Introduction
Prostate cancer (PCa) is the most frequently
diagnosed cancer and the second leading cause of cancer death among
men in developed countries (1).
Most patients are initially responsive to androgen-deprivation
therapy (ADT); however, PCa can eventually become resistant to ADT
and progress to castration-resistant prostate cancer (CRPC).
Currently, CRPC is difficult to treat, and most clinical trials for
advanced PCa have shown limited benefits, with disease progression
and metastasis to the bone or other sites (2,3).
Therefore, understanding the molecular mechanisms of CRPC and the
metastatic pathways underlying PCa using currently available
genomic approaches would help to elucidate for this disease.
The discovery of non-coding RNAs (ncRNAs) in the
human genome was an important conceptual breakthrough in the
post-genome sequencing era (4).
Further improving our understanding of ncRNAs is necessary for
continued progress in cancer research. MicroRNAs (miRNAs) are
endogenous small ncRNA molecules (19–22 bases in length) that
regulate protein-coding gene expression by repressing translation
or cleaving RNA transcripts in a sequence-specific manner (5). Numerous studies have shown that
miRNAs are aberrantly expressed in many human cancers and that they
play significant roles in the initiation, development and
metastasis of those cancers (6–9).
Moreover, normal regulatory mechanisms can be disrupted by the
aberrant expression of tumor-suppressive or oncogenic miRNAs in
cancer cells (10,11). Therefore, identification of
aberrantly expressed miRNAs is an important first step toward
elucidating miRNA-mediated oncogenic pathways.
Based on these data, we have sought to elucidate the
miRNA expression signatures of PCa clinical specimens and have
investigated the specific roles of miRNAs in PCa oncogenesis using
differentially expressed miRNAs (12). Recently, we demonstrated that
several miRNAs are downregulated in PCa and that miR-1/133a
and miR-143/145 clusters function as tumor suppressors,
targeting several oncogenic genes (13,14).
Data from our analysis of the miRNA signature of PCa showed that
miR-29s (including miR-29a/b/c) are significantly
downregulated in cancer tissues, suggesting that miR-29s may act as
tumor suppressors.
The aim of the present study was to investigate the
functional significance of miR-29s and to identify the
molecular pathways and targets regulated by these miRNAs in PCa
cells. Our data demonstrated that restoration of mature
miR-29s inhibited cancer cell proliferation. Moreover, gene
expression data and in silico database analysis showed that
laminin γ1 (LAMC1), which is involved in the ‘focal
adhesion’ pathway, was a potential target of miR-29-mediated
regulation. Silencing of the LAMC1 gene significantly
inhibited cell migration and invasion in cancer cells. Thus, our
study revealed that tumor-suppressive miR-29s regulated
cancer pathways and cancer-related target molecules, providing new
insights into the potential mechanisms of PCa oncogenesis and
metastasis.
Materials and methods
Clinical prostate specimens
Clinical specimens were obtained from patients at
the Teikyo University Chiba Medical Center from 2012 to 2013. All
patients had elevated levels of prostate-specific antigen (PSA) and
had undergone transrectal prostate needle biopsy. Non-cancerous
prostate tissue (n=33) was obtained from patients who were negative
for malignancy without indurations on the prostate. Prostate cancer
tissues (n=37) contained 90–100% malignant cells in the biopsy
cores.
The patient backgrounds and clinicopathological
characteristics are summarized in Table 1A and B. Before tissue collection, all patients
provided written informed consent of tissue donation for research
purposes. The protocol was approved by the Institutional Review
Board of Teikyo University.
 | A, Patient characteristics used for
miR-29s expression |
A, Patient characteristics used for
miR-29s expression
| No. | Age (yrs.) | PSA (ng/ml) | Gleason score | TNM classification
|
|---|
| T | N | M |
|---|
| PCa | | | | | | |
| 1 | 67 | 244 | 4+4 | 4 | 1 | 1 |
| 2 | 70 | 395 | 4+4 | 3a | 1 | 1 |
| 3 | 83 | 49.9 | 4+5 | 3a | 0 | 0 |
| 4 | 68 | 212 | 4+4 | 3b | 1 | 0 |
| 5 | 80 | 589 | 4+4 | 3b | 1 | 0 |
| 6 | 72 | 2530 | 4+5 | 3b | 0 | 1 |
| 7 | 76 | 12.5 | 4+5 | 3b | 1 | 1 |
| 8 | 67 | 153 | 4+4 | 4 | 1 | 1 |
| 9 | 82 | 808.8 | 4+4 | 4 | 1 | 1 |
| 10 | 88 | 50.5 | 4+4 | 3a | 0 | 0 |
| 11 | 69 | 3.45 | 4+3 | 3a | 0 | 0 |
| 12 | 64 | 486 | 4+5 | 4 | 1 | 1 |
| 13 | 74 | 60.8 | 5+5 | 4 | 1 | 1 |
| 14 | 63 | 49.6 | 4+4 | 3b | 1 | 0 |
| 15 | 69 | 1060 | 4+5 | 4 | 1 | 1 |
| 16 | 67 | 34.9 | 4+4 | 3b | 1 | 1 |
| 17 | 64 | 7.23 | 4+4 | 3b | 1 | 0 |
| 18 | 79 | 3750 | 4+4 | 3a | 0 | 1 |
| 19 | 78 | 1400 | 4+4 | 3a | 0 | 1 |
| 20 | 77 | 2640 | 4+4 | 4 | 1 | 1 |
| 21 | 69 | 554 | 4+4 | 3b | 1 | 1 |
| 22 | 82 | 59.5 | 4+5 | 3b | 0 | 0 |
| 23 | 70 | 1030 | 4+4 | 3b | 1 | 1 |
| 24 | 78 | 892 | 4+5 | 4 | 1 | 1 |
| 25 | 86 | 24.2 | 4+4 | 3b | 0 | 0 |
| 26 | 67 | 64.6 | 4+5 | 3b | 0 | 1 |
| 27 | 71 | 63.8 | 4+3 | 3a | 0 | 0 |
|
| Non-PCa | | | | | | |
| 28 | 69 | 12.2 | | | | |
| 29 | 66 | 11.9 | | | | |
| 30 | 85 | 10.1 | | | | |
| 31 | 55 | 11.2 | | | | |
| 32 | 67 | 22 | | | | |
| 33 | 66 | 7.33 | | | | |
| 34 | 69 | 7.92 | | | | |
| 35 | 68 | 7.24 | | | | |
| 36 | 53 | 4.33 | | | | |
| 37 | 62 | 5.14 | | | | |
| 38 | 63 | 10 | | | | |
| 39 | 62 | 5.11 | | | | |
