Introduction
Loss of heterozygosity of chromosome 17 is a
frequent occurrence in epithelial ovarian cancer (EOC) and
particularly in high-grade serous carcinomas (HGSC), which is one
of the most common histotypes of EOCs (1–6).
This has been largely attributed to the inactivation of the tumour
suppressor gene TP53, which is located at 17p13.1, as it is
the most frequently mutated gene in HGSCs, however, other genes
involved in tumour suppressor pathways have been proposed (1–4,7–14).
For example, our group recently identified 158 underexpressed
chromosome 17 genes in a transcriptome analysis of HGSCs, which
included FKBP10, a gene that maps to 17q21.1. Interestingly,
FKBP10 was among the genes upregulated in a genetically
modified EOC cell line rendered non-tumourigenic as a consequence
of an unique gene complementation assay involving chromosome
transfer, and thus may be one of a number of genes
transcriptionally reprogrammed as a consequence of tumour
suppression (15,16).
FKBP10 encodes FKBP65, a 65 kDa FK506 binding
protein that is a member of the FKBP-type peptidylprolyl cis/trans
isomerase family (17,18). This protein localizes to the
endoplasmic reticulum and acts as a molecular chaperone and binding
partner of type I collagen (19–21).
The interaction with type I collagen is interesting as
COL1A1, which is a gene that maps to 17q21.33, was also
found significantly underexpressed in our chromosome 17
transcriptome analyses of HGSCs (14). FKBP65 is expressed in developing
tissues and re-expressed in adult tissues following injury
(19,20). In the mouse, the only tissues that
continue to express FKBP65 are reproductive tissues (ovary, uterus
and mammary glands) during phases of growth and remodelling (E.C.
Davis, unpublished data). Although somatic mutations inactivating
FKBP10 have not been identified, such as in recent reports
of genome-wide exomic sequencing analyses of HGSCs by The Cancer
Genome Atlas Research Network (5)
and other cancer types (22),
germline FKBP10 mutations have been described in association
with autosomal-recessive osteogenesis imperfecta and Bruck
syndrome, which are connective tissue disorders characterized by
defects in type I collagen (23,24).
Though epithelial malignancies in patients with osteogenesis
imperfecta are rare, there is at least one case report of a
32-year-old who developed a low-grade serous ovarian carcinoma with
stage IIIb disease (25).
The localization of FKBP10 to chromosome 17
and its interesting expression profile in the murine adult normal
ovary, HGSCs and our genetically modified EOC cell line, prompted
our further investigation of this gene in ovarian cancer samples.
In this study, we investigated the expression profile of
FKBP10 in HGSCs, and its expression in a set of
well-characterized EOC cell lines that differ in their growth
characteristics and tumourigenic potential. We also investigated
protein expression by immunohistochemistry analysis of a tissue
microarray and related the expression profile to patient outcome.
COLIA1 gene and protein expression was also investigated
given its purported interaction with FKBP65.
Materials and methods
Tissue specimens, cell lines, and
clinical information
The HGSCs, primary cultures of normal ovarian
surface epithelial cells (NOSE), and EOC cell lines (OV90, TOV112D,
TOV81D, TOV21G, TOV1946, OV1946 and TOV2223) examined for gene
expression have been described previously (1,14,26).
Briefly, the HGSC samples were from chemotherapy naïve patients and
the cell lines were derived from long-term passages of malignant
ovarian ascites from an undifferentiated adeno-carcinoma (OV-90),
high-grade endometrioid adenocarcinoma (TOV112D), serous carcinomas
(TOV81D, TOV1946, OV1946 and TOV2223) and a clear cell carcinoma
(TOV21G), where TOV1946 and OV1946 were derived from malignant
ovarian ascites (OV1946) or tumour (TOV1946) from the same
patient.
The HGSC (n=196) cases represented in the tissue
array were derived from archival blocks of paraffin-embedded
tissues samples as previously described (27). The tumour grade and disease stage
(Table I) were designated
according to the International Federation of Gynaecology and
Obstetrics. Disease-free interval, defined as time to doubling of
the upper normal limit of the serum cancer antigen marker CA-125 or
the detection of a new lesion by ultrasound or CT-scan imaging, and
overall survival, defined according to the Response Evaluation
Criteria in Solid Tumors (28)
were extracted from the Système d’Archivage des Données en
Oncologie (Table I). Normal ovary
and fallopian tube tissues used in immunohistochemistry analyses
were retrieved from archival paraffin-embedded samples. Normal
ovarian tissues were obtained from ovarectomy (age 40) and
hysterectomy (age 45) cases due to a benign uterine tumour. The
fallopian tube tissues were adjacent normal tissues collected from
women diagnosed with serous ovarian cancer. All samples and related
clinical information were obtained with informed written consent at
the Centre Hospitalier de l’ Universite de Montreal (CHUM) -
Hôpital Notre Dame.
 | Table IDescription of HGSC tissue array. |
Table I
Description of HGSC tissue array.
| Patient
characteristic | All cases | G2 | G3 |
|---|
| No. of cases | 196 | 30 | 166 |
| Mean age of
diagnosis, years (range) | 62 (34–89) | 60 (42–82) | 62 (34–89) |
| Stage I | 15 | 2 | 13 |
| Stage II | 19 | 2 | 17 |
| Stage III | 139 | 23 | 116 |
| Stage IV | 23 | 3 | 20 |
| Mean disease free
interval, months (range) | 22 (0–134) | 15 (0–57) | 23 (0–134) |
| No. of censured
patients | 52 | 6 | 46 |
| No. of non-censured
patients | 144 | 24 | 120 |
| Mean overall
survival, months (range) | 35 (0–134) | 28 (0–102) | 36 (0–134) |
| No. of censured
patients | 110 | 19 | 91 |
| No. of non-censured
patients | 86 | 11 | 75 |
Gene expression analyses
Gene expression was assessed by semi-quantitative
RT-PCR analysis using cDNA synthesized from total RNA prepared as
previously described (3,13,15,29).
Primers were designed using Primer3 software (30) based on genomic structures of
FKBP10 and COL1A1 available from the March 2006 human
reference sequence (NCBI Build 36.1/hg18) assembly (31) and alignment of reference sequences
of each gene, NM_021939 and NM_000088.3, respectively. The
FKBP10 forward primer is 5′-GTGGAACAAGGAAGA CACC-3′, and the
reverse primer is 5′-CTTCCTTCTCTCTCC AGGAC-3′, yielding a 238 base
pair product. The COL1A1 forward primer is
5′-GTGCTCCTGTATTGCTG-3′, and the reverse primer is
5′-CTCGCTTTCCTTCCTCTC-3′, yielding a 207 base pair product. The
RT-PCR-based assays were performed essentially as previously
described (32). RT-PCR of
18S RNA was performed to assess RNA quality. Primer
sequences for 18S were reported previously (3).
Immunohistochemistry analysis of tissue
arrays
FKBP65 and COL1A1 protein expression was assessed by
immunohisto-chemistry analysis using a tissue array containing 0.6
mm cores derived from paraffin-embedded tissue blocks representing
196 HGSC cases selected based on a review of hematoxylin and
eosin-stained slides prepared as described previously (27). The array also contained 11 normal
fallopian tube samples. Immunohistochemistry analysis was performed
on the tissue array and sections prepared from two normal ovary
paraffin-embedded tissue blocks. Five-micron sections were mounted
onto frosted plus slides, deparaffinized in Citrisolv (Fisher
Scientific) for 90 min and then rehydrated in an ethanol gradient.
Before primary antibody incubation, antigen retrieval was performed
using 0.05% Tween-20 in 10 mM sodium citrate (pH 6.0) at 90°C for
20 min. The slides were washed with 0.1% Triton X-100 in
Tris-buffered saline (TBS) [0.5 M Tris, 1.5 M NaCl, (pH 7.4)]. The
slides were then washed and blocked with TBS containing 0.1% bovine
serum albumin (TBS/BSA) for 3×10 min and then incubated with Ultra
V Block (LabVision Corp., Fremont, CA, USA) for 7 min. After
washing again in TBS/BSA, the slides were incubated overnight at
4°C with either a polyclonal FKBP65 antibody, raised against a
synthetic peptide to the C-terminus of the mouse FKBP65 (33), or a polyclonal type I collagen
antibody (Calbiochem), diluted at 1:300 and 1:1000, respectively.
Slides were incubated in Value Primary Antibody Enhancer (LabVision
Corp.) for 20 min before incubation in AP Value Polymer (LabVision
Corp.) for 30 min at room temperature. Staining was visualized
using FastRed (LabVision Corp.) and slides were counterstained with
hematoxylin for 30 sec. The tissue arrays were scanned with an
Aperio ScanScope XT Digital Slide Scanner and images were viewed at
high resolution using Aperio ImageScope Software version 11.02. Two
observers examined the images independently and scored them based
on staining intensity ranked as absent, low, moderate or high, for
both the epithelial and stromal tissue components of each core. The
inter-observer correlation coefficients were 0.792 for FKBP65 and
0.747 for COL1A1. The inter-observer correlation coefficient was
calculated using SPSS software version 16.0 (SPSS Inc., Chicago,
IL, USA), where the minimum threshold was 0.7.
Statistical analyses
Spearman analysis was used to evaluate the
correlation of FKBP65 and COL1A1 staining intensities. The
association between tumour grade and staining intensity was
evaluated using an independent sample t-test. The association
between disease stage and staining intensity was evaluated using a
one-way ANOVA. The relationship between staining intensity and
disease-free interval or overall survival was evaluated using the
log-rank test and visualized by Kaplan-Meier survival curve
analysis. All statistical analyses were performed with SPSS
software version 16.0 (SPSS Inc.), p-values <0.05 were
considered significant.
Results
Gene expression analyses of FKBP10 and
COL1A1
A previous gene expression microarray analysis of
chromo-some 17 genes identified FKBP10 and COL1A1
among the 158 genes underexpressed in HGSCs as compared with NOSEs
(14). To verify these
observations, we investigated the expression of these genes by
performing RT-PCR analyses on the samples used in the microarray
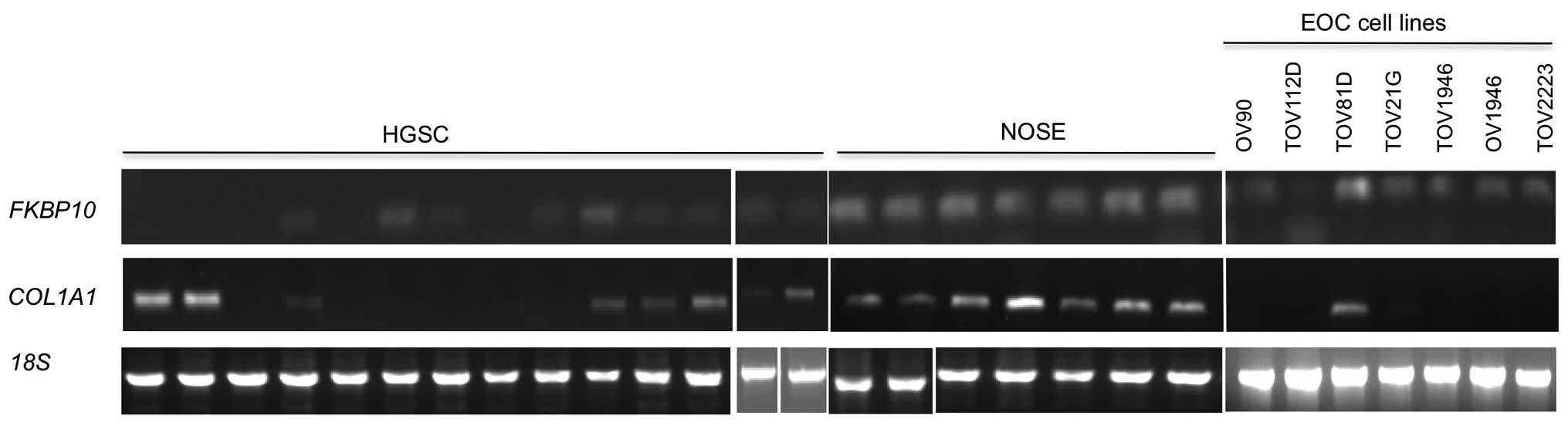
analyses. As shown in Fig. 1,
FKBP10 and COL1A1 expression was clearly detectable
in all NOSEs. In contrast, FKBP10 expression in the HGSCs
was undetectable or expressed at levels lower than that observed in
the NOSEs. COL1A1 expression was more variable, ranging from
clearly detectable levels comparable to those observed in NOSEs in
some samples, to undetectable or lower levels of expression
relative to the NOSEs (Fig. 1).
