Introduction
Colorectal cancer (CRC) is the most prevalent
malignant neoplasm within the gastrointestinal tract, affecting
1.36 million individuals globally and accounting for ~10% of all
cancer cases worldwide (1).
Previous research studies have indicated that colon cancer results
in ~700,000 deaths each year, ranking it fourth in
cancer-associated deaths globally (2). While chemotherapeutic agents
significantly improve outcomes in advanced-stage CRC, their
efficacy is limited to only 15-25% of the patient population
(3,4). Consequently, the identification of
novel oncogenic mechanisms is essential for enhancing our
understanding of CRC pathophysiology and for developing novel
therapeutic approaches.
In recent years, advancements in the understanding
of tumor development have highlighted the involvement of autophagy
in CRC (5,6). Autophagy, a cellular
self-degradation mechanism that maintains a consistent energy
supply, acts as a quality control system during the early stages of
tumorigenesis by preventing tissue damage and genomic instability,
thereby inhibiting cancer progression (7-9).
Within the tumor microenvironment (TME), increased autophagic flux
generally facilitates tumor cell survival and proliferation
(10). Furthermore, the concept
of 'regulated cell death' encompasses the following three distinct
mechanisms: Apoptosis, autophagy-dependent cell death and necrosis.
The induction of cell death in cancer cells is fundamental to
numerous cancer treatment strategies (11). Biological agents that trigger
autophagy-dependent cell death in cancer cells are recognized as
valuable tools in cancer therapy.
In the previous study conducted by our group,
acetylated RNA immunoprecipitation and sequencing (acRIP-seq) of
CRC was performed indicating that YWHAH drives progression via ac4C
modification (12). Subsequent
bioinformatic analysis indicated that YWHAH could modulate
autophagy and apoptosis in CRC via the MAPK/ERK axis. 14-3-3
proteins, including YWHAH, are conserved regulators found in all
eukaryotes. YWHAH can interact with target proteins, thereby
modulating protein-protein interactions and the functionality of
their ligands. This is achieved via specific mechanisms, such as
induction of structural modifications, masking or unmasking of
functional sites and alteration of the intracellular localization
of the ligands (13,14). YWHAH exhibits high specificity
for various ligands and plays significant roles in numerous
physiological processes (13,14).
In the present study, the expression levels of YWHAH
in CRC were investigated, together with its relationship with
autophagy and associated mechanisms, thereby providing insights
into CRC pathogenesis.
Materials and methods
Tissue specimens
Tissue specimens from patients with primary CRC
treated at the Second Affiliated Hospital of Xuzhou Medical
University were collected from January 2018 to December 2023. Among
the 85 patients, there were 43 males and 42 females, with ages
ranging from 35 to 69 years and a median age of 56 years. The
Ethics Committee of Xuzhou Medical University approved the use of
these clinical samples (approval no. 2023100505) and all patients
provided written informed consent. The animal experiment of the
present study was approved by the Animal Ethics Committee of Xuzhou
Medical University (approval no. 2023110512; Xuzhou, China).
Small interfering (si)-RNA, plasmid
synthesis and transfection
The CRC cell lines LOVO and HCT116, were provided
from the Chinese Academy of Sciences' Cell Bank. LOVO and HCT116
cells were cultured in DMEM (cat. no. G4515-500ML; Wuhan Servicebio
Technology Co., Ltd.). The media were supplemented with 10% fetal
bovine serum (FBS; Wuhan Servicebio Technology Co., Ltd.),
streptomycin and penicillin (×100). YWHAH-targeting siRNAs and a
negative control (NC) were synthesized by Ruijie Bio (https://www.11467.com/shanghai/co/936727.htm). LOVO
and HCT116 cells were transfected using Endo-Free Plasmid Midi Kit
(Omega Corporation) with YWHAH siRNAs (15 pmol) or NC-siRNA at 37°C
for 24-48 h, and the effects were assessed using reverse
transcription-quantitative PCR (RT-qPCR). The siRNA sequences are
provided in Table SI. The
verification of knockdown and overexpression efficiency is shown in
Fig. S1, and siR-2 was selected
for subsequent studies.
RT-qPCR
The CRC cell and tissue RNAs were extracted using
TRIzol® (Thermo Fisher Scientific, Inc.). Reverse
transcription reaction was used to convert the RNA to cDNA and
YWHAH primers synthesized by Takara Bio USA, Inc according to the
manufacturer's protocol. The cDNA was chilled for subsequent use.
qPCR was conducted on a LongGene Q2000B, starting hybridization for
2 min at 50°C and subsequent amplification for 40 cycles at 95°C
for 2 min, 95°C for 15 sec (denaturation), and 60°C for 60 sec
(extension). Primer sequences for qPCR experiments are provided in
Table SII. The
2−∆∆Cq method was applied for mRNA analysis and GAPDH
was used as the reference gene.
Cell Counting Kit-8 (CCK-8) assays
CCK-8 assays were conducted using a kit (Beyotime
Institute of Biotechnology). CRC cells (5×103
cells/well) were incubated in 96-well plates. Following reagent
treatments, the medium in each well was substituted with 100
μl CCK-8 solution (comprising 90 μl fresh medium and
10 μl CCK-8 reagent). The color reaction starts at 37°C and
was allowed to proceed for 2 h to maintain consistency. The
measurement of optical density at 450 nm was performed using a
spectrophotometer.
Flow cytometry
During Bisphenol A (BPA, 99%; MilliporeSigma)
treatment, 5×105 CRC cells per well were incubated in
6-well plates and cultured overnight; they were harvested 48 h
later and resuspended in binding buffer. The cell suspension was
incubated away from light for 20 min with Annexin V-fluorescein
isothiocyanate and propidium iodide (Beyotime Institute of
Biotechnology). Late apoptosis was subsequently assessed via flow
cytometry (CellQuestPro software; BD FACSCalibur; BD
Biosciences).
Western blotting
The proteins were extracted using RIPA buffer
(Beyotime Institute of Biotechnology) with protease inhibitors,
incubated at 4°C for 30 min and centrifuged (12,000 × g, 20 min, at
4°C). The protein concentration levels in the supernatants were
assessed by BCA assays and 30 μg samples were
electrophoresed using 8% SDS-PAGE gels prior to their transfer to
nitrocellulose membrane. Following blocking (5% skimmed milk) at
37°C for 30 min, the membranes were incubated overnight at 4°C with
primary antibodies (Table SIII)
at a ratio of 1:5,000 according to the antibody instructions (GAPDH
was diluted at a ratio of 1:5,000), followed by incubation for 30
min at room temperature with a Goat Anti-Rabbit IgG H&L
secondary antibody (cat. no. 111-035-003; Jackson ImmunoResearch
Laboratories, Inc.), which was diluted and prepared according to
the antibody instructions. The protein bands were detected and
visualized using the ChemiDocTM XRS+ system (Bio-Rad
Laboratories, Inc.) and Ultra-sensitive ECL chemiluminescence kit
(Beyotime Institute of Biotechnology). Image software (V.1.8.0;
National Institutes of Health) was used for densitometric
analysis.
RFP-GFP-hLC3 dual fluorescence
microscopy
A two-color fluorescence fusion protein, namely
RFP-GFP-hLC3, was used to monitor autophagy. The fusion protein
emits green and red fluorescence signals upon localization to
autophagosomes, resulting in yellow signals in merged images during
autophagy induction. The cells were plated onto glass cover slips
(1×1.5 cm) in Corning 6-well plates overnight to adhere. A total of
6 h after transfection with siRNA and plasmids, cells were
subsequently transfected with tandem RFP-GFP-hLC3 adenoviruses
[pGMLV-CMV-RFP-GFP-hLC3-Puro Lentivirus, Genomeditech (Shanghai)
Co., Ltd.] according to the manufacturer's instructions. Following
the 24-h incubation, the cells were rinsed with PBS at room
temperature, fixed in 4% paraformaldehyde at room temperature for
10-20 min, counterstained with 4',6-diamidino-2-phenylindole and
subsequently covered with coverslips. Confocal images were obtained
with the Olympus FV1000 laser scanning confocal fluorescence
microscope. Quantification of autophagosomes with green and red
fluorescence was performed using ImageJ software (V.1.8.0, National
institutes of Health) by counting the LC3 puncta per cell.
Animal models
To establish the model, the cells in the logarithmic
growth phase were selected; the old culture medium was discarded
from the dish, followed by washing once with PBS. The cells were
subsequently digested with 0.25% trypsin for 1-2 min and FBS was
added promptly to terminate digestion and prevent over-digestion.
Following low-speed centrifugation (1,000 × g, 5 min) to collect
the cells, they were washed twice with PBS, resuspended in
serum-free medium, pipetted evenly, counted and adjusted to a
density of 3×107 cells/ml with sterile PBS. A total of
24 4-6-week-old healthy SPF-grade female BALB/c-nu nude mice with
similar body weights SPF-grade female BALB/c-nu nude mice were
randomly selected from the breeding cage, and 100-μl tumor
cells (3×106 cells) were subcutaneously incubated at the
axilla of the right forelimb, with the injection site pressed with
a sterile cotton swab to terminate bleeding prior to returning the
mice to a sterilized cage for continuous feeding. The temperature
was controlled at 20-26°C, and the humidity maintained between
40-70%. For the light/dark cycle, a 12/12-h light/dark cycle was
adopted. The air exchange rate was 10-15-fold per h, and clean,
special breeders provided food and sterile drinking water. The
daily observations of the mental state of the mice, their activity,
diet and defecation were conducted, while their body weights were
measured weekly with an electronic balance and the longest and
shortest diameters of the subcutaneous xenografts were obtained
with a vernier caliper to calculate the volume using the following
formula: V (mm3)=longest diameter (mm) × shortest
diameter × shortest diameter × 0.5. This equation was used for
plotting the growth curve with time on the abscissa and tumor
volume on the ordinate (The maximum volume of a single tumor did
not exceed 1,000 mm3). A total of 5 weeks later, the
nude mice were weighed, euthanized by cervical dislocation and the
xenografts were completely removed. The excised tumors were washed
with sterile PBS, images were captured by experimental group on
white paper, weighed and the data were recorded prior to storage at
−80°C.
Transwell detect cell migration and
invasion
Cells were collected and gently blown into a fresh
complete medium to obtain a uniform suspension. The cells were
counted and spread onto 6-well plates in a 37°C 5% CO2
incubator overnight before transfection. After 24 h, the cells were
collected for analysis. Pancreatic enzyme digestion was performed
at 1,000 rpm for 5 min, then the supernatant was removed. Cells
were resuspended in 5-ml sterile PBS, a small sample was counted
using a hemocytometer at 1,000 rpm, and cells were collected again
after 5 min. In total, 3 ml of serum-free medium was added to
adjust cell density to 2×105/ml. Cells were added to
sterile 24-well plates with 500-μl complete medium per
well.; cells were transferred to 8-μm Transwell chambers
using forceps and gently inserted into the plate. The cell
suspension was mixed and 200 μl were added to the Transwell
chamber. Incubation lasted for 48 h, then cells were rinsed twice
with PBS. In total, 500 μl 4% PFA was added at room
temperature for 20 min. PFA was removed, washing three times with
PBS, and cells were stained with crystal violet for 20 min. Washing
three times with PBS was performed to remove unbound dye. The
Transwell chamber surface was gently wiped with a cotton swab to
remove residual dye, then rinsed once with PBS and allowed to
air-dry. Chambers were placed directly on a microscope slide and
images of purple-stained cells were captured.
The MAPK/ERK signaling pathway inhibitor PD98059 was
purchased from Beyotime Institute of Biotechnology, while its
agonist U-46619 and the autophagy inducer RAPA were obtained from
MedChemExpress. Additionally, 3-methoxyamphetamine (3-MA) was
purchased from OriLeaf Biotechnology. Based on the concentration
screening results, 2 μmol/l rapamycin, 5 mmol/l 3-MA, 20
μM PD98059, and 2 μM U46619 were selected for the
experiment.
Immunohistochemical assay
The tissues were treated with 4% paraformaldehyde at
room temperature for 24 h, rinsed in running water and dehydrated
using gradient alcohols. Dehydrated tissues were embedded in wax
using an embedding machine, cooled at −20°C, trimmed and sectioned
at 4-μm thickness slices. Following incubation at 40°C for
1-3 min (on a water surface to flatten), the sections were
retrieved with glass slides, incubated at 60°C and stored. For
immunohistochemical staining, the sections were incubated at 62°C
for 1 h, the wax was removed and rehydrated prior to antigen
retrieval in citrate buffer (pH 6.0) with microwave treatment
(high-power for 1 min, medium-low for 9 min). Following cooling,
the sections were washed with PBS and treated with 3%
H2O in methanol to block endogenous peroxidase activity,
followed by PBS washing. Blocking was performed with diluted sheep
serum (1:10) (ZSGM-BIO) for 1 h at 37°C. Primary antibody
incubation with anti-rabbit YWHAH (1:1,000) was carried out
overnight at 4°C, followed by 30 min at room temperature, and
washing with PBST (1X PBS + 0.05% Tween-20). An HRP-labeled
secondary antibody (1:1,000) was added for 30 min at 37°C, followed
by PBST washing. DAB color development lasted for 1 min; it was
terminated with water and examined under a microscope.
Counterstaining with hematoxylin for 30 sec was followed by washing
and examination. The sections underwent dehydration, clearing and
mounting with neutral gum.
Online database
The RNA-sequencing expression (level 3) profiles and
the corresponding clinical information for CRC were downloaded from
The Cancer Genome Atlas (https://portal.gdc.cancer.gov/analysis_page?app=Downloads).
Statistical analysis
The data were subjected to analysis using SPSS
version 21.0 (IBM Corp.) or GraphPad Prism version 10.1.2
(Dotmatics). The nominal and absolute values were assessed
employing either paired t-tests or one-way ANOVA. All analyses were
conducted utilizing R version 4.0.3 (https://www.r-project.org/), along with the
appropriate R packages. P<0.05 was considered to indicate a
statistically significant difference. Data are expressed as the
mean ± standard deviation (SD). Error bars in all figures indicate
SD from three biological replicates.
Results
Association between YWHAH and clinical
pathological characteristics of CRC
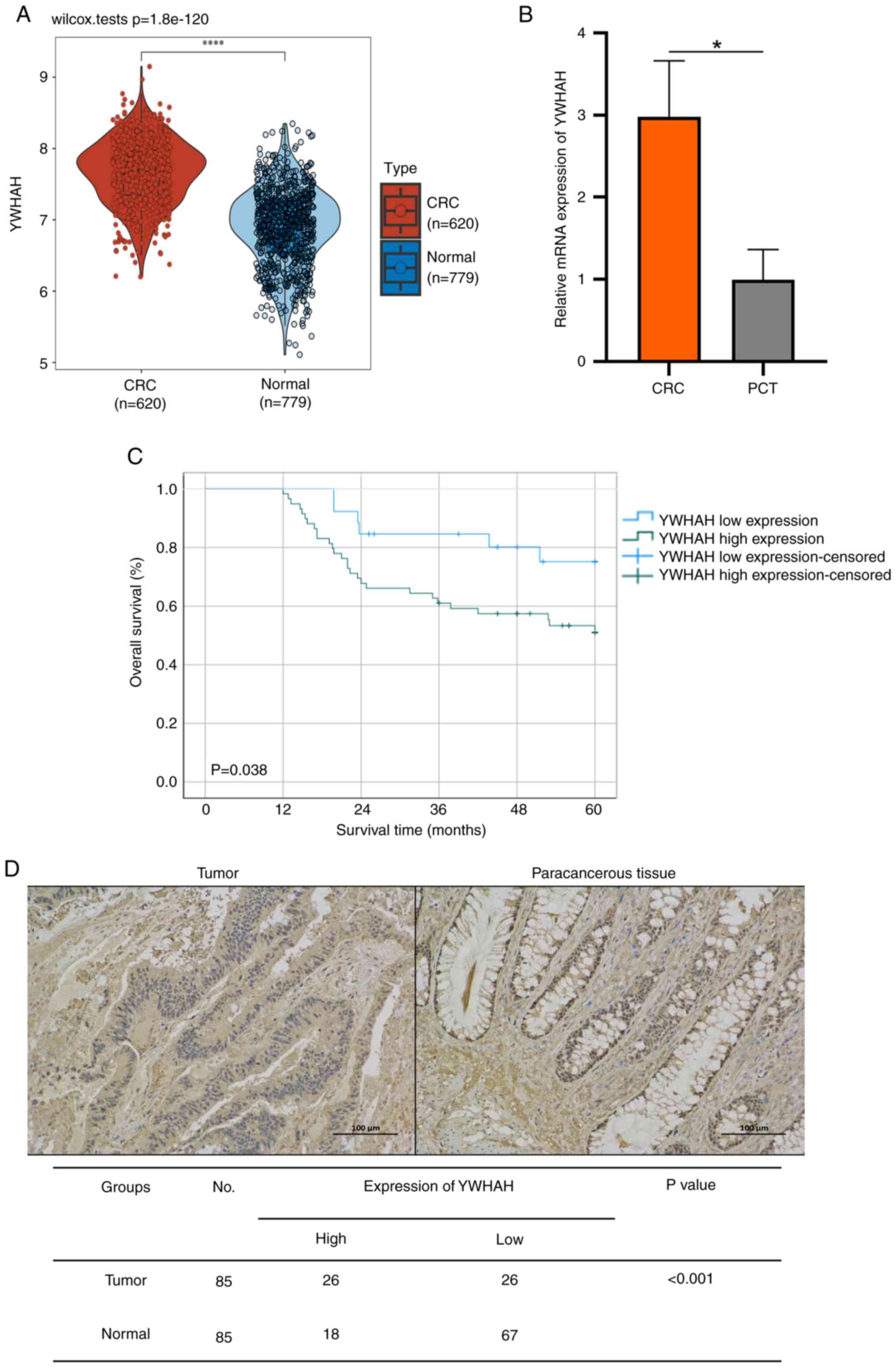
In a previous study using acRIP-seq, the involvement
of YWHAH in CRC was established. The analysis with the R software
markedly raised YWHAH levels in tumors relative to normal tissues
(Fig. 1A). RT-qPCR confirmed
this result in CRC tumor and paracancerous tissue (PCT) (Fig. 1B). Immunohistochemical analysis
was performed on 85 CRC tissue pairs (tumor and PCT), revealing
notably high YWHAH levels in tumors (P<0.001; Fig. 1D) and a strong association with
poor overall survival (P=0.038; Fig.
