Introduction
Pancreatic cancer is the most lethal common
malignancy, with estimated 43,920 new cases and 37,390 deaths
occurring in the United States in 2012 (1). Despite the standardization of
surgical techniques and advances in systemic treatments, <5% of
patients survive 5 years after diagnosis; and this survival rate
has remained unchanged for 40 years (2). Furthermore, <20% of patients are
diagnosed with localized, potentially curable tumors at
presentation; while 80–85% of patients present with an inoperable
disease and rapidly succumb to this malignancy (3). In addition, pancreatic cancer
responds poorly to most chemotherapeutic agents (3). Hence, there is an urgent need for a
better understanding of the molecular mechanisms that contribute to
pancreatic cancer development and progression as well as for new
potential diagnostic and prognostic tumor markers.
Epithelial-mesenchymal transition (EMT) plays an
important role in human physiology and pathophysiology in processes
such as organ development, wound healing, organ fibrosis and cancer
progression (4–6). This process is accompanied by
dramatic changes in cellular morphology, the loss and remodeling of
cell-cell and cell-matrix adhesions and the gain of migratory and
invasive capabilities (4–7). In pancreatic cancer, induction of EMT
leads to acquisition of invasive, metastatic properties as well as
chemoresistance (8–10). Therefore, EMT might be an important
mechanism involved in pancreatic cancer progression and might
contribute to its poor prognosis. All these findings suggest that
characterization of EMT effectors is likely to yield new insights
into metastasis and novel avenues for treatment of pancreatic
cancer.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that post-transcriptionally regulate gene expression by
pairing with complementary nucleotide sequences in the
3′-untranslated region (3′-UTR) of target mRNAs (11). Several previous studies have
revealed that miRNAs play an important role in EMT and repress
target mRNAs through translational downregulation and deadenylation
(12–14).
Carcinoembryonic antigen-related cell adhesion
molecule 6 (CEACAM6; 19q13.2) is a glycosylphosphatidylinositol
(GPI)-linked immunoglobulin superfamily member. There is
accumulating evidence that CEACAM6 is overexpressed in several
epithelial carcinomas including colon, breast, non-small cell lung
cancer and intrahepatic cholangiocarcinoma (15–19).
In addition, it is involved in many crucial cellular events such as
migration, invasion and tumorigenicity (20,21).
Recent studies have suggested that CEACAM6 plays important roles in
pancreatic cancer development and progression. Indeed,
adenocarcinoma gene expression profiling studies have shown a 20-
to 25-fold higher expression of CEACAM6 compared to normal
pancreatic ductal epithelial cells (22). Moreover, deregulated overexpression
of CEACAM6 has been shown to inhibit differentiation and anoikis
(20). Conversely, knockdown of
CEACAM6 has been shown to reverse anoikis resistance and inhibit
the metastatic potential in pancreatic cancer mouse xenograft
models in vivo by enhancing caspase-3-mediated apoptosis
(21). In addition, CEACAM6 gene
silencing markedly increased sensitivity to gemcitabine-mediated
cytotoxicity (23).
Nevertheless, there are no previous studies on the
role of CEACAM6 in pancreatic cancer EMT and the mechanisms
regulating CEACAM6 expression in tumor progression still remain to
be elucidated.
In the present study, we demonstrated that CEACAM6
is an important regulator of pancreatic cancer EMT, migration and
invasion in vitro and metastasis in vivo.
Furthermore, we showed that CEACAM6 might be a miR-29a/b/c target
gene in the pancreatic cancer cell line CFPAC-1.
Materials and methods
Cell culture
Human pancreatic cancer cell lines CFPAC-1 and
PANC-1 were purchased from Shanghai Cell Bank (Shanghai, China) and
cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS)
(Sigma, St. Louis, MO, USA), 2 mM glutamine, 100 μg/ml penicillin
and 100 μg/ml streptomycin. All cells were incubated at 37°C in a
humidified chamber supplemented with 5% CO2.
Immunohistochemistry
Pancreatic cancer tissue samples were obtained from
99 patients undergoing a pancreatectomy for pancreatic cancer at
the First Affiliated Hospital of Nanjing Medical University between
2008 and 2010 and were confirmed by a pathologist. All patients
provided informed consent for their participation in the study,
which was approved by the Ethics Committee of Nanjing Medical
University, China.
For the immunohistochemistry analysis, 4-μm thick
paraffin-embedded tissue sections were deparaffinized in xylene,
rehydrated in graded alcohol and blocked in methanol containing 3%
hydrogen peroxide. The slides were covered with a blocking solution
for 1 h at room temperature and incubated with mouse anti-human
CEACAM6 monoclonal antibody (Abcam, Cambridge, MA, USA) or mouse
anti-human E-cadherin monoclonal antibody (Abcam) for 2 h at 37°C.
After rinsing with phosphate-buffered saline (PBS; pH 7.4)
solution, sections were treated with a goat anti-mouse secondary
antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at
37°C. Next, the slides were incubated with 3,3-diaminobenzidine
(DAB) solution for 10 min and then counterstained with hematoxylin.
CEACAM6 and E-cadherin expression were quantified using Image-Pro
Plus version 6.0 (Media Cybernetics, Inc., Bethesda, MD, USA).
Generation of stable cell lines
For the generation of stable cell lines in our
study, CEACAM6 lentiviral constructs were amplified using PrimeSTAR
HS DNA Polymerase (Takara, DR010A, Dalian, China) and ligated into
the Lv-CMV-EGFP vector. The shRNAs for human CEACAM6 were designed
in our lab and constructed in pLKO.1-puro vectors. Three shRNA
plasmids were constructed against different CEACAM6 coding sequence
(CDS) regions and a scrambled sequence was made as a negative
control. All plasmids were verified by sequencing (Invitrogen).
After infection with lentivirus, cells were tested for CEACAM6 gene
overexpression or knockdown efficiency. One construct with ≥80%
knockdown efficiency was selected and used in further studies. The
shRNA sequences used in knockdown studies were as follows:
shCEACAM6 (sense: 5′-GCCCCAGAAUCGUAUUGGUTT-3′ and antisense:
5′-ACCAAUACGAUUCUGGGGCTT-3′) and shCont (sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense:
5′-ACGUGACACGUUCGGAGAATT-3′).
