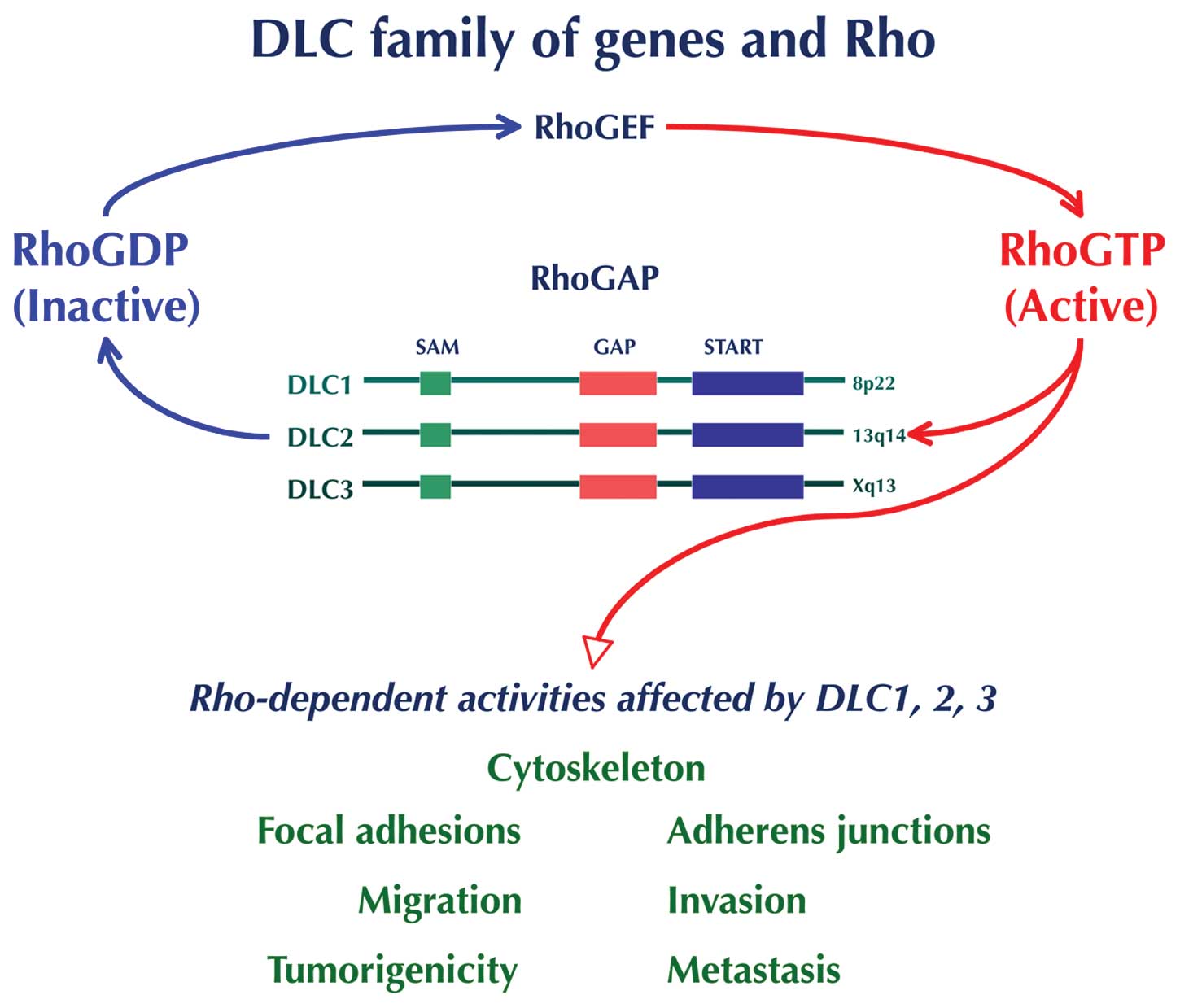
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bishop JM: The molecular genetics of
cancer. Science. 235:305–311. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rowley JD: Ph1-positive leukaemia,
including chronic myelogenous leukaemia. Clin Haematol. 9:55–86.
1980.PubMed/NCBI
|
|
4
|
Kraus MH, Popescu NC, Amsbaugh SC and King
CR: Overexpression of the EGF receptor-related protooncogene erbB-2
in human mammary tumor cell lines by different molecular
mechanisms. EMBO J. 6:605–610. 1987.PubMed/NCBI
|
|
5
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gore SD, Baylin S, Sugar E, Carraway H,
Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, Rudek
MA, Zhao M, Smith BD, Manning J, Jiemjit A, Dover G, Mays A,
Zwiebel J, Murgo A, Weng LJ and Herman JG: Combined DNA
methyltransferase and histone deacetylase inhibition in the
treatment of myeloid neoplasms. Cancer Res. 66:6361–6369. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Richon VM, Garcia-Vargas J and Hardwick
JS: Development of vorinostat: current applications and future
perspectives for cancer therapy. Cancer Lett. 280:201–210. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellullar carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Altekruse SF, McGlynn KA and Reichman ME:
Hepatocellular carcinima incidence, mortality and survival trends
in United States from 1975 to 2005. J Clin Oncol. 27:1485–1491.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones RG and Thompson CB: Tumor suppressor
and cell metabolism: a recipe for cancer growth. Genes Dev.
