Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer in males and the second in females worldwide, and
is the fourth leading cause of cancer death in males and the third
in females worldwide (1). Although
several molecular target therapies have been developed for CRCs,
the prognosis of this disease in advanced stages is still poor.
Therefore, personalized therapies using biomarkers and new
molecular therapies are expected to improve current therapies. For
example, cetuximab, a therapeutic antibody to an epidermal growth
factor receptor (EGFR), was reported to be effective in CRC
treatment. The randomized phase III study (CRYSTAL: Cetuximab
Combined with Irinotecan in First-Line Therapy for Metastatic
Colorectal Cancer) and randomized phase II study (OPUS: Oxaliplatin
and Cetuximab in First-Line Treatment of metastatic CRC)
demonstrated that addition of cetuximab to first-line chemotherapy
in patients with KRAS wild-type CRC significantly improved the
treatment outcome compared with chemotherapy alone. However, the
benefit of cetuximab was limited to patients with KRAS wild-type
tumors. The overall survival (OS) rate in these trials was still
less than two years, and adverse events such as skin reactions,
infusion-related reactions and diarrhea were more frequent with
cetuximab plus chemotherapy than with chemotherapy alone (2,3).
Therefore, there is a need for personalized therapies that are more
effective for CRCs including the KRAS mutant, but with fewer
adverse events.
Oncoantigens are proteins with oncogenic functions
that are overexpressed in malignant cells of various origins and in
normal reproductive tissues such as the testis, ovary, and placenta
(4,5). Oncoantigens are considered a
candidate biomarker and therapeutic target for cancer therapy. We
performed genome-wide gene expression analyses and subsequent
tissue microarray analyses of solid tumor tissues using a cDNA
microarray containing 25,000 genes or expressed sequence tags
(ESTs) (6–10). To date, we have isolated several
oncoantigens involved in development and/or progression of cancer
(11–42). These data revealed that cell
division cycle-associated protein 1 (CDCA1) was
overexpressed in cancer tissues including CRC and lung cancer
tissues. In our previous reports, CDCA1 proteins were detected in
many lung cancers with varying histologic types and were associated
with a poorer prognosis for patients with non-small cell lung
carcinomas (NSCLC). Knockdown of CDCA1 expression with siRNA
significantly suppressed growth of NSCLC cell lines (16). In addition, the
HLA-A0201-restricted peptides derived from CDCA1 induced
peptide-specific cytotoxic T lymphocytes (CTLs), suggesting that
CDCA1 is a likely target for molecular therapy and/or immunotherapy
(39). CDCA1 mRNA
expression was also observed in CRCs and tissues of gastric
cancers, and was correlated with cancer growth (43). Moreover, CDCA1 was associated with
a decrease in progression-free survival of multiple myeloma
patients, and a decrease in probability of biochemical-free
survival in localized prostate cancer (44).
CDCA1 plays a role in regulating mitosis. CDCA1, and
its binding partner KNTC2, are members of the Ndc80 complex, which
comprises the two Ndc80 sub-complexes (KNTC2)-Nuf2 (CDCA1) and
Spc24-Spc25 (46). The CDCA1-KNTC2
complex is highly conserved in prokaryotic and eukaryotic cells,
and plays an important role in kinetochore functions and the
spindle checkpoint (46). Although
some reports describe CDCA1 expression in human cancers, no
report has revealed the function of CDCA1 in colon cancer
growth/survival in detail, or the clinical or prognostic value of
CDCA1 protein as a tissue biomarker for various colon cancers.
We present evidence that CDCA1 plays a significant
role in the malignant potential of CRC, and is a promising
diagnostic and prognostic biomarker, as well as a therapeutic
target for treating CRC.
Materials and methods
Colorectal cancer cell lines and tissue
samples
The human CRC cell lines, Caco-2, CCD-841, COLO205,
LoVo, HCT116, HT-29, SW48, SW480, SW620 and SW948 were purchased
from American Type Culture Collection (ATCC; Rockville, MD, USA).
All cells were grown in monolayers in the appropriate medium
(Table I), supplemented with 10%
fetal bovine serum (FBS) (Nichirei Biosciences, Tokyo, Japan), and
were maintained at 37°C in atmospheres of humidified air. Eight CRC
tissue samples and adjacent normal colorectal tissue samples were
obtained from patients undergoing surgery at Shiga University of
Medical Science Hospital. In addition, we obtained 434 CRC and
adjacent normal colorectal tissue samples for immunostaining on
tissue microarrays from CRC patients without distant metastases who
underwent surgery at Kanagawa Cancer Center Hospital. Individual
institutional ethics committees approved this study and the use of
all clinical materials.
 | Table I.The human CRC cell lines. |
Table I.
The human CRC cell lines.
| Cell line | Medium |
|---|
| Caco-2 | Eagle’s minimum
essential medium |
| CCD-841 | Dulbecco’s modified
Eagle’s medium |
| COLO205 | RPMI-1640
medium |
| LoVo | F-12K medium |
| HCT116 | McCoy’s 5a medium
modified |
| HT-29 | McCoy’s 5a medium
modified |
| SW48 | Leibovitz’s L-15
medium |
| SW480 | Leibovitz’s L-15
medium |
| SW620 | Leibovitz’s L-15
medium |
| SW948 | Leibovitz’s L-15
medium |
Semi-quantitative reverse
transcription-PCR
Total RNAs were extracted from cultured cells and
clinical tissues using Maxwell 16 LEV simplyRNA Purification kit
(Promega, Madison, WI, USA) according to the manufacturer’s
protocol. The RNAs from cultured cells and clinical tissues, as
well as commercially available mRNAs from normal human colon and
rectum samples (Agilent Technologies, Santa Clara, CA, USA) were
reversely transcribed using ReverTra Ace® qPCR RT kit
(Toyobo, Osaka, Japan). Semi-quantitative reverse transcription-PCR
(RT-PCR) experiments were carried out with the following
synthesized CDCA1-specific primers or with β-actin
(ACTB)-specific primers as an internal control: CDCA1,
5′-GAGAAACTGAAGTCCCAGGAAAT-3′ and 5′-CTGATACTTCCATTCGCTTCAAC-3′;
ACTB, 5′-GCC ACCCCACTTCTCTCTAA-3′ and 5′-CACGAAGGCTCAT
CATTCAA-3′. RT-PCR reactions were optimized for the number of
cycles to ensure product intensity within the logarithmic phase of
amplification.
Western blot analysis
Cells were lysed in Pierce RIPA buffer (Thermo
Scientific, Waltham, MA, USA) that included 1% protease inhibitor
cocktail (Thermo Scientific). After homogenization, the cell
lysates were incubated on ice for 30 min and centrifuged at 14,000
rpm for 15 min to separate the supernatant from cellular debris.
The amount of total protein was estimated using a Quick Start
Bradford Protein Assay kit (Bio-Rad, Hercules, CA, USA), and the
proteins were then mixed with SDS sample buffer and incubated at
37°C for 15 min before loading them into a 12% SDS-PAGE gel. After
electrophoresis, the proteins were transferred onto an Amersham
Hybond-P PVDF Membrane (GE Healthcare, Buckinghamshire, UK).
Membranes were blocked using 4% Block Ace (Dainippon
Pharmaceutical, Osaka, Japan), and incubated with anti-Nuf2 (alias
CDCA1) antibody (catalog no. ab96147; Abcam, Cambridge, MA, USA)
and mouse anti-β-actin antibody (catalog no. 8H10D10; Cell
Signaling Technology, Danvers, MA, USA). In the final step, the
membranes were incubated with enhanced chemiluminescence (ECL)
anti-rabbit IgG, horseradish peroxidase (HRP)-linked whole
antibody, ECL anti-mouse IgG and HRP-linked whole antibody (GE
Healthcare). Protein bands were visualized using ECL detection
reagents (GE Healthcare).
Immunocytochemistry
Cultured cells were washed twice with PBS(-), fixed
in 4% formaldehyde solution for 30 min at room temperature and
rendered permeable by a 3-min treatment with PBS(-) containing 0.1%
Triton X-100. Cells were covered with CAS Block (Invitrogen,
Carlsbad, CA, USA) for 7 min to block non-specific binding before
the primary antibody reaction. Then the cells were incubated with
anti-Nuf2 antibody. The immune complexes were stained with Alexa
Fluor 488 goat anti-rabbit IgG (Life Technologies, Grand Island,
NY, USA) and mounted with Vectashield mounting medium with DAPI
(Vector Laboratories, Burlingame, CA, USA), and viewed using a
motorized inverted microscope IX81 (Olympus, Tokyo, Japan).
Immunohistochemistry and tissue
microarray analysis
To investigate the significance of CDCA1
overexpression in clinical CRCs, we stained tissue sections using
EnVision+ kit/horseradish peroxidase (HRP;
DakoCytomation, Glostrup, Denmark). Anti-Nuf2 antibody was added
after blocking of endogenous peroxidase and proteins and each
section was incubated with HRP-labeled anti-rabbit IgG as the
secondary antibody. Substrate-chromogen was added and the specimens
were counterstained with hematoxylin and eosin (HE). Tumor tissue
microarrays were constructed according to previously published
procedures, using formalin-fixed CRCs that were surgically resected
(11–13). Tissue areas selected for sampling
were determined by the visual alignment with the corresponding
HE-stained sections on slides. Three, four or five tissue cores
(diameter, 0.6 mm; height, 3–4 mm) taken from donor tumor blocks
were placed into recipient paraffin blocks using a tissue
microarrayer (Beecher Instruments, Sun Prairie, WI, USA). A core of
normal tissue was punched from each case. Sections (5-μm
thick) of the resulting microarray block were used for
immunohistochemical analysis. Positivity and staining intensity
were recorded as absent, weak or strongly positive.
Statistical analysis
We used contingency tables to correlate
clinicopathological variables, such as gender, age, histologic
type, and pathologic tumor-node-metastasis (TNM) stage, with CDCA1
protein expression levels determined by tissue microarray analysis.
Survival curves were calculated from the surgery date to the
CRC-related time of death or to the last follow-up observation.
Kaplan-Meier curves were calculated for each relevant variable and
for CDCA1 expression; differences in survival times among patient
subgroups were analyzed using the log-rank test. Univariate
analysis was performed using the Cox proportional-hazard regression
model to determine associations between clinicopathological
variables and mortality. We first analyzed associations between
death and possible prognostic factors including age, gender,
pathologic tumor classification and pathologic node classification,
taking into consideration one factor at a time. Then, a
multivariate Cox analysis was applied on backward (stepwise)
procedures that always forced strong CDCA1 expression into the
model, along with any or all variables that satisfied an entry
level P-value of <0.05. As the model continued to add factors,
independent factors did not exceed an exit level of P<0.05.
Effect of small-interfering RNA against
CDCA1 on CRC cell growth
To evaluate the biological functions of CDCA1 in CRC
cells, we used siRNA oligonucleotides (Sigma-Aldrich Japan, Tokyo,
Japan). The sequences targeting each gene were as follows:
si-CDCA1-1, 5′-AAUGCCAGACAAGAAGUG GUG-3′; si-CDCA1-2,
5′-AAGAUGCUGCUGAAAGGG AGA-3′; si-EGFP, 5′-GAAGCAGCACGACUUCUUC-3′;
and si-LUC, 5′-CGUACGCGGAAUACUUCGA-3′. CRC cells, SW480 or SW948,
were transfected with either of siRNAs (si-CDCA1-1 or siCDCA1-2)
using Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer’s instructions. Cell viability was measured using
triplicate 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) assays (Cell Counting kit-8 solution; Dojindo
Laboratories, Kumamoto, Japan) after the transfection. Flow
cytometric analysis was performed using CycleTest Plus DNA reagent
kit and FACSVerse system (BD Immunocytometry Systems, San Jose, CA,
USA). Caspase 3/7 assay (Caspase-Glo 3/7 assay; Promega) and TUNEL
assay (DeadEnd Fluorometric TUNEL System; Promega) were performed
according to the manufacturer’s instructions. Endogenous CDCA1
protein expression was detected using western blotting.