Gene expression of FKBP10 and COL1A1 was also
investigated in cell lines established as long-term passages from
chemotherapy naïve EOC samples (1,26).
FKBP10 expression was detectable in all EOC cell lines with
the highest level of expression observed in TOV81D (Fig. 1). In contrast, COL1A1
expression was detectable in only TOV81D (Fig. 1), which is a non-tumourigenic EOC
cell line.
Immunohistochemical staining of FKBP65
and COL1A1
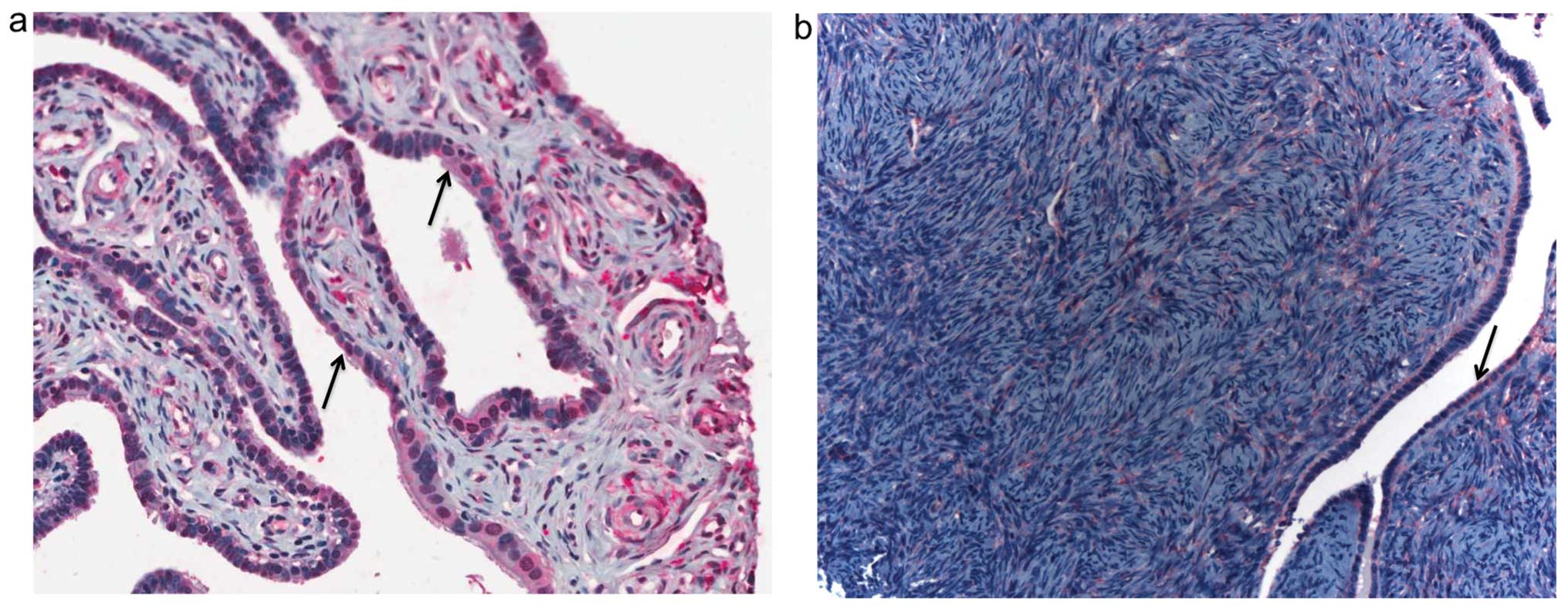
Immunohistochemistry analysis was performed to
characterize FKBP65 protein expression in normal ovary and
fallopian tube as the expression profile in human tissues purported
to be the origins of HGSCs have not previously been described.
Staining was evident in both the epithelial and stromal cells of
the tissues (Fig. 2).