23:537–548. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Keck CL, Zimonjic DB, Yuan BZ,
Thorgeirsson SS and Popescu NC: Nonrandom breakpoints of unbalanced
chromosome translocations in human hepatocellular carcinoma cell
lines. Cancer Genet Cytogenet. 111:37–44. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zimonjic DB, Keck CL, Thorgeirsson SS and
Popescu NC: Novel recurrent genetic imbalances in human
hepatocellular carcinoma cell lines identified by comparative
genomic hybridization. Hepatology. 29:1208–1214. 1999. View Article : Google Scholar
|
|
13
|
Popescu NC: Genetic alterations in cancer
as a result of breakage at fragile sites. Cancer Lett. 192:1–17.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
15
|
Simon D, Knowles B and Weith A:
Abnormalities of chromosome 1 and loss of heterozygosity on 1p in
primary hepatomas. Oncogene. 6:765–770. 1991.PubMed/NCBI
|
|
16
|
Yeh SH, Chen PJ, Chen HL, Lai MY, Wang CC
and Chen DS: Frequent genetic alterations at the distal region of
chromosome 1p in human hepatocellular carcinomas. Cancer Res.
54:4188–4192. 1994.PubMed/NCBI
|
|
17
|
Woo HG, Park ES, Lee JS, Lee YH, Ishikawa
T, Kim YJ and Thorgeirsson SS: Identification of potential driver
genes in human liver carcinoma by genomewide screening. Cancer Res.
69:4059–4066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan BZ, Keck-Waggoner C, Zimonjic DB,
Thorgeirsson SS and Popescu NC: Alterations of FHIT gene in human
hepatocellular carcinoma. Cancer Res. 60:1049–1053. 2000.PubMed/NCBI
|
|
19
|
Imreh S, Klein G and Zabarovsky ER: Search
for unknown tumor-antagonizing genes. Genes Chromosomes Cancer.
38:307–321. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Popescu NC, Klein G and Imreh S:
The interferon-alpha responsive gene TMEM7 suppresses cell
proliferation and is downregulated in human hepatocellular
carcinoma. Cancer Genet Cytogenet. 177:6–15. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zimonjic DB, Zhou X, Lee JS,
Ullmannova-Benson V, Tripathi V, Thorgeirsson SS and Popescu NC:
Acquired genetic and functional alterations associated with
transforming growthfactor beta type I resistance in Hep3B human
hepatocellular carcinoma cell line. J Cell Mol Med. 13:3985–3992.
2009. View Article : Google Scholar
|
|
22
|
Ludes-Meyers JH, Bednarek AK, Popescu NC,
Bedford M and Aldaz CM: WWOX, the common chromosomal fragile site,
FRA16D, cancer gene. Cytogenet Genome Res. 100:101–110. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park SW, Ludes-Meyers J, Zimonjic DB,
Durkin ME, Popescu NC and Aldaz CM: Frequent downregulation and
loss of WWOX gene expression in human hepatocellular carcinoma. Br
J Cancer. 91:753–759. 2004.PubMed/NCBI
|
|
24
|
Yuan BZ, Zhou X, Zimonjic DB, Durkin ME
and Popescu NC: Amplification and overexpression of the EMS 1
oncogene, a possible prognostic marker, in human hepatocellular
carcinoma. J Mol Diagn. 5:48–53. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zimonjic DB, Durkin ME, Keck-Waggoner CL,
Park SW, Thorgeirsson SS and Popescu NC: SMAD5 gene expression, re
arrangements, copy number, and amplification at fragile site FRA5C
in human hepatocellular carcinoma. Neoplasia. 5:390–396. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Emi M, Fujiwara Y, Ohata H, Tsuda H,
Hirohashi S, Koike M, Miyaki M, Monden M and Nakamura Y: Allelic
loss at chromosome band 8p21.3-p22 is associated with progression
of hepatocellular carcinoma. Genes Chromosomes Cancer. 7:152–157.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pineau P, Nagai H, Prigent S, Wei Y,
Gyapay G, Weissenbach J, Tiollais P, Buendia MA and Dejean A:
Identification of three distinct regions of allelic deletions on
the short arm of chromosome 8 in hepatocellular carcinoma.
Oncogene. 18:3127–3134. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin LX, Tang ZY, Sham JS, Ma ZC, Ye SL,
Zhou XD, Wu ZQ, Trent JM and Guan XY: The association of chromosome
8p deletion and tumor metastasis in human hepatocellular carcinoma.
Cancer Res. 59:5662–5665. 1999.PubMed/NCBI
|
|
29
|
Chan KL, Lee JM, Guan XY, Fan ST and Ng
IO: High-density allelotyping of chromosome 8p in hepatocellular
carcinoma and clinicopathologic correlation. Cancer. 94:3179–3185.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kahng YS, Lee YS, Kim BK, Park WS, Lee JY
and Kang CS: Loss of heterozygosity of chromosome 8p and 11p in the
dysplastic nodule and hepatocellular carcinoma. J Gastroenterol
Hepatol. 18:430–436. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pang JZ, Qin LX, Ren N, Hei ZY, Ye QH, Jia
WD, Sun BS, Lin GL, Liu DY, Liu YK and Tang ZY: Loss of
heterozygosity at D8S298 is a predictor for long-term survival of
patients with tumor-node-metastasis stage I of hepatocellular
carcinoma. Clin Cancer Res. 13:7363–7369. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yam JW, Wong CM and Ng IO: Molecular and
functional genetics in hepatocellular carcinoma. Front Biosci
(Schol Ed). 2:117–134. 2010. View
Article : Google Scholar
|
|
33
|
Birnbaum D, Adélaïde J, Popovici C,
Charafe-Jauffret E, Mozziconacci MJ and Chaffanet M: Chromosome arm
8p and cancer: a fragile hypothesis. Lancet Oncol. 4:639–642. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Popescu NC: Fragile sites and cancer genes
on the short arm of chromosome 8. Lancet Oncol. 5:772004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fujiwara Y, Ohata H, Kuroki T, Koyama K,
Tsuchiya E, Monden M and Nakamura Y: Isolation of a candidate tumor
suppressor gene on chromosome 8p21.3-p22 that is homologous to an
extracellular domain of the PDGF receptor beta gene. Oncogene.