Real-time quantitative reverse
transcription PCR (qRT-PCR)
For the real-time quantitative RT-PCR analysis,
total RNA was extracted using TRIzol reagent (Invitrogen) and cDNA
was synthesized using the PrimeScript RT kit (Takara). Real-time
quantitative reverse transcription PCR (qRT-PCR) was performed
using a FastStart Universal SYBR Green Master (Rox) (Roche, USA)
and ABI PRISM 7500 Sequence Detection System (Applied Biosystems,
Life Technologies Corp., CA, USA). The relative expression of mRNA
was examined as the inverse log of the ΔΔCt and normalized to the
reference gene, GAPDH. Primers for qPCR were synthesized by
Invitrogen (Shanghai, China) and the sequences were as follows:
CEACAM6 sense: 5′-AGAAGCTAGCAGAGACCATGGGACCC-3′, antisense:
5′-AAATTCTAGAGGGCTGCTATATCAGAGCC-3′. GAPDH sense:
5′-TCACCCACACTGTGCCCATCTACGA-3′, antisense:
5′-CAGCGGAACCGCTCATTGCCAATGG-3′. The other primers are available
upon request.
The miR-29a/b/c level was quantified by qRT-PCR
using a TaqMan probe (Applied Biosystems, Foster City, CA, USA),
with RNU6B small nuclear RNA as an internal reference. Their
relative levels were analyzed in triplicate on an ABI PRISM 7900
Sequence Detection System (Applied Biosystems), according to the
manufacturer’s protocol.
Western blot analysis
For the western blot analysis, cells were lysed
using a RIPA buffer with 1% phenylmethanesulfonyl fluoride (PMSF).
Protein concentration was measured using a BCA kit (Keygen,
Nanjing, China). Equal amounts of protein (30 μg) were resolved
with 10% SDS-PAGE and transferred to polyvinylidene difluoride
(PVDF) membranes (Millipore, Bedford, MA, USA). Membranes were
probed with primary antibodies for 12 h at 4°C and then incubated
with secondary antibodies for 2 h at room temperature. CEACAM6
(1:250), E-cadherin (1:1,000), vimentin (1:500) and ZEB1 (1:100)
antibodies were from Abcam and the ZEB2 (1:200) antibody was from
Santa Cruz Biotechnology. The goat anti-rabbit and goat anti-mouse
secondary antibodies were from Beyotime (Nantong, China). GAPDH
antibody (1:500) (Beyotime) was used as an internal control.
Electrochemiluminescence was performed with a ChemiImager 5500
imaging system (Alpha Innotech Co., San Leandro, CA, USA).
Target prediction and microRNA
transfection
Three online programs, TargetScan (http://www.targetscan.org), Microcosm Targets
(http://www.ebi.ac.uk) and microRNA (http://www.microrna.org) were used for predicting
miRNAs that might target CEACAM6. CFPAC-1 cells overexpressing
CEACAM6 were used for target miRNA verification. The miRNA mimics
were designed and synthesized by Genepharma (Shanghai, China). The
miR-29a/b/c mimics and the negative control were as follows:
miR-29a sense: 5′-UAGCACCAUCUGAAAUCGGUUA-3′ and antisense:
5′-ACCGAUUUCAGAUGGUGCUAUU-3′; miR-29b sense:
5′-UAGCACCAUUUGAAAUCAGUGUU-3′ and antisense:
5′-CACUGAUUUCAAAUGGUGCUAUU-3′; miR-29c sense:
5′-UAGCACCAUUUGAAAUCGGUUA-3′ and antisense:
5′-ACCGAUUUCAAAUGGUGCUAUU-3′; negative control sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense:
5′-ACGUGACACGUUCGGAGAATT-3′.
MicroRNA transfection was performed using
Lipofectamine 2000 (Invitrogen). In brief, CPFAC-1 cells were grown
in 6-well plates to 50% confluency before transfection. Total RNA
and proteins were extracted at 48 h post-transfection and used for
qRT-PCR and western blot analysis, respectively.
Luciferase reporter assay
For the luciferase reporter assay, reporter plasmids
were constructed by ligating 60-bp synthetic oligonucleotides
(Invitrogen) containing putative miRNA binding sites from the human
CEACAM6 3′-UTR or their mutant versions into XbaI-FseI sites of the
pGL3-control vector (Promega, Madison, WI, USA). Cells were plated
at 1.5×105 cells/well in 24-well plates 24 h before
transfection. Cells were transfected with 200 ng of luciferase
reporter plasmid plus 80 ng of pRL-TK (Promega) in combination with
60 pmol of the microRNA mimics or negative control using
Lipofectamine 2000 (Invitrogen). Luciferase activity was measured
48 h after transfection using the Dual-Luciferase Reporter Assay
System (Promega). Firefly luciferase activity was normalized to
renilla luciferase activity for each transfected well.
Cell proliferation assay
Cell proliferation was assessed by the MTT assay.
Cells were plated at 1,000 cells/well on 96-well plates. Twenty
microliters of MTT (5 mg/ml) was added to each well and plates were
incubated for 4 h at 37°C, then 200 μl of DMSO was added to each
well and plates were agitated for 15 min. The optical density (OD)
value of each well was determined by measuring the absorbance,
respectively, at 492 and 620 nm (reference). Survival percentage
(%) was calculated relative to the control.
Cell migration and invasion assay
The cell migration assay was performed using 6.5-mm
chambers with 8-μm pores (Corning, Corning, NY, USA). In brief,
cells were seeded in the upper chambers in serum-free DMEM
(1×104 cells in 200 μl) and 600 μl of 10% FBS-DMEM was
added into the lower wells. After 24 h at 37°C, cells migrating to
the bottom of the membrane were stained with 0.1% crystal violet in
methanol. Images of three random ×10 magnification fields were
captured from each membrane and the number of migratory cells was
counted. For the cell invasion assay, similar inserts coated with
Matrigel were used to determine the invasive potential of the
cells. All experiments were done in triplicate.
Orthotopic pancreatic tumor xenograft
model
Athymic nude mice (BALB/cA-nu (nu/nu))
(4–6-week-old) were purchased from the Nanjing Medical University
Animal Center (Nanjing, China). Mice were anesthetized with 2.5%
avertin and the injection site was cleaned with 70% ethanol. A 1-cm
incision was made in the left subcostal region and the pancreas was
exposed. A solution of 1×106 PANC-1-CEACAM6 or
PANC-1-Cont cells in 30 μl of PBS was injected into the tail of the
pancreas (ten mice per group). The peritoneum and skin were closed
with a 4-/T0 surgical suture. Four weeks post-inoculation, all
surviving mice were sacrificed and evaluated macroscopically for
the presence of orthotopic tumors and metastases in the liver.
Tumor volumes were determined by the formula: tumor volume
(mm3) = [length (mm)] × [width (mm)]2 × 0.52
(24).