Results
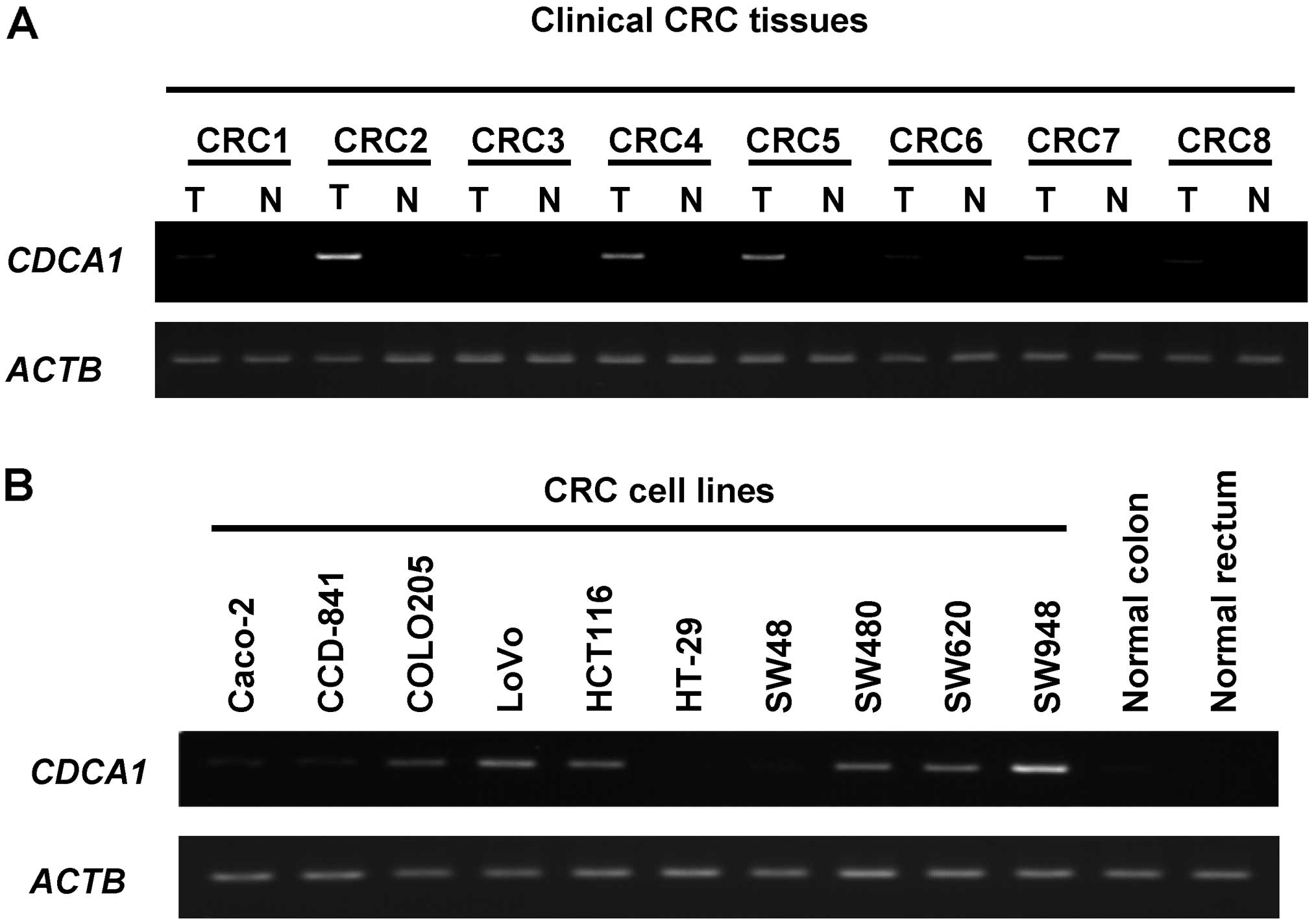
CDCA1 gene expression in CRC tissues and
cell lines
We previously demonstrated using gene expression
profile analysis that CDCA1 was overexpressed in cancer
tissues including CRC tissues (data not shown). We also checked
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/; ID: 88360759) and
found that CDCA1 gene expression in colorectal cancer
tissues was higher in all of 17 clinical CRC tissues than in their
corresponding normal tissues. Because our original gene expression
profile database and the publicly available database revealed high
CDCA1 expression levels in clinical CRCs, we performed a
semi-quantitative RT-PCR analysis of CDCA1 gene expression
in both the cancer and corresponding normal colorectal tissues
isolated from 8 CRC patients and the 10 CRC cell lines.
CDCA1 gene was highly expressed in 4 of the 8 CRC tissues
and in 6 of the 10 CRC cell lines, but was not detected in any
normal colorectal tissues (Fig.
1).
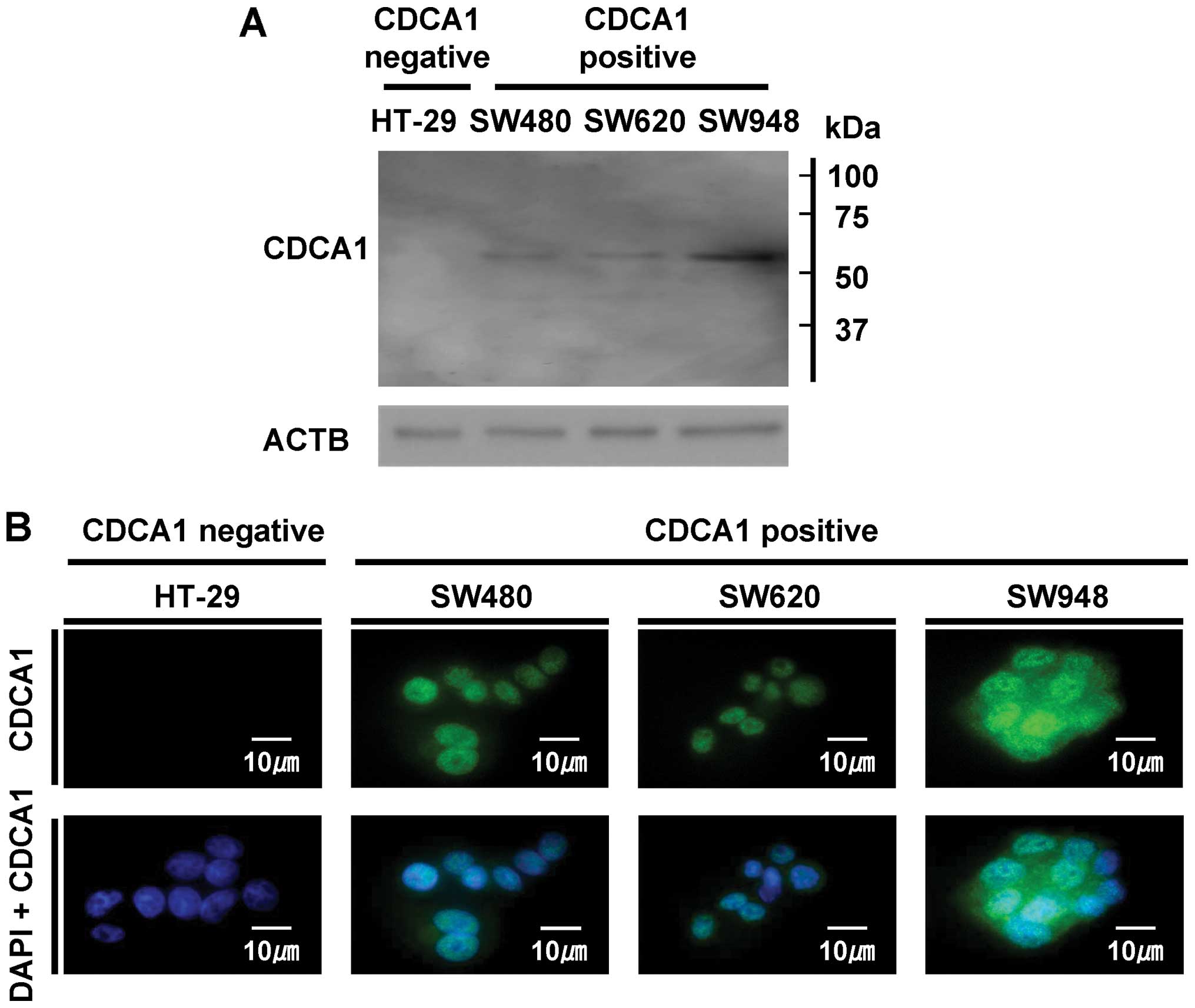
CDCA1 protein expression and its
subcellular localization in CRC cells
To determine the CDCA1 protein expression and its
subcellular localization in CRC cells, we performed western
blotting and immunofluorescence analyses using anti-CDCA1 antibody,
CDCA1-positive CRC cells (SW480, SW620 and SW948), and
CDCA1-negative HT-29 cells. The band was detected using
western blotting in CDCA1-positive SW480, SW620 and SW948
cells, whereas no signal was detected in CDCA1-negative
HT-29 cells (Fig. 2A). In
addition, through an immunofluorescence analysis, we detected CDCA1
protein primarily in the nucleus and cytoplasm of
CDCA1-positive SW480, SW620 and SW948 cells, but not in
CDCA1-negative HT-29 cells (Fig. 2B).
