Introduction
microRNAs (miRNAs) are approximately 22 nucleotides
long and act as negative regulators of gene expression by
inhibiting mRNA translation or promoting mRNA degradation (1). Hundreds of miRNAs have been
identified in the human genome; bioinformatics and cloning studies
have estimated that miRNAs may regulate 30% of all human genes.
miRNAs can be expressed in a tissue-specific and developmental
stage-specific manner, and can be involved in the regulation of
embryonic stem cell fate (2,3).
They have attracted significant attention given their important
roles in cell-cycle, apoptosis and carcinogenesis (4). Thus, deregulation of miRNA expression
may lead to a variety of disorders, including human cancer.
Emerging evidence indicates that several miRNAs are deregulated in
human malignancies and it has been proposed that some miRNAs may
have oncogenic or tumor suppressor functions (5). For instance, the miR-17–92 cluster is
markedly overexpressed in B-cell lymphomas and miR-124 has been
documented as a tumor suppressor capable of inhibiting cell
proliferation of glioma (6,7).
Gastric cancer is one of the most common
malignancies and the second leading cause of cancer-related
mortality worldwide. It is estimated that 360,000 individuals
succumb to gastric cancer each year in China. Despite recent
advances in combining surgery, radiotherapy and chemotherapy no
effective targeting therapy is available for gastric cancer, mainly
due to a lack of complete understanding of the molecular mechanisms
underlying gastric cancer development (8). Some miRNAs have been reported to be
associated with the initiation and progression of gastric cancer.
One such miRNA, miR-106a, was found aberrantly expressed in gastric
carcinoma tissue and was able to promote gastric carcinogenesis.
The Let-7 miRNAs family as a class of tumor suppressor genes showed
low levels in gastric carcinoma tissue (9,10).
These suggest that miRNAs are involved in gastric carcinogenesis
via a variety of patterns and pathways.
Cho et al screened the expression pattern of
miRNA-372 in several human gastric cancer cell lines and found that
it is expressed only in the AGS cell line. miR-372 also supports
the growth of the gastric cancer cell line (11). Belair et al reported that
Helicobacter pylori downregulates the miR-371-372-373
cluster in proliferating gastric epithelial cells to achieve cell
cycle arrest (12). The growth
advantages that AGS cells gained from the high expression of the
miR-371-372-373 cluster confer to them characteristics of gastric
stem cell/progenitors rather than gastric mucosa differentiated
epithelium. However, the molecular details of miR-372 involvement
in gastric cancer development remain unclear. In the present study,
we found that miR-372 was upregulated in AGS cells when compared to
the undifferentiated human gastric carcinoma cell line HGC-27.
Notably, overexpression of miR-372 improved HGC-27 cell growth, but
inhibition of miR-372 using miR-372 inhibitor suppressed
proliferation and increased apoptosis of AGS cells in vitro.
In HGC-27 and AGS cells, we confirmed that TNFAIP1 was a target of
miR-372. TNFAIP1 is an immediate-early response gene of endothelium
induced by TNFα and IL-6 (13). It
may play roles in DNA synthesis, DNA repair, cell apoptosis and
human diseases (14,15). TNFAIP1 was highly expressed in
normal cell lines while it was lowly expressed in cancer cell lines
(16). The overexpression of
TNFAIP1 could accelerate the apoptosis of HeLa cells (17). Moreover, TNFAIP1 and KCTD10
suppressed the transcriptional activities of NFκB (18). We found that TNFAIP1 links miR-372
to NFκB activation, leading to increased cell survival by
transactivating anti-apoptotic genes downstream of NFκB.
Collectively, our data indicate that an oncogenic role of miR-372
may be through control of cell growth and cell apoptosis by
downregulating the TNFAIP1 and positively regulating NFκB
expression.
Materials and methods
Cell culture and transfection
The human gastric carcinoma cell lines (AGS, HGC-27)
and HEK293, Cos7 cell lines were obtained from the Cell Bank of the
Chinese Academy of Sciences (China). Cells were cultured in DMEM
(Gibco) or F12 (Gibco) media that was supplemented with 10% fetal
bovine serum, 100 μg/ml of streptomycin and 100 IU/ml of
penicillin, and maintained at 37°C in a humidified atmosphere with
5% CO2. The miRNA mimics and inhibitors that were used
in this study were purchased from GenePharma (Shanghai, China). All
transfections were carried out using Lipofectamine 2000
(Invitrogen, USA), according to the manufacturer’s
instructions.
MTT assay
The AGS cells were seeded in 24-cell well plates at
16,000 cells/well. Twenty-four hours after transfection, the cells
were incubated with 80 μl MTT at 37°C for another 4 h. Then
the medium was removed and the precipitated formazan was dissolved
in 400 μl DMSO. After shaking for 15 min, the absorbance at
420 nm was detected using a microplate spectrophotometer.
FACS assays
Cells were harvested at 600 g for 3 min, washed
twice in PBS at room temperature and resuspended in appropriate
PBS. Then, cells were resuspended in 1X FACS banding buffer
containing Annexin-V and propidium iodide and analyzed using a FACS
flow cytometer (BD Biosciences). A total of 20,000 cells were
counted for each sample.
Plasmid construction
The 3′UTR of TNFAIP1 was amplified from HeLa cDNA
using RT-PCR and was inserted into the 3′-end of the firefly
luciferase gene of the dual-luciferase miRNA target expression
vector pmirGLO (Promega Corp., Madison, WI, USA) between the
SacI and Xbal sites. Similarly, 3′UTR mutants, which
contained mutated miR-372 binding sites, were cloned to the pmirGLO
between the same sites. The primer sequences used for RT-PCR
amplification are shown in Table
I. All primers were synthesized by Shanghai Sangon Biotech
(Sangon Biotech, China). Sequences that were inserted into the
plasmid were verified by DNA sequencing. pCMV and pLuc-NFκB vectors
were purchased from Clontech.
 | Table I.The oligonucleotides used in this
study. |
Table I.