10:891–895. 1995.PubMed/NCBI
|
|
36
|
Yuan BZ, Miller MJ, Keck CL, Zimonjic DB,
Thorgeirsson SS and Popescu NC: Cloning, characterization, and
chromosomal localization of a gene frequently deleted in human
liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res.
58:2196–2199. 1998.PubMed/NCBI
|
|
37
|
Yan J, Yu Y, Wang N, Chang Y, Ying H, Liu
W, He J, Li S, Jiang W, Li Y, Liu H, Wang H and Xu Y:
LFIRE-1/HFREP-1, a liver-specific gene, is frequently downregulated
and has growth suppressor activity in hepatocellular carcinoma.
Oncogene. 23:1939–1949. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shih YL, Shyu RY, Hsieh CB, Lai HC, Liu
KY, Chu TY and Lin YW: Promoter methylation of the secreted
frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular
carcinoma. Cancer. 107:579–590. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang J, Zhang YL, Teng XM, Lin Y, Zheng
DL, Yang PY and Han ZG: Down-regulation of SFRP1 as a putative
tumor suppressor gene can contribute to human hepatocellular
carcinoma. BMC Cancer. 7:1262007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lei KF, Wang YF, Zhu XQ, Lu PC, Sun BS,
Jia HL, Ren N, Ye QH, Sun HC, Wang L, Tang ZY and Qin LX:
Identification of MSRA gene on chromosome 8p as a candidate
metastasis suppressor for human hepatitis B virus-positive
hepatocellular carcinoma. BMC Cancer. 7:1722007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang J, Zheng DL, Qin FS, Cheng N, Chen
H, Wan BB, Wang YP, Xiao HS and Han ZG: Genetic and epigenetic
silencing of SCARA5 may contribute to human hepatocellular
carcinoma by activating FAK signaling. J Clin Invest. 120:223–241.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Finch PW, He X, Kelley MJ, Uren A,
Schaudies P, Popescu NC, Rudicoff S, Aaronson SA, Varmus HE and
Rubin JS: Purification and molecular cloning of a secreted,
Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA.
94:6670–6675. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rubin JS, Barshishat-Kupper M,
Feroze-Merzoug F and Xi ZF: Secreted WNT antagonists as tumor
suppressors: pro and con. Front Biosci. 11:2093–2105. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saini S, Liu J, Yamamura S, Majid S,
Kawakami K, Hirata H and Dahiya R: Functional significance of
secreted Frizzled-related protein 1 in metastatic renal cell
carcinomas. Cancer Res. 69:6815–6822. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kawamoto K, Hirata H, Kikuno N, Tanaka Y,
Nakagawa M and Dahiya R: DNA methylation and histone modifications
cause silencing of Wnt antagonist gene in human renal cell
carcinoma cell lines. Int J Cancer. 123:535–542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Thompson MD and Monga SP: WNT/beta-catenin
signaling in liver health and disease. Hepatology. 45:1298–1305.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Takigawa Y and Brown AM: Wnt signaling in
liver cancer. Curr Drug Targets. 9:1013–1024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ching YP, Wong CM, Chan SF, Leung TH, Ng
DC, Jin DY and Ng IO: Deleted in liver cancer (DLC) 2 encodes a
RhoGAP protein with growth suppressor function and is
underexpressed in hepatocellular carcinoma. J Biol Chem.
278:10824–10830. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Durkin EM, Ullmannova V, Guan M and
Popescu NC: Deleted in liver cancer 3(DLC-3), a novel
RhoGTPase-activating protein, is downregulated in cancer and
inhibits tumor cell growth. Oncogene. 26:4580–4589. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Durkin ME, Yuan BZ, Zhou X, Zimonjic DB,
Lowy DR, Thorgeirsson SS and Popescu NC: DLC-1: a Rho
GTPase-activating protein and tumor suppressor. J Cell Mol Med.
11:1185–1207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Low JS, Tao Q, Ng KM, Goh HK, Shu XS, Woo
WL, Ambinder RF, Srivastava G, Shamay M, Chan AT, Popescu NC and
Hsieh WS: A novel isoform of the 8p22 tumor suppressor gene DLC1
suppresses tumor growth and is frequently silenced in multiple
common tumors. Oncogene. 30:1923–1935. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liao YC and Lo SH: Deleted in liver
cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J
Biochem Cell Biol. 40:843–847. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xue W, Krasnitz A, Lucito R, Sordella R,
Vanaelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers
S, Zender L and Lowe SW: DLC1 is a chromosome 8p tumor suppressor
whose loss promotes hepatocellular carcinoma. Genes Dev.
22:1439–1444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lahoz A and Hall A: DLC1: a significant
GAP in the cancer genome. Genes Dev. 22:1724–1730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vigil D, Cherfils J, Rossman KL and Der
CJ: Ras superfamily GEFs and GAPs: validated and tractable targets
for cancer therapy? Nat Rev Cancer. 12:842–857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Durkin ME, Avner MR, Huh CG, Yuan BZ,
Thorgeirsson SS and Popescu NC: DLC-1, a Rho GTPase-activating
protein with tumor suppressor function, is essential for embryonic
development. FEBS Lett. 579:1191–1196. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hers I, Wherlock M, Homma Y, Yagisawa H
and Tavaré JM: Identification of p122RhoGAP (deleted in liver
cancer-1) Serine 322 as a substrate for protein kinase B and
ribosomal S6 kinase in insulin-stimulated cells. J Biol Chem.
