dispersion phase of the cytoplasm and can maintain the structure of
various organelles and the activity of enzymes to a certain extent
(25-28).
Sucrose density gradient centrifugation is a classic
method for extracting mitochondria by separating cellular fractions
of different densities (29). It
involves three main processes: Tissue homogenization, fractionation
and analysis (30-32). Homogenization refers to the
disruption of cells or tissues in a homogenizer by adding sucrose
at a low temperature to form a homogenate containing various
organelles and other substances (33). Fractionation is the sequential
settling of particles of different densities and sizes in the
sample by centrifugation at different speeds. Analysis refers to
the use of biochemical methods to identify the morphological
function of the separated components; it is conducted using the
Janus green live dyeing method, which is easy to operate and stable
in performance. However, at high concentrations, sucrose has a high
viscosity and high osmotic pressure, which can easily cause
repeated shrinkage and mitochondrial expansion. Compared with
sucrose, the price of commonly used density gradient media
(including Percoll, Nycodenz and OptiPrep) is generally higher, but
the morphology of the extracted mitochondria is generally complete.
Percoll has a low diffusion constant, the gradient formed is very
stable and it does not penetrate the biofilm; as such, it minimizes
organelle rupture and is often used to isolate platelet
mitochondria (12,34,35). Nycodenz is increasingly widely
used owing to its high density, low viscosity and lack of effect on
osmotic pressure (36-38). The yield of intact mitochondria is
significantly higher in Nycodenz gradients containing sorbitol as
an osmotic stabilizer instead of sucrose (37,38). As a dimer of Nycodenz, OptiPrep
has the advantage of forming automatic gradients in a short period
of time (39-42). Additionally, some researchers use
streptavidin magnetic beads to separate Arabidopsis
mitochondria. After the tissues are lysed, they are mixed with
anti-mitochondrial outer membrane protein 22 (TOM22) magnetic beads
and the mixed samples placed in the sorting column. Only
mitochondria remain on the sorting column after washing, followed
by elution, isolating the complete mitochondria in less than 30 min
with a success rate, purity and integrity significantly higher than
the density gradient centrifugation (43-47). Therefore, the magnetic bead method
can be used to extract mitochondria in tissues with fewer
mitochondria. As such, this approach will probably become
increasingly common in mitochondrial extraction and purification
(48-50). In conclusion, among the current
mitochondrial extraction and purification methods, the magnetic
bead method has the best effect on eliminating impurities such as
microsomes and peroxisomes and the mitochondrial purity obtained by
the differential centrifugation method is the lowest and the effect
on eliminating these impurities is the worst.
Mitochondria are organelles with a complex
bi-membrane structure that regulate the entry and output of
proteins, lipids, solutes and metabolite products and protect the
cytoplasm from harmful mitochondrial products (51-53). Mitochondria can engulf abnormal
mitochondria and remove excess harmful mitochondrial products to
protect the body. This process is called mitophagy (54-56). Most mitochondria are spherical,
rod-shaped, or tubular; however, mitochondrial morphology varies
widely among tissues and cells depending on the energy requirements
of cells and the location of mitochondria within the cell (53,57). For example, mitochondria are
spherical at synaptic terminals, whereas they appear as highly
elongated rods in axons. In senescent and functionally impaired
cells, mitochondrial morphology is significantly different from
that in normal cells and they can be irregularly shaped (53,58,59). Therefore, morphological changes
can be used in the initial assessment of mitochondrial
function.
After over 50 years since its development, electron
microscopy (EM) has become the central tool for observing
organelles in eukaryotic cells and is the gold standard for
observing mitochondrial structure (60). It can reveal mitochondrial
swelling, rupture and other abnormalities of damaged mitochondria.
However, it cannot clearly distinguish mitochondria from other
membranous structures and is occasionally confusing. In the 1980s,
atomic force microscopy, as an emerging observation method, could
study the surface structure and properties of substances by
detecting the extremely weak interatomic interaction between the
surface of the sample to be tested and a miniature force-sensitive
element. Due to the characteristics of resolution and real-time
imaging, changes such as the formation of mitochondrial swelling
can also be observed under liquid conditions but are significantly
affected by the probe; thus, the application range is small
(61-64)
The recently developed AiryScan microscope (Zeiss
AG) can acquire images at high speed with high sensitivity to
effectively observe the kinetic processes of mitochondrial fission,
fusion and autophagy (65-67).
In addition, both wide-field fluorescence microscopy and
high-resolution confocal laser scanning microscopy can be used for
imaging analyses of morphological changes in mitochondria with
higher specificity than that of EM, but the dynamic changes of the
mitochondria cannot be observed (68-76).
In most cases, microscopy can be used to observe and
analyze two-dimensional mitochondrial morphologies and quantities.
However, although this method is suitable for analyzing adherent
cells with flat morphology, it is not suitable for thicker cells
(77-83). Three-dimensional confocal
microscopy can be used to observe mitochondrial morphology by
observing specifically labeled mitochondrial proteins at the 3D
level (84-87). In addition, after labeling
mitochondria with specific dyes, mitochondrial morphology can be
visualized using a combination of immunofluorescent staining and
computer images (58,88,89).
Mitochondrial membrane potential (MMP) refers to the
negative potential difference between the two sides of the inner
mitochondrial membrane. It is a sensitive indicator for evaluating
mitochondrial function (90-93). It is closely associated with
cellular homeostasis and is most commonly used to determine the
metabolic state of mitochondria (93-98).
Fluorescent dye probes used for flow cytometry are
now commonly used in MMP assays. For example, rhodamine 123, a
specific stain developed in the 1980s, is widely used in flow
cytometry and MMP assays. In normal cells, rhodamine 123 can
selectively enter the mitochondrial matrix depending on MMP and can
emit bright yellow-green fluorescence; when cells undergo apoptosis
or necrosis, the mitochondrial membrane permeability transition
pore (mPTP) is abnormally opened and MMP is unbalanced. Rhodamine
123 is released from mitochondria, resulting in a significant
decrease in the yellow-green fluorescence intensity in
mitochondria, which reflects the changes in MMP (50,99-102). 5,5',6,6'-Tetrachloro-1,1',3,
3'-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) has higher
sensitivity than that of rhodamine 123. At low MMP levels, JC-1
exists as a monomer and produces green fluorescence; at high MMP
levels, JC-1 aggregates in the mitochondrial matrix and forms
polymeric JC-1. This can be used for qualitative and quantitative
analyses of MMP by fluorescence microscopy or flow cytometry
(50,96,101,103-108). Tetramethyl rhodamine methyl
ester (TMRM) and tetramethyl rhodamine ethyl ester (TMRE), like
JC-1, are specific dyes that have recently become common tools for
measuring MMP (109-112). TMRM can be excited at 488 nm,
showing red-orange fluorescence and its fluorescence intensity has
a linear relationship with MMP. Compared to rhodamine 123 and JC-1,
these two dyes are very soluble, have short loading times (15-20
min) and have extremely low cytotoxicity, requiring micromolar
inhibition of mitochondrial function. With staining concentrations
in the range of 0.5-30 nM (the concentration of JC-1 needs to be
>0.1 µM), the accumulation in mitochondria is limited to
the change of membrane potential and the sensitivity is extremely
high; this is markedly suitable for quantitative analysis of
mitochondrial membrane potential and quantitative flow cytometry
(113-118). However, in quantitative flow
cytometry studies, the data must be corrected for the signal of
MitoTracker Green FM, a dye that is not dependent on mitochondrial
membrane potential. It is worth noting that the above fluorescent
probes for measuring MMP are applicable to most tissues and cells,
including plant cells and bacteria.
Fluorescence resonance energy transfer (FRET) is a
non-radiative energy transition that transfers energy from the
excited state of the donor to the excited state of the acceptor
through intermolecular electric dipole interactions (119,120). This process does not involve
photons, so it is non-radiative. This analytical method has the
advantages of rapidity, sensitivity and simplicity. Fluorescence
resonance energy transfer molecular pairs (FRET Pairs) have been
designed and synthesized to monitor MMPs (121). The FRET donor molecule (FixD) is
constructed by attaching a benzyl chloride group to a fluorophore
with green fluorescence emission. FixD can be attached to and fixed
in mitochondria by sulfhydryl groups of mitochondrial proteins. The
FRET acceptor (LA) is a mitochondrial membrane potential-dependent
probe with green absorption and deep red fluorescence emission.
When MMP is at a normal level, both FixD and LA target
mitochondria. When FixD has an excitation wavelength of 405 nm,
FRET occurs between FixD and LA, allowing green fluorescence to be
detected but not deep red LA fluorescence emissions. When MMP is
gradually reduced, LA will gradually fall off from mitochondria.
While FixD is still fixed in mitochondria, the distance between the
molecules gradually blocks the occurrence of FRET between FixD and
LA molecules, allowing deep red fluorescence emission to be
detected gradually. The decrease and the gradual increase of green
fluorescence emission can be used to monitor the dynamic changes of
MMPs (122), providing new ideas
for the development of novel MMP fluorescent probes and real-time
in situ studies of MMPs in living organisms, tissues and
cells (123,124).
MMP varies greatly among sites on the mitochondrial
membrane; therefore, accurate measurement of MMP requires further
study (125). In recent years,
low concentrations of a hemicyanine derivative (TPP-CY) have been
used to monitor trace changes in MMP at the subcellular level
during apoptosis with very high sensitivity (125). This approach is a potentially
useful tool for evaluating cell health.
Among organelles, mitochondria consume the most
oxygen in cells and this oxygen consumption often reflects
mitochondrial function (126-128). In the heart, mitochondrial
oxygen consumption can be measured to determine cardiac
mitochondrial function, providing an indicator of cardiac function
(129-131). In children, mitochondrial
dysfunction causes mitochondrial heart disease with hypertrophic
myocardial infarction as the primary symptom; however, the exact
mechanism and etiology remain to be investigated (129,132).
Oxygen electrode polarography is a common method for
determining mitochondrial oxygen consumption and refers to the
incubation of mitochondria in an oxygen-consuming medium in a
magnetically stirred incubator at 30°C. Briefly, rotenone is used
to inhibit complex I in the electron transport chain, followed by
the addition of succinate to measure mitochondrial state IV
respiration (non-phosphorylating respiration). State III
respiration is measured by incubating mitochondria in the presence
of succinate and adenosine diphosphate. The respiratory control
ratio (RCR) is the ratio of the state III respiration rate to state
IV respiration rate, with a normal value of 3-10 (133-135). A low RCR indicates impaired
mitochondrial ATP synthesis and mitochondrial dysfunction and a
high RCR indicates vigorous cellular activity and accelerated
metabolism (127,136,137).
In addition, the hippocampal analyzer can measure
the changes in oxygen and pH levels through sensors and then
automatically calculate the rate and detect the cellular oxygen
consumption rate (OCR) and extracellular phosphorylation rate
(ECAR) in real time to characterize the metabolic status of cells.
Where OCR is caused by mitochondrial electron transfer, ECAR is
derived from lactic acid fermentation (glycolytic acidification)
and carbon dioxide produced by mitochondria (mitochondrial
acidification) (138-140).
OCR is used to study mitochondrial oxidative
phosphorylation function, with pMoles/min as the readout type
(141). Generally, basal
respiration in a normal state is measured first and then oligomycin
is added to inhibit ATP synthase. This is a significant decrease in
OCR, leaving only proton leakage (142). The oxygen consumption rate is
caused by proton leakage and the reduced section is the oxygen
consumption rate (ATP production) of oxidative phosphorylation.
With the addition of the uncoupling agent FCCP, electron transport
loses the constraints of the proton gradient and proceeds at a
maximum rate (143). Therefore,
the OCR increases sharply, reaching the maximum oxygen consumption
(maximal respiration); the difference between this value and the
basal respiration is termed the spare respiratory capacity.
Finally, adding an electron transport inhibitor, such as antimycin
A, completely inhibits electron transport and reduces the oxygen
consumption rate to a minimum (144).
ECAR is often used to study metabolic conditions
such as glycolysis, with mpH/min as the readout type (139,140,142). The basal value before adding
glucose is non-catalytic acid production, such as mitochondrial
acidification caused by carbon dioxide produced by mitochondrial
respiration. Glucose is then added and the elevated value
represents glycolysis. After the addition of oligomycin, the
production of acid increases because oxidative phosphorylation is
inhibited and the cells are forced to use lactic acid fermentation
for energy. The value at this time is called glycolytic capacity
and the difference from glycolysis is termed glycolytic reserve
(140,142,143). Last added is 2-deoxyglucose, a
competitive hexokinase inhibitor that can block glycolysis, so the
curve should return to the basic value following its addition
(144-146).
However, the direct measurement of glycolysis by
ECAR is somewhat biased since the addition of glucose enhances
glycolysis and oxidative phosphorylation. This will lead to
increased mitochondrial acidification, causing the calculated
amount of glycolysis to be high (147-149).
It is worth noting that during the measurement
process of the hippocampal analyzer, the interference of phenol red
should be avoided because it causes errors in the measurement
results (141,150,151), but the specific reasons remain
to be elucidated. In conclusion, the hippocampal analyzer can
monitor OCR and ECAR to obtain multiple other parameters in a
single analysis, including basal respiration, ATP-related
respiration, maximal respiration, spare respiratory capacity and
non-mitochondrial oxygen consumption, all of which can provide
information on the mechanism of mitochondrial dysfunction (152,153).
mPTP is a class of protein complexes between the
inner and outer mitochondrial membranes that permit the passage of
substances with a molecular weight of <1.5 kDa and serve as the
structural basis for transitions in mitochondrial permeability
(188-191). Additionally, mPTP is very
sensitive to changes in intracellular and extracellular ion
concentrations and serves an important role in signal transduction
systems. It is currently hypothesized that the abnormal opening of
mPTP is closely associated with abnormal changes in Ca2+
concentrations, oxidative stress and mitochondrial DNA (mtDNA)
mutations (154,188,189,192,193). By contrast, MMP and
mitochondrial Ca2+ concentrations are the principal
drivers of mPTP opening, resulting in the release of cytochrome c
and other substances associated with cell death into the cytosol
(191,192,194-197). This leads to mitochondrial
swelling and reduced mitochondrial respiratory chain activity,
which can cause various diseases, such as neurodegenerative
diseases and cancers (190,198-200). Furthermore, studies have shown
that PINK1 can inhibit mPTP opening by downregulating intracellular
ROS levels, suggesting that mitochondrial autophagy serves a
regulatory role in mPTP opening (191-193). Various methods have been
developed for detecting mPTP, such as the patch-clamp,
spectrophotometric and active substance labeling methods. The
patch-clamp method is the earliest, originating in 1976 and can
reflect ion channel activity by recording ion channel currents to
evaluate mitochondrial function (188,189,201). As the magnification of AFM is as
high as 1 billion times, the opening of mPTP can be directly
observed, which can serve a guiding role in the abnormal opening of
mPTP (202-205). Fully automated patch-clamp
techniques have recently emerged; these are simple in operation and
have greatly improved efficiency but are only applicable to the
detection of cells in suspension. Compared to the active substance
labeling and patch-clamp methods, spectrophotometry is simpler and
more commonly used.
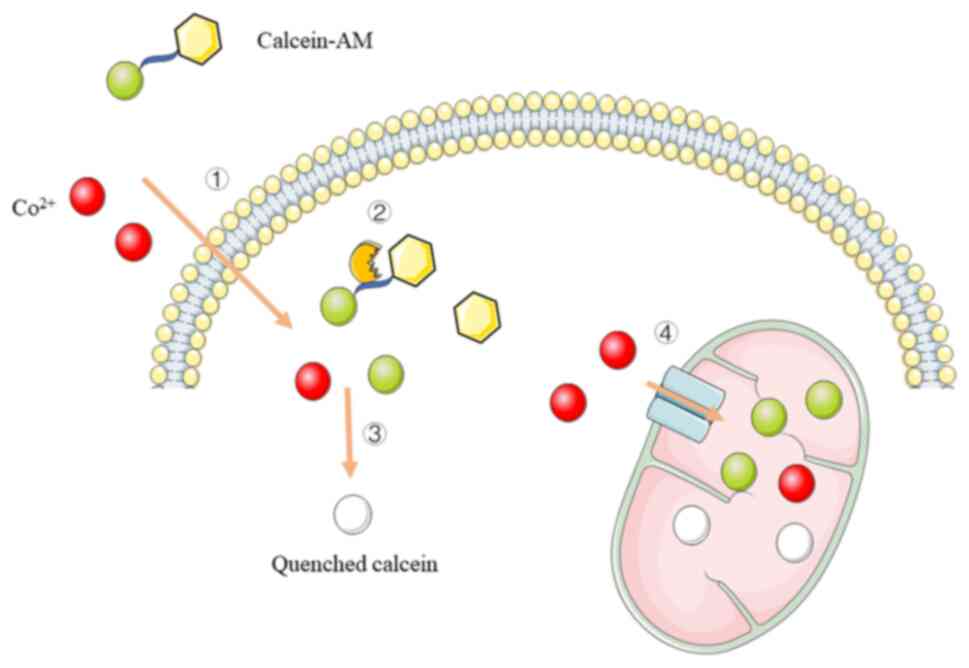
The calcein-cobalt fluorescent probe technique is an
emerging technique for the detection of mPTP and is simple in
operation and highly sensitive (Fig.
2). Calcein-AM (190,194,
198,206,207), in which the acetylmethoxy methyl
ester (AM) group enhances the hydrophobicity of the stain for easy
penetration of the living cell membrane, is used to fluorescently
label living cells. Next, calcein-AM is cleaved by intracellular
esterases to yield highly fluorescent and polar calcein (208-210). When cells are incubated with
calcein and Co2+, both enter the cytoplasm; however,
calcein is further captured by mitochondria (211,212). Calcein that accumulates in the
mitochondria exhibits fluorescent staining, whereas calcein
remaining in the cytoplasm or released from the mitochondria into
the cytoplasm is rapidly quenched by Co2+ (213-219). Under normal physiological
conditions, mPTP opens transiently and calcein that enters the
cytoplasm from the mitochondria is rapidly quenched. In
pathological states, such as calcium overload and oxidative stress,
mPTP can appear to be continuously open and Co2+ in the
cytoplasm can enter the mitochondria to quench the calcein
fluorescence, resulting in a gradual decrease in fluorescence
intensity in the mitochondria, thus indicating the degree of mPTP
opening (195,196,220-222).
ATP is often considered the primary energy currency
of cells and is primarily derived from the mitochondria (137,223-228). It serves major roles in material
transport, energy conversion and information transfer. Mitochondria
are sensitive to external environmental stimuli, such as hypoxia,
oxidative stress, toxic substances and high glucose. Once
mitochondria are damaged, ATP production decreases and free radical
production increases, which affects a number of cellular processes
and contributes to the development of a number of diseases, such as
Parkinson's disease, cancer, cardiovascular disease and endocrine
dysfunction (224-227). Therefore, ATP levels are a key
indicator of the status of cellular energy metabolism and
mitochondrial function.
Analyzing ATP levels requires freshly extracted
mitochondria, as mitochondria must remain intact and in a coupled
state (229). Several techniques
are available to measure mitochondrial ATP levels, including
chromatography, electrophoresis, high-performance liquid
chromatography (HPLC) and enzymatic analysis (225,227,229-232). Chromatography and
electrophoresis are chemical methods that were developed in the 18
and 19th centuries and have gradually improved. Classic liquid
chromatography uses a large-diameter glass tube column and a
difference in liquid levels at room temperature and atmospheric
pressure to force the mobile phase (231,232). However, this technique has low
column efficiency and is very time-consuming (often requiring
several hours). HPLC was developed based on classic liquid
chromatography following the introduction of gas chromatography
theory in the late 1960s. The differences between HPLC and classic
liquid chromatography include a faster analysis speed, smaller and
more uniform particles as packing material and high column
efficiency of the small particles. However, this causes high
resistance and requires high pressure to force the mobile phase;
therefore, this technique is also known as high-speed liquid
chromatography (233-235). HPLC can be used to determine
differences in cellular energy substances in different states, is
easy to operate and has high sensitivity (233-236). The enzymatic method is based on
spectrophotometry, where ADP production is assessed by measuring
the absorbance of NAD+ in phosphoenolpyruvate (237-240). Fluorescence analysis techniques
have been improved in recent years and are commonly used to
determine mitochondrial ATP synthesis activity (241-244). For example, in the
luciferin-luciferase luminescence method, luciferin is rapidly
oxidized under the action of luciferase, producing green
fluorescence and the amount of luminescence is linearly correlated
with the level of ATP (245,246). This is a fast and accurate
method; however, fluorescein is an amphiphilic molecule whose
carboxyl group is charged at physiological pH and thus does not
easily cross the cell membrane (244-246). A novel synthetic fluorescent
probe called Mito-Rh can specifically identify ATP in mitochondria
with high sensitivity and a detection range of 0.1-10 mM. In
another method, the level of ATP can be determined directly by
measuring the amount of inorganic phosphate based on the principle
that ATP gives rise to ADP and inorganic phosphate (225). In addition, FRET can also be
used to detect the level of ATP synthesis after labeling the ATP
synthase subunit. When CFP and YFP are labeled on ATP synthase
subunits, when the ATP synthase activity is enhanced, the
interaction between the subunits is enhanced, the shortened
distance between the subunits brings CFP and YFP closer to each
other and FRET occurs and CFP excites YFP to emit yellow
fluorescence. The lower the green fluorescence intensity received
by the detector, the higher the ATP synthase activity and the
higher the ATP level. When ATP synthase activity is low, the
interaction between subunits is weakened, FRET hardly occurs and
CFP is excited at this time and the cell emits green light.