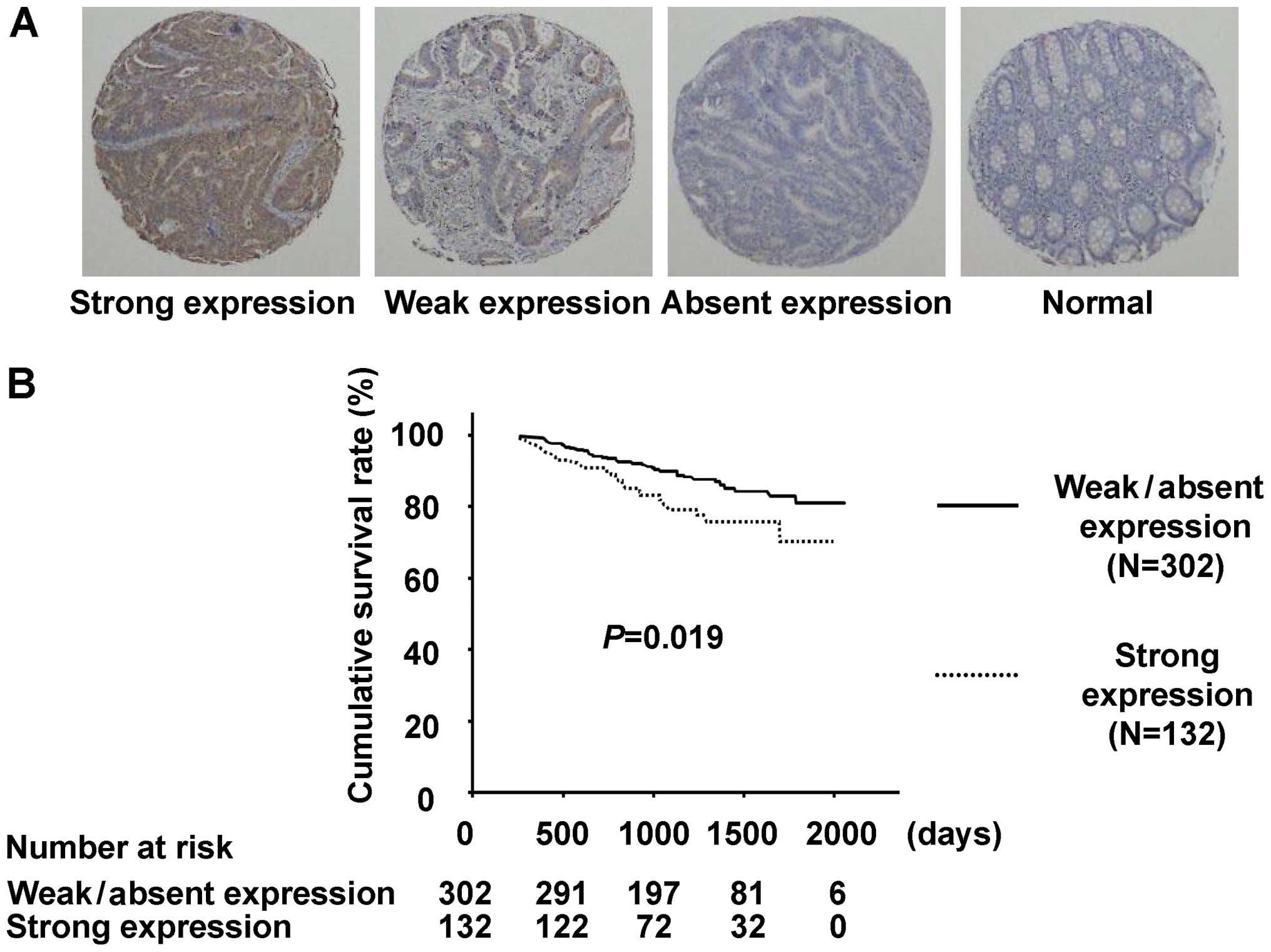
Association of CDCA1 overexpression with
poor clinical outcomes for CRC patients
To verify the biological and clinicopathological
significance of CDCA1 in clinical CRCs, we examined CDCA1 protein
expression with immunohistochemical analysis using anti-CDCA1
antibody and tissue microarrays for the 434 CRC cases without
distant metastases that underwent surgical resection. CDCA1
staining was observed primarily in the cell nucleus and cytoplasm
of tumor cells, but was hardly detectable in surrounding normal
tissues (Fig. 3A). We classified a
CDCA1 expression pattern in the tissue array ranging from
absent/weak to strong. Positive staining was observed in 240 of the
434 (55.3%) CRC cases, whereas no staining was observed in adjacent
normal tissues. Of the 434 CRC cases examined, CDCA1 was strongly
stained in 132 (30.4%), weakly stained in 108 (24.9%) and unstained
in 194 (44.7%; Table II). CRC
patients whose tumors showed strong CDCA1 expression had shorter
survival compared with those with weak/absent CDCA1 expression
(P=0.019 by the log-rank test; Fig.
3B). We also applied univariate analysis to evaluate
associations between patient prognosis and other factors, including
gender (male versus female), age (<65 years versus ≥65 years),
histologic type (tub1, tub2, pap versus por1, por2, sig, others),
pT factor (Tis, T1, T2 versus T3, T4), pN factor (N0, N1 versus
N2), and CDCA1 status (weak, absent versus strong). Among these
variables, CDCA1 status (P=0.020) and advanced pT stage
(P<0.001) were significantly associated with poorer prognosis
(Table III). In multivariate
analysis of the prognostic factors, strong CDCA1 expression
(P=0.008) and higher pT stage (P<0.001) were identified as
independent prognostic factors (Table
III).
 | Table II.Association between CDCA1 positivity
in colorectal cancer tissues and patient characteristics
(n=434). |
Table II.