1C). YWHAH expression was significantly linked to CRC tissue
differentiation (P=0.024), tumor node metastasis (TNM) stage
(P=0.011), N stage (P=0.028) and vascular invasion (P=0.04;
Table I).
 | Table IThe relationship between YWHAH
protein expression and clinicopathological parameters in patients
with CRC. |
Table I
The relationship between YWHAH
protein expression and clinicopathological parameters in patients
with CRC.
| Clinicopathological
characteristics | Total (n=85) | Expression level of
YWHAH
| P-value |
|---|
| High | Low |
|---|
| CRC | 85 | 59 (69.4%) | 26 (30.6%) | |
| Age, years | | | | 0.806 |
| ≥60 | 48 | 36 | 12 | |
| <60 | 37 | 23 | 14 | |
| Sex | | | | 0.174 |
| Female | 42 | 29 | 13 | |
| Male | 43 | 30 | 13 | |
| Tumor size | | | | 0.335 |
| ≥5 cm | 36 | 22 | 14 | |
| <5 cm | 49 | 37 | 12 | |
| G stage | | | | 0.024 |
| G1 | 33 | 22 | 11 | |
| G2 | 34 | 24 | 10 | |
| G3 | 18 | 13 | 5 | |
| T stage | | | | 0.673 |
| T1/T2 | 17 | 11 | 6 | |
| T3/T4 | 68 | 48 | 20 | |
| TNM stage | | | | 0.011 |
| I/II | 49 | 35 | 14 | |
| III/IV | 36 | 24 | 12 | |
| N stage | | | | 0.028 |
| N0 | 49 | 28 | 21 | |
|
N1+2 | 36 | 31 | 5 | |
| Perineural
invasion | | | | 0.3725 |
| No | 56 | 40 | 16 | |
| Yes | 29 | 19 | 10 | |
| Vascular
invasion | | | | 0.04 |
| No | 53 | 34 | 19 | |
| Yes | 32 | 25 | 7 | |
These studies are in consistency with previous
research, suggesting that YWHAH is a major factor in CRC
tumorigenesis and could function as a biomarker and a target for
therapy.
Cell and animal experiments for the
verification of YWHAH on CRC proliferation
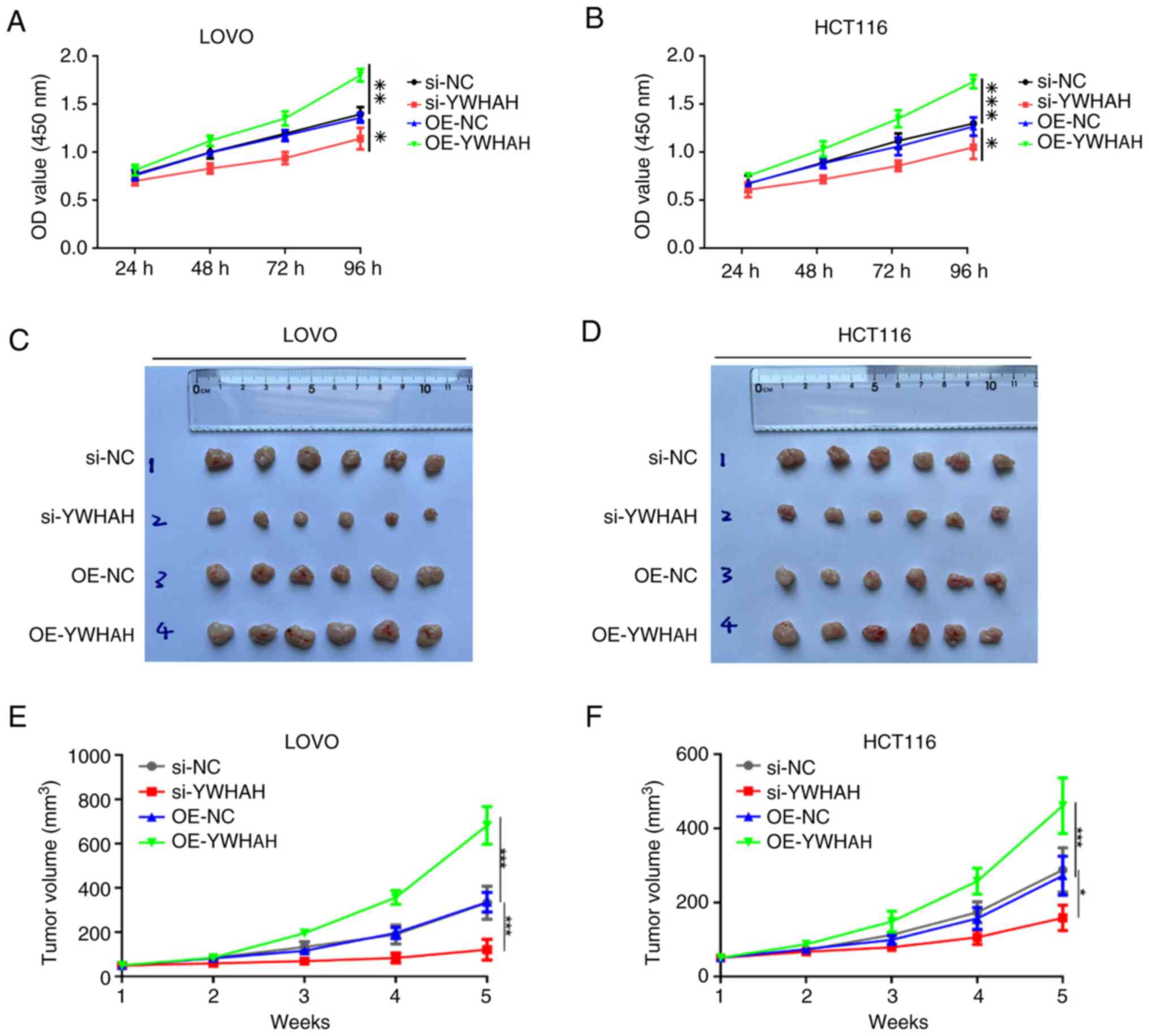
In our previous study, colony formation assays were
conducted, and the results showed that YWHAH overexpression
markedly increased colony numbers in both cell lines (12). In the present study, the CCK-8
assay was performed to assess the influence of varying YWHAH
expression levels on the proliferative capacity of CRC cells. As
illustrated in Fig. 2A and B,
YWHAH overexpression significantly enhanced the proliferative
ability of LOVO cells, whereas YWHAH suppression markedly
diminished their proliferative capacity (P<0.05). Similar
results were observed in HCT116 cells.
To assess the oncogenic potential of YWHAH in
vivo, subcutaneous xenograft models in nude mice were performed
utilizing cells transfected with si-NC, si-YWHAH, overexpression
(OE)-NC and OE-YWHAH constructs. Tumor growth was monitored on a
weekly basis and growth curves were subsequently generated.
Following a period of 5 weeks, the mice were euthanized, and the
tumors were excised to measure their volumes. The findings
indicated that tumors in the YWHAH overexpression group exhibited a
significantly larger volume compared with those in the OE-NC group.
Conversely, knockdown of YWHAH expression resulted in a
significantly reduced tumor volume compared with the si-NC group
(Fig. 2C-F). These in
vitro and in vivo results collectively suggest that
YWHAH facilitates the proliferation of CRC cells.
Impact of YWHAH on the
epithelial-to-mesenchymal transition (EMT) of CRC cells
The authors' previous study found that knockdown of
YWHAH expression levels significantly lowered the numbers of
migratory and invasive cells in both cell lines (P<0.01). In
contrast to these observations, YWHAH overexpression exhibited the
opposite effect, increasing both these processes. These results
suggested that YWHAH positively regulated the migratory and
invasive capabilities of CRC cells (12).
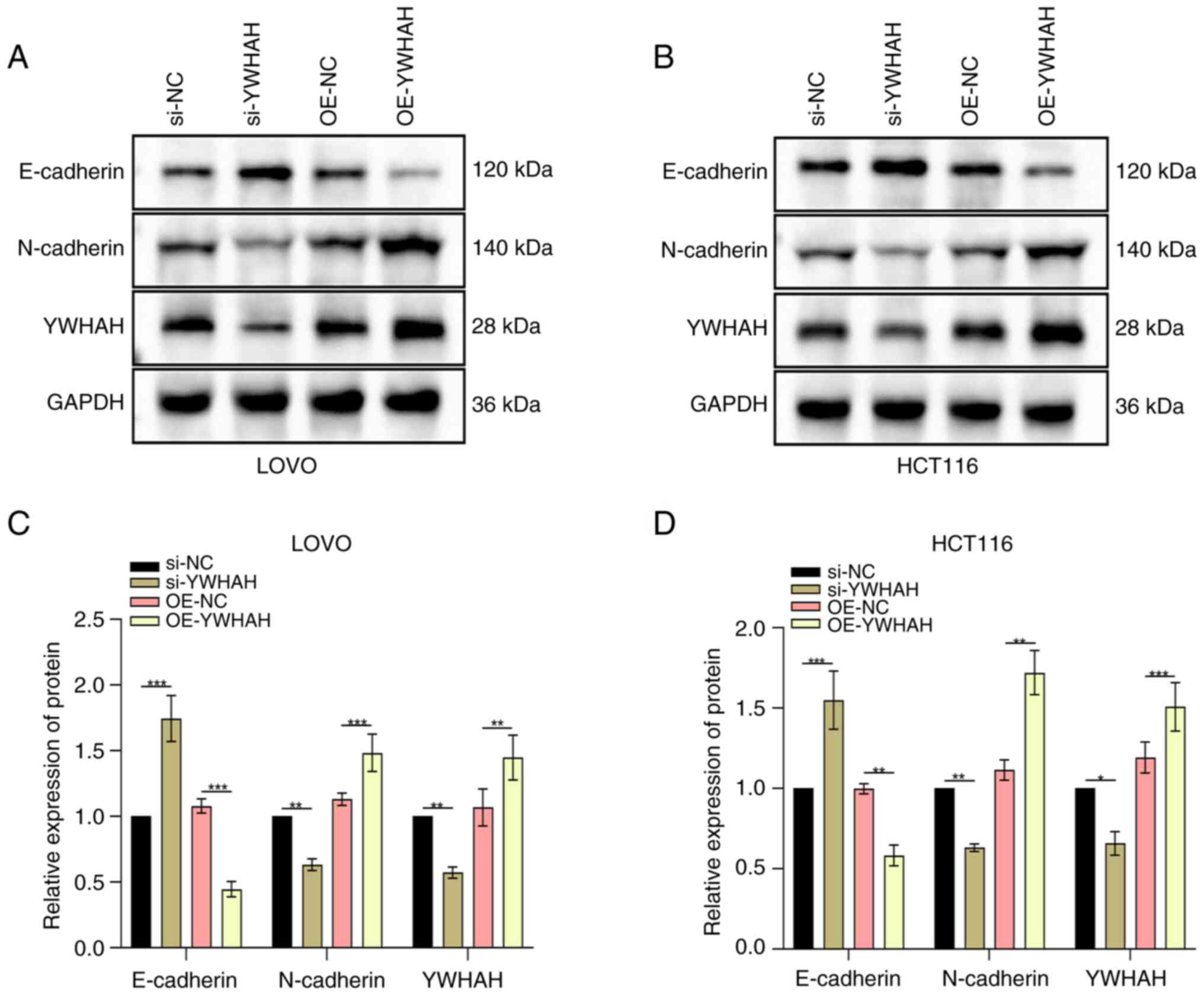
To further assess the influence of YWHAH on the EMT
of CRC cells, western blotting was utilized to assess the changes
in EMT markers. These indicated a decrease in E-cadherin and an
increase in N-cadherin levels in cells overexpressing YWHAH.
Conversely, in knockdown cells, E-cadherin was elevated, while
N-cadherin levels declined (Fig.
3A-D).
Association between YWHAH and apoptosis
in CRC cells
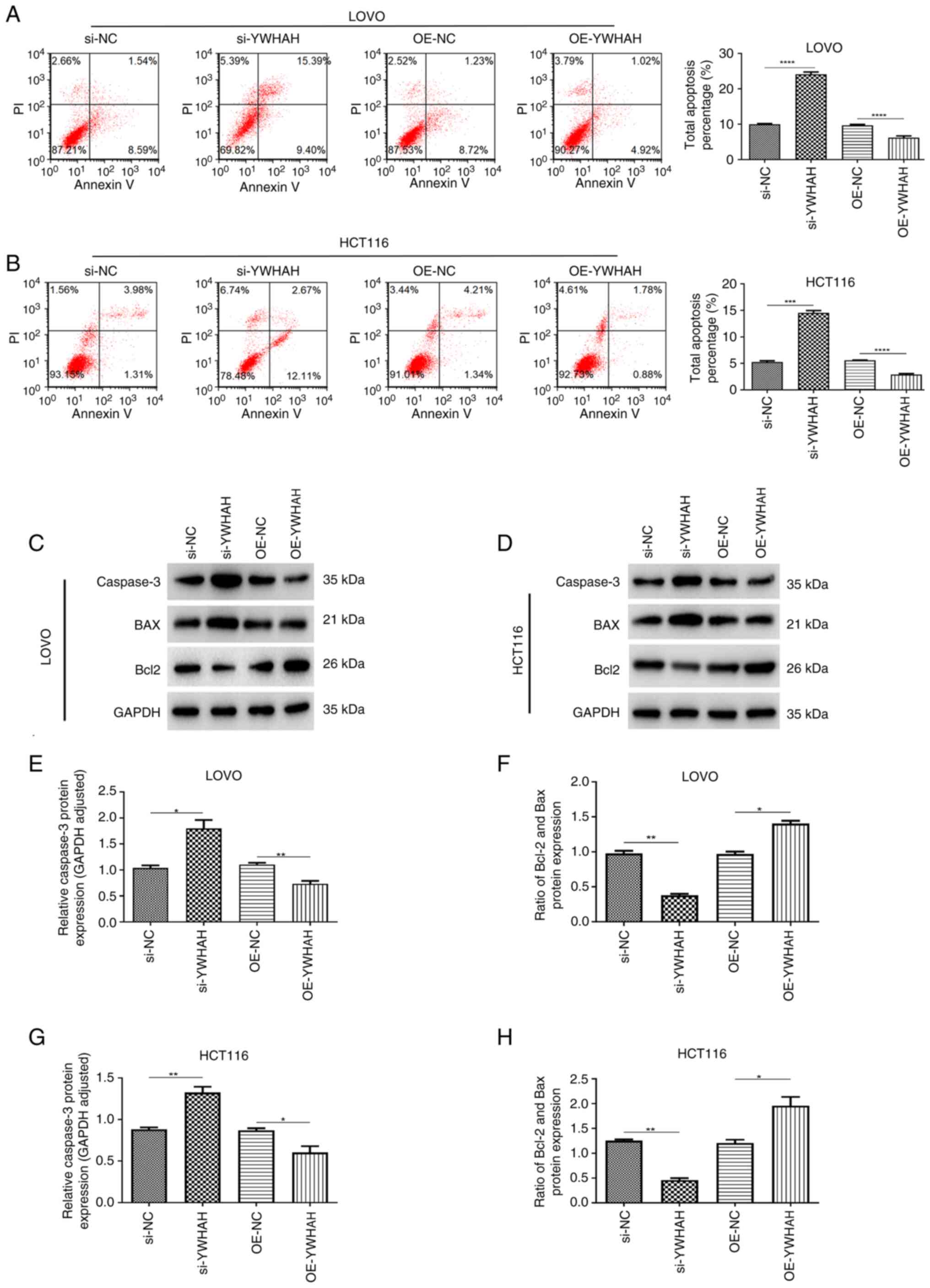
The induction of apoptosis is closely associated
with tumor suppression. Flow cytometry was used to assess the
function of YWHAH in modulating the apoptotic response in CRC
cells. As illustrated in Fig. 4,
knockdown of YWHAH expression via siRNA (si-YWHAH) significantly
increased the apoptotic rate in both cell lines, while OE-YWHAH
resulted in reduced apoptotic rates in these cell lines (Fig. 4A and B). Western blot analysis
further revealed that knockdown of YWHAH expression led to an
upregulation of caspase 3 and a decrease of Bcl-2/BAX protein
levels in LOVO and HCT116 cell lines. Conversely, YWHAH
overexpression significantly inhibited the expression of caspase 3
whilst it enhanced the Bcl-2/BAX protein in both cell lines
(Fig. 4C-H). Collectively, these
findings suggested that YWHAH overexpression suppresses apoptosis
in CRC cells.
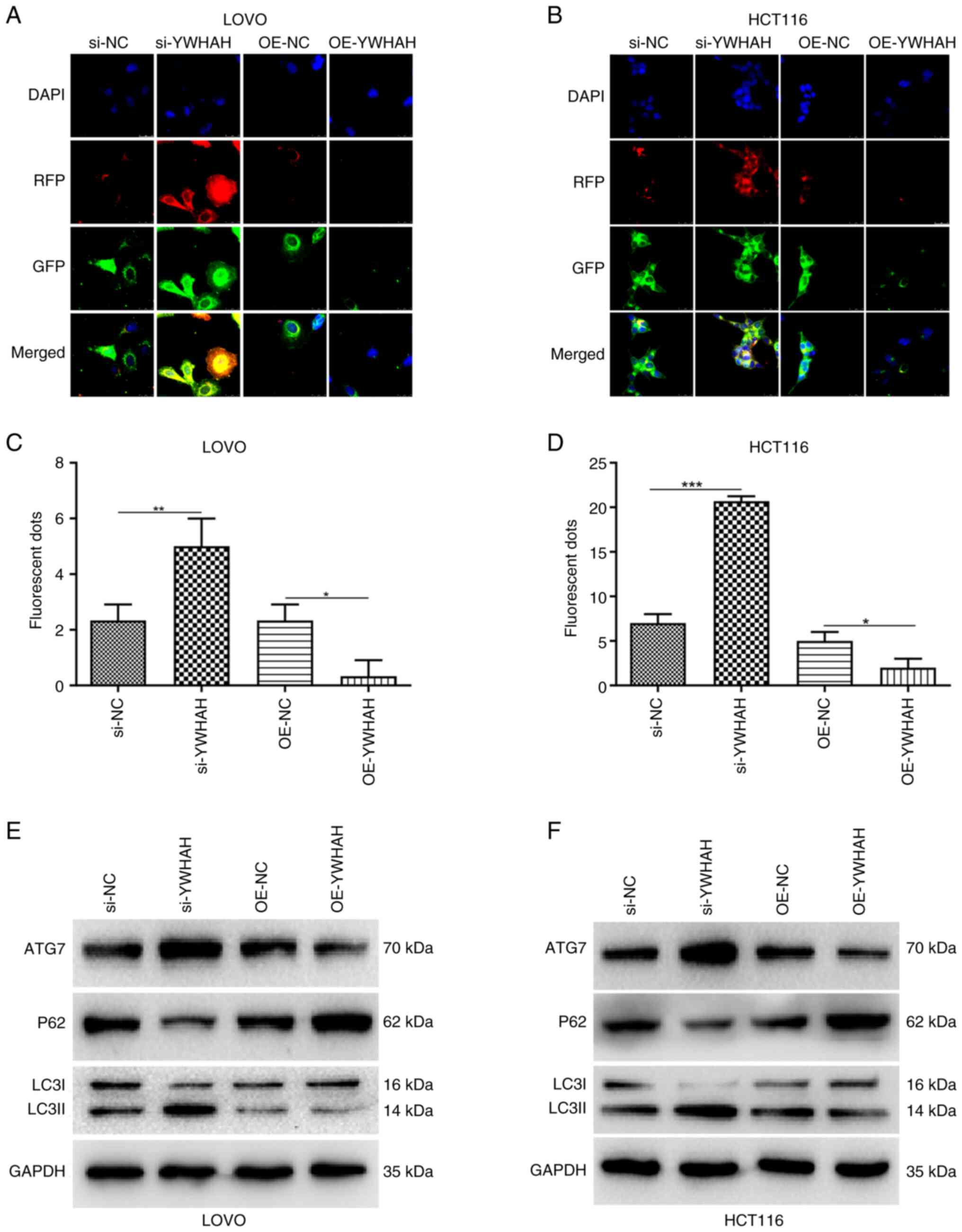
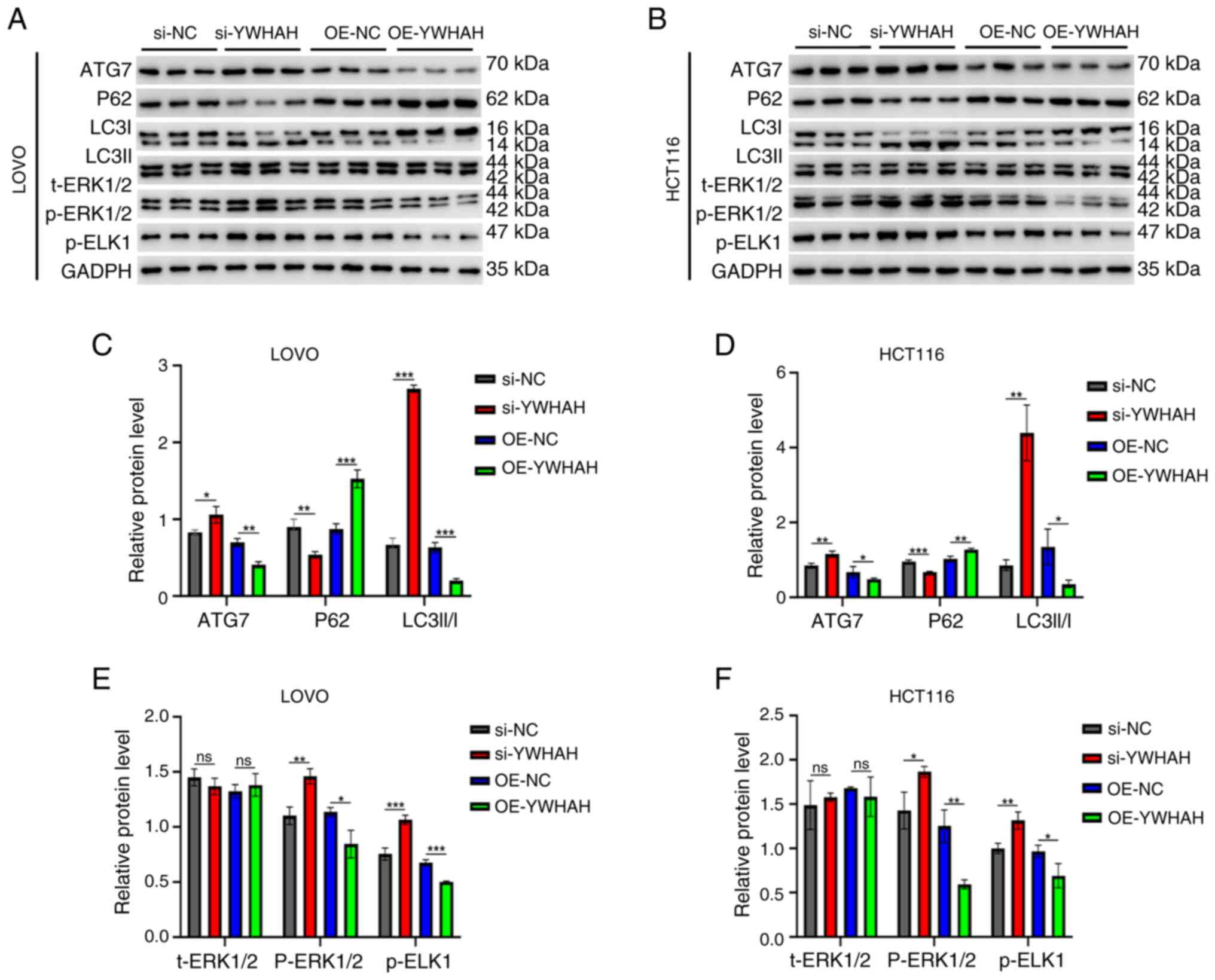
Association between YWHAH and autophagy
in CRC cells
A dual-labeled LC3B construct enables the distinct
visualization of autophagosomes and autolysosomes, capitalizing on
the divergent pH stabilities of its two fluorescent components.
Specifically, within the acidic environment of lysosomes, the
fluorescent signals from the red (mRFP) and green (GFP) proteins
undergo differential quenching. By utilizing the mRFP-GFP-LC3 dual
fluorescence system, a more profound understanding of the role of
YWHAH in autophagy was obtained. Fluorescence microscopy indicated
significantly increased numbers of fluorescent puncta in
YWHAH-knockdown CRC cells compared with those of the controls.
