Introduction
Glioblastoma multiforme (GBM) is the most common,
lethal and aggressive type of primary brain tumor with a median
survival of 9–12 months. Despite advances in the basic
understanding of cancer biology and therapeutic advances in other
neoplasms, the poor prognosis for GBM has not improved in the last
four decades and its treatment remains ineffective and essentially
palliative (1,2). Though the exact molecular mechanisms
for glioma genesis remain unclear, recent studies have reported
that there were several microRNA (miRNA or miR) abnormalities in
human gliomas, including miR-451 (3). miRNAs are now being used as a new
type of molecules of interest to elucidate tumorigenesis and novel
therapeutic approaches could be developed by targeting miRNAs that
are altered in glioma (4).
miRNAs are a class of small (~22 nucleotides)
non-coding RNAs that function as negative regulators of gene
expression at the post-transcriptional level by binding to
complimentary sequences in, mainly, the 3′-untranslated regions
(3′-UTRs) of specific mRNAs (5–11).
These miRNAs play important roles in apoptosis, proliferation,
differentiation, development, and metabolism (12–14).
In particular, miRNAs show altered expression in tumors in relation
to normal tissues and miRNA aberrations may be important in tumor
progression (15). hsa-miR-451 is
located on chromosome 17q11.2, a region known to be amplified in
certain types of cancers, in close proximity to ERBB2 (17q12)
(16,17). Studies have shown that miR-451
inhibited cell growth (3),
proliferation, and invasion and enhance apoptosis (18). The identification of target genes
associated with altered miRNA expression might accurately elucidate
the role of miRNAs in cancer biology (15).
In this study, we confirmed the hypo-expression of
miR-451 in gliomas using quantitative real-time PCR and fluorescent
in situ hybridization. The calcium binding protein 39 gene
(CAB39) predicted by bioinformatics analysis as a target gene of
the miR-451, was validated by fluorescent reporter assay. We
further analyzed the signaling pathway miR-451 might regulate in
human glioma and found that miR-451 modulated the expression of
multiple downstream molecules such as LKB1, AMPK, PI3K and AKT,
suggesting that miR-451 may act as a tumor-suppressor factor and
regulate the PI3K/AKT pathway through LKB1 and AMPK.
Materials and methods
Patients and samples
Tissue specimens and clinical information were
obtained as part of an approved study by the Institutional Review
Board at the Tianjin Medical University, China. Forty-six human
glioma tissues were collected with patient consent at the time of
operation, grading of tumors was carried out with WHO criteria
(World Health Organization, 2007). The tissue samples included: 3
grade I tumors (3 hairy cell astrocytoma); 8 grade II tumors (1
protoplasmic astrocytomas, 6 fibrocytic astrocytomas and 1 mixed
oligoastrocytomas); 15 grade III gliomas (all of these tumors were
anaplastic astrocytomas); and 15 grade IV glioblastomas (GBMs).
Five normal brain tissues were obtained from patients with
traumatic brain injury and brain tumors for internal decompression.
Immediately after surgery, samples were snap-frozen and stored in
liquid nitrogen.
A glioma tissue microarray was purchased from
Shaanxi Chaoying Biotechnology (Xi’an, China). Pathologic grades of
tumors on the microarray were defined according to the 2007 WHO
criteria as follows: 4 grade I tumors (4 Pilocytic astrocytoma), 18
grade II tumors (15 astrocytoma, 2 oligoastrocytoma and 1
oligodendroglioma), 14 grade III tumors (10 anaplastic astrocytoma,
3 anaplastic oligoastrocytoma and 1 anaplastic oligodendroglioma),
39 grade IV tumors (all glioblastomas); 5 were normal brain tissue.
Each dot represented a tissue spot from one individual specimen,
selected and pathologically confirmed. The array dot diameter was
1.5 mm. All microarrays were stored in the dark at 4°C.
In situ hybridization
Using sense locked nucleic acid (LNA)-modified
oligonucleotide probes, in situ hybridization was performed
with an in situ hybridization kit (Boster Biological
Technology, Ltd., Wuhan, China). The sequences of the LNA/DNA
oligonucleotides contained locked nucleic acids at five consecutive
centrally located bases (indicated by the underline) as shown:
HSA-miR-451 5′-TTGAG TCATT
ACCAT TGCCA AA-3′. The glioma tissue microarrays were
deproteinated, and then prehybridized for 2 h in hybridization
liquid in a humidified chamber (50% formamide, 5 × SSC). The probes
(miR-451 10 ng) were added to the sections on the microarray and
incubated overnight at 40°C in a water bath. After washing with PBS
3 times, the same volume of anti-digoxigenin-rhodamine and
streptavidin-FITC solution was added and incubated for 2 h at room
temperature in the dark. Nuclei were counterstained with a DAPI
karyotyping kit (Genmed, USA). After washing with PBS 3 times,
sections were sealed and detected under a fluorescence microscope
with an OptiGrid system and analyzed by IPP6.1 (Olympus, Tokyo,
Japan).