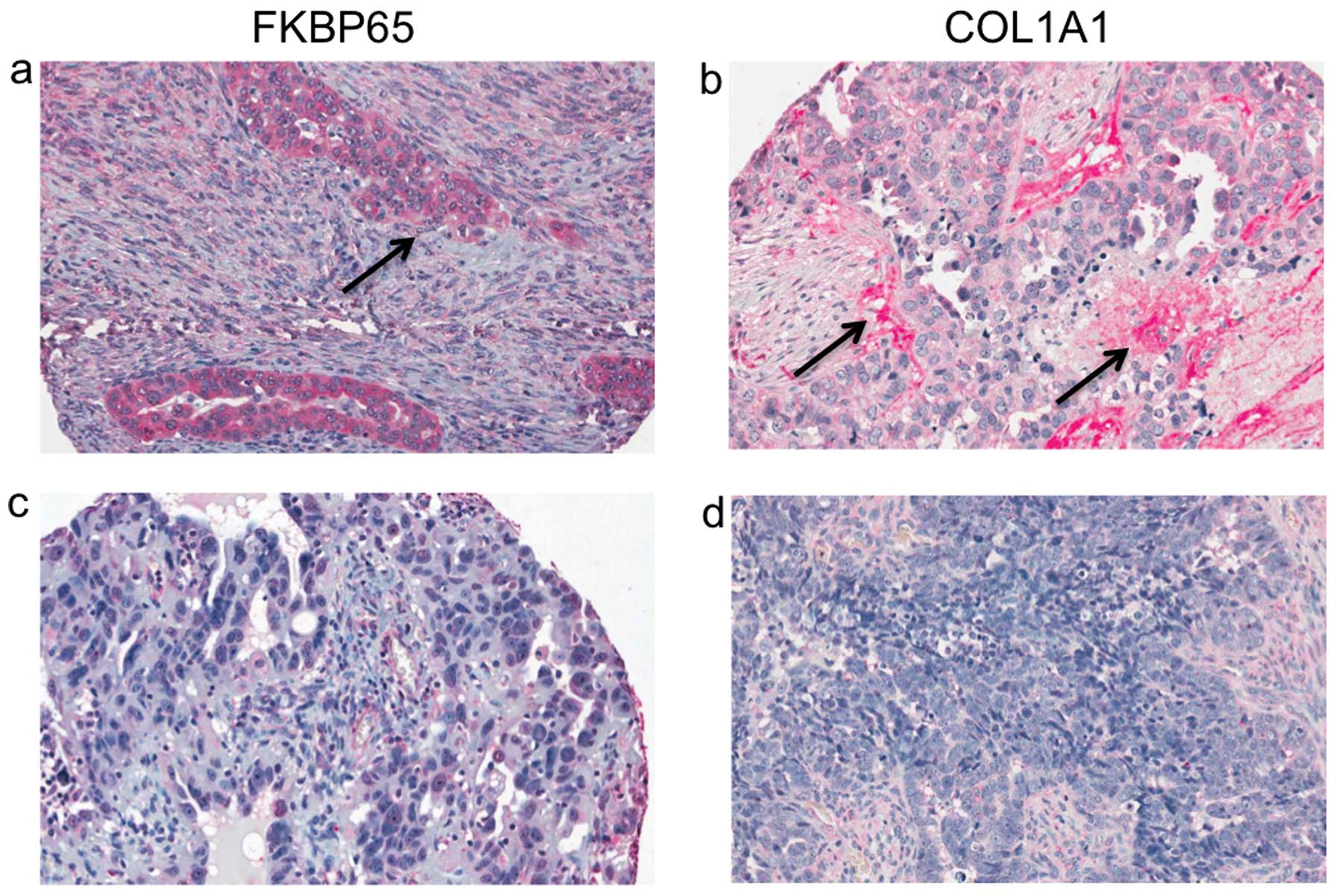
FKBP65 and COL1A1 protein expression in HGSCs was
investigated by immunohistochemistry analysis using a tissue array
containing 196 cores from tumour samples (Table I). Staining was localized to the
cytoplasm for both FKBP65 and COL1A1 and was observed in both the
epithelial and stromal cell components of the tumour samples
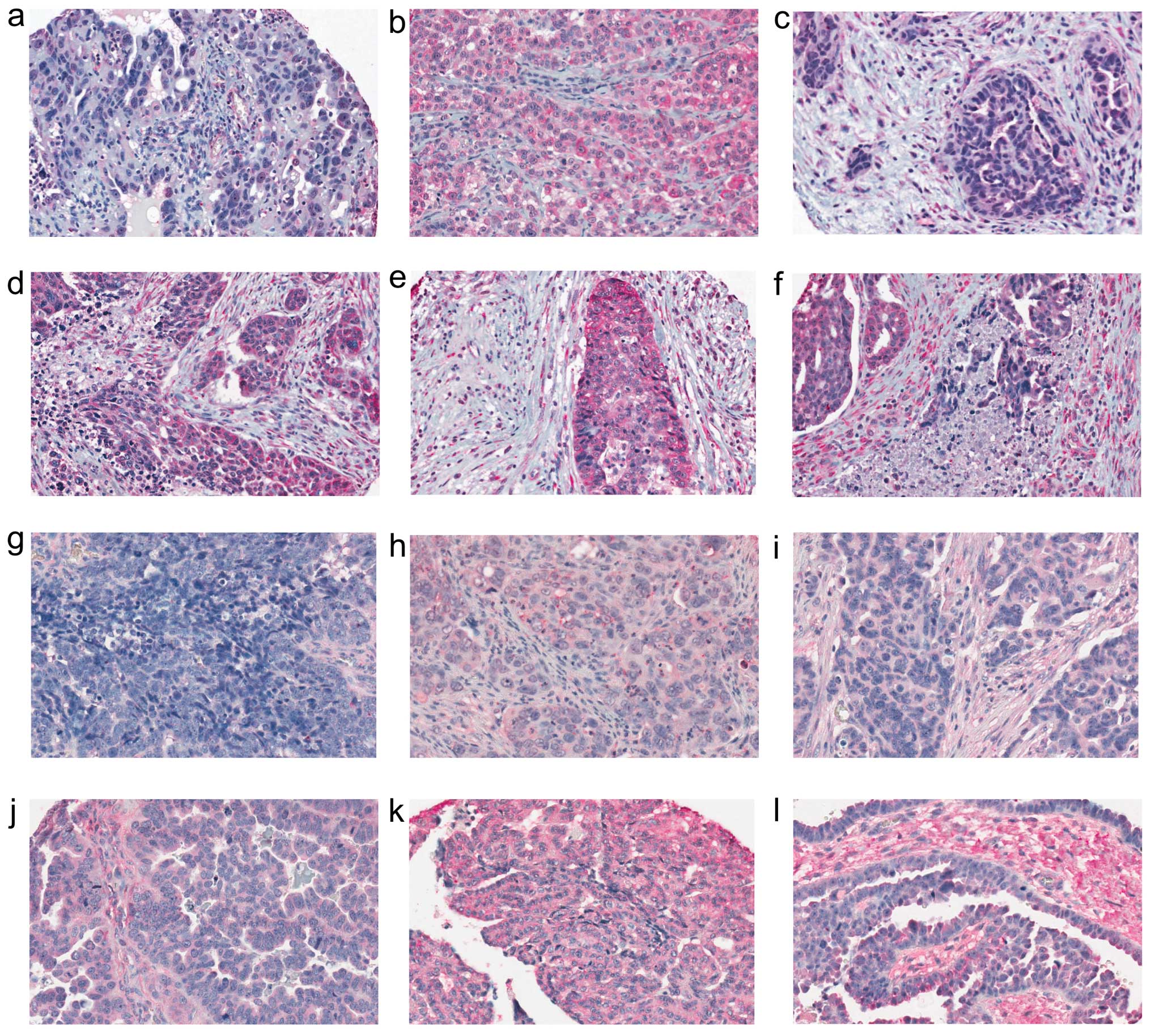
(Fig. 3). To characterize the
expression pattern, intensity of staining was scored as absent,
low, moderate or high for both the epithelial and stromal cell
components where possible, as scoring was not possible for some of
the samples due to the type of cells present or the quality of core
(Fig. 4). The majority of the
epithelial cell components of the tumour samples scored as either
low or moderate for FKBP65 expression, and less than 10% of samples
scored as either absent or high (Table
II). A somewhat similar staining intensity pattern for the
epithelial component of the tumours was also observed with COL1A1,
although there were more cases that scored with low intensity
levels (Table II). In contrast,
FKBP65 expression patterns for the stromal components of the tumour
were almost equally distributed among all staining intensity
categories, with the largest number of samples (32.3%) exhibiting
moderate staining intensity (Table
II). Although the largest number of samples (44.5%) also
exhibited moderate staining of COL1A1 in the stromal component of
the tumour, there were fewer samples that had an absent intensity
score (Table II). There was a
significant correlation between FKBP65 and COL1A1 staining
intensity in the epithelial cell component (p<0.001), but not in
the stromal component (p=0.101) of the tumour samples analysed.