The oligonucleotides used in this
study.
| Name | | Sequence (5′-3′) |
|---|
| PmirGLO-TNFAIP1 | Forward | CGAGCTCGTGCTGCCTGGGTCTCTGC |
| Reverse | GCTCTAGAGCAGCTGCTCTGTCGGATGTTT |
| TNFAIP1-mut1 | Forward | CCTGCTCATGTCTGGAGAC |
| Reverse | CAGACATGAGCAGGGGCAA |
| TNFAIP1-F2 | Forward |
TGTGCAGAAGGGCTACTGC |
| TNFAIP1-R2 | Reverse |
GCAGTAGCCCTTCTGCACA |
| TNFAIP1-mut2 | Forward | ATTCTCATGTACATGACAATAAG |
| Reverse | TCAGTACAACATGAGTTAAAGAA |
| TNFAIP1-CDS | Forward |
AGTCGACGATGTCAGGGGACACCTGT |
| Reverse |
GGGTACCTCAGTCACGATGAGTGGA |
| RT-TNFAIP1 | Forward |
GCACTTTGGGCACCATTTTGA |
| Reverse |
CGGTTCTGAGGGAGGGTGAT |
| U6 | Forward |
TGCGGGTGCTCGCTTCGGCAGC |
| miR-372 | Forward |
GCCCGCAAAGTGCTGCGACAT |
| Reverse
(universal) |
CCAGTGCAGGGTCCGAGGT |
| β-actin | Forward |
CCTGTACGCCAACACAGTGC |
| Reverse |
ATACTCCTGCTTGCTGATCC |
Luciferase assay
The dual-luciferase reporter plasmids were
co-transfected with miRNA mimics (GenePharma) into HEK293 cells or
AGS cells. At 24 h post-transfection, cells were assayed for
luciferase activity using the Dual-Glo Luciferase assay system
(Promega), according to the manufacturer’s instructions. The
firefly luciferase activities were normalized to Renilla luciferase
activity. The firefly luciferase activity of the cells that were
transfected with miRNA mimics was represented as the percentage of
activity relative to that of the cells that were transfected with
negative control miRNA mimics. For each transfection, the
luciferase activity was averaged from three replicates (19).
RNA extraction and real-time RT-PCR
analysis
Total RNA was extracted using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions. For
miRNA detection, 2 μg of small RNA was reverse transcribed
to cDNA using miRNA First-Strand cDNA Synthesis kit (Invitrogen)
according to the manufacturer’s instructions. Quantitative
real-time PCR (qRT-PCR) analysis for miR-372 was performed in
triplicate with the SYBR Green PCR Master Mix (Perkin-Elmer,
Applied Biosystems) according to the manufacturer’s instructions.
U6 RNA was used to normalize expression. To detect the target
genes, 2 μg of total RNA was reverse transcribed to cDNA
using oligo(dT) primers and Moloney murine leukemia virus reverse
transcriptase (Promega). qRT-PCR was used to determine the
expression levels of TNFAIP1 using the primers presented in
Table I. β-actin levels were used
to normalize expression. Data analysis was performed using the
2−ΔΔCt method (20).
Immunofluorescent staining
Cells were cultured on glass coverslips in the
12-cell well. The cells were fixed using 4% paraformaldehyde,
permeabilized and incubated with the primary antibodies and then
with the secondary antibodies. The primary antibodies used were
rabbit polyclonal anti-TNFAIP1, and mouse monoclonal anti-NFκB
(P65) antibodies, while the secondary antibodies were Alexa Fluor
488 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse antibodies
(Molecular Probes). Hoechst 33258 (Sigma) was used to stain the
nuclei. Fluorescence on the processed slips was analyzed using a
fluorescent microscope (ZESS).
Western blot analysis
At 24 h posttransfection, cells were harvested and
lysed in RIPA buffer [50 mM Tris-HCl (pH 7.2), 150 mM NaCl, 1%
(v/v) Triton X-100, 1% (w/v) sodium deoxycholate, 0.1% (w/v) SDS]
with protease inhibitors. Proteins were separated on 10%
SDS-polyacrylamide gel and transferred to PVDF membranes. A
PageRuler prestained protein ladder was used as a molecular marker.
Antibodies to TNFAIP1, NFκB, caspase-8, PARP, Bcl-2 or the
endogenous control β-actin were incubated with the blot overnight
at 4°C. The secondary antibody was added after the membrane was
washed three times with TBST. The protein was detected using a
HRP-conjugated secondary antibody and a Chemilucent ECL Detection
system (Millipore, Billerica, MA, USA).
Statistical analysis
Results are presented as the means ± standard
deviation from at least three separate experiments. The differences
among groups were analyzed using the double-sided Student’s t-test
and P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-372 regulates growth of the gastric
cancer cell line
It is known that miR-372 is involved in the
tumorigenesis of the testicular germ cell tumor of adolescents and
adults. Cho et al(11)
analyzed the miR-372 expression profile of several gastric cancer
cell lines and only found the AGS cell line expressing miR-372.
They also showed miR-372 plays an important role in gastric
adenocarcinoma carcinogenesis. To further investigate the
relationship between miR-372 and gastric adenocarcinoma, we
introduced HGC-27, which is an undifferentiation gastric cancer
cell to our experiment. The relative expression of miR-372 measured
using real-time PCR demonstrated miR-372 expression levels in AGS
are markedly higher than in HGC-27 cells, in which miR-372 was
almost undetected (data not shown).
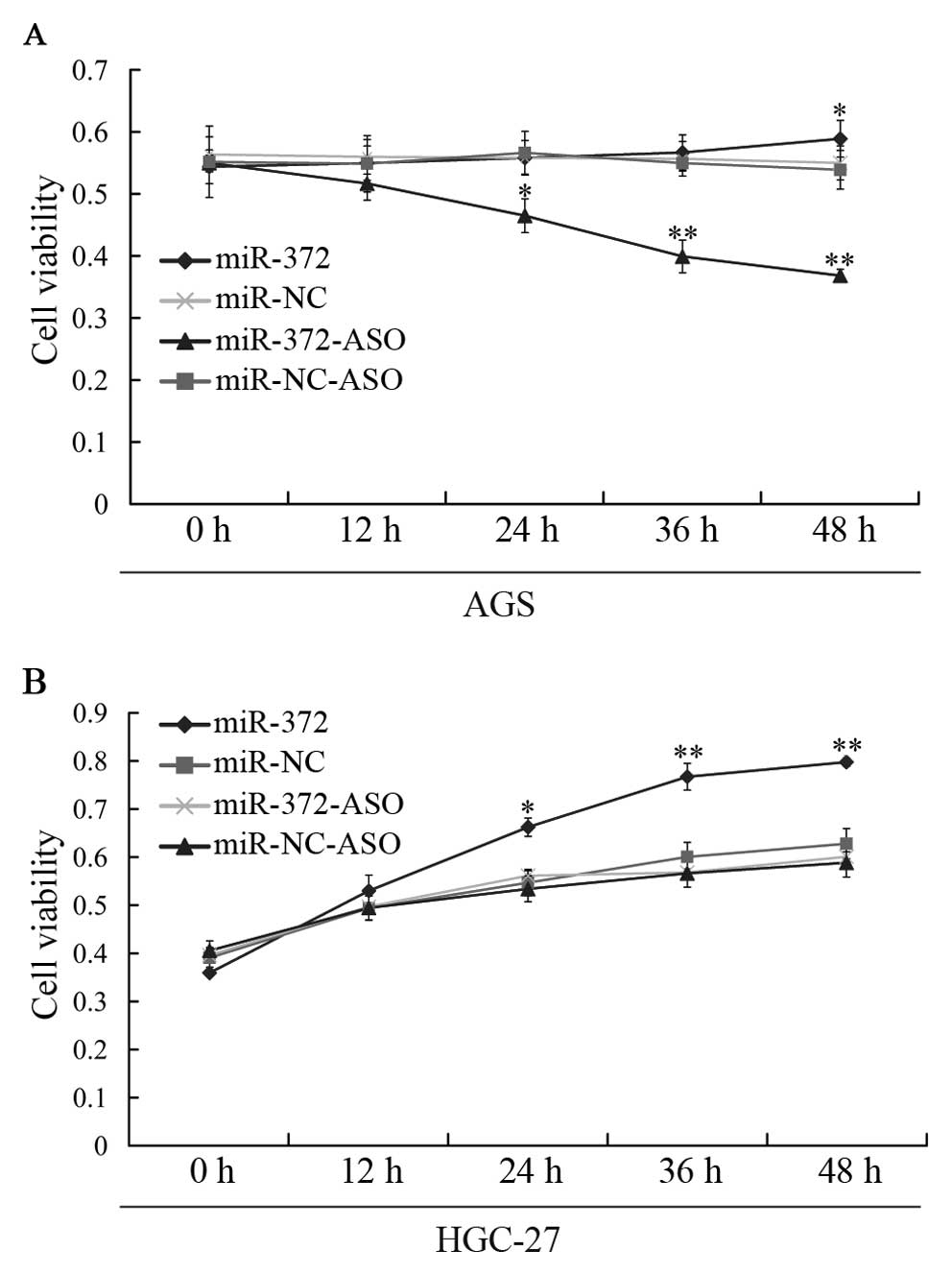
Next, we studied the growth of gastric cancer cells
affected by miR-372 using MTT assay. We found that the growth rate
of AGS cells transfected with the miR-372 inhibitor (miR-372-ASO)
was lower than that of cells transfected with miR-NC, miR-NC-ASO
and miR-372 mimics (Fig. 1A).