Statistical analysis
All experiments were repeated in triplicate. All
values were expressed as mean ± standard deviation (SD).
Statistical significance was determined using the Student’s t-test,
Kaplan-Meier survival analysis, log-rank test and Spearman
correlation using SPSS 17.0 (Chicago, IL, USA). P<0.05 were
considered as statistically significant.
Results
CEACAM6 expression in pancreatic cancer
is correlated with clinicopathological characteristics and the EMT
marker E-cadherin
In this study, we examined the expression of CEACAM6
in 99 pancreatic tumor tissue samples by immunohistochemistry.
Positive CEACAM6 immunohistochemical reaction was localized to the
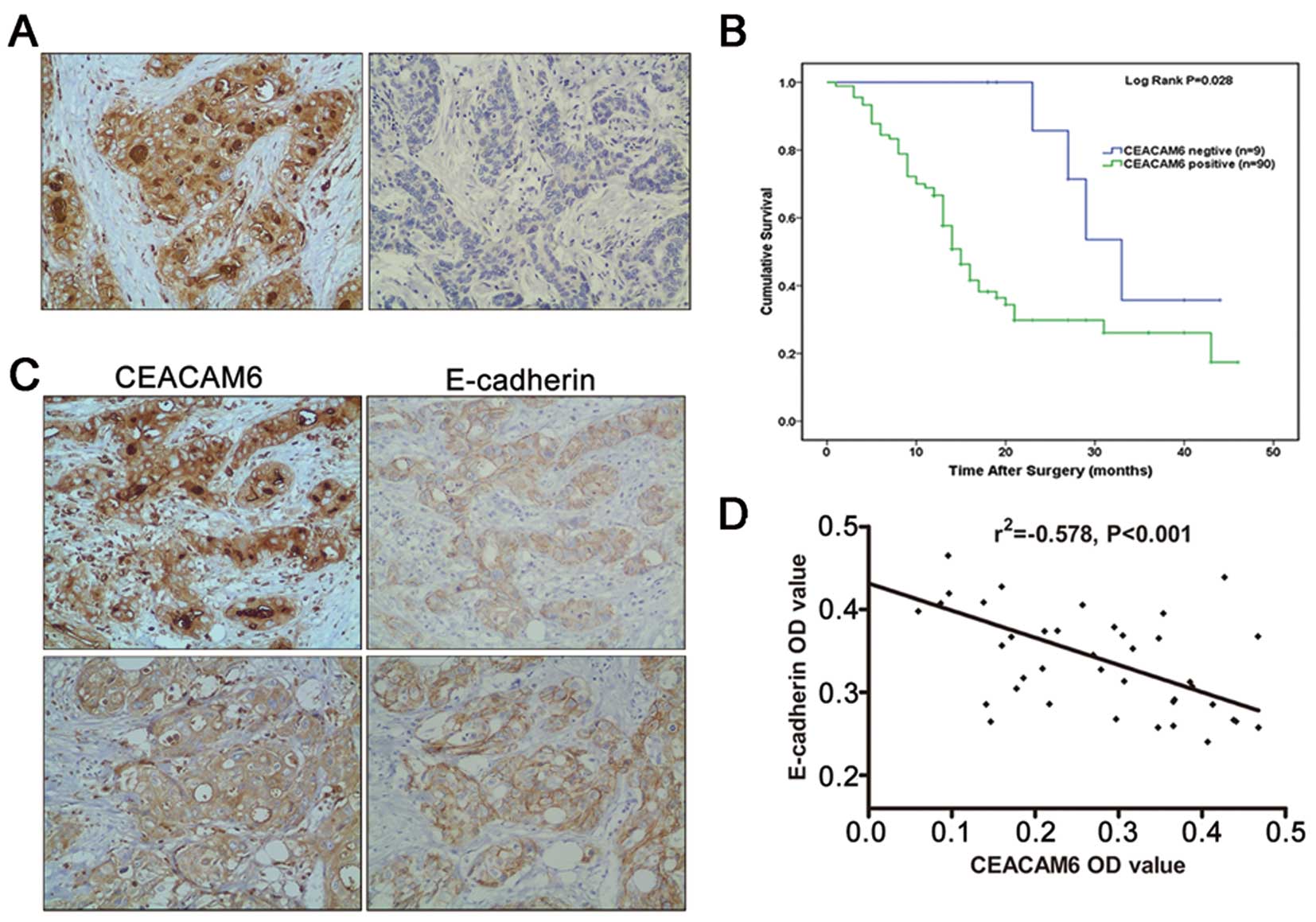
membrane and cytoplasm of tumor cells (Fig. 1A) and was detected in 90.9% (90/99)
of samples. Furthermore, we examined the correlation between
CEACAM6 expression and the clinicopathological characteristics of
patients. The results of this analysis are summarized in Table I.
 | Table IAssociation between CEACAM6
imunohistochemical expression and clinicopathological
characteristics of pancreatic cancer patients. |
Table I
Association between CEACAM6
imunohistochemical expression and clinicopathological
characteristics of pancreatic cancer patients.
| CEACAM6
expression | |
|---|
|
| |
|---|
| Characteristic | Positive | Negative | P-value |
|---|
| Gender | | | 0.563 |
| Male | 51 | 6 | |
| Female | 39 | 3 | |
| Age (years) | | | 0.703 |
| ≤60.8 | 46 | 4 | |
| >60.8 | 44 | 5 | |
| Size (cm) | | | 0.898 |
| ≤3.78 | 52 | 5 | |
| >3.78 | 38 | 4 | |
|
Differentiation | | | 0.041 |
| Poor | 8 | 0 | |
| Moderate | 75 | 6 | |
| Well | 7 | 3 | |
| Positive lymph
nodes | | | 0.019 |
| No | 43 | 8 | |
| Yes | 47 | 1 | |
| Perineural
invasion | | | 0.295 |
| No | 25 | 4 | |
| Yes | 65 | 5 | |
| Stage T1/T2/T3 | | | 0.060 |
| T1 | 19 | 5 | |
| T2 | 40 | 3 | |
| T3 | 31 | 1 | |
| Location | | | 0.295 |
| Head | 65 | 5 | |
| Body and
limbs | 25 | 4 | |
In brief, CEACAM6 expression correlated with tumor
differentiation and positive lymph node status (P<0.05);
however, no correlation of CEACAM6 expression with patients’ age,
gender, tumor location, tumor size, perineural invasion, or T stage
was observed (P>0.05). Furthermore, according to the
Kaplan-Meier test, patients with CEACAM6-negative tumors had
significantly longer overall survival, compared with those with
CEACAM6-positive tumors (P<0.05) (Fig. 1B).