In addition, a multi-color ATP indicator has
appeared in recent years. Different from the previous indicators
that can only specifically detect intracellular ATP, the
multi-color ATP indicator is based on a single fluorescent protein
indicator with red, green and blue colors (247-249). Alternatively, it can
simultaneously detect ATP in different organelles in the same cell
and simultaneously detect ATP dynamics in the mitochondria of
mammalian, plant and even worm cells and will have an assured role
in promoting energy metabolism research in the future (225,226).
The mitochondrial respiratory chain, with functions
in energy production, the regulation of cell death and calcium
metabolism (183,250-253), is located on the inner
mitochondrial membrane and consists of five complexes.
Mitochondrial respiratory chain complex I (NADH oxidase) and
mitochondrial respiratory chain complex II (succinate
dehydrogenase) are the major elements for electrons entering the
mitochondrial electron transport chain (ETC). Complex I oxidizes
NADH and transfers electrons to coenzyme Q (254-257). Complex II transfers electrons
from succinate to coenzyme Q, a process that does not involve
proton transport (258-260). Mitochondrial respiratory chain
complex III (cytochrome c reductase) is an essential protein for
mitochondrial oxidative phosphorylation, the gatekeeper of the
mitochondrial respiratory chain and a major source of third
reactive oxygen species. Complex III transfers electrons from
coenzyme Q to cytochrome c while using the released energy to pump
protons into the intermembrane space. The mitochondrial respiratory
chain complex IV (cytochrome c oxidase) is the terminal electron
acceptor of the mitochondrial electron transport chain. Complex IV
transfers electrons from cytochrome c to oxygen, half the number of
protons is synthesized into water and the other half is pumped into
the intermembrane space. Mitochondrial respiratory chain complex V
and the above four complexes complete oxidative phosphorylation to
generate ATP, which is called ATP synthase, also known as
F1F0-ATPase (254,260-265). The energy released by complex V
through the electron transport chain during respiration or
photosynthesis is first converted into a transmembrane proton
(H+) gradient and the proton then flows along the proton
gradient and passes through ATP synthase to enable ADP+Pi to
synthesize ATP (266-269). It is also hypothesized that
abnormalities in mitochondrial complexes are closely associated
with mitochondrial encephalopathy, mitochondrial liver disease and
mitochondrial nephropathy (265). It should be noted that the
mitochondrial respiratory chain complex is closely related to the
occurrence of tumors (251,270-272). Therefore, mitochondrial complex
inhibitors may be used as a new treatment for tumors (252,253,260,273). Therefore, the accurate detection
of mitochondrial complexes is essential and spectrophotometric
assays remain the first-line technique for detecting the activity
of mitochondrial respiratory chain complexes I-V (266,274,275).
Mitochondrial respiratory chain function can also
be determined by RCR, which reflects both mitochondrial integrity
and mitochondrial oxidative respiratory chain function (256,265,267,301).
As the central organelle for cellular oxidative
phosphorylation, mitochondria are the principal site of ROS
production (3,302-305). Under physiological conditions,
the intracellular antioxidant defense system is in equilibrium with
oxygen radicals. The levels of intracellular ROS, including
superoxide radicals, hydrogen peroxide and its downstream products
(peroxides and hydroxyl radicals), are maintained at low
physiological ranges. Under pathological conditions, the balance
between the intracellular antioxidant system and oxygen radicals is
disrupted. When intracellular ROS levels are too high,
mitochondrial structure and function are impaired and cytochrome c
is released through mPTP, resulting in damage to mitochondrial
enzymes, lipids and nucleic acids as well as oxidative stress
(303,306-310). ROS can also attack mitochondrial
DNA (mtDNA) to produce oxidative damage, resulting in reduced
mitochondrial ATP synthesis and MMP damage. Therefore, the
functional status of mitochondria can be determined by measuring
ROS levels (311-313).
Common methods for detecting ROS include the
chemical reaction method, selective electrode method,
spectrophotometry and direct detection by kits. ROS shows high
reactivity and can react with different compounds to produce
various products, which can be analyzed quantitatively or
qualitatively. The chemical reaction method is characterized by
high sensitivity, low cost and simple operation; however, it has
poor specificity and measurement results are easily affected by
some redox reactions or enzyme-catalyzed reactions.
Tetranitromethane, nitrotetrazolium blue chloride (NBT), cytochrome
c, epinephrine and reduced coenzyme I are commonly used for
spectrophotometric methods; these react with superoxide anion
radicals to produce ferrous cytochromes with a specific absorbance
(detectable at a wavelength of 550 nm), which can be used to
directly measure ROS levels (307,314-317). The NBT assay is highly sensitive
and is commonly used for the histochemical localization of oxygen
radicals; however, it is difficult to measure dynamic changes in
oxygen radicals in cells or aqueous systems. Cytochrome c has
oxidative activity and can be used to detect the production of
oxygen radicals. However, cytochrome c is easily reduced by other
reducing agents and is therefore limited for the accurate
localization of oxygen radicals. In the last decade, a number of
ROS kits have been developed to detect intracellular or
mitochondrial ROS (mtROS) levels directly. Intracellular ROS are
usually measured using the fluorescent probe DCHF-DA, which is
non-fluorescent and can freely cross the cell membrane. After
DCHF-DA enters cells, it is hydrolyzed by intracellular esterases
to generate DCHF, which cannot enter or exit the cell membrane,
thus allowing the probe to easily label the cell. In the presence
of ROS, DCHF is oxidized to produce the fluorescent substance DCF,
whose fluorescence intensity is directly proportional to
intracellular ROS levels. mtROS is usually measured using the
fluorescent probe MitoSOX, which is highly specific to
mitochondrial ROS and is characterized by simple operation, low
background signals, wide linear range and high detection
efficiency; however, it requires the immediate imaging of assay
results and protection from light to prevent fluorescence
quenching. Prior to the widespread use of kits, ROS levels were
indirectly measured by detecting products of oxidative damage.
Levels of malondialdehyde (MDA) reflect the degree of lipid
peroxidation in the body and can be measured using the
thiobarbituric acid (TBA) chemical colorimetric method.
Condensation under acidic conditions generates the MDA-TBA complex,
a red product with a maximum absorption peak at 535 nm, which can
be used to indirectly determine the MDA content by
spectrophotometry, indicating ROS levels. However, this technique
has poor sensitivity and is prone to contamination. Fluorescent
protein-based ROS detection methods are designed by combining
fluorescent proteins and prokaryotic redox-sensitive proteins
(318,319). The recombinant proteins are
introduced into cells via plasmids or adenoviruses and target
organelles to detect intracellular redox status (320,321). Redox-dependent fluorescence
spectral changes of recombinant proteins are achieved through
structural changes of disulfide bonds and part of the backbone
under oxidative conditions (319,321).
Electron spin resonance (ESR) technology has
emerged in recent years. Also known as electron paramagnetic
resonance (EPR), its principle is similar to nuclear magnetic
resonance (322-325). The sample is controlled in a
fixed frequency microwave and the applied magnetic field is then
changed so that the electron energy level difference is the same as
the microwave energy (326,327). Unpaired electrons can move
between the two energy levels and the net absorption energy of the
microwave can be measured to obtain the ESR spectrum. Due to the
high reactivity and short lifespan of ROS, the ESR signal is not
easy to detect directly. The combination of ESR and spin traps can
make up for this defect. The spin-electron trapping agent reacts
with free radicals to generate relatively stable free radical
addition products that are easily detected by ESR, which is then
determined by ESR technology. This powerful and reliable technique
can unambiguously measure the presence of free radicals in
biological samples. ROS is the most direct and effective method for
detecting free radicals and is widely used in physics, chemistry
and biomedicine (328-331).
Human mitochondria carry a small circular
double-stranded genome of 16569 bp known as mtDNA, which encodes
mitochondrial 16S and 12S ribosomal RNA, 22 mitochondrial tRNA
molecules and 13 respiratory chain proteins. Each organism contains
only one type of mtDNA and mutations such as the conversion,
inversion, insertion, or deletion of one or several bases of mtDNA,
resulting in more than one type of mtDNA within an individual, are
referred to as mtDNA heterogeneity (332-335). Owing to the lack of protective
histones and effective DNA repair systems, the mutation frequency
of mtDNA is ~10 times higher than that of nuclear DNA (336-339). Moreover, mutated mtDNA gradually
accumulates and can cause irreversible damage to the nervous,
cardiovascular, respiratory and reproductive systems after reaching
a certain threshold (60-80%). In addition to these diseases,
studies have also shown that mtDNA mutations are closely associated
with the development of infertility (308,339-342). mtDNA dysfunction can be both
quantitative (e.g., mtDNA copy number variation and deletions) and
qualitative (e.g., strand breaks, point mutations and oxidative
damage) (343-345).
mtDNA can be released from the cell as circulating
free mitochondrial DNA (CCF-mtDNA) via extracellular vesicles (EVs)
(346,347). CCF-mtDNA can serve as a
damage-associated molecular pattern leading to the activation of
inflammatory pathways, a process closely associated with TLR9.
Numerous reports have shown that elevated levels of CCF-mtDNA are
associated with various TLR9-dependent pathologies, such as
rheumatoid arthritis, atherosclerosis, hypertension, acute liver
injury and nonalcoholic steatohepatitis (48,348).
Unrepaired depurinated/depyrimidinated sites (AP
sites) in mtDNA lead to the misbinding of nucleotides, which can
have serious downstream effects (372-374). Therefore, the rapid and accurate
quantification of AP sites in mtDNA is crucial for the real-time
assessment of mtDNA oxidative damage. Researchers have used a
specific fluorescent probe (BTBM-CN2) for the real-time detection
of mtDNA (375-378). At ~20 sec after contact with AP
sites, red fluorescence is detectable at 598 nm and after ~100 sec,
green fluorescence is detectable at 480 nm. More AP sites result in
green fluorescence with greater intensity and duration and the
degree of mtDNA damage can be quantified based on the time of
appearance and intensity of fluorescence at 480 nm. Doxorubicin
(Dox), a common anticancer drug, not only causes damage to the
nuclear DNA of cells but can also be rapidly inserted into the
mtDNA of living cells, causing the aggregation of mtDNA nucleoids
and changing the distribution of nuclear proteins (375-382). Therefore, after Dox induces
mtDNA damage, morphological changes of mtDNA can be tracked in real
time using the two-photon fluorescent probe CNQ, which emits red
fluorescence and is localized to mtDNA. When incubated with Dox,
dynamic changes in mtDNA can be observed, providing a new method
for studying mtDNA damage in real time (383,384).
In addition to primary mitochondrial disease caused
by mtDNA damage, mitochondrial dysfunction occurs in a number of
infectious and non-infectious diseases (262,385,386), such as inflammation,
neurodegeneration, diabetes, obesity and cardiovascular disease and
several therapies targeting mitochondria have been developed
(Table II). Mitochondrial
transplantation and mitochondrial replacement can fundamentally
address the inadequate energy supply in pathological states and
have been applied in clinical settings for the treatment of
pediatric congenital heart disease (385).
Leber hereditary optic neuropathy (LHON), the most
common primary mitochondrial disease, is a maternally-inherited
bilateral-blinding optic neuropathy mainly caused by mtDNA
mutations, including m.3460G>A (MT-ND1), m.11778G>A (MT-ND4)
and m.14484T>C (MT-ND6), of which m.11778G>A is the most
common mutation (387,388). These mutations can affect the
mitochondrial respiratory chain complex I of retinal ganglion
cells, impair mitochondrial function and increase the production of
reactive oxygen species, leading to apoptosis and optic nerve
degeneration and atrophy, which further leads to rapidly
progressive loss of binocular vision (389-391). Treatment of LHON is mostly based
on ectopic expression, that is, intravitreal injection of
adeno-associated viral vectors with mitochondrial targeting
sequences and then guiding the translated protein into mitochondria
to restore mitochondrial function, which has been successfully and
safely applied to cell models. Transplant into an inducible LHON
animal model that preserves retinal ganglion cells and visual
function (392,393).
The mitochondrial diseases associated with mtDNA
deletion mainly include chronic progressive external
ophthalmoplegia (CPEO), Kearns-Sayre syndrome (KSS) and Pearson
syndrome. CPEO is mostly associated with m.3243A>G(MT-TL1)
deletion, which manifests as progressive paralysis of the ocular
muscles, resulting in ocular movement disorders and ptosis, which
usually appear in late childhood or early adulthood (394,395). KSS is a more severe syndrome
than CPEO and is mostly associated with m.8993T>G (APT6)
deletion. Its main clinical manifestations are progressive external
ophthalmoplegia and retinitis pigmentosa, usually occurring before
the age of 20 (396-399). Other symptoms may include mild
skeletal muscle weakness, hearing loss, cognitive impaired
cognitive function and diabetes. Pearson's syndrome is a syndrome
caused by sideroblastic anemia and pancreatic exocrine
insufficiency. There are very few cases (~100 cases worldwide) that
may be related to the deletion of ATPase 6 and 8. Most patients die
during infancy; however, a minority of patients who survive into
adulthood tend to develop symptoms of KSS syndrome. Due to the
double-membrane structure of mitochondria and the inability of
foreign nucleic acids to recombine on endogenous mtDNA (168,400,401), there is currently no effective
method to directly import nucleic acids into mitochondria and the
localization of proteins to mitochondria is a routine practice in
the treatment of mitochondrial diseases. In principle, expression
of mitochondrial-targeted DNases that specifically recognize
mutated sequences can remove mutated mtDNA, or at least reduce its
abundance in a heterogeneous background. Restriction endonucleases,
zinc finger nucleases and transcription activator-like effector
nucleases have been tested and proven effective; these specific
enzymes can be used to eliminate aberrant mtDNA and thereby reduce
the rate of aberrant mtDNA in cells (402-406).
In addition, mitochondrial neurogastrointestinal
encephalomyopathy, a rare mitochondrial disease, is often
associated with TYMP gene mutations, manifesting as splanchnic
neuropathy and marked motor impairment, often combined with CPEO,
sensorimotor polyneuropathy and white matter encephalopathy
(407-409). With advances in gene editing
technology, CRISPR/Cas9 has been proposed for the treatment of
mitochondrial diseases, aiming to eliminate abnormal mtDNA
sequences through the principles of bacterial immunology (410,411).
To treat primary mitochondrial diseases, gene
therapy based on ectopic expression is still the first choice;
however, the application of viral vectors in live animals to
correct any gene mutation still has the following significant
problems: High cost (390,412-415), carcinogenicity and
immunogenicity. Non-viral vector-mediated in situ
mitochondrial gene therapy may be a promising approach to overcome
the bottleneck of existing gene therapy LHON, such as
liposome-based nanoparticles, which require further investigation
(416-421).
Mesenchymal stem cell-derived EVs are a promising
nanotherapeutic strategy to effectively attenuate mitochondrial
damage and the inflammatory response by promoting mitochondrial
transcription factor A expression and preventing mtDNA damage and
leakage from target cells (422).
Oxidative stress caused by mitochondrial
dysfunction is one of the etiologies of metabolic disease and is a
potential target for the treatment of metabolic and
neurodegenerative disorders (55,168,423-426). A number of antioxidants, such as
vitamin E, ubiquinone, N-acetylcysteine, glutathione and
melatonin, can effectively scavenge mitochondrial ROS and regulate
redox processes, thus alleviating or curing disease. Antibiotics
(e.g., tetracyclines and actinomycins), drugs (e.g., creatine and
ursodeoxycholic acid) and exercise can significantly improve
oxidative stress and balance mitochondrial fission and fusion, thus
increasing the number of mitochondria, contributing to the
treatment of cancer (400-406,426-442). SS31 and mitoTEMPO are novel
mitochondrial-targeted antioxidants that have a scavenging effect
on ROS (443-446). In addition, SS31 accumulates in
the mitochondrial membrane to protect and restore the mitochondrial
structure without affecting healthy mitochondria (162,447-453). Thus, SS31 and mitoTEMPO have
protective effects on a variety of diseases, including heart and
kidney-related diseases, as well as sepsis and diabetes, which have
been demonstrated in a variety of animal models (454-457). The use of nanomaterials for
mitochondrial targeting therapy has become a recent focus of
research. Nanomaterials are materials with at least one of three
spatial dimensions at the nanometer scale (1-100 nm). They are a
new generation of materials composed of nanoparticles with sizes
between atoms, molecules and macroscopic systems and are widely
used in the medical field owing to their large specific surface
area and excellent biocompatibility. Ideally, medical nanomaterials
should remain quiescent in normal tissues but accumulate precisely
and act in mitochondria under pathophysiological conditions
(404,458,459). Delocalized lipophilic cations
(DLCs), such as triphenylphosphine (TPP) and
mitochondria-penetrating peptides (MPPs), serve a major role in
mitochondria-targeted therapies. DLCs can accumulate specifically
in the mitochondria of tumor cells and increase their MMP, leading
to altered mitochondrial membrane permeability and inducing
apoptosis (56,130,400,403,428,458-470). Studies have shown that graphene
has a large specific surface area, good targeting and high
biocompatibility, making it a promising nanodelivery system
(441,471-473). Mitochondrial biogenesis is
driven by PCG-1α, which can increase the number of mitochondria in
the cell and thus meet the evolving energy demands of the cell,
alleviating ATP deficiency in patients with mitochondrial diseases.
Promoting mitochondrial biogenesis is also an important component
of mitochondrial therapeutics (474). Resveratrol,
5-aminoimidazole-4-carboxamide riboside, epicatechin and RTA-408
have significant pro-mitochondrial biogenesis effects; the
treatment of mice with these drugs enhances the expression of
mitochondrial electron transport chain proteins and mitochondrial
transcription factors and increases the abundance of mitochondrial
cristae (54,401,402,405,406,441, 471-478).
As the powerhouses of the cell, mitochondria are at
the center of cellular oxidative phosphorylation and are critical
for growth and development as well as the development of a number
of diseases. Mitochondrial abnormalities can cause disturbances in
the intracellular environment and can lead to a variety of
diseases, such as mitochondrial heart disease, mitochondrial
encephalopathy, mitochondrial myopathy and even various pathologies
of the reproductive and respiratory systems. Therefore, the
accurate detection of mitochondrial abnormalities is essential for
clinical guidance.
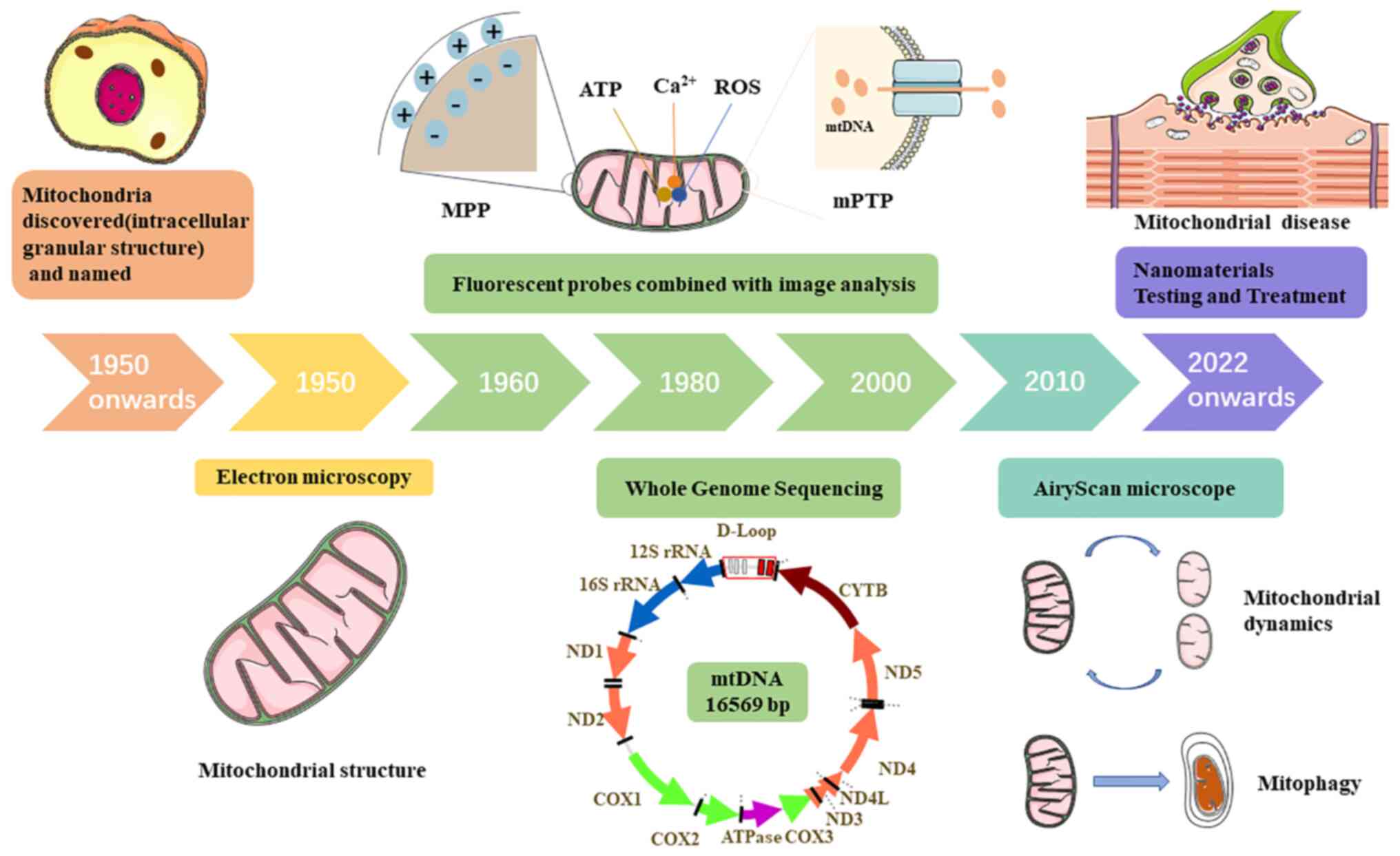
Since the beginning of the last century, a number
of methods for mitochondrial research have been developed (Fig. 3), from the discovery of
mitochondria as intracellular granular structures to the
observation of mitochondrial microstructures via EM and the use of
fluorescent probes to detect physiological indicators within
mitochondria. The application of these methods has provided
theoretical foundations for the detection and treatment of
mitochondrial diseases. Accordingly, the treatment of mitochondrial
diseases has gradually evolved from drug-based therapy to
multidisciplinary combination therapies, such as the use of
nanomaterials to precisely transport therapeutic drugs into
mitochondria for targeted drug delivery, substantially improving
therapeutic efficiency. However, the methods by which therapeutic
efficacy is achieved still warrant investigation. The combined
application of biomedicine and material science may be a promising
means of detection and treatment. Notably, the specific molecular
mechanism underlying the pathogenesis of the mitochondrial disease
remains unclear. Current monitoring and treatment strategies cannot
completely cure mitochondrial disease but only alleviate symptoms
or slow disease progression. Therefore, methods for detection and
treatment that are specific to the molecular mechanisms are needed.