Analysis of hsa-miR-451 candidate target
genes
Previous studies in our laboratory have shown a
negative correlation between miR-451 and the expressions of AKT1
and c-Myc (18). We further
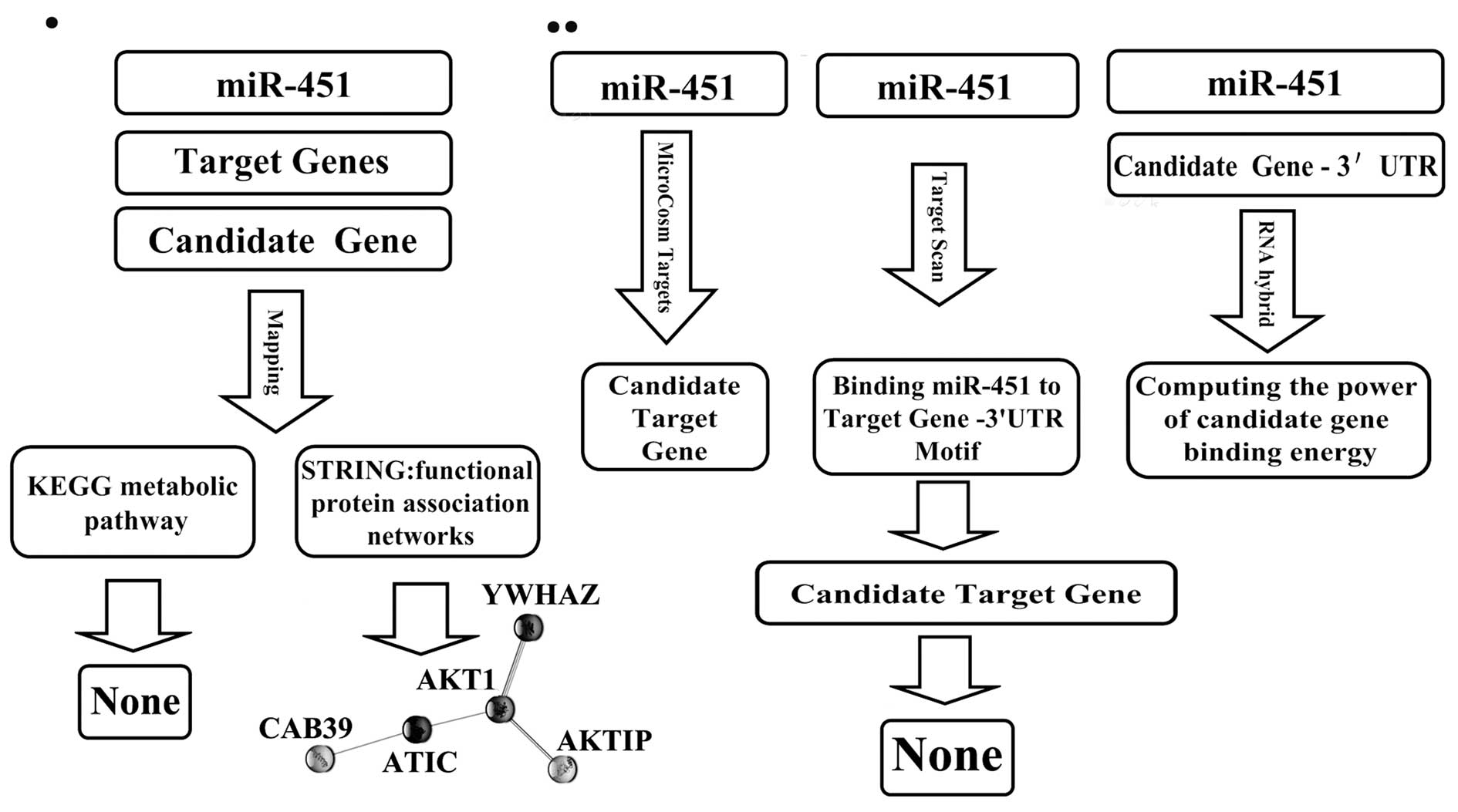
analyzed our data using several databases (Fig. 1). First, we used TargetScan 5.1
(http://www.targetscan.org) to search for
candidate miR-451 target genes and found 14 (YTHDF2, ZNF644,
CUGBP2, C11orf30, FMNL3, FBXO33, AKTIP, VAPA, RKHD2, SAMD4B, OSR1,
TTN, CAB39, YWHAZ). Next, we used STRING, the functional protein
association networks database (http://www.bork.embl-heidelberg.de/STRING/) (19), to explore possible interactions
between the 14 candidate miR-451 target genes and AKT1 and c-Myc.
To exploit the possible interactions that were identified between
the miR-451 target genes and AKT1 and c-Myc we used the KEGG
pathway database (http://www.genome.jp/kegg/) (20). For a second prediction of possible
hsa-miR-451 (human miR-451) gene targets we used MicroCosm Targets
(http://microrna.sanger.ac.uk/) (21) and investigated whether AKT1 and
c-Myc were included in the prediction. Finally, to further evaluate
the possibility of AKT1 and c-Myc as hsa-miR-451 target genes we
used RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid)
(22).
Cell culture and transfection
The human GBM cell lines LN229, U87 and U251 were
purchased from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences, Shanghai, China. The human
glioblastoma cell line A172 was gifted by Professor Jinhuan Wang
(Tianjin First Central Hospital, China). The cells were maintained
in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA)
supplemented with 12% fetal bovine serum (Invitrogen, Carlsbad,
USA), and incubated at 37°C with 5% CO2. Transfections
with hsa-miR-451 mimics were performed in serum-free medium 24 h
after plating, with Lipofectamine 2000 (Invitrogen). The
oligonucleotide sequence of the hsa-miR-451 mimics was:
5′-AAACCGUUACCAUUACUGAGUU-3′. A scrambled siRNA sequence
(5′-TTCTCCGAACGTGTCACGT-3′) was used as the negative control (Gima
Biol Engineering Inc., Shanghai, China). Cells were then cultured
in complete medium 6 h later.
Quantative real-time PCR analysis
Total RNA of the GBM cells (U251, LN229, A172, U87),
as well as of the 46 snap-frozen human glioma tissues was harvested
using TRIzol (Invitrogen) following the manufacturer’s protocol.
The concentration and purity of RNA were determined using
NanoDrop® ND-1000. Total RNA (2 μg) was used for cDNA
synthesis by reverse transcription using M-MLV Reverse
Transcriptase (Promega) according to the manufacturer’s protocol.
Expressions of mature miR-451 were quantified by miR-qRT PCR using
the Hairpin-it™ miRNA qPCR Quantitation kit (GenePharma Co. Ltd.).
All PCR reactions were performed using standard PCR conditions:
stage 1: 95°C for 3 min (1 cycle); stage 2: 95°C for 12 sec,
followed by 62°C for 40 sec; stage 3: from 62°C up to 95°C,
followed by 0.2°C for 2 sec (1 cycle). U6 was used as the internal
control. Data are shown as fold change and analyzed initially using
Opticon Monitor Analysis Software V2.02 software (MJ Research,
Waltham, MA, USA).