Additionally, we examined the expression of the EMT
marker E-cadherin by immunohistochemistry and correlated it to
CEACAM6 expression. A positive immunohistochemical reaction for
E-cadherin was observed mainly on membranes of normal glands and
cancer cells (Fig. 1C). Pearson
correlative analysis indicated significantly negative correlations
between CEACAM6 and E-cadherin expression (P<0.01) (Fig. 1D).
CEACAM6 promotes EMT in pancreatic cancer
cells
To determine the potential role of CEACAM6 in
regulating EMT in pancreatic cancer, we analyzed the influence of
CEACAM6 overexpression and silencing in PANC-1 and CFPAC-1
pancreatic cancer cell lines, respectively.
To analyze the influence of CEACAM6 overexpression,
we transfected the CEACAM6 expression vector Lv-CMV-EGFP-CEACAM6 or
the control vector Lv-CMV-EGFP into PANC-1 cells, which typically
express low levels of CEACAM6. The overexpression of CEACAM6 in
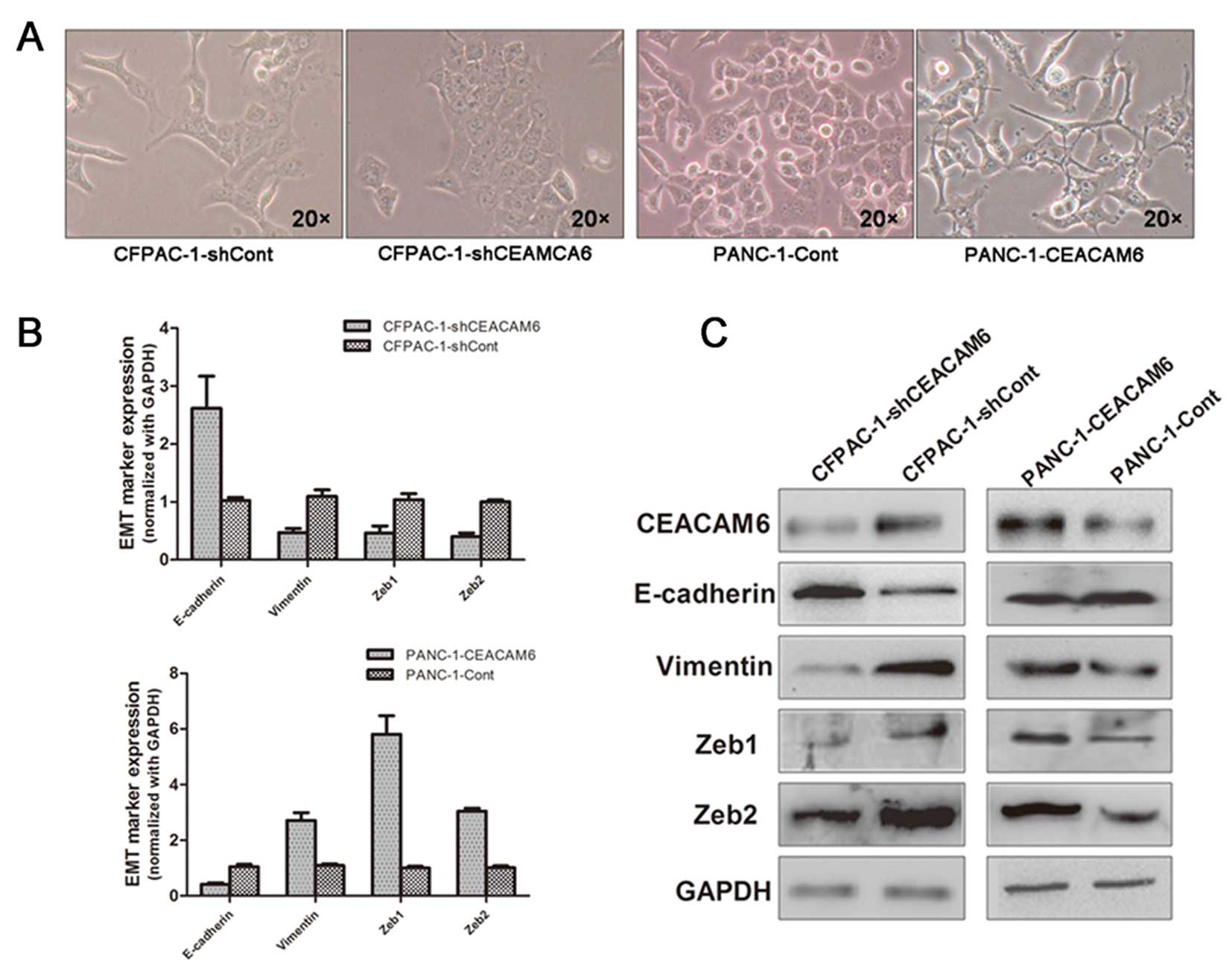
PANC-1 cells induced loose cell contact and spindle-shaped
morphology reminiscent of EMT, while cells transfected with the
control vector maintained the cobblestone-like morphology (Fig. 2A). Next, we observed that elevated
expression of CEACAM6 significantly increased the expression of the
mesenchymal marker vimentin but decreased the expression of the
epithelial marker E-cadherin (Fig. 2B
and C, respectively).
Furthermore, in the silencing experiment, we
transfected the pLKO.1-puro-shCAECAM6 vector or the control vector
pLKO.1-puro-shScramble into CFPAC-1 cells, which typically express
high levels of CEACAM6. Knockdown of CEACAM6 in CFPAC-1 cells led
to typical transition from mesenchymal to epithelial morphology and
a concomitant decrease in vimentin and increase in E-cadherin
expression, as evidenced by both qRT-PCR and western blot analysis.
Collectively, these findings indicate that altered CEACAM6
expression affects pancreatic cancer cell EMT in vitro.
Furthermore, we examined the levels of the known EMT
activators ZEB1 and ZEB2 in relation to CEACAM6 overexpression or
knockdown in pancreatic cancer cell lines. ZEB1 and ZEB2 expression
was significantly increased in PANC-1 cells overexpressing CEACAM6;
whereas in CFPAC-1 cells transfected with CEACAM6, the expression
of the silencing vectors ZEB1 and ZEB2 was repressed (Fig. 2B and C, respectively). These
results suggest a potential role of ZEB1 and ZEB2 in
CEACAM6-regulated EMT.