Using multi-omics and artificial intelligence, artificial
mitochondrial models can be established through molecular
co-assembly technology and mitochondria-targeted drugs can be
screened to conduct in-depth discussions on abnormal mitochondria,
which may elucidate the pathogenesis of mitochondrial diseases at
the molecular level and provide new treatments for mitochondrial
diseases.
Data sharing is not applicable to this article, as
no data sets were generated or analyzed during the current
study.
YY wrote the first draft of this review. HS
provided valuable comments on this first draft. Both authors read
and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the Foundation of Liaoning
Education Department (grant no. JCZR2020014), the Liaoning Province
Key R&D Program (grant no. 2020JH2/10300141) and the 345 Talent
Project of Shengjing Hospital.
|
1
|
Akbari M, Kirkwood TBL and Bohr VA:
Mitochondria in the signaling pathways that control longevity and
health span. Ageing Res Rev. 54:1009402019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bock FJ and Tait SWG: Mitochondria as
multifaceted regulators of cell death. Nat Rev Mol Cell Biol.
21:85–100. 2020. View Article : Google Scholar
|
|
3
|
Chakrabarty RP and Chandel NS:
Mitochondria as signaling organelles control mammalian stem cell
fate. Cell Stem Cell. 28:394–408. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hood DA, Memme JM, Oliveira AN and Triolo
M: Maintenance of skeletal muscle mitochondria in health, exercise,
and aging. Annu Rev Physiol. 81:19–41. 2019. View Article : Google Scholar
|
|
5
|
Li L, Conradson DM, Bharat V, Kim MJ,
Hsieh CH, Minhas PS, Papakyrikos AM, Durairaj AS, Ludlam A,
Andreasson KI, et al: A mitochondrial membrane-bridging machinery
mediates signal transduction of intramitochondrial oxidation. Nat
Metab. 3:1242–1258. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martínez-Reyes I and Chandel NS:
Mitochondrial TCA cycle metabolites control physiology and disease.
Nat Commun. 11:1022020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim Hong HT, Bich Phuong TT, Thu Thuy NT,
Wheatley MD and Cushman JC: Simultaneous chloroplast, mitochondria
isolation and mitochondrial protein preparation for two-dimensional
electrophoresis analysis of ice plant leaves under well watered and
water-deficit stressed treatments. Protein Expr Purif. 155:86–94.
2019. View Article : Google Scholar
|
|
8
|
Boussardon C and Keech O: Cell
type-specific isolation of mitochondria in Arabidopsis. Methods Mol
Biol. 2363:13–23. 2022. View Article : Google Scholar
|
|
9
|
Elekofehinti OO, Kamdem JP, Saliu TP,
Famusiwa CD, Boligon A and Teixeira Rocha JB: Improvement of
mitochondrial function by Tapinanthus globifer (A.Rich.) Tiegh.
Against hepatotoxic agent in isolated rat's liver mitochondria. J
Ethnopharmacol. 242:1120262019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gäbelein CG, Feng Q, Sarajlic E, Zambelli
T, Guillaume-Gentil O, Kornmann B and Vorholt JA: Mitochondria
transplantation between living cells. PLoS Biol. 20:e30015762022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee D, Lee YH, Lee KH, Lee BS, Alishir A,
Ko YJ, Kang KS and Kim KH: Aviculin isolated from lespedeza cuneata
induce apoptosis in breast cancer cells through
mitochondria-mediated caspase activation pathway. Molecules.
25:17082020. View Article : Google Scholar :
|
|
12
|
Léger JL, Jougleux JL, Savadogo F, Pichaud
N and Boudreau LH: Rapid isolation and purification of functional
platelet mitochondria using a discontinuous percoll gradient.
Platelets. 31:258–264. 2020. View Article : Google Scholar
|
|
13
|
Léger JL, Pichaud N and Boudreau LH:
Purification of functional platelet mitochondria using a
discontinuous percoll gradient. Methods Mol Biol. 2276:57–66. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liao PC, Bergamini C, Fato R, Pon LA and
Pallotti F: Isolation of mitochondria from cells and tissues.
Methods Cell Biol. 155:3–31. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin YT, Chen ST, Chang JC, Teoh RJ, Liu CS
and Wang GJ: Green extraction of healthy and additive free
mitochondria with a conventional centrifuge. Lab Chip.
19:3862–3869. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Long Q, Huang L, Huang K and Yang Q:
Assessing mitochondrial bioenergetics in isolated mitochondria from
mouse heart tissues using oroboros 2k-oxygraph. Methods Mol Biol.
1966:237–246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rahman MH, Xiao Q, Zhao S, Wei AC and Ho
YP: Extraction of functional mitochondria based on membrane
stiffness. Methods Mol Biol. 2276:343–355. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramezani M, Samiei F and Pourahmad J:
Anti-glioma effect of pseudosynanceia melanostigma venom on
isolated mitochondria from glioblastoma cells. Asian Pac J Cancer
Prev. 22:2295–2302. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruzzenente B and Metodiev MD: Linear
density sucrose gradients to study mitoribosomal biogenesis in
tissue-specific knockout mice. Methods Mol Biol. 2224:47–60. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Cao L, Li Y, Liu H, Zhang M, Ma H,
Wang B, Yuan X and Liu Q: Gracillin isolated from reineckia carnea
induces apoptosis of A549 cells via the mitochondrial pathway. Drug
Des Devel Ther. 15:233–243. 2021. View Article : Google Scholar :
|
|
21
|
Chandra K, Kumar V, Werner SE and Odom TW:
Separation of stabilized MOPS gold nanostars by density gradient
centrifugation. ACS Omega. 2:4878–4884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen BY, Sung CW, Chen C, Cheng CM, Lin
DP, Huang CT and Hsu MY: Advances in exosomes technology. Clin Chim
Acta. 493:14–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Écija-Arenas Á, Román-Pizarro V and
Fernández-Romero JM: Luminescence continuous flow system for
monitoring the efficiency of hybrid liposomes separation using
multiphase density gradient centrifugation. Talanta.
222:1215322021. View Article : Google Scholar
|
|
24
|
Hu P, Fabyanic E, Kwon DY, Tang S, Zhou Z
and Wu H: Dissecting cell-type composition and activity-dependent
transcriptional state in mammalian brains by massively parallel
single-nucleus RNA-Seq. Mol Cell. 68:1006–1015.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jerri HA, Sheehan WP, Snyder CE and
Velegol D: Prolonging density gradient stability. Langmuir.
26:4725–4731. 2010. View Article : Google Scholar
|
|
26
|
Johnson ME, Montoro Bustos AR and
Winchester MR: Practical utilization of spICP-MS to study sucrose
density gradient centrifugation for the separation of
nanoparticles. Anal Bioanal Chem. 408:7629–7640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pužar Dominkuš P, Stenovec M, Sitar S,
Lasič E, Zorec R, Plemenitaš A, Žagar E, Kreft M and Lenassi M:
PKH26 labeling of extracellular vesicles: Characterization and
cellular internalization of contaminating PKH26 nanoparticles.
Biochim Biophys Acta Biomembr. 1860:1350–1361. 2018. View Article : Google Scholar
|
|
28
|
Wang J, Shen T, Huang X, Kumar GR, Chen X,
Zeng Z, Zhang R, Chen R, Li T, Zhang T, et al: Serum hepatitis B
virus RNA is encapsidated pregenome RNA that may be associated with
persistence of viral infection and rebound. J Hepatol. 65:700–710.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sugiura A, Nagashima S, Tokuyama T, Amo T,
Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, et
al: MITOL regulates endoplasmic reticulum-mitochondria contacts via
Mitofusin2. Mol Cell. 51:20–34. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong B, Cheng J, Qiao Y, Zhou R, He Y and
Yeung ES: Separation of nanorods by density gradient
centrifugation. J Chromatogr A. 1218:3823–3829. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng X, Xu K, Zhou B, Chen T, Huang Y, Li
Q, Wen F, Ge W, Wang J, Yu S, et al: A circulating extracellular
vesicles-based novel screening tool for colorectal cancer revealed
by shotgun and data-independent acquisition mass spectrometry. J
Extracell Vesicles. 9:17502022020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu J, Liu B, Wang Z, Wang D, Ni H, Zhang
L and Wang Y: Exosomes from nicotine-stimulated macrophages
accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC
migration and proliferation. Theranostics. 9:6901–6919. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qattan AT, Mulvey C, Crawford M, Natale DA
and Godovac-Zimmermann J: Quantitative organelle proteomics of
MCF-7 breast cancer cells reveals multiple subcellular locations
for proteins in cellular functional processes. J Proteome Res.
9:495–508. 2010. View Article : Google Scholar
|
|
34
|
Hassani M, Hellebrekers P, Chen N, van
Aalst C, Bongers S, Hietbrink F, Koenderman L and Vrisekoop N: On
the origin of low-density neutrophils. J Leukoc Biol. 107:809–818.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi W, Wang Y, Zhang C, Jin H, Zeng Z, Wei
L, Tian Y, Zhang D and Sun G: Isolation and purification of immune
cells from the liver. Int Immunopharmacol. 85:1066322020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grist TM, Canon CL, Fishman EK, Kohi MP
and Mossa-Basha M: Short-, mid-, and long-term strategies to manage
the shortage of iohexol. Radiology. 304:289–293. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang S, Su M, Liu B, Liu R, Zheng H, Qiu
W and Zhang Z: Evaluation of blood induced influence for
high-definition intravascular ultrasound (HD-IVUS). IEEE Trans
Ultrason Ferroelectr Freq Control. 69:98–105. 2022. View Article : Google Scholar
|
|
38
|
Warwick J and Holness J: Measurement of
glomerular filtration rate. Semin Nucl Med. 52:453–466. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elgamal S, Cocucci E, Sass EJ, Mo XM,
Blissett AR, Calomeni EP, Rogers KA, Woyach JA, Bhat SA, Muthusamy
N, et al: Optimizing extracellular vesicles' isolation from chronic
lymphocytic leukemia patient plasma and cell line supernatant. JCI
Insight. 6:e1379372021. View Article : Google Scholar
|
|
40
|
Inoue T, Kusumoto S, Iio E, Ogawa S,
Suzuki T, Yagi S, Kaneko A, Matsuura K, Aoyagi K and Tanaka Y:
Clinical efficacy of a novel, high-sensitivity HBcrAg assay in the
management of chronic hepatitis B and HBV reactivation. J Hepatol.
75:302–310. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tóth EÁ, Turiák L, Visnovitz T, Cserép C,
Mázló A, Sódar BW, Försönits AI, Petővári G, Sebestyén A, Komlósi
Z, et al: Formation of a protein corona on the surface of
extracellular vesicles in blood plasma. J Extracell Vesicles.
10:e121402021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Veerman RE, Teeuwen L, Czarnewski P,
Güclüler Akpinar G, Sandberg A, Cao X, Pernemalm M, Orre LM,
Gabrielsson S and Eldh M: Molecular evaluation of five different
isolation methods for extracellular vesicles reveals different
clinical applicability and subcellular origin. J Extracell
Vesicles. 10:e121282021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cartuche L, Reyes-Batlle M, Sifaoui I,
Arberas-Jiménez I, Piñero JE, Fernández JJ, Lorenzo-Morales J and
Díaz-Marrero AR: Antiamoebic activities of indolocarbazole
metabolites isolated from streptomyces sanyensis cultures. Mar
Drugs. 17:5882019. View Article : Google Scholar :
|
|
44
|
Jiang S, Zhang E, Ruan H, Ma J, Zhao X,
Zhu Y, Xiu X, Han N, Li J, Zhang H, et al: Actinomycin V induces
apoptosis associated with mitochondrial and PI3K/AKT pathways in
human CRC cells. Mar Drugs. 19:5992021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li K, Liang Z, Chen W, Luo X, Fang W, Liao
S, Lin X, Yang B, Wang J, Tang L, et al: Iakyricidins A-D,
antiproliferative piericidin analogues bearing a carbonyl group or
cyclic skeleton from streptomyces iakyrus SCSIO NS104. J Org Chem.
84:12626–12631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu L, Zhu H, Wu W, Shen Y, Lin X, Wu Y,
Liu L, Tang J, Zhou Y, Sun F and Lin HW: Neoantimycin F, a
streptomyces-derived natural product induces mitochondria-related
apoptotic death in human non-small cell lung cancer cells. Front
Pharmacol. 10:10422019. View Article : Google Scholar :
|
|
47
|
Rawat PS, Jaiswal A, Khurana A, Bhatti JS
and Navik U: Doxorubicin-induced cardiotoxicity: An update on the
molecular mechanism and novel therapeutic strategies for effective
management. Biomed Pharmacother. 139:1117082021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal
T, Junger W, Brohi K, Itagaki K and Hauser CJ: Circulating
mitochondrial DAMPs cause inflammatory responses to injury. Nature.
464:104–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Deng X, Liu J, Liu L, Sun X, Huang J and
Dong J: Drp1-mediated mitochondrial fission contributes to
baicalein-induced apoptosis and autophagy in lung cancer via
activation of AMPK signaling pathway. Int J Biol Sci. 16:1403–1416.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ma ZJ, Lu L, Yang JJ, Wang XX, Su G, Wang
ZL, Chen GH, Sun HM, Wang MY and Yang Y: Lariciresinol induces
apoptosis in HepG2 cells via mitochondrial-mediated apoptosis
pathway. Eur J Pharmacol. 821:1–10. 2018. View Article : Google Scholar
|
|
51
|
Ke H, Dass S, Morrisey JM, Mather MW and
Vaidya AB: The mitochondrial ribosomal protein L13 is critical for
the structural and functional integrity of the mitochondrion in
plasmodium falciparum. J Biol Chem. 293:8128–8137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Galvan DL, Green NH and Danesh FR: The
hallmarks of mitochondrial dysfunction in chronic kidney disease.
Kidney Int. 92:1051–1057. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wiemerslage L and Lee D: Quantification of
mitochondrial morphology in neurites of dopaminergic neurons using
multiple parameters. J Neurosci Methods. 262:56–65. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Labarta E, de Los Santos MJ, Escribá MJ,
Pellicer A and Herraiz S: Mitochondria as a tool for oocyte
rejuvenation. Fertil Steril. 111:219–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li WQ, Wang Z, Hao S, He H, Wan Y, Zhu C,
Sun LP, Cheng G and Zheng SY: Mitochondria-targeting polydopamine
nanoparticles to deliver doxorubicin for overcoming drug
resistance. ACS Appl Mater Interfaces. 9:16793–16802. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin Y, Liu J, Bai R, Shi J, Zhu X, Liu J,
Guo J, Zhang W, Liu H and Liu Z: Mitochondria-inspired
nanoparticles with microenvironment-adapting capacities for
on-demand drug delivery after ischemic injury. ACS Nano.
14:11846–11859. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Smith GM and Gallo G: The role of
mitochondria in axon development and regeneration. Dev Neurobiol.
78:221–237. 2018. View Article : Google Scholar :
|
|
58
|
Bastian C, Day J, Politano S, Quinn J,
Brunet S and Baltan S: Preserving mitochondrial structure and
motility promotes recovery of white matter after ischemia.
Neuromolecular Med. 21:484–492. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bhargava P and Schnellmann RG:
Mitochondrial energetics in the kidney. Nat Rev Nephrol.
13:629–646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Granata C, Jamnick NA and Bishop DJ:
Training-induced changes in mitochondrial content and respiratory
function in human skeletal muscle. Sports Med. 48:1809–1828. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hammond K, Ryadnov MG and Hoogenboom BW:
Atomic force microscopy to elucidate how peptides disrupt
membranes. Biochim Biophys Acta Biomembr. 1863:1834472021.
View Article : Google Scholar
|
|
62
|
Heath GR, Kots E, Robertson JL, Lansky S,
Khelashvili G, Weinstein H and Scheuring S: Localization atomic
force microscopy. Nature. 594:385–390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Müller DJ, Dumitru AC, Lo Giudice C, Gaub
HE, Hinterdorfer P, Hummer G, De Yoreo JJ, Dufrêne YF and Alsteens
D: Atomic force microscopy-based force spectroscopy and
multiparametric imaging of biomolecular and cellular systems. Chem
Rev. 121:11701–11725. 2021. View Article : Google Scholar
|
|
64
|
Vogt N: Atomic force microscopy in
super-resolution. Nat Methods. 18:8592021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kolossov VL, Sivaguru M, Huff J, Luby K,
Kanakaraju K and Gaskins HR: Airyscan super-resolution microscopy
of mitochondrial morphology and dynamics in living tumor cells.
Microsc Res Tech. 81:115–128. 2018. View Article : Google Scholar
|
|
66
|
Rocha EM, De Miranda B and Sanders LH:
Alpha-synuclein: Pathology, mitochondrial dysfunction and
neuroinflammation in Parkinson's disease. Neurobiol Dis.
109:249–257. 2018. View Article : Google Scholar
|
|
67
|
Szymański J, Janikiewicz J, Michalska B,
Patalas-Krawczyk P, Perrone M, Ziółkowski W, Duszyński J, Pinton P,
Dobrzyń A and Więckowski MR: Interaction of mitochondria with the
endoplasmic reticulum and plasma membrane in calcium homeostasis,
lipid trafficking and mitochondrial structure. Int J Mol Sci.
18:15762017. View Article : Google Scholar
|
|
68
|
Adam N, Beattie TL and Riabowol K:
Fluorescence microscopy methods for examining telomeres during cell
aging. Ageing Res Rev. 68:1013202021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huang L, Chen H, Luo Y, Rivenson Y and
Ozcan A: Recurrent neural network-based volumetric fluorescence
microscopy. Light Sci Appl. 10:622021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Thiele JC, Helmerich DA, Oleksiievets N,
Tsukanov R, Butkevich E, Sauer M, Nevskyi O and Enderlein J:
Confocal fluorescence-lifetime single-molecule localization
microscopy. ACS Nano. 14:14190–14200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang Y, Zong H, Zong C, Tan Y, Zhang M,
Zhan Y and Cheng JX: Fluorescence-detected mid-infrared
photothermal microscopy. J Am Chem Soc. 143:11490–11499. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Alexander JF, Seua AV, Arroyo LD, Ray PR,
Wangzhou A, Heiβ-Lückemann L, Schedlowski M, Price TJ, Kavelaars A
and Heijnen CJ: Nasal administration of mitochondria reverses
chemotherapy-induced cognitive deficits. Theranostics.
11:3109–3130. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Dumitru AC, Stommen A, Koehler M, Cloos
AS, Yang J, Leclercqz A, Tyteca D and Alsteens D: Probing PIEZO1
localization upon activation using high-resolution atomic force and
confocal microscopy. Nano Lett. 21:4950–4958. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wu Y, Han X, Su Y, Glidewell M, Daniels
JS, Liu J, Sengupta T, Rey-Suarez I, Fischer R, Patel A, et al:
Multiview confocal super-resolution microscopy. Nature.
600:279–284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yordanov S, Neuhaus K, Hartmann R,
Díaz-Pascual F, Vidakovic L, Singh PK and Drescher K:
Single-objective high-resolution confocal light sheet fluorescence
microscopy for standard biological sample geometries. Biomed Opt
Express. 12:3372–3391. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhao Y, Raghuram A, Kim HK, Hielscher AH,
Robinson JT and Veeraraghavan A: High resolution, deep imaging
using confocal time-of-flight diffuse optical tomography. IEEE
Trans Pattern Anal Mach Intell. 43:2206–2219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Dalecká M, Sabó J, Backová L, Rösel D,
Brábek J, Benda A and Tolde O: Invadopodia structure in 3D
environment resolved by near-infrared branding protocol combining
correlative confocal and FIB-SEM microscopy. Int J Mol Sci.
22:78052021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Guo R, Barnea I and Shaked NT:
Limited-angle tomographic phase microscopy utilizing confocal
scanning fluorescence microscopy. Biomed Opt Express. 12:1869–1881.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lamers MM, van der Vaart J, Knoops K,
Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D,
Bezstarosti K, Koopman CD, et al: An organoid-derived
bronchioalveolar model for SARS-CoV-2 infection of human alveolar
type II-like cells. EMBO J. 40:e1059122021. View Article : Google Scholar
|
|
80
|
Messal HA, Almagro J, Zaw Thin M, Tedeschi
A, Ciccarelli A, Blackie L, Anderson KI, Miguel-Aliaga I, van
Rheenen J and Behrens A: Antigen retrieval and clearing for
whole-organ immunofluorescence by FLASH. Nat Protoc. 16:239–262.
2021. View Article : Google Scholar
|
|
81
|
Miyashita L, Foley G, Gill I, Gillmore G,
Grigg J and Wertheim D: Confocal microscopy 3D imaging of diesel
particulate matter. Environ Sci Pollut Res Int. 28:30384–30389.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Restall BS, Kedarisetti P, Haven NJM,
Martell MT and Zemp RJ: Multimodal 3D photoacoustic remote sensing
and confocal fluorescence microscopy imaging. J Biomed Opt.
26:0965012021. View Article : Google Scholar :
|
|
83
|
Rodriguez-Gallardo S, Kurokawa K,
Sabido-Bozo S, Cortes-Gomez A, Perez-Linero AM, Aguilera-Romero A,
Lopez S, Waga M, Nakano A and Muñiz M: Assay for dual cargo sorting
into endoplasmic reticulum exit sites imaged by 3D super-resolution
confocal live imaging microscopy (SCLIM). PLoS One.
16:e02581112021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Durand MJ, Ait-Aissa K, Levchenko V,
Staruschenko A, Gutterman DD and Beyer AM: Visualization and
quantification of mitochondrial structure in the endothelium of
intact arteries. Cardiovasc Res. 115:1546–1556. 2019. View Article : Google Scholar :
|
|
85
|
Bartolák-Suki E and Suki B: Tuning
mitochondrial structure and function to criticality by
fluctuation-driven mechanotransduction. Sci Rep. 10:4072020.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chandhok G, Lazarou M and Neumann B:
Structure, function, and regulation of mitofusin-2 in health and
disease. Biol Rev Camb Philos Soc. 93:933–949. 2018. View Article : Google Scholar
|
|
87
|
Kowaltowski AJ, Menezes-Filho SL, Assali
EA, Gonçalves IG, Cabral-Costa JV, Abreu P, Miller N, Nolasco P,
Laurindo FRM, Bruni-Cardoso A and Shirihai OS: Mitochondrial
morphology regulates organellar Ca2+ uptake and changes
cellular Ca2+ homeostasis. FASEB J. 33:13176–13188.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Csordás G, Weaver D and Hajnóczky G:
Endoplasmic reticulum-mitochondrial contactology: Structure and
signaling functions. Trends Cell Biol. 28:523–540. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xie LL, Shi F, Tan Z, Li Y, Bode AM and
Cao Y: Mitochondrial network structure homeostasis and cell death.
Cancer Sci. 109:3686–3694. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Correia-Álvarez E, Keating JE, Glish G,
Tarran R and Sassano MF: Reactive oxygen species, mitochondrial
membrane potential, and cellular membrane potential are predictors
of E-liquid induced cellular toxicity. Nicotine Tob Res. 22(Suppl
1): S4–S13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhou Y, Long Q, Wu H, Li W, Qi J, Wu Y,
Xiang G, Tang H, Yang L, Chen K, et al: Topology-dependent,
bifurcated mitochondrial quality control under starvation.
Autophagy. 16:562–574. 2020. View Article : Google Scholar :
|
|
92
|
Du R, Bei H, Jia L, Huang C, Chen Q, Wang
J, Wu F, Chen J and Bo H: A low-cost, accurate method for detecting
reticulocytes at different maturation stages based on changes in
the mitochondrial membrane potential. J Pharmacol Toxicol Methods.
101:1066642020. View Article : Google Scholar
|
|
93
|
Ganta KK, Mandal A and Chaubey B:
Depolarization of mitochondrial membrane potential is the initial
event in non-nucleoside reverse transcriptase inhibitor efavirenz
induced cytotoxicity. Cell Biol Toxicol. 33:69–82. 2017. View Article : Google Scholar
|
|
94
|
Dreier DA, Denslow ND and Martyniuk CJ:
Computational in vitro toxicology uncovers chemical structures
impairing mitochondrial membrane potential. J Chem Inf Model.
59:702–712. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lee JY, Lim W, Ham J, Kim J, You S and
Song G: Ivermectin induces apoptosis of porcine trophectoderm and
uterine luminal epithelial cells through loss of mitochondrial
membrane potential, mitochondrial calcium ion overload, and
reactive oxygen species generation. Pestic Biochem Physiol.
159:144–153. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tao L, Liu X, Da W, Tao Z and Zhu Y:
Pycnogenol achieves neuroprotective effects in rats with spinal
cord injury by stabilizing the mitochondrial membrane potential.
Neurol Res. 42:597–604. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Haider SZ, Mohanraj N, Markandeya YS,
Joshi PG and Mehta B: Picture perfect: Imaging mitochondrial
membrane potential changes in retina slices with minimal stray
fluorescence. Exp Eye Res. 202:1083182021. View Article : Google Scholar
|
|
98
|
Zhang G, Yang W, Zou P, Jiang F, Zeng Y,
Chen Q, Sun L, Yang H, Zhou N, Wang X, et al: Mitochondrial
functionality modifies human sperm acrosin activity, acrosome
reaction capability and chromatin integrity. Hum Reprod. 34:3–11.
2019. View Article : Google Scholar
|
|
99
|
Sakthivel R, Malar DS and Devi KP: Phytol
shows anti-angiogenic activity and induces apoptosis in A549 cells
by depolarizing the mitochondrial membrane potential. Biomed
Pharmacother. 105:742–752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Alyasin A, Momeni HR and Mahdieh M:
Aquaporin3 expression and the potential role of aquaporins in
motility and mitochondrial membrane potential in human spermatozoa.
Andrologia. 52:e135882020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Alpert NM, Guehl N, Ptaszek L,
Pelletier-Galarneau M, Ruskin J, Mansour MC, Wooten D, Ma C,
Takahashi K, Zhou Y, et al: Quantitative in vivo mapping of
myocardial mitochondrial membrane potential. PLoS One.
13:e01909682018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kuwahara Y, Roudkenar MH, Suzuki M,
Urushihara Y and Fukumoto M, Saito Y and Fukumoto M: The
Involvement of mitochondrial membrane potential in cross-resistance
between radiation and docetaxel. Int J Radiat Oncol Biol Phys.
96:556–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Marcondes NA, Terra SR, Lasta CS, Hlavac
NRC, Dalmolin ML, Lacerda LA, Faulhaber GAM and González FHD:
Comparison of JC-1 and MitoTracker probes for mitochondrial
viability assessment in stored canine platelet concentrates: A flow
cytometry study. Cytometry A. 95:214–218. 2019. View Article : Google Scholar
|
|
104
|
Poznanski RR, Cacha LA, Ali J, Rizvi ZH,
Yupapin P, Salleh SH and Bandyopadhyay A: Induced mitochondrial
membrane potential for modeling solitonic conduction of
electrotonic signals. PLoS One. 12:e01836772017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Georgakopoulos ND, Wells G and Campanella
M: The pharmacological regulation of cellular mitophagy. Nat Chem
Biol. 13:136–146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bikas A, Jensen K, Patel A, Costello J,
Kaltsas G, Hoperia V, Wartofsky L, Burman K and Vasko V: Mitotane
induces mitochondrial membrane depolarization and apoptosis in
thyroid cancer cells. Int J Oncol. 55:7–20. 2019.PubMed/NCBI
|
|
107
|
Gloria A, Wegher L, Carluccio A, Valorz C,
Robbe D and Contri A: Factors affecting staining to discriminate
between bull sperm with greater and lesser mitochondrial membrane
potential. Anim Reprod Sci. 189:51–59. 2018. View Article : Google Scholar
|
|
108
|
Saraf KK, Kumaresan A, Chhillar S, Nayak
S, Lathika S, Datta TK, Gahlot SC, Karan P, Verma K and Mohanty TK:
Spermatozoa with high mitochondrial membrane potential and low
tyrosine phosphorylation preferentially bind to oviduct explants in
the water buffalo (Bubalus bubalis). Anim Reprod Sci. 180:30–36.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Cano M, Datta S, Wang L, Liu T,
Flores-Bellver M, Sachdeva M, Sinha D and Handa JT: Nrf2 deficiency
decreases NADPH from impaired IDH shuttle and pentose phosphate
pathway in retinal pigmented epithelial cells to magnify oxidative
stress-induced mitochondrial dysfunction. Aging Cell.
20:e134442021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
El Manaa W, Duplan E, Goiran T, Lauritzen
I, Vaillant Beuchot L, Lacas-Gervais S, Morais VA, You H, Qi L and
Salazar M: et al Transcription- and phosphorylation-dependent
control of a functional interplay between XBP1s and PINK1 governs
mitophagy and potentially impacts Parkinson disease
pathophysiology. Autophagy. 17:4363–4385. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Franco-Iborra S, Plaza-Zabala A, Montpeyo
M, Sebastian D, Vila M and Martinez-Vicente M: Mutant HTT
(huntingtin) impairs mitophagy in a cellular model of Huntington
disease. Autophagy. 17:672–689. 2021. View Article : Google Scholar :
|
|
112
|
Hamilton K, Krause K, Badr A, Daily K,
Estfanous S, Eltobgy M, Khweek AA, Anne MNK, Carafice C, Baetzhold
D, et al: Defective immunometabolism pathways in cystic fibrosis
macrophages. J Cyst Fibros. 20:664–672. 2021. View Article : Google Scholar :
|
|
113
|
Rabinovich-Nikitin I, Rasouli M, Reitz CJ,
Posen I, Margulets V, Dhingra R, Khatua TN, Thliveris JA, Martino
TA and Kirshenbaum LA: Mitochondrial autophagy and cell survival is
regulated by the circadian clock gene in cardiac myocytes during
ischemic stress. Autophagy. 17:3794–3812. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Rovini A, Heslop K, Hunt EG, Morris ME,
Fang D, Gooz M, Gerencser AA and Maldonado EN: Quantitative
analysis of mitochondrial membrane potential heterogeneity in
unsynchronized and synchronized cancer cells. FASEB J.
35:e211482021. View Article : Google Scholar
|
|
115
|
Samuvel DJ, Li L, Krishnasamy Y, Gooz M,
Takemoto K, Woster PM, Lemasters JJ and Zhong Z: Mitochondrial
depolarization after acute ethanol treatment drives mitophagy in
living mice. Autophagy. 1–15. 2022.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Wang Q and Hutt KJ: Evaluation of
mitochondria in mouse oocytes following cisplatin exposure. J
Ovarian Res. 14:652021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yazdankhah M, Ghosh S, Shang P, Stepicheva
N, Hose S, Liu H, Chamling X, Tian S, Sullivan MLG, Calderon MJ, et
al: BNIP3L-mediated mitophagy is required for mitochondrial
remodeling during the differentiation of optic nerve
oligodendrocytes. Autophagy. 17:3140–3159. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Young VC and Artigas P: Displacement of
the Na+/K+ pump's transmembrane domains
demonstrates conserved conformational changes in P-type 2 ATPases.
Proc Natl Acad Sci USA. 118:e20193171182021. View Article : Google Scholar
|
|
119
|
Cui Y, Duan W, Jin Y, Wo F, Xi F and Wu J:
Graphene quantum dot-decorated luminescent porous silicon dressing
for theranostics of diabetic wounds. Acta Biomater. 131:544–554.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kambe Y and Yamaoka T: Initial immune
response to a FRET-based MMP sensor-immobilized silk fibroin
hydrogel in vivo. Acta Biomater. 130:199–210. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Feng R, Guo L, Fang J, Jia Y, Wang X, Wei
Q and Yu X: Construction of the FRET pairs for the visualization of
mitochondria membrane potential in dual emission colors. Anal Chem.
91:3704–3709. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lee H, Kim SJ, Shin H and Kim YP:
Collagen-immobilized extracellular FRET reporter for visualizing
protease activity secreted by living cells. ACS Sens. 5:655–664.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Liu L, Chu H, Yang J, Sun Y, Ma P and Song
D: Construction of a magnetic-fluorescent-plasmonic nanosensor for
the determination of MMP-2 activity based on SERS-fluorescence
dual-mode signals. Biosens Bioelectron. 212:1143892022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhan Y, Ling S, Huang H, Zhang Y, Chen G,
Huang S, Li C, Guo W and Wang Q: Rapid unperturbed-tissue analysis
for intraoperative cancer diagnosis using an enzyme-activated
NIR-II nanoprobe. Angew Chem Int Ed Engl. 60:2637–2642. 2021.
View Article : Google Scholar
|
|
125
|
Wang C, Wang G, Li X, Wang K, Fan J, Jiang
K, Guo Y and Zhang H: Highly sensitive fluorescence molecular
switch for the ratio monitoring of trace change of mitochondrial
membrane potential. Anal Chem. 89:11514–11519. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Rao M, Jaber BL and Balakrishnan VS:
Chronic kidney disease and acquired mitochondrial myopathy. Curr
Opin Nephrol Hypertens. 27:113–120. 2018. View Article : Google Scholar
|
|
127
|
Zhu SC, Chen C, Wu YN, Ahmed M, Kitmitto
A, Greenstein AS, Kim SJ, Shao YF and Zhang YH: Cardiac complex II
activity is enhanced by fat and mediates greater mitochondrial
oxygen consumption following hypoxic re-oxygenation. Pflugers Arch.
472:367–374. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kurhaluk N, Lukash O, Nosar V,
Portnychenko A, Portnichenko V, Wszedybyl-Winklewska M and
Winklewski PJ: Liver mitochondrial respiratory plasticity and
oxygen uptake evoked by cobalt chloride in rats with low and high
resistance to extreme hypobaric hypoxia. Can J Physiol Pharmacol.
97:392–399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Acetoze G, Champagne J, Ramsey JJ and
Rossow HA: Liver mitochondrial oxygen consumption and efficiency of
milk production in lactating Holstein cows supplemented with
copper, manganese and zinc. J Anim Physiol Anim Nutr (Berl).
102:e787–e797. 2018. View Article : Google Scholar
|
|
130
|
Kalyanaraman B, Cheng G, Hardy M, Ouari O,
Lopez M, Joseph J, Zielonka J and Dwinell MB: A review of the
basics of mitochondrial bioenergetics, metabolism, and related
signaling pathways in cancer cells: Therapeutic targeting of tumor
mitochondria with lipophilic cationic compounds. Redox Biol.
14:316–327. 2018. View Article : Google Scholar
|
|
131
|
Banh RS, Iorio C, Marcotte R, Xu Y,
Cojocari D, Rahman AA, Pawling J, Zhang W, Sinha A, Rose CM, et al:
PTP1B controls non-mitochondrial oxygen consumption by regulating
RNF213 to promote tumour survival during hypoxia. Nat Cell Biol.
18:803–813. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Campos JC, Queliconi BB, Bozi LHM, Bechara
LRG, Dourado PMM, Andres AM, Jannig PR, Gomes KMS, Zambelli VO,
Rocha-Resende C, et al: Exercise reestablishes autophagic flux and
mitochondrial quality control in heart failure. Autophagy.
13:1304–1317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Rossow HA, Acetoze G, Champagne J and
Ramsey JJ: Measuring liver mitochondrial oxygen consumption and
proton leak kinetics to estimate mitochondrial respiration in
holstein dairy cattle. J Vis Exp. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Morimoto N, Hashimoto S, Yamanaka M,
Nakano T, Satoh M, Nakaoka Y, Iwata H, Fukui A, Morimoto Y and
Shibahara H: Mitochondrial oxygen consumption rate of human embryos
declines with maternal age. J Assist Reprod Genet. 37:1815–1821.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Darr CR, Cortopassi GA, Datta S, Varner DD
and Meyers SA: Mitochondrial oxygen consumption is a unique
indicator of stallion spermatozoal health and varies with
cryopreservation media. Theriogenology. 86:1382–1392. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Müller ME, Vikstrom S, König M,
Schlichting R, Zarfl C, Zwiener C and Escher BI: Mitochondrial
toxicity of selected micropollutants, their mixtures, and surface
water samples measured by the oxygen consumption rate in cells.
Environ Toxicol Chem. 38:1000–1011. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Thomas LW and Ashcroft M: Exploring the
molecular interface between hypoxia-inducible factor signalling and
mitochondria. Cell Mol Life Sci. 76:1759–1777. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Espinosa JA, Pohan G, Arkin MR and
Markossian S: Real-time assessment of mitochondrial toxicity in
HepG2 cells using the Seahorse extracellular flux analyzer. Curr
Protoc. 1:e752021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Fu Y, Wang D, Wang H, Cai M, Li C, Zhang
X, Chen H, Hu Y, Zhang X, Ying M, et al: TSPO deficiency induces
mitochondrial dysfunction, leading to hypoxia, angiogenesis, and a
growth-promoting metabolic shift toward glycolysis in glioblastoma.
Neuro Oncol. 22:240–252. 2020.
|
|
140
|
Gu X, Ma Y, Liu Y and Wan Q: Measurement
of mitochondrial respiration in adherent cells by Seahorse XF96
cell mito stress Test. STAR Protoc. 2:1002452021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Eagleson KL, Villaneuva M, Southern RM and
Levitt P: Proteomic and mitochondrial adaptations to early-life
stress are distinct in juveniles and adults. Neurobiol Stress.
13:1002512020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Maremanda KP, Sundar IK and Rahman I: Role
of inner mitochondrial protein OPA1 in mitochondrial dysfunction by
tobacco smoking and in the pathogenesis of COPD. Redox Biol.
45:1020552021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Nishida M, Yamashita N, Ogawa T, Koseki K,
Warabi E, Ohue T, Komatsu M, Matsushita H, Kakimi K, Kawakami E, et
al: Mitochondrial reactive oxygen species trigger
metformin-dependent antitumor immunity via activation of
Nrf2/mTORC1/p62 axis in tumor-infiltrating CD8T lymphocytes. J
Immunother Cancer. 9:e0029542021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Nishida Y, Nawaz A, Kado T, Takikawa A,
Igarashi Y, Onogi Y, Wada T, Sasaoka T, Yamamoto S, Sasahara M, et
al: Astaxanthin stimulates mitochondrial biogenesis in insulin
resistant muscle via activation of AMPK pathway. J Cachexia
Sarcopenia Muscle. 11:241–258. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Sabogal-Guáqueta AM, Hobbie F, Keerthi A,
Oun A, Kortholt A, Boddeke E and Dolga A: Linalool attenuates
oxidative stress and mitochondrial dysfunction mediated by
glutamate and NMDA toxicity. Biomed Pharmacother. 118:1092952019.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Tian T, Zhang Y, Wu T, Yang L, Chen C, Li
N, Li Y, Xu S, Fu Z, Cui X, et al: miRNA profiling in the
hippocampus of attention-deficit/hyperactivity disorder rats. J
Cell Biochem. 120:3621–3629. 2019. View Article : Google Scholar
|
|
147
|
Ooi K, Hu L, Feng Y, Han C, Ren X, Qian X,
Huang H, Chen S, Shi Q, Lin H, et al: Sigma-1 receptor activation
suppresses microglia M1 polarization via regulating endoplasmic
reticulum-mitochondria contact and mitochondrial functions in
stress-induced hypertension rats. Mol Neurobiol. 58:6625–6646.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Shetty T, Park B and Corson TW:
Measurement of mitochondrial respiration in the murine retina using
a Seahorse extracellular flux analyzer. STAR Protoc. 2:1005332021.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wang SH, Zhu XL, Wang F, Chen SX, Chen ZT,
Qiu Q, Liu WH, Wu MX, Deng BQ, Xie Y, et al: LncRNA H19 governs
mitophagy and restores mitochondrial respiration in the heart
through Pink1/Parkin signaling during obesity. Cell Death Dis.
12:5572021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Andersen JV, Jakobsen E, Waagepetersen HS
and Aldana BI: Distinct differences in rates of oxygen consumption
and ATP synthesis of regionally isolated non-synaptic mouse brain
mitochondria. J Neurosci Res. 97:961–974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Hubbard WB, Joseph B, Spry M, Vekaria HJ,
Saatman KE and Sullivan PG: Acute mitochondrial impairment
underlies prolonged cellular dysfunction after repeated mild
traumatic brain injuries. J Neurotrauma. 36:1252–1263. 2019.
View Article : Google Scholar
|
|
152
|
McAlpin BR, Mahalingam R, Singh AK,
Dharmaraj S, Chrisikos TT, Boukelmoune N, Kavelaars A and Heijnen
CJ: HDAC6 inhibition reverses long-term doxorubicin-induced
cognitive dysfunction by restoring microglia homeostasis and
synaptic integrity. Theranostics. 12:603–619. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Raut S, Patel R and Al-Ahmad AJ: Presence
of a mutation in PSEN1 or PSEN2 gene is associated with an impaired
brain endothelial cell phenotype in vitro. Fluids Barriers CNS.
18:32021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Algieri C, Trombetti F, Pagliarani A,
Ventrella V and Nesci S: The mitochondrial
F1FO -ATPase exploits the dithiol redox state
to modulate the permeability transition pore. Arch Biochem Biophys.
712:1090272021. View Article : Google Scholar
|
|
155
|
Sun C, Liu X, Wang B, Wang Z, Liu Y, Di C,
Si J, Li H, Wu Q, Xu D, et al: Endocytosis-mediated mitochondrial
transplantation: Transferring normal human astrocytic mitochondria
into glioma cells rescues aerobic respiration and enhances
radiosensitivity. Theranostics. 9:3595–3607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Sun JY, Zhao SJ, Wang HB, Hou YJ, Mi QJ,
Yang MF, Yuan H, Ni QB, Sun BL and Zhang ZY: Ifenprodil improves
long-term neurologic deficits through antagonizing
glutamate-induced excitotoxicity after experimental subarachnoid
hemorrhage. Transl Stroke Res. 12:1067–1080. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Boyman L, Karbowski M and Lederer WJ:
Regulation of mitochondrial ATP production: Ca2+
signaling and quality control. Trends Mol Med. 26:21–39. 2020.