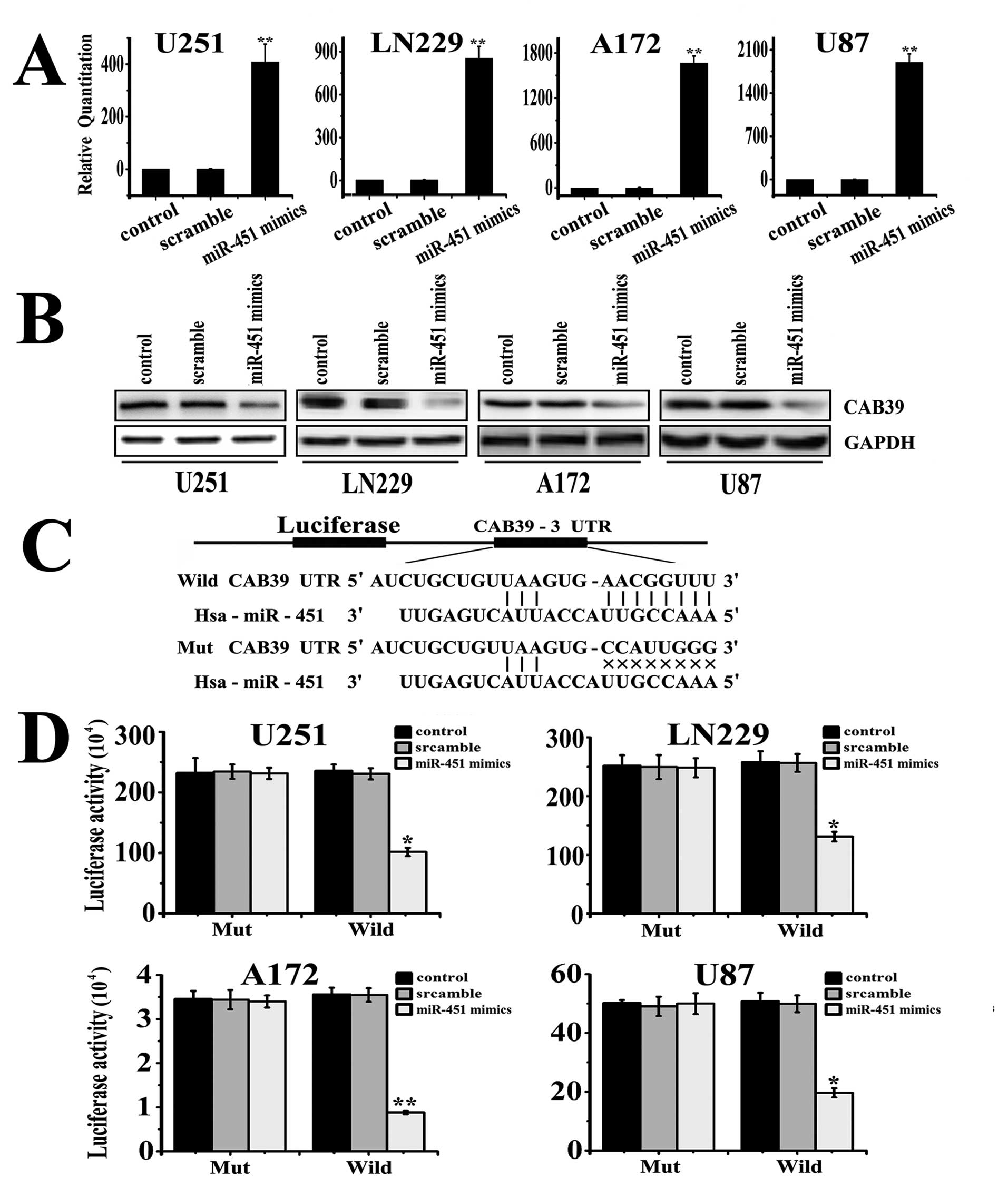
Luciferase activity assay
The pGL3-CAB39-3′UTR-Subcloning and
pGL3-CAB39-3′UTR-Mut plasmids were purchased from GenScript
(Nanjing, China). The cDNA was cloned into the
XbaI/XbaI site of the pGL3-control vector downstream
of the luciferase gene, to generate pGL3-CAB39 vectors with the
following oligonucleotide sequences (Fig. 3C): The gene CAB39 3′UTR-Wild:
3′-TCTAGATGTTAGCTATTCAG CATCAGGCACTCTTATTGATTCATGAGGAACACTGC
TAATCTGCTGTTAAGTGAACGGTTTTTCATTTTACCCT
TTTGTTTTTCAGTCCAGGTTGGAGATCGTAGCTGCTG CTGCTTGCACACTCTAGAA-5′; The
gene CAB39 3′UTR-Mut: 3′-TGTTAGCTATTCAGCATCAGGCACTCTTATTGA
TTCATGAGGAACATTACTGCTAATCTGCTGTTAAGTGC
CATTGGGTTCATTTTACCCTTTTGTTTTTCAGTCCAGG
TTGGAGATCGTAGCTGCTGCTGCTTGCACACC-5′.
For the luciferase reporter assay, the U251, LN229,
U87 and A172 cells were cultured in 96-well plates (2000 cells per
well), transfected with 5 pmol of the hsa-miR-451 mimic
oligonucleotide with Lipofectamine 2000. 24 h after transfection,
the cells were transfected again, this time with 0.2 μg of either
the pGL3-CAB39-3-UTR-Subcloning plasmids or the
pGL3-CAB39-3′UTR-Mut plasmids with Lipofectamine 2000. After 48 h
of this transfection, luciferase activity was measured using the
Luciferase Assay System (Promega).