However, the growth rate of HGC-27 cells was elevated when
transfected with miR-372 mimics, compared with the growth rate of
cells treated with miR-372-ASO, miR-NC-ASO and miR-NC (Fig. 1B). Our data indicate that miR-372
could affect the growth of the gastric cancer cell line.
miR-372 affects apoptosis of AGS
cells
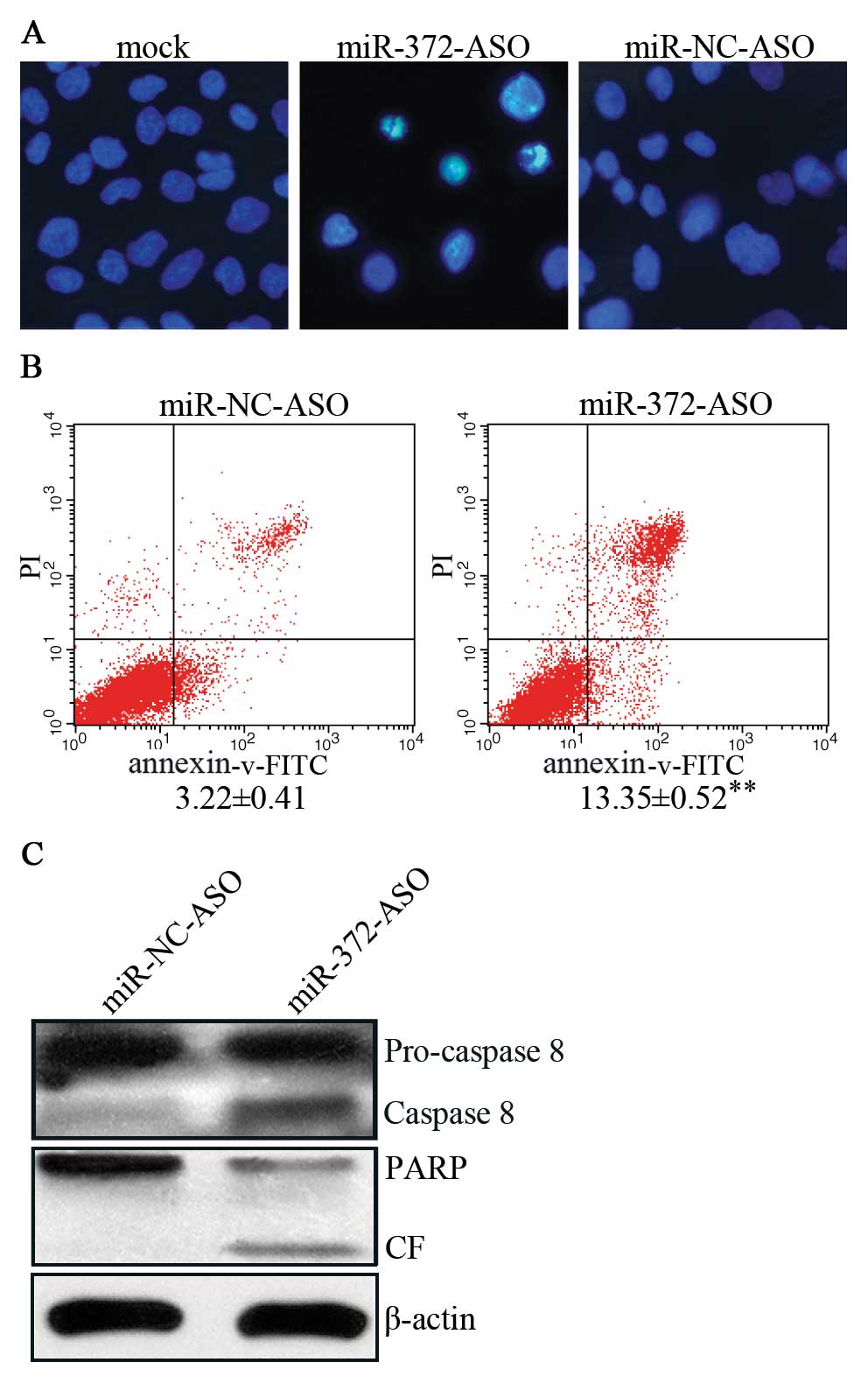
Next, we focused on AGS cells with aberrant
expression of miR-372. To determine whether miR-372 regulates
apoptosis of AGS, we conducted Hoechst 33258 staining assay. The
results indicated that suppression of miR-372 induced nuclear foci
compared with mock and miR-NC-ASO (Fig. 2A). Moreover, we detected cell
apoptosis in AGS cells using an Annexin-V-FITC apoptosis detection
kit and found that apoptosis rates were elevated by ∼4-fold in
miR-372-ASO vs. miR-NC-ASO-treated cells (Fig. 2B). Further experiments revealed
that miR-372-ASO-treated AGS cells induced the expression of PARP
and cleaved caspase-8 (Fig. 2C),
members of a general apoptosis pathway. In brief, these results
indicate that miR-372 regulates the caspase-8 apoptosis pathway and
inhibits the apoptosis of AGS cells.
TNFAIP1 is a target of miR-372 in AGS
cells
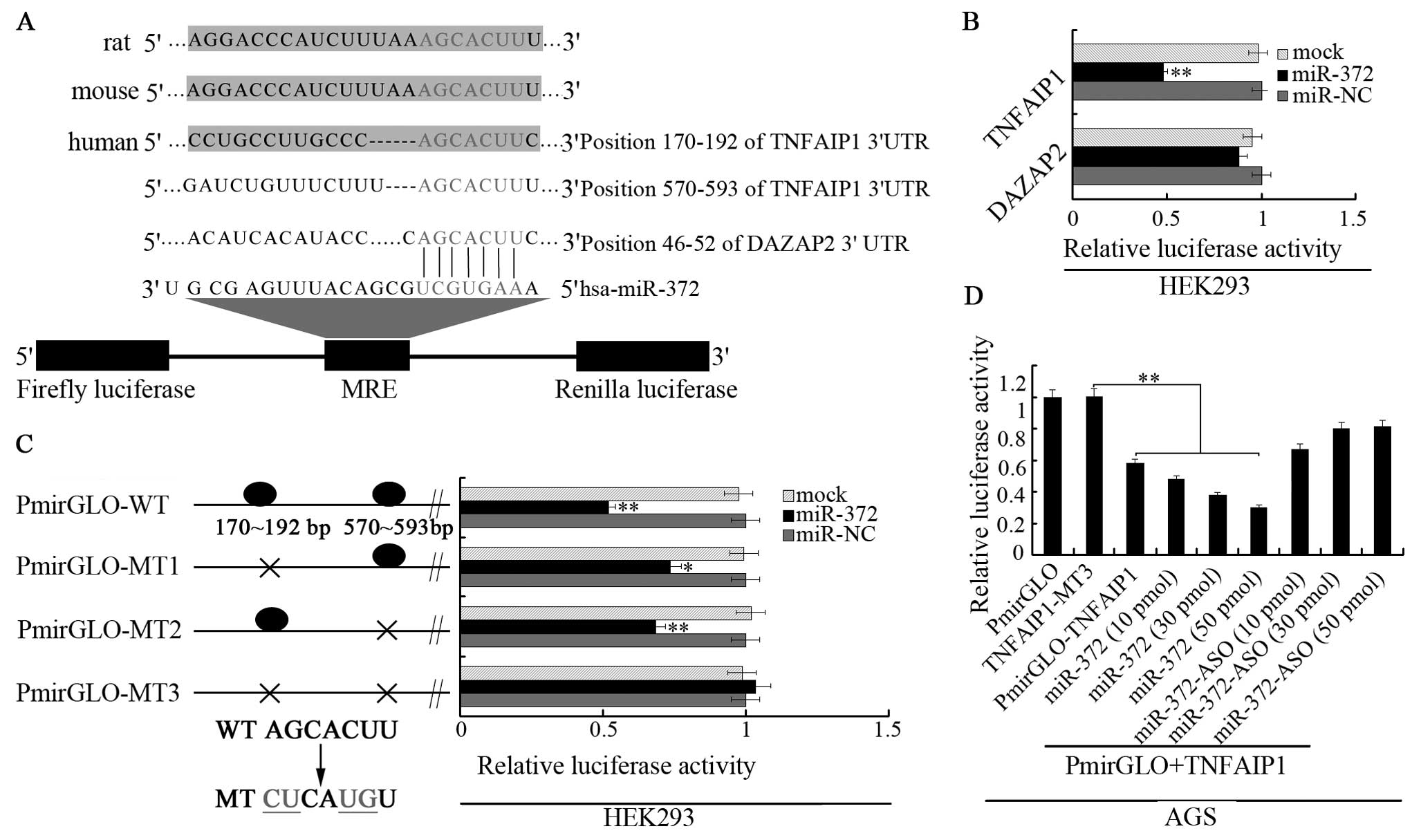
To understand the mechanisms by which miR-372
increases AGS cell growth and inhibits apoptosis, we used several
computational methods to help identify miR-372 targets in humans.
We used three open access programs (TargetScan, miRanda and picTar)
to predict targets of miR-372. We found TNFAIP1 and DAZAP2 have a
relative higher score and have potential to be target genes of
miR-372. We then cloned the TNFAIP1 3′UTR and DAZAP2 3′UTR target
sites into the PmirGLO plasmid (the predicted binding sites for
miR-372 are shown in Fig. 3A). The
luciferase reporter assay which was performed in HEK293 cells
indicated that the activity of the reporter containing the 3′UTR of
the TNFAIP1 gene was decreased following treatment with miR-372
mimics, whereas the reporter containing the 3′UTR of DAZAP2 genes
was not obviously altered (Fig.
3B). We also constructed three mutant vectors of the 3′UTR of
TNFAIP1 as shown in Fig. 3C. Two
single site mutation vectors, a whole site mutation vector and the
luciferase reporter assay were performed with the mutated construct
in the HEK293 cells. The results demonstrated that the luciferase
activity was elevated in all mutated vectors and the relative
luciferase activity of two single site mutation vectors increased
less compared with the whole site mutation vector. Further
luciferase assay was conducted in AGS cells (Fig. 3D). miR-372 showed a significant
reduction of luciferase activity, with either TNFAIP1 luciferase
vector or co-transfected PmirGLO-MT3 with miR-372 mimics (three
gradients). The reduction of luciferase activity by miR-372 mimics
could be rescued by co-transfection of miR-372 inhibitor (three
gradients) with PmirGLO-MT3. Thus, these results demonstrate that
miR-372 can direct bind the two presumptive sites of TNFAIP1
3′UTR.