Functional role of CEACAM6 in pancreatic
cancer cell proliferation, migration and invasion in vitro
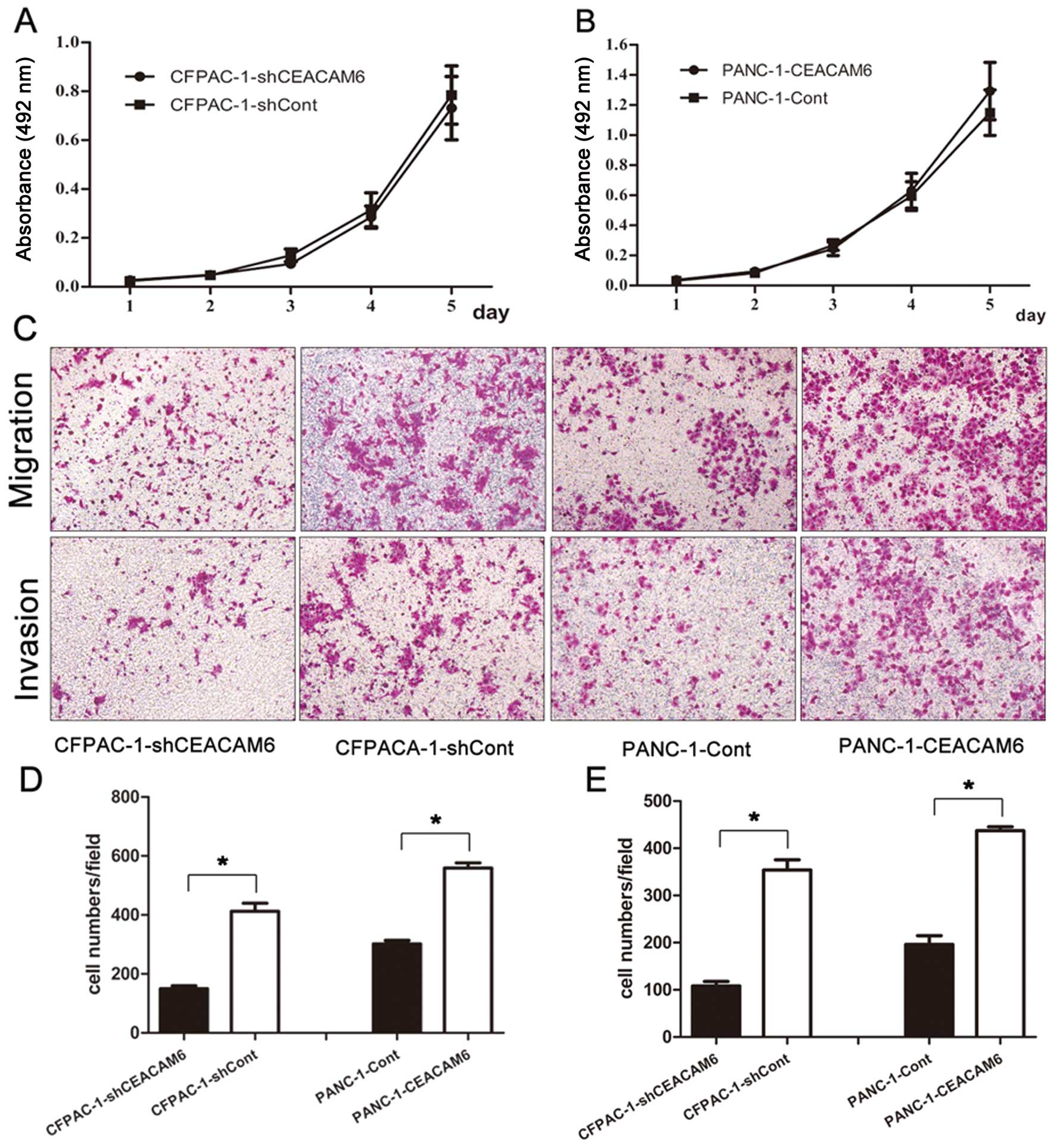
CEACAM6 knockdown and overexpression did not
markedly affect the proliferative ability of CFPAC-1 and PANC-1
cell lines (CFPACA-1-shCEACAM6 vs. CFPAC-1-shCont: 0.731±0.129 vs.
0.785±0.119, P=0.626; PANC-1-CEACAM6 vs. PANC-1-Cont: 1.293±0.190
vs. 1.149±0.150, P=0.364) (Fig. 3A and
B, respectively). Although the results were not consistent with
the impact of altered CEACAM6 expression on proliferation of
pancreatic cancer cells in vitro, the data showed that
overexpression of CEACAM6 promoted the migration (PANC-1-CEACAM6
vs. PANC-1-Cont: 559.1±51.9 vs. 301.6±36.2, P<0.01) and invasion
(PANC-1-CEACAM6 vs. PANC-1-Cont: 437.2±25.1 vs. 196.2±56.2,
P<0.01) abilities of PANC-1 cells (Fig. 3D and E, respectively), whereas
knockdown of CEACAM6 attenuated cell migration (CFPACA-1-shCEACAM6
vs. CFPAC-1-shCont: 149.7±30.3 vs. 412.2±83.1, P<0.01) and
invasion (CFPACA-1-shCEACAM6 vs. CFPAC-1-shCont: 108.2±27.9 vs.
354.1±64.0, P<0.01) in CFPAC-1 cells (Fig. 3C, D and E, respectively).