These observations indicated that the disruption of YWHAH
expression could enhance autophagy. Furthermore, the red
fluorescent puncta were substantially more abundant than the green
ones and their co-localization resulted in a predominantly red
region, corroborating the efficient progression of autophagy.
Conversely, the opposite effects were observed in YWHAH-OE CRC
cells (Fig. 5A-D). These
findings were subsequently validated by western blot analysis
(Fig. 5E-F). Specifically,
knockdown of YWHAH expression via siRNA led to an upregulation of
autophagy related 7 (ATG7) and LC3II/I protein expression in both
LOVO and HCT116 cells, while it resulted in a downregulation of P62
protein expression. In contrast to these observations, YWHAH
overexpression significantly suppressed the levels of ATG7 and the
LC3II/I ratio in both cell lines, while increasing P62 expression
specifically in LOVO cells. In addition, YWHAH overexpression
exhibited a tendency to increase P62 expression in the HCT116 cell
line.
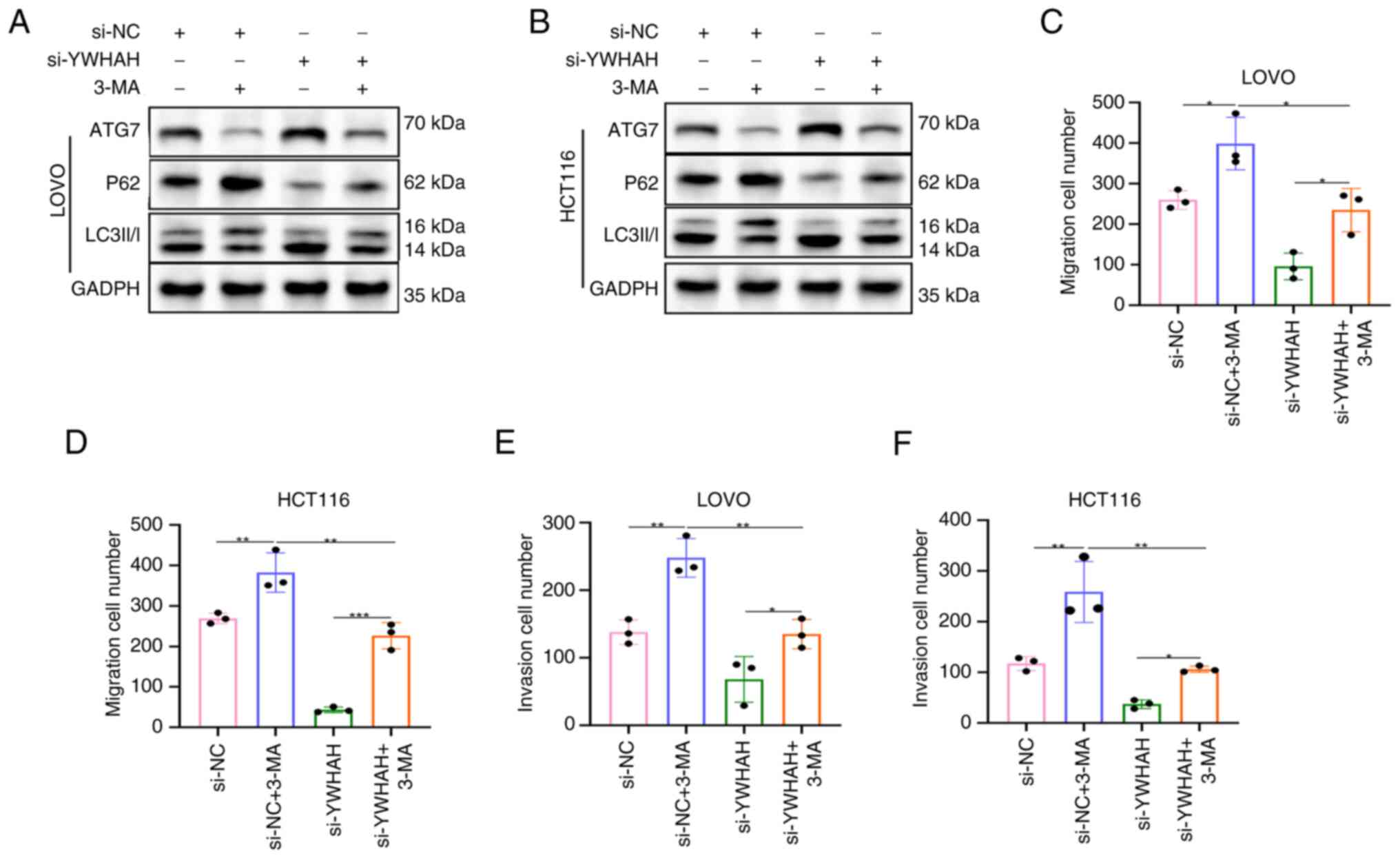
Inhibition of autophagy reverses the
suppressive effects of si-YWHAH on CRC cell migration and
invasion
It has been found that YWHAH inhibits autophagy in
CRC cells. To explore if this affects cell migration and invasion,
experiments altering autophagic flux were conducted. Initially,
autophagy was inhibited and the data indicated that si-YWHAH
increased ATG7 and LC3II/I levels, while it decreased those of P62,
confirming that it induced autophagy. In contrast to these
observations, the addition of 3-methoxyamphetamine (3-MA) to both
si-NC and si-YWHAH groups reduced ATG7 and LC3II/I levels, while it
increased P62 levels (Fig. 6A and
B), indicating that 3-MA suppressed autophagy in these
cells.