View Article : Google Scholar
|
|
158
|
Bravo-Sagua R, Parra V, López-Crisosto C,
Díaz P, Quest AF and Lavandero S: Calcium transport and signaling
in mitochondria. Compr Physiol. 7:623–634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Marchi S, Patergnani S, Missiroli S,
Morciano G, Rimessi A, Wieckowski MR, Giorgi C and Pinton P:
Mitochondrial and endoplasmic reticulum calcium homeostasis and
cell death. Cell Calcium. 69:62–72. 2018. View Article : Google Scholar
|
|
160
|
Chow J, Rahman J, Achermann JC, Dattani MT
and Rahman S: Mitochondrial disease and endocrine dysfunction. Nat
Rev Endocrinol. 13:92–104. 2017. View Article : Google Scholar
|
|
161
|
Cieluch A, Uruska A and
Zozulinska-Ziolkiewicz D: Can we prevent mitochondrial dysfunction
and diabetic cardiomyopathy in type 1 diabetes mellitus?
Pathophysiology and treatment options. Int J Mol Sci. 21:28522020.
View Article : Google Scholar :
|
|
162
|
Ding XW, Robinson M, Li R, Aldhowayan H,
Geetha T and Babu JR: Mitochondrial dysfunction and beneficial
effects of mitochondria-targeted small peptide SS-31 in diabetes
mellitus and Alzheimer's disease. Pharmacol Res. 171:1057832021.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Fisher JJ, Vanderpeet CL, Bartho LA,
McKeating DR, Cuffe JSM, Holland OJ and Perkins AV: Mitochondrial
dysfunction in placental trophoblast cells experiencing gestational
diabetes mellitus. J Physiol. 599:1291–1305. 2021. View Article : Google Scholar
|
|
164
|
Jelenik T and Roden M: Mitochondrial
plasticity in obesity and diabetes mellitus. Antioxid Redox Signal.
19:258–268. 2013. View Article : Google Scholar :
|
|
165
|
Rovira-Llopis S, Bañuls C, Diaz-Morales N,
Hernandez-Mijares A, Rocha M and Victor VM: Mitochondrial dynamics
in type 2 diabetes: Pathophysiological implications. Redox Biol.
11:637–645. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Zhao H, Li T, Wang K, Zhao F, Chen J, Xu
G, Zhao J, Li T, Chen L, Li L, et al: AMPK-mediated activation of
MCU stimulates mitochondrial Ca2+ entry to promote
mitotic progression. Nat Cell Biol. 21:476–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Calvo-Rodriguez M, Hou SS, Snyder AC,
Kharitonova EK, Russ AN, Das S, Fan Z, Muzikansky A, Garcia-Alloza
M, Serrano-Pozo A, et al: Increased mitochondrial calcium levels
associated with neuronal death in a mouse model of Alzheimer's
disease. Nat Commun. 11:21462020. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Bhatti JS, Bhatti GK and Reddy PH:
Mitochondrial dysfunction and oxidative stress in metabolic
disorders-a step towards mitochondria based therapeutic strategies.
Biochim Biophys Acta Mol Basis Dis. 1863:1066–1077. 2017.
View Article : Google Scholar
|
|
169
|
Guo Q, Bi J, Wang H and Zhang X:
Mycobacterium tuberculosis ESX-1-secreted substrate protein EspC
promotes mycobacterial survival through endoplasmic reticulum
stress-mediated apoptosis. Emerg Microbes Infect. 10:19–36. 2021.
View Article : Google Scholar :
|
|
170
|
Galla L, Vajente N, Pendin D, Pizzo P,
Pozzan T and Greotti E: Generation and characterization of a new
FRET-Based Ca2+ sensor targeted to the nucleus. Int J
Mol Sci. 22:99452021. View Article : Google Scholar
|
|
171
|
Isshiki M, Nishimoto M, Mizuno R and
Fujita T: FRET-based sensor analysis reveals caveolae are spatially
distinct Ca2+ stores in endothelial cells. Cell Calcium.
54:395–403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Laskaratou D, Fernández GS, Coucke Q, Fron
E, Rocha S, Hofkens J, Hendrix J and Mizuno H: Quantification of
FRET-induced angular displacement by monitoring sensitized acceptor
anisotropy using a dim fluorescent donor. Nat Commun. 12:25412021.
View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Nagai T, Ibata K, Park ES, Kubota M,
Mikoshiba K and Miyawaki A: A variant of yellow fluorescent protein
with fast and efficient maturation for cell-biological
applications. Nat Biotechnol. 20:87–90. 2002. View Article : Google Scholar
|
|
174
|
Ucar H, Watanabe S, Noguchi J, Morimoto Y,
Iino Y, Yagishita S, Takahashi N and Kasai H: Mechanical actions of
dendritic-spine enlargement on presynaptic exocytosis. Nature.
600:686–689. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Yoon S, Pan Y, Shung K and Wang Y:
FRET-based Ca2+ biosensor single cell imaging interrogated by
high-frequency ultrasound. Sensors (Basel). 20. pp. 49982020,
View Article : Google Scholar
|
|
176
|
Chen J, Qiu M, Zhang S, Li B, Li D, Huang
X, Qian Z, Zhao J, Wang Z and Tang D: A calcium phosphate drug
carrier loading with 5-fluorouracil achieving a synergistic effect
for pancreatic cancer therapy. J Colloid Interface Sci.
605:263–273. 2022. View Article : Google Scholar
|
|
177
|
Fan Y and Simmen T: Mechanistic
connections between endoplasmic reticulum (ER) Redox Control And
Mitochondrial Metabolism. Cells. 8:10712019. View Article : Google Scholar :
|
|
178
|
Shoshan-Barmatz V, Nahon-Crystal E,
Shteinfer-Kuzmine A and Gupta R: VDAC1, mitochondrial dysfunction,
and Alzheimer's disease. Pharmacol Res. 131:87–101. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Country MW and Jonz MG: Mitochondrial KATP
channels stabilize intracellular Ca2+ during hypoxia in retinal
horizontal cells of goldfish (Carassius auratus). J Exp Biol.
224:jeb2426342021. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Davidson SM, Padró T, Bollini S, Vilahur
G, Duncker DJ, Evans PC, Guzik T, Hoefer IE, Waltenberger J, Wojta
J and Weber C: Progress in cardiac research: From rebooting cardiac
regeneration to a complete cell atlas of the heart. Cardiovasc Res.
117:2161–2174. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Leduc-Gaudet JP, Hussain SNA, Barreiro E
and Gouspillou G: Mitochondrial dynamics and mitophagy in skeletal
muscle health and aging. Int J Mol Sci. 22:81792021. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Li S, Chen J, Liu M, Chen Y, Wu Y, Li Q,
Ma T, Gao J, Xia Y, Fan M, et al: Protective effect of HINT2 on
mitochondrial function via repressing MCU complex activation
attenuates cardiac microvascular ischemia-reperfusion injury. Basic
Res Cardiol. 116:652021. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Mollazadeh H, Tavana E, Fanni G, Bo S,
Banach M, Pirro M, von Haehling S, Jamialahmadi T and Sahebkar A:
Effects of statins on mitochondrial pathways. J Cachexia Sarcopenia
Muscle. 12:237–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Nakamura T, Ogawa M, Kojima K, Takayanagi
S, Ishihara S, Hattori K, Naguro I and Ichijo H: The mitochondrial
Ca2+ uptake regulator, MICU1, is involved in cold
stress-induced ferroptosis. EMBO Rep. 22:e515322021. View Article : Google Scholar
|
|
185
|
Chen M, Mu L, Wang S, Cao X, Liang S, Wang
Y, She G, Yang J, Wang Y and Shi W: A single silicon nanowire-based
ratiometric biosensor for Ca2+ at various locations in a
neuron. ACS Chem Neurosci. 11:1283–1290. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Jiang Y, Fang Y, Ye Y, Xu X, Wang B, Gu J,
Aschner M, Chen J and Lu R: Anti-cancer effects of
3,3'-diindolylmethane on human hepatocellular carcinoma cells is
enhanced by calcium ionophore: The role of cytosolic
Ca2+ and p38 MAPK. Front Pharmacol. 10:11672019.
View Article : Google Scholar
|
|
187
|
Mata-Martínez E, Sánchez-Tusie AA, Darszon
A, Mayorga LS, Treviño CL and De Blas GA: Epac activation induces
an extracellular Ca2+-independent Ca2+ wave
that triggers acrosome reaction in human spermatozoa. Andrology.
9:1227–1241. 2021. View Article : Google Scholar
|
|
188
|
Wacquier B, Combettes L and Dupont G: Dual
dynamics of mitochondrial permeability transition pore opening. Sci
Rep. 10:39242020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Nesci S, Trombetti F, Ventrella V and
Pagliarani A: From the Ca2+-activated
F1FO-ATPase to the mitochondrial permeability
transition pore: An overview. Biochimie. 152:85–93. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Cui Y, Pan M, Ma J, Song X, Cao W and
Zhang P: Recent progress in the use of mitochondrial membrane
permeability transition pore in mitochondrial dysfunction-related
disease therapies. Mol Cell Biochem. 476:493–506. 2021. View Article : Google Scholar
|
|
191
|
Chinopoulos C: Mitochondrial permeability
transition pore: Back to the drawing board. Neurochem Int.
117:49–54. 2018. View Article : Google Scholar
|
|
192
|
Briston T, Selwood DL, Szabadkai G and
Duchen MR: Mitochondrial permeability transition: A molecular
lesion with multiple drug targets. Trends Pharmacol Sci. 40:50–70.
2019. View Article : Google Scholar
|
|
193
|
Rottenberg H and Hoek JB: The path from
mitochondrial ROS to aging runs through the mitochondrial
permeability transition pore. Aging Cell. 16:943–955. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhou B, Kreuzer J, Kumsta C, Wu L, Kamer
KJ, Cedillo L, Zhang Y, Li S, Kacergis MC, Webster CM, et al:
Mitochondrial permeability uncouples elevated autophagy and
lifespan extension. Cell. 177:299–314.e16. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Baines CP and Gutiérrez-Aguilar M: The
still uncertain identity of the channel-forming unit(s) of the
mitochondrial permeability transition pore. Cell Calcium.
73:121–130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Ying Z, Xiang G, Zheng L, Tang H, Duan L,
Lin X, Zhao Q, Chen K, Wu Y, Xing G, et al: Short-term
mitochondrial permeability transition pore opening modulates
histone lysine methylation at the early phase of somatic cell
reprogramming. Cell Metab. 28:935–945.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Burke PJ: Mitochondria, bioenergetics and
apoptosis in cancer. Trends Cancer. 3:857–870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Pérez MJ, Ponce DP, Aranguiz A, Behrens MI
and Quintanilla RA: Mitochondrial permeability transition pore
contributes to mitochondrial dysfunction in fibroblasts of patients
with sporadic Alzheimer's disease. Redox Biol. 19:290–300. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Kalani K, Yan SF and Yan SS: Mitochondrial
permeability transition pore: A potential drug target for
neurodegeneration. Drug Discov Today. 23:1983–1989. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Naryzhnaya NV, Maslov LN and Oeltgen PR:
Pharmacology of mitochondrial permeability transition pore
inhibitors. Drug Dev Res. 80:1013–1030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Shah SS, Lannon H, Dias L, Zhang JY, Alper
SL, Pollak MR and Friedman DJ: APOL1 kidney risk variants induce
cell death via mitochondrial translocation and opening of the
mitochondrial permeability transition pore. J Am Soc Nephrol.
30:2355–2368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Gao G, Wang Z, Lu L, Duan C, Wang X and
Yang H: Morphological analysis of mitochondria for evaluating the
toxicity of α-synuclein in transgenic mice and isolated
preparations by atomic force microscopy. Biomed Pharmacother.
96:1380–1388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Ghosh P, Bhoumik A, Saha S, Mukherjee S,
Azmi S, Ghosh JK and Dungdung SR: Spermicidal efficacy of VRP, a
synthetic cationic antimicrobial peptide, inducing apoptosis and
membrane disruption. J Cell Physiol. 233:1041–1050. 2018.
View Article : Google Scholar
|
|
204
|
Jiang S, Zu Y, Wang Z, Zhang Y and Fu Y:
Involvement of mitochondrial permeability transition pore opening
in 7-xylosyl-10-deacetylpaclitaxel-induced apoptosis. Planta Med.
77:1005–1012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Tricaud N, Gautier B, Berthelot J,
Gonzalez S and Van Hameren G: Traumatic and diabetic schwann cell
demyelination is triggered by a transient mitochondrial calcium
release through voltage dependent anion channel 1. Biomedicines.
10:14472022. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Mukherjee R, Mareninova OA, Odinokova IV,
Huang W, Murphy J, Chvanov M, Javed MA, Wen L, Booth DM, Cane MC,
et al: Mechanism of mitochondrial permeability transition pore
induction and damage in the pancreas: Inhibition prevents acute
pancreatitis by protecting production of ATP. Gut. 65:1333–1346.
2016. View Article : Google Scholar
|
|
207
|
Urbani A, Giorgio V, Carrer A, Franchin C,
Arrigoni G, Jiko C, Abe K, Maeda S, Shinzawa-Itoh K, Bogers JFM, et
al: Purified F-ATP synthase forms a Ca2+-dependent
high-conductance channel matching the mitochondrial permeability
transition pore. Nat Commun. 10:43412019. View Article : Google Scholar
|
|
208
|
Aqawi M, Sionov RV, Gallily R, Friedman M
and Steinberg D: Anti-bacterial properties of cannabigerol toward
streptococcus mutans. Front Microbiol. 12:6564712021. View Article : Google Scholar :
|
|
209
|
Asperti M, Bellini S, Grillo E, Gryzik M,
Cantamessa L, Ronca R, Maccarinelli F, Salvi A, De Petro G, Arosio
P, et al: H-ferritin suppression and pronounced mitochondrial
respiration make hepatocellular carcinoma cells sensitive to
RSL3-induced ferroptosis. Free Radic Biol Med. 169:294–303. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Daniyal M, Liu Y, Yang Y, Xiao F, Fan J,
Yu H, Qiu Y, Liu B, Wang W and Yuhui Q: Anti-gastric cancer
activity and mechanism of natural compound 'Heilaohulignan C'
isolated from Kadsura coccinea. Phytother Res. 35:3977–3987. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Datki Z, Acs E, Balazs E, Sovany T, Csoka
I, Zsuga K, Kalman J and Galik-Olah Z: Exogenic production of
bioactive filamentous biopolymer by monogonant rotifers. Ecotoxicol
Environ Saf. 208:1116662021. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Ge Y, Wang C, Zhang W, Lai S, Wang D and
Wang L: Coassembly behavior and rheological properties of a
β-hairpin peptide with dicarboxylates. Langmuir. 37:11657–11664.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
He A, Wang L, Wang Q, Luan W and Qi F:
Protective effects of micronized fat against ultraviolet B-induced
photoaging. Plast Reconstr Surg. 145:712–720. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Jiang Q, Su DY, Wang ZZ, Liu C, Sun YN,
Cheng H, Li XM and Yan B: Retina as a window to cerebral
dysfunction following studies with circRNA signature during
neurodegeneration. Theranostics. 11:1814–1827. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Kirk NM, Vieson MD, Selting KA and
Reinhart JM: Cytotoxicity of cultured canine primary hepatocytes
exposed to itraconazole is decreased by pre-treatment with
glutathione. Front Vet Sci. 8:6217322021. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Lan HY, An P, Liu QP, Chen YY, Yu YY, Luan
X, Tang JY and Zhang H: Aidi injection induces apoptosis of
hepatocellular carcinoma cells through the mitochondrial pathway. J
Ethnopharmacol. 274:1140732021. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Li C, Sun G, Chen B, Xu L, Ye Y, He J, Bao
Z, Zhao P, Miao Z, Zhao L, et al: Nuclear receptor coactivator
4-mediated ferritinophagy contributes to cerebral ischemia-induced
ferroptosis in ischemic stroke. Pharmacol Res. 174:1059332021.
View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Liu X, Xing S, Xu Y, Chen R, Lin C and Guo
L: 3-Amino-1,2,4-triazole-derived graphitic carbon nitride for
photodynamic therapy. Spectrochim Acta A Mol Biomol Spectrosc.
250:1193632021. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Suo L, Liu C, Zhang QY, Yao MD, Ma Y, Yao
J, Jiang Q and Yan B: METTL3-mediated N
6-methyladenosine modification governs pericyte
dysfunction during diabetes-induced retinal vascular complication.
Theranostics. 12:277–289. 2022. View Article : Google Scholar :
|
|
220
|
Panel M, Ruiz I, Brillet R, Lafdil F,
Teixeira-Clerc F, Nguyen CT, Calderaro J, Gelin M, Allemand F,
Guichou JF, et al: Small-molecule inhibitors of cyclophilins block
opening of the mitochondrial permeability transition pore and
protect mice from hepatic ischemia/reperfusion injury.
Gastroenterology. 157:1368–1382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Winquist RJ and Gribkoff VK: Targeting
putative components of the mitochondrial permeability transition
pore for novel therapeutics. Biochem Pharmacol. 177:1139952020.
View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Yu CH, Davidson S, Harapas CR, Hilton JB,
Mlodzianoski MJ, Laohamonthonkul P, Louis C, Low RRJ, Moecking J,
De Nardo D, et al: TDP-43 triggers mitochondrial DNA release via
mPTP to activate cGAS/STING in ALS. Cell. 183:636–649.e18. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Wu S and Zou MH: AMPK, mitochondrial
function, and cardiovascular disease. Int J Mol Sci. 21:49872020.
View Article : Google Scholar :
|
|
224
|
Lee P, Chandel NS and Simon MC: Cellular
adaptation to hypoxia through hypoxia inducible factors and beyond.
Nat Rev Mol Cell Biol. 21:268–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Tan KY, Li CY, Li YF, Fei J, Yang B, Fu YJ
and Li F: Real-time monitoring ATP in mitochondrion of living
cells: A specific fluorescent probe for ATP by dual recognition
sites. Anal Chem. 89:1749–1756. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Arai S, Kriszt R, Harada K, Looi LS,
Matsuda S, Wongso D, Suo S, Ishiura S, Tseng YH, Raghunath M, et
al: RGB-color intensiometric indicators to visualize spatiotemporal
dynamics of ATP in single cells. Angew Chem Int Ed Engl.
57:10873–10878. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Potter M, Newport E and Morten KJ: The
Warburg effect: 80 Years on. Biochem Soc Trans. 44:1499–1505. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Nesci S, Pagliarani A, Algieri C and
Trombetti F: Mitochondrial F-type ATP synthase: multiple enzyme
functions revealed by the membrane-embedded FO
structure. Crit Rev Biochem Mol Biol. 55:309–321. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Schönfeld P and Wojtczak L: Short- and
medium-chain fatty acids in energy metabolism: The cellular
perspective. J Lipid Res. 57:943–954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Chistiakov DA, Shkurat TP, Melnichenko AA,
Grechko AV and Orekhov AN: The role of mitochondrial dysfunction in
cardiovascular disease: A brief review. Ann Med. 50:121–127. 2018.
View Article : Google Scholar
|
|
231
|
Costa R, Peruzzo R, Bachmann M, Montà GD,
Vicario M, Santinon G, Mattarei A, Moro E, Quintana-Cabrera R,
Scorrano L, et al: Impaired mitochondrial ATP production
downregulates Wnt signaling via ER stress induction. Cell Rep.
28:1949–1960.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Rambold AS and Pearce EL: Mitochondrial
dynamics at the interface of immune cell metabolism and function.
Trends Immunol. 39:6–18. 2018. View Article : Google Scholar
|
|
233
|
Roger AJ, Muñoz-Gómez SA and Kamikawa R:
The origin and diversification of mitochondria. Curr Biol.
27:R1177–R1192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Guntur AR, Gerencser AA, Le PT, DeMambro
VE, Bornstein SA, Mookerjee SA, Maridas DE, Clemmons DE, Brand MD
and Rosen CJ: Osteoblast-like MC3T3-E1 cells prefer glycolysis for
ATP production but adipocyte-like 3T3-L1 cells prefer oxidative
phosphorylation. J Bone Miner Res. 33:1052–1065. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Depaoli MR, Karsten F,
Madreiter-Sokolowski CT, Klec C, Gottschalk B, Bischof H, Eroglu E,
Waldeck-Weiermair M, Simmen T, Graier WF and Malli R: Real-time
imaging of mitochondrial ATP dynamics reveals the metabolic setting
of single cells. Cell Rep. 25:501–512.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Hampl V, Čepička I and Eliáš M: Was the
mitochondrion necessary to start eukaryogenesis? Trends Microbiol.
27:96–104. 2019. View Article : Google Scholar
|
|
237
|
Beamer E, Conte G and Engel T: ATP release
during seizures-a critical evaluation of the evidence. Brain Res
Bull. 151:65–73. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Buckel W, Hetzel M and Kim J: ATP-driven
electron transfer in enzymatic radical reactions. Curr Opin Chem
Biol. 8:462–467. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Chen H and Zhang YPJ: Enzymatic
regeneration and conservation of ATP: Challenges and opportunities.
Crit Rev Biotechnol. 41:16–33. 2021. View Article : Google Scholar
|
|
240
|
Dorr BM and Fuerst DE: Enzymatic amidation
for industrial applications. Curr Opin Chem Biol. 43:127–133. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Finley D and Prado MA: The proteasome and
its network: Engineering for adaptability. Cold Spring Harb
Perspect Biol. 12:a0339852020. View Article : Google Scholar
|
|
242
|
Hammler D, Marx A and Zumbusch A:
Fluorescencelifetime-sensitive probes for monitoring ATP cleavage.
