Introduction
Glioma is the most common primary brain tumor
affecting yearly 3–5/100,000 and occurring mainly in adults >45
years old (1–3). The ability of glioma to invade and
infiltrate diffusely contiguous brain tissue limits the complete
surgical resection and the efficacy of standard therapies (4). Glioblastoma multiforme (GBM), a grade
IV astrocytoma as currently defined by the World Health
Organization (WHO) classification (5), is the most common and the most lethal
form of brain tumor. The therapeutic approach against this tumor
consists in surgical resection followed by radiation and
chemotherapy with temozolomide (TMZ) (6,7). At
present, such treatment can only slightly modify the patient’s
outcome. In fact, the tumor typically recurs after an average of
only 6.9 months, resulting in a median survival rate of <1 year
following diagnosis (8). Permanent
cell lines represent important tools to study the behaviour of
human tumors, such as their growth and metabolism, drug sensitivity
and resistance, as well as genomic and expression profiles
(9–11). Furthermore, some cultivated cancer
cells can originate a novel tumor when transplanted into nude mice,
providing an experimental system to test potential novel
therapeutic drugs (12). Here we
report the establishment and the characterization, by cytogenetic
and molecular approaches, of a novel cell line termed as ANGM-CSS,
derived from a patient with GBM.
Materials and methods
Patient history
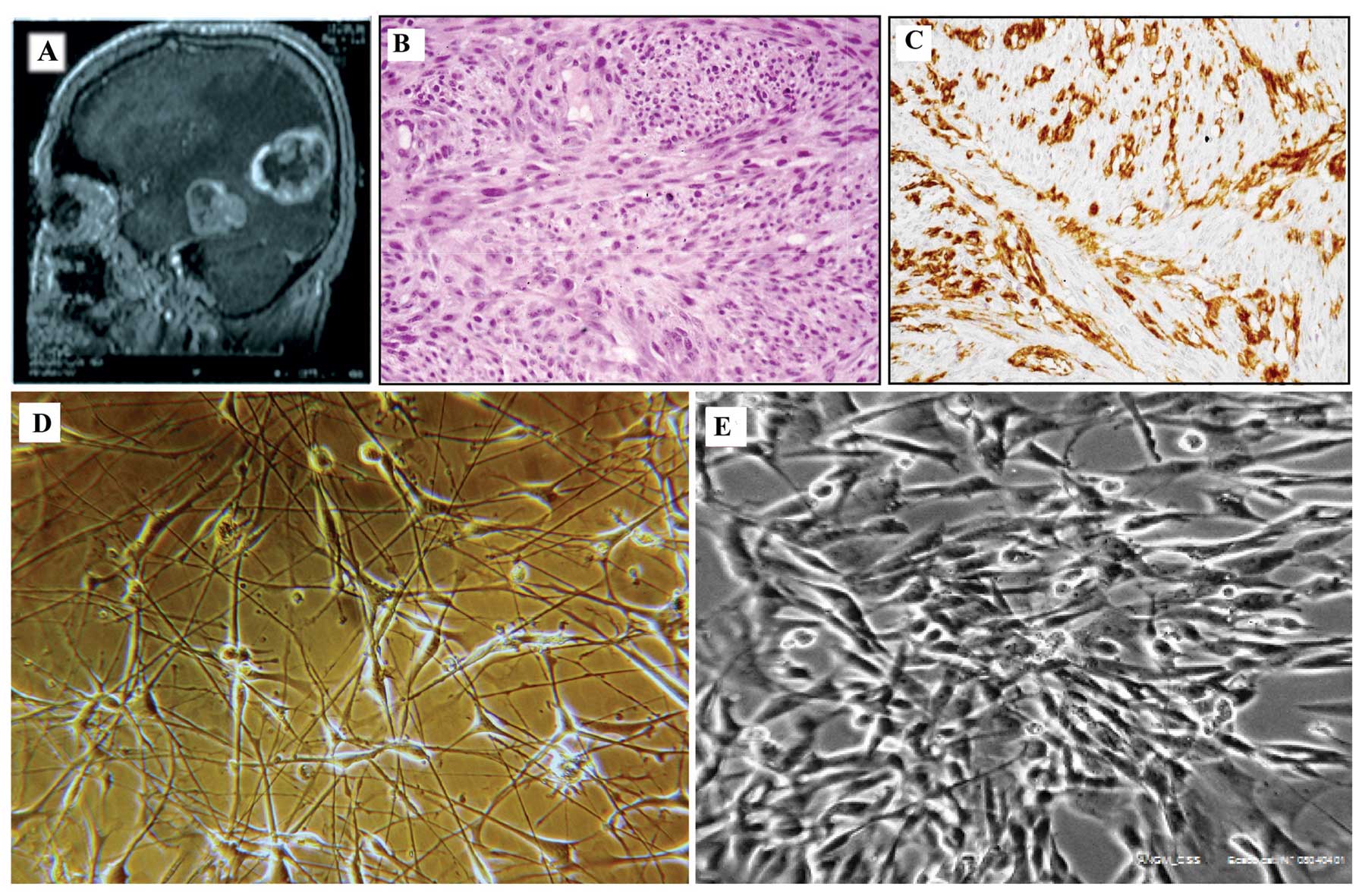
The patient was a 56-year-old male, surgically
treated to remove a large primary tumor localized in the left
temporo-occipital lobe with invasion of the ventricular cornus
(Fig. 1A). Hematoxylin and eosin
staining (Fig. 1B) and
immunohistochemical examination for GFAP, α-SMA and HMB45 were
performed as part of the routine assessment of tumor
type/phenotype. Immunostaining demonstrated a weak positivity for
GFAP (Fig. 1C) but was negative
for α-SMA and HMB45 (data not shown). The final diagnosis was GBM
with spindle, mitotically active cells showing a fascicular growth
pattern. After surgery, the patient was treated with temozolomide
at a daily dose of 75 mg/m2 of body surface in
association with fractionated radiotherapy (60 Gy) for 6–7 weeks.