The Transwell assay indicated that the si-NC + 3-MA
group exhibited a significantly higher number of migratory and
invasive cells than the si-NC group, suggesting increased migration
and invasion following inhibition of autophagy. In the si-YWHAH +
3-MA group, cell migratory and invasive activities were lower than
those noted in the si-NC + 3-MA group, yet higher than those of the
si-YWHAH group, indicating that 3-MA reversed the inhibitory
effects of si-YWHAH on CRC cell migration and invasion (Figs 6C-F and S2). This confirmed that inhibition of
autophagy could counteract the suppressive impact of knockdown of
YWHAH expression on these processes.
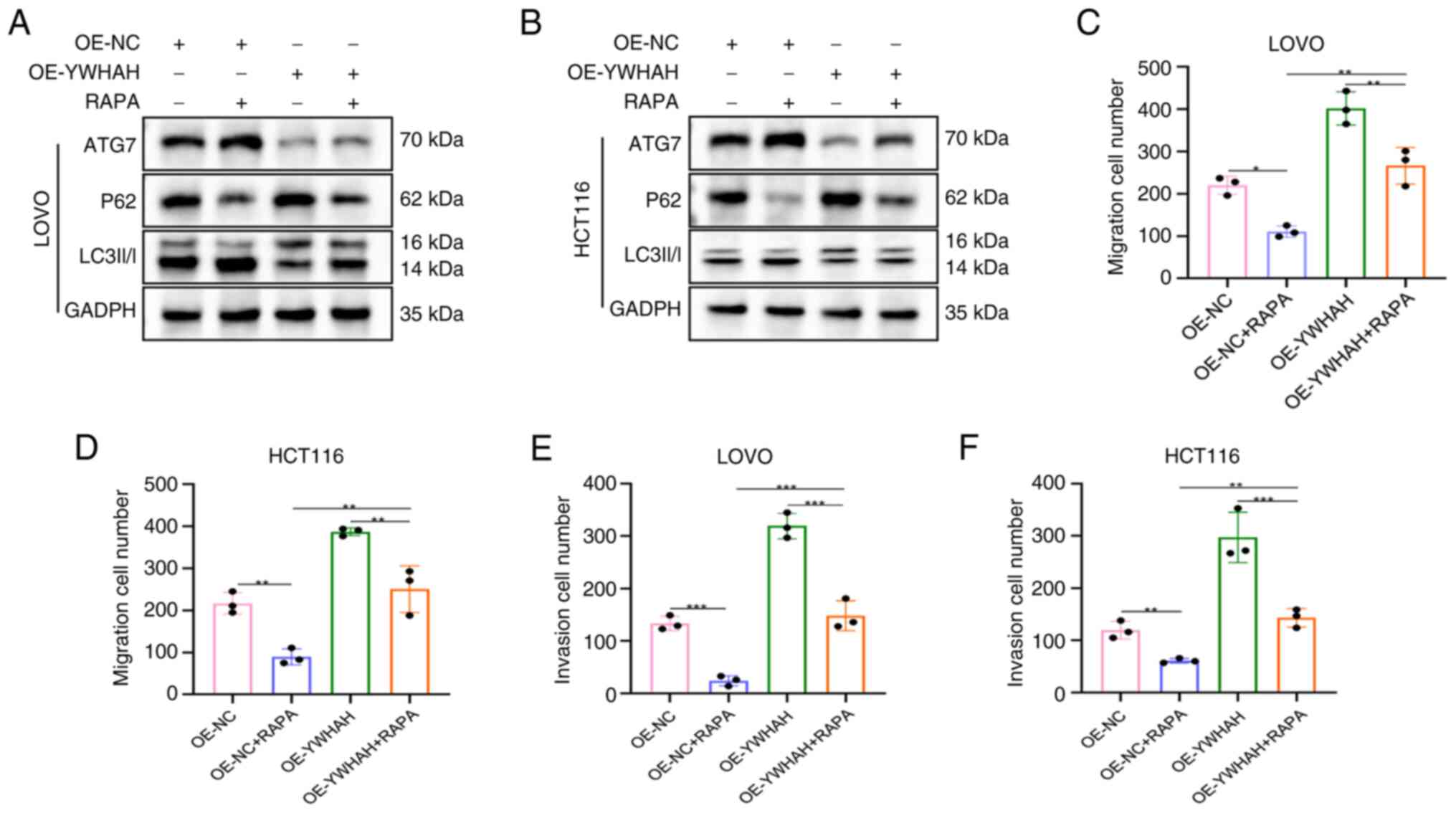
Activation of autophagy reverses the
promoting effect of OE-YWHAH on CRC cell migration and
invasion
An in-depth analysis of migration and invasion in
CRC cells was performed with stable knockdown and overexpression
models of YWHAH by introducing the autophagy inducer Rapamycin
(RAPA). In the OE-YWHAH group, the levels of ATG7 and LC3II/I were
markedly reduced compared with those of OE-NC cells, while P62
levels were notably elevated, suggesting that OE-YWHAH inhibits
autophagy in CRC cells, corroborating previous research findings.
In contrast to these observations, OE-NC + RAPA cells exhibited a
marked rise in ATG7 and LC3II/I levels together with reduced P62
levels compared with those noted in OE-NC cells. Similarly,
OE-YWHAH + RAPA cells demonstrated markedly elevated ATG7 and
LC3II/I levels and decreased P62 levels compared with OE-YWHAH
cells, indicating that RAPA effectively activated autophagy in CRC
cells (Fig. 7A and B).
Furthermore, the results from the Transwell migration and invasion
assays revealed that the number of migratory and invasive cells in
the si-NC + RAPA cells was significantly decreased compared with
that of OE-NC cells (Figs. 7C-F
and S3), suggesting that
autophagy activation led to a relative reduction in migratory and
invasive activities.
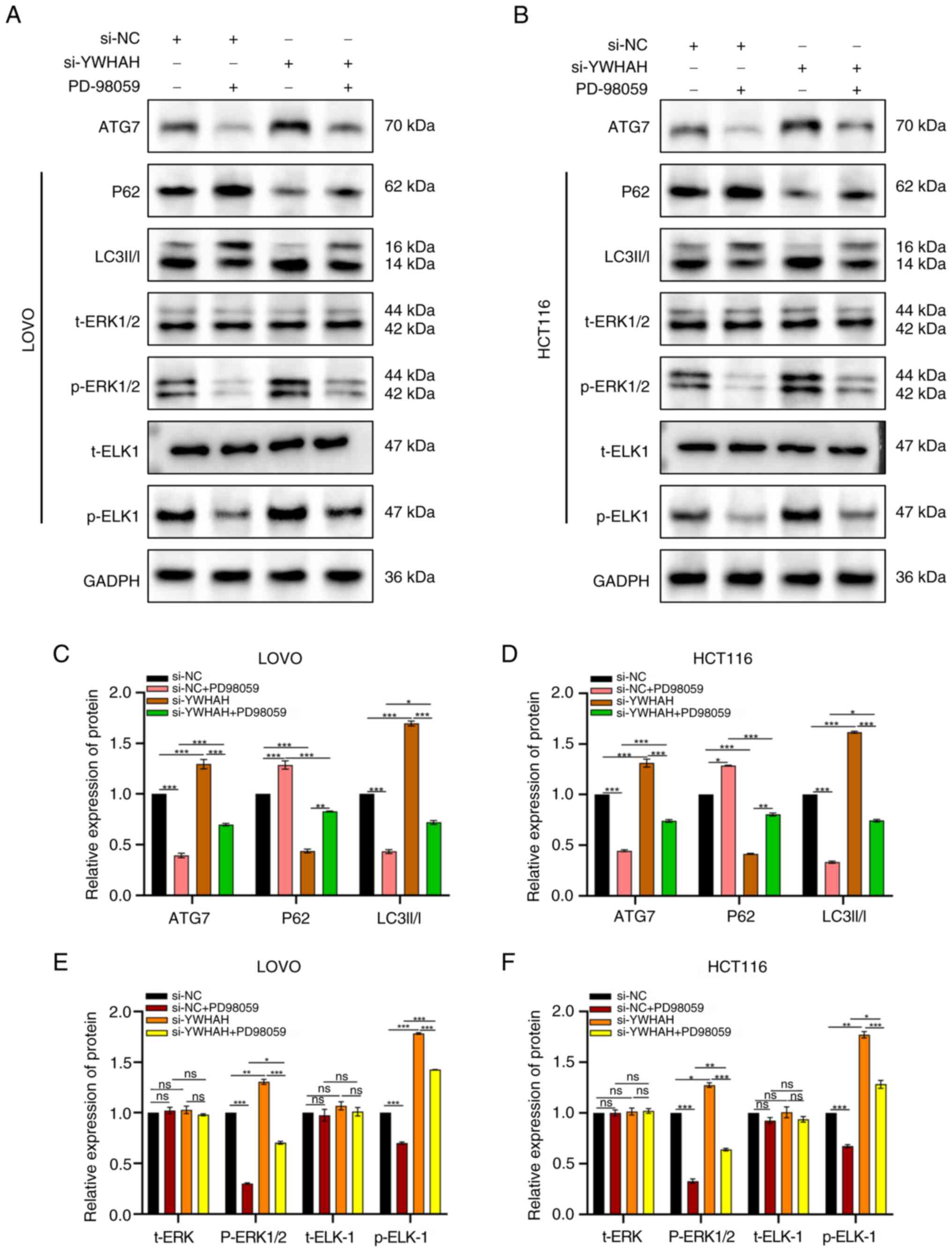
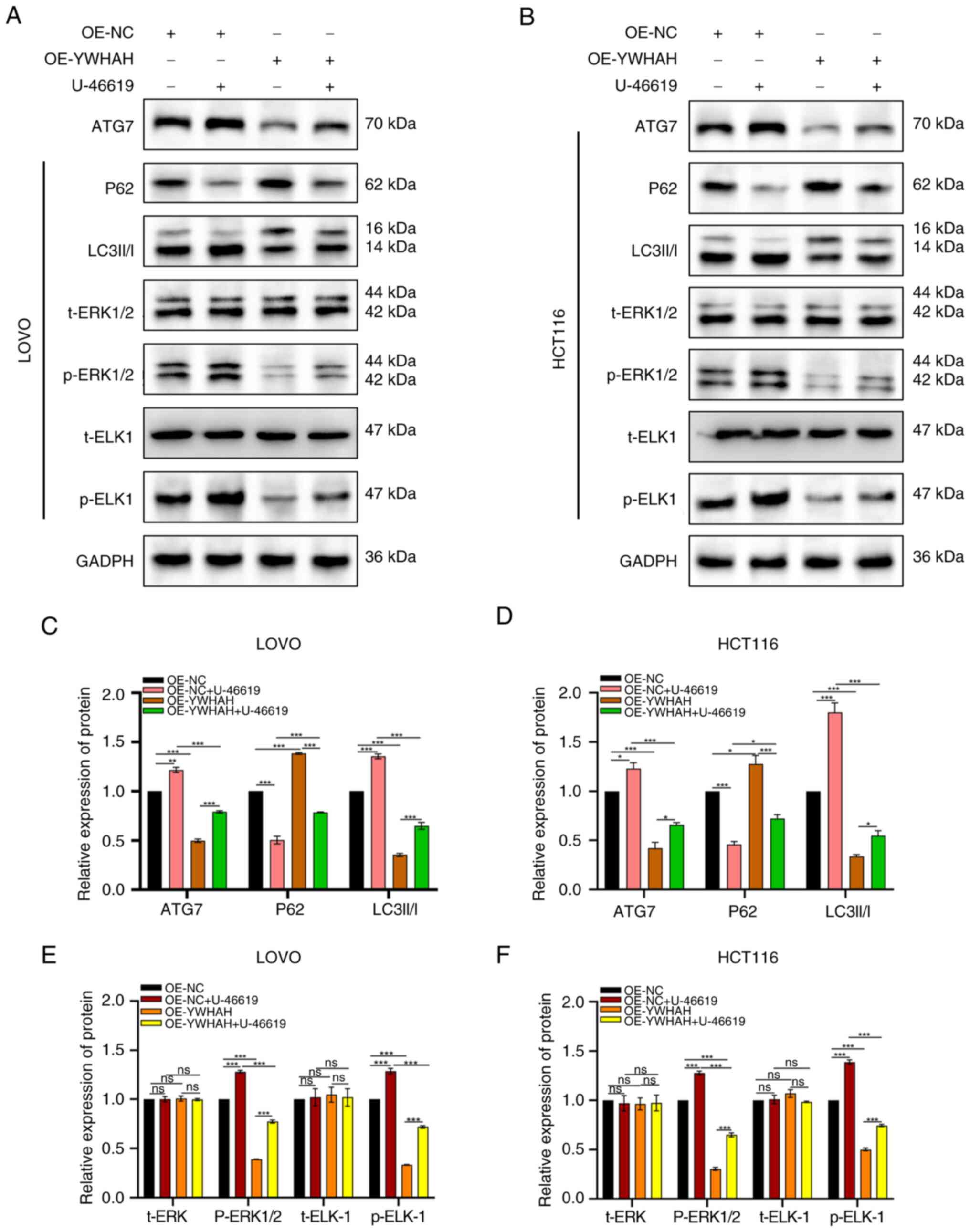
YWHAH induces autophagy via the MAPK/ERK
signaling pathway
In subsequent studies, the MAPK/ERK signaling
pathway inhibitor PD98059 and the agonist U-46619 were used to
evaluate the levels of the related pathway proteins ERK1/2,
phosphorylated (p)-ERK1/2, and p-ETS Like-1 protein (ELK)-1 by
western blotting. The aim of these experiments was to assess
whether YWHAH exhibited regulatory effects on the ERK signaling
pathway.
Previous studies have demonstrated that YWHAH
overexpression inhibits autophagy, whereas its knockdown promotes
autophagy. The present study explored the ERK pathway-related
protein expression following alteration of the YWHAH levels. The
results revealed that si-YWHAH increased p-ERK1/2 and p-ELK-1
expression levels, activating the MAPK/ERK pathway, while OE-YWHAH
decreased their expression levels, suppressing the pathway. In
addition, si-YWHAH reversed the inhibitory effect of PD98059 on the
pathway, as p-ERK1/2 and p-ELK-1 levels were higher in si-YWHAH +
PD98059 cells compared with those of the si-NC + PD98059 cells
(Fig. 8). Subsequent
investigations indicated that p-ERK1/2 and p-ELK-1 levels were
significantly elevated in OE-NC + U46619 cells compared with those
of the OE-NC cells. They were also higher in OE-YWHAH + U46619
cells compared with those of the OE-YWHAH cells; however, they were
lower than those noted in the OE-NC + U46619 cells (Fig. 9). This suggested that YWHAH
overexpression could counteract the activation of the signaling
pathway by U46619. The protein expression levels were statistically
significant, with the exception of t-ERK. Overall, these results