Association between CDCA1 positivity
in colorectal cancer tissues and patient characteristics
(n=434).
| n=434 | CDCA1 Strong
positive n=132 | CDCA1 Weak positive
n=108 | CDCA1 Absent
n=194 | P-value (Strong
versus weak, absent) |
|---|
| Gender | | | | | |
| Male | 249 | 74 | 73 | 102 | 0.752 |
| Female | 185 | 58 | 35 | 92 | |
| Age (years) | | | | | |
| <65 | 215 | 74 | 54 | 87 | 0.077 |
| ≥65 | 219 | 58 | 54 | 107 | |
| Histologic
type | | | | | |
| tub1, tub2,
pap | 385 | 116 | 99 | 170 | 0.743 |
| por1, por2, sig,
others | 49 | 16 | 9 | 24 | |
| pT factor | | | | | |
| Tis+T1+T2 | 132 | 43 | 32 | 57 | 0.571 |
| T3+T4 | 302 | 89 | 76 | 137 | |
| pN factor | | | | | |
| N0+N1 | 364 | 108 | 89 | 167 | 0.479 |
| N2 | 70 | 24 | 19 | 27 | |
 | Table III.Cox’s proportional hazards model
analysis of prognostic factors in patient with colorectal
cancers. |
Table III.
Cox’s proportional hazards model
analysis of prognostic factors in patient with colorectal
cancers.
| Variables | Hazards ratio (95%
CI) |
Unfavorable/favorable | P-value |
|---|
| Univariate
analysis | | | |
| CDCA1 | 1.813
(1.097–2.997) | Strong/weak,
absent | 0.020a |
| Gender | 0.775
(0.464–1.295) | Male/female | 0.330 |
| Age (years) | 1.189
(0.725–1.952) | ≥65/<65 | 0.493 |
| Histologic
type | 0.643
(0.258–1.604) | tub1, tub2,
pap/por1, por2, sig, others | 0.344 |
| pT factor | 14.222
(3.477–58.166) |
T3+T4/Tis+T1+T2 | <0.001a |
| pN factor | 1.299
(0.706–2.393) | N2/N0+N1 | 0.401 |
| Multivariate
analysis | | | |
| CDCA1 | 1.977
(1.195–3.269) | Strong/weak,
absent | 0.008a |
| pT factor | 14.877
(3.636–60.874) |
T3+T4/Tis+T1+T2 | <0.001a |
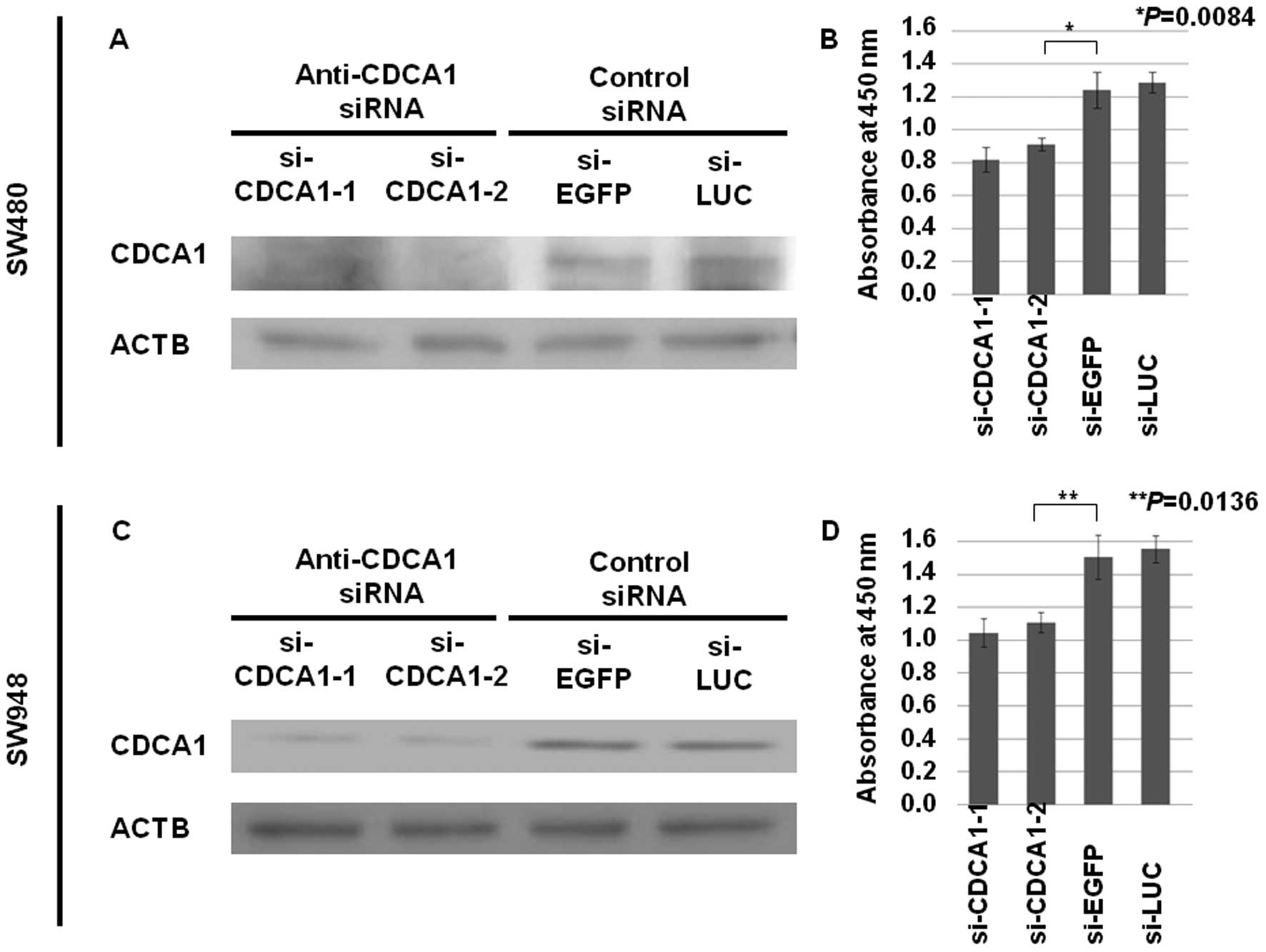
Growth-inhibitory effects of siRNAs
against CDCA1
To assess the role of CDCA1 in CRC cell growth
and/or survival, we knocked down CDCA1 expression in the
CDCA1-positive CRC cell lines SW480 and SW948 using two siRNAs
against CDCA1 (si-CDCA1-1 and -2), along with two control siRNAs
(siRNAs for EGFP and LUC). Transfection of si-CDCA1s into CRC cells
reduced CDCA1 protein levels, and significantly reduced cell
viability (Fig. 4). These results
indicate that CDCA1 is indispensable for growth and survival of CRC
cells.