Chemistry. 24:15329–15335. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Ishida A, Yamada Y and Kamidate T:
Colorimetric method for enzymatic screening assay of ATP using
Fe(III)-xylenol orange complex formation. Anal Bioanal Chem.
392:987–994. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Midelfort CF and Rose IA: A stereochemical
method for detection of ATP terminal phosphate transfer in
enzymatic reactions. Glutamine synthetase J Biol Chem.
251:5881–5887. 1976. View Article : Google Scholar
|
|
245
|
Ušaj M, Moretto L, Vemula V, Salhotra A
and Månsson A: Single molecule turnover of fluorescent ATP by
myosin and actomyosin unveil elusive enzymatic mechanisms. Commun
Biol. 4:642021. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Vasta JD, Corona CR, Wilkinson J, Zimprich
CA, Hartnett JR, Ingold MR, Zimmerman K, Machleidt T, Kirkland TA,
Huwiler KG, et al: Quantitative, wide-spectrum kinase profiling in
live cells for assessing the effect of cellular ATP on target
engagement. Cell Chem Biol. 25:206–214.e11. 2018. View Article : Google Scholar :
|
|
247
|
Klier PEZ, Martin JG and Miller EW:
Imaging reversible mitochondrial membrane potential dynamics with a
masked rhodamine voltage reporter. J Am Chem Soc. 143:4095–4099.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Mita M, Sugawara I, Harada K, Ito M,
Takizawa M, Ishida K, Ueda H, Kitaguchi T and Tsuboi T: Development
of red genetically encoded biosensor for visualization of
intracellular glucose dynamics. Cell Chem Biol. 29:98–108.e4. 2022.
View Article : Google Scholar
|
|
249
|
Murata O, Shindo Y, Ikeda Y, Iwasawa N,
Citterio D, Oka K and Hiruta Y: Near-infrared fluorescent probes
for imaging of intracellular Mg2+ and application to
multi-color imaging of Mg2+, ATP, and mitochondrial
membrane potential. Anal Chem. 92:966–974. 2020. View Article : Google Scholar
|
|
250
|
Billingham LK, Stoolman JS, Vasan K,
Rodriguez AE, Poor TA, Szibor M, Jacobs HT, Reczek CR, Rashidi A,
Zhang P, et al: Mitochondrial electron transport chain is necessary
for NLRP3 inflammasome activation. Nat Immunol. 23:692–704. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Fernström J, Mellon SH, McGill MA, Picard
M, Reus VI, Hough CM, Lin J, Epel ES, Wolkowitz OM and Lindqvist D:
Blood-based mitochondrial respiratory chain function in major
depression. Transl Psychiatry. 11:5932021. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Spinelli JB, Rosen PC, Sprenger HG,
Puszynska AM, Mann JL, Roessler JM, Cangelosi AL, Henne A, Condon
KJ, Zhang T, et al: Fumarate is a terminal electron acceptor in the
mammalian electron transport chain. Science. 374:1227–1237. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Vercellino I and Sazanov LA: The assembly,
regulation and function of the mitochondrial respiratory chain. Nat
Rev Mol Cell Biol. 23:141–161. 2022. View Article : Google Scholar
|
|
254
|
Colaço HG, Barros A, Neves-Costa A, Seixas
E, Pedroso D, Velho T, Willmann KL, Faisca P, Grabmann G, Yi HS, et
al: Tetracycline antibiotics induce host-dependent disease
tolerance to infection. Immunity. 54:53–67.e7. 2021. View Article : Google Scholar :
|
|
255
|
Dennerlein S, Wang C and Rehling P:
Plasticity of mitochondrial translation. Trends Cell Biol.
27:712–721. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Diebold LP, Gil HJ, Gao P, Martinez CA,
Weinberg SE and Chandel NS: Mitochondrial complex III is necessary
for endothelial cell proliferation during angiogenesis. Nat Metab.
1:158–171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Flønes IH, Ricken G, Klotz S, Lang A,
Ströbel T, Dölle C, Kovacs GG and Tzoulis C: Mitochondrial
respiratory chain deficiency correlates with the severity of
neuropathology in sporadic Creutzfeldt-Jakob disease. Acta
Neuropathol Commun. 8:502020. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Manczak M, Kandimalla R, Yin X and Reddy
PH: Mitochondrial division inhibitor 1 reduces dynamin-related
protein 1 and mitochondrial fission activity. Hum Mol Genet.
28:177–199. 2019. View Article : Google Scholar :
|
|
259
|
Markevich NI, Galimova MH and Markevich
LN: Hysteresis and bistability in the succinate-CoQ reductase
activity and reactive oxygen species production in the
mitochondrial respiratory complex II. Redox Biol. 37:1016302020.
View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Mazat JP, Devin A and Ransac S: Modelling
mitochondrial ROS production by the respiratory chain. Cell Mol
Life Sci. 77:455–465. 2020. View Article : Google Scholar
|
|
261
|
Timón-Gómez A, Garlich J, Stuart RA,
Ugalde C and Barrientos A: Distinct roles of mitochondrial HIGD1A
and HIGD2A in respiratory complex and supercomplex biogenesis. Cell
Rep. 31:1076072020. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Grünewald A, Kumar KR and Sue CM: New
insights into the complex role of mitochondria in Parkinson's
disease. Prog Neurobiol. 177:73–93. 2019. View Article : Google Scholar
|
|
263
|
Ansó E, Weinberg SE, Diebold LP, Thompson
BJ, Malinge S, Schumacker PT, Liu X, Zhang Y, Shao Z, Steadman M,
et al: The mitochondrial respiratory chain is essential for
haematopoietic stem cell function. Nat Cell Biol. 19:614–625. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Wang HW, Zhu SQ, Liu J, Miao CY, Zhang Y
and Zhou BH: Fluoride-induced renal dysfunction via respiratory
chain complex abnormal expression and fusion elevation in mice.
Chemosphere. 238:1246072020. View Article : Google Scholar
|
|
265
|
Weiland D, Brachvogel B, Hornig-Do HT,
Neuhaus JFG, Holzer T, Tobin DJ, Niessen CM, Wiesner RJ and Baris
OR: Imbalance of mitochondrial respiratory chain complexes in the
epidermis induces severe skin inflammation. J Invest Dermatol.
138:132–140. 2018. View Article : Google Scholar
|
|
266
|
Weinberg SE, Singer BD, Steinert EM,
Martinez CA, Mehta MM, Martínez-Reyes I, Gao P, Helmin KA,
Abdala-Valencia H, Sena LA, et al: Mitochondrial complex III is
essential for suppressive function of regulatory T cells. Nature.
565:495–499. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
267
|
Wu M, Gu J, Zong S, Guo R, Liu T and Yang
M: Research journey of respirasome. Protein Cell. 11:318–338. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Yamada S, Ozaki H and Noguchi K: The
mitochondrial respiratory chain maintains the photosynthetic
electron flow in Arabidopsis thaliana leaves under high-light
stress. Plant Cell Physiol. 61:283–295. 2020. View Article : Google Scholar
|
|
269
|
Yamashita K, Miyazaki T, Fukuda Y,
Mitsuyama J, Saijo T, Shimamura S, Yamamoto K, Imamura Y, Izumikawa
K, Yanagihara K, et al: The novel arylamidine T-2307 selectively
disrupts yeast mitochondrial function by inhibiting respiratory
chain complexes. Antimicrob Agents Chemother. 63:e00374–19. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Fernandez-Vizarra E and Zeviani M:
Mitochondrial disorders of the OXPHOS system. FEBS Lett.
595:1062–1106. 2021. View Article : Google Scholar
|
|
271
|
Hernansanz-Agustín P, Choya-Foces C,
Carregal-Romero S, Ramos E, Oliva T, Villa-Piña T, Moreno L,
Izquierdo-Álvarez A, Cabrera-García JD, Cortés A, et al:
Na+ controls hypoxic signalling by the mitochondrial
respiratory chain. Nature. 586:287–291. 2020. View Article : Google Scholar
|
|
272
|
Kobayashi A, Azuma K, Ikeda K and Inoue S:
Mechanisms underlying the regulation of mitochondrial respiratory
chain complexes by nuclear steroid receptors. Int J Mol Sci.
21:66832020. View Article : Google Scholar :
|
|
273
|
Martínez-Reyes I, Cardona LR, Kong H,
Vasan K, McElroy GS, Werner M, Kihshen H, Reczek CR, Weinberg SE,
Gao P, et al: Mitochondrial ubiquinol oxidation is necessary for
tumour growth. Nature. 585:288–292. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Castellana S, Biagini T, Petrizzelli F,
Parca L, Panzironi N, Caputo V, Vescovi AL, Carella M and Mazza T:
MitImpact 3: Modeling the residue interaction network of the
respiratory chain subunits. Nucleic Acids Res. 49(D1): D1282–D1288.
2021. View Article : Google Scholar :
|
|
275
|
Wang M, Ren X, Wang L, Lu X, Han L, Zhang
X and Feng J: A functional analysis of mitochondrial respiratory
chain cytochrome bc1 complex in gaeumannomyces tritici
by RNA silencing as a possible target of carabrone. Mol Plant
Pathol. 21:1529–1544. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
276
|
Mirali S, Botham A, Voisin V, Xu C,
St-Germain J, Sharon D, Hoff FW, Qiu Y, Hurren R, Gronda M, et al:
The mitochondrial peptidase, neurolysin, regulates respiratory
chain supercomplex formation and is necessary for AML viability.
Sci Transl Med. 12:eaaz82642020. View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Heyman E, Daussin F, Wieczorek V, Caiazzo
R, Matran R, Berthon P, Aucouturier J, Berthoin S, Descatoire A,
Leclair E, et al: Muscle oxygen supply and use in type 1 diabetes,
from ambient air to the mitochondrial respiratory chain: Is there a
limiting step? Diabetes Care. 43:209–218. 2020. View Article : Google Scholar
|
|
278
|
Lobo-Jarne T, Pérez-Pérez R, Fontanesi F,
Timón-Gómez A, Wittig I, Peñas A, Serrano-Lorenzo P,
García-Consuegra I, Arenas J, Martín MA, et al: Multiple pathways
coordinate assembly of human mitochondrial complex IV and
stabilization of respiratory supercomplexes. EMBO J.
39:e1039122020. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Mohanraj K, Wasilewski M, Benincá C,
Cysewski D, Poznanski J, Sakowska P, Bugajska Z, Deckers M,
Dennerlein S, Fernandez-Vizarra E, et al: Inhibition of proteasome
rescues a pathogenic variant of respiratory chain assembly factor
COA7. EMBO Mol Med. 11:e95612019. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Formosa LE, Dibley MG, Stroud DA and Ryan
MT: Building a complex complex: Assembly of mitochondrial
respiratory chain complex I. Semin Cell Dev Biol. 76:154–162. 2018.
View Article : Google Scholar
|
|
281
|
Gao M, Yi J, Zhu J, Minikes AM, Monian P,
Thompson CB and Jiang X: Role of mitochondria in ferroptosis. Mol
Cell. 73:354–363.e3. 2019. View Article : Google Scholar :
|
|
282
|
Maclean AE, Hertle AP, Ligas J, Bock R,
Balk J and Meyer EH: Absence of complex I is associated with
diminished respiratory chain function in european mistletoe. Curr
Biol. 28:1614–1619.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Senkler J, Rugen N, Eubel H, Hegermann J
and Braun HP: Absence of complex I implicates rearrangement of the
respiratory chain in European mistletoe. Curr Biol.
28:1606–1613.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
284
|
Signes A and Fernandez-Vizarra E: Assembly
of mammalian oxidative phosphorylation complexes I-V and
supercomplexes. Essays Biochem. 62:255–270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
285
|
Kazak L, Chouchani ET, Stavrovskaya IG, Lu
GZ, Jedrychowski MP, Egan DF, Kumari M, Kong X, Erickson BK, Szpyt
J, et al: UCP1 deficiency causes brown fat respiratory chain
depletion and sensitizes mitochondria to calcium overload-induced
dysfunction. Proc Natl Acad Sci USA. 114:7981–7986. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
286
|
Kozlov AV, Lancaster JR Jr, Meszaros AT
and Weidinger A: Mitochondria-meditated pathways of organ failure
upon inflammation. Redox Biol. 13:170–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Letts JA and Sazanov LA: Clarifying the
supercomplex: The higher-order organization of the mitochondrial
electron transport chain. Nat Struct Mol Biol. 24:800–808. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Guo R, Zong S, Wu M, Gu J and Yang M:
Architecture of Human Mitochondrial Respiratory Megacomplex
I2III2IV2. Cell.
170:1247–1257.e12. 2017. View Article : Google Scholar
|
|
289
|
Jian C, Xu F, Hou T, Sun T, Li J, Cheng H
and Wang X: Deficiency of PHB complex impairs respiratory
supercomplex formation and activates mitochondrial flashes. J Cell
Sci. 130:2620–2630. 2017.PubMed/NCBI
|
|
290
|
Ndi M, Marin-Buera L, Salvatori R, Singh
AP and Ott M: Biogenesis of the bc1 complex of the
mitochondrial respiratory chain. J Mol Biol. 430:3892–3905. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Priesnitz C and Becker T: Pathways to
balance mitochondrial translation and protein import. Genes Dev.
32:1285–1296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Lin KH, Xie A, Rutter JC, Ahn YR,
Lloyd-Cowden JM, Nichols AG, Soderquist RS, Koves TR, Muoio DM,
MacIver NJ, et al: Systematic dissection of the metabolic-apoptotic
interface in AML reveals heme biosynthesis to be a regulator of
drug sensitivity. Cell Metab. 29:1217–1231.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Lobo-Jarne T and Ugalde C: Respiratory
chain supercomplexes: Structures, function and biogenesis. Semin
Cell Dev Biol. 76:179–190. 2018. View Article : Google Scholar :
|
|
294
|
Tsai YL, Coady TH, Lu L, Zheng D, Alland
I, Tian B, Shneider NA and Manley JL: ALS/FTD-associated protein
FUS induces mitochondrial dysfunction by preferentially
sequestering respiratory chain complex mRNAs. Genes Dev.
34:785–805. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Balsa E, Soustek MS, Thomas A, Cogliati S,
García-Poyatos C, Martín-García E, Jedrychowski M, Gygi SP,
Enriquez JA and Puigserver P: ER and nutrient stress promote
assembly of respiratory chain supercomplexes through the PERK-eIF2α
axis. Mol Cell. 74:877–890e6. 2019. View Article : Google Scholar
|
|
296
|
Chinopoulos C: Acute sources of
mitochondrial NAD+ during respiratory chain dysfunction.
Exp Neurol. 327:1132182020. View Article : Google Scholar
|
|
297
|
Cogliati S, Lorenzi I, Rigoni G, Caicci F
and Soriano ME: Regulation of mitochondrial electron transport
chain assembly. J Mol Biol. 430:4849–4873. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Nagao T, Shintani Y, Hayashi T, Kioka H,
Kato H, Nishida Y, Yamazaki S, Tsukamoto O, Yashirogi S, Yazawa I,
et al: Higd1a improves respiratory function in the models of
mitochondrial disorder. FASEB J. 34:1859–1871. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
299
|
Vankayala R and Hwang KC:
Near-infrared-light-activatable nanomaterial-mediated
phototheranostic nanomedicines: An emerging paradigm for cancer
treatment. Adv Mater. 30:e17063202018. View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Wang S, Zhang Z, Wei S, He F, Li Z, Wang
HH, Huang Y and Nie Z: Near-infrared light-controllable MXene
hydrogel for tunable on-demand release of therapeutic proteins.
Acta Biomater. 130:138–148. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
301
|
Zhang S, Weinberg S, DeBerge M, Gainullina
A, Schipma M, Kinchen JM, Ben-Sahra I, Gius DR, Yvan-Charvet L,
Chandel NS, et al: Efferocytosis fuels requirements of fatty acid
oxidation and the electron transport chain to polarize macrophages
for tissue repair. Cell Metab. 29:443–456.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
302
|
Luo X, Gong X, Su L, Lin H, Yang Z, Yan X
and Gao J: Activatable mitochondria-targeting organoarsenic
prodrugs for bioenergetic cancer therapy. Angew Chem Int Ed Engl.
60:1403–1410. 2021. View Article : Google Scholar
|
|
303
|
Jiang H, Zhang XW, Liao QL, Wu WT, Liu YL
and Huang WH: Electrochemical monitoring of paclitaxel-induced ROS
release from mitochondria inside single cells. Small.
15:e19017872019. View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Kaplan P, Tatarkova Z, Sivonova MK, Racay
P and Lehotsky J: Homocysteine and mitochondria in cardiovascular
and cerebrovascular systems. Int J Mol Sci. 21:76982020. View Article : Google Scholar :
|
|
305
|
Koch RE, Josefson CC and Hill GE:
Mitochondrial function, ornamentation, and immunocompetence. Biol
Rev Camb Philos Soc. 92:1459–1474. 2017. View Article : Google Scholar
|
|
306
|
Zhang L, Wang X, Cueto R, Effi C, Zhang Y,
Tan H, Qin X, Ji Y, Yang X and Wang H: Biochemical basis and
metabolic interplay of redox regulation. Redox Biol. 26:1012842019.
View Article : Google Scholar : PubMed/NCBI
|
|
307
|
Zhu X, Liu G, Bu Y, Zhang J, Wang L, Tian
Y, Yu J, Wu Z and Zhou H: In situ monitoring of mitochondria
regulating cell viability by the RNA-specific fluorescent
photosensitizer. Anal Chem. 92:10815–10821. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
308
|
Blanco FJ, Valdes AM and Rego-Pérez I:
Mitochondrial DNA variation and the pathogenesis of osteoarthritis
phenotypes. Nat Rev Rheumatol. 14:327–340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
Fuhrmann DC and Brüne B: Mitochondrial
composition and function under the control of hypoxia. Redox Biol.
12:208–215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Lee JH and Paull TT: Mitochondria at the
crossroads of ATM-mediated stress signaling and regulation of
reactive oxygen species. Redox Biol. 32:1015112020. View Article : Google Scholar : PubMed/NCBI
|
|
311
|
Madreiter-Sokolowski CT, Thomas C and
Ristow M: Interrelation between ROS and Ca2+ in aging
and age-related diseases. Redox Biol. 36:1016782020. View Article : Google Scholar
|
|
312
|
Angelova PR, Esteras N and Abramov AY:
Mitochondria and lipid peroxidation in the mechanism of
neurodegeneration: Finding ways for prevention. Med Res Rev.
41:770–784. 2021. View Article : Google Scholar
|
|
313
|
van der Reest J, Nardini Cecchino G,
Haigis MC and Kordowitzki P: Mitochondria: Their relevance during
oocyte ageing. Ageing Res Rev. 70:1013782021. View Article : Google Scholar : PubMed/NCBI
|
|
314
|
Martins WK, Santos NF, Rocha CS, Bacellar
IOL, Tsubone TM, Viotto AC, Matsukuma AY, Abrantes ABP, Siani P,
Dias LG and Baptista MS: Parallel damage in mitochondria and
lysosomes is an efficient way to photoinduce cell death. Autophagy.
15:259–279. 2019. View Article : Google Scholar :
|
|
315
|
Kleih M, Böpple K, Dong M, Gaißler A,
Heine S, Olayioye MA, Aulitzky WE and Essmann F: Direct impact of
cisplatin on mitochondria induces ROS production that dictates cell
fate of ovarian cancer cells. Cell Death Dis. 10:8512019.
View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Sidlauskaite E, Gibson JW, Megson IL,
Whitfield PD, Tovmasyan A, Batinic-Haberle I, Murphy MP, Moult PR
and Cobley JN: Mitochondrial ROS cause motor deficits induced by
synaptic inactivity: Implications for synapse pruning. Redox Biol.
16:344–351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Yang J, Chen Z, Liu N and Chen Y:
Ribosomal protein L10 in mitochondria serves as a regulator for ROS
level in pancreatic cancer cells. Redox Biol. 19:158–165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
318
|
Erard M, Dupré-Crochet S and Nüße O:
Biosensors for spatiotemporal detection of reactive oxygen species
in cells and tissues. Am J Physiol Regul Integr Comp Physiol.
314:R667–R683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Jiang X, Wang L, Carroll SL, Chen J, Wang
MC and Wang J: Challenges and opportunities for small-molecule
fluorescent probes in redox biology applications. Antioxid Redox
Signal. 29:518–540. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
320
|
Ortega-Villasante C, Burén S, Barón-Sola
Á, Martínez F and Hernández LE: In vivo ROS and redox potential
fluorescent detection in plants: Present approaches and future
perspectives. Methods. 109:92–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Ortega-Villasante C, Burén S,
Blázquez-Castro A, Barón-Sola Á and Hernández LE: Fluorescent in
vivo imaging of reactive oxygen species and redox potential in
plants. Free Radic Biol Med. 122:202–220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Dragišić Maksimović J, Mojović M, Vučinić
Ž and Maksimović V: Spatial distribution of apoplastic
antioxidative constituents in maize root. Physiol Plant.