 | Table IIDistribution of staining intensities
for FKBP65 and COL1A1 in HGSC samples. |
Table II
Distribution of staining intensities
for FKBP65 and COL1A1 in HGSC samples.
| Protein | Cell type | Total no. of
samples scored | Staining intensity
score (%) |
|---|
|
|---|
| Absent | Low | Moderate | High |
|---|
| FKBP65 | Epithelial | 193 | 2 (1.0) | 107 (55.4) | 69 (35.8) | 15 (7.8) |
| Stromal | 195 | 46 (23.6) | 40 (20.5) | 63 (32.3) | 46 (23.6) |
| COL1A1 | Epithelial | 191 | 2 (1.0) | 134 (70.2) | 53 (27.7) | 2 (1.0) |
| Stromal | 191 | 9 (4.7) | 51 (26.7) | 85 (44.5) | 46 (24.1) |
Protein expression and disease stage,
disease-free interval or overall survival
The staining intensity patterns of FKBP65 and COL1A1
were characterized with respect to disease stage, though the
majority of samples (83%) were from cases with advanced stage
III/IV disease (Table I). There
were no statistically significant differences (data not shown) in
the distribution of staining intensity patterns for the proteins
assayed for either the epithelial or stromal cell components of the
tumour samples and disease stage (Table III).
 | Table IIIDistribution of staining intensities
for FKBP65 and COL1A1 by stage. |
Table III
Distribution of staining intensities
for FKBP65 and COL1A1 by stage.
| Protein | Cell type | Stage | Total no. of
samples scored | Staining intensity
score (%) |
|---|
|
|---|
| Absent | Low | Moderate | High |
|---|
| FKBP65 | Epithelial | I | 15 | 0 | 9 (60) | 5 (33.3) | 1 (6.6) |
| II | 18 | 0 | 12 (66.6) | 5 (27.7) | 1 (5.5) |
| III | 137 | 2 (1.5) | 75 (54.7) | 52 (38.0) | 8 (5.8) |
| IV | 23 | 0 | 11 (97.8) | 7 (30.4) | 5 (21.7) |
| Stromal | I | 15 | 5 (33.3) | 2 (13.3) | 4 (26.6) | 4 (26.6) |
| II | 19 | 0 | 6 (31.6) | 8 (42.1) | 5 (26.3) |
| III | 138 | 37 (26.8) | 27 (19.6) | 42 (30.4) | 32 (23.2) |
| IV | 23 | 4 (17.4) | 5 (21.7) | 9 (39.1) | 5 (21.7) |
| COL1A1 | Epithelial | I | 15 | 0 | 10 (66.6) | 5 (33.3) | 0 |
| II | 19 | 0 | 15 (78.9) | 4 (21.1) | 0 |
| III | 135 | 1 (0.7) | 98 (72.6) | 36 (26.7) | 0 |
| IV | 22 | 1 (4.5) | 11 (50) | 8 (36.4) | 2 (9.1) |
| Stromal | I | 15 | 1 (6.6) | 2 (13.3) | 6 (40) | 6 (40) |
| II | 18 | 0 | 5(27.8) | 7 (38.9) | 6 (33.3) |
| III | 136 | 6 (4.4) | 37(27.2) | 65 (47.8) | 28(20.6) |
| IV | 22 | 2 (9.1) | 7 (31.8) | 7 (31.8) | 6 (27.3) |
The staining intensities for FKBP65 and COL1A1 were
also evaluated with respect to disease-free interval and overall
survival. No significant relationships were observed with staining
intensities of COL1A1 for the epithelial or stromal components of
the tumour samples and either of these clinical parameters (data
not shown). Significant relationships were also not found with
staining intensities of FKBP65 for the epithelial or stromal
components of the tumour samples and disease-free interval (data
not shown). In contrast, there was a significant association
between prolonged overall survival and the presence of FKBP65
protein in the epithelial component when evaluated for each
staining category (p=0.005) or when cases with no staining are
compared with those with low, moderate and high staining combined
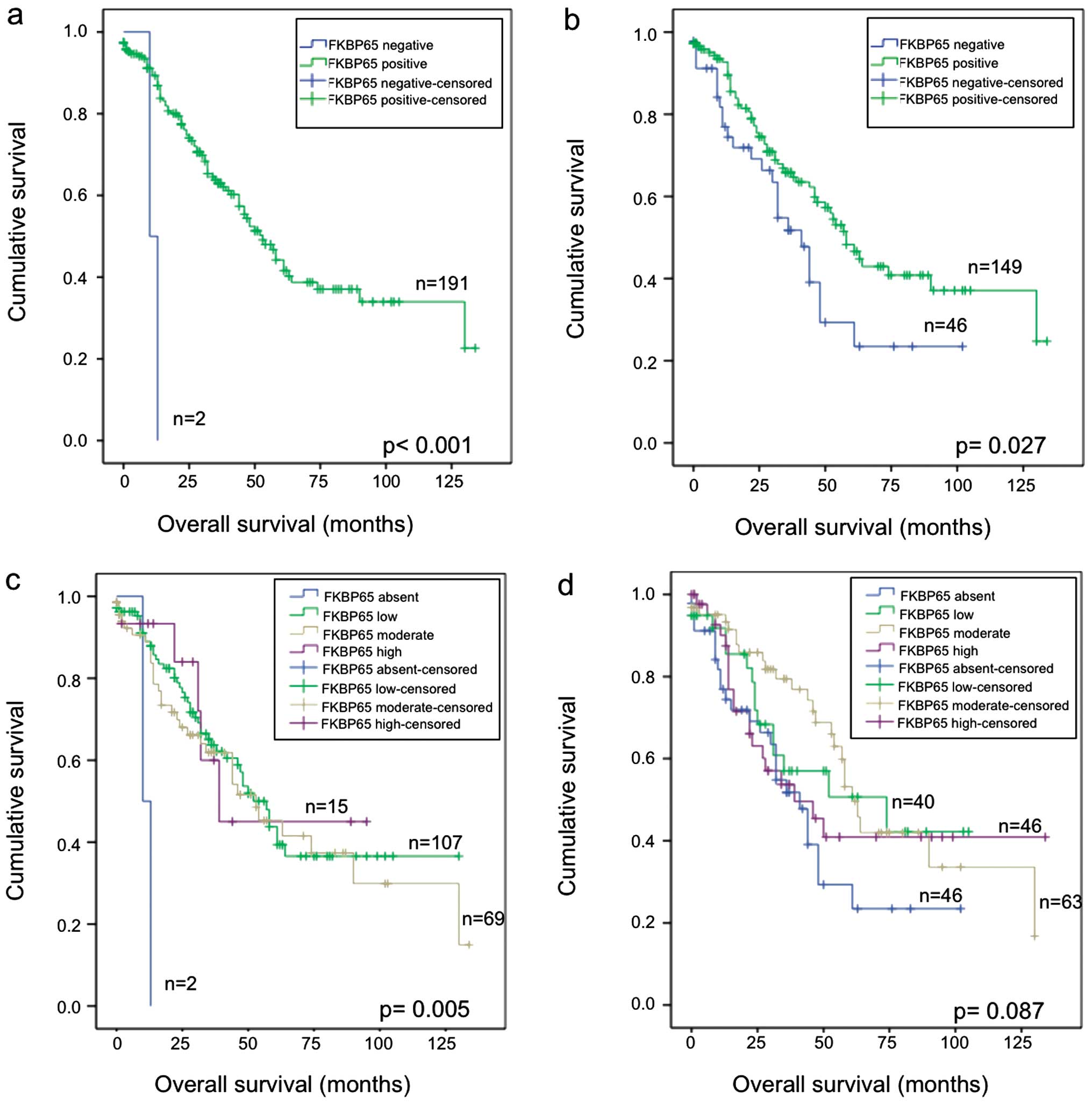
(p<0.001) (Fig. 5a and c).
Although there are only two samples with no detectable staining in
the epithelial component of the tumour, it is interesting that the
cases with the highest staining levels for FKBP65 where among the
cases which had the longest overall survival (Fig. 5c). A similar analysis of FKBP65
staining found a significant association (p=0.027) with prolonged
overall survival when the samples were analysed for the absence or
presence of staining in the stromal component (Fig. 5b). Although there was no
significant difference with overall survival and staining based on
each staining intensity category (p=0.087), it is interesting that
the cases scored absent for staining in the stromal component were
among those with the poorest overall survival (Fig. 5d).
Discussion
In this study we have verified that FKBP10
expression is absent or low in HGSC and the results are consistent
with the low frequency of high intensity staining patterns of
FKBP65 in tumour samples. FKBP65 expression in the epithelial cells
of normal ovarian surface and the distal fimbrae of the fallopian
tube is interesting considering that both of these tissues have
been proposed as the progenitor cell type for HGSC (34). During the course of this study,
decreased expression of FKBP65 in a study of 57 EOC samples of
different histological subtypes, which included HGSCs, was reported
independently (35). These
observations suggest the possibility that FKBP65 expression is also
important in the biology of the other histological subtypes of EOC.
This notion is supported by the observation that FKBP10 was
underexpressed in most of our EOC cell lines, which were derived
from epithelial ovarian tumour samples that differed in their
histology.
Our results from the FKBP10 expression assays
of the EOC cell lines also suggest that underexpression might be
associated with tumourigenicity. Although our assays were
semi-quanti tative, the highest level of gene expression was
observed in TOV81D, the only cell line in our series of EOC cell
lines tested that is unable to form tumours in mouse tumour
xenograft assays and lacks in vitro growth pheno-types
characteristics of tumourigenic cell lines (26,36).
Our expression results are consistent with previously published
studies from our group where, by semi-quantitative RT-PCR and gene
expression microarray analyses, TOV81D exhibited gene expression
profiles that resemble those of NOSEs (3,14,16,37–39).
Notable also is the underexpression of FKBP10 observed in
the tumourigenic OV90 cell line, as this gene was induced in OV90
cell line hybrids that were rendered nontumourigenic as a
consequence of the transfer chromosome 3 in genetic complementation
assays aimed at identifying genes implicated in tumour suppressor
pathways (15,16). Thus, decreased expression of
FKBP10 may be important in tumour suppressor pathways.