miR-372 regulates TNFAIP1 expression at
both the mRNA and protein levels
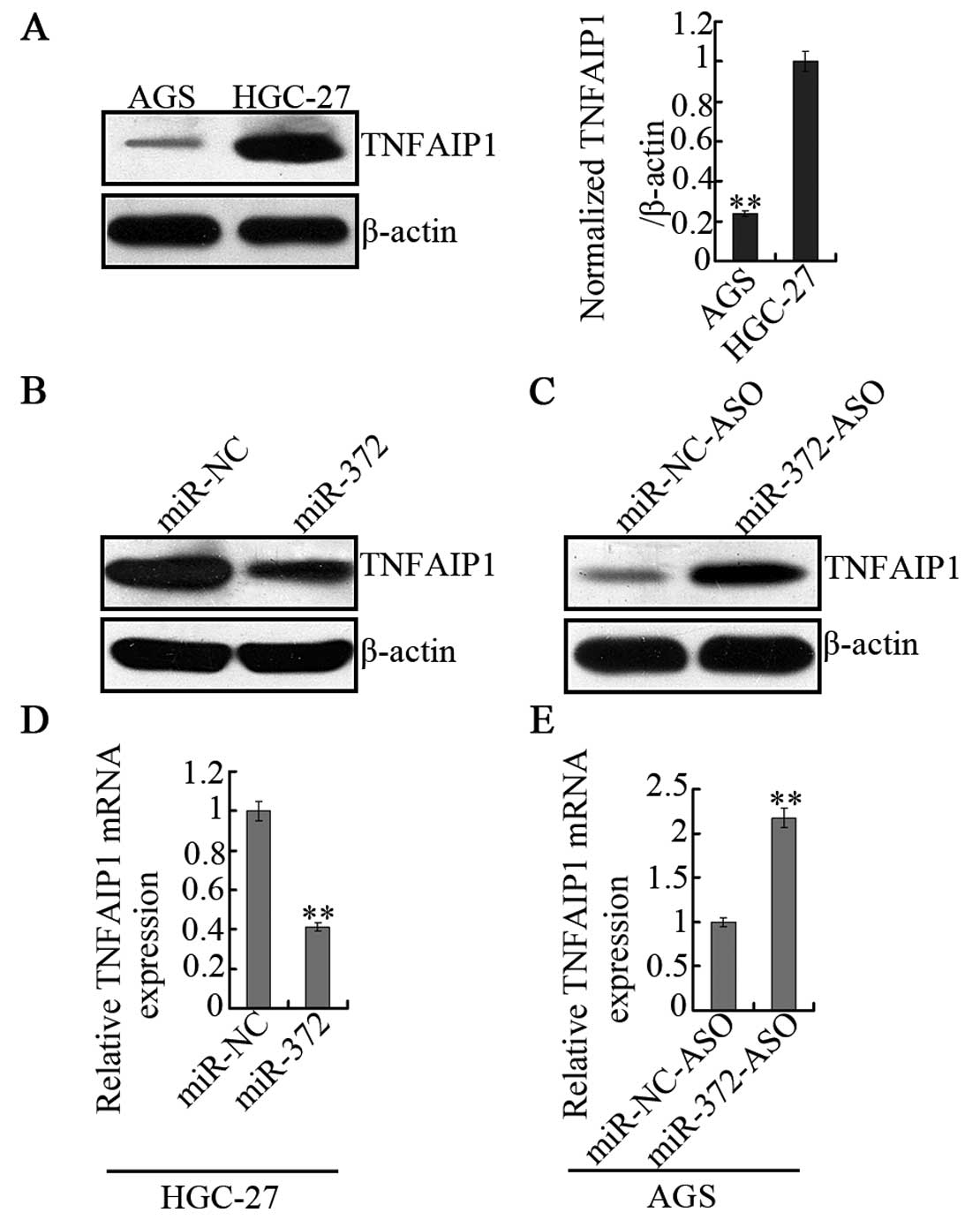
To determine whether miR-372 can regulate TNFAIP1
expression in vivo, the background protein levels of AGS and
HGC-27 cells were also detected (Fig.
4A). This result was consistent with the hypothesis that AGS
cells are characterized by high miR-372 expression and lower
TNFAIP1 protein levels, while in HGC-27 cells, miR-372 is
undetected and TNFAIP1 protein levels are higher. Furthermore,
overexpression of miR-372 mimics or negative control (miR-NC) was
performed in HGC-27 cells. The result was that overexpression of
miR-372 strongly decreased expression of TNFAIP1 at both the
protein (Fig. 4B) and the mRNA
level (Fig. 4D). However, when
transfected with miR-372 inhibitors (miR-372-ASO) in AGS cells,
which is 2′Ome chemically modified, single stranded nucleic acid
designed to specifically bind to and inhibit endogenous miR-372
molecules, the observations indicated that TNFAIP1 is negatively
regulated by miR-372 both at the protein (Fig. 4C) and mRNA levels (Fig. 4E). These results demonstrate that
miR-372 negatively regulates TNFAIP1 expression.
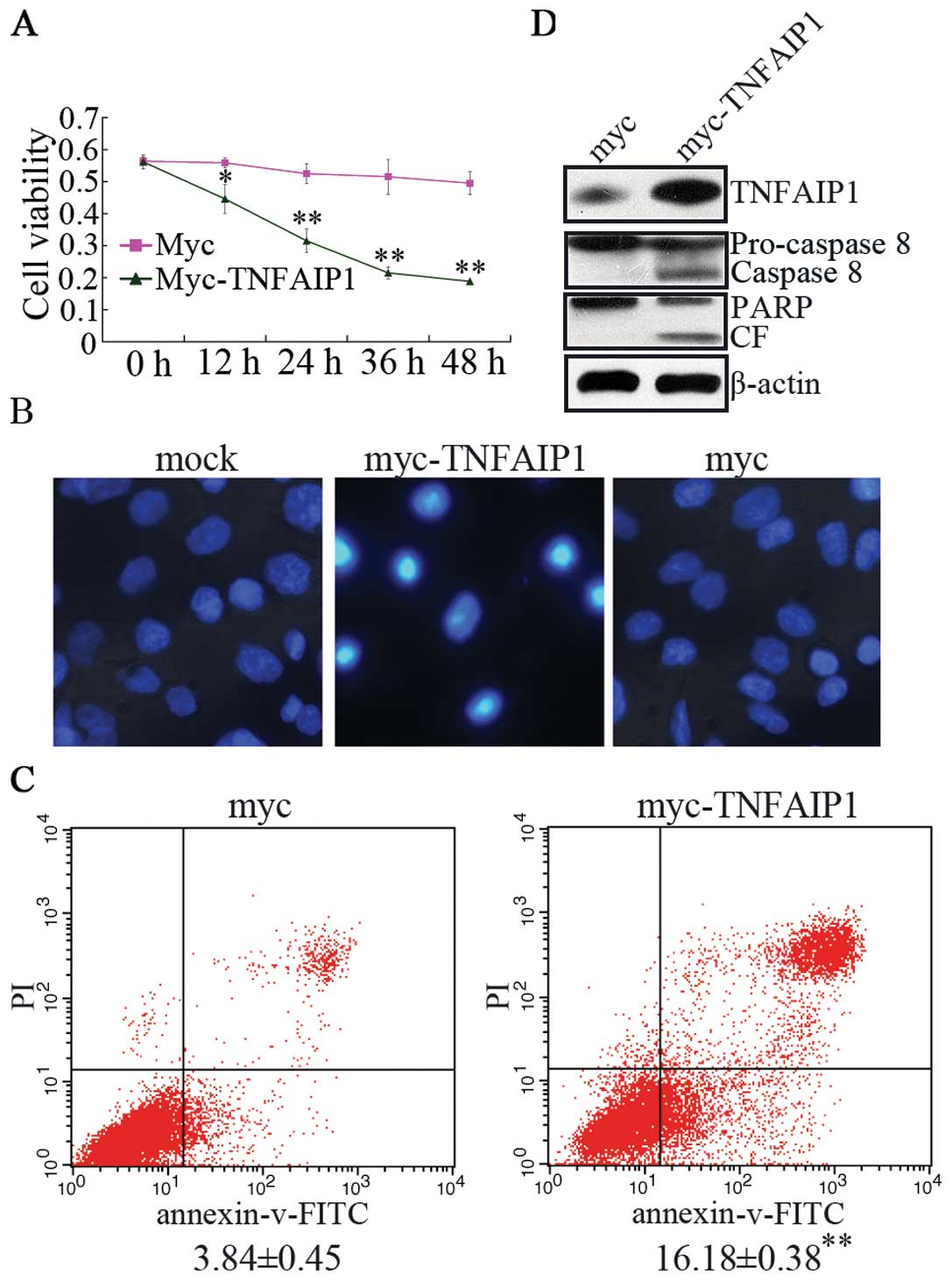
The effect of TNFAIP1 overexpression is
consistent with miR-372-ASO treated in AGS cells
To further investigate the effect of TNFAIP1 in AGS
cells, we constructed TNFAIP1 expression vector pCMV-TNFAIP1, which
did not contain 5′ and 3′UTR of TNFAIP1. Then, pCMV-TNFAIP1 and
pCMV vectors were transfected with AGS cells to study cellular
mechanisms such as cell activity and apoptosis. This experiment
indicated that independently overexpression of TNFAIP1 suppressed
cell viability (Fig. 5A) and
increased the apoptosis of AGS cells (Fig. 5B and C). We also examined the
cleavage of caspase-8 and the subsequent proteolytic cleavage of
PARP in AGS cells transfected with pCMV-TNFAIP1 (Fig. 5D). Western blotting assay revealed
that cleavage of apical pro-caspase-8 and PARP into the
characteristic activate fragments. In brief, these results
demonstrated that TNFAIP1 produces the same effect as the miR-372
inhibitor treated in AGS cells and indicates that miR-372 regulates
cell growth, apoptosis of AGS cells by targeting TNFAIP1.