173:818–828. 2021. View Article : Google Scholar
|
|
323
|
Emoto MC, Sato-Akaba H, Hamaue N,
Kawanishi K, Koshino H, Shimohama S and Fujii HG: Early detection
of redox imbalance in the APPswe/PS1dE9 mouse model of Alzheimer's
disease by in vivo electron paramagnetic resonance imaging. Free
Radic Biol Med. 172:9–18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
324
|
Gotham JP, Li R, Tipple TE, Lancaster JR
Jr, Liu T and Li Q: Quantitation of spin probe-detectable oxidants
in cells using electron paramagnetic resonance spectroscopy: To
probe or to trap? Free Radic Biol Med. 154:84–94. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
325
|
He L, Li MX, Chen F, Yang SS, Ding J, Ding
L and Ren NQ: Novel coagulation waste-based Fe-containing
carbonaceous catalyst as peroxymonosulfate activator for pollutants
degradation: Role of ROS and electron transfer pathway. J Hazard
Mater. 417:1261132021. View Article : Google Scholar : PubMed/NCBI
|
|
326
|
Hinoshita M, Abe T, Sato A, Maeda Y and
Takeyoshi M: Development of a new photosafety test method based on
singlet oxygen generation detected using electron spin resonance. J
Appl Toxicol. 41:247–255. 2021. View Article : Google Scholar
|
|
327
|
Matsumoto KI, Ueno M, Shoji Y and
Nakanishi I: Heavy-ion beam-induced reactive oxygen species and
redox reactions. Free Radic Res. 55:450–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Mendoza C, Désert A, Khrouz L, Páez CA,
Parola S and Heinrichs B: Heterogeneous singlet oxygen generation:
In-operando visible light EPR spectroscopy. Environ Sci Pollut Res
Int. 28:25124–25129. 2021. View Article : Google Scholar
|
|
329
|
Okazaki Y, Ishidzu Y, Ito F, Tanaka H,
Hori M and Toyokuni S: L-Dehydroascorbate efficiently degrades
non-thermal plasma-induced hydrogen peroxide. Arch Biochem Biophys.
700:1087622021. View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Prasad A, Manoharan RR, Sedlářová M and
Pospíšil P: Free radical-mediated protein radical formation in
differentiating monocytes. Int J Mol Sci. 22:99632021. View Article : Google Scholar : PubMed/NCBI
|
|
331
|
Yamaguchi M, Ma T, Tadaki D, Hirano-Iwata
A, Watanabe Y, Kanetaka H, Fujimori H, Takemoto E and Niwano M:
Bactericidal activity of bulk nanobubbles through active oxygen
species generation. Langmuir. Aug 2–2021.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
332
|
Zhang K, Deng R, Teng X, Li Y, Sun Y, Ren
X and Li J: Direct visualization of single-nucleotide variation in
mtDNA using a CRISPR/Cas9-mediated proximity ligation assay. J Am
Chem Soc. 140:11293–11301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
333
|
Moriyama M, Koshiba T and Ichinohe T:
Influenza A virus M2 protein triggers mitochondrial DNA-mediated
antiviral immune responses. Nat Commun. 10:46242019. View Article : Google Scholar : PubMed/NCBI
|
|
334
|
Baumann K: mtDNA robs nuclear dNTPs. Nat
Rev Mol Cell Biol. 20:6632019. View Article : Google Scholar : PubMed/NCBI
|
|
335
|
Lazo S, Noren Hooten N, Green J, Eitan E,
Mode NA, Liu QR, Zonderman AB, Ezike N, Mattson MP, Ghosh P and
Evans MK: Mitochondrial DNA in extracellular vesicles declines with
age. Aging Cell. 20:e132832021. View Article : Google Scholar :
|
|
336
|
Li D, Du X, Guo X, Zhan L, Li X, Yin C,
Chen C, Li M, Li B, Yang H and Xing J: Site-specific selection
reveals selective constraints and functionality of tumor somatic
mtDNA mutations. J Exp Clin Cancer Res. 36:1682017. View Article : Google Scholar : PubMed/NCBI
|
|
337
|
Medeiros TC and Graef M: Autophagy
determines mtDNA copy number dynamics during starvation. Autophagy.
15:178–179. 2019. View Article : Google Scholar
|
|
338
|
Fontana GA and Gahlon HL: Mechanisms of
replication and repair in mitochondrial DNA deletion formation.
Nucleic Acids Res. 48:11244–11258. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
339
|
Wanrooij PH, Tran P, Thompson LJ, Carvalho
G, Sharma S, Kreisel K, Navarrete C, Feldberg AL, Watt DL, Nilsson
AK, et al: Elimination of rNMPs from mitochondrial DNA has no
effect on its stability. Proc Natl Acad Sci USA. 117:14306–14313.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
340
|
Wei W and Chinnery PF: Inheritance of
mitochondrial DNA in humans: Implications for rare and common
diseases. J Intern Med. 287:634–644. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
341
|
Ignatenko O, Chilov D, Paetau I, de Miguel
E, Jackson CB, Capin G, Paetau A, Terzioglu M, Euro L and
Suomalainen A: Loss of mtDNA activates astrocytes and leads to
spongiotic encephalopathy. Nat Commun. 9:702018. View Article : Google Scholar : PubMed/NCBI
|
|
342
|
Kasahara T and Kato T: What can
mitochondrial DNA analysis tell us about mood disorders? Biol
Psychiatry. 83:731–738. 2018. View Article : Google Scholar
|
|
343
|
Larsson NG and Wedell A: Mitochondria in
human disease. J Intern Med. 287:589–591. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
344
|
Bagge EK, Fujimori-Tonou N,
Kubota-Sakashita M, Kasahara T and Kato T: Unbiased PCR-free
spatio-temporal mapping of the mtDNA mutation spectrum reveals
brain region-specific responses to replication instability. BMC
Biol. 18:1502020. View Article : Google Scholar : PubMed/NCBI
|
|
345
|
Chiang JL, Shukla P, Pagidas K, Ahmed NS,
Karri S, Gunn DD, Hurd WW and Singh KK: Mitochondria in ovarian
aging and reproductive longevity. Ageing Res Rev. 63:1011682020.
View Article : Google Scholar : PubMed/NCBI
|
|
346
|
Li H, Slone J, Fei L and Huang T:
Mitochondrial DNA variants and common diseases: A mathematical
model for the diversity of age-related mtDNA mutations. Cells.
8:6082019. View Article : Google Scholar :
|
|
347
|
Nissanka N and Moraes CT: Mitochondrial
DNA heteroplasmy in disease and targeted nuclease-based therapeutic
approaches. EMBO Rep. 21:e496122020. View Article : Google Scholar : PubMed/NCBI
|
|
348
|
West AP and Shadel GS: Mitochondrial DNA
in innate immune responses and inflammatory pathology. Nat Rev
Immunol. 17:363–375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
349
|
Asfaram S, Fakhar M, Mohebali M, Ziaei
Hezarjaribi H, Mardani A, Ghezelbash B, Akhoundi B, Zarei Z and
Moazeni M: A convenient and sensitive kDNA-PCR for screening of
leishmania infantum latent infection among blood donors in a highly
endemic focus, northwestern Iran. Acta Parasitol. 67:842–850. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
350
|
Semerikov VL, Semerikova SA, Khrunyk YY
and Putintseva YA: Sequence capture of mitochondrial genome with
PCR-generated baits provides new insights into the biogeography of
the genus abies mill. Plants (Basel). 11. pp. 7622022, View Article : Google Scholar
|
|
351
|
Tay E, Chen SC, Green W, Lopez R and
Halliday CL: Development of a real-time PCR assay to identify and
distinguish between cryptococcus neoformans and cryptococcus gattii
species complexes. J Fungi (Basel). 8:4622022. View Article : Google Scholar
|
|
352
|
Wang J, Balciuniene J, Diaz-Miranda MA,
McCormick EM, Aref-Eshghi E, Muir AM, Cao K, Troiani J, Moseley A,
Fan Z, et al: Advanced approach for comprehensive mtDNA genome
testing in mitochondrial disease. Mol Genet Metab. 135:93–101.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
353
|
Yang Z, Slone J and Huang T:
Next-generation sequencing to characterize mitochondrial genomic
DNA heteroplasmy. Curr Protoc. 2:e4122022. View Article : Google Scholar : PubMed/NCBI
|
|
354
|
Allouche J, Rachmin I, Adhikari K, Pardo
LM, Lee JH, McConnell AM, Kato S, Fan S, Kawakami A, Suita Y, et
al: NNT mediates redox-dependent pigmentation via a UVB- and
MITF-independent mechanism. Cell. 184:4268–4283.e20. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
355
|
Cornman RS, McKenna JE Jr and Fike JA:
Composition and distribution of fish environmental DNA in an
adirondack watershed. PeerJ. 9:e105392021. View Article : Google Scholar : PubMed/NCBI
|
|
356
|
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S,
Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu
YP, Acevedo-Arozena A, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy (4th
edition)1. Autophagy. 17:1–382. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
357
|
Matsui H, Ito J, Matsui N, Uechi T,
Onodera O and Kakita A: Cytosolic dsDNA of mitochondrial origin
induces cytotoxicity and neurodegeneration in cellular and
zebrafish models of Parkinson's disease. Nat Commun. 12:31012021.
View Article : Google Scholar : PubMed/NCBI
|
|
358
|
Rhie A, McCarthy SA, Fedrigo O, Damas J,
Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J,
et al: Towards complete and error-free genome assemblies of all
vertebrate species. Nature. 592:737–746. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
359
|
Rossmann MP, Hoi K, Chan V, Abraham BJ,
Yang S, Mullahoo J, Papanastasiou M, Wang Y, Elia I, Perlin JR, et
al: Cell-specific transcriptional control of mitochondrial
metabolism by TIF1γ drives erythropoiesis. Science. 372:716–721.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
360
|
Wiessner M, Maroofian R, Ni MY, Pedroni A,
Müller JS, Stucka R, Beetz C, Efthymiou S, Santorelli FM, Alfares
AA, et al: Biallelic variants in HPDL cause pure and complicated
hereditary spastic paraplegia. Brain. 144:1422–1434. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
361
|
Wong HH, Seet SH, Maier M, Gurel A,
Traspas RM, Lee C, Zhang S, Talim B, Loh AYT, Chia CY, et al: Loss
of C2orf69 defines a fatal autoinflammatory syndrome in humans and
zebrafish that evokes a glycogen-storage-associated
mitochondriopathy. Am J Hum Genet. 108:1301–1317. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
362
|
Zhang DG, Zhao T, Hogstrand C, Ye HM, Xu
XJ and Luo Z: Oxidized fish oils increased lipid deposition via
oxidative stress-mediated mitochondrial dysfunction and the
CREB1-Bcl2-Beclin1 pathway in the liver tissues and hepatocytes of
yellow catfish. Food Chem. 360:1298142021. View Article : Google Scholar : PubMed/NCBI
|
|
363
|
Borsche M, König IR, Delcambre S, Petrucci
S, Balck A, Brüggemann N, Zimprich A, Wasner K, Pereira SL, Avenali
M, et al: Mitochondrial damage-associated inflammation highlights
biomarkers in PRKN/PINK1 parkinsonism. Brain. 143:3041–3051. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
364
|
Fernström J, Ohlsson L, Asp M, Lavant E,
Holck A, Grudet C, Westrin Å and Lindqvist D: Plasma circulating
cell-free mitochondrial DNA in depressive disorders. PLoS One.
16:e02595912021. View Article : Google Scholar : PubMed/NCBI
|
|
365
|
Gonçalves VF, Mendes-Silva AP, Koyama E,
Vieira E, Kennedy JL and Diniz B: Increased levels of circulating
cell-free mtDNA in plasma of late life depression subjects. J
Psychiatr Res. 139:25–29. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
366
|
Liu Y, Zhou K, Guo S, Wang Y, Ji X, Yuan
Q, Su L, Guo X, Gu X and Xing J: NGS-based accurate and efficient
detection of circulating cell-free mitochondrial DNA in cancer
patients. Mol Ther Nucleic Acids. 23:657–666. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
367
|
Maresca A, Del Dotto V, Romagnoli M, La
Morgia C, Di Vito L, Capristo M, Valentino ML and Carelli V;
ER-MITO Study Group: Expanding and validating the biomarkers for
mitochondrial diseases. J Mol Med (Berl). 98:1467–1478. 2020.
View Article : Google Scholar
|
|
368
|
Nie S, Lu J, Wang L and Gao M:
Pro-inflammatory role of cell-free mitochondrial DNA in
cardiovascular diseases. IUBMB Life. 72:1879–1890. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
369
|
Valenti D, Vacca RA, Moro L and Atlante A:
Mitochondria can cross cell boundaries: An overview of the
biological relevance, pathophysiological implications and
therapeutic perspectives of intercellular mitochondrial transfer.
Int J Mol Sci. 22:83122021. View Article : Google Scholar : PubMed/NCBI
|
|
370
|
Zhong XY, Guo Y and Fan Z: Increased level
of free-circulating MtDNA in maintenance hemodialysis patients:
Possible role in systemic inflammation. J Clin Lab Anal.
36:e245582022. View Article : Google Scholar : PubMed/NCBI
|
|
371
|
Zhou G, Li Y, Li S, Liu H, Xu F, Lai X,
Zhang Q, Xu J and Wan S: Circulating cell-free mtDNA content as a
non-invasive prognostic biomarker in HCC patients receiving TACE
and traditional Chinese medicine. Front Genet. 12:7194512021.
View Article : Google Scholar : PubMed/NCBI
|
|
372
|
Angelova PR, Andruska KM, Midei MG,
Barilani M, Atwal P, Tucher O, Milner P, Heerinckx F and Shchepinov
MS: RT001 in progressive supranuclear palsy-clinical and in-vitro
observations. Antioxidants (Basel). 10. pp. 10212021, View Article : Google Scholar
|
|
373
|
Bjørklund G, Tinkov AA, Hosnedlová B,
Kizek R, Ajsuvakova OP, Chirumbolo S, Skalnaya MG, Peana M, Dadar
M, El-Ansary A, et al: The role of glutathione redox imbalance in
autism spectrum disorder: A review. Free Radic Biol Med.
160:149–162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
374
|
Blotto BL, Lyra ML, Cardoso MCS, Trefaut
Rodrigues M, R Dias I, Marciano-Jr E, Dal Vechio F, Orrico VGD,
Brandão RA, Lopes de Assis C, et al: The phylogeny of the
casque-headed treefrogs (Hylidae: Hylinae: Lophyohylini).
Cladistics. 37:36–72. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
375
|
Langton AK, Ayer J, Griffiths TW, Rashdan
E, Naidoo K, Caley MP, Birch-Machin MA, O'Toole EA, Watson REB and
Griffiths CEM: Distinctive clinical and histological
characteristics of atrophic and hypertrophic facial photoageing. J
Eur Acad Dermatol Venereol. 35:762–768. 2021. View Article : Google Scholar :
|
|
376
|
Luo ZL, Sun HY, Wu XB, Cheng L and Ren JD:
Epigallocatechin-3-gallate attenuates acute pancreatitis induced
lung injury by targeting mitochondrial reactive oxygen species
triggered NLRP3 inflammasome activation. Food Funct. 12:5658–5667.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
377
|
Rebelo AP, Eidhof I, Cintra VP,
Guillot-Noel L, Pereira CV, Timmann D, Traschütz A, Schöls L,
Coarelli G, Durr A, et al: Biallelic loss-of-function variations in
PRDX3 cause cerebellar ataxia. Brain. 144:1467–1481. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
378
|
Wu HC, Rérolle D, Berthier C, Hleihel R,
Sakamoto T, Quentin S, Benhenda S, Morganti C, Wu C, Conte L, et
al: Actinomycin D targets NPM1c-primed mitochondria to restore
PML-driven senescence in AML therapy. Cancer Discov. 11:3198–3213.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
379
|
Feng B, Wang K, Liu J, Mao G, Cui J, Xuan
X, Jiang K and Zhang H: Ultrasensitive apurinic/apyrimidinic
site-specific ratio fluorescent rotor for real-time highly
selective evaluation of mtDNA oxidative damage in living cells.
Anal Chem. 91:13962–13969. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
380
|
Dabravolski SA, Nikiforov NG, Zhuravlev
AD, Orekhov NA, Grechko AV and Orekhov AN: Role of the mtDNA
mutations and mitophagy in inflammaging. Int J Mol Sci.
23:13232022. View Article : Google Scholar : PubMed/NCBI
|
|
381
|
Hamel Y, Mauvais FX, Madrange M, Renard P,
Lebreton C, Nemazanyy I, Pellé O, Goudin N, Tang X, Rodero MP, et
al: Compromised mitochondrial quality control triggers
lipin1-related rhabdomyolysis. Cell Rep Med. 2:1003702021.
View Article : Google Scholar : PubMed/NCBI
|
|
382
|
Karshovska E, Wei Y, Subramanian P,
Mohibullah R, Geißler C, Baatsch I, Popal A, Corbalán Campos J,
Exner N and Schober A: HIF-1α (hypoxia-inducible factor-1α)
promotes macrophage necroptosis by regulating miR-210 and miR-383.
Arterioscler Thromb Vasc Biol. 40:583–596. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
383
|
Gao F, Li L, Fan J, Cao J, Li Y, Chen L
and Peng X: An off-on two-photon carbazole-based fluorescent probe:
Highly targeting and super-resolution imaging of mtDNA. Anal Chem.
91:3336–3341. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
384
|
Grady JP, Pickett SJ, Ng YS, Alston CL,
Blakely EL, Hardy SA, Feeney CL, Bright AA, Schaefer AM, Gorman GS,
et al: mtDNA heteroplasmy level and copy number indicate disease
burden in m.3243A>G mitochondrial disease. EMBO Mol Med.
10:e82622018. View Article : Google Scholar :
|
|
385
|
Bozi LHM, Campos JC, Zambelli VO, Ferreira
ND and Ferreira JCB: Mitochondrially-targeted treatment strategies.
Mol Aspects Med. 71:1008362020. View Article : Google Scholar
|
|
386
|
Jing X, Yang F, Shao C, Wei K, Xie M, Shen
H and Shu Y: Role of hypoxia in cancer therapy by regulating the
tumor microenvironment. Mol Cancer. 18:1572019. View Article : Google Scholar : PubMed/NCBI
|
|
387
|
Amore G, Romagnoli M, Carbonelli M,
Barboni P, Carelli V and La Morgia C: Therapeutic options in
hereditary optic neuropathies. Drugs. 81:57–86. 2021. View Article : Google Scholar :
|
|
388
|
Chen JJ and Bhatti MT: Gene therapy for
leber hereditary optic neuropathy: Is vision truly RESCUED?
Ophthalmology. 128:661–662. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
389
|
Mejia-Vergara AJ, Seleme N, Sadun AA and
Karanjia R: Pathophysiology of conversion to symptomatic leber
hereditary optic neuropathy and therapeutic implications: A review.
Curr Neurol Neurosci Rep. 20:112020. View Article : Google Scholar : PubMed/NCBI
|
|
390
|
Newman NJ, Yu-Wai-Man P, Carelli V, Moster
ML, Biousse V, Vignal-Clermont C, Sergott RC, Klopstock T, Sadun
AA, Barboni P, et al: Efficacy and safety of intravitreal gene
therapy for leber hereditary optic neuropathy treated within 6
months of disease onset. Ophthalmology. 128:649–660. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
391
|
Stenton SL, Sheremet NL, Catarino CB,
Andreeva NA, Assouline Z, Barboni P, Barel O, Berutti R, Bychkov I,
Caporali L, et al: Impaired complex I repair causes recessive
leber's hereditary optic neuropathy. J Clin Invest.
131:e1382672021. View Article : Google Scholar
|
|
392
|
Wang L, Ding H, Chen BT, Fan K, Tian Q,
Long M, Liang M, Shi D, Yu C and Qin W: Occult primary white matter
impairment in leber hereditary optic neuropathy. Eur J Neurol.
28:2871–2881. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
393
|
Yu-Wai-Man P, Newman NJ, Carelli V, Moster
ML, Biousse V, Sadun AA, Klopstock T, Vignal-Clermont C, Sergott
RC, Rudolph G, et al: Bilateral visual improvement with unilateral
gene therapy injection for leber hereditary optic neuropathy. Sci
Transl Med. 12:eaaz74232020. View Article : Google Scholar : PubMed/NCBI
|
|
394
|
Heighton JN, Brady LI, Sadikovic B, Bulman
DE and Tarnopolsky MA: Genotypes of chronic progressive external
ophthalmoplegia in a large adult-onset cohort. Mitochondrion.
49:227–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
395
|
Wu Y, Kang L, Wu HL, Hou Y and Wang ZX:
Optical coherence tomography findings in chronic progressive
external ophthalmoplegia. Chin Med J (Engl). 132:1202–1207. 2019.
View Article : Google Scholar
|
|
396
|
Del Monte F, Angelini F, Villar AM and
Gabbarini F: The arrhythmic risk in Kearns-Sayre syndrome: Still
many questions unanswered. Europace. 23:980–981. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
397
|
Di Mambro C, Tamborrino PP and Drago F:
The arrhythmic risk in Kearns-Sayre syndrome: Still many questions
unanswered-Authors' reply. Europace. 23:981–982. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
398
|
Di Nora C, Paldino A, Miani D, Finato N,
Pizzolitto S, De Maglio G, Vendramin I, Sponga S, Nalli C, Sinagra
G and Livi U: Heart transplantation in Kearns-Sayre syndrome.
Transplantation. 103:e393–e394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
399
|
Nguyen MTB, Micieli J and Margolin E:
Teaching neuroImages: Kearns-Sayre syndrome. Neurology.