Overexpression of CEACAM6 enhances
metastatic ability of PANC-1 cells in vivo
To assess the significance of CEACAM6 expression
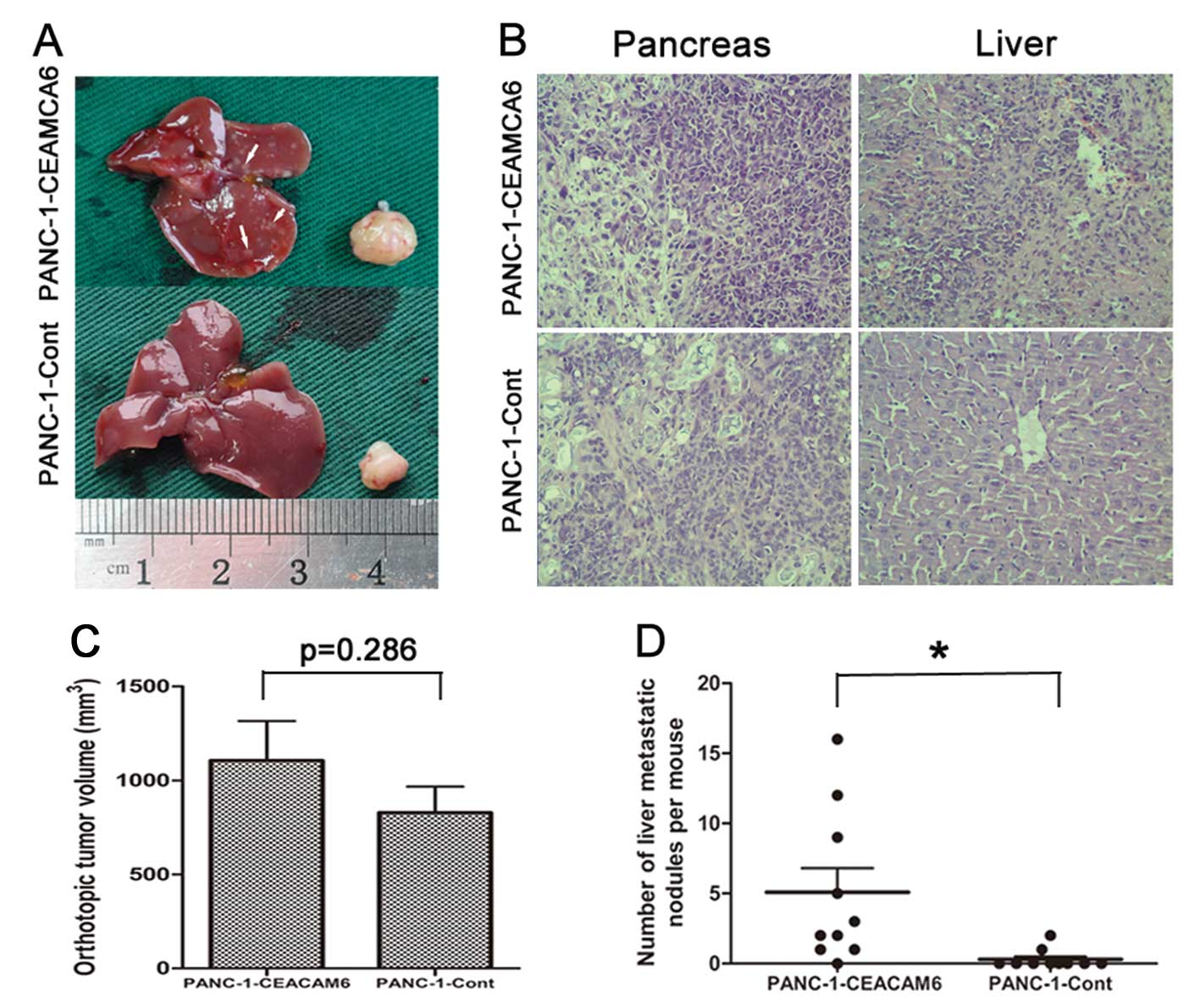
in vivo, PANC-1-CEACAM6 cells were orthotopically injected
into the pancreas of nude mice, while PANC-1-Cont cells were used
as a control. Four weeks after injection, mice were sacrificed and
tumor volume and metastatic liver nodules were counted and
confirmed histologically (Fig. 4A and
B, respectively).
The tumor volume showed no significant difference
between the two groups (PANC-1-CEACAM6 vs. PANC-1-Cont:
1105.5±666.7 mm3 vs. 828.5±439.2 mm3,
P=0.286) (Fig. 4C). Nevertheless,
a statistically significant difference in the mean metastatic liver
nodule number in PANC-1-CEACAM6 and PANC-1-Cont groups was observed
(5.10 and 0.30, respectively, P<0.05) (Fig. 4D).
Modulating effect of miR-29a/b/c on
CEACAM6 expression
Using bioinformatic tools (TargetScan, Microcosm
Targets and microRNA), we predicted that miR-29a/b/c might be the
most potent regulator of the CEACAM6 gene. Therefore, we decided to
test our hypothesis in CFPAC-1 cells using a constructed reporter
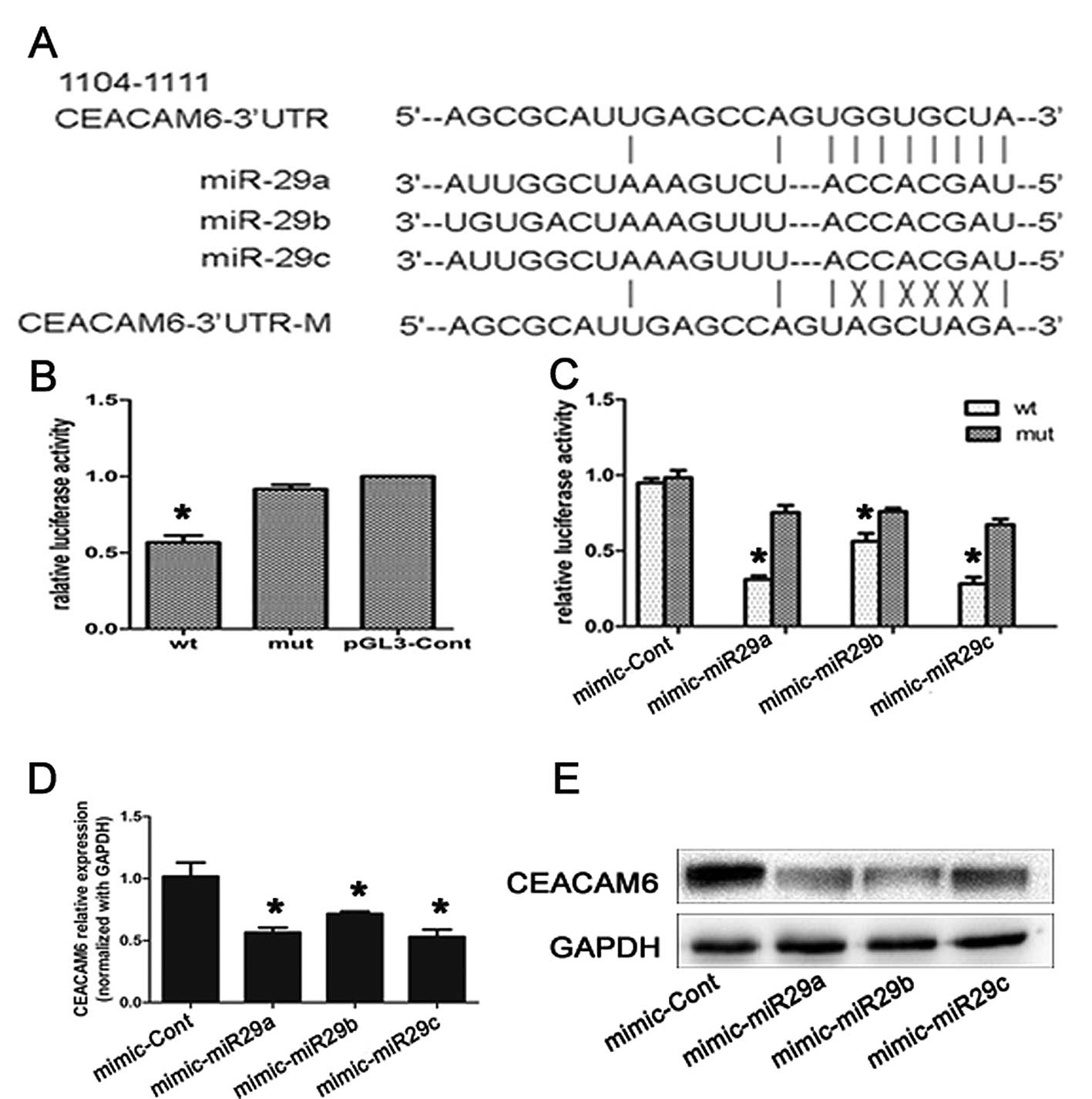
plasmid carrying the CEACAM6 wild-type and mutant-type 3′-UTR
region (Fig. 5A).