We observed a significant association between FKBP65
staining intensity in the epithelial cells of HGSC samples and
overall survival. An association between high FKBP65 expression and
prolonged survival was also observed in the independent study of 57
EOC samples by Henriksen et al, although the finding was not
significant (35). In their
association analyses the small sample size and possibly the
inclusion of different histological subtypes, which are known to
exhibit differences in outcome (40), may have affected the interpretation
of results. In our analyses, though the samples with no FKBP65
staining exhibited the poorest outcome, they represented only two
HGSC cases in our cohort. Notable, however, is that the samples
exhibiting the highest staining intensity were among the HGSC cases
with the longest overall survival. Interestingly, the staining
intensities of the stromal component of the tumour samples were
also associated with overall survival. There is mounting evidence
that ovarian cancer progression involves reciprocal communication
between malignant epithelial cells and the adjacent stromal
microenvironment possibly relating to malignant
epithelium-activated fibroblasts (41). Although not fully explored in the
context of ovarian cancer, gene expression profiles of stromal
cells have been found to be predictive of disease outcome in breast
cancer (42). The expression
profile of COL1A1 was similar to FKBP65 as demonstrated by the
significant positive correlation of staining intensities by
immunohistochemistry of HGSCs. The similarities in expression
profiles have been reported previously, and this is consistent with
FKBP65 being a type I collagen chaperone (19,21).
In conclusion, we found FKBP65 underexpressed in a
proportion of HGSCs, and its expression correlated with that of
COL1A1 in epithelial cells. We also reported on the interesting
finding that absence of FKBP65 staining was associated with poorer
overall survival warranting replication of our study with larger
cohorts. The interesting FKBP10 expression profiles in our
EOC cell lines that differ in their tumourigenic potential could
also be explored to further elucidate the molecular pathways that
involve FKBP65.
Acknowledgements
We thank Jason Madore for his
technical assistance. P.M.W. is a recipient of a Doctoral Research
Award (DRA) from the Canadian Institute of Health Research (CIHR).
The Research Institute of the McGill University Health Centre and
the Centre de Recherche du Centre Hospitalier de l’ Université de
Montréal receive support from the Fonds de Recherche du Québec -
Santé. Clinical specimens were provided by the Banque de tissus et
de données of the Réseau de Recherche sur le Cancer of the Fonds de
Recherche du Québec - Santé which is affiliated with the Canadian
Tumour Repository Network. This research was supported by grants
from the CIHR to P.N.T., D.M.P. and A.-M.M.-M.
References
|
1
|
Dion F, Mes-Masson AM, Seymour RJ,
Provencher D and Tonin PN: Allelotyping defines minimal imbalance
at chromosomal region 17q25 in non-serous epithelial ovarian
cancers. Oncogene. 19:1466–1472. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Foulkes WD, Black DM, Stamp GW, Solomon E
and Trowsdale J: Very frequent loss of heterozygosity throughout
chromosome 17 in sporadic ovarian carcinoma. Int J Cancer.
54:220–225. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Presneau N, Dewar K, Forgetta V,
Provencher D, Mes-Masson AM and Tonin PN: Loss of heterozygosity
and transcriptome analyses of a 1.2 Mb candidate ovarian cancer
tumor suppressor locus region at 17q25.1–q25.2. Mol Carcinog.
43:141–154. 2005.PubMed/NCBI
|
|
4
|
Wojnarowicz PM, Provencher DM, Mes-Masson
AM and Tonin PN: Chromosome 17q25 genes, RHBDF2 and CYGB, in
ovarian cancer. Int J Oncol. 40:18656–1880. 2012.PubMed/NCBI
|
|
5
|
TCGA: Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar
|
|
6
|
Gorringe KL, George J, Anglesio MS, et al:
Copy number analysis identifies novel interactions between genomic
loci in ovarian cancer. PloS One. 5:e114082010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pieretti M, Cavalieri C, Conway PS,
Gallion HH, Powell DE and Turker MS: Genetic alterations
distinguish different types of ovarian tumors. Int J Cancer.
64:434–440. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sangha N, Wu R, Kuick R, et al:
Neurofibromin 1 (NF1) defects are common in human ovarian serous
carcinomas and co-occur with TP53 mutations. Neoplasia.
10:1362–1372. 2008.PubMed/NCBI
|
|
9
|
Feng Q, Deftereos G, Hawes SE, et al: DNA
hypermethylation, Her-2/neu overexpression and p53 mutations in
ovarian carcinoma. Gynecol Oncol. 111:320–329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rathi A, Virmani AK, Schorge JO, et al:
Methylation profiles of sporadic ovarian tumors and nonmalignant
ovaries from high-risk women. Clin Cancer Res. 8:3324–3331.
2002.PubMed/NCBI
|
|
11
|
Pergolizzi R, Appierto V, Crosti M, et al:
Role of retinoic acid receptor overexpression in sensitivity to
fenretinide and tumorigenicity of human ovarian carcinoma cells.
Int J Cancer. 81:829–834. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bruening W, Prowse AH, Schultz DC,
Holgado-Madruga M, Wong A and Godwin AK: Expression of OVCA1, a
candidate tumor suppressor, is reduced in tumors and inhibits
growth of ovarian cancer cells. Cancer Res. 59:4973–4983.
1999.PubMed/NCBI
|
|
13
|
Presneau N, Mes-Masson AM, Ge B,
Provencher D, Hudson TJ and Tonin PN: Patterns of expression of
chromosome 17 genes in primary cultures of normal ovarian surface
epithelia and epithelial ovarian cancer cell lines. Oncogene.
22:1568–1579. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wojnarowicz PM, Breznan A, Arcand SL, et
al: Construction of a chromosome 17 transcriptome in serous ovarian
cancer identifies differentially expressed genes. Int J Gynecol
Cancer. 18:963–975. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cody NA, Ouellet V, Manderson EN, et al:
Transfer of chromosome 3 fragments suppresses tumorigenicity of an
ovarian cancer cell line monoallelic for chromosome 3p. Oncogene.
26:618–632. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Quinn MC, Filali-Mouhim A, Provencher DM,
Mes-Masson AM and Tonin PN: Reprogramming of the transcriptome in a
novel chromosome 3 transfer tumor suppressor ovarian cancer cell
line model affected molecular networks that are characteristic of
ovarian cancer. Mol Carcinog. 48:648–661. 2009. View Article : Google Scholar
|
|
17
|
Coss MC, Winterstein D, Sowder RC II and
Simek SL: Molecular cloning, DNA sequence analysis, and biochemical
characterization of a novel 65-kDa FK506-binding protein (FKBP65).
J Biol Chem. 270:29336–29341. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng B, MacDonald JR, Bann JG, et al:
Chicken FK506-binding protein, FKBP65, a member of the FKBP family
of peptidylprolyl cis-trans isomerases, is only partially inhibited
by FK506. Biochem J. 330:109–114. 1998.PubMed/NCBI
|
|
19
|
Patterson CE, Abrams WR, Wolter NE,
Rosenbloom J and Davis EC: Developmental regulation and coordinate
reexpression of FKBP65 with extracellular matrix proteins after
lung injury suggest a specialized function for this endoplasmic
reticulum immunophilin. Cell Stress Chaperones. 10:285–295. 2005.
View Article : Google Scholar
|
|
20
|
Patterson CE, Schaub T, Coleman EJ and
Davis EC: Developmental regulation of FKBP65. An ER-localized
extra-cellular matrix binding-protein. Mol Biol Cell. 11:3925–3935.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ishikawa Y, Vranka J, Wirz J, Nagata K and
Bachinger HP: The rough endoplasmic reticulum-resident
FK506-binding protein FKBP65 is a molecular chaperone that
interacts with collagens. J Biol Chem. 283:31584–31590. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Forbes SA, Bindal N, Bamford S, et al:
COSMIC: mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acid Res. 39:D945–D950. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alanay Y, Avaygan H, Camacho N, et al:
Mutations in the gene encoding the RER protein FKBP65 cause
autosomal-recessive osteogenesis imperfecta. Am J Hum Genet.
86:551–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kelley BP, Malfait F, Bonafe L, et al:
Mutations in FKBP10 cause recessive osteogenesis imperfecta and
Bruck syndrome. J Bone Miner Res. 26:666–672. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nishida T, Oda T, Sugiyama T, Izumi S and
Yakushiji M: Concurrent ovarian serous carcinoma and osteogenesis
imperfecta. Arch Gynecol Obstet. 253:153–156. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ouellet V, Zietarska M, Portelance L, et
al: Characterization of three new serous epithelial ovarian cancer
cell lines. BMC Cancer. 8:1522008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le Page C, Marineau A, Bonza PK, et al:
BTN3A2 expression in epithelial ovarian cancer is associated with
higher tumor infiltrating T cells and a better prognosis. PloS One.
7:e385412012.PubMed/NCBI
|
|
28
|
Therasse P, Arbuck SG, Eisenhauer EA, et
al: New guidelines to evaluate the response to treatment in solid
tumors. European Organization for Research and Treatment of Cancer,
National Cancer Institute of the United States, National Cancer
Institute of Canada. J Natl Cancer Inst. 92:205–216. 2000.
View Article : Google Scholar
|
|
29
|
Ouellet V, Provencher DM, Maugard CM, et
al: Discrimination between serous low malignant potential and
invasive epithelial ovarian tumors using molecular profiling.
Oncogene. 24:4672–4687. 2005. View Article : Google Scholar
|
|
30
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.PubMed/NCBI
|
|
31
|
Karolchik D, Baertsch R, Diekhans M, et
al: The UCSC Genome Browser Database. Nucleic Acid Res. 31:51–54.
2003. View Article : Google Scholar
|
|
32
|
Arcand SL, Maugard CM, Ghadirian P, et al:
Germline TP53 mutations in BRCA1 and BRCA2 mutation-negative French
Canadian breast cancer families. Breast Cancer Res Treat.
108:399–408. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Davis EC, Broekelmann TJ, Ozawa Y and
Mecham RP: Identification of tropoelastin as a ligand for the 65-kD
FK506-binding protein, FKBP65, in the secretory pathway. J Cell
Biol. 140:295–303. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salvador S, Gilks B, Kobel M, Huntsman D,
Rosen B and Miller D: The fallopian tube: primary site of most
pelvic high-grade serous carcinomas. Int J Gynecol Cancer.
19:58–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Henriksen R, Sorensen FB, Orntoft TF and
Birkenkamp-Demtroder K: Expression of FK506 binding protein 65
(FKBP65) is decreased in epithelial ovarian cancer cells compared
to benign tumor cells and to ovarian epithelium. Tumour Biol.
32:671–676. 2011. View Article : Google Scholar
|
|
36
|
Provencher DM, Lounis H, Champoux L, et
al: Characterization of four novel epithelial ovarian cancer cell
lines. In Vitro Cell Dev Biol Anim. 36:357–361. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Birch AH, Quinn MC, Filali-Mouhim A,
Provencher DM, Mes-Masson AM and Tonin PN: Transcriptome analysis
of serous ovarian cancers identifies differentially expressed
chromosome 3 genes. Mol Carcinog. 47:56–65. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cody NA, Shen Z, Ripeau JS, et al:
Characterization of the 3p12.3-pcen region associated with tumor
suppression in a novel ovarian cancer cell line model genetically
modified by chromosome 3 fragment transfer. Mol Carcinog.
48:1077–1092. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Manderson EN, Birch AH, Shen Z, Mes-Masson
AM, Provencher D and Tonin PN: Molecular genetic analysis of a cell
adhesion molecule with homology to L1CAM, contactin 6, and
contactin 4 candidate chromosome 3p26pter tumor suppressor genes in
ovarian cancer. Int J Gynecol Cancer. 19:513–525. 2009. View Article : Google Scholar
|
|
40
|
Kobel M, Kalloger SE, Boyd N, et al:
Ovarian carcinoma subtypes are different diseases: implications for
biomarker studies. PLoS Med. 5:e2322008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schauer IG, Sood AK, Mok S and Liu J:
Cancer-associated fibroblasts and their putative role in
potentiating the initiation and development of epithelial ovarian
cancer. Neoplasia. 13:393–405. 2011.PubMed/NCBI
|
|
42
|
Finak G, Bertos N, Pepin F, et al: Stromal
gene expression predicts clinical outcome in breast cancer. Nat
Med. 14:518–527. 2008. View
Article : Google Scholar : PubMed/NCBI
|