Western blot analysis
Cells were harvested 48 h after transfection, rinsed
three times with ice-cold PBS and then extracted with 1% Nonidet
P-40 lysis buffer (20 mM Tris, pH 8.0, 137 mM NaCl, 1% Nonidet
P-40, 10% glycerol, 1 mM CaCl2, 1 mM MgCl2, 1
mM phenylmethylsulfonyl fluoride, 1 mM sodium fluoride, 1 mM sodium
orthovanadate, and a protease inhibitor mixture). Homogenates were
clarified by centrifugation at 20,000 × g for 15 min at 4°C, and
protein concentrations were measured by Nanodrop spectrophotometer
(Gene, USA). Protein lysates (50 μg) from each sample were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) on 10% SDS polyacrylamide gel. The
separated proteins were transferred to polyvinylidene difluoride
(PVDF) membranes (Millipore, Billerica, MA). The membrane was
incubated with primary antibodies against CAB39, LKB1, AMPK,
p-AMPK, PI3K (p110α) and p-AKT1/2/3 (1:1000 dilution, Santa Cruz,
USA), followed by incubation with an HRP-conjugated secondary
antibody (1:1000 dilution, Zhongshan Bio Corp, Beijing, China). The
specific protein was detected using a SuperSignal protein detection
kit (Pierce, USA). After washing with stripping buffer, the
membrane was re-probed with antibody against GAPDH (1:1000
dilution, Santa Cruz).
Subcutaneous tumor assay
BALB/c-A 6-week-old nude mice were purchased from
the animal center of the Cancer Institute of Chinese Academy of
Medical Science and housed individually in ventilated microisolator
cages with water and food. All experimental procedures were carried
out according to the regulations and internal biosafety and
bioethics guidelines of Tianjin Medical University and the Tianjin
Municipal Science and Technology Commission. The LN229 subcutaneous
tumor xenograft model was established as previously described
(23). When the tumors were
approximately 5 mm in length, the mice were randomly divided into 3
groups (10 mice per group): the LN229 control group, the LN229
scramble PBS-treated group, and the LN229 miR-451-treated group. A
mixture of 5 μl oligonucleotides containing scramble miR-451 mimics
and 10 μl Lipofectamine was injected into the xenograft tumor model
in a multi-site injection manner. The mice in the LN229 control
group received 10 μl of PBS only. Treatment was conducted every
four days, until the end of the experiment. The tumor volume was
measured with a caliper every 3 days using the formula, volume =
length × width2/2. At the end of a 21-day observation
period, the mice were sacrificed and the tumor tissues were removed
for formalin fixation and preparation of paraffin-embedded sections
for immunohistochemical analysis.
Immunohistochemistry analysis
The paraffin-embedded tissue sections were examined
for CAB39, AMPK, p-AMPK, LKB1, PI3K, p-AKT, and GAPDH expression,
while the glioma tissue microarrays were examined for CAB39
expression, and H&E staining. Sections were dewaxed, treated
with 3% H2O2 for 10 min, and incubated with
appropriate primary antibodies (1:100; Santa Cruz Biotechnology)
overnight at 4°C. They were then treated with biotinylated
secondary antibody (1:100) for 1 h at room temperature, followed by
incubation with avidin-biotin complex (ABC)-peroxidase for a
further 1 h. After washing with Tris buffer, the sections were
incubated with 3,3′-diaminobenzidine (DAB) for 5 min, rinsed in
water, counterstained with hematoxylin, and visualized using a
light microscope.
Statistical analysis
SPSS10.0 was used for statistical analysis. One-way
analysis of variance (ANOVA) and χ2 test was used to
analyze the significance between groups. The LSD method of multiple
comparisons with parental and control vector groups was used when
the probability for ANOVA was statistically significant.
Statistical significance was determined at p<0.05.