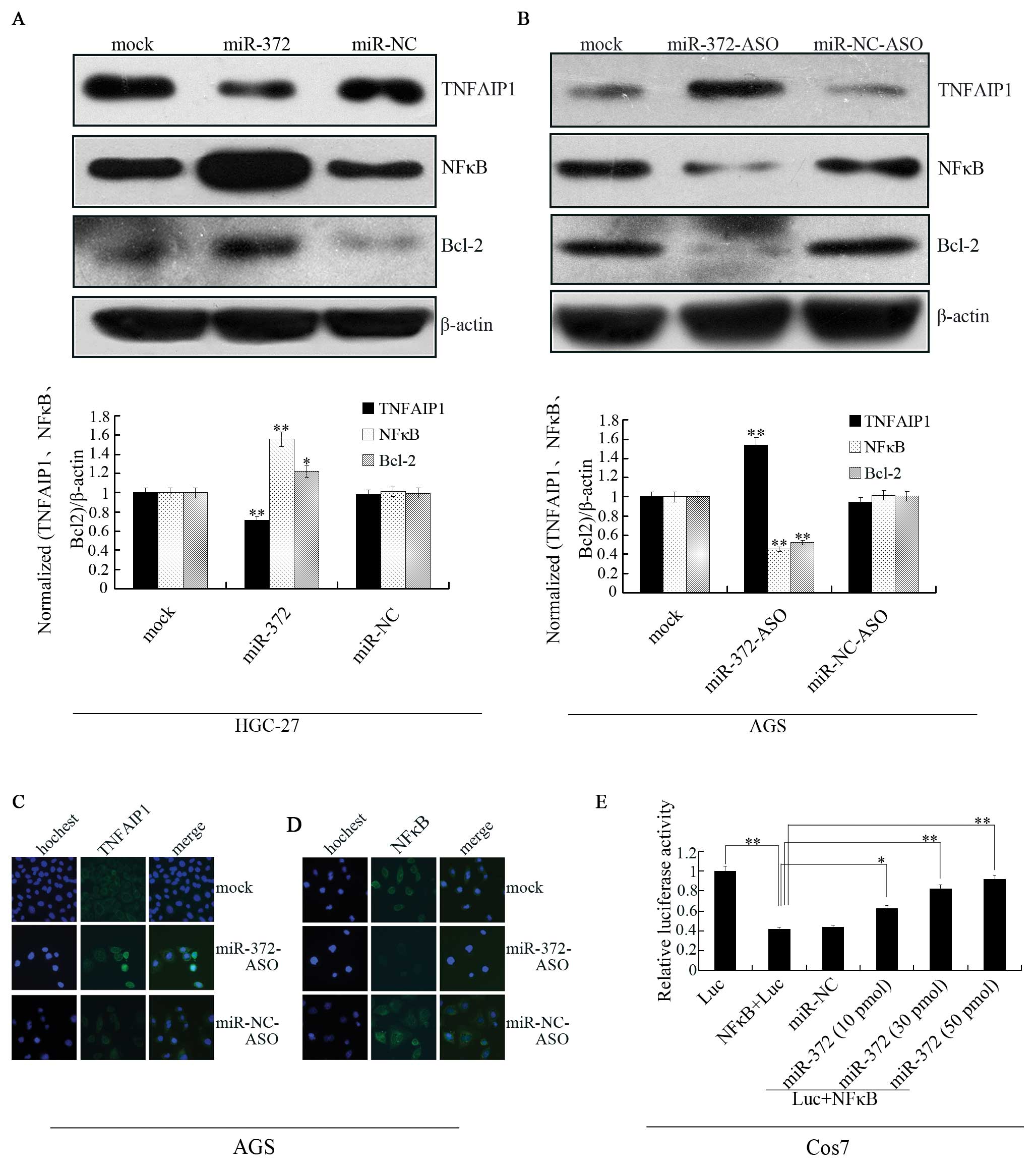
miR-372 regulates the expression of NFκB
(p65) through TNFAIP1
TNFAIP1 can be induced by TNFα and IL6 at the same
time. The TLR family and TNFα receptor family possess numerous
biological functions such as cell apoptosis, cell proliferation and
inflammatory response and, mostly, these physiological functions
are performed by activating the transcriptional activity of NFκB
and then regulating downstream gene expression and TLR and TNFRs
signal pathways are precisely regulated by other negative
regulators in return. However, TNFAIP1 is a protein that can be
induced by the tumor necrosis factor α (TNFα). Several microRNAs,
such as Let-7, miR-218 and miR-199a, are related to NFκB signaling
(21). Thus, we hypothesized that
miR-372 is also involved in NFκB signaling by TNFAIP1.
To further investigate the regulation network of
miR-372, we detected TNFAIP1 and NFκB (p65) in miR-372 mimic
transfected cells (HGC-27) (Fig.
6A) and miR-372-ASO transfected cells (AGS) (Fig. 6B). In this experiment, TNFAIP1 was
inhibited by miR-372 and NFκB presented striking upregulation in
the HGC-27 cells. On the contrary, along with TNFAIP1 expression
improved by suppression of miR-372 in the AGS cells, NFκB showed
downregulation and BCL-2 protein levels, which were reported to be
regulated by TNFAIP1 in HeLa cells, also decreased (22). Immunofluorescence staining also
revealed an increase in TNFAIP1 protein expression (Fig. 6C) and a reduction of NFκB (p65)
levels in AGS cells treated with miR-372-ASO (Fig. 6D). Furthermore, we detected NFκB
luciferase activity alteration in Cos7 cells, in which TNFAIP1
expression was markedly high. We transiently transfected with
luciferase empty vector (Luc) or Luc+NFκB and transiently
co-transfected Luc+NFκB with miR-372 to Cos7 cells (Fig. 6E). The results demonstrated the
luciferase activity of NFκB decreased compared to the luciferase
vector (Luc) and can be reversed by the treatment of miR-372.
Therefore, these findings reveal that miR-372 can regulate NFκB
signal by affecting TNFAIP1.
Discussion
At present, the role of miRNAs in a variety of human
malignant neoplasms has gained significant attention, with
intensive investigations into the potential of inhibiting miRNAs as
a novel approach to treatment. However, how these miRNA molecules
contribute to the pathogenesis of cancer remains largely unknown. A
detailed study on the biological functions of the miRNA-mRNA
interaction network is therefore urgently required.
The miR-372 gene is located on genomic chromosome at
19q13.42. The functions of miR-372 have yet to be fully explored
and its relevance to carcinogenesis remains unclear. Previous
studies showed that miR-372 enhances cell proliferation, stimulates
cell cycle progression, or decreases apoptosis in testicular tumor
germ cells and in gastric cancer cells. By contrast, miR-372
suppresses cell growth in human cervical cancer (23). Considering that the gene expression
profile is tumor-specific and that the influence of miRNA
alterations might strongly depend on tumor context, these results
are expected.
We analyzed the proliferation rate of gastric cancer
cells transfected with miR-372 or anti-miR-372 to identify its
oncogenic property. Our results showed that the proliferation
capacity was repressed after downregulation of miR-372 in gastric
cancer AGS cell proliferation, whereas the over-expression of
miR-372 promoted gastric cancer HGC-27 cell survival rate,
indicating that miR-372 might be a potential growth factor for
gastric cancer cells. Furthermore, we found that miR-372 decreases
apoptosis in gastric cancer cells.
Since the impact of cancer miRNAs on cancer biology
depends on the functions of the downstream targets they suppress,
we need to discover the targets of each miRNA. It is well known
that miRNAs can regulate the gene expression of multiple targets to
perform pleiotropic functions (24,25).
According to PicTar, miR-372 has 571 predicted targets in humans.
We found, through several databases, numerous important candidate
targets, among which TNFAIP1 is a direct target of miR-372 in the
control of gastric cancer cell proliferation (26,27).
TNFAIP1 is a protein which can be induced by TNFα and IL-6. It also
relates to DNA synthesis, DNA repair, cell apoptosis and human
diseases. In the present study, using a luciferase reporter system
and expression assay, we confirmed that TNFAIP1 is negatively
regulated by miR-372. When we examined the relationship between
TNFAIP1 mRNA and miR-372 in gastric cancer cell lines, an inverse
correlation between miR-372 and the TNFAIP1 protein expression was
noted by western blot analysis and immunofluorescence (Fig. 4C). TNFAIP1 was shown to be
expressed at a high level in normal cells and was downregulated in
several cancer-derived cell lines. miR-372 overexpression conferred
cell resistance to apoptosis, and the increased resistance
correlated with the level of TNFAIP1. Conversely, reducing miR-372
expression by miRNA inhibitors greatly sensitized cells to
apoptosis. miR-372 may play a key role in tumorigenesis by
conferring cells resistance to apoptosis.
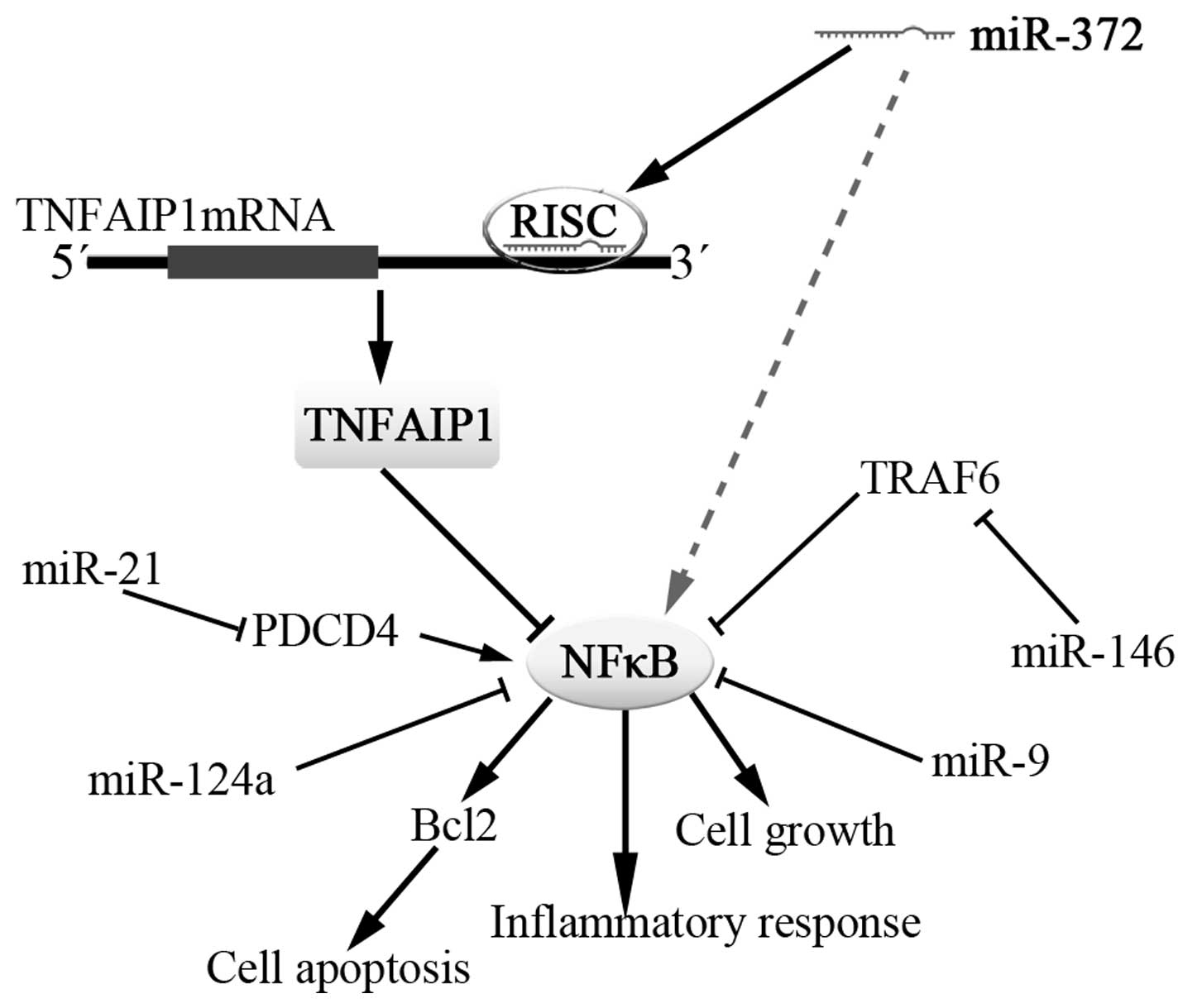
In recent years, several miRNAs have been found to
be involved in NFκB signaling by targeting NFκB regulators or
effectors and participating in positive or negative feedback loops.