As shown in Fig. 5B and
C, miR-29a/b/c significantly suppressed luciferase activity
when the wild-type 3′-UTR of CEACAM6 was present (P<0.05). To
verify that miR-29a/b/c acts as a negative regulator of CEACAM6
translation, we transfected CFPAC-1 cells with miR-29a/b/c mimics
and tested the endogenous CEACAM6 mRNA and protein expression
levels by qRT-PCR and western blot analysis, respectively. CEACAM6
mRNA levels decreased 48 h after miR-29a/b/c transfection (Fig. 5D, P<0.05). Additionally, western
blot analysis showed that 48 h after transfection, overexpression
of miR-29a/b/c resulted in a significant decrease in CEACAM6
protein level (Fig. 5E). These
results collectively suggest that miR-29a/b/c may, at least in
part, be responsible for the regulation of CEACAM6 expression in
vitro.
Discussion
Pancreatic cancer is the tenth most common cancer
and the fourth most common cause of cancer mortality worldwide
(1). In the past few decades,
great efforts have been made to elucidate the molecular mechanisms
underlying its tumorigenicity, invasion and metastasis in order to
find new potential diagnostic and prognostic markers for early
detection as well as to develop new targeted anticancer therapies.
Nevertheless, the detailed mechanisms of pancreatic cancer
development and progression to metastasis still remain obscure.
Previous studies have shown that CEACAM6 is
overexpressed in many carcinomas, including pancreatic cancer
(15,18,19,25).
It has been suggested that CEACAM6 overexpression is associated
with greater resistance to anoikis and high cellular invasion
potential in vitro as well as higher metastatic potential
in vivo(21,26–28).
The reason why CEACAM6 overexpression is associated with aggressive
biological behavior of cancer cells has not been fully
clarified.
In the present study, we found that CEACAM6 was
highly expressed in most pancreatic cancer tissue samples and this
expression was closely associated with poor prognosis in pancreatic
cancer patients. In addition, we have for the first time
demonstrated that CEACAM6 directly impacts EMT, migration, invasion
and metastasis of pancreatic cancer cells. More importantly, our
study is the first to show that miR-29a/b/c can regulate CEACAM6 at
the post-transcriptional level.
Emerging evidence suggests that EMT is associated
with the loss of epithelial and gain of mesenchymal
characteristics, resulting in an increased invasive, metastatic and
chemo-resistance potential of tumor cells and thus having an
important role in cancer progression and prognosis (29,30).
In the present study, we found that CEACAM6 is highly expressed in
most pancreatic tumor tissues. Clinicopathological analysis
revealed that expression of CEACAM6 protein was significantly
related to tumor differentiation and lymph node metastasis. Our
results are in agreement with those of a previous study by Duxbury
et al in which the expression of CEACAM6 correlated with
tumor grade and positive lymph node status (25). In addition, the observed cell
morphology, molecular biomarkers and biological behavior found in
our study were consistent with EMT characteristics. Moreover, we
demonstrated that elevated CEACAM6 expression could contribute to
EMT phenotype acquisition characterized by the typical mesenchymal
morphology, through its influence on upregulation of the
mesenchymal cell marker vimentin and downregulation of the
epithelial cell marker E-cadherin. Conversely, decreased CEACAM6
expression in our study was associated with the reversal of EMT
through downregulation of vimentin and upregulation of E-cadherin.
Furthermore, these results are consistent with the observed
clinical data that showed a significantly negative correlation
between CEACAM6 and E-cadherin expression in 40 pancreatic cancer
tissues.
ZEB1 and ZEB2, two members of the ZEB family, are
important regulators of EMT and are implicated in the tumorigenesis
of many human cancers (12,31).
We found that ZEB1 and ZEB2 expression was significantly increased
in PANC-1 cells overexpressing CEACAM6. On the contrary, ZEB1 and
ZEB2 expression was repressed in CFPAC-1 cells in which the CEACAM6
was silenced. Based on these findings, we can speculate on the
possible role of CEACAM6 in EMT regulation through its effects on
ZEB1 and ZEB2.
The functional study of the role of CEACAM6 in
pancreatic cancer cell lines demonstrated that PANC-1 cells, which
typically express low levels of CEACAM6 when transfected with
CEACAM6 gene, have greater migratory and invasive abilities
compared to control-transfected cells. Furthermore, RNA
interference-mediated gene suppression of CEACAM6 in the
overexpressing pancreatic cancer cell line CFPAC-1 showed marked
reduction in migration and invasion capabilities of transfected
cells. These findings are consistent with our CEACAM6
immunohistochemistry results as well as in vivo experiments
on nude mouse models. In brief, the expression of CEACAM6 in our
study was associated with lymph node metastasis in pancreatic
cancer patients. Moreover, CEACAM6 overexpression in PANC-1 cells
enhanced their ability to form liver metastasis in nude mouse
models. Nevertheless, the proliferation ability of pancreatic
cancer cells was not affected with either the overexpression or
knockdown of CEACAM6 in vitro. This result is further
supported by our findings that CEACAM6 overexpression does not
influence the orthotopic tumor volume in nude mouse models.
Recent studies in colon cancer, cholangiocarcinoma,
hepatocellular carcinoma (HCC) and lung cancer have suggested that
miR-29 may have a significant role in tumor biology (32–35).