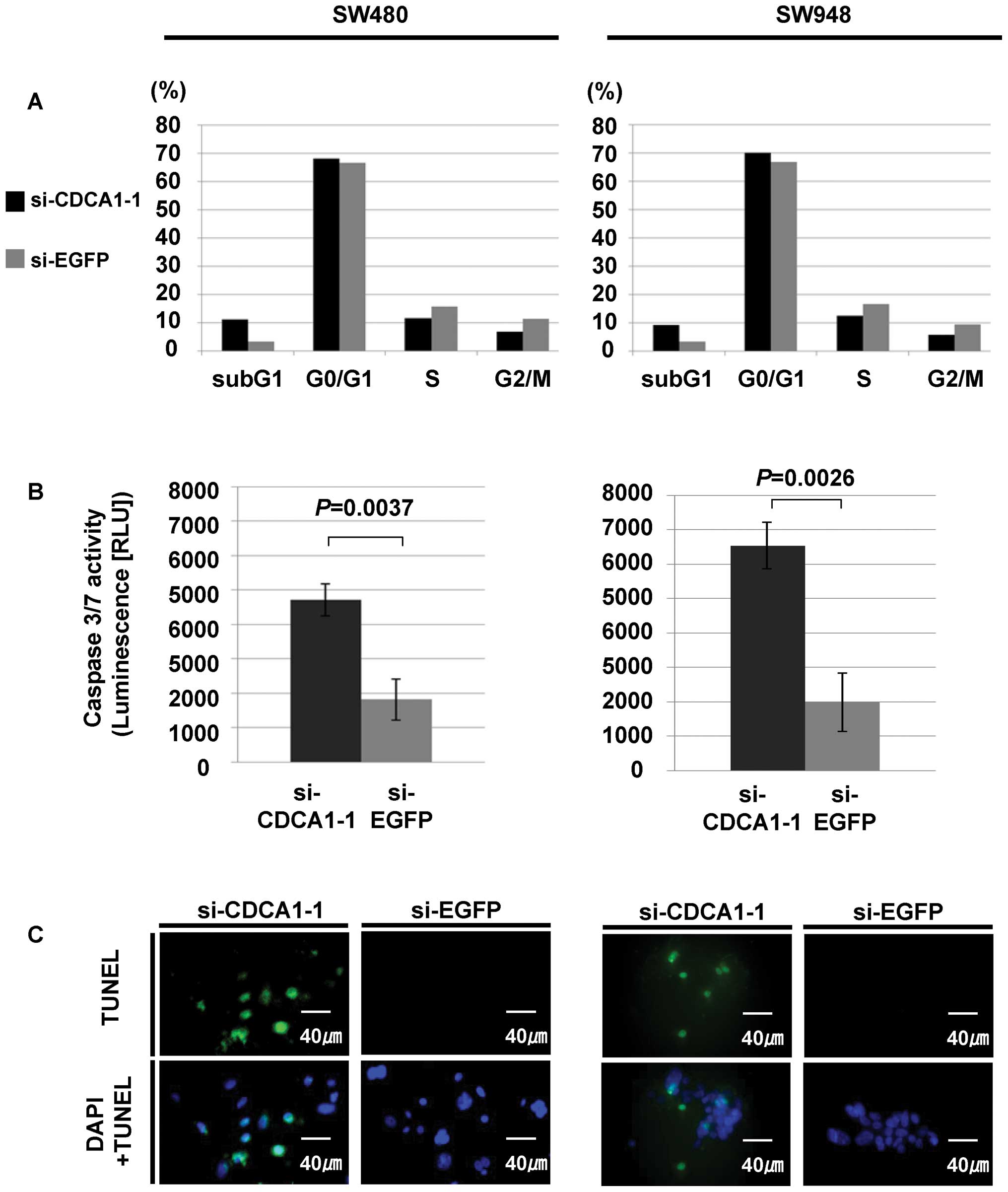
To further investigate the mechanisms of CRC cell
growth suppression caused by reduced CDCA1 expression, we measured
the DNA contents using flow cytometric analysis after CDCA1
(si-CDCA1-1) siRNA transfection into CRC cells. We observed an
increased sub-G1 fraction in SW480 and SW948 cells transfected with
si-CDCA1-1, compared with those transfected with control siRNAs
(Fig. 5A). In addition, caspase
3/7 assay detected an increase in caspase 3/7 activity, whereas
TUNEL assay showed an increase in TUNEL staining in SW480 and SW948
cells transfected with siRNAs for CDCA1, compared with those
transfected with control siRNAs. These results independently
demonstrated activation of caspase cascades and subsequent DNA
fragmentation in CRC cells transfected with siRNAs for CDCA1
(Fig. 5B and C).
Discussion
Significant advances in the development of
molecular-targeting drugs for cancer therapy have been achieved in
the last two decades. However, the number of patients that respond
to the presently available treatments is limited, and a subset of
the patients suffers from severe adverse reactions without clinical
benefits. Therefore, it is critical to develop new anticancer
agents that are highly specific to malignant cells and have a
minimum risk of adverse side effects.
We previously reported that CDCA1 overexpression
plays a key role in the proliferation of lung cancer. CDCA1 is one
of the highly conserved components of nuclear division cycle
complex and is categorized as an oncoantigen that can induce
peptide-specific CTLs against solid tumors (16,39).
In the present study, we demonstrated that CDCA1 expression is also
elevated in many CRC tissues. Similar to previous findings in lung
cancer cells, knocking down CDCA1 with siRNA inhibited CRC cell
growth, which suggests increased CDCA1 expression is necessary for
CRC cell proliferation and/or survival. CDCA1 protein functions at
kinetochores for stable microtubule attachment and stable
kinetochore localization of centromere-associated protein E
(CENP-E) in the cervical cancer cell line HeLa (45–47).
When CDCA1 expression was reduced in these cells, kinetochores
failed to form attachments with spindle microtubules, which were
followed by aberrant chromosome segregation, a prolonged mitotic
blockade, and cell death (45–47).
This aberrant exit from mitosis has characteristics of both
apoptosis and catastrophe (45).