indicated that YWHAH negatively regulated the ERK axis in CRC
cells.
To further investigate the relationship between the
regulation of autophagy by YWHAH in CRC and the involvement of the
MAPK/ERK signaling pathway, the expression levels of
autophagy-related proteins were investigated following inhibition
and activation of the ERK signaling pathway.
Western blotting indicated that in si-YWHAH cells,
ATG7 and LC3II/I levels were significantly elevated, while P62
levels were lower compared with those noted in si-NC cells
(Fig. 8). The introduction of
the inhibitor PD98059 to the si-YWHAH group decreased the levels of
ATG7 and LC3II/I, while it increased those of P62, indicating
reduced autophagy and reversal of the activation effect of si-YWHAH
on autophagy in CRC cells (Fig.
8). Conversely, in the OE-YWHAH group, ATG7 and LC3II/I levels
were lower, and P62 expression was higher. The addition of the
activator U-46619 increased ATG7 and LC3II/I levels and reduced P62
expression (Fig. 9), suggesting
enhanced autophagy and reversal of the inhibitory effect of
OE-YWHAH on autophagy in these cells. In summary, these results
indicated that in CRC cells, YWHAH regulated autophagy in a manner
dependent on the MAPK/ERK axis.
YWHAH regulates autophagy in CRC tissues
via the MAPK/ERK axis in vivo
To evaluate if YWHAH inhibits autophagy in CRC via
the MAPK/ERK pathway, the expression levels of autophagic and
signaling proteins were investigated in nude mouse tumors. In the
si-YWHAH group, the expression levels of the autophagic markers
ATG7 and LC3II/I were increased, while the levels of P62 were
reduced, indicating enhanced autophagy. Conversely, in OE-YWHAH
cells, the expression levels of ATG7 and LC3II/I were lowered,
while those of P62 were increased, suggesting reduced autophagy. In
addition, p-ERK1/2 and p-ELK-1 levels were increased in the
si-YWHAH group and decreased in the OE-YWHAH group (Fig. 10A-F). These results confirmed
that YWHAH inhibited autophagy in CRC via the MAPK/ERK pathway.
 | Figure 10YWHAH inhibits autophagy through
MAPK/ERK signaling pathway in vivo. (A and B) Western
blotting was used to evaluate the protein expression of ATG7, P62,
LC3II/I, t-ERK1/2, p-ERK1/2 and p-ELK-1 when the nude mice were
injected with CRC cells. (C-F) Semi-quantitative analysis of
autophagy related proteins and MAPK/ERK signaling related proteins
when the nude mice were injected with CRC cells.
*P<0.05, **P<0.01 and
***P<0.001. Data are presented as the mean ± standard
deviation (n=3 independent experiments). YWHAH, tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein Eta;
t-, total; p-, phosphorylated; CRC, colorectal cancer; ATG7,
autophagy related 7; OE, overexpression; si-, small interfering;
NC, negative control; ns, not significant (P>0.05). |
Discussion
The 14-3-3 protein, YWHAH, is integral to processes,
such as cell signaling, apoptosis, cell cycle modulation,
transcription and the malignant transformation of cells (15-18). In a previous investigation,
ac4C-acetylation sequencing was employed to identify YWHAH; the
data substantiated that acetylation significantly influenced the
stability of YWHAH (12). In the
present study, a comprehensive examination of the role of YWHAH in
CRC was performed; moreover, its association with the clinical
pathological characteristics was investigated. By using various
cellular and animal experiments, the effects of YWHAH were examined
on cellular proliferation, migration, invasion and autophagy of CRC
cells. These findings enhance the understanding of the involvement
of YWHAH in CRC progression and offer novel insights for future
therapeutic approaches.
CRC development is a multi-stage process marked by
gene expression changes and phenotypic shifts that activate
signaling pathways, enhancing malignancy via inhibition of
apoptosis, increased angiogenesis and EMT (19-22). These factors collectively drive
CRC initiation and progression. Tumor cell migration and invasion
are key to malignancy, with autophagy playing a complex role by
affecting cell survival, proliferation, migration and invasion via
intracellular homeostasis (23-26). BMP9, for instance, promotes
autophagy and inhibits breast cancer cell migration via the
c-Myc/SNHG3/mTOR pathway, suggesting similar mechanisms in CRC
(27). CD133 influences CRC cell
migration and invasion by modulating autophagy, while interactions
between inflammatory factors and autophagy in the TME may also aid
CRC metastasis (27). Elevated
levels of YWHAH have been observed in CRC tissues; this protein was
linked to tissue differentiation, lymph node metastasis, vascular
invasion and TNM staging. In cells, YWHAH overexpression boosted
CRC cell proliferation, migration and invasion, while knockdown of
its expression inhibited these activities. In vivo, YWHAH
overexpression increased tumor size and weight, whereas knockdown
of its expression reduced them. YWHAH also promoted CRC cell
invasion and metastasis by regulating EMT. Autophagy involves
pathways such as the PI3K/Akt/mTOR and MAPK family members, such as
ERK, which exhibit a complex interaction with autophagy (27-31).
In the present study, it was revealed that YWHAH
influences the MAPK/ERK pathway activation, regulating autophagy in
CRC cells. Overexpression of YWHAH enhances CRC cell migration and
invasion by negatively affecting autophagy and mediating the
MAPK/ERK pathway. However, the small sample size of the tissues
used may limit the findings and other mechanisms may also play a
role in the oncogenic effects of YWHAH. Current models may not
fully capture human CRC complexity. Future research should examine
the interactions of YWHAH with other pathways, validate findings in
larger studies and develop models that mimic human CRC more
efficiently. The exploration of the targeted therapies of YWHAH in
combination with the existing treatments can improve the outcomes
of CRC.
In summary, the current data indicated that YWHAH
plays a pivotal role in the progression of CRC by modulating
apoptosis and autophagy via the MAPK/ERK signaling pathway.
Targeting YWHAH or its downstream pathways can provide novel
therapeutic strategies for the treatment of CRC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QL was responsible for conceptualizing, developing
methods, conducting formal analysis, managing resources, validating
findings, curating data, drafting the original manuscript, and
reviewing and editing the final version. YW contributed to
conceptualization, resource allocation, and reviewing and editing
of the writing. ZY contributed to the methodology, formal analysis,
resources, and writing, reviewing and editing of the manuscript. PZ
developed methodology. JW conducted formal analysis and data
validation. CZ performed formal analysis and data curation. ZS
developed methodology and validated data. CX contributed to
conceptualization, investigation, data validation, and writing,
reviewing and editing of the manuscript. All authors read and
approved the final version of the manuscript. QLi and PZ confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
All procedures conducted in studies involving human
participants adhered to the ethical standards set by the Ethics
Committee of the Second Affiliated Hospital of Xuzhou Medical
University (approval no. 2023100505), as well as the principles
outlined in the 1964 Declaration of Helsinki and its subsequent
amendments concerning research involving human subjects. Written
informed consent was provided by all participants or their legal
representatives. All animal experiments received approval (approval
no. 2023110512) from the Ethics Committee of Xuzhou Medical
University (Xuzhou, China) for the use of animals and were
performed in compliance with the National Institutes of Health
Guidelines for the Care and Use of Laboratory Animals. All
institutional and national guidelines for the care and use of
laboratory animals were strictly followed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein Eta
|
|
CRC
|
colorectal cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
3-MA
|
3-methoxyamphetamine
|
|
RAPA
|
rapamycin
|
|
acRIP-seq
|
acetylated RNA immunoprecipitation and
sequencing
|
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jiangsu Provincial Key
Laboratory of Tumor Biotherapy (grant no. XZSYSKF2023033).
References
|
1
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI
|
|
3
|
Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh
Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M and Saad A:
Colorectal cancer epidemiology: Recent trends and impact on
outcomes. Curr Drug Targets. 22:998–1009. 2021. View Article : Google Scholar
|
|
4
|
Gonzalez NS, Ros Montana FJ, Illescas DG,
Argota IB, Ballabrera FS and Elez Fernandez ME: New insights into
adjuvant therapy for localized colon cancer. Hematol Oncol Clin
North Am. 36:507–520. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Long J, He Q, Yin Y, Lei X, Li Z and Zhu
W: The effect of miRNA and autophagy on colorectal cancer. Cell
Prolif. 53:e129002020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orlandi G, Roncucci L, Carnevale G and
Sena P: Different roles of apoptosis and autophagy in the
development of human colorectal cancer. Int J Mol Sci.
24:102012023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang D, He J, Dong J, Wu S, Liu S, Zhu H
and Xu T: UM-6 induces autophagy and apoptosis via the Hippo-YAP
signaling pathway in cervical cancer. Cancer Lett. 519:2–19. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiao YN, Wu LN, Xue D, Liu XJ, Tian ZH,
Jiang ST, Han SY and Li PP: Marsdenia tenacissima extract induces
apoptosis and suppresses autophagy through ERK activation in lung
cancer cells. Cancer Cell Int. 18:1492018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pohl C and Dikic I: Cellular quality
control by the ubiquitin-proteasome system and autophagy. Science.
366:818–822. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Green DR: The coming decade of cell death
research: Five riddles. Cell. 177:1094–1107. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Q, Yuan Z, Wang Y, Zhai P, Wang J,
Zhang C, Shao Z and Xing C: Unveiling YWHAH: A potential
therapeutic target for overcoming CD8(+) T cell exhaustion in
colorectal cancer. Int Immunopharmacol. 135:1123172024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kacirova M, Novacek J, Man P, Obsilova V
and Obsil T: Structural basis for the 14-3-3 protein-dependent
inhibition of phosducin function. Biophys J. 112:1339–1349. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Obsil T and Obsilova V: Structural basis
of 14-3-3 protein functions. Semin Cell Dev Biol. 22:663–672. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Darling DL, Yingling J and Wynshaw-Boris
A: Role of 14-3-3 proteins in eukaryotic signaling and development.
Curr Top Dev Biol. 68:281–315. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen NM, Meyer D, Meyer L, Chand S,
Jagadesan S, Miravite M, Guda C, Yelamanchili SV and Pendyala G:
Identification of YWHAH as a novel brain-derived extracellular
vesicle marker post long-term midazolam exposure during early
development. Cells. 12:9662023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Y, Yang M and Huang W: 14-3-3 σ: A
potential biomolecule for cancer therapy. Clin Chim Acta.
511:50–58. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Munier CC, Ottmann C and Perry MWD: 14-3-3
modulation of the inflammatory response. Pharmacol Res.
163:1052362021. View Article : Google Scholar
|
|
19
|
Malki A, ElRuz RA, Gupta I, Allouch A,
Vranic S and Al Moustafa AE: Molecular Mechanisms of colon cancer
progression and metastasis: Recent insights and advancements. Int J
Mol Sci. 22:1302020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nicolini A and Ferrari P: Involvement of
tumor immune microenvironment metabolic reprogramming in colorectal
cancer progression, immune escape, and response to immunotherapy.
Front Immunol. 15:13537872024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shin AE, Giancotti FG and Rustgi AK:
Metastatic colorectal cancer: Mechanisms and emerging therapeutics.
Trends Pharmacol Sci. 44:222–236. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Kornmann M and Traub B: Role of
epithelial to mesenchymal transition in colorectal cancer. Int J
Mol Sci. 24:148152023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Yang Y, Qi X, Cui P, Kang Y, Liu
H, Wei Z and Wang H: SLC14A1 and TGF-β signaling: A feedback loop
driving EMT and colorectal cancer metachronous liver metastasis. J
Exp Clin Cancer Res. 43:2082024. View Article : Google Scholar
|
|
24
|
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L,
Xu Y, He J, Lan J, Ou Q, et al: Tumor-derived lactate promotes
resistance to bevacizumab treatment by facilitating autophagy
enhancer protein RUBCNL expression through histone H3 lysine 18
lactylation (H3K18la) in colorectal cancer. Autophagy. 20:114–130.
2024. View Article : Google Scholar :
|
|
25
|
Xie Q, Liu Y and Li X: The interaction
mechanism between autophagy and apoptosis in colon cancer. Transl
Oncol. 13:1008712020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Foerster EG, Mukherjee T, Cabral-Fernandes
L, Rocha JDB, Girardin SE and Philpott DJ: How autophagy controls
the intestinal epithelial barrier. Autophagy. 18:86–103. 2022.
View Article : Google Scholar :
|
|
27
|
Adebayo AS, Agbaje K, Adesina SK and
Olajubutu O: Colorectal cancer: Disease process, current treatment
options, and future perspectives. Pharmaceutics. 15:26202023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma L, Zhang R, Li D, Qiao T and Guo X:
Fluoride regulates chondrocyte proliferation and autophagy via
PI3K/AKT/mTOR signaling pathway. Chem Biol Interact.
349:1096592021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang K, Luan L, Li X, Sun X and Yin J: ERK
inhibition in glioblastoma is associated with autophagy activation
and tumorigenesis suppression. J Neurooncol. 156:123–137. 2022.
View Article : Google Scholar
|
|
30
|
Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y and
Hu LL: ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med.
19:1997–2007. 2020.PubMed/NCBI
|
|
31
|
Lee S, Rauch J and Kolch W: Targeting MAPK
signaling in cancer: Mechanisms of drug resistance and sensitivity.
Int J Mol Sci. 21:11022020. View Article : Google Scholar : PubMed/NCBI
|