The patient died 18 months after surgery.
Establishment of primary culture
After surgical removal, upon institutional Ethics
Committee approval, the tumor specimen was placed immediately in
DMEM-F12 medium without serum, repeatedly washed with
phosphate-buffered saline (PBS, Invitrogen, Carlsbad, CA, USA) and
then placed in a 30-mm Petri dish. The specimen was cut in 1–2 mm
or tinier fragments and transferred in a poly-D-lysine treated
flask with a small amount of Dulbecco’s modified Eagle’s medium/F12
medium (D-MEM/F12, Invitrogen) (1:1, v/v) supplemented with 10%
fetal bovine serum (FBS, Invitrogen), 100 U/ml penicillin and 100
μg/ml streptomycin (PenStrep, Invitrogen) in order to allow
the fragments to adhere to the surface of the flask. Primary
culture was incubated at 37°C in 5% CO2 humidified
atmosphere, and 5 ml of complete medium was added 24 h later. After
one week, the primary culture was washed with PBS to remove
non-adherent fragments and fresh medium prewarmed at 37°C was
added. These procedures were repeated every 3 days until primary
culture reached local confluence. Then cells were treated with
0.05% trypsin (Invitrogen) and 0.02% EDTA (Invitrogen), washed with
PBS and transferred into a T-75 flask without biocoat and
containing complete medium DMEM/F12. Next, the culture was serially
transferred by using the same procedures once or twice a week.
Every 10 passages, one amount of cells was counted with Z1-Coulter
(IL-Laboratories). Cells (50×106) were frozen in medium
with 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen.
Cells were propagated by serial passages (split ratio 1:3) along 2
years, until 105th passage. Growth curves were established at the
32nd passages by seeding 1×105 cells into three 35-mm
culture dishes. Triplicate dishes were harvested and counted daily
with Z1-Coulter (IL-Laboratories). The cell number was determined
as the average number of cells ± SD in each time interval.
Immunophenotypical characterization of
ANGM-CSS cell line
Immunofluorescence analysis was performed at
different passages to establish GFAP, nestin, CD133 and vimentin
localization in the ANGM cell cultures. After trypsin treatment,
105 cells were seeded on 20×20 mm coverslips, rinsed
twice in PBS, and fixed in PBS containing 4% formaldehyde (pH
7.2–7.4) for 10 min at room temperature (RT), washed for 10 min
three times with PBS, permeabilized with PBS/0.2% Triton X-100 (MP
Biomedical) and blocked for 30 min with PBS containing bovine serum
albumin (BSA). After three washings with PBS, the cells were
incubated with a primary antibody against CD133 (AP2010b, purified
rabbit, ABGENT), nestin (sc-23927, mouse monoclonal
IgG1, Santa Cruz Biotechnology), vimentin (sc-6260,
mouse monoclonal IgG1, Santa Cruz Biotechnology), GFAP
(sc-58766, mouse monoclonal IgG1, Santa Cruz
Biotechnology) diluted 1:300 in BSA for 1 h at RT. After extensive
washing in PBS, the coverslips were incubated with the secondary
antibodies for 30 min at room temperature. A goat anti-mouse
IgG-FITC conjugated (sc-2010, 1:200, Santa Cruz Biotechnology) for
vimentin, nestin and GFAP, and a goat anti-rabbit Ig-G FITC
conjugated (sc-2012, 1:200, Santa Cruz Biotechnology) for CD-133
were used as secondary antibodies. Nuclei were then washed only
once with PBS and stained with DAPI (1 mg/ml). The fluorescent
staining was visualized using Nikon E1000 microscopy.
Cytogenetic analysis
ANGM-CSS cells were subcultured at passages 5, 32,
67, 86 and incubated overnight with Colcemid (0.05 mg/ml,
Invitrogen). Cells were dispersed with 0.5% trypsin (Invitrogen)
and 0.02 EDTA (Invitrogen), then washed with 1X PBS and treated
with 0.075 M KCl hypotonic solution and fetal bovine serum (1:1,
v/v) for 20 min at 37°C. The cells were then fixed with
methanol:acetic acid (3:1, v/v) solution and stored for 1 h at
−20°C. Cell suspension were dropped on ice glass slides and stained
in Giemsa stain after banding with GAG-acid solution at 55°C.
Karyotyping was performed by use of the Genikon software (Nikon
Italia, Firenze, Italy) and described in accordance to the
International System for Human Cytogenetic Nomenclature (ISCN
2009).
SNP array analysis
SNP array experiments and data analysis were
performed using the Genome-Wide Human SNP 6.0 array (Affymetrix,
Santa Clara, CA, USA) as previously described (13).
Multicolor fluorescence in situ
hybridization (M-FISH)
M-FISH analysis of ANGM-CSS cells was performed
using the commercially available 24-colour SpectraVysion probe
(Abbott), according to the manufacturer’s instructions. Metaphase
images were captured using a Leica DM-RXA2 epifluorescence
microscope equipped with an 8-position automated filter wheel and a
cooled CCD camera (Princeton Instruments). Six fluorescent images
per metaphase were captured using filter combinations specific for
SpectrumGold, SpectrumAqua, SpectrumGreen, FRed, Red, and DAPI.
Images were processed using the Leica CW4000 M-FISH software.
FISH
BAC clones for FISH analysis were selected according
to the March 2006 release of the UCSC Human Genome Browser
(http://genome.ucsc.edu) (data not shown). FISH
experiments were carried out as previously described (14).
Quantitative reverse transcription
polymerase chain reaction (RTq-PCR)
Expression analysis of EGFR and MET
genes was performed comparing the mRNA levels in the ANGM-CSS cell
line and normal human astrocytes (NHA) (Lonza Walkersville, MD,
USA) by QRT-PCR on 7700 sequence detection system (Applied
Biosystems, Foster City, CA, USA) using MGB TaqMan chemistry and
the 2−ΔCt relative method for relative quantification
(15). For the analysis were used
the following TaqMan® Gene Expression Assays (Applied
Biosystems): MET (Hs01565580_m1) and EGFR
(Hs01076092_m1). The human large ribosomal protein transcript
(Human RPLP09, Applied Biosystems) was used as endogenous
control.
Quantitative methylation-specific PCR
(QMSP) for the MGMT gene
DNA extracted from ANGM-CSS underwent bisulfite
treatment and subsequent DNA purification using the Epitect
Bisulfate kit (Qiagen Sci, MD, USA) according to the manufacturer’s
instructions. Bisulphite converted DNA was used as template for
fluorescence-based real-time QMSP. Real-time PCR experiments for
MGMT were performed as previously described (16).
TP53 and KRAS mutation analysis
PCR amplifications of KRAS gene exon 2 and
TP53 gene exons 4–8 were performed as previously described
(17). Amplification reactions
were performed in a GeneAmp PCR System 9700 (Perkin-Elmer, Foster
City, CA, USA) in a final reaction volume of 25 μl
containing 100 ng of genomic DNA template, 0.25 nM dNTPs, 20 pmol
of each primers, 1 U HotMaster Taq polymerase (Eppendorf), in 1X
PCR reaction buffer. All PCR products were purified using
GFX™ PCR DNA and Gel Band Purification kit (GE
Healthcare, Buckinghamshire, UK) and sequenced. Sequencing
reactions were performed in 10 μl of final volume using 3
pmol of primer, 4–6 ng of DNA template and 1 μl of Big Dye
Terminator Ready Reaction mix v.1.1 (Applied Biosystems).
Sequencing reactions were loaded on an ABI 3100 capillary sequencer
(Applied Biosystems) and by the Sequencing Analysis software v.3.7
(PE Applied Biosystems).
FIG-ROS1 and FGFR3-TACC3 fusion gene
detection
Total RNA was isolated from ANGM-CSS cell line using
the TRIzol reagent (InvitrogenTM Life Technologies,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
The Agilent 2100 Bioanalyzer was used to measure the quantity,
integrity and purity of total RNA. RNA (1 μg) was reverse
transcribed by High Capacity cDNA Reverse Transcription kit (Life
Technologies) according to the manufacturer’s instructions. To
detect the possible presence of FIG-ROS1 fusion transcript,
reverse transcriptase (RT)-PCR experiments were performed using
primers previously reported (18).
The detection of the FGFR3-TACC3 fusion gene was performed
on the basis of the results previously described (19) and using the primers TACC3_Ex5_f
(CTTGAACTCTGCCAGCACCT) and FGFR3_Ex16_r GTGGGCAAACACGGAGTC.
Briefly, for both chimeric genes, 2 μl cDNA was used as
template in a final volume of 50 μl containing 10X PCR
buffer, 0.25 mM of each dNTP, 0.5 μM of each forward and
reverse primer, 0.5 U HotMaster Taq DNA polymerase (5Prime). The
PCRs were run on a GeneAmp PCR System 9700 (Applied Biosystem) with
the cycling profile of initial denaturation for 2 min at 94°C
followed by 35 cycles of 1 min at 94°C, 1 min at 56°C and 2 min at
72°C, with a final extension for 10 min at 72°C. For the
experiments for FIG-ROS1 detection, the cDNA of U118MG cell
line was used as positive control (18–20).
The PCR product (20 μl) was analyzed by electrophoresis
through a 1% agarose gel containing ethidium bromide for
staining.
Murine xenograft model
Six female, 6-week old athymic nude mice
(Crl:CD1-NU-Foxn1nu from Charles River
Laboratories, Italy) were used for transplantation studies in
accordance with national and institutional guidelines and were kept
under specific pathogen-free (SPF) conditions. The mice were
inoculated subcutaneously with 200 μl of cell suspension at
four different concentrations (from a minimum concentration of
5×105 cells in 200 μl of mixture PBS/Matrigel
1:1, to a maximum concentration of 10×106 cells in 200
μl of mixture PBS/Matrigel 1:1). Mice were bilaterally
inoculated on the flank (three injections for each concentration),
while a control mouse was inoculated with Matrigel. The tumor
diameter was measured weekly with a digital caliper and the tumor
volume (mm3) was calculated as previously described
(21). All mice used in the
experiment were monitored daily for signs of suffering and to
identify cases of spontaneous death.
Results
The primary culture grew initially slowly, reaching
cell confluence three weeks after surgical removal. Analysis by
phase contrast microscopy showed a mixed population with
dendritic-like and spindle cells (Fig.
1D). From the 26th passage, the cells showed a spindle shape
with large nuclei. Cells were propagated until the 105th passage,
growing continuously for >2 years, without morphological
changes. The doubling time was calculated at the 32nd passage. One
day after subculturing, the cells entered an exponential growth
phase, where the doubling time was ∼60 h. Immunofluorescence
analysis showed a decrease in GFAP immunoreactivity, starting from
the 17th passage to completely disappear during further serial
passages in vitro. The cells were persistently positive for
vimentin and nestin, while only a small fraction of the ANGM-CSS
cell population showed a CD133 staining (data not shown).
Cytogenetic and molecular analyses were performed at the 32nd, at
the 70th and at the last passages. Metaphase spreads from long-term
cultured cells were treated by conventional cytogenetic methods for
karyotype analysis and thirty-two metaphases were analyzed. The
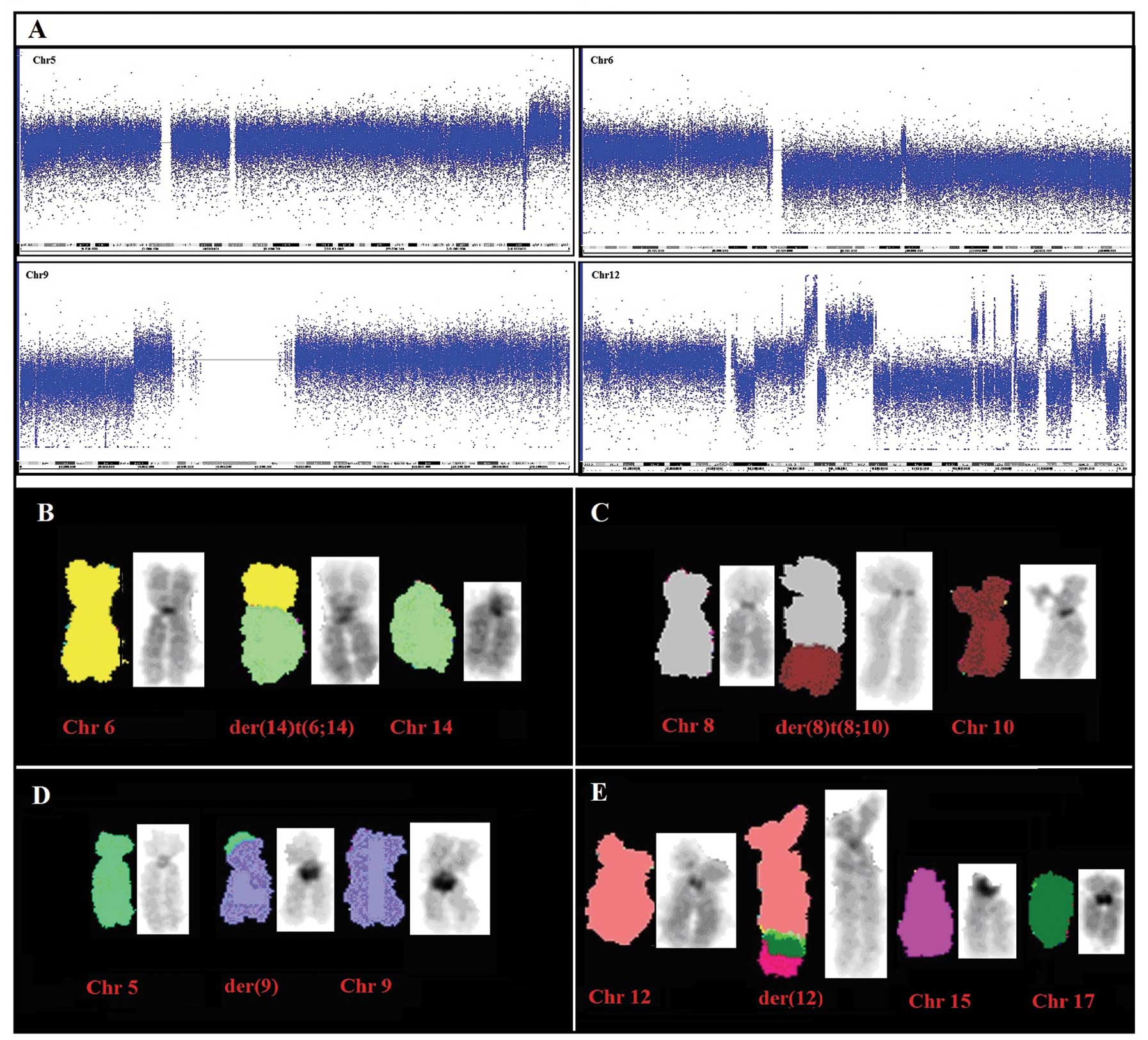
cytogenetic analysis showed a strong karyotypic complexity and
heterogeneity; the chromosome number was near-tetraploid and in
addition to whole chromosome losses (chromosomes 10, 14 and 21) and
gains (chromosomes 7, 20 and 19), two clonal chromosomal
translocations were observed: a t(6;14) (p12;q11.2), and a t(8;10)
(q24.2;q21.1). Moreover, the recurrent deletion of the long arm of
chromosome 6 was detected and one marker chromosome apparently
composed of chromosome 12 material [mar (12)] was found to be recurrent in 100% of
the cell population (Fig. 2).
According to the 2009 recommendations of the International System
for Human Cytogenetic Nomenclature (22), the karyotype was described as
follows:
88∼91,XXYY,−6,+7,del(9)(p21.1)×2,−10,−12,+der(14)t(6;14)(p12;q11.2)×2,−17,+19,+20,−21,+marx2[p32].
To better define genomic gains and losses, SNP array
analysis was performed on DNA extracted at the same passages. The
SNP analysis confirmed the high genomic complexity, showing, in
addition to the losses and gains of whole chromosomes detected by
chromosome banding, the deletion of the 6q, copy number alterations
at 5q (gain of a 14.6 Mb region from 5q34 to 5qter) and 9p (loss of
a 28.7-Mb segment from 9p24.3 to 9p21.1); in addition, chromosome
12 showed copy number changes from 10 to 1 along its long arm
(Fig. 3A), similarly to regions of
chromosomes 7, 16 and 17. The overall SNP array results are listed
in Table II.
 | Table II.SNP array results. |
Table II.
SNP array results.
| Chromosome | Gain
(chromosome/cytoband) | Loss
(chromosome/cytoband) |
|---|
| 2 | | 2p11.2 |
| 3 | | 3q13.31–3q13.32,
3q13.31 |
| 4 | | 4q34.3 |
| 5 | 5q34–5q35.3, | 5q34, 5q34 |
| 6 | | 6q16.3–6q27,
6q11.1–6q16.2 |
| 7 | 7,
7p22.3–7q36.3 | |
| 9 | | 9p24.2,
9p24.3-9p24.2, 9p24.2-9p21.1 |
| 10 | | 10, 10q23.1,
10p15.3–10q23.1, 10q23.1–10q26.3 |
| 12 | 12q24.23–12q24.31,
12q24.32, 12q22–12q23.1, 12q14.1–12q21.1, 12q24.12–12q24.13,
12q23.1, 12q23.1–12q23.2, 12q13.2–12q13.3, 12q24.31, 12q24.13,
12q23.3, 12q13.3–12q14.1 | 12q12, 12q23.1,
12q21.1, 12q23.1, 12q23.1, 12q23.2–12q23.3, 12q21.1–12q22,
12q23.3–12q24.12, 12q24.13–12q24.23, 12q14.1,
12q24.32–12q24.33 |
| 14 | 14q11.2 | 14 |
| 17 | 17p13.1, 17p13.2,
17p13.3 |
17q11.1–17q21.31 |
| 19 | 19,
19p13.3–19q13.43 | |
| 20 | 20,
20p13–20q13.33 | |
| 21 | | 21 |
| X | | Xp22.33–Xq28 |
M-FISH analysis confirmed the presence of the
unbalanced translocation der(14)t(6;14)(p12;q11.2) and detected two
additional unbalanced translocations: der(8)t(8;10)(q24.2;q21.1)
and der(9)t(5;9)(q34;p21), in agreement with the SNP array data and
clarified that the additional material on der(12), already detected
by G-banding and SNP-array CGH, originated from amplified material
from chromosomes 7, 12 and 17 (Fig.
3B–E). Chromosome aberrations detected by both SNP array and
M-FISH analyses, were further validated by FISH experiments with
locus-specific BAC probes. Interestingly, the long arm of the
der(12) was shown to be almost entirely composed by amplified
chromosome 12 sequences, together with chromosomes 7, 16 and 17
material (Fig. 4). Moreover, we
finely mapped the breakpoints of all unbalanced translocations
(data not shown). Then, combining classical cytogenetic, molecular
cytogenetic and SNP array data for ANGM-CCS cell line, it was
possible to define the karyotype as:
88∼91,XXYY,−6,+7,+add(9)(p21.1)ishdup(5)(q34)×2,+der(8)t(8;10)(q24.2;q21.1),−10,+der(12)ishins(7;17;16)(12
qter→12q24::7?::17?::16?::12q24.3)×2,+der(14)t(6;14)(p12;q11.2)×2,−17,+19,+20[cp32].
ANGM-CSS showed an increased expression of EGFR and
MET as compared to a normal human astrocytes (NHA) cell line
(data not shown). The relative mRNA expression value for the
MET and EGFR in the tumor cell line was, respectively
4.65 and 1.45. ANGM-CSS did not show methylation of the MGMT
promoter or pathogenic mutations in the hotspot regions (exons 5–8)
of the TP53 gene, neither were mutations detected in codons
12 and 13 of the KRAS gene. No FIG-ROS1 or
FGFR3-TACC3 chimeric gene was detected. Six weeks after the
subcutaneous injection of ANGM-CSS cells in athymic nude mice, all
animals showed growth of a macroscopically visible tumor. Mice were
sacrified eight weeks after injections and the tumors were
immediately excised. One fragment was used for serial
transplantation in other mice whereas the other section was used to
start a new culture. The newly injected mice developed tumors two
weeks after inoculation. The morphology of the cells grown after
heterotransplantation did not differ from the initial culture
(Fig. 1E). The human origin of the
tumor cells was confirmed by chromosome analysis that revealed the
same karyotype (data not shown).
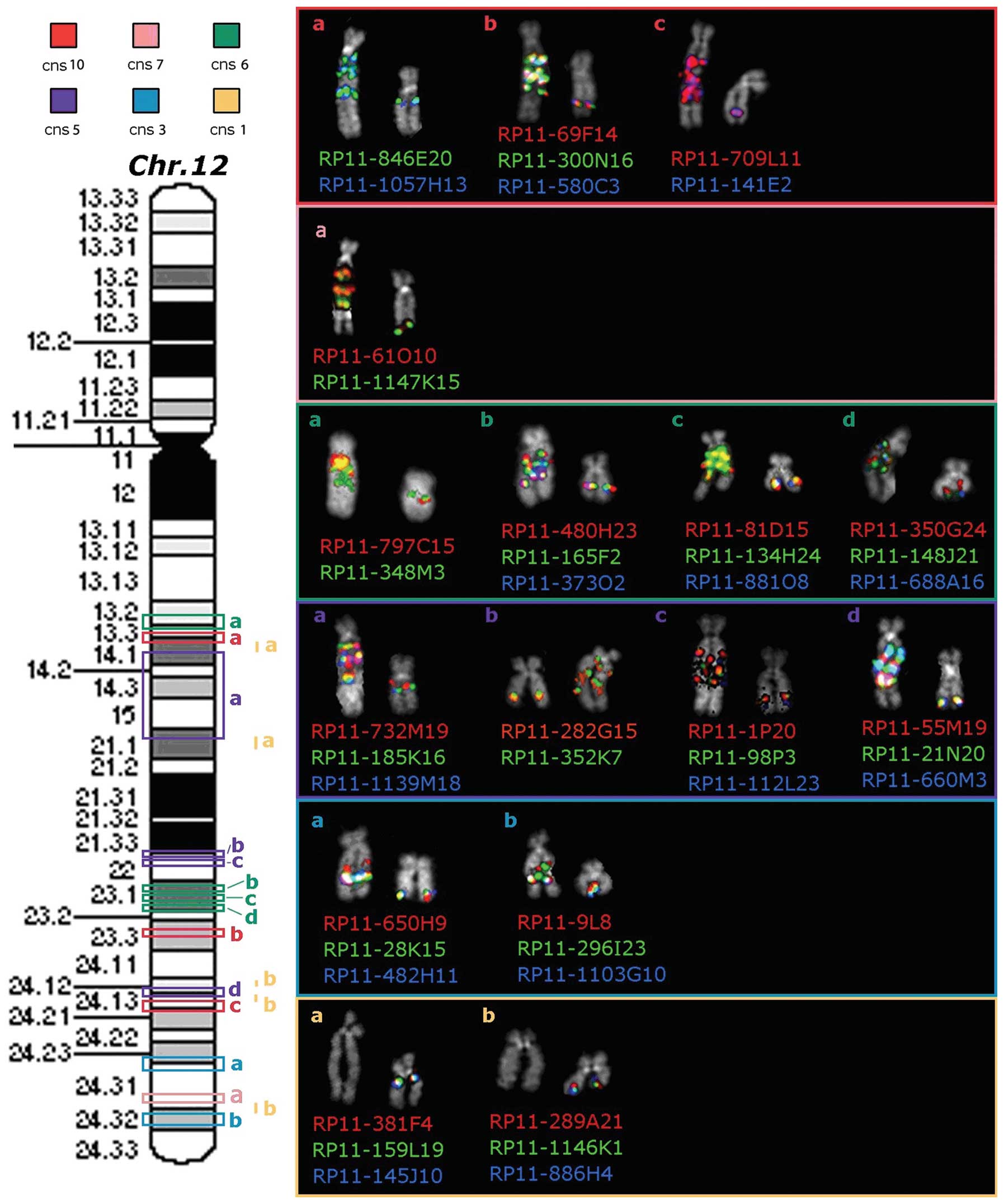 | Figure 4.Map of the chromosome 12
locus-specific BAC probes used in FISH experiments (on the left)
and partial metaphases showing the results obtained on both the
normal and rearranged chromosomes 12 (on the right). Red, pink,
green, violet and light blue rectangles on the chromosome 12
ideogram correspond to amplified sequences with copy number state
(cns) of 10, 7, 6, 5 and 3, respectively. Conversely, yellow bars
at the right side of the ideogram correspond to heterozygously
deleted sequences, with cns of 1. Different chromosome 12 regions,
with the same cns, are indicated, from the centromere to the
telomere, by lowercase letters (a–d) in the same color. FISH
results obtained with BAC probes, corresponding to each
amplified/deleted region, are represented on the right side of the
figure. |
Discussion
We successfully established a novel cell line, named
ANGM-CSS, from a patient with GBM. Cell line immunoreactivity for
GFAP showed a decrease after serial passages in culture, as
previously described (23–27); whereas, the immunohistochemical
positivity for vimentin and nestin was persistent. The
immunohistochemical results recapitulate the phenotype of the GMB
cell lines, according to data from literature (26–28).
We provided a detailed characterization of ANGM-CSS by use of
cytogenetic and molecular approaches. As expected, the cytogenetic
analysis of ANGM-CSS showed a very complex karyotype characterized
by several chromo-some aberrations, in line with the data described
in previous studies concerning human glioma cell lines (29–31).
Gliomas have been extensively analyzed by genetic techniques. They
typically show highly complex karyotypes (31). The most common numerical
chromosomal changes include losses of 9p, chr10, chr17 and chr22
and gain of chromosomes chr7 and chr20 (31–35).
Other structural abnormalities were also reported in chromosomal
arms 1p, 6q, 9p, 9q and 13q (31).
Similary to the latter data, G-banding, molecular cytogenetic and
SNP array analysis performed on ANGM-CSS disclosed a
near-tetraploid karyotype harbouring numerous copy number changes,
including gain of whole chromosomes 7, 20 and 19 and loss of whole
chromosomes 10, 14 and 21, in addition to gains of sub-regions in
5q, 7 and 17 and loss of 6q and 9p. Moreover, three structural
chromosome rearrangements were identified in ANGM-CSS: a
t(8;10)(q24.2;q21.1), a t(6;14)(p12;q11.2) and an add(9)(p21.1) ish
dup(5)(q34). In summary, the genetic analyses indicate that the
ANGM-CSS cell line recapitulates the key properties of GBM. The
Cancer Genome Atlas Research Network reported in GBM frequent
genetic alterations in three critical pathways: the RTK/RAS/PI3K,
the p53 and the RB signalling pathways (36). ANGM-CSS presents a profound
deregulation of proliferation and survival due to disruption of the
RTK/RAS/PI3K pathway due to the amplification of the EGFR
and MET genes and the homozygous deletion of NF1 and
PTEN genes, as detected by SNP array analysis (Table I). Quantitative RT-PCR analysis
confirmed an increased expression at the mRNA level of the
EGFR and MET genes. The cell line under study does
not show the presence of FIG-ROS1 and FGFR3-TACC3
chimeric transcripts, already described for U118MG glioblastoma
cell line and for primary tumors, respectively (19,20).
 | Table I.Comparison between the genetic
abnormalities reported by the Cancer Genome Atlas (TCGC) research
network at gene loci critically involved in gliomas and gains and
losses detected in ANGM-CSS cell line by SNP array
analysis.a |
Table I.
Comparison between the genetic
abnormalities reported by the Cancer Genome Atlas (TCGC) research
network at gene loci critically involved in gliomas and gains and
losses detected in ANGM-CSS cell line by SNP array
analysis.a
| Gene | Locus | TCGAb | ANGM-CSS |
|---|
| Genes involved in
RTK/RAS/PI-3K signaling | | | |
| EGFR | 7p12.3-p12.1 | Amplified | 45% amplified |
| ERBB2 | 17q21.1 | Mutated | 8% homozygous
deletion |
| PDGFRA | 4q12 | Amplified | 13% no change |
| MET | 7q31 | Amplified | 4% amplified |
| NF1 | 17q11.2 | Homozygous
deletion | 18% homozygous
deletion |
| RAS | 6p21.3 | Mutated | 2% no change |
| PI3K | 3q26.3 | Mutated | 15% no change |
| PTEN | 10q23.31 | Homozygous
deletion | 36% homozygous
deletion |
| AKT | 14q32.3 | Amplified | 2% no change |
| FOXO | 6q21 | Mutated | 1% homozygous
deletion |
| TP53
regulation | | | |
| CDKN2A | 9p21 | Homozygous
deletion | 49% deleted
Hom |
| MDM2 | 12q14.3–q15 | Amplified | 14% amplified |
| MDM4 | 1q32 | Amplified | 7% no change |
| TP53 | 17p13.1 | Homozygous
deletion | 35% no change |
| RB signaling | | | |
| CDKN2A | 9p21 | Homozygous
deletion | 52% homozygous
deletion |
| CDKN2B | 9p21 | Homozygous
deletion | 47% homozygous
deletion |
| CDKN2C | 1p32 | Homozygous
deletion | 2% no change |
| CDK4 | 12q14 | Amplified | 18% amplified |
| CCND2 | 12p13 | Amplified | 2% amplified |
| CDK6 | 7q21–22 | Amplified | 1% amplified |
| RB1 | 13q14.1–q14.2 | Homozygous
deletion | 11% homozygous
deletion |
While no mutation or deletions were detected in the
TP53 gene, ANGM-CSS was characterized by amplification of
the MDM2 gene and homozygous deletion of the CDKN2A
gene (Table I). The MDM2
gene encodes for a protein involved in the degradation of the p53
protein and its expression is negatively regulated by the ARF
protein, encoded by CDKN2A, leading to an abnormal
regulation of apoptosis and senescence mediated by p53 (37). The Rb pathway was affected by the
amplification of CDK4 and CDK6 and the homozygous
deletion of the CDKN2A/CDKN2B genes. All these alterations
affect the G1/S phase progression. Another feature of the ANGM-CSS
is the absence of methylation in the promoter region of the
MGMT gene. Alkylating agents induce cell death by forming
cross-links between adjacent DNA strands through the alkylation of
the O6 position of guanine. MGMT promoter
hypermethylation with consequent loss of MGMT protein expression
reduces the DNA repair activity of glioma cells overcoming
resistance to alkylating agents. The absence of MGMT
promoter hypermethylation in ANGM-CSS leads to a transcriptionally
active MGMT which rapidly removes the alkyl adducts
preventing the formation of cross-links thereby causing resistance
to alkylating drugs (38,39). ANGM-CSS cell also gave rise to
tumors in vivo. All six SCID mice that were injected with
tumor cells developed solid tumors, demonstrating that this cell
line is capable of being propagated in animal models, which may aid
in the development of test systems for new therapies.
Acknowledgements
This study was supported by Italian
Health Ministry.
References
|
1.
|
Furnari FB, Fenton T, Bachoo RM, et al:
Malignant astrocytic glioma: genetics, biology, and paths to
treatment. Genes Dev. 21:2683–26710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Porter K, McCarthy B, Berbaum M, et al:
Conditional survival of all primary brain tumor patients by age,
behavior, and histology. Neuroepidemiology. 36:230–239. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Preusser M, de Ribaupierre S, Wöhrer A, et
al: Current concepts and management of glioblastoma. Ann Neurol.
70:9–21. 2011. View Article : Google Scholar
|
|
4.
|
Cheng L, Wu Q, Guryanova O, et al:
Elevated invasive potential of glioblastoma stem cells. Biochem
Biophys Res Commun. 406:643–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Stupp R, Mason WP, van den Bent MJ, et al:
Radiotherapy plus concomitant and adjuvant temozolomide for
glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Stupp R, Hegi ME, Mason WP, et al: Effects
of radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009.
|
|
8.
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bakir A, Gezen F, Yildiz O, et al:
Establishment and characterization of a human glioblastoma
multiforme cell line. Cancer Genet Cytogenet. 103:46–51. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Nuki Y, Uchinokura S, Miyata S, et al:
Establishment and characterization of a new human glioblastoma cell
line, NYGM. Hum Cell. 17:145–150. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Wang J, Wang X, Jiang S, et al:
Establishment of a new human glioblastoma multiforme cell line
(WJ1) and its partial characterization. Cell Mol Neurobiol.