Recent investigations also demonstrated that knocking down
centrosomal proteins such as aurora A and ninein in HeLa cells led
to aberrant spindle formation and subsequent cell death, which are
accompanied by several features of apoptosis (48). Therefore, we presume that apoptosis
of CRC cells induced by CDCA1 expression inhibition by siRNA could
result from a similar mechanism. However, further studies are
needed to clarify the relationship between CDCA1 suppression and
mitotic catastrophe or apoptosis. Many proteins that regulate
mitosis are aberrantly expressed in human tumor cells when compared
with their normal counterparts, and some of these function as
oncogenes, such as aurora kinase and polo-like kinase (49,50).
Some of these proteins are potential targets for anticancer agents.
For example, highly conserved aurora kinases are critical mitotic
regulators (49). Several aurora
kinase inhibitors, including ZM447439, Hesperadin and VX-680 have
been described as anticancer drugs (49). CDCA1 could serve as a valuable
target for molecular-targeted therapies, as well as peptide vaccine
immunotherapy for CRC.
In addition, we demonstrated that CDCA1 was highly
expressed in 55.3% of surgically resected samples obtained from CRC
patients and this overexpression was associated with a poorer
prognosis. A publicly available microarray database (http://www.prognoscan.org/) revealed a significant
correlation between high CDCA1 expression and a reduced OS
period for CRC patients (dataset no. GSE17536; P=0.028054), which
independently supports our data that CDCA1 expression has
prognostic value for CRC patients. CDCA1 positivity in CRC tissues
could provide a clinical prognostic indicator that warrants
intensive follow-up in patients and/or addition of adjuvant
chemotherapy after surgical treatments.
To examine the mechanisms of CDCA1 activation and
overexpression in CRCs, we searched previous publications and
databases for CDCA1 including the CGH and genome sequencing
(http://cancer.sanger.ac.uk/cosmic/gene/) databases.
Missense mutation was indicated in 6 of the 652 CRCs (0.92%), but
no CDCA1 gene amplification or translocation was reported in
CRCs. Therefore, we speculate that overexpression of CDCA1 may be
primarily caused by epigenetic mechanism. Further analysis of
CDCA1, including screening using functional assays for an
activating mutation or epigenetic regulating mechanisms of CDCA1
may further clarify the oncogenic function of CDCA1.
In conclusion, our data suggest that CDCA1
contributes to the viability and malignant potential of CRC cells,
and is a clinically promising prognostic biomarker in addition to a
potential molecular target for treating CRC.
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Van Cutsem E, Köhne CH, Hitre E, Zaluski
J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G,
Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M,
Nippgen J and Rougier P: Cetuximab and chemotherapy as initial
treatment for metastatic colorectal cancer. N Engl J Med.
360:1408–1417. 2009.PubMed/NCBI
|
|
3.
|
Bokemeyer C, Van Cutsem E, Rougier P,
Ciardiello F, Heeger S, Schlichting M, Celik I and Köhne CH:
Addition of cetuximab to chemotherapy as first-line treatment for
KRAS wild-type metastatic colorectal cancer: pooled analysis of the
CRYSTAL and OPUS randomised clinical trials. Eur J Cancer.
48:1466–1475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Daigo Y and Nakamura Y: From cancer
genomics to thoracic oncology: discovery of new biomarkers and
therapeutic targets for lung and esophageal carcinoma. Gen Thorac
Cardiovasc Surg. 56:43–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Kikuchi T, Daigo Y, Katagiri T, et al:
Expression profiles of non-small cell lung cancers on cDNA
microarrays: identification of genes for prediction of lymph-node
metastasis and sensitivity to anti-cancer drugs. Oncogene.
22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kakiuchi S, Daigo Y, Ishikawa N, et al:
Prediction of sensitivity of advanced non-small cell lung cancers
to gefitinib (Iressa, ZD1839). Hum Mol Genet. 13:3029–3043. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Kikuchi T, Daigo Y, Ishikawa N, et al:
Expression profiles of metastatic brain tumor from lung
adenocarcinomas on cDNA microarray. Int J Oncol. 28:799–805.
2006.PubMed/NCBI
|
|
9.
|
Taniwaki M, Daigo Y, Ishikawa N, et al:
Gene expression profiles of small-cell lung cancers: molecular
signatures of lung cancer. Int J Oncol. 29:567–575. 2006.PubMed/NCBI
|
|
10.
|
Yamabuki T, Daigo Y, Kato T, et al:
Genome-wide gene expression profile analysis of esophageal squamous
cell carcinomas. Int J Oncol. 28:1375–1384. 2006.PubMed/NCBI
|
|
11.
|
Suzuki C, Daigo Y, Kikuchi T, Katagiri T
and Nakamura Y: Identification of COX17 as a therapeutic target for
non-small cell lung cancer. Cancer Res. 63:7038–7041.
2003.PubMed/NCBI
|
|
12.
|
Kato T, Daigo Y, Hayama S, et al: A novel
human tRNA-dihydrouridine synthase involved in pulmonary
carcinogenesis. Cancer Res. 65:5638–5646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Furukawa C, Daigo Y, Ishikawa N, et al:
Plakophilin 3 oncogene as prognostic marker and therapeutic target
for lung cancer. Cancer Res. 65:7102–7110. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Suzuki C, Daigo Y, Ishikawa N, et al: ANLN
plays a critical role in human lung carcinogenesis through the
activation of RHOA and by involvement in the phosphoinositide
3-kinase/AKT pathway. Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Takahashi K, Furukawa C, Takano A, et al:
The neuromedin U-growth hormone secretagogue receptor
1b/neurotensin receptor 1 oncogenic signaling pathway as a
therapeutic target for lung cancer. Cancer Res. 66:9408–9419. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Hayama S, Daigo Y, Kato T, et al:
Activation of CDCA1-KNTC2, members of centromere protein complex,
involved in pulmonary carcinogenesis. Cancer Res. 66:10339–10348.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kato T, Hayama S, Yamabuki T, et al:
Increased expression of IGF-II mRNA-binding protein 1 is associated
with the tumor progression in patients with lung cancer. Clin
Cancer Res. 13:434–442. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hayama S, Daigo Y, Yamabuki T, et al:
Phosphorylation and activation of cell division cycle associated 8
by aurora kinase B plays a significant role in human lung
carcinogenesis. Cancer Res. 67:4113–4122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Taniwaki M, Takano A, Ishikawa N, et al:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Kato T, Sato N, Hayama S, et al:
Activation of holliday junction recognizing protein involved in the
chromosomal stability and immortality of cancer cells. Cancer Res.