Results
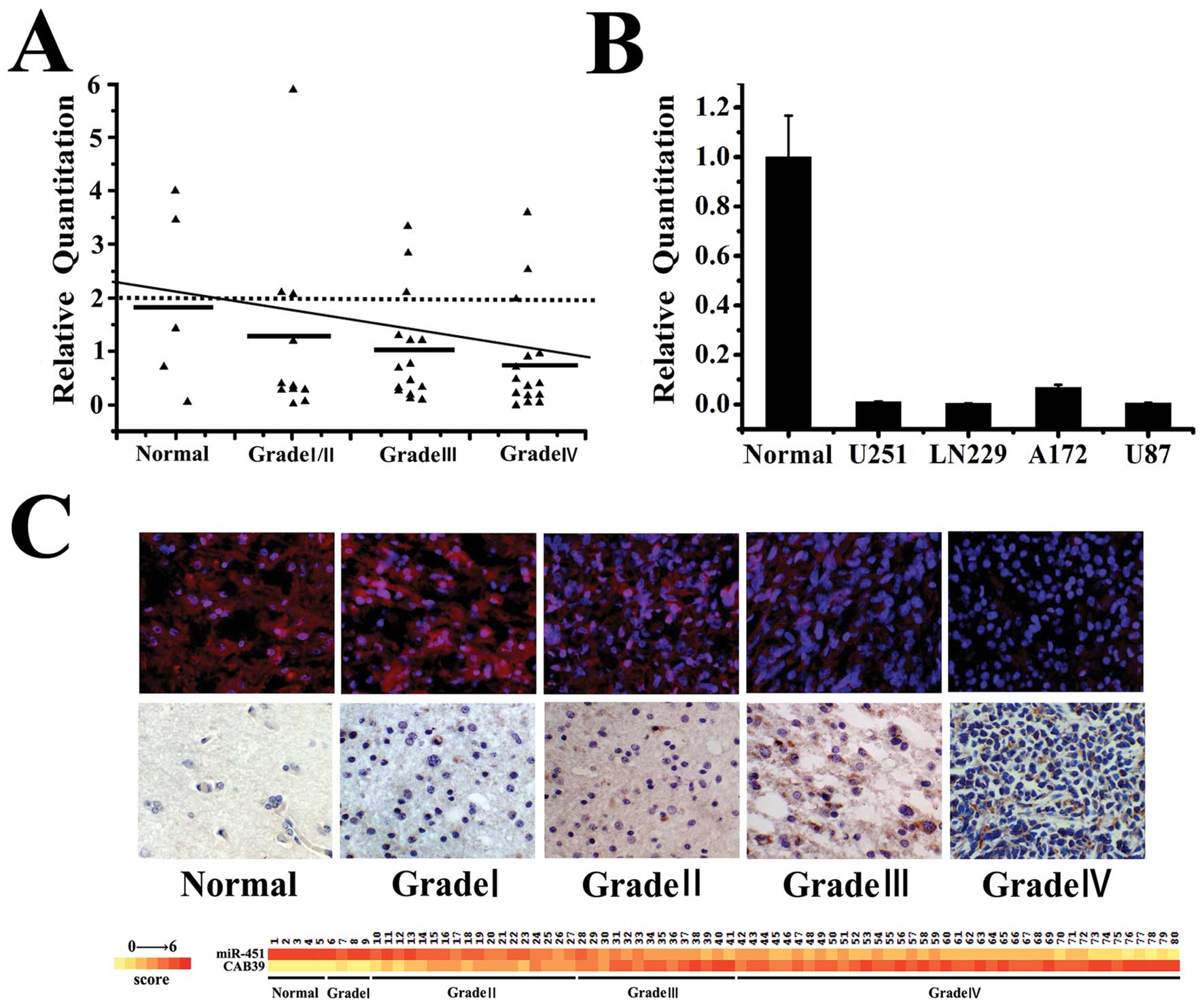
miR-451 expression was negatively
correlated with the WHO grades of gliomas
To further study the biological role of miR-451 in
human glioma tissues, we examined miR-451 expression in normal
brain tissues, glioma tissues and glioma cell lines by quantitative
RT-PCR. As shown (Fig. 2A and B),
miR-451 expression decreased with the increasing WHO grades of
glioma tissues. In situ hybridization analysis revealed that
miR-451 was expressed in gliomas and its total positive rate was
94.67% (71/75). The expression of miR-451 decreased markedly in
high grade gliomas (WHO grades III and IV) compared to its
expression in low grade gliomas (WHO grades I and II). Indeed,
22/22 low grade gliomas exhibited detectable levels of miR-451,
while in 4/53 high grade gliomas miR-451 was at detectable levels
(p<0.05) (Fig. 2C).
CAB39 is a target gene of miR-451 in
glioma
Although the reported under-expression of miR-451 in
some types of tumors suggested a role in cancer development, the
underlying mechanism is still unclear because little is known about
the miR-451 target genes. Therefore, the identification of
miR-451-regulated targets is a necessary step to understand how
miR-451 functions. We used a three-step consequential approach to
identify miR-451 target genes. First, target genes were predicted
by bioinformatics analysis, then, predicted genes were tested with
Western blotting and finally, potential target genes were validated
by fluorescent reporter assay (25).
miR-451 target gene(s)
identification
Bioinformatics analysis failed to identify AKT1
and/or c-Myc as hsa-miR-451 target genes. However, using the STRING
proteins functional association network database, AKT1 was shown to
have a direct association with AKTIP and YWHAZ and an indirect
association with CAB39, all three of these genes were predicted to
be hsa-miR-451 target genes (Fig.
1). CAB39-AMPK-mTOR-AKT1 was inferred in KEGG pathway database,
with CAB39-AMPK-mTOR documented while AKT1 not. It is possible that
hsa-miR-451 interacts indirectly with AKT1, through its target
genes. Both the STRING and KEGG databases indicated that CAB39 was
the preferential candidate target gene of hsa-miR-451.
miR-451 target gene confirmation
Quantitative real-time PCR showed that miR-451
expression increased in U251, LN229, A172, and U87 cells by 340.14,
849.22, 1680.88 and 2033.85-fold respectively after transfection
with the miR-451 mimics, compared to its expression in the control
and scramble treated cells (Fig.
3A). The transfected cells were used in subsequent Western blot
assays and CAB39 was found to be significantly downregulated in
cells treated with the hsa-miR-451 mimic oligonucleotide. Finally,
as shown in Fig. 3B, luciferase
activity was significantly decreased in cells co-transfected with
hsa-miR-451 and pGL3-CAB39-3′UTR-wild plasmid cells compared to its
activity in the scramble, negative control and pGL3-CAB39-3′UTR-Mut
plasmid-treated cells (p=0.0011, Fig.
3D). Together, these data demonstrated that CAB39 is a target
gene of the miR-451 in glioma.
Previously we have shown that miR-451 expression was
negatively correlated with glioma WHO grades in quantitative
real-time PCR and in situ hybridization. Here, we found a
similar phenomenon between CAB39 expression and glioma WHO grades
(Fig. 2C), using
immunohistochemistry. CAB39 was expressed in gliomas and its total
positive rate was 96% (72/75). The levels of CAB39 increased
markedly in high grade gliomas (WHO grades III and IV) in
comparison to low grade gliomas (WHO grades I and II). Indeed,
53/53 high grade gliomas exhibited detectable levels of CAB39,
while 3/22 low grade gliomas the protein was at undetectable levels
(p<0.05).