281:4762–4770. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Murakami R, Osanai T, Tomita H, Sasaki S,
Maruyama A, Itoh K, Homma Y and Okumura K: p122 protein enhances
intra-cellular calcium increase to acetylcholine: its possible role
in the pathogenesis of coronary spastic angina. Arterioscler Thromb
Vasc Biol. 30:1968–1975. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu J, Li Y, Fan X, Zhang C, Wang Y and
Zhao Z: Analysis of gene expression profile of periodontal ligament
cells subjected to cyclic compressive force. DNA Cell Biol.
30:865–873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ng IO, Liang ZD, Cao L and Lee TK: DLC1 is
deleted in primary hepatocellular carcinoma and exerts inhibitory
effects on the proliferation of hepatoma cell lines with deleted
DLC1. Cancer Res. 60:6581–6584. 2000.PubMed/NCBI
|
|
61
|
Park SW, Durkin ME, Thorgeirsson SS and
Popescu NC: DNA variants of DLC-1, a candidate tumor suppressor
gene in human hepatocellular carcinoma. Int J Oncol. 23:133–137.
2003.PubMed/NCBI
|
|
62
|
Liao YC, Shih YP and Lo SH: Mutations in
the focal adhesion targeting region of deleted in liver cancer-1
attenuate their expression and function. Cancer Res. 68:7718–7722.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A,
Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya
T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP,
Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR,
Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G,
Vogelstein B, Velculescu VE and Kinzler KW: Core signaling pathways
in human pancreatic cancers revealed by global genomic analyses.
Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yachida S, Jones S, Bozic I, Antal T,
Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA,
Velculescu VE, Kinzler KW, Vogelstein B and Iacobuzio- Donahue CA:
Distant metastasis occurs late during the genetic evolution of
pancreatic cancer. Nature. 467:1114–1117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dong X, Zhou G, Zhai Y, Zhang H, Yang H,
Zhi L, Zhang X, Chu J and He F: Association of DLC1 gene
polymorphism with susceptibility to hepatocellular carcinoma in
Chinese hepatitis B virus carriers. Cancer Epidemiol. 33:265–270.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Teodoridis JM, Hardie C and Brown R: CpG
island methylator phenotype (CIMP) in cancer: causes and
implications. Cancer Lett. 268:177–186. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yuan BZ, Durkin ME and Popescu NC:
Promoter hypermethylation of DLC-1, a candidate tumor suppressor
gene, in several common human cancers. Cancer Genet Cytogenet.
140:113–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wong CM, Lee JM, Ching YP, Jin DY and Ng
IO: Genetic and epigenetic alterations of DLC-1 gene in
hepatocellular carcinoma. Cancer Res. 63:7646–7651. 2003.PubMed/NCBI
|
|
69
|
Ko FC, Yeung YS, Wong CM, Chan LK, Poon
RT, Ng IO and Yam JW: Deleted in liver cancer 1 isoforms are
distinctly expressed in human tissues, functionally different and
under differential transcriptional regulation in hepatocellular
carcinoma. Liver Int. 30:139–148. 2010. View Article : Google Scholar
|
|
70
|
Croce MC: Causes and consequences of
microRNA dysregulation in cancer. Nature Rev Genet. 10:704–714.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Banaudha K, Kaliszewski M, Korolnek T,
Florea L, Yeung ML, Kuan KT and Kumar A: MicroRNA silencing of
tumor suppressor DLC-1 promotes efficient hepatitis C virus
replication in primary human hepatocytes. Hepatology. 53:53–61.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wong CM, Yam JW, Ching YP, Yau TO, Leung
TH, Jin DY and Ng IO: Rho GTPase-activating protein deleted in
liver cancer suppresses cell proliferation and invasion in
hepatocellular carcinoma. Cancer Res. 65:8861–8868. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Healy KD, Hodgson L, Kim TY, Shutes AT,
Maddileti S, Juliano RL, Hahn KM, Harden TK, Bang YJ and Der CJ:
DLC1 suppresses non-small lung cancer growth and invasion by
RhoGAP-dependent and independent mechanisms. Mol Carcinog.
47:326–337. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kim TY, Lee JW, Kim HP, Jong HS, Kim TY,
Jung M and Bang YJ: DLC-1, a GTPase-activating protein for Rho, is
associated with cell proliferation, morphology and migration in
human hepatocellular carcinoma. Biochem Biophys Res Commun.
355:72–77. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Qian X, Li G, Asmussen HK, Asnaghi L, Vass
WC, Braverman R, Yamada KM, Popescu NC, Papageorge AG and Lowy DR:
Oncogenic inhibition by a deleted in liver cancer gene requires
cooperation between tensin binding and Rho-specific
GTPase-activating protein activities. Proc Natl Acad Sci USA.
104:9012–9017. 2007. View Article : Google Scholar
|
|
76
|
Guan M, Tripathi V, Zhou X and Popescu NC:
Adenovirus-mediated restoration of the expression of the tumor
suppressor gene DLC1 inhibits the proliferation and tumorigenicity
of aggressive, androgen-independent human prostate cancer cell
lines: Prospects for gene therapy. Cancer Gene Ther. 15:371–381.
2008. View Article : Google Scholar
|
|
77
|
Zhou X, Zimonjic DB, Park SW, Yang XY,
Durkin ME and Popescu NC: DLC1 suppresses distant dissemination of
human hepatocellular carcinoma cells in nude mice through reduction
of RhoA GTPase activity, actin cytoskeletal disruption and
down-regulation of genes involved in metastasis. Int J Oncol.