Indeed, Xiong et al have shown that miR-29 expression was
reduced in the majority of hepatocellular carcinomas included in
their study and its downregulation was significantly associated
with poor disease-free survival in HCC patients (36).
In our study, miR-29a/b/c overexpression induced a
significant downregulation of the CEACAM6 protein and mRNA levels
in vitro. In addition, the overexpression of miR-29a/b/c was
associated with suppression of luciferase-CEACAM6-3′-UTR activity,
indicating that CEACAM6 is a direct target of miR-29a/b/c.
In conclusion, our results suggest that CEACAM6
plays an important role in the progression and metastasis of human
pancreatic cancer by promoting EMT via the ZEB1/ZEB2 pathway. In
addition, we have for the first time shown that miR-29a/b/c can
regulate CEACAM6 at the post-transcriptional level. Therefore, we
conclude that targeting these signaling pathways may be a feasible
and effective approach for treatment of pancreatic cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (NO. NFSC 30972912).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar
|
|
3
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar
|
|
4
|
Savagner P: The epithelial-mesenchymal
transition (EMT) phenomenon. Ann Oncol. 21(Suppl 7): vii89–92.
2010.
|
|
5
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iwatsuki M, Mimori K, Yokobori T, et al:
Epithelial-mesenchymal transition in cancer development and its
clinical significance. Cancer Sci. 101:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rhim AD, Mirek ET, Aiello NM, et al: EMT
and dissemination precede pancreatic tumor formation. Cell.
148:349–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cano CE, Motoo Y and Iovanna JL:
Epithelial-to-mesenchymal transition in pancreatic adenocarcinoma.
ScientificWorldJournal. 10:1947–1957. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krantz SB, Shields MA, Dangi-Garimella S,
Munshi HG and Bentrem DJ: Contribution of epithelial-to-mesenchymal
transition and cancer stem cells to pancreatic cancer progression.
J Surg Res. 173:105–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim T, Veronese A, Pichiorri F, et al: p53
regulates epithelial-mesenchymal transition through microRNAs
targeting ZEB1 and ZEB2. J Exp Med. 208:875–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tellez CS, Juri DE, Do K, et al: EMT and
stem cell-like properties associated with miR-205 and miR-200
epigenetic silencing are early manifestations during
carcinogen-induced transformation of human lung epithelial cells.
Cancer Res. 71:3087–3097. 2011. View Article : Google Scholar
|
|
14
|
Li Y, Van den Boom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jantscheff P, Terracciano L, Lowy A, et
al: Expression of CEACAM6 in resectable colorectal cancer: a factor
of independent prognostic significance. J Clin Oncol. 21:3638–3646.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maraqa L, Cummings M, Peter MB, et al:
Carcinoembryonic antigen cell adhesion molecule 6 predicts breast
cancer recurrence following adjuvant tamoxifen. Clin Cancer Res.
14:405–411. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poola I, Shokrani B, Bhatnagar R, DeWitty
RL, Yue Q and Bonney G: Expression of carcinoembryonic antigen cell
adhesion molecule 6 oncoprotein in atypical ductal hyperplastic
tissues is associated with the development of invasive breast
cancer. Clin Cancer Res. 12:4773–4783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singer BB, Scheffrahn I, Kammerer R,
Suttorp N, Ergun S and Slevogt H: Deregulation of the CEACAM
expression pattern causes undifferentiated cell growth in human
lung adenocarcinoma cells. PLoS One. 5:e87472010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ieta K, Tanaka F, Utsunomiya T, Kuwano H
and Mori M: CEACAM6 gene expression in intrahepatic
cholangiocarcinoma. Br J Cancer. 95:532–540. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Strickland LA, Ross J, Williams S, et al:
Preclinical evaluation of carcinoembryonic cell adhesion molecule
(CEACAM) 6 as potential therapy target for pancreatic
adenocarcinoma. J Pathol. 218:380–390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duxbury MS, Ito H, Zinner MJ, Ashley SW
and Whang EE: CEACAM6 gene silencing impairs anoikis resistance and
in vivo metastatic ability of pancreatic adenocarcinoma
cells. Oncogene. 23:465–473. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iacobuzio-Donahue CA, Maitra A, Olsen M,
et al: Exploration of global gene expression patterns in pancreatic
adenocarcinoma using cDNA microarrays. Am J Pathol. 162:1151–1162.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duxbury MS, Ito H, Benoit E, Waseem T,
Ashley SW and Whang EE: A novel role for carcinoembryonic
antigen-related cell adhesion molecule 6 as a determinant of
gemcitabine chemoresistance in pancreatic adenocarcinoma cells.
Cancer Res. 64:3987–3993. 2004. View Article : Google Scholar
|
|
24
|
Fu X, Tao L, Li M, Fisher WE and Zhang X:
Effective treatment of pancreatic cancer xenografts with a
conditionally replicating virus derived from type 2 herpes simplex
virus. Clin Cancer Res. 12:3152–3157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duxbury MS, Matros E, Clancy T, et al:
CEACAM6 is a novel biomarker in pancreatic adenocarcinoma and PanIN
lesions. Ann Surg. 241:491–496. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duxbury MS, Ito H, Benoit E, Zinner MJ,
Ashley SW and Whang EE: Overexpression of CEACAM6 promotes
insulin-like growth factor I-induced pancreatic adenocarcinoma
cellular invasiveness. Oncogene. 23:5834–5842. 2004. View Article : Google Scholar
|
|
27
|
Duxbury MS, Ito H, Benoit E, Ashley SW and
Whang EE: CEACAM6 is a determinant of pancreatic adenocarcinoma
cellular invasiveness. Br J Cancer. 91:1384–1390. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lewis-Wambi JS, Cunliffe HE, Kim HR,
Willis AL and Jordan VC: Overexpression of CEACAM6 promotes
migration and invasion of oestrogen-deprived breast cancer cells.
Eur J Cancer. 44:1770–1779. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
the importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eastham AM, Spencer H, Soncin F, Ritson S,
Merry CL, Stern PL and Ward CM: Epithelial-mesenchymal transition
events during human embryonic stem cell differentiation. Cancer
Res. 67:11254–11262. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leshem O, Madar S, Kogan-Sakin I, et al:
TMPRSS2/ERG promotes epithelial to mesenchymal transition through
the ZEB1/ZEB2 axis in a prostate cancer model. PLoS One.
6:e216502011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cummins JM, He Y, Leary RJ, et al: The
colorectal microRNAome. Proc Natl Acad Sci USA. 103:3687–3692.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: mir-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Braconi C, Kogure T, Valeri N, et al:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.PubMed/NCBI
|