In this study, we found that miR-372 is also associated with NFκB.
When miR-372 overexpression was performed in HGC-27 cells, NFκB
expression was increased with the inhibition of TNFAIP1. While
miR-372 was inhibited in AGS cells, NFκB presented downregulation
accompanied by increased TNFAIP1. Furthermore, NFκB transcription
luciferase assay in Cos7 cells indicated the increase of luciferase
activity related to the expression of TNFAIP1. Therefore, TNFAIP1
regulated by miR-372 may finally have an effect on NFκB signaling
and the miRNAs which converged with the NFκB network may be
enlarged. The oncogenic properties of miR-372 may partly derive
from increased NFκB (Fig. 7).
Furthermore, although we have shown that miR-372 can target TNFAIP1
and affect NFκB signaling, our laboratory recently demonstrated
that TNFAIP1 suppressed the transcriptional activities of NFκB.
However, the detailed action between TNFAIP1 and NFκB may need
further study as well as the reason why miR-372 was overexpressed
in AGS cells. Our findings demonstrate that miR-372 enhances cell
proliferation, or decreases apoptosis in gastric cancer cells via
directly regulating the expression of TNFAIP1, suggesting that
miR-372 can serve as a potential target in the treatment of gastric
cancer. We have shown that miR-372 affects NFκB and that these
effects may be achieved by regulating the expression of TNFAIP1.
This may, in part, be the reason for miR-372 possessing oncogenic
properties.
Acknowledgements
This study was supported by the
National Natural Science Foun dation of China (grant nos. 81071696,
31071150 and 30900827), the Hunan Provincial Innovation Foundation
for Postgraduate (CX2012B227) and the Project of Chang Sha Science
and Technology Plan (K1109006-31).
References
|
1.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Berezikov E, Thuemmler F, Van Laake LW, et
al: Diversity of microRNAs in human and chimpanzee brain. Nat
Genet. 38:1375–1377. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Wienholds E and Plasterk RHA: MicroRNA
function in animal development. FEBS Lett. 579:5911–5922. 2005.
View Article : Google Scholar
|
|
4.
|
Carleton M, Cleary MA and Linsley PS:
MicroRNAs and cell cycle regulation. Cell Cycle. 6:2127–2132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Inomata M, Tagawa H, Guo YM, Kameoka Y,
Takahashi N and Sawada K: MicroRNA-17-92 down-regulates expression
of distinct targets in different B-cell lymphoma subtypes. Blood.
113:3962009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Silber J, Lim D, Petritsch C, et al:
miR-124 and miR-137 inhibit proliferation of glioblastoma
multiforme cells and induce differentiation of brain tumor stem
cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Gao C, Zhang Z, Liu W, Xiao S, Gu W and Lu
H: Reduced microRNA-218 expression is associated with high nuclear
factor kappa B activation in gastric cancer. Cancer. 116:41–49.
2010.PubMed/NCBI
|
|
9.
|
Motoyama K, Inoue H, Nakamura Y, Uetake H,
Sugihara K and Mori M: Clinical significance of high mobility group
A2 in human gastric cancer and its relationship to let-7 microRNA
family. Clin Cancer Res. 14:2334–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Xiao B, Guo J, Miao Y, et al: Detection of
miR-106a in gastric carcinoma and its clinical significance. Clin
Chim Acta. 400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Cho WJ, Shin JM, Kim JS, et al: miR-372
regulates cell cycle and apoptosis of ags human gastric cancer cell
line through direct regulation of LATS2. Mol Cells. 28:521–527.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Belair C, Baud J, Chabas S, et al:
Helicobacter pylori interferes with an embryonic stem cell
micro RNA cluster to block cell cycle progression. Silence. 2:1–16.
2011. View Article : Google Scholar
|
|
13.
|
Wolf F, Marks R, Sarma V, et al:
Characterization of a novel tumor necrosis factor-alpha-induced
endothelial primary response gene. J Biol Chem. 267:1317–1326.
1992.PubMed/NCBI
|
|
14.
|
Link CD, Taft A, Kapulkin V, et al: Gene
expression analysis in a transgenic Caenorhabditis elegans
Alzheimer’s disease model. Neurobiol Aging. 24:397–413. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Yang L, Liu N, Hu X, et al: CK2
phosphorylates TNFAIP1 to affect its subcellular localization and
interaction with PCNA. Mol Biol Rep. 37:2967–2973. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Yang LP, Zhou AD, Li H, et al: Expression
profile in the cell lines of human TNFAIP1 gene. Yi Chuan.
28:918–922. 2006.PubMed/NCBI
|
|
17.
|
Kim DM, Chung KS, Choi SJ, et al: RhoB
induces apoptosis via direct interaction with TNFAIP1 in HeLa
cells. Int J Cancer. 125:2520–2527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hu X, Yan F, Wang F, et al: TNFAIP1
interacts with KCTD10 to promote the degradation of KCTD10 proteins
and inhibit the transcriptional activities of NF-kappaB and AP-1.
Mol Biol Rep. 39:9911–9919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ma X, Becker Buscaglia LE, Barker JR and
Li Y: MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 3:159–166.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Hu W, Xie J, Zhao J, Xu Y, Yang S and Ni
W: Involvement of Bcl-2 family in apoptosis and signal pathways
induced by cigarette smoke extract in the human airway smooth
muscle cells. DNA Cell Biol. 28:13–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Tian RQ, Wang XH, Hou LJ, et al:
MicroRNA-372 is down-regulated and targets cyclin-dependent kinase
2 (CDK2) and cyclin A1 in human cervical cancer, which may
contribute to tumorigenesis. J Biol Chem. 286:25556–25563. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Alvarez JP, Pekker I, Goldshmidt A, Blum
E, Amsellem Z and Eshed Y: Endogenous and synthetic microRNAs
stimulate simultaneous, efficient, and localized regulation of
multiple targets in diverse species. Plant Cell. 18:1134–1151.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Valastyan S, Reinhardt F, Benaich N, et
al: A pleiotropically acting microRNA, miR-31, inhibits breast
cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Gillis AJ, Stoop HJ, Hersmus R, et al:
High-throughput microRNAome analysis in human germ cell tumours. J
Pathol. 213:319–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Subramanyam D, Lamouille S, Judson RL, et
al: Multiple targets of miR-302 and miR-372 promote reprogramming
of human fibroblasts to induced pluripotent stem cells. Nat
Biotechnol. 29:443–448. 2011. View
Article : Google Scholar : PubMed/NCBI
|