32:1258–1291. 2008.
|
|
78
|
Holeiter G, Heering J, Erlmann P, Schmid
S, Jähne R and Olayioye MA: Deleted in liver cancer 1 controls
migration through a Dia1-dependent signaling pathway. Cancer Res.
68:8743–8751. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Erlmann P, Schmid S, Horenkamp FA, Geyer
M, Pomorski TG and Olayioye M: DLC1 activation requires lipid
interation through a polybasic region preceding the RhoGap domain.
Mol Biol Cell. 20:4400–4411. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhong D, Zhang J, Yang S, Soh UJ,
Buschdorf JP, Zhou YT, Yang D and Low BC: The SAM domain of the
RhoGAP DLC1 binds EF1A1 to regulate cell migration. J Cell Sci.
122:414–424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar
|
|
82
|
Gómez del Pulgar T, Benitah SA, Valerón
PF, Espina C and Lacal JC: Rho GTPase expression in tumourigenesis:
evidence for a significant link. Bioessays. 27:602–613.
2005.PubMed/NCBI
|
|
83
|
Jaffe AB and Hall A: Rho GTPases in
transformation and metastasis. Adv Cancer Res. 84:57–80. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ridley AJ: Rho proteins and cancer. Breast
Cancer Res Treat. 84:13–19. 2004. View Article : Google Scholar
|
|
85
|
Grise F, Bidaud A and Moreau V: Rho
GTPases in hepatocellular carcinoma. Biochim Biophys Acta.
1795:137–151. 2009.PubMed/NCBI
|
|
86
|
Roessler S, Long EL, Budhu A, Chen Y, Zhao
X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, Tang ZY, He P, Hunter KW,
Thorgeirsson SS, Meltzer PS and Wang XW: Integrative genomic
identification of genes on 8p associated with hepatocellular
carcinoma progression and patient survival. Gastroenterology. Dec
24–2011.(Epub ahead of print).
|
|
87
|
Pihur V and Datta S and Datta S: Finding
common genes in multiple cancer types through meta-analysis of
microarray experiments: a rank aggregation approach. Genomics.
92:400–403. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhou X, Thorgeirsson SS and Popescu NC:
Restoration of DLC-1 gene expression induces apoptosis and inhibits
both cell growth and tumorigenicity in human hepatocellular
carcinoma cells. Oncogene. 23:1308–1313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kawai K, Yamaga M, Iwamae Y, Kiyota M,
Kamata H, Hirata H, Homma Y and Yagisawa H: A PLCdelta1-binding
protein, p122RhoGAP, is localized in focal adhesions. Biochem Soc
Trans. 32:1107–1109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kim TY, Vigil D, Der CJ and Juliano RL:
Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in
regulation of the cytoskeleton and cell motility. Cancer Metastasis
Rev. 28:77–83. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yuan BZ, Jefferson AM, Millecchia L,
Popescu NC and Reynolds SH: Morphological changes and nuclear
translocation of DLC1 tumor suppressor protein precede apoptosis in
human non-small cell lung carcinoma cells. Exp Cell Res.
313:3868–3880. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Goodison S, Yuan J, Sloan D, Kim R, Li C,
Popescu NC and Urquidi V: The RhoGAP protein DLC-1 functions as a
metastasis suppressor in breast cancer cells. Cancer Res.
65:6042–6053. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ko FC, Chan LK, Tung EK, Lowe SW, Ng IO
and Yam JW: Akt phosphorylation of deleted in liver cancer 1
abrogates its suppression of liver cancer tumorigenesis and
metastasis. Gastroenterology. 139:1397–1407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yam JW, Ko FC, Chan CY, Jin DY and Ng IO:
Interaction of deleted in liver cancer 1 with tensin2 in caveolae
and implications in tumor suppression. Cancer Res. 66:8367–8372.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liao YC, Si L, deVere White RW and Lo SH:
The phosphotyrosine-independent interaction of DLC-1 and the SH2
domain of cten regulates focal adhesion localization and growth
suppression activity of DLC-1. J Cell Biol. 176:43–49. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hall EH, Daugherty AE, Choi CK, Horwitz AF
and Brautigan DL: Tensin1 requires protein phosphatase-1alpha in
addition to RhoGAP DLC-1 to control cell polarization, migration,
and invasion. J Biol Chem. 284:34713–34722. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chan LK, Ko FC, Ng IO and Yam JW: Deleted
in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2
PTB domain interaction and is required for tumor-suppressive
function. PLoS One. 4:e55722009. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hafizi S, Sernstad E, Swinny JD, Gomez MF
and Dahlbäck B: Individual domains of Tensin2 exhibit distinct
subcellular localisations and migratory effects. Int J Biochem Cell
Biol. 42:52–61. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Clark K, Howe JD, Pullar CE, Green JA,
Artym VV, Yamada KM and Critchley DR: Tensin 2 modulates cell
contractility in 3D collagen gels through the RhoGAP DLC1. J Cell
Biochem. 109:808–817. 2010.PubMed/NCBI
|
|
100
|
Kawai K, Kitamura SY, Maehira K, Seike J
and Yagisawa H: START-GAP1/DLC1 is localized in focal adhesions
through interaction with the PTB domain of tensin2. Adv Enzyme
Regul. 50:202–215. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Du X, Qian X, Papageorge A, Vass WC,
Braverman R and Lowy DR: Complex formation between DLC START domain
and Cav1 contributes to the tumor suppressor function of DLC1. Proc
Am Assoc Cancer Res. 52:5232011.
|
|
102
|
Yang XY, Guan M, Vigil D, Der CJ, Lowy DR
and Popescu NC: p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor
protein and inhibits its RhoA GTPase and growth-suppressing
activities. Oncogene. 28:1401–1409. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tripathi V, Zimonjic DB and Popescu NC:
DLC1 and α-catenin protein interaction enhances DLC1 antioncogenic
activity by stabilizing adherens junctions and suppressing NFκB
signaling. Proc Am Assoc Cancer Res. 52:9622011.
|
|
104
|
Yang X, Popescu NC and Zimonjic DB: DLC1
interaction with S100A10 mediates inhibtion of in vitro cell
invasion and tumorigenicity of lung cancer cells through a
RhoGAP-indpendent mechanism. Cancer Res. 71:2916–2925.
2011.PubMed/NCBI
|
|
105
|
Scholz RP, Gustafsson JO, Hoffmann P,
Jaiswal M, Ahmadian MR, Eisler S, Erlmann P, Schmid S, Hausser A
and Olayioye MA: The tumor suppressor protein DLC1 is regulated by
PKD-mediated GAP domain phosphorylation. Exp Cell Res. 317:496–503.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tompa P: Intrinsically unstructured
proteins. Trends Biochem Sci. 27:527–533. 2002. View Article : Google Scholar
|
|
107
|
Hermeking H: The 14-3-3 cancer connection.
Nat Rev Cancer. 3:931–943. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Scholz RP, Regner J, Theil A, Erlmann P,
Holeiter G, Jähne R, Schmid S, Hausser A and Olayioye MA: DLC1
interacts with 14-3-3 proteins to inhibit RhoGAP activity and block
nucleocytoplasmic shuttling. J Cell Sci. 122:92–102. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wuestefeld T and Zender L: DLC1 and liver
cancer: the Akt connection. Gastroenterology. 139:1093–1096. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Oliveira AM, Ross JS and Fletcher JA:
Tumor suppressor genes in breast cancer: the gatekeepers and the
caretakers. Am J Clin Pathol. 124:S16–S28. 2005.PubMed/NCBI
|
|
111
|
Meyer N and Penn LZ: Reflecting on 25
years with MYC. Nat Rev Cancer. 8:976–990. 2008.PubMed/NCBI
|
|
112
|
Varmus H: Retroviruses. Science.
240:1427–1435. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Peters G: Oncogenes at viral integration
sites. Cell Growth Differ. 1:503–510. 1990.PubMed/NCBI
|
|
114
|
Popescu NC and Zimonjic DB:
Chromosome-mediated alterations of the MYC gene in human cancer. J
Cell Mol Med. 6:151–159. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Payne GS, Bishop JM and Varmus HE:
Multiple arrangements of viral DNA and an activated host oncogene
in bursal lymphomas. Nature. 295:209–214. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Popescu NC, Zimonjic DB and DiPaolo JA:
Viral integration, fragile sites and proto-oncogenes in human
neoplasia. Hum Genet. 84:383–386. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu J, Kaur G, Zhawar VK, Zimonjic DB,
Popescu NC, Kandpal R and Athwal RS: Role of SV40 integration site
at chromosomal interval 1q21.1 in immortalized CRL2504 cells.
Cancer Res. 69:7819–7825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Weinberg RA: Integrated genomes of animal
viruses. Annu Rev Biochem. 49:197–226. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bester AC, Schwartz M, Schmidt M, Garrigue
A, Hacein-Bey-Abina S, Cavazzana-Calvo M, Ben-Porat N, Von Kalle C,
Fischer A and Kerem B: Fragile sites are preferential targets for
integrations of MLV vectors in gene therapy. Gene Ther.
13:1057–1059. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Croce CM and Nowell PC: Molecular basis of
human B cell neoplasia. Blood. 65:1–7. 1985.
|
|
121
|
Zimonjic DB, Keck-Waggoner C and Popescu
NC: Novel genomic imbalances and chromosome translocations
involving c-myc gene in Burkitt’s lymphoma. Leukemia. 15:1582–1588.
2001.PubMed/NCBI
|
|
122
|
Alitalo KM and Schwab M: Oncogene
amplification in tumor cells. Adv Cancer Res. 47:235–281. 1986.
View Article : Google Scholar
|
|
123
|
Alitalo K, Schwab M, Lin CC, Varmus HE and
Bishop JM: Homogeneously staining chromosomal regions contain
amplified copies of an abundantly expressed cellular oncogene
(c-myc) in malignant neuroendocrine cells from a human colon
carcinoma. Proc Natl Acad Sci USA. 80:1707–1711. 1983. View Article : Google Scholar
|
|
124
|
Zimonjic DB, Keck-Waggoner CL, Yuan BZ,
Kraus MH and Popescu NC: Profile of genetic alterations and
tumorigenicity of human breast cancer cells. Int J Oncol.
16:221–230. 2000.PubMed/NCBI
|
|
125
|
Elenbaas B, Spirio L, Koerner F, Fleming
MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC and Weinberg RA:
Human breast cancer cells generated by oncogenic transformation of
primary mammary epithelial cells. Genes Dev. 15:50–65. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Kaposi-Novak P, Libbrecht L, Woo HG, Lee
YH, Sears NC, Coulouarn C, Conner EA, Factor VM, Roskams T and
Thorgeirsson SS: Central role of c-Myc during malignant conversion
in human hepatocarcinogenesis. Cancer Res. 69:2775–2782. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: from genes to environment. Nat Rev Cancer.
6:674–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Schlaeger C, Longerich T, Schiller C,
Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R,
Lichter P, Schirmacher P and Radlwimmer B: Etiology-dependent
molecular mechanisms in human hepatocarcinogenesis. Hepatology.
47:511–520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wei Y, Ponzetto A, Tiollais P and Buendia
MA: Multiple rearrangements and activated expression of c-myc
induced by woodchuck hepatitis virus integration in a primary liver
tumour. Res Virol. 143:89–96. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Tokino T and Matsubara K: Chromosomal
sites for hepatitis B virus integration in human hepatocellu lar
carcinoma. J Virol. 65:6761–6764. 1991.PubMed/NCBI
|
|
131
|
Yunis JJ, Soreng AL and Bowe AE: Fragile
sites are targets of diverse mutagens and carcinogens. Oncogene.
1:59–69. 1987.PubMed/NCBI
|
|
132
|
Yang L, He J, Chen L and Wang G: Hepatitis
B virus X protein upregulates expression of SMYD3 and C-MYC in
HepG2 cells. Med Oncol. 26:445–451. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Santoni-Rugiu E, Nagy P, Jensen MR, Factor
VM and Thorgeirsson SS: Evolution of neoplastic development in the
liver of transgenic mice co-expressing c-myc and transforming
growth factor. Am J Pathol. 149:407–428. 1996.PubMed/NCBI
|
|
134
|
Sargent LM, Sanderson ND and Thorgeirsson
SS: Ploidy and karyotypic alterations associated with early events
in the development of hepatocarcinogenesis in transgenic mice
harboring c-myc and transforming growth factor alpha transgenes.
Cancer Res. 56:2137–2142. 1996.
|
|
135
|
Factor VM, Laskowska D, Jensen MR, Woitach
JT, Popescu NC and Thorgeirsson SS: Vitamin E reduces chromosomal
damage and inhibits hepatic tumor formation in a transgenic mouse
model. Proc Natl Acad Sci USA. 97:2196–2201. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Sargent LM, Zhou X, Keck CL, Sanderson ND,
Zimonjic DB, Popescu NC and Thorgeirsson SS: Nonrandom cytogenetic
alterations in hepatocellular carcinoma from transgenic mice
overexpressing c-Myc and transforming growth factor-alpha in the
liver. Am J Path. 154:1047–1055. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Grisham JW: Interspecies comparison of
liver carcinogenesis: implications for cancer risk assessment.
Carcinogenesis. 18:59–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Grisham JW: Molecular genetic alterations
in primary hepatocellular meoplasm: hepatocellular adenoma,
hepatocellular carcinoma, and hepatoblastoma. The Molecular Basis
of Human Cancer. Coleman WB and Tsongalis GT: Humana Press; Totowa,
New Jersey, NJ: pp. 259–346. 2001
|
|
139
|
Durkin ME, Keck-Waggoner CL, Popescu NC
and Thorgeirsson SS: Integration of a c-myc transgene results in
disruption of the mouse Gtf2ird1 gene, the homologue of the human
GTF2IRD1 gene hemizygously deleted in Williams-Beuren syndrome.
Genomics. 73:20–27. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Tassabehji M, Hammond P, Karmiloff-Smith
A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T,
Metcalfe K, Rucka A, Stewart H, Read AP, Maconochie M and Donnai D:
GTF2IRD1 in craniofacial development of humans and mice. Science.
310:1184–1187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zimonjic DB, Ullmannova-Benson V, Factor
VM, Thorgeirsson SS and Popescu NC: Recurrent and nonrandom DNA
copy number and chromosome alterations in Myc transgenic mouse
model for hepatocellular carcinogenesis: implications for human
disease. Cancer Genet Cytogenet. 191:17–26. 2009. View Article : Google Scholar
|
|
142
|
Zimonjic DB, Zhang H, Shan Z, Factor VM,
Trent J, Thorgeirsson SS and Popescu NC: DNA amplification
associated with double minutes originating from chromosome 19 in
mouse hepatocellular carcinoma. Cytogenet Cell Genet. 93:114–116.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Murakami H, Sanderson ND, Nagy P, Marino
PA, Merlino G and Thorgeirsson SS: Transgenic mouse model for
synergistic effects of nuclear oncogenes and growth factors in
tumorigenesis: interaction of c-myc and transforming growth factor
alpha in hepatic oncogenesis. Cancer Res. 53:1719–1723. 1993.
|
|
144
|
Shachaf CM, Kopelman AM, Arvanitis C,
Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B,
Cardiff RD, Yang Q, Bishop JM, Contag CH and Felsher DW: MYC
inactivation uncovers pluripotent differentiation and tumour
dormancy in hepatocellular cancer. Nature. 431:1112–1117. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Zender L, Spector MS, Xue W, Flemming P,
Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D,
Lucito R, Powers S and Lowe SW: Identification and validation of
oncogenes in liver cancer using an integrative oncogenomic
approach. Cell. 125:1253–1267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Roth JA and Grammer SF: Tumor suppressor
gene therapy. Tumor Supressor Genes: Regulation, Function and
Medicinal Applications. El-Deiry WS: 2. Humana Press; Totowa, NJ:
pp. 577–597. 2003, View Article : Google Scholar
|
|
147
|
Weinstein IB: Cancer. Addiction to
oncogenes - the Achilles heel of cancer. Science. 297:63–64. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Sharma SV and Settleman J: Oncogene
addiction: setting the stage for molecularly targeted cancer
therapy. Genes Dev. 21:3214–3231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang L, Zhang J, Xie L, Xie X, Guo Q, Lv
J, Gao Z, Qian Z, Yin X, Zheng L, Zhu G, Ji Q and Ren Z: Molecular
characterization of hepatocellular carcinoma (HCC) patient derived
explant models. Proc Am Assoc Cancer Res. 52:5772011.
|
|
150
|
Wong CC, Wong CM, Au SL and Ng IO:
RhoGTPases and Rho-effectors in hepatocellular carcinoma
metastasis: ROCK N’ Rho move it. Liver Int. 30:642–656.
2010.PubMed/NCBI
|
|
151
|
Takamura M, Sakamoto M, Genda T, Ichida T,
Asakura H and Hirohashi S: Inhibition of intrahepatic metastasis of
human hepatocellular carcinoma by Rho-associated protein kinase
inhibitor Y-27632. Hepatology. 33:577–581. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Ogawa T, Tashiro H, Miyata Y, Ushitora Y,
Fudaba Y, Kobayashi T, Arihiro K, Okajima M and Asahara T:
Rho-associated kinase inhibitor reduces tumor recurrence after
liver transplantation in a rat hepatoma model. Am J Transplant.
7:347–355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Nakajima M, Hayashi K, Egi Y, Katayama K,
Amano Y, Uehata M, Ohtsuki M, Fujii A, Oshita K, Kataoka H, Chiba
K, Goto N and Kondo T: Effect of Wf-536, a novel ROCK inhibitor,
against metastasis of B16 melanoma. Cancer Chemother Pharmacol.
52:319–324. 2003. View Article : Google Scholar
|
|
154
|
McHenry PR and Vargo-Gogola T: Pleiotropic
functions of Rho GTPase signaling: a Trojan horse or Achilles’ heel
for breast cancer treatment? Curr Drug Targets. 11:1043–1058.
2010.
|
|
155
|
Ullmannova V and Popescu NC: Inhibition of
cell proliferation, induction of apoptosis, reactivation of DLC1,
and modulation of other gene expression by dietary flavone in
breast cancer cell lines. Cancer Detect Prev. 31:110–118. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Pang X, Yi T, Yi Z, Cho S G, Qu W, Pinkaew
D, Fujise K and Liu M: Morelloflavone, a biflavonoid, inhibits
tumor angiogenesis by targeting rho GTPases and extracellular
signal-regulated kinase signaling pathways. Cancer Res. 69:518–525.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Liu H, Dong A, Gao C, Tan C, Xie Z, Zu X,
Qu L and Jiang Y: New synthetic flavone derivatives induce
apoptosis of hepatocarcinoma cells. Bioorg Med Chem. 18:6322–6328.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Yoshizumi T, Ohta T, Ninomiya I, Terada I,
Fushida S, Fujimura T, Nishimura G, Shimizu K, Yi S and Miwa K:
Thiazolidinedione, a peroxisome proliferator-activated
receptor-gamma ligand, inhibits growth and metastasis of HT-29
human colon cancer cells through differentiation-promoting effects.
Int J Oncol. 25:631–639. 2004.
|
|
159
|
Zhou X, Yang XY and Popescu NC:
Synergistic antineoplastic effect of DLC1 tumor suppressor protein
and histone deacetylase inhibitor, suberoylanilide hydroxamic acid
(SAHA), on prostate and liver cancer cells: perspectives for
therapeutics. Int J Oncol. 36:999–1005. 2010.
|
|
160
|
Chung GE, Yoon JH, Lee JH, Kim HY, Myung
SJ, Yu SJ, Lee SH, Lee SM, Kim YJ and Lee HS: Ursodeoxycholic
acid-induced inhibition of DLC1 protein degradation leads to
suppression of hepatocellular carcinoma cell growth. Oncol Rep.
25:1739–1746. 2011.PubMed/NCBI
|
|
161
|
Murphy DJ, Junttila MR, Pouyet L, Karnezis
A, Shchors K, Bui DA, Brown-Swigart L, Johnson L and Evan GI:
Distinct thresholds govern Myc’s biological output in vivo. Cancer
Cell. 14:447–457. 2008.PubMed/NCBI
|
|
162
|
Larsson LG and Henricksson MA: The Yin and
Yang functions of the Myc oncoprotein in cancer development and as
targets for therapy. Exp Cell Res. 316:1429–1437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Lin CP, Liu CR, Lee CN, Chan TS and Liu
HE: Targeting c-Myc as a novel approach for hepatocellular
carcinoma. World J Hepatol. 2:16–20. 2010.PubMed/NCBI
|
|
164
|
Brooks TA and Hurley LH: Targeting MYC
expression through G-quardruplexes. Genes Cancer. 1:641–649. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR,
Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici
M, Voliotis D and Bruix J: SHARP Investigators Study Group:
Sorafenib in advanced hepatocellular carcinoma. N Engl J Med.
359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Llovet J M and Bruix J: Molecular targeted
therapies in hepatocellular carcinoma. Hepatology. 48:1312–1327.
2008.
|
|
167
|
Cao Z, Fan-Minogue H, Bellovin DI,
Yevtodiyenko A, Arzeno J, Yang Q, Gambhir SS and Felsher DW: MYC
phosphorylation, activation, and tumorigenic potential in
hepatocellular carcinoma are regulated by HMG-CoA reductase. Cance
Res. 71:2286–2297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Kawata S, Yamasaki E, Nagase T, Inui Y,
Ito N, Matsuda Y, Inada M, Tamura S, Noda S, Imai Y and Matsuzawa
Y: Effect of pravastatin on survival in patients with advanced
hepatocellular carcinoma. A randomized controlled trial. Br J
Cancer. 84:886–891. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Homma Y and Emori Y: A dual functional
signal mediator showing RhoGAP and phospholipase C-delta
stimulating activities. EMBO J. 14:286–291. 1995.PubMed/NCBI
|
|
170
|
Ponting CP and Aravind L: START: a
lipid-binding domain in StAR, HD-ZIP and signalling proteins.
Trends Biochem Sci. 24:130–132. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Wang H, Han H, Mousses S and Von Hoff DD:
Targeting loss-of-function mutations in tumor-suppressor genes as a
strategy for development of cancer therapeutic agents. Semin Oncol.
33:513–520. 2006. View Article : Google Scholar : PubMed/NCBI
|