92:e519–e520. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
400
|
Ashton TM, McKenna WG, Kunz-Schughart LA
and Higgins GS: Oxidative phosphorylation as an emerging target in
cancer therapy. Clin Cancer Res. 24:2482–2490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
401
|
Bonora M, Wieckowski MR, Sinclair DA,
Kroemer G, Pinton P and Galluzzi L: Targeting mitochondria for
cardiovascular disorders: Therapeutic potential and obstacles. Nat
Rev Cardiol. 16:33–55. 2019. View Article : Google Scholar :
|
|
402
|
Ni K, Lan G, Veroneau SS, Duan X, Song Y
and Lin W: Nanoscale metal-organic frameworks for
mitochondria-targeted radiotherapy-radiodynamic therapy. Nat
Commun. 9:43212018. View Article : Google Scholar : PubMed/NCBI
|
|
403
|
Porporato PE, Filigheddu N, Pedro JMB,
Kroemer G and Galluzzi L: Mitochondrial metabolism and cancer. Cell
Res. 28:265–280. 2018. View Article : Google Scholar :
|
|
404
|
Qi T, Chen B, Wang Z, Du H, Liu D, Yin Q,
Liu B, Zhang Q and Wang Y: A pH-activatable nanoparticle for
dual-stage precisely mitochondria-targeted photodynamic anticancer
therapy. Biomaterials. 213:1192192019. View Article : Google Scholar : PubMed/NCBI
|
|
405
|
Ramachandra CJA, Hernandez-Resendiz S,
Crespo-Avilan GE, Lin YH and Hausenloy DJ: Mitochondria in acute
myocardial infarction and cardioprotection. EBioMedicine.
57:1028842020. View Article : Google Scholar : PubMed/NCBI
|
|
406
|
Soukas AA, Hao H and Wu L: Metformin as
anti-aging therapy: Is it for everyone? Trends Endocrinol Metab.
30:745–755. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
407
|
Bonora E, Chakrabarty S, Kellaris G,
Tsutsumi M, Bianco F, Bergamini C, Ullah F, Isidori F, Liparulo I,
Diquigiovanni C, et al: Biallelic variants in LIG3 cause a novel
mitochondrial neurogastrointestinal encephalomyopathy. Brain.
144:1451–1466. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
408
|
D'Angelo R, Boschetti E, Amore G, Costa R,
Pugliese A, Caporali L, Gramegna LL, Papa V, Vizioli L, Capristo M,
et al: Liver transplantation in mitochondrial neurogastrointestinal
encephalomyopathy (MNGIE): Clinical long-term follow-up and
pathogenic implications. J Neurol. 267:3702–3710. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
409
|
Hirano M, Carelli V, De Giorgio R, Pironi
L, Accarino A, Cenacchi G, D'Alessandro R, Filosto M, Martí R,
Nonino F, et al: Mitochondrial neurogastrointestinal
encephalomyopathy (MNGIE): Position paper on diagnosis, prognosis,
and treatment by the MNGIE international network. J Inherit Metab
Dis. 44:376–387. 2021. View Article : Google Scholar :
|
|
410
|
Kripps K, Nakayuenyongsuk W, Shayota BJ,
Berquist W, Gomez-Ospina N, Esquivel CO, Concepcion W, Sampson JB,
Cristin DJ, Jackson WE, et al: Successful liver transplantation in
mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). Mol
Genet Metab. 130:58–64. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
411
|
Parés M, Fornaguera C, Vila-Julià F, Oh S,
Fan SHY, Tam YK, Comes N, Vidal F, Martí R, Borrós S and Barquinero
J: Preclinical assessment of a gene-editing approach in a mouse
model of mitochondrial neurogastrointestinal encephalomyopathy. Hum
Gene Ther. 32:1210–1223. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
412
|
Jackson CB, Turnbull DM, Minczuk M and
Gammage PA: Therapeutic manipulation of mtDNA heteroplasmy: A
shifting perspective. Trends Mol Med. 26:698–709. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
413
|
Jiang Z and Shen H: Mitochondria: Emerging
therapeutic strategies for oocyte rescue. Reprod Sci. 29:711–722.
2022. View Article : Google Scholar
|
|
414
|
Mok BY, de Moraes MH, Zeng J, Bosch DE,
Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, et
al: A bacterial cytidine deaminase toxin enables CRISPR-free
mitochondrial base editing. Nature. 583:631–637. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
415
|
Ng YS, Bindoff LA, Gorman GS, Klopstock T,
Kornblum C, Mancuso M, McFarland R, Sue C M, Suomalainen A, Taylor
RW, et al: Mitochondrial disease in adults: Recent advances and
future promise. Lancet Neurol. 20:573–584. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
416
|
Fang H, Yao S, Chen Q, Liu C, Cai Y, Geng
S, Bai Y, Tian Z, Zacharias AL, Takebe T, et al: De novo-designed
near-infrared nanoaggregates for super-resolution monitoring of
lysosomes in cells, in whole organoids, and in vivo. ACS Nano.
13:14426–14436. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
417
|
Gong X, Pu X, Wang J, Yang L, Cui Y, Li L,
Sun X, Liu J, Bai J and Wang Y: Enhancing of nanocatalyst-driven
chemodynaminc therapy for endometrial cancer cells through
inhibition of PINK1/parkin-mediated mitophagy. Int J Nanomedicine.
16:6661–6679. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
418
|
González LF, Bevilacqua LE and Naves R:
Nanotechnology-based drug delivery strategies to repair the
mitochondrial function in neuroinflammatory and neurodegenerative
diseases. Pharmaceutics. 13:20552021. View Article : Google Scholar : PubMed/NCBI
|
|
419
|
Gu X, Kwok RTK, Lam JWY and Tang BZ:
AIEgens for biological process monitoring and disease theranostics.
Biomaterials. 146:115–135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
420
|
He C, Jiang S, Yao H, Zhang L, Yang C,
Jiang S, Ruan F, Zhan D, Liu G, Lin Z, et al: High-content analysis
for mitophagy response to nanoparticles: A potential sensitive
biomarker for nanosafety assessment. Nanomedicine. 15:59–69. 2019.
View Article : Google Scholar
|
|
421
|
He G, Pan X, Liu X, Zhu Y, Ma Y, Du C, Liu
X and Mao C: HIF-1α-mediated mitophagy determines ZnO
nanoparticle-induced human osteosarcoma cell death both in vitro
and in vivo. ACS Appl Mater Interfaces. 12:48296–48309. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
422
|
Zhao M, Liu S, Wang C, Wang Y, Wan M, Liu
F, Gong M, Yuan Y, Chen Y, Cheng J, et al: Mesenchymal stem
cell-derived extracellular vesicles attenuate mitochondrial damage
and inflammation by stabilizing mitochondrial DNA. ACS Nano.
15:1519–1538. 2021. View Article : Google Scholar
|
|
423
|
Macdonald R, Barnes K, Hastings C and
Mortiboys H: Mitochondrial abnormalities in Parkinson's disease and
Alzheimer's disease: Can mitochondria be targeted therapeutically?
Biochem Soc Trans. 46:891–909. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
424
|
Tan DX, Manchester LC, Liu X,
Rosales-Corral SA, Acuna-Castroviejo D and Reiter RJ: Mitochondria
and chloroplasts as the original sites of melatonin synthesis: A
hypothesis related to melatonin's primary function and evolution in
eukaryotes. J Pineal Res. 54:127–138. 2013. View Article : Google Scholar
|
|
425
|
Lee JH, Park A, Oh KJ, Lee SC, Kim WK and
Bae KH: The role of adipose tissue mitochondria: Regulation of
mitochondrial function for the treatment of metabolic diseases. Int
J Mol Sci. 20:49242019. View Article : Google Scholar :
|
|
426
|
Wallace DC: Mitochondrial genetic
medicine. Nat Genet. 50:1642–1649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
427
|
Strobbe D and Campanella M: Anxiolytic
therapy: A paradigm of successful mitochondrial pharmacology.
Trends Pharmacol Sci. 39:437–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
428
|
Wang XQ, Peng M, Li CX, Zhang Y, Zhang M,
Tang Y, Liu MD, Xie BR and Zhang XZ: Real-time imaging of free
radicals for mitochondria-targeting hypoxic tumor therapy. Nano
Lett. 18:6804–6811. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
429
|
Kim HK, Noh YH, Nilius B, Ko KS, Rhee BD,
Kim N and Han J: Current and upcoming mitochondrial targets for
cancer therapy. Semin Cancer Biol. 47:154–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
430
|
Lleonart ME, Grodzicki R, Graifer DM and
Lyakhovich A: Mitochondrial dysfunction and potential anticancer
therapy. Med Res Rev. 37:1275–1298. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
431
|
Tian J, Huang B, Cui Z, Wang P, Chen S,
Yang G and Zhang W: Mitochondria-targeting and ROS-sensitive smart
nanoscale supramolecular organic framework for combinational
amplified photodynamic therapy and chemotherapy. Acta Biomater.
130:447–459. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
432
|
Kim HJ, Maiti P and Barrientos A:
Mitochondrial ribosomes in cancer. Semin Cancer Biol. 47:67–81.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
433
|
Chen WW, Freinkman E and Sabatini DM:
Rapid immunopurification of mitochondria for metabolite profiling
and absolute quantification of matrix metabolites. Nat Protoc.
12:2215–2231. 2017. View Article : Google Scholar
|
|
434
|
Jung HS, Lee JH, Kim K, Koo S, Verwilst P,
Sessler JL, Kang C and Kim JS: A mitochondria-targeted
cryptocyanine-based photothermogenic photosensitizer. J Am Chem
Soc. 139:9972–9978. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
435
|
Roth KG, Mambetsariev I, Kulkarni P and
Salgia R: The mitochondrion as an emerging therapeutic target in
cancer. Trends Mol Med. 26:119–134. 2020. View Article : Google Scholar :
|
|
436
|
Nash GT, Luo T, Lan G, Ni K, Kaufmann M
and Lin W: Nanoscale metal-organic layer isolates phthalocyanines
for efficient mitochondria-targeted photodynamic therapy. J Am Chem
Soc. 143:2194–2199. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
437
|
Russell OM, Gorman GS, Lightowlers RN and
Turnbull DM: Mitochondrial diseases: Hope for the future. Cell.
181:168–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
438
|
Saeb-Parsy K, Martin JL, Summers DM,
Watson CJE, Krieg T and Murphy MP: Mitochondria as therapeutic
targets in transplantation. Trends Mol Med. 27:185–198. 2021.
View Article : Google Scholar
|
|
439
|
Kelly B and Pearce EL: Amino assets: How
amino acids support immunity. Cell Metab. 32:154–175. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
440
|
Rahman J and Rahman S: Mitochondrial
medicine in the omics era. Lancet. 391:2560–2574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
441
|
Tabish TA and Narayan RJ:
Mitochondria-targeted graphene for advanced cancer therapeutics.
Acta Biomater. 129:43–56. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
442
|
Yuan P, Deng FA, Liu YB, Zheng RR, Rao XN,
Qiu XZ, Zhang DW, Yu XY, Cheng H and Li SY: Mitochondria targeted
O2 economizer to alleviate tumor hypoxia for enhanced
photodynamic therapy. Adv Healthc Mater. 10:e21001982021.
View Article : Google Scholar
|
|
443
|
Ballarò R, Lopalco P, Audrito V, Beltrà M,
Pin F, Angelini R, Costelli P, Corcelli A, Bonetto A, Szeto HH, et
al: Targeting mitochondria by SS-31 ameliorates the whole body
energy status in cancer- and chemotherapy-induced cachexia. Cancers
(Basel). 13. pp. 8502021, View Article : Google Scholar
|
|
444
|
Bhatti JS, Tamarai K, Kandimalla R,
Manczak M, Yin X, Ramasubramanian B, Sawant N, Pradeepkiran JA,
Vijayan M, Kumar S and Reddy PH: Protective effects of a
mitochondria-targeted small peptide SS31 against
hyperglycemia-induced mitochondrial abnormalities in the liver
tissues of diabetic mice, Tallyho/JngJ mice. Mitochondrion.
58:49–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
445
|
Deng HF, Yue LX, Wang NN, Zhou YQ, Zhou W,
Liu X, Ni YH, Huang CS, Qiu LZ, Liu H, et al: Mitochondrial iron
overload-mediated inhibition of Nrf2-HO-1/GPX4 assisted ALI-induced
nephrotoxicity. Front Pharmacol. 11:6245292021. View Article : Google Scholar : PubMed/NCBI
|
|
446
|
Le Gal K, Wiel C, Ibrahim MX, Henricsson
M, Sayin VI and Bergo MO: Mitochondria-targeted antioxidants MitoQ
and MitoTEMPO Do not influence BRAF-driven malignant melanoma and
KRAS-driven lung cancer progression in mice. Antioxidants (Basel).
10:1632021. View Article : Google Scholar
|
|
447
|
Bhatti JS, Thamarai K, Kandimalla R,
Manczak M, Yin X, Kumar S, Vijayan M and Reddy PH:
Mitochondria-targeted small peptide, SS31 ameliorates diabetes
induced mitochondrial dynamics in male TallyHO/JngJ mice. Mol
Neurobiol. 58:795–808. 2021. View Article : Google Scholar :
|
|
448
|
Grosser JA, Fehrman RL, Keefe D, Redmon M
and Nickells RW: The effects of a mitochondrial targeted peptide
(elamipretide/SS31) on BAX recruitment and activation during
apoptosis. BMC Res Notes. 14:1982021. View Article : Google Scholar : PubMed/NCBI
|
|
449
|
He Y, Chen Z, Zhang R, Quan Z, Xu Y, He B
and Ren Y: Mitochondrial-targeted antioxidant peptide SS31 prevents
RPE cell death under oxidative stress. Biomed Res Int.
2022:61803492022. View Article : Google Scholar : PubMed/NCBI
|
|
450
|
He Y, Quan Z, Zhang R, He B, Xu Y, Chen Z,
Ren Y and Li K: Preparation of targeted mitochondrion
nanoscale-release peptides and their efficiency on eukaryotic
cells. J Biomed Nanotechnol. 17:1679–1689. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
451
|
He Y, Zhang R, Quan Z, He B, Xu Y, Chen Z,
Ren Y and Liu X: Synthesis, characterization, and specific
localization of mitochondrial-targeted antioxidant peptide SS31
probe. Biomed Res Int. 2021:99156992021. View Article : Google Scholar : PubMed/NCBI
|
|
452
|
Sun M, Ma J, Ye J, Fan H, Le J and Zhu J:
Protective effect of mitochondria-targeted antioxidant peptide
SS-31 in sepsis-induced acute kidney injury. Zhonghua Wei Zhong
Bing Ji Jiu Yi Xue. 33:1418–1422. 2021.In Chinese.
|
|
453
|
Zhu Y, Luo M, Bai X, Li J, Nie P, Li B and
Luo P: SS-31, a mitochondria-targeting peptide, ameliorates kidney
disease. Oxid Med Cell Longev. 2022:12955092022. View Article : Google Scholar : PubMed/NCBI
|
|
454
|
Olgar Y, Billur D, Tuncay E and Turan B:
MitoTEMPO provides an antiarrhythmic effect in aged-rats through
attenuation of mitochondrial reactive oxygen species. Exp Gerontol.
136:1109612020. View Article : Google Scholar : PubMed/NCBI
|
|
455
|
Tuncer S, Akkoca A, Celen MC and Dalkilic
N: Can MitoTEMPO protect rat sciatic nerve against
ischemia-reperfusion injury? Naunyn Schmiedebergs Arch Pharmacol.
394:545–553. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
456
|
Vrijsen S, Besora-Casals L, van Veen S,
Zielich J, Van den Haute C, Hamouda NN, Fischer C, Ghesquière B,
Tournev I, Agostinis P, et al: ATP13A2-mediated endo-lysosomal
polyamine export counters mitochondrial oxidative stress. Proc Natl
Acad Sci USA. 117:31198–31207. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
457
|
Wang Y, Zhao Y, Wang Z, Sun R, Zou B, Li
R, Liu D, Lin M, Zhou J, Ning S, et al: Peroxiredoxin 3 inhibits
acetaminophen-induced liver pyroptosis through the regulation of
mitochondrial ROS. Front Immunol. 12:6527822021. View Article : Google Scholar : PubMed/NCBI
|
|
458
|
Gao J, Zhan J and Yang Z:
Enzyme-instructed self-assembly (EISA) and hydrogelation of
peptides. Adv Mater. 32:e18057982020. View Article : Google Scholar
|
|
459
|
Liu C, Liu B, Zhao J, Di Z, Chen D, Gu Z,
Li L and Zhao Y: Nd3+-sensitized upconversion
metal-organic frameworks for mitochondria-targeted amplified
photodynamic therapy. Angew Chem Int Ed Engl. 59:2634–2638. 2020.
View Article : Google Scholar
|
|
460
|
Lu M, Qu A, Li S, Sun M, Xu L, Kuang H and
Xu C: Mitochondria-targeting plasmonic spiky nanorods increase the
elimination of aging cells in vivo. Angew Chem Int Ed Engl.
59:8698–8705. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
461
|
Li C, Zhang W, Liu S, Hu X and Xie Z:
Mitochondria-targeting organic nanoparticles for enhanced
photodynamic/photothermal therapy. ACS Appl Mater Interfaces.
12:30077–30084. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
462
|
Zhang CX, Cheng Y, Liu DZ, Liu M, Cui H,
Zhang BL, Mei QB and Zhou SY: Mitochondria-targeted cyclosporin A
delivery system to treat myocardial ischemia reperfusion injury of
rats. J Nanobiotechnology. 17:182019. View Article : Google Scholar : PubMed/NCBI
|
|
463
|
Sun J, Zhang J, Tian J, Virzì GM, Digvijay
K, Cueto L, Yin Y, Rosner MH and Ronco C: Mitochondria in
sepsis-induced AKI. J Am Soc Nephrol. 30:1151–1161. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
464
|
Yang G, Chen C, Zhu Y, Liu Z, Xue Y, Zhong
S, Wang C, Gao Y and Zhang W: GSH-activatable NIR nanoplatform with
mitochondria targeting for enhancing tumor-specific therapy. ACS
Appl Mater Interfaces. 11:44961–44969. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
465
|
Gabandé-Rodríguez E, Gómez de Las Heras MM
and Mittelbrunn M: Control of inflammation by calorie restriction
mimetics: On the crossroad of autophagy and mitochondria. Cells.
9:822019. View Article : Google Scholar
|
|
466
|
Cho H, Cho YY, Shim MS, Lee JY, Lee HS and
Kang HC: Mitochondria-targeted drug delivery in cancers. Biochim
Biophys Acta Mol Basis Dis. 1866:1658082020. View Article : Google Scholar : PubMed/NCBI
|
|
467
|
Liu K, Zhou Z, Pan M and Zhang L: Stem
cell-derived mitochondria transplantation: A promising therapy for
mitochondrial encephalomyopathy. CNS Neurosci Ther. 27:733–742.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
468
|
Deng Y, Jia F, Chen X, Jin Q and Ji J: ATP
suppression by pH-activated mitochondria-targeted delivery of
nitric oxide nanoplatform for drug resistance reversal and
metastasis inhibition. Small. 16:e20017472020. View Article : Google Scholar : PubMed/NCBI
|
|
469
|
Gao C, Wang Y, Sun J, Han Y, Gong W, Li Y,
Feng Y, Wang H, Yang M, Li Z, et al: Neuronal mitochondria-targeted
delivery of curcumin by biomimetic engineered nanosystems in
Alzheimer's disease mice. Acta Biomater. 108:285–299. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
470
|
Andrieux P, Chevillard C, Cunha-Neto E and
Nunes JPS: Mitochondria as a cellular hub in infection and
inflammation. Int J Mol Sci. 22:113382021. View Article : Google Scholar : PubMed/NCBI
|
|
471
|
Zeng WN, Yu QP, Wang D, Liu JL, Yang QJ,
Zhou ZK and Zeng YP: Mitochondria-targeting graphene oxide
nanocomposites for fluorescence imaging-guided synergistic
phototherapy of drug-resistant osteosarcoma. J Nanobiotechnology.
19:792021. View Article : Google Scholar : PubMed/NCBI
|
|
472
|
Nam HY, Hong JA, Choi J, Shin S, Cho SK,
Seo J and Lee J: Mitochondria-targeting peptoids. Bioconjug Chem.
29:1669–1676. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
473
|
El-Hattab AW, Zarante AM, Almannai M and
Scaglia F: Therapies for mitochondrial diseases and current
clinical trials. Mol Genet Metab. 122:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
474
|
Han Y, Chu X, Cui L, Fu S, Gao C, Li Y and
Sun B: Neuronal mitochondria-targeted therapy for Alzheimer's
disease by systemic delivery of resveratrol using dual-modified
novel biomimetic nanosystems. Drug Deliv. 27:502–518. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
475
|
Mohammadinejad R, Moosavi MA, Tavakol S,
Vardar DÖ, Hosseini A, Rahmati M, Dini L, Hussain S, Mandegary A
and Klionsky DJ: Necrotic, apoptotic and autophagic cell fates
triggered by nanoparticles. Autophagy. 15:4–33. 2019. View Article : Google Scholar
|
|
476
|
Vincent AE, Turnbull DM, Eisner V,
Hajnóczky G and Picard M: Mitochondrial nanotunnels. Trends Cell
Biol. 27:787–799. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
477
|
Wu T, Liang X, Liu X, Li Y, Wang Y, Kong L
and Tang M: Induction of ferroptosis in response to graphene
quantum dots through mitochondrial oxidative stress in microglia.
Part Fibre Toxicol. 17:302020. View Article : Google Scholar : PubMed/NCBI
|
|
478
|
Wang H, Shi W, Zeng D, Huang Q, Xie J, Wen
H, Li J, Yu X, Qin L and Zhou Y: pH-activated,
mitochondria-targeted, and redox-responsive delivery of paclitaxel
nanomicelles to overcome drug resistance and suppress metastasis in
lung cancer. J Nanobiotechnology. 19:1522021. View Article : Google Scholar : PubMed/NCBI
|