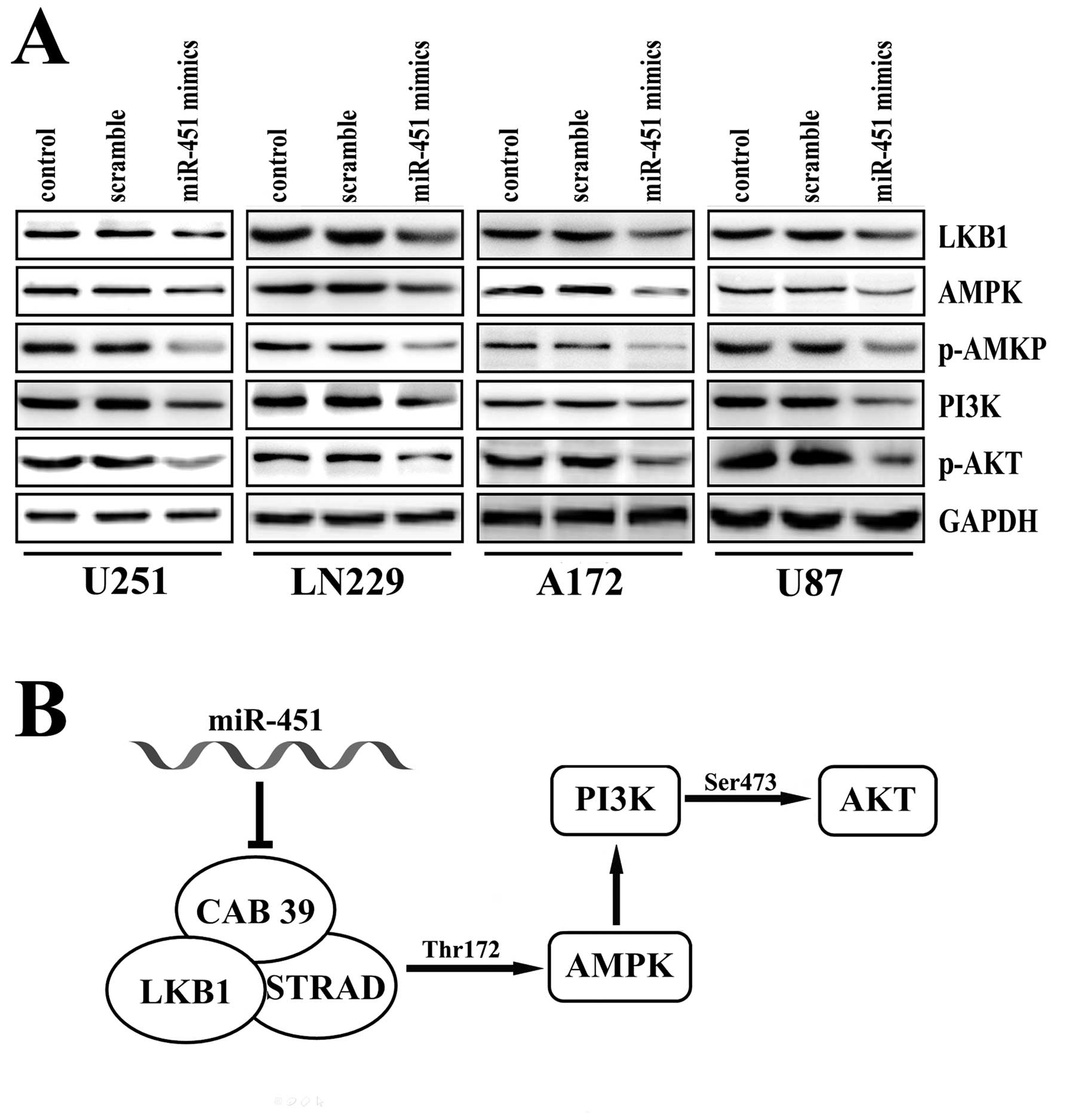
miR-451 regulated PI3K/ATK pathway
factors in human glioma in vitro
Previous data from our laboratory showed that
miR-451 had a significant impact on cell proliferation, invasion
and apoptosis in human glioblastoma cell lines, possibly by
regulating AKT expression (18).
AKT1 plays a critical role in controlling a diversity of cellular
functions, such as protein synthesis, cell cycle, cell survival and
apoptosis (26,27). We, therefore, investigated
AKT-related pathways. As shown in Fig.
4B, obvious activation of phosphorylated-AKT was observed in
U251, LN229, A172 and U87 cells after transfection with the miR-451
mimics. Consistently, over-expression of miR-451 led to a marked
down-regulation of LKB1, AMPK, p-AMPK, and PI3K, all of which are
involved in the pathway upstream of AKT (Fig. 4A). These data suggest that the
tumor suppressor activity of miR-451 in glioblastoma cells likely
acts through its regulation of the PI3K/AKT pathway.
miR-451 inhibited the growth of LN229
glioblastoma cells in vivo
To further understand the anti-tumor effect of
miR-451 and its role in the signaling pathway in vivo, we
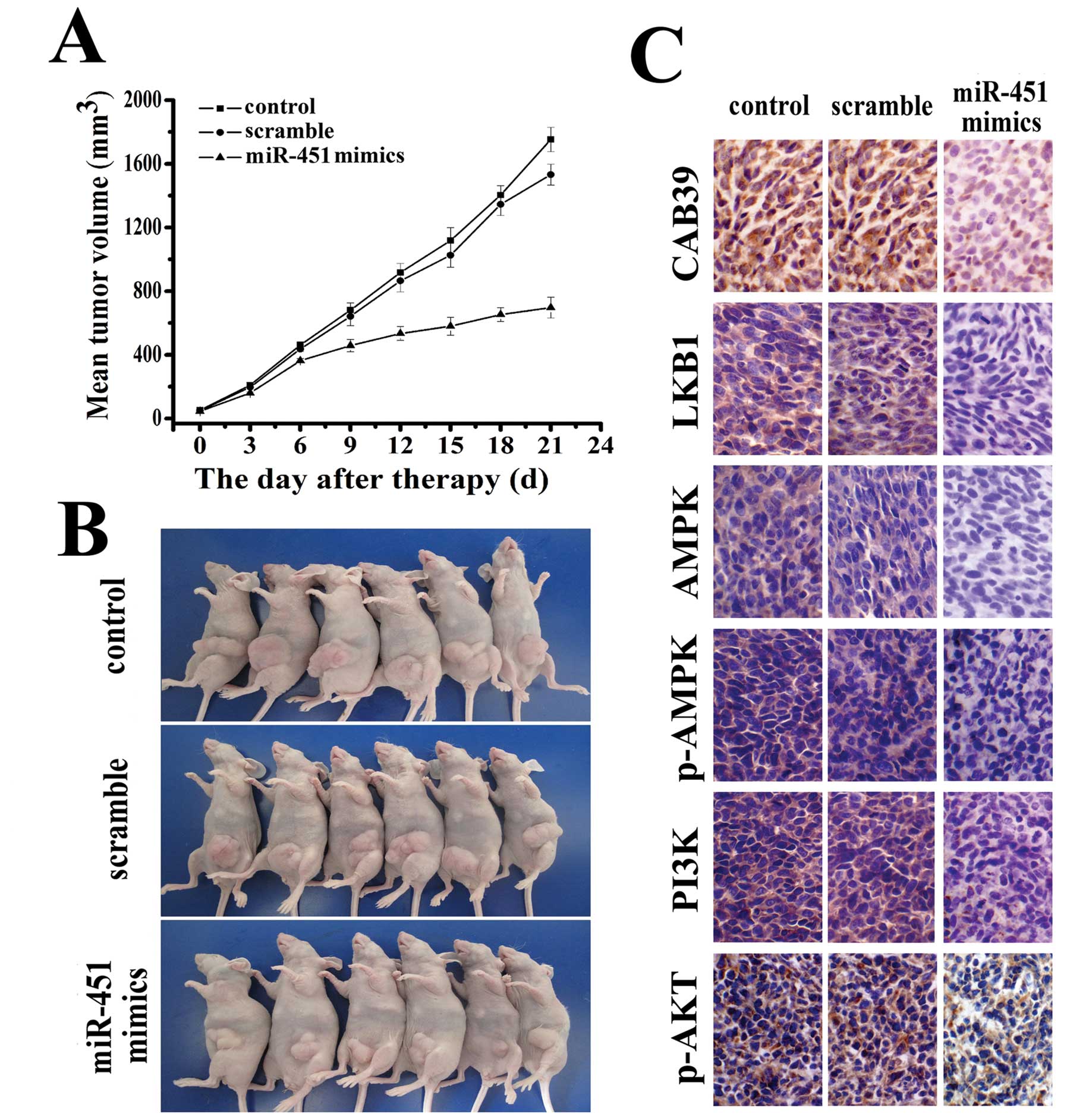
employed an LN229 xenograft glioma mouse model. The mean volume of
the tumors used in this study prior to treatment was 56±20.35
mm3 (Fig. 5B). During
the first 3 days of observation following intratumoral
administration of miR-451, tumors in both the control and treated
groups grew slowly with no marked differences in tumor size between
them. Tumors in the group treated with miR-451 maintained a slow
growth rate throughout the experiment while tumors in the control
group began to grow faster. On day 12, tumors of the mice in the
miR-451 treated group started to show statistically significant
differences in size compared to tumors of the mice in the control
group (p<0.05). At the termination of the study, the difference
in tumor mass between the miR-451 treated group and the control
group was marked (p<0.01). No difference in tumor volume was
observed between the control and PBS-treated groups (Fig. 5A). To determine whether
intratumoral miR-451 administration affected the expression of
factors in the PI3K/AKT signaling pathway, the expressions of
CAB39, LKB1, AMPK, p-AMPK, PI3K and p-AKT were tested and were
found to show significant downregulation in an
immunohistopathological examination (Fig. 5C).
Discussion
miRNA expression profiling studies revealed that a
number of miRNAs were dysregulated in human glioblastoma. miRNA-451
was one of them. A tumor suppressive role of miR-451 was shown in
gliomas in vitro (18), but
whether or not miR-451 actually participated in gliomagenesis still
needs further investigation. Expression alteration can be a useful
index of such activity. We used a high throughput experiment,
microarray in situ hybridization examination, and further