| 40 | 74 | 8.3 | | | | |
| 41 | 65 | 4.3 | | | | |
| 42 | 72 | 5.43 | | | | |
| 43 | 66 | 5.35 | | | | |
| 44 | 67 | 5 | | | | |
| 45 | 66 | 19.5 | | | | |
| 46 | 63 | 12.9 | | | | |
| 47 | 53 | 6.69 | | | | |
| 48 | 63 | 5.3 | | | | |
| 49 | 66 | 7.73 | | | | |
| 50 | 57 | 7.3 | | | | |
 | B, Patient characteristics used for
LAMC1 expression |
B, Patient characteristics used for
LAMC1 expression
| No. | Age (yrs.) | PSA (ng/ml) | Gleason score | TNM classification
|
|---|
| T | N | M |
|---|
| PCa | | | | | | |
| 1 | 78 | 989 | 4+4 | 4 | 1 | 1 |
| 2 | 69 | 26 | 3+4 | 3b | 0 | 0 |
| 3 | 78 | 19.3 | 4+4 | 3a | 0 | 0 |
| 4 | 73 | 478 | 4+3 | 3b | 0 | 1 |
| 5 | 72 | 102 | 4+4 | 3a | 0 | 0 |
| 6 | 65 | 212 | 4+4 | 4 | 1 | 1 |
| 7 | 81 | 11.4 | 4+4 | 2 | 0 | 0 |
| 8 | 78 | 121 | 4+5 | 3b | 1 | 0 |
| 9 | 79 | 633 | 4+5 | 3b | 1 | 0 |
| 10 | 58 | 482 | 4+4 | 3b | 1 | 0 |
|
| Non-PCa | | | | | | |
| 11 | 60 | 5.6 | | | | |
| 12 | 56 | 8.4 | | | | |
| 13 | 61 | 8.6 | | | | |
| 14 | 62 | 35.5 | | | | |
| 15 | 73 | 6.0 | | | | |
| 16 | 57 | 5.2 | | | | |
| 17 | 64 | 4.4 | | | | |
| 18 | 60 | 5.7 | | | | |
| 19 | 63 | 11.4 | | | | |
| 20 | 65 | 13.2 | | | | |
Cell culture and RNA extraction
Human prostate cancer cells (PC3 and DU145) were
obtained from the American Type Culture Collection (Manassas, VA,
USA) and maintained in RPMI-1640 medium supplemented with 10% fetal
bovine serum in a humidified atmosphere of 5% CO2 and
95% air at 37°C.
Total RNA was isolated using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
protocol. RNA quality was confirmed using an Agilent 2100
Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Quantitative real-time reverse
transcription polymerase chain reaction (RT-PCR)
The procedure for PCR quantification was carried out
as previously described (12–14).
TaqMan probes and primers for LAMC1 (P/N: Hs00267056_m1; Applied
Biosystems, Foster City, CA, USA) and for GUSB (the internal
control; P/N: Hs00939627_m1; Applied Biosystems) were
assay-on-demand gene expression products. The expression levels of
miR-29a (Assay ID: 002112), miR-29b (Assay ID:
000413) and miR-29c (Assay ID: 000587) were analyzed by
TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay; Applied
Biosystems) and normalized to the expression of RNU48 (Assay
ID: 001006). All reactions were performed in triplicate, and each
assay included negative control reactions that lacked cDNA.
Transfection with mature miRNA and
small-interfering RNA (siRNA)
The following mature miRNA species were used in this
study: mirVana miRNA mimic for hsa-miR-29a-3p (product ID:
MC12499; Applied Biosystems), hsa-miR-29b-3p (product ID:
MC10103), and hsa-miR-29c-3p (product ID: MC10518). The
following si-RNAs were used: Stealth Select RNAi si-RNA,
si-LAMC1 (P/N: HSS105959, HSS180591; Invitrogen), and
negative control miRNA/siRNA (P/N: AM17111; Applied Biosystems).
RNAs were incubated with OPTI-MEM (Invitrogen) and Lipofectamine
RNAiMax reagent (Invitrogen). The transfection efficiencies of
miRNA in PC3 and DU145 cells were confirmed based on downregulation
of TWF1 (PTK9) mRNA following transfection with
miR-1 as previously reported (15).
Cell proliferation, migration and
invasion assays
To investigate the functional significance of
miR-29s, we performed cell proliferation, migration and
invasion assays using PC3 and DU145 cells. The experimental
procedures were performed as described in our previous studies
(12–16).
Western blotting
Cells were harvested 72 h after transfection, and
lysates were prepared. Fifty micrograms of protein from each
lysates was separated on Mini-Protean TGX gels (Bio-Rad, Hercules,
CA, USA) and transferred to PVDF membranes. Immunoblotting was
performed with mouse anti-LAMC1 antibodies (1:250; HSA001909;
Sigma-Aldrich, St. Louis, MO, USA), and anti-GAPDH antibodies
(1:1000; ab8245, Abcam, Cambridge, UK) were used as an internal
loading control.
Genome-wide gene expression and in silico
analysis for the identification of genes regulated by miR-29s
To identify target genes of miR-29s, we used
in silico analysis and genome-wide gene expression analysis.
First, we screened genes using TargetScan release 6.2 (http://www.targetscan.org/). These genes were then
categorized into KEGG (Kyoto Encyclopedia of Genes and Genomes)
pathways using GeneCodis analysis (http://genecodis.cnb.csic.es/). To identify
upregulated genes in PCa, we analyzed a publicly available gene
expression data set in GEO (accession no.: GSE29079).
Plasmid construction and dual-luciferase
reporter assay
Partial sequences of the LAMC1
3′-untranslated region (UTR) of putative miR-29s binding
sites (180 bp) were inserted between the XhoI-PmeI
restriction sites in the 3′-UTR of the hRluc gene in the
psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). PC3 cells
were transfected with 5 ng of the vector and 10 nM miR-29s
using Lipofectamine 2000 (Invitrogen). The activities of firefly
and Renilla luciferases in cell lysates were determined with
a dual-luciferase assay system (E1910; Promega). Normalized data
were calculated as the ratio of Renilla/firefly luciferase
activities as previously described (17).
Statistical analysis
The relationships between 2 groups and the numerical
values obtained by real-time RT-PCR were analyzed using paired
t-tests. The relationships among 3 variables and numerical values
were analyzed using the Bonferroni-adjusted Mann-Whitney U test.
All analyses were performed using Expert StatView software (version
4, SAS Institute Inc., Cary, NC, USA).
Results
Expression levels of miR-29s
(miR-29a/b/c) in PCa specimens
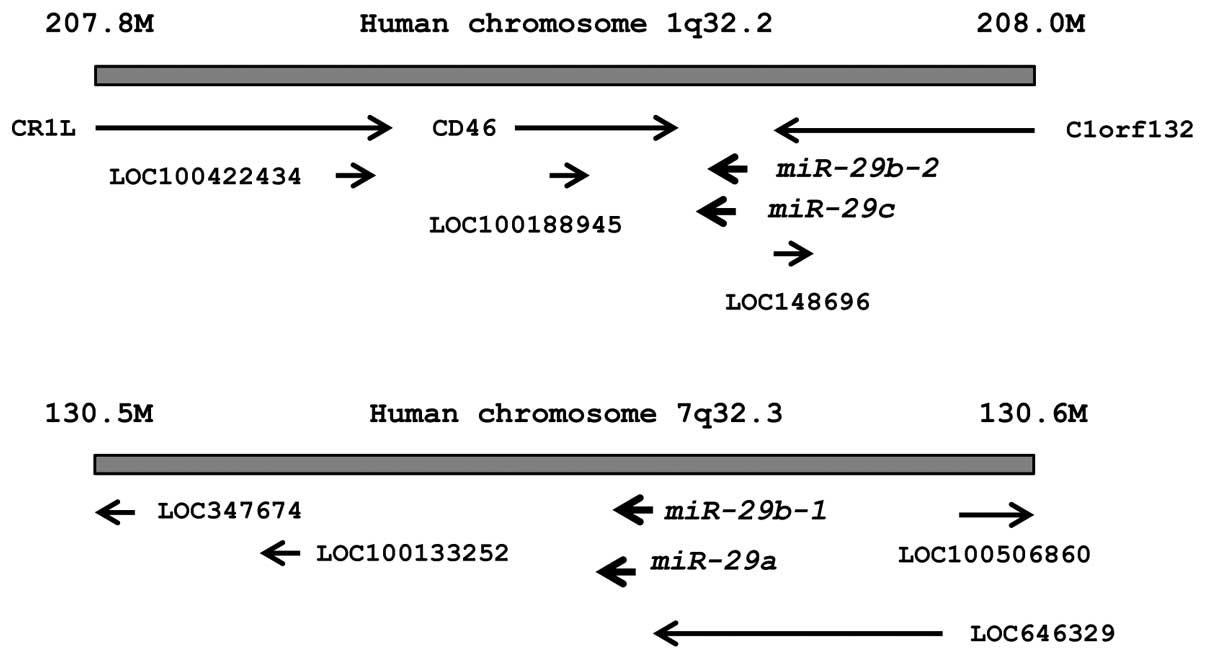
The chromosomal locations of miR-29s in the
human genome are shown in Fig. 1.
These miRNAs were clustered at 2 different human genomic loci;
miR-29b-1 and miR-29a were located at 7q32, and
miR-29b-2 and miR-29c were located at 1q32.
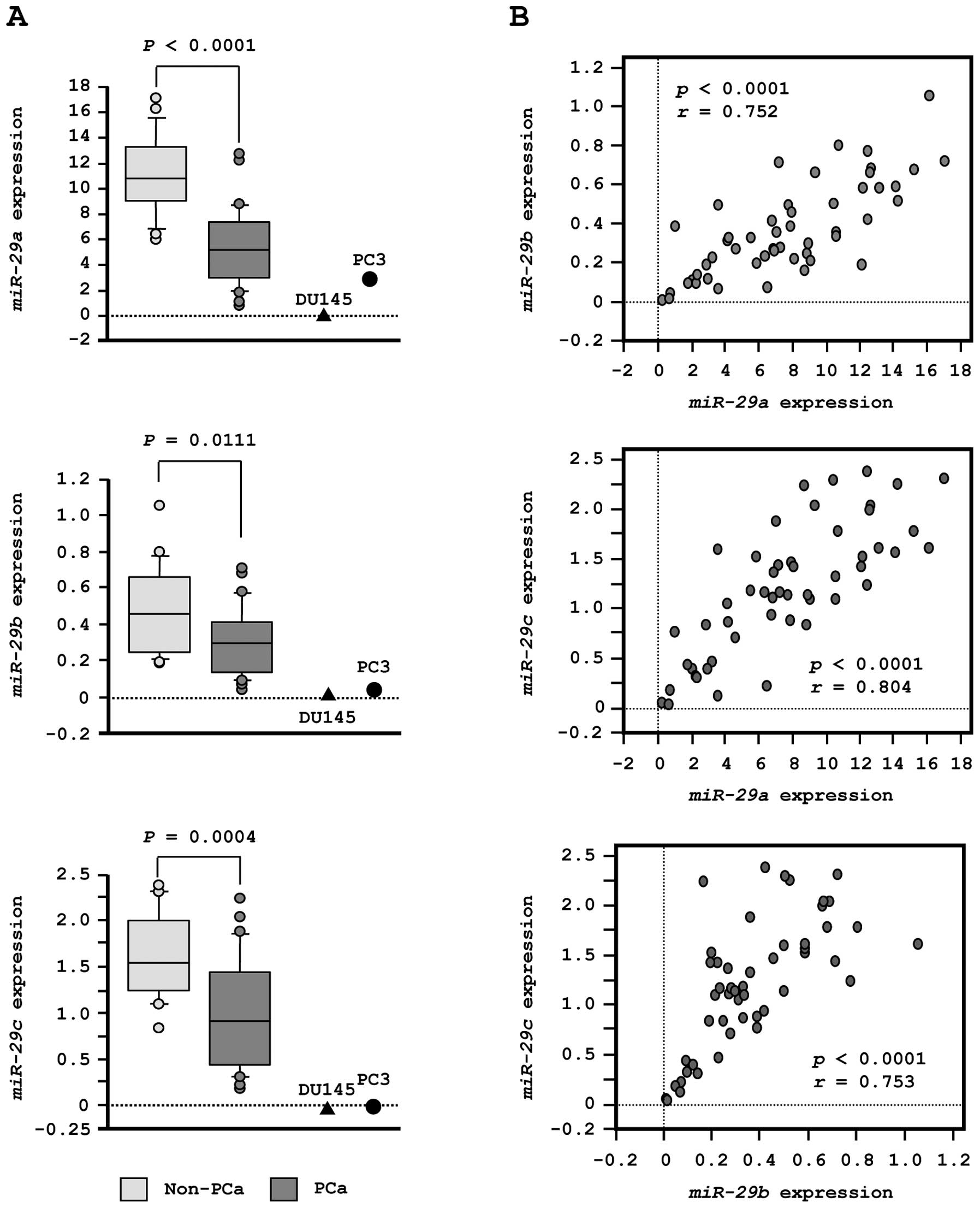
First, we evaluated the expression levels of
miR-29s (miR-29a/b/c) in normal prostate tissues (n=23) and
PCa tissues (n=27). In patients from whom normal prostate tissues
were collected, the median PSA level was 7.3 ng/ml (range, 4.3–22
ng/ml). In contrast, in patients from whom PCa tissues were
collected, PSA levels were quite high, with a median of 212 ng/ml
(range, 3.5–3750 ng/ml). Twenty-one PCa patients had progressive
disease with N1 or M1 according to TNM classification (Table IA).
To validate our previous miRNA profiling results in
PCa, we evaluated expression of miR-29s in clinical PCa
specimens. The expression levels of miR-29a, miR-29b and
miR-29c were significantly lower in tumor tissues than in
corresponding non-cancerous tissues (P<0.0001, P= 0.0111 and P=
0.0004, respectively; Fig. 2A).
Spearman’s rank test showed a positive correlation between the
expression of miR-29a and that of miR-29b (R=0.752
and P<0.0001, Fig. 2B). The
expression of miR-29a was positively correlated with that of
miR-29c (R= 0.804 and P<0.0001, Fig. 2B). Similarly, the expression of
miR-29b was positively correlated with that of
miR-29c (R=0.753 and P<0.0001, Fig. 2B).
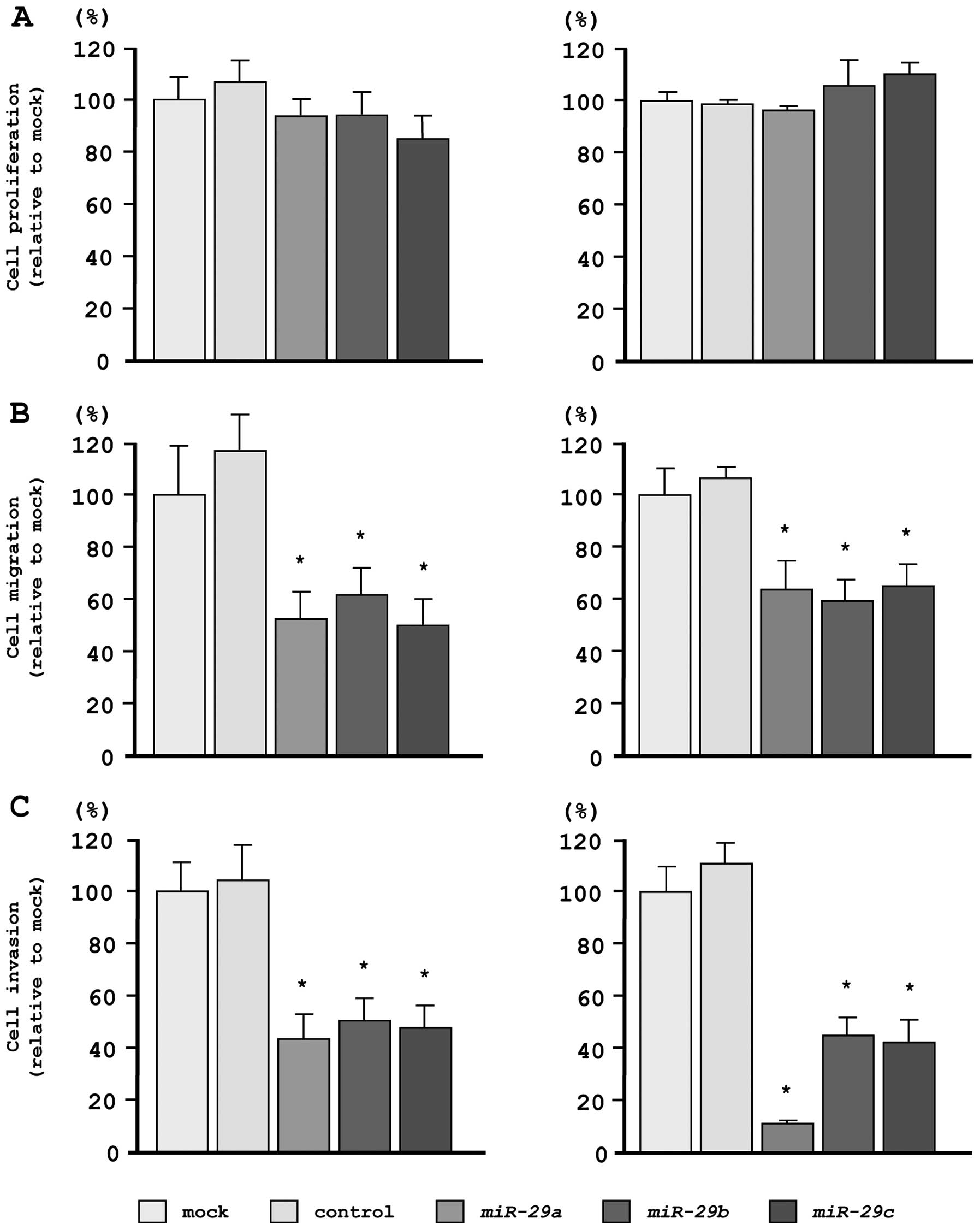
Effects of restoring miR-29s expression
levels on cell proliferation, migration, and invasion in PC3 and
DU145 PCa cells
To investigate the functional effects of
miR-29s, we performed gain-of-function studies using miRNA
transfection in PC3 and DU145 cell lines.
As observed using XTT assays, cell proliferation was
not inhibited in miR-29s transfectants, as compared with
mock- or miR-control-transfected cells (Fig. 3A). However, cell migration activity
was significantly inhibited in miR-29s transfectants, as
compared with mock- or miR-control-transfected cells (Fig. 3B). Moreover, in Matrigel invasion
assays, transfection with miR-29s significantly inhibited
cell invasion as compared with mock- or miR-control-transfected
cells (Fig. 3C).
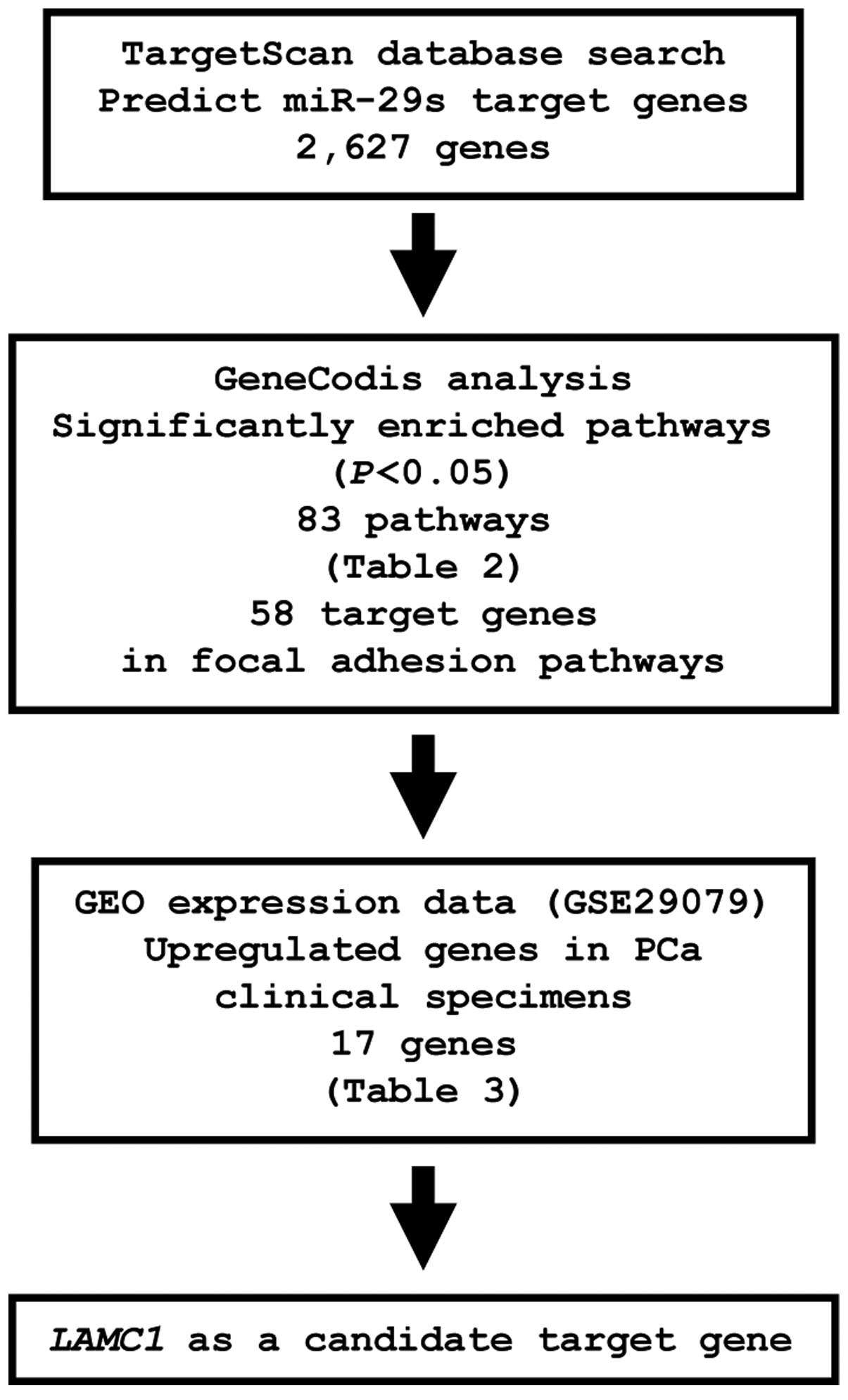
Identification of candidate genes
targeted by miR-29s
To identify genes targeted by miR-29s, we
analyzed a combination of in silico and gene expression data
in PCa. First, we screened miR-29s-targeted genes using the
TargetScan database and identified 2,627 genes, which were then
categorized into KEGG pathways using GeneCodis analysis, yielding
83 pathways identified as significantly enriched (Table II). Among these pathways, we
focused on the focal adhesion pathway because this pathway has been
implicated in cancer cell migration and invasion. A total of 58
genes were identified in this pathway. The gene set was then
analyzed with available gene expression data from the GEO
(accession no.: GSE29079), and genes upregulated in PCa were chosen
(Fig. 4).
 | Table II.Significantly enriched pathways among
predicted miR-29s target genes (top 10 pathways). |
Table II.
Significantly enriched pathways among
predicted miR-29s target genes (top 10 pathways).
| No. of genes | P-value | Annotations |
|---|
| 77 | 8.47E-18 | (KEGG) 05200:
Pathways in cancer |
| 58 | 1.13E-17 | (KEGG) 04510: Focal
adhesion |
| 34 | 3.41E-15 | (KEGG) 05222: Small
cell lung cancer |
| 30 | 9.11E-12 | (KEGG) 04512:
ECM-receptor interaction |
| 46 | 6.31E-11 | (KEGG) 04144:
Endocytosis |
| 32 | 6.86E-11 | (KEGG) 05146:
Amoebiasis |
| 33 | 2.52E-09 | (KEGG) 05145:
Toxoplasmosis |
| 25 | 6.83E-09 | (KEGG) 04974:
Protein digestion and absorption |
| 30 | 4.35E-07 | (KEGG) 04360: Axon
guidance |
| 29 | 7.07E-07 | (KEGG) 04722:
Neurotrophin signaling pathway |
From this selection, 17 candidate genes were
identified as miR-29s targets (Table III). Among these candidates, we
focused on the LAMC1 gene and examined LAMC1 function
and characteristics in further analyses.
 | Table III.Candidate target genes for
miR-29s in focal adhesion pathway. |
Table III.
Candidate target genes for
miR-29s in focal adhesion pathway.
| Entrez gene ID | Gene symbol | Gene name | Location | GEO expression data
| Conserved
sites | Poorly conserved
sites |
|---|
| Fold change | Log FC |
|---|
| 1277 | COL1A1 | Collagen, type I,
α1 | 17q21.33 | 2.35 | 1.23 | 3 | 0 |
| 1278 | COL1A2 | Collagen, type I,
α2 | 7q22.1 | 1.74 | 0.80 | 2 | 0 |
| 3480 | IGF1R | Insulin-like growth
factor 1 receptor | 15q26.3 | 1.69 | 0.71 | 0 | 1 |
| 1281 | COL3A1 | Collagen, type III,
α1 | 2q31 | 1.67 | 0.74 | 2 | 0 |
| 3915 | LAMC1 | Laminin, γ1
(formerly LAMB2) | 1q31 | 1.63 | 0.71 | 1 | 0 |
| 7058 | THBS2 | Thrombospondin
2 | 6q27 | 1.54 | 0.62 | 0 | 1 |
| 7057 | THBS1 | Thrombospondin
1 | 15q15 | 1.53 | 0.61 | 0 | 1 |
| 5159 | PDGFRB | Platelet-derived
growth factor receptor, β polypeptide | 5q33.1 | 1.30 | 0.38 | 1 | 0 |
| 8503 | PIK3R3 |
Phosphoinositide-3-kinase, regulatory
subunit 3 (γ) | 1p34.1 | 1.28 | 0.36 | 1 | 0 |
| 1282 | COL4A1 | collagen, type IV,
α1 | 13q34 | 1.26 | 0.33 | 2 | 0 |
| 2889 | RAPGEF1 | Rap guanine
nucleotide exchange factor (GEF) 1 | 9q34.3 | 1.26 | 0.33 | 0 | 1 |
| 1293 | COL6A3 | Collagen, type VI,
α3 | 2q37 | 1.25 | 0.32 | 1 | 0 |
| 1290 | COL5A2 | Collagen, type V,
α2 | 2q14-q32 | 1.19 | 0.25 | 2 | 0 |
| 22801 | ITGA11 | Integrin, α11 | 15q23 | 1.19 | 0.25 | 1 | 0 |
| 208 | AKT2 | v-akt murine
thymoma viral oncogene | 19q13.1-q13.2 | 1.15 | 0.21 | 0 | 4 |
| 1284 | COL4A2 | homolog 2 collagen,
type IV, α2 | 13q34 | 1.15 | 0.21 | 1 | 0 |
| 1399 | CRKL | v-crk sarcoma virus
CT10 oncogene homolog (avian)-like | 22q11.21 | 1.11 | 0.15 | 0 | 1 |
LAMC1 is a direct target of miR-29s in
PCa cells
We performed quantitative real-time RT-PCR and
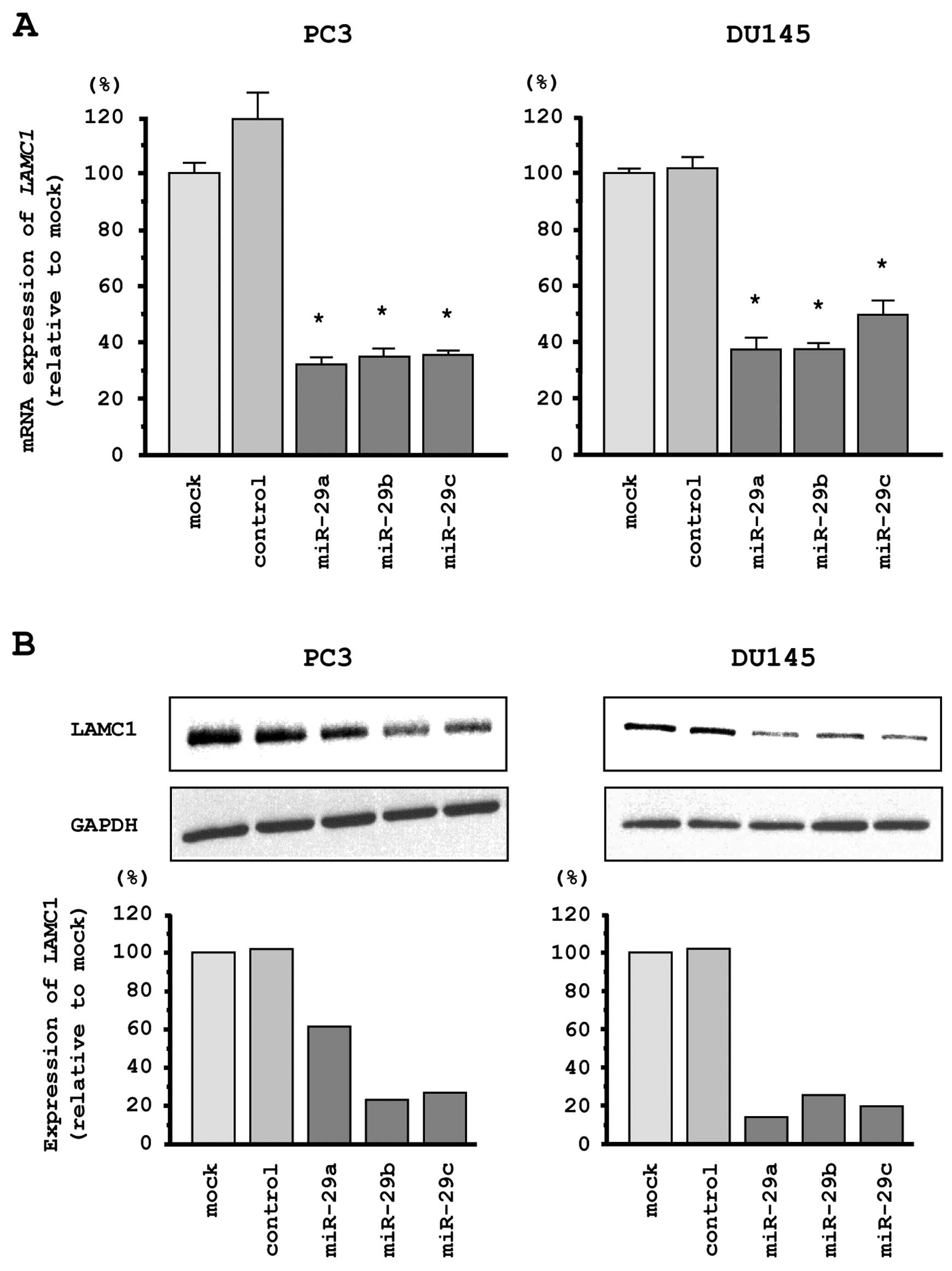
western blotting in PC3 and DU145 cells to investigate whether
restoration of miR-29s altered LAMC1 gene and LAMC1
protein expression. The mRNA and protein expression levels of
LAMC1/LAMC1 were significantly repressed in miR-29s
transfectants as compared with mock- or miR-control transfected
cells (Fig. 5A and B).
Therefore, we next performed luciferase reporter
assays in PC3 cells to determine whether LAMC1 mRNA had
target sites for miR-29s. The TargetScan database predicted
that the putative miR-29s binding site existed in the
3′-untranslated region (UTR) of LAMC1 (position 1463–1470).
We used vectors encoding a partial wild-type sequence of the 3′-UTR
of LAMC1 mRNA, including predicted miR-29s target
sites. We found that the luminescence intensity was significantly
reduced by transfection of miR-29s with the vector carrying
the wild-type 3′-UTR of LAMC1 (Fig. 5C).
Effects of downregulating LAMC1 on cell
proliferation, migration and invasion in PCa cell lines
To investigate the functional role of LAMC1
in PCa cells, we performed loss-of-function studies using
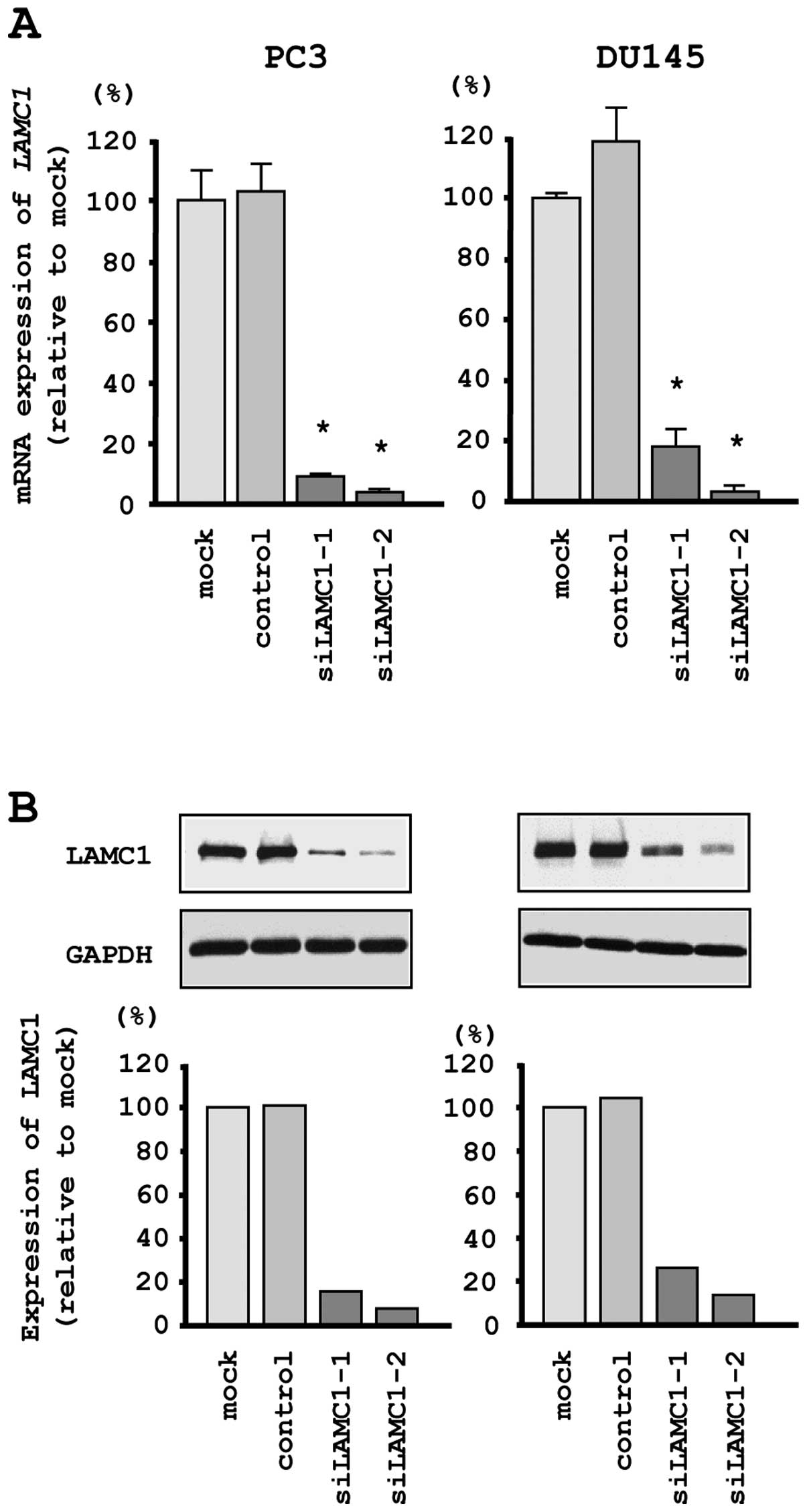
si-LAMC1 transfectants. First, we evaluated the knockdown
efficiency of si-LAMC1 transfection in PC3 and DU145 cells.
Quantitative real-time RT-PCR and western blotting indicated that
the siRNA effectively downregulated LAMC1/LAMC1 expression
in both cell lines (Fig. 6A and
B).
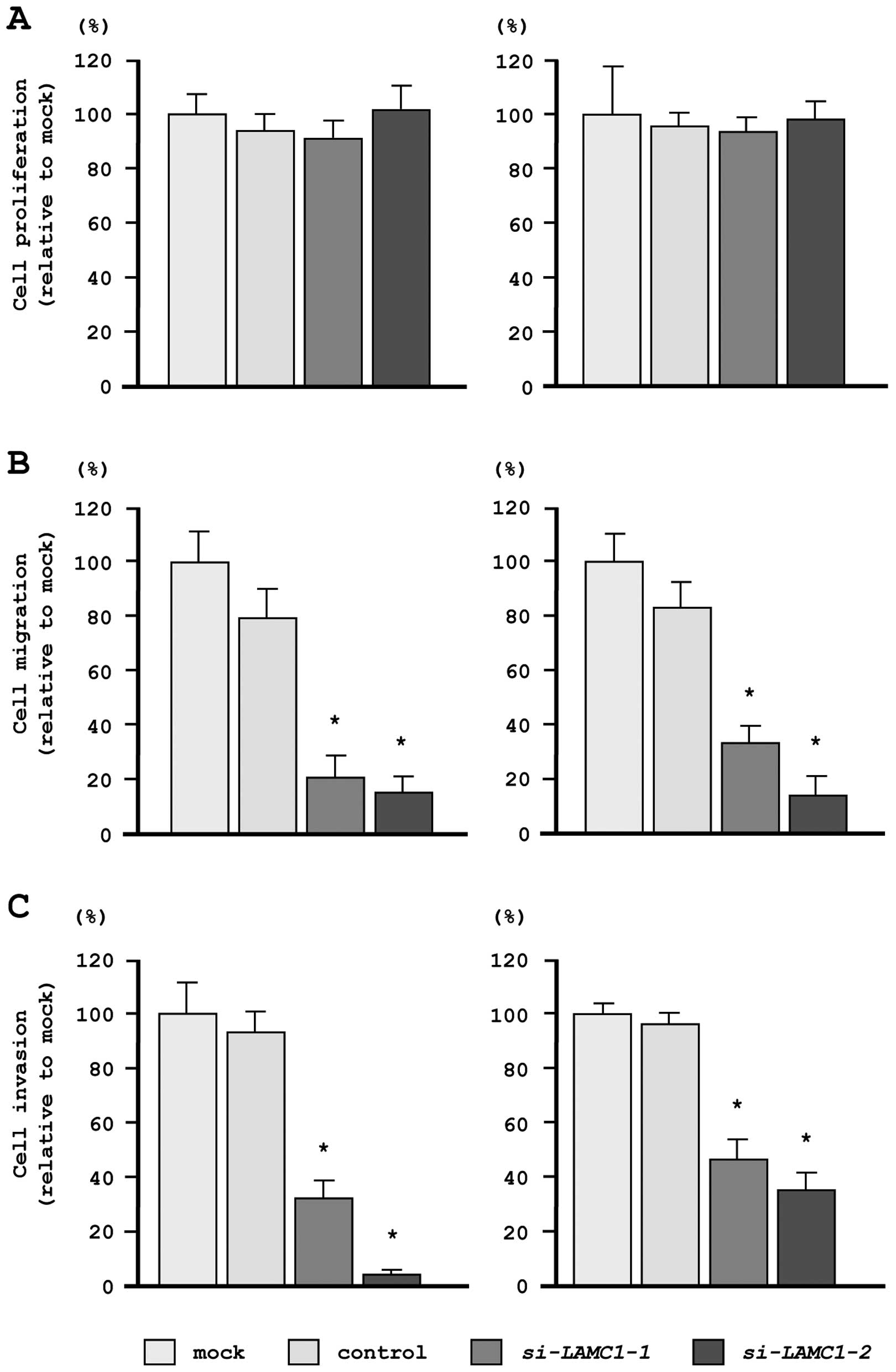
In our functional analyses, XTT assays demonstrated
that cell proliferation was not inhibited in si-LAMC1
transfectants, as compared with mock- or miR-control-transfected
cells (Fig. 7A). In contrast,
transfection with si-LAMC1 inhibited both cell migration and
invasion, as compared with mock- or miR-control-transfected cells
(Fig. 7B and C), similar to the
results observed for restoration of miR-29s.
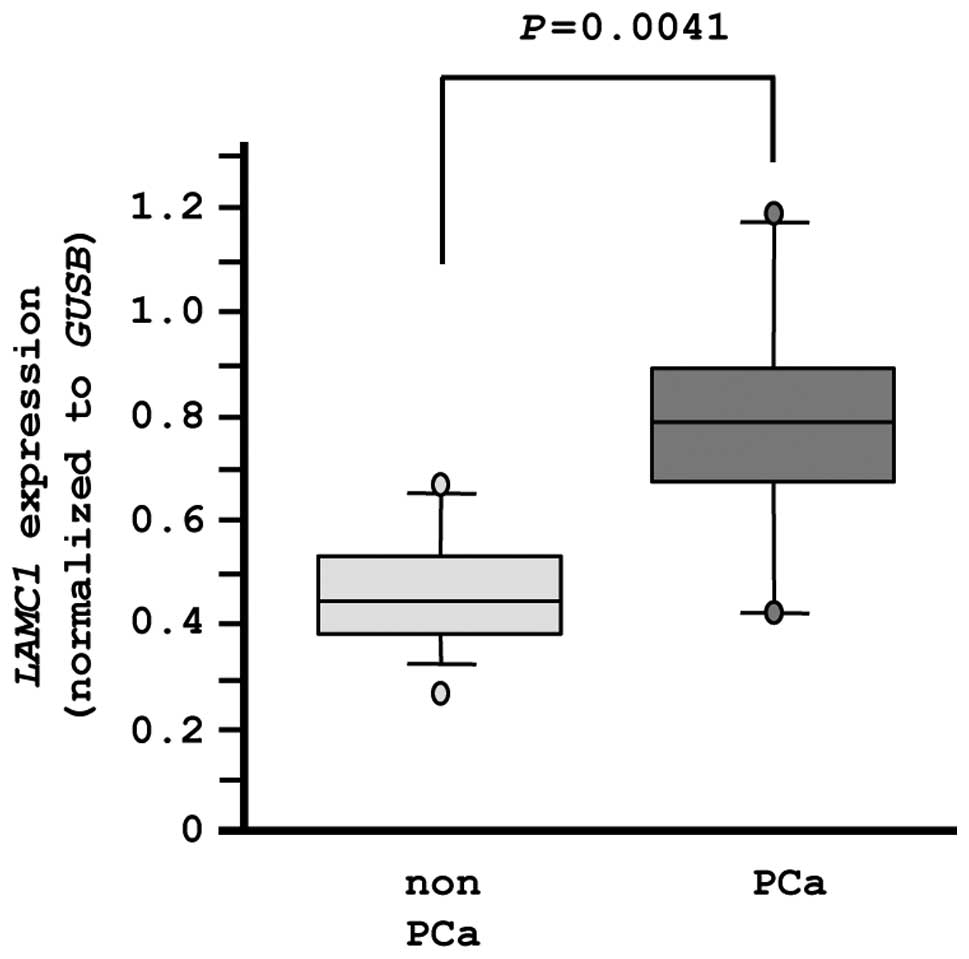
LAMC1 expression in PCa specimens
Finally, we evaluated the mRNA expression levels of
LAMC1 in normal prostate tissues (n=10) and PCa tissues
(n=10). The expression of LAMC1 was significantly higher in
PCa tissues compared with normal tissues (P=0.0041), as
demonstrated by RT-PCR (Fig. 8).
In this analysis, an independent set of clinical specimens was used
(Table IB).
Discussion
Emerging evidence has demonstrated that aberrantly
expressed miRNAs upset the tightly regulated miRNA/protein-coding
gