Introduction
Disruption in apoptosis is involved in cancer
development and progression, where insufficient apoptosis
contributes to the process of carcinogenesis and acquirement of
therapeutic resistance (1,2). Apoptosis is initiated and executed by
proteases known as caspases whose activation is believed to be
required for apoptosis to occur in most instances (3), whereas the inhibitor of apoptosis
(IAP) family of proteins is believed to inhibit apoptosis as potent
caspase inhibitors (4).
Livin-β, also known as melanoma inhibitor of
apoptosis protein (ML-IAP), has been identified as a member of the
IAP family (5–7). Livin-β contains a single BIR
(baculoviral IAP repeat) domain as well a carboxyl-terminal RING
domain. It was supposed to suppress downstream caspase-3 and -7 and
protect cells from proapoptotic stimuli. An alternative splicing
variant α isoform was also identified, which is almost identical
except for a 54-bp truncation in exon 6 (8). It seems the two isoforms do not
differ significantly in their biological activities.
The expression of Livin-β was detectable in a panel
of tumor types, including melanoma, leukemias, bladder cancer,
breast cancer, cervical cancer, colon cancer, nasopharyngeal cancer
and different forms of lung cancer (9–12).
Our previous study indicated the expression profile of Livin-β was
restricted to melanoma, but not melanocytic naevus (13). We also found Livin-β was hardly
detectable in astrocytoma (14).
Taken together, these observations implicate Livin-β may play a
role for the variant malignant phenotype of tumors.
Noteworthy, Livin-β undergoes site-specific cleavage
in cells by effector caspase-3 and -7 and a truncated fragment
(tLivin) without its N-terminal 52 amino acids was produced
(15). However, the biological
consequence of the cleavage remains largely ignored. Here, we show
ectopic expression of the truncated fragment tLivin was able to
induce the activation of caspase-3 and this fragment was further
cleaved into a protective C-terminal fragment in cells.
Materials and methods
Plasmid construction
The full-length cDNA of Livin-β was amplified from
total RNA isolated from fresh tissue of a metastatic malignant
melanoma by RT-PCR with the following primers: 5′-AAT AGA TCT CAT
GGG ACC TAA AGA CAG TGCC-3′ (sense) and 5′-AAT GGA TCC GGA CAG GAA
GGT GCG-3′ (antisense). When the product of the expected size was
obtained, it was inserted into UA cloning vector pDrive (Qiagen,
Valencia, CA, USA) and then subcloned into pcDNA 3.1 (Invitrogen,
Calsbad, CA, USA) to obtain the expression plasmid. The construct
myc-tLivin expressing Livin-β lack of N-terminal 52 amino acids
with myc tag at its N-terminus was generated by cloning the PCR
product into pCMV-myc vector (Clontech, Palo Alto, CA, USA). To
create an expression plasmid for the full-length ML-IAP with a
C-terminal green fluorescence protein (GFP) fusion, PCR
amplification was performed with Pfu polymerase (Fermentas, St.
Leon Rot, Germany) using the UA clone as a template. GFPliv was
then generated by cloning the PCR product into the BamHI and
XhoI sites of pEGFP-N1 (Clontech). GFP-tLivin and GFPlivC
were constructed by the same strategy. To create a vector
expressing full-length ML-IAP with an N-terminal hexahistidine tag,
the sequence encompassing Livin-β was generated by PCR and cloned
into the BamHI and HindIII sites of PQE30 (Qiagen).
The construct expressing Livin-β lacking N-terminal 52 amino acids
was made in the same manner. The expression construct for caspase-3
contained a hexahistidine tag at the C terminus of the full-length
protein was kindly provided by Professor G.S. Salvesen. All the
constructs were verified by sequencing.
Site-directed mutagenesis
Site-directed mutagenesis of tLivin was conducted to
generate a D220A substitution mutation by over-lap PCR with the
following mutagenic primers: 5′-GCG CCT CCA CAG CCC TGG CTC-3′ and
5′-AGC CAG GGC TGT GGA GGCG-3′, combined with standard primers
matching the regions of flanking the coding DNA in expression
vector PQEtLivin. The DNA with expected point mutatin was inserted
into the TA cloning plasmid pMD18-T (Takara). After the mutant
construct was confirmed by sequencing, the DNA was subcloned into
PQE30 for expression in bacteria.
Cell culture, transfection and western
blotting
Human A549 lung adenocarcinoma cells, human
embryonic kidney 293 cells and colon cancer HCT116 cells harboring
wild-type P53 were purchased from the American Type Culture
Collection. HCT116 cells with P53 knock-out were donated by
Professor Vogelstein (Howard Hughes Medical Institute). The cells
were maintained in Dulbecco’ modified Eagle’s medium (Invitrogen)
supplemented with 10% FBS and antibiotics at 37°C.
For transfection of cells, a standard lipofectin
method was employed. Cells were seeded in 6-well plates or 96-well
plates. Lipofectamine 2000 (Invitrogen) was used at a ration of 2.5
μg/μl in input plasmid and lipofectin-DNA complexes
were incubated with cells for 6 h at 37°C.
Forty-eight hours after transfection, cells were
harvested in lysis buffer [50 mM Tris-buffered HCl, 1% NP-40, 1%
SDS, 100 mM EDTA, 100 mM PMSF, 0.5% Triton X-100 and 1 mM
proteinase inhibitor cocktail (Sigma)] and protein were quantified
using Bradford protein assay reagent (Bio-Rad). Total proteins were
resolved by SDS polyacrylamide (Bio-Rad) gel electrophoresis.
Proteins were electroblotted to PVDF membrane (Amersham Biosciences
Ltd., Little Chalfont, UK) and then incubated with block solution
(5% non-fat milk, 0.1% Tween-20, in TBS) at room temperature for 1
h. Following blocking, the membrane was incubated with an
appropriate primary antibody and then incubated with a
corresponding horseradish peroxidase conjugated or fluorescein
isothiocyanate (0 FITC)-tagged secondary antibody. The blots were
developed by an enhanced ECL method (Pierce, Englewood Cliffs, NJ,
USA). When FITC-tagged secondary antibody was used, the blot was
subjected to Tyhoon Trio fluorescence scanner (Amersham
Biosciences).
Reagents and antibodies
Cytotoxic drug cisplatin was purchased from Qilu
Pharmaceutical Inc. (Shangdong, China). Stock preparation of the
reagents was stored at −20°C. The pan-caspase inhibitor z-VAD-fmk
(Promega, Madison, WI, USA) was prepared in DMSO at a concentration
of 20 μM. The fluorogenic substrate z-DEVD-AFC and
colorimetric substrate z-DEVD-pNA for caspase-3 were obtained from
Molecular Probes.
Primary antibodies used in this study included the
following: anti-His mAb (Sigma), anti-myc mAb (Clontech),
anti-GAPDH mAb (Kangcheng, Shanghai, China), anti-Livin polyclonal
antibody (R&D, Minneapolis, MN, USA), anti-caspase-3
(full-length and active fragment) polyclonal antibody (Santa Cruz),
anti-XIAP polyclonal antibody (ProteinTech Group Inc., Chicago, IL,
USA), anti-caspase-8 polyclonal antibody (Santa Cruz), anti-cIAP2
polyclonal antibody (Santa Cruz) and anti-GFP polyclonal antibody
(Novus, St. Louis, MO, USA).
MTT assay
Survival of cells after treatment was quantified by
a modified MTT assay (Promega). Briefly, cells were seeded in
96-well tissue culture plate, in 100-μl culture medium. Then
the cells were either transfected with different plasmids or
treated with cisplatin. MTT labeling solution (15 μl) was
added to each well and the cells were further incubated at 37°C for
4 h. Solubilization solution (100 μl) was added to each well
for incubation overnight. The absorbance was measured at a
wavelength of 570 nm with a microplate reader (Bio-Rad). Data were
the average of 5-wells. Untreated cells served as the indicator of
100% cell viability.
Expression and purification of
proteins
For production of recombinant Livin-β, full-length
cDNA of Livin-β was cloned into the plasmid PQE30 (Qiagen). The
plasmids were introduced into Escherichia coli strain JM109.
The His6-tagged protein was prepared from the inclusion body upon
induction with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG)
at 37°C for 4 h. Purified protein was isolated using Ni-chelating
column (Amersham Biosciences) on the AKTA chromato graphy system
(Amersham Biosciences) and eluting with an imidazole gradient from
0–200 mM in 50 mM Tris, 300 mM Na2HPO4, pH
8.0. Coomassie blue staining analysis after SDS-PAGE revealed
purity of >90%. Recombinant Livin-β protein was renatured by
dialyzing against the buffer containing decreased urea
concentration. Its biological activity was confirmed in an in
vitro recombinant caspase-3 activity inhibition assay.
Recombinant tLivin protein was generated in
Escherichia coli strain XL-1 blue, induced with 1 mM IPTG at
30°C for 4 h. Then the bacteria were lysed and soluble fraction was
collected and subjected to Nickel column purification. For the
tLivin mutant, the protein was expressed in the same way. Also, the
mutant protein was purified with Nickel charged magnetic beads
(Toyobo, Osaka, Japan) according to the manufacturer’s
instructions. Expression and purification of active form of
caspase-3 were described previously (16). These proteins were quantified by
BCA method.
Preparation and purification of
anti-Livin-β polyclonal antibody
Anti-Livin-β sera were raised in rabbits with
purified Livin-β protein as immunogen. The immunization protocol
was as follows: the rabbit was immunized subcutaneously with 300
μg protein in complete and incomplete Freund’s adjuvant 6
times at one-week interval. Antisera titers were monitored by ELISA
where the plate was coated with purified protein. The sera were
harvested and polyclonal antibody was purified with an affinity
blue-gel (Bio-Rad) on the chromatography system. The purified
antibody was assayed for its ability to recognize recombinant and
endogenous ML-IAP protein in western blot analysis.
DEVDase activity
The assay was performed on a SpectraMAX M5 plate
reader coupled with SoftMax software (Molecular Devices, Sunnyvale,
CA, USA) operating in the kinetic mode at 37°C. DEVDase acitivity
was determined by using colorimetric pNA substrates (maximal
absorbance at 405 nm) or fluorogenic AFC substrate (excitation
wavelength 400 nm and emission wavelength 505 nm). Assay buffer was
50 mM HEPES, pH 7.4; 100 mM NaCl, 0.1% Chaps, 10 mM dithiothreitol
and 10% sucrose. Data were recorded every 30 sec for various
periods of time as appropriate for each assay.
Apoptosis assay
Cells were transfected with the indicated GFP fusion
plasmid to allow quantitation of transfection efficiency. Green
(transfected) cells were then scored as either apoptotic or viable
according to standard morphological criteria. Apoptotic cells were
typically retracted from the substratum, shrunk and exhibited
dramatic membrane blebbing along with the formation of numerous
apoptotic bodies. A minimum of 300 transfected cells per treatment
was evaluated. The images of GFP expression were captured by an
Axiovert 200 inverted fluorescence microscope (Zeiss, Oberkochen,
Germany).
Preparation of cytosolic fraction
Cytosolic extracts from 293 cells with different
treatments were harvested by gentle scraping into
phosphate-buffered saline at 4°C, pelleted by centrifugation and
subsequently washed once in the same buffer. The cell pellet was
resuspended in HEB (20 mM Pipes, 10 mM KCl, 5 mM EDTA, 2 mM
MgCl2, 1 mM dithiothreitol, pH 7.4) at 4°C and pelleted
by centrifugation at 1,000 × g. The cell pellet was then
resuspended in an equal volume of HEB and allowed to swell on ice
for 30 min. The cells were then cracked by passing through a
24-gauge needle and pelleted by centrifugation at 16,000 × g for 30
min and the supernatant (cytosolic extract) recovered. The
resultant pellets were collected as nuclear fraction.
Cell cycle analysis
Cells were trypsinized and prepared for cell cycle
analysis using the Vindelov protocol with propidium iodine (PI) for
DNA staining (17). Cell cycle
analysis was performed using a FACscan (Becton-Dickinsson, San
Jose, CA, USA). The data were analyzed and calculated by ModFit LT
for Mac V3.1 (Verity Software House, Inc., Topsham, ME, USA).
γH2AX immunostaining
Cells grown on the slides were fixed in 4%
paraformaldehyde and permeabilized with 0.1% Triton X-100. Then
γH2AX Ab (Abcam) diluted in Antibody Diluent (Dako) was applied and
incubated at 4°C overnight. Alexa fluor 488 conjugated anti-mouse
Ab was applied, followed by DAPI incubation. The ProLong Gold
AntiFade Reagent was dripped onto the slides. Finally, the
fluorescence was observed under a fluorescence microscope (Axiovert
200).
Co-immunoprecipitation
The co-immunoprecipitation was performed with the
c-Myc tag co-IP kit (Thermo Scientific, Rockford, IL, USA).
Briefly, cell were transfected with myc-tLivin as described above.
Lysates were immobilized with anti-c-Myc agarose overnight at 4°C.
The agarose was washed twice with TBST and then the c-Myc tagged
protein was eluted with elution buffer. The c-Myc tagged tLivin was
probed with an anti-Livin antibody as described.
Statistical analysis
Statistical analysis was performed by SPSS 13.0 (IBM
software). The 2-side Student’s t-test for independent samples was
used for the analysis of all data. P-values <0.05 were
considered to be statistically significant.
Results
Truncated Livin exerted a pro-apoptotic
effect
We introduced the expression plasmid for the
truncated Livin (tLivin) into the A549 lung adenocarcinoma cells.
Two days after transfection, A549 cells treated with tLivin
manifested as retracted from the substratum, shrunken and with
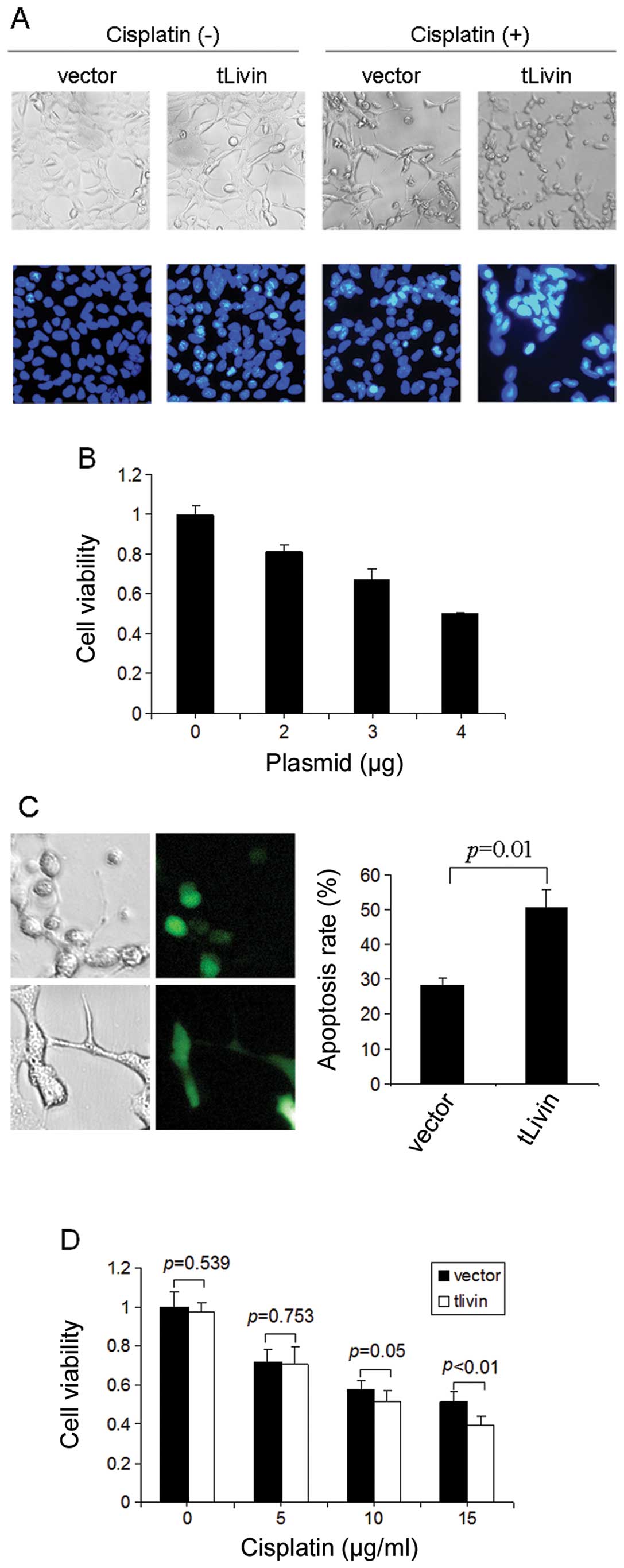
membrane blebbing, typical features of cell apoptosis (Fig. 1A). We also used the Hoechst 33258
staining to check for apoptosis after treatment with tLivin.
Consistently, tLivin induced apoptosis in A549 cells, which was
characterized by chromatin condensation and fragmentation (Fig. 1A). To further evaluate the effects
of tLivin, an MTT assay was performed to determine cell viability.
The treatment reduced the cell viability compared with empty vector
in a dose-dependent manner. The higher dose of tLivin plasmid led
to less cell viability compared with the same amount of empty
vector (Fig. 1B). We also
constructed a tLivin and GFP fusion plasmid (GFP-tLivin) and used
this plasmid to evaluate the pro-apoptotic efficiency of tLivin. In
this assay, ectopic expression of tLivin led to ∼30% of apoptosis
while the apoptosis rate of control GFP treated cells was <10%
(Fig. 1C). Besides, we also
observed that the expression of tLivin sensitized A549 cells to
cytotoxic chemotherapeutic agent cisplatin, assayed morphologically
or by fluorescence staining (Fig.
1A). In addition, we found the synergistic effect of tLivin
with cisplatin was more significant with the higher dose of
cisplatin than the lower dose (Fig.
1D). The dose of 15 mg/ml of cisplatin was chosen for the
following experiments.
Truncated Livin synergizes with cisplatin
through caspase-3 mediated apoptosis
A synergistic effect of tLivin combined with
cisplatin on A549 cells was observed and we sought to explore the
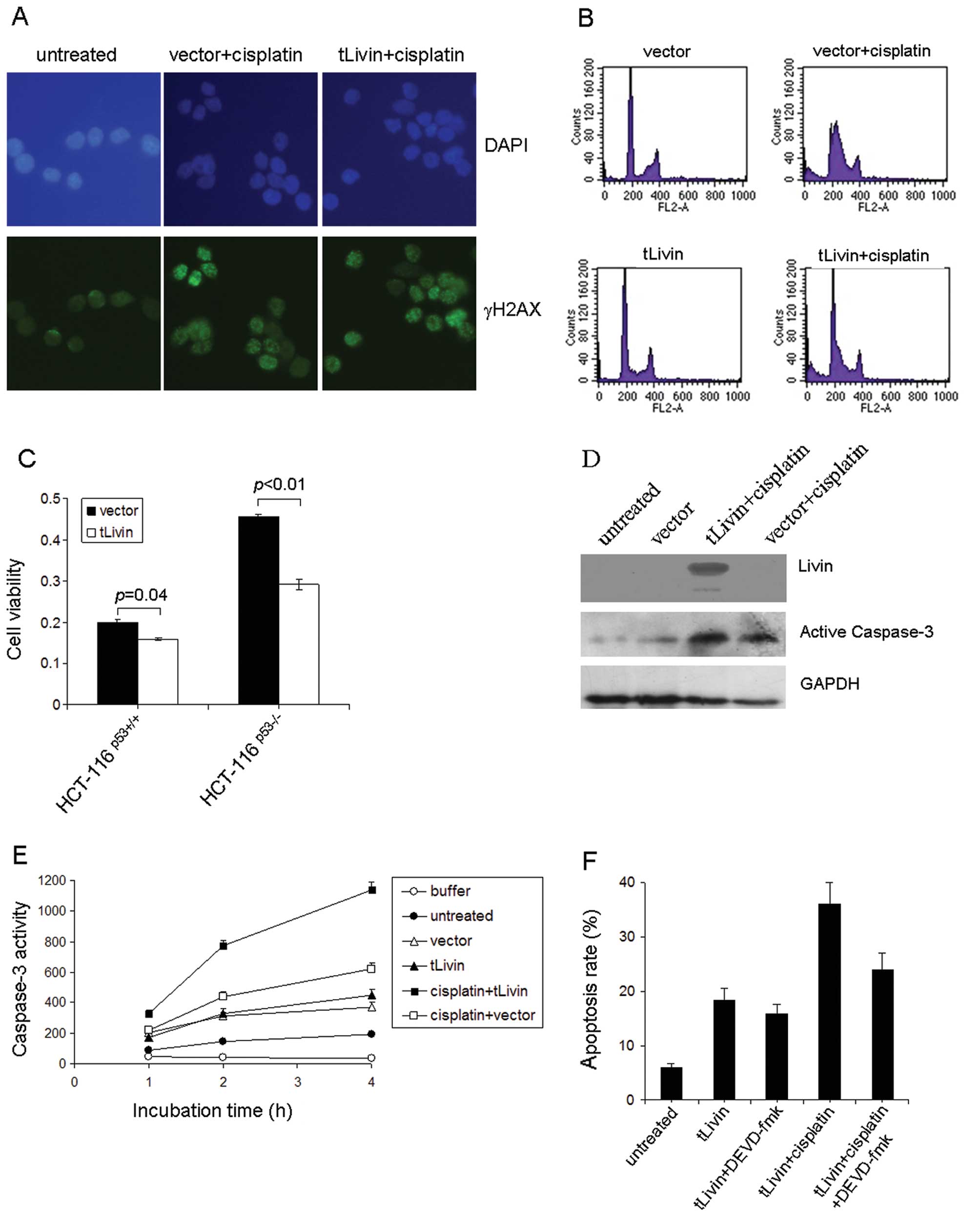
possible mechanism. The DNA break repair capacity was determined by
immunostaining for γH2AX after cisplatin treatment. No difference
was noticed between tLivin-treated or vector-treated cells
(Fig. 2A). Next the cell cycle
distribution of A549 cells with or without cisplatin treatment was
examined by flow cytometry. The cells transfected with tLivin had
even less changes in cell cycle distribution after cisplatin
treatment although they were more vulnerable to cisplatin (Fig. 2B). Therefore, the synergistic
effects of tLivin could not be attributed to the cell cycle
re-distribution. The impact of P53 status on the effects of tLivin
was also checked. To this end, human colon cancer HCT116 cells with
wild-type (HCT116P53+/+) or null TP53
(HCT116P53−/−) were transfected with tLivin and treated
with cisplatin. In both cell types, tLivin transfection led to less
viability than vector transfection (Fig. 2C).
Then the active form of caspase-3 was examined by
Western blotting. The cells transfected with tLivin showed a
stronger signal of the 17-kDa form of caspase-3 which indicated a
higher activity of caspase-3 in this setting (Fig. 2D). A DEVDase activity assay was
further conducted to confirm the observation. In this experiment,
the highest DEVDase activity was observed in the tLivin transfected
cells, followed by vector transfected ones (Fig. 2E). The cytotoxic effect of tLivin
was possibly attributed to apoptosis, therefore an inhibitor of
caspase-3 was used and found to markedly reduce cell death by
tLivin or tLivin combined with cisplatin (Fig. 2F).
Truncated Livin was mainly localized in
cytoplasm
It was reported that Livin-β was localized in the
cytoplasm by using an immunostaining method. We wanted to explore
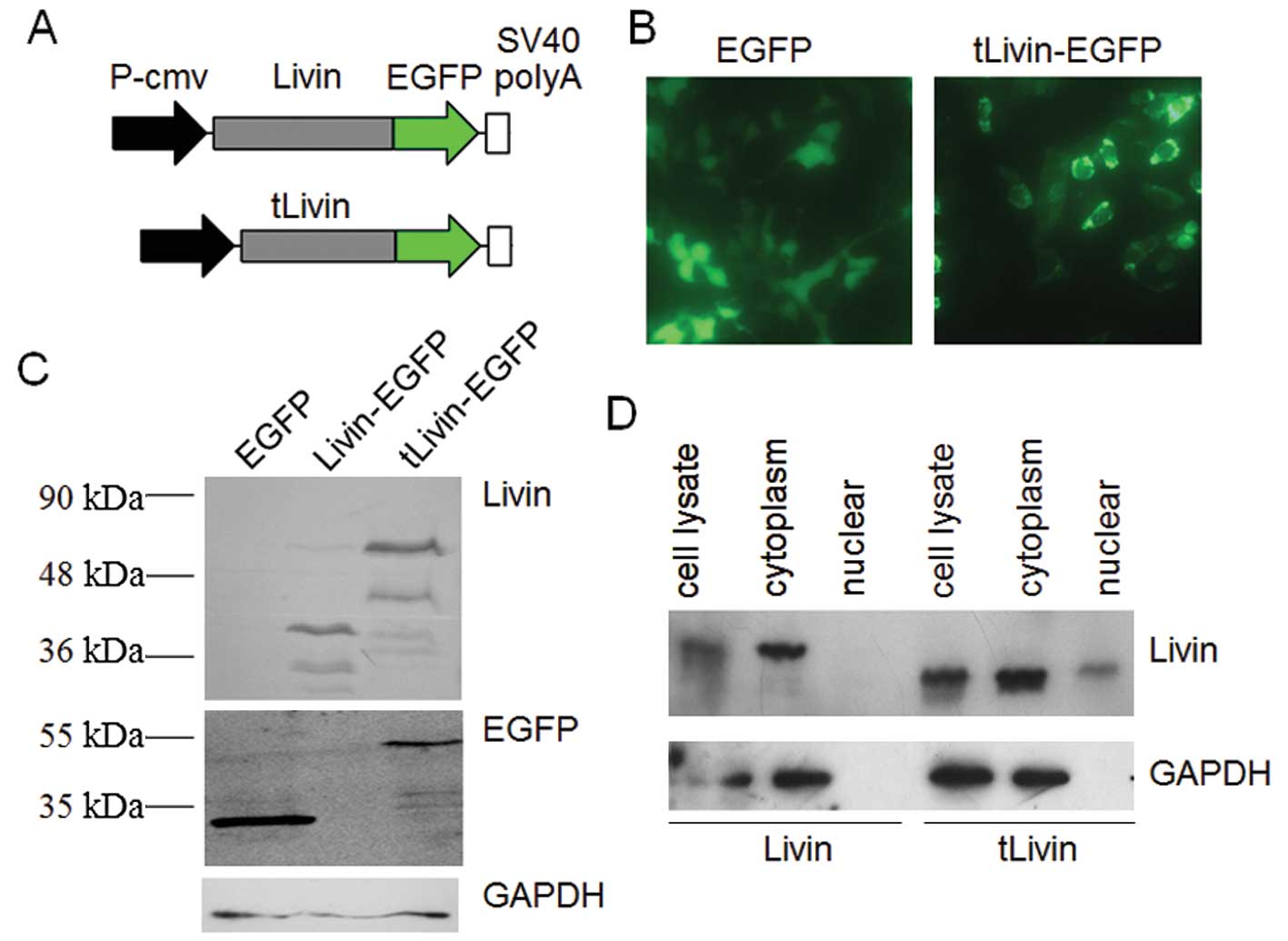
the subcellular localization of tLivin. Here, the expression
plasmid for GFP-fused tLivin (GFP-tLivin) was constructed to study
its distribution in living cells (Fig.
3A). Also the recombinant GFP-Livin-β vector was prepared in
parallel. We first transfected A549 cells with the constructed
plasmid or GFP plasmid. Forty-eight hours later, the living cells
were examined by fluorescence microscope. GFP protein displayed an
evenly diffused localization throughout the whole cells transfected
with pEGFP-N1. GFP-tLivin was mainly found to localize in the
cytoplasm (Fig. 3B). Western blot
analysis confirmed properly expressed GFP and GFP-tLivin (Fig. 3C). However, the GFP-Livin-β vector
failed to show any fluorescence with our repeated efforts (data not
shown). In Western blotting only fragments of GFP-Livin-β fusion
protein were detected (Fig. 3C).
The exact mechanism underlying the improper expression of
GFP-Livin-β remains elusive. To further investigate the subcellular
localization of tLivin, we carried our subcellular fractionation
experiments in Livin or tLivin transfected cells. Livin-β was
almost completely located in the cytoplasm. In parallel, tLivin was
mainly detected in the cytoplasmic fraction (Fig. 3D).
Truncated Livin was cleaved into a
smaller fragment in cells
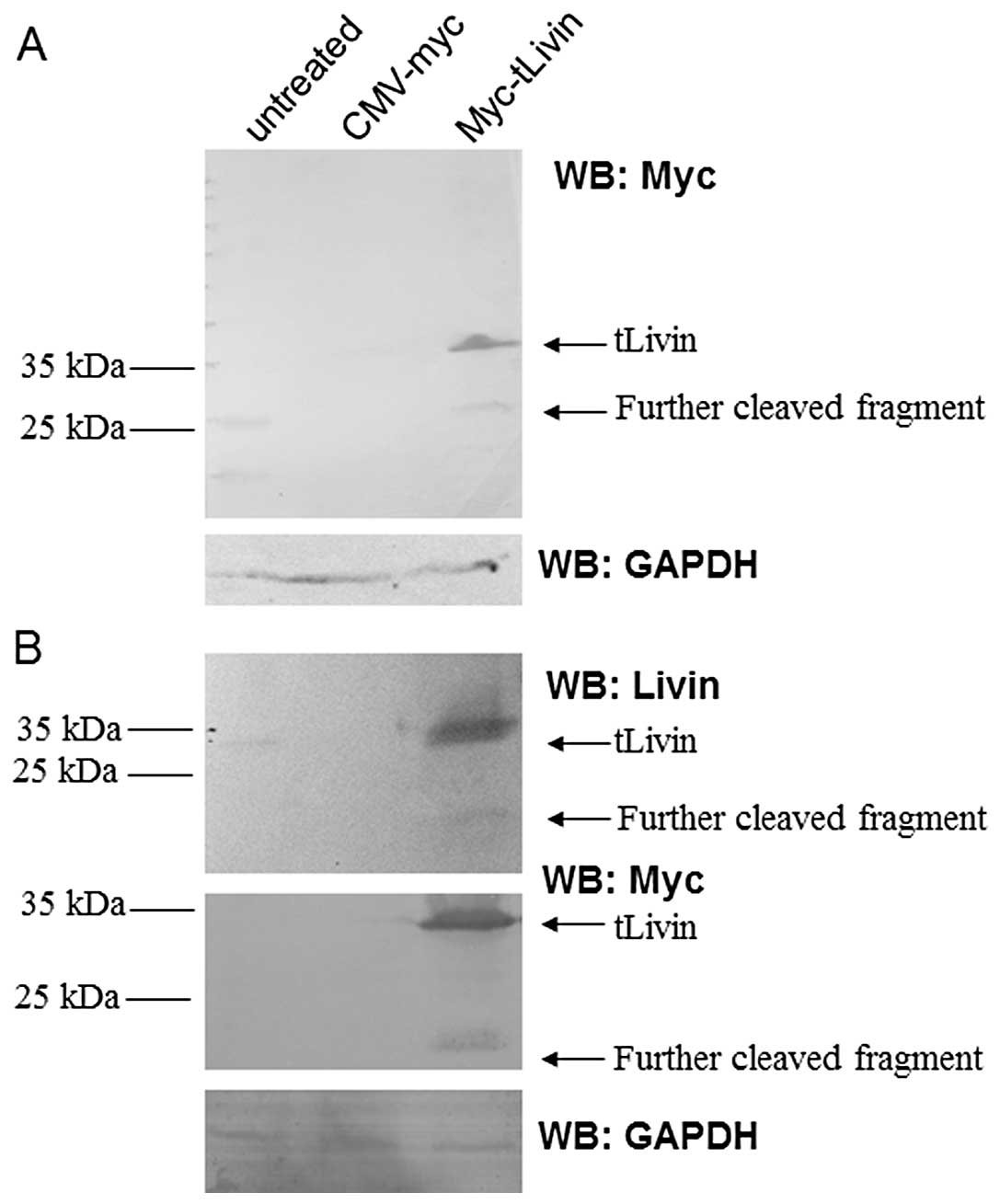
To confirm the expression of tLivin after
transfection, lysates prepared from A549 cells transfected with
myc-tLivin were immunoblotted with a monoclonal antibody against
Myc tag. The 37-kDa myc-tLivin protein band was readily detected in
cells transfected with the recombinant expression plasmid. In
addition to full-length Myc-tLivin protein, an immunoreactive band
of ∼22 kDa was also detected after transfection. The additional
band was also detected by the antibody against Livin (Fig. 4A), suggesting that tLivin may be
cleaved after expression. In order to further confirm this
observation, similar results were obtained in 293 cells. To exclude
the possibility of non-specific immunoreactivity in western blot
analysis, the lysates were also probed with a rabbit polyclonal
antibody generated against recombinant Livin-β. Once more, a
similar cleavage was observed (Fig.
4B).
Cleavage of tLivin is dependent on
caspase-3 activation
We demonstrated that overexpression of tLivin led to
apoptosis with activation of caspase-3 and tLivin mainly localized
in the cytoplasm, the same compartment with caspase-3. tLivin was
cleaved to smaller fragments. Based on these observations, we
wondered whether tLivin cleavage was mediated with caspase-3. To
confirm this hypothesis, we used the caspase inhibitor zVAD-fmk to
treat A549 cells. We found pre-treatment with zVAD-fmk abrogated
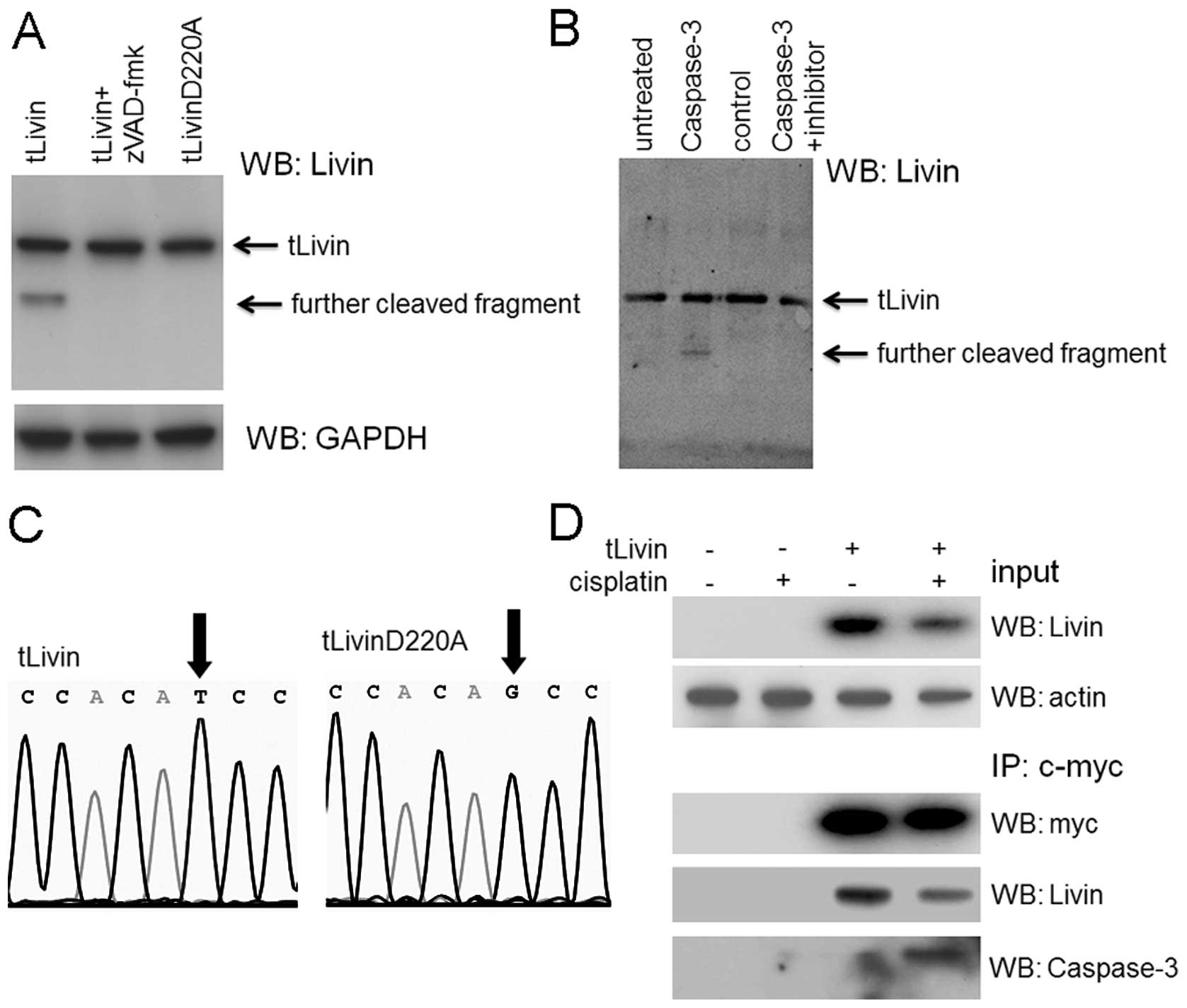
the cleavage of tLivin (Fig. 5A).
We also performed a co-IP experiment where c-Myc tagged tLivin
overexpressed by transfection was precipitated with anti-c-Myc
antibody agarose. The precipitated tLivin was confirmed by the
anti-Livin antibody, besides the band recognized by anti-caspase-3
antibody at ∼17-kDa size, corresponding to the cleaved caspase-3
fragment (Fig. 5D).
In an attempt to reconstitute the cleavage reaction
in vitro, we prepared and purified tLivin recombinant
protein from bacteria expression system. The tLivin protein was
incubated with pre-activated caspase-3 protein for 1 h at 37°C and
then the reaction mixture was resolved by SDS-PAGE and probed with
anti-livin antibody. In this in vitro reaction, tLivin was
cleaved into a smaller fragment, similar to that observed in cell
lysates (Fig. 5B). However, tLivin
protein incubated with a protein control E-cadherin failed to
cleave.
Next, the amino sequence of tLivin was scanned and
one possible caspase-3 substrate tetra-peptide (217GARD220) was
recognized. To map the cleavage site of tLivin, a site-directed
mutagenesis was performed where the glutid acid in position 220 was
mutated to alanine (Fig. 5C). The
resulting protein, tLivin D220A, was also incubated with active
form of caspase-3. However, this point mutation abrogated the
cleavage potential of tLivin (Fig.
5A).
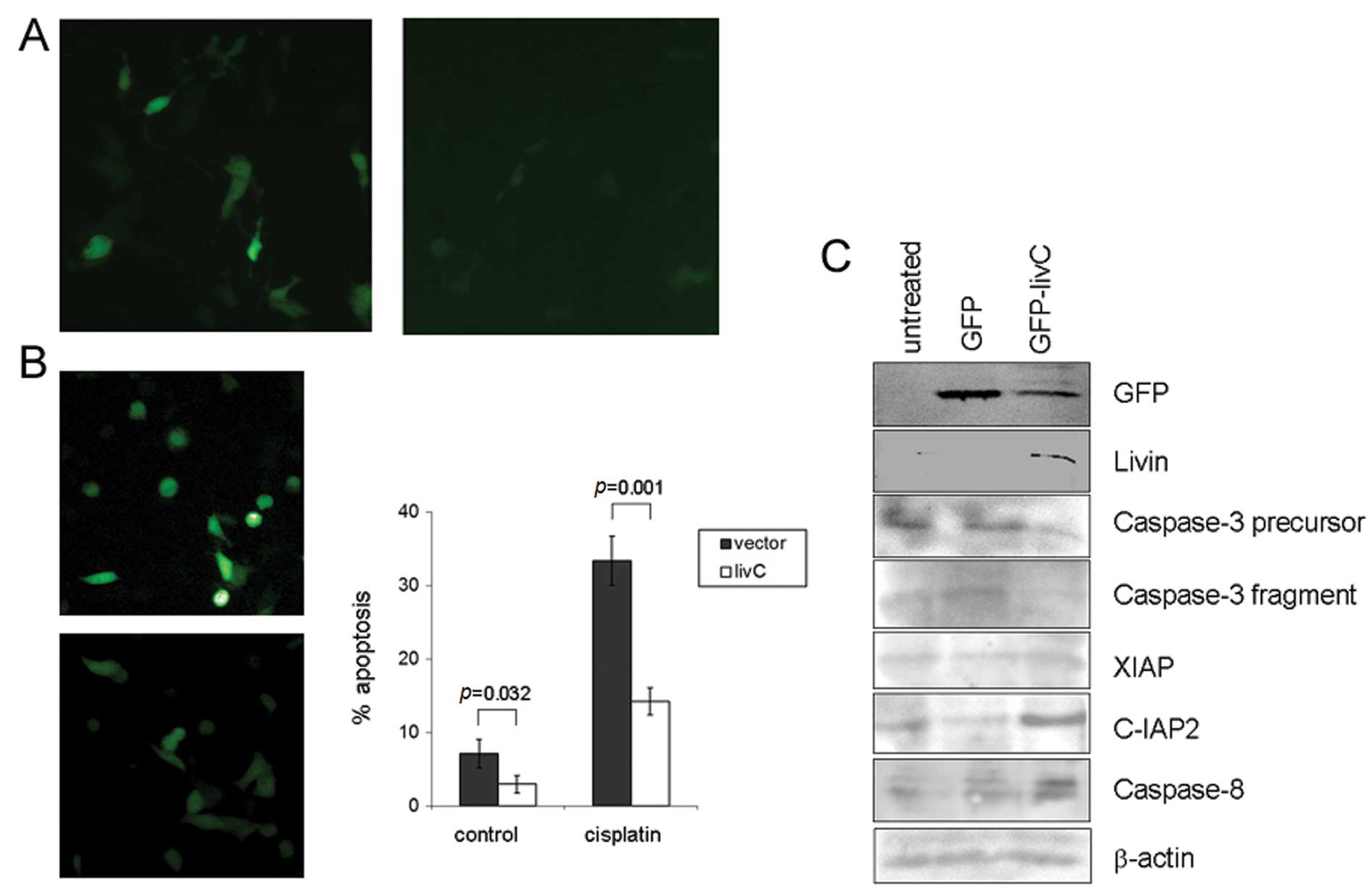
The cleaved C-terminal fragment of tLivin was an
anti-apoptotic factor. Because the RING finger of XIAP mediated
ubiquitination of caspases, we hypothesized the C-terminus of
tLivin had anti-apoptotic function. To confirm this hypothesis, we
transfected A549 cells with a construct expressing the C-terminus
of tLivin (amino acids 221–280, livC) fused with GFP and found it
could antagonize the apoptotic effect of cisplatin (Fig. 6B). We also observed a significant
decrease in the expression intensity of GFP, which was fused to the
N-terminus of livC (Fig. 6A). To
further explore the possible mechanism for this effect of livC, the
cell lysates were probed with a panel of antibodies. We found the
protein content of caspase-3 decreased in cells transfected with
livC, both of full-length and cleaved active form of caspase-3
(Fig. 6C). These data may help to
explain the anti-apoptotic effect of livC in cells.
Discussion
Key mediators in apoptosis regulation are of intense
biological interest. Livin-β underwent site-specific cleavage in
cells upon apoptosis stimulation and a truncated fragment (tLivin)
without its N-terminal 52 amino acids was produced (15). In the present study, we constructed
an expression plasmid for the truncated fragment of Livin-β
(tLivin) and validated its pro-apoptotic effect in lung
adenocarcinoma cells. Interestingly, we observed tLivin was further
cleaved into a smaller fragment in cells and the cleavage was
supposed to be mediated by caspases. Accordingly, the treatment
with the caspase inhibitor or introduction of a site-directed
mutagenesis of 220Asp to 220Ala abrogated the cleavage. Further,
the cleavage was reconstituted in vitro with the purified
recombinant tLivin protein and active caspase-3 protein, a reaction
that was abrogated by zVAD-fmk. We further constructed the
expression plasmid for the cleaved C-terminal fragment (livC) and
found it exerted an anti-apoptotic effect.
The first 52 amino acids was removed from Livin-β
and a truncated form (tLivin) with pro-apoptotic effects was
produced thereafter (15). The
cleavage was proposed to be mediated by caspase-3 or another
non-canonical caspase (18). Our
study observed the pro-apoptotic fragment tLivin activates caspases
and is cleaved into even smaller fragments by the latter. The
C-terminal fragment produced by the second cleavage was with
anti-apoptotic activity which was possibly attributed to its
ubiquitination activity. Cells may use this negative feed-back
mechanism to avoid the magnification of cell death signal. Thus,
our study reveals a complex interaction between Livin-β and
caspases and helps to dissect the full functional spectrum of
Livin-β in cells.
Nachimias et al(15) reported that the Livin-β was cleaved
in vivo by caspase-3, however, in their experiments the
site-directed mutagenesis of D220A failed to abrogate the cleavage.
In our study the mutant of tLivinD220A lost the potential of being
cleaved by active form of caspase-3. This difference, we thought,
was due to the different recognition efficiency of substrate
sequence. In fact, the optimal sequence for caspase-3 recognition
was DEVD (19). The sequence
neighboring amino acid 52 of Livin-β was DVHD, which was similar to
the original sequence to a greater extent than that of neighboring
amino acid 220, GARD. This possibly explained the observed cleavage
in our experiment where the optimal sequence had been removed.
We found the C-terminus of Livin-β, containing the
RING motif, exerted anti-apoptotic effects on cells. Similarly, the
function of RING motif has been discussed for other members of IAP
family. Suzuki et al reported the RING motif in XIAP
promotes proteasomal degradation of caspase-3 and enhances the
anti-apoptotic effect of XIAP (20). On the contrary, Silke et al
failed to observe such an effect (21). In their study, the RING motif in
XIAP mediated the degradation of other IAPs and actually promoted
cell death. From these reports and others (22), one may speculate the RING motif
probably have variant impact on cells, depending on different cell
type and stimuli. Our results were supported by a report that RING
motif in Livin-β enhanced cell survival and this was possibly
through the degradation of Smac/DIABLO (23).
Livin-β was reported to exert anti-apoptotic effects
against detrimental stimuli (5–7),
however controversies remain on the mechanism underlying its
anti-apoptotic activity. It was initially reported to suppress
downstream effector caspase-3 and-7 through its BIR domain
(5–7). However, later experiments failed to
prove this notion. Vucic et al reported Livin merely weakly
inhibited caspase-9 activity with inhibition constant Ki
∼3–5 μM. Also, it was argued that Livin might regulate
apoptosis by sequestering SMAC from XIAP (24). Another report awaiting for further
confirmation stated that inhibition of apoptosis by Livin might
involve the TAK1/JNK1 pathway (25). In our study, we found that the
C-terminal fragment potently inhibits apoptosis induced by
cisplatin. We propose it was attributed to the decreased expression
level of caspase-3 which was possibly related to the ubiquitination
property of the RING domain, probably the anti-apoptotic effect of
Livin-β was also partially attributed to the RING domain.
Previous reports indicated Livin-β was a cytoplasmic
protein and tLivin was also localized in the cytoplasm fraction
(6,26). These reports were in good agreement
with the present study. Interestingly, we also found the fragment
containing only the RING distributed diffusely throughout the whole
cell. The findings argue for the region flanking of the BIR domain
to be critical for proper subcellular localization of ML-IAP and
tLivin. Although lacking of data to demonstrate the relationship
between the subcellular localization and the different biological
properties of fragments of Livin-β, similar studies have been
proposed for other key molecules in apoptosis regulation. For
example, Smac/DIABLO located in the mitochondria and was released
into the cytoplasm during apoptosis (27). The different nuclear and
cytoplasmic localization pattern of Survivin determined its
different action in apoptosis (14). We might reason that the proper
localization of Livin-β was important for its function in
vivo.
Livin-β has been proposed as critical for
therapeutic resistance in tumors (28). In addition, the prognostic value of
Livin-β was examined in tumor samples retrospectively at the mRNA
or protein level (9–12). The Livin-β expression was
associated with poor prognosis at least in lung, bladder and
nasopharyngeal cancer. Efforts were devoted to interfere with the
activity of Livin-β in order to improve the prognosis of cancer
patients with Livin-β expression. siRNA or small peptide were
developed capable of efficiently disrupting the function of Livin-β
in cells, rendering cells vulnerable to treatment (29,30).
Cancer gene therapy and cancer vaccine based on Livin-β were also
tested in preclinical models (31,32).
All these studies highlight the value of Livin-β as a therapeutic
target. Because tLivin has pro-apoptotic effects, its potential
therapeutic usage has been proposed (33). However, the present study provides
a deeper insight into this scenario. We suggest the C-terminal
fragment of tLivin has protective effects on cells and impedes the
therapeutic benefits. More efforts are needed to fulfill the
maximum therapeutic potential of tLivin.
In conclusion, the present study focused on the
fragment of Livin-β without its N-terminal 52 amino acids tLivin.
It was shown that tLivin localized in the cytoplasm and it was
further cleaved by caspase-3 to an anti-apoptotic factor. Our study
may contribute to the elucidation of functional spectrum of
Livin-β.
Acknowledgements
The authors thank Professor G.S.
Salvesen for providing the expression plasmid for caspase-3 and
Professor B. Vogelstein for the HCT-116p53−/− cells.
This study was supported by grants from National Natural Science
Fund of China (81272684, 30901756) and Doctoral Fund of Ministry of
Education of China (20090181120100).
References
|
1
|
Ghobrial IM, Witzig TE and Adjei AA:
Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin.
55:178–194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhivotovsky B and Orrenius S:
Carcinogenesis and apoptosis: paradigms and paradoxes.
Carcinogenesis. 27:1939–1945. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taylor RC, Cullen SP and Martin SJ:
Apoptosis: controlled demolition at the cellular level. Nat Rev Mol
Cell Biol. 9:231–241. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Srinivasula SM and Ashwell JD: IAPs:
what’s in a name? Mol Cell. 30:123–135. 2008.
|
|
5
|
Kasof GM and Gomes BC: Livin, a novel
inhibitor of apoptosis protein family member. J Biol Chem.
276:3238–3246. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vucic D, Stennicke HR, Pisabarro MT, et
al: ML-IAP, a novel inhibitor of apoptosis that is preferentially
expressed in human melanomas. Curr Biol. 10:1359–1366. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin JH, Deng G, Huang Q and Morser J:
KIAP, a novel member of the inhibitor of apoptosis protein family.
Biochem Biophys Res Commun. 279:820–831. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashhab Y, Alian A, Polliack A, et al: Two
splicing variants of a new inhibitor of apoptosis gene with
different biological properties and tissue distribution pattern.
FEBS Lett. 495:56–60. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanabe H, Yagihashi A, Tsuji N, et al:
Expression of survivin mRNA and livin mRNA in non-small-cell lung
cancer. Lung Cancer. 46:299–304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gazzaniga P, Gradilone A, Giuliani L, et
al: Expression and prognostic significance of Livin, Survivin and
other apoptosis-related genes in the progression of superficial
bladder cancer. Ann Oncol. 14:85–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choi J, Hwang YK, Sung KW, et al:
Expression of Livin, an anti-apoptotic protein, is an independent
favorable prognostic factor in childhood acute lymphoblastic
leukemia. Blood. 109:471–47. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiang Y, Yao H, Wang S, et al: Prognostic
value of Survivin and Livin in nasopharyngeal carcinoma.
Laryngoscope. 116:126–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong J, Chen N, Zhou Q, et al: Melanoma
inhibitor of apoptosis protein is expressed differentially in
melanoma and melanocytic naevus, but similarly in primary and
metastatic melanomas. J Clin Pathol. 58:1081–1085. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Chen N, Wang X, et al: Apoptosis
and proliferation markers in diffusely infiltrating astrocytomas:
profiling of 17 molecules. J Neuropathol Exp Neurol. 65:905–913.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nachmias B, Ashhab Y, Bucholtz V, et al:
Caspase-mediated cleavage converts Livin from an antiapoptotic to a
proapoptotic factor: implications for drug-resistant melanoma.
Cancer Res. 63:6340–6349. 2003.PubMed/NCBI
|
|
16
|
Zhou Q, Krebs JF, Snipas SJ, et al:
Interaction of the baculovirus anti-apoptotic protein p35 with
caspases. Specificity, kinetics and characterization of the
caspase/p35 complex. Biochemistry. 37:10757–1065. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vindeløv LL, Christensen IJ and Nissen N:
Standardization of high-resolution flow cytometric DNA analysis by
the simultaneous use of chicken and trout red blood cells as
internal reference standards. Cytometry. 3:328–331. 1983.PubMed/NCBI
|
|
18
|
Yan H, Brouha B, Liu T, et al: Proteolytic
cleavage of Livin (ML-IAP) in apoptotic melanoma cells potentially
mediated by a non-canonical caspase. J Dermatol Sci. 43:189–200.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Timmer JC and Salvesen GS: Caspase
substrates. Cell Death Differ. 14:66–72. 2007. View Article : Google Scholar
|
|
20
|
Suzuki Y, Nakabayashi Y and Takahashi R:
Ubiquitin-protein ligase activity of X-linked inhibitor of
apoptosis protein promotes proteasomal degradation of caspase-3 and
enhances its anti-apoptotic effect in Fas-induced cell death. Proc
Natl Acad Sci USA. 98:8662–8667. 2001. View Article : Google Scholar
|
|
21
|
Silke J, Kratina T, Chu D, et al:
Determination of cell survival by RING-mediated regulation of
inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci
USA. 102:16182–16187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang H, Joazeiro CA, Bonfoco E, et al:
The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein
ligase and promotes in vitro monoubiquitination of caspases 3 and
7. J Biol Chem. 275:26661–26664. 2000.PubMed/NCBI
|
|
23
|
Ma L, Huang Y, Song Z, et al: Livin
promotes Smac/DIABLO degradation by ubiquitin-proteasome pathway.
Cell Death Differ. 13:2079–2088. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vucic D, Franklin MC, Wallweber HJ, et al:
Engineering ML-IAP to produce an extraordinarily potent caspase 9
inhibitor: implications for Smac-dependent anti-apoptotic activity
of ML-IAP. Biochem J. 385:11–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sanna MG, da Silva Correia J, Ducrey O, et
al: IAP suppression of apoptosis involves distinct mechanisms: the
TAK1/JNK1 signaling cascade and caspase inhibition. Mol Cell Biol.
22:1754–1766. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nachmias B, Lazar I, Elmalech M, et al:
Subcellular localization determines the delicate balance between
the anti- and proapoptotic activity of Livin. Apoptosis.
12:1129–1142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang QH and Du C: Smac/DIABLO selectively
reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and
livin in HeLa cells. J Biol Chem. 279:16963–16970. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Crnkovic-Mertens I, Muley T, Meister M, et
al: The anti-apoptotic livin gene is an important determinant for
the apoptotic resistance of non-small cell lung cancer cells. Lung
Cancer. 54:135–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crnkovic-Mertens I, Bulkescher J, Mensger
C, et al: Isolation of peptides blocking the function of
anti-apoptotic Livin protein. Cell Mol Life Sci. 67:1895–1905.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Crnkovic-Mertens I, Hoppe-Seyler F and
Butz K: Induction of apoptosis in tumor cells by siRNA-mediated
silencing of the livin/ML-IAP/KIAP gene. Oncogene. 22:8330–8336.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang R, Lin F, Wang X, et al: Silencing
Livin gene expression to inhibit proliferation and enhance
chemosensitivity in tumor cells. Cancer Gene Ther. 15:402–412.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie J, Xiong L, Tao X, et al: Antitumor
effects of murine bone marrow-derived dendritic cells infected with
xenogeneic livin alpha recombinant adenoviral vectors against Lewis
lung carcinoma. Lung Cancer. 68:338–345. 2010. View Article : Google Scholar
|
|
33
|
Wang L, Zhang Q, Liu B, et al: Challenge
and promise: roles for Livin in progression and therapy of cancer.
Mol Cancer Ther. 7:3661–3669. 2008. View Article : Google Scholar : PubMed/NCBI
|