validated the result using the more quantitative and more sensitive
quantitative real-time PCR test on clinical samples. miR-451 was
down-regulated in glioma tissues and a significant negative
correlation was revealed between miR-451 expression and glioma WHO
grades.
Fluorescent reporter assay is generally accepted as
a gold standard to determine miRNA targets. Using this assay, we
demonstrated that CAB39 was a target gene of miR-451. CAB39 (MO25)
is an armadillo repeat scaffolding-like protein with two isoforms,
CAB39α and CAB39β (28,29). Structural studies have revealed
that the CAB39α forms an extended α-helical repeat rod-like
structure, distantly related to the armadillo repeat domain
(30). CAB39 is a component of the
trimeric LKB1-STRAD-MO25 complex (29,31)
and its role is to stabilize the binding of STRAD to LKB1 and
re-localize LKB1 from the nucleus to the cytoplasm (29,32).
LKB1, a member of the serine/threonine kinase family, regulates
cell polarity and functions as a tumor suppressor. Mutations in
this widely expressed protein kinase in humans result in a disorder
termed Peutz-Jeghers syndrome (PJS), which predisposes the sufferer
to a wide spectrum of benign and malignant tumors (33,34).
LKB1 is activated through its interaction with STRAD (37) and MO25 (29). AMP-activated protein kinase (AMPK)
is a sensor of cellular energy charge which regulates physiological
processes that consume or regenerate ATP to restore the energy
charge in the cells (35). Studies
in mammalian cells demonstrated that LKB1 complexed to STRAD and
MO25 activated AMPK by phosphorylating Thr172, and that the STRAD
and MO25 subunits enhanced phosphorylation of AMPK by over 100-fold
(36,37). The phosphatidylinositol-3′ kinase
(PI3K) family plays complex and extensive roles in many aspects of
cell biology and metabolism (38).
Signaling through PI3Ks is central to cell survival, growth and
proliferation which, as a consequence, is frequently activated in
many human cancers, including glioblastoma (39). Data from Luo et al provided
new evidence that AMPK activated Akt by regulating PI3K (40).
To verify the significance of LKB1/AMPK, PI3K/AKT
signaling in glioma, we performed Western blotting in human glioma
cell lines and immunohistochemistry in a tumor xenograft mouse
model. Consistently, over-expression of miR-451 led to a marked
downregulation of factors upstream of AKT. This evidence, both
in vitro and in vivo, implied that miR-451 can
suppress cell proliferation in human glioma through the LKB1/AMPK
and PI3K/AKT pathway. However, more evidence still needs to be
found. Increasing the expression of miR-451 might be a useful
therapeutic strategy for treating glioma in the future.
Acknowledgements
This study was supported by the China National
Natural Scientific Fund (Grant no. 81172406), the Tianjin Science
and Technology Committee (Grant no. 09JCZDJC20500) and the
Technology Fund of the Tianjin Public Health Bureau (Grant no.