27:831–843. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Fomchenko EI and Holland EC: Mouse models
of brain tumors and their applications in preclinical trials. Clin
Cancer Res. 12:5288–5297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Palumbo O, Palumbo P, Palladino T, et al:
A novel deletion in 2q24.1q24.2 in a girl with mental retardation
and generalized hypotonia: a case report. Mol Cytogenet. 5:12012.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Macchia G, Trombetta D, Möller E, et al:
FOSL1 as a candidate target gene for 11q12 rearrangements in
desmoplastic fibroblastoma. Lab Invest. 92:735–743. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Scintu M, Vitale R, Prencipe M, et al:
Genomic instability and increased expression of BUB1B and MAD2L1
genes in ductal breast carcinoma. Cancer Lett. 254:298–307. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Parrella P, la Torre A, Copetti M, et al:
High specificity of quantitative methylation-specific PCR analysis
for MGMT promoter hypermethylation detection in gliomas. J Biomed
Biotechnol. 2009:5316922009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Hibi K, Robinson CR, Booker S, et al:
Molecular detection of genetic alterations in the serum of
colorectal cancer patients. Cancer Res. 58:1405–1407.
1998.PubMed/NCBI
|
|
18.
|
Gu TL, Deng X, Huang F, et al: Survey of
tyrosine kinase signaling reveals ROS kinase fusions in human
cholangiocarcinoma. PLoS One. 6:e156402011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Singh D, Chan JM, Zoppoli P, et al:
Transforming fusions of FGFR and TACC genes in human glioblastoma.
Science. 337:1231–1235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Charest A, Lane K, McMahon K, et al:
Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma
with an interstitial del(6) (q21q21). Genes Chromosomes Cancer.
37:58–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Burfeind P, Chernicky CL, Rininsland F, et
al: Antisense RNA to the type I insulin-like growth factor receptor
suppresses tumor growth and prevents invasion by rat prostate
cancer cells in vivo. Proc Natl Acad Sci USA. 93:7263–7268. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Shaffer LG, Slovak ML and Campbell LJ:
ISCN 2009: An International System for Human Cytogenetic
Nomenclature. S. Karger; Basel: 2009
|
|
23.
|
Lolait SJ, Harmer JH, Auteri G, et al:
Expression of glial fibrillary acidic protein, actin, fibronectin
and factor VIII antigen in human astrocytomas. Pathology.
15:373–378. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Lipsky RH and Silverman SJ: Effects of
mycophenolic acid on detection of glial filaments in human and rat
astrocytoma cultures. Cancer Res. 47:4900–4904. 1987.PubMed/NCBI
|
|
25.
|
Bocchini V, Casalone R, Collini P, et al:
Changes in glial fibrillary acidic protein and karyotype during
culturing of two cell lines established from human glioblastoma
multiforme. Cell Tissue Res. 265:73–81. 1991. View Article : Google Scholar
|
|
26.
|
Veselska R, Kuglik P, Cejpek P, et al:
Nestin expression in the cell lines derived from glioblastoma
multiforme. BMC Cancer. 6:322003. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Jiang Y and Uhrbom L: On the origin of
glioma. Ups J Med Sci. 117:113–121. 2012. View Article : Google Scholar
|
|
28.
|
Krupkova O, Loja T, Zambo I, et al: Nestin
expression in human tumors and tumor cell lines. Neoplasma.
57:291–298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Bigner DD, Bigner SH, Ponten J, et al:
Heterogeneity of Genotypic and phenotypic characteristics of
fifteen permanent cell lines derived from human gliomas. J
Neuropathol Exp Neurol. 40:201–229. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Roversi G, Pfundt R, Moroni RF, et al:
Identification of novel genomic markers related to progression to
glioblastoma through genomic profiling of 25 primary glioma cell
lines. Oncogene. 25:1571–1583. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Dahlback HS, Brandal P, Meling TR, et al:
Genomic aberrations in 80 cases of primary glioblastoma multiforme:
Pathogenetic heterogeneity and putative cytogenetic pathways. Genes
Chromosomes Cancer. 48:908–924. 2009. View Article : Google Scholar
|
|
32.
|
Rey JA, Bello MJ, de Campos JM, et al:
Chromosomal patterns in human malignant astrocytomas. Cancer Genet
Cytogenet. 29:201–221. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Bigner SH, Mark J and Bigner DD:
Cytogenetics of human brain tumors. Cancer Genet Cytogenet.
47:141–154. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kruse CA, Mitchell DH,
Kleinschmidt-DeMasters BK, et al: Characterization of a continuous
human glioma cell line DBTRG-05MG: growth kinetics, karyotype,
receptor expression, and tumor suppressor gene analyses. In Vitro
Cell Dev Biol. 28A:609–614. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Gjerset RA, Fakhrai H, Shawler DL, et al:
Characterization of a new human glioblastoma cell line that
expresses mutant p53 and lacks activation of the PDGF pathway. In
Vitro Cell Dev Biol Anim. 31:207–214. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Uhrinova S, Uhrin D, Powers H, et al:
Structure of free MDM2 N-terminal domain reveals conformational
adjustments that accompany p53-binding. J Mol Biol. 350:587–598.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Esteller M, Garcia-Foncillas J, Andion E,
et al: Inactivation of the DNA-repair gene MGMT and the clinical
response of gliomas to alkylating agents. N Engl J Med.
343:1350–1354. 2000. View Article : Google Scholar
|
|
39.
|
Hegi ME, Diserens AC, Gorlia T, et al:
MGMT gene silencing and benefit from temozolomide in glioblastoma.
N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|