67:8544–8553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kato T, Sato N, Takano A, et al:
Activation of placenta-specific transcription factor distal-less
homeobox 5 predicts clinical outcome in primary lung cancer
patients. Clin Cancer Res. 14:2363–2370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Dunleavy EM, Roche D, Tagami H, et al:
HJURP is a cell-cycle-dependent maintenance and deposition factor
of CENP-A at centromeres. Cell. 137:485–497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Hirata D, Yamabuki T, Miki D, et al:
Involvement of epithelial cell transforming sequence 2 (ECT2)
oncoantigen in lung and esophageal cancer progression. Clin Cancer
Res. 15:256–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sato N, Koinuma J, Fujita M, et al:
Activation of WD repeat and high-mobility group box DNA binding
protein 1 in pulmonary and esophageal carcinogenesis. Clin Cancer
Res. 16:226–239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Nguyen MH, Koinuma J, Ueda K, et al:
Phosphorylation and activation of cell division cycle associated 5
by mitogen-activated protein kinase play a crucial role in human
lung carcinogenesis. Cancer Res. 70:5337–5347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Aragaki M, Takahashi K, Akiyama H, et al:
Characterization of a cleavage stimulation factor, 3′ pre-RNA,
subunit 2, 64 kDa (CSTF2) as a therapeutic target for lung cancer.
Clin Cancer Res. 7:5889–5900. 2011.
|
|
27.
|
Masuda K, Takano A, Oshita H, et al:
Chondrolectin is a novel diagnostic biomarker and a therapeutic
target for lung cancer. Clin Cancer Res. 17:7712–7722. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Fujitomo T, Daigo Y, Matsuda K, Ueda K and
Nakamura Y: Critical function for nuclear envelope protein TMEM209
in human pulmonary carcinogenesis. Cancer Res. 72:4110–4118. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Oshita H, Nishino R, Takano A, et al:
RASEF is a novel diagnostic biomarker and a therapeutic target for
lung cancer. Mol Cancer Res. 11:937–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ishikawa N, Daigo Y, Yasui W, et al: ADAM8
as a novel serological and histochemical marker for lung cancer.
Clin Cancer Res. 10:8363–8370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Ishikawa N, Daigo Y, Takano A, et al:
Increases of amphiregulin and transforming growth factor-alpha in
serum as predictors of poor response to gefitinib among patients
with advanced non-small cell lung cancers. Cancer Res.
65:9176–9184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Yamabuki T, Takano A, Hayama S, et al:
Dickkopf-1 as a novel serologic and prognostic biomarker for lung
and esophageal carcinomas. Cancer Res. 67:2517–2525. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Ishikawa N, Takano A, Yasui W, et al:
Cancer-testis antigen lymphocyte antigen 6 complex locus K is a
serologic biomarker and a therapeutic target for lung and
esophageal carcinomas. Cancer Res. 67:11601–11611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Takano A, Ishikawa N, Nishino R, et al:
Identification of nectin-4 oncoprotein as a diagnostic and
therapeutic target for lung cancer. Cancer Res. 69:6694–6703. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Sato N, Yamabuki T, Takano A, et al: Wnt
inhibitor Dickkopf-1 as a target for passive cancer immunotherapy.
Cancer Res. 70:5326–5336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Nishino R, Takano A, Oshita H, et al:
Identification of Epstein-Barr virus-induced gene 3 as a novel
serum and tissue biomarker and a therapeutic target for lung
cancer. Clin Cancer Res. 17:6272–6286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Suda T, Tsunoda T, Daigo Y, Nakamura Y and
Tahara H: Identification of human leukocyte antigen-A24-restricted
epitope peptides derived from gene products upregulated in lung and
esophageal cancers as novel targets for immunotherapy. Cancer Sci.
98:1803–1808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Mizukami Y, Kono K, Daigo Y, et al:
Detection of novel cancer-testis antigen-specific T-cell responses
in TIL, regional lymph nodes, and PBL in patients with esophageal
squamous cell carcinoma. Cancer Sci. 99:1448–1454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Harao M, Hirata S, Irie A, et al:
HLA-A2-restricted CTL epitopes of a novel lung cancer-associated
cancer testis antigen, cell division cycle associated 1, can induce
tumor-reactive CTL. Int J Cancer. 123:2616–2625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kono K, Mizukami Y, Daigo Y, et al:
Vaccination with multiple peptides derived from novel cancer-testis
antigens can induce specific T-cell responses and clinical
responses in advanced esophageal cancer. Cancer Sci. 100:1502–1509.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Yokomine K, Senju S, Nakatsura T, et al:
The forkhead box M1 transcription factor, as a possible
immunotherapeutic tumor-associated antigen. Int J Cancer.
126:2153–2163. 2010.
|
|
42.
|
Tomita Y, Imai K, Senju S, et al: A novel
tumor-associated antigen, cell division cycle 45-like can induce
cytotoxic T lymphocytes reactive to tumor cells. Cancer Sci.
102:697–705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Kaneko N, Miura K, Gu Z, Karasawa H,
Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H,
Shibata C, Sasaki I and Horii A: siRNA-mediated knockdown against
CDCA1 and KNTC2, both frequently overexpressed in colorectal and
gastric cancers, suppresses cell proliferation and induces
apoptosis. Biochem Biophys Res Commun. 390:1235–1240. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
van Duin M, Broyl A, de Knegt Y,
Goldschmidt H, Richardson PG, Hop WC, van der Holt B,
Joseph-Pietras D, Mulligan G, Neuwirth R, Sahota SS and Sonneveld
P: Cancer testis antigens in newly diagnosed and relapse multiple
myeloma: prognostic markers and potential targets for
immunotherapy. Haematologica. 96:1662–1669. 2011.PubMed/NCBI
|
|
45.
|
DeLuca JG, Moree B, Hickey JM, Kilmartin
JV and Salmon ED: hNuf2 inhibition blocks stable
kinetochore-microtubule attachment and induces mitotic cell death
in HeLa cells. J Cell Biol. 159:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
DeLuca JG, Howell BJ, Canman JC, Hickey
JM, Fang G and Salmon ED: Nuf2 and Hec1 are required for retention
of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr
Biol. 13:2103–2109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Liu D, Ding X, Du J, Cai X, Huang Y, Ward
T, Shaw A, Yang Y, Hu R, Jin C and Yao X: Human NUF2 interacts with
centromere-associated protein E and is essential for a stable
spindle microtubule-kinetochore attachment. J Biol Chem.
282:21415–21424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Kimura M, Yoshioka T, Saio M, Banno Y,
Nagaoka H and Okano Y: Mitotic catastrophe and cell death induced
by depletion of centrosomal proteins. Cell Death Dis. 4:e6032013.
View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Keen N and Taylor S: Aurora-kinase
inhibitors as anticancer agents. Nat Rev Cancer. 4:927–936. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Jang YJ, Kim YS and Kim WH: Oncogenic
effect of Polo-like kinase 1 expression in human gastric
carcinomas. Int J Oncol. 29:589–594. 2006.PubMed/NCBI
|