09KZ112).
References
|
1
|
Furnari FB, Fenton T, Bachoo RM, et al:
Malignant astrocytic glioma: genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007.
|
|
2
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: the avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010.
|
|
3
|
Gal H, Pandi G, Kanner AA, et al: MIR-451
and imatinib mesylate inhibit tumor growth of glioblastoma stem
cells. Biochem Biophys Res Commun. 376:86–90. 2008.
|
|
4
|
Zhang CZ, Zhang JX, Zhang AL, et al:
MiR-221 and miR-222 target PUMA to induce cell survival in
glioblastoma. Mol Cancer. 9:2292010.
|
|
5
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009.
|
|
6
|
Moriyama T, Ohuchida K, Mizumoto K, et al:
MicroRNA-21 modulates biological functions of pancreatic cancer
cells including their proliferation, invasion, and chemoresistance.
Mol Cancer Ther. 8:1067–1074. 2009.
|
|
7
|
Selbach M, Schwanhausser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008.
|
|
8
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008.
|
|
9
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009.
|
|
10
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.
|
|
11
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.
|
|
13
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
|
|
14
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006.
|
|
15
|
Bandres E, Agirre X, Ramirez N, Zarate R
and Garcia-Foncillas J: MicroRNAs as cancer players: potential
clinical and biological effects. DNA Cell Biol. 26:273–282.
2007.
|
|
16
|
Mahlamaki EH, Barlund M, Tanner M, et al:
Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in
pancreatic cancer. Genes Chromosomes Cancer. 35:353–358. 2002.
|
|
17
|
Varis A, Wolf M, Monni O, et al: Targets
of gene amplification and overexpression at 17q in gastric cancer.
Cancer Res. 62:2625–2629. 2002.
|
|
18
|
Nan Y, Han L, Zhang A, et al: MiRNA-451
plays a role as tumor suppressor in human glioma cells. Brain Res.
1359:14–21. 2010.
|
|
19
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: a database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261.
2003.
|
|
20
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.
|
|
21
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008.
|
|
22
|
Kruger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34:W451–W454. 2006.
|
|
23
|
Kang CS, Zhang ZY, Jia ZF, et al:
Suppression of EGFR expression by antisense or small interference
RNA inhibits U251 glioma cell growth in vitro and in vivo. Cancer
Gene Ther. 13:530–538. 2006.
|
|
24
|
Zhang J, Han L, Zhang A, et al: AKT2
expression is associated with glioma malignant progression and
required for cell survival and invasion. Oncol Rep. 24:65–72.
2010.
|
|
25
|
Wan HY, Guo LM, Liu T, Liu M, Li X and
Tang H: Regulation of the transcription factor NF-kappaB1 by
microRNA-9 in human gastric adenocarcinoma. Mol Cancer.
9:162010.
|
|
26
|
Brazil DP, Park J and Hemmings BA: PKB
binding proteins. Getting in on the Akt. Cell. 111:293–303.
2002.
|
|
27
|
Dufour G, Demers MJ, Gagne D, et al: Human
intestinal epithelial cell survival and anoikis. Differentiation
state-distinct regulation and roles of protein kinase B/Akt
isoforms. J Biol Chem. 279:44113–44122. 2004.
|
|
28
|
Alessi DR, Sakamoto K and Bayascas JR:
LKB1-dependent signaling pathways. Annu Rev Biochem. 75:137–163.
2006.
|
|
29
|
Boudeau J, Baas AF, Deak M, et al:
MO25alpha/beta interact with STRADalpha/beta enhancing their
ability to bind, activate and localize LKB1 in the cytoplasm. EMBO
J. 22:5102–5114. 2003.
|
|
30
|
Milburn CC, Boudeau J, Deak M, Alessi DR
and van Aalten DM: Crystal structure of MO25 alpha in complex with
the C terminus of the pseudo kinase STE20-related adaptor. Nat
Struct Mol Biol. 11:193–200. 2004.
|
|
31
|
Brajenovic M, Joberty G, Kuster B,
Bouwmeester T and Drewes G: Comprehensive proteomic analysis of
human Par protein complexes reveals an interconnected protein
network. J Biol Chem. 279:12804–12811. 2004.
|
|
32
|
Baas AF, Boudeau J, Sapkota GP, et al:
Activation of the tumour suppressor kinase LKB1 by the STE20-like
pseudokinase STRAD. EMBO J. 22:3062–3072. 2003.
|
|
33
|
Hemminki A, Markie D, Tomlinson I, et al:
A serine/threonine kinase gene defective in Peutz-Jeghers syndrome.
Nature. 391:184–187. 1998.
|
|
34
|
Jenne DE, Reimann H, Nezu J, et al:
Peutz-Jeghers syndrome is caused by mutations in a novel serine
threonine kinase. Nat Genet. 18:38–43. 1998.
|
|
35
|
Hardie DG, Scott JW, Pan DA and Hudson ER:
Management of cellular energy by the AMP-activated protein kinase
system. FEBS Lett. 546:113–120. 2003.
|
|
36
|
Hawley SA, Boudeau J, Reid JL, et al:
Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and
MO25 alpha/beta are upstream kinases in the AMP-activated protein
kinase cascade. J Biol. 2:282003.
|
|
37
|
Woods A, Johnstone SR, Dickerson K, et al:
LKB1 is the upstream kinase in the AMP-activated protein kinase
cascade. Curr Biol. 13:2004–2008. 2003.
|
|
38
|
Vanhaesebroeck B, Leevers SJ, Ahmadi K, et
al: Synthesis and function of 3-phosphorylated inositol lipids.
Annu Rev Biochem. 70:535–602. 2001.
|
|
39
|
Cheng CK, Fan QW and Weiss WA: PI3K
signaling in glioma-animal models and therapeutic challenges. Brain
Pathol. 19:112–120. 2009.
|
|
40
|
Tao R, Gong J, Luo X, et al: AMPK exerts
dual regulatory effects on the PI3K pathway. J Mol Signal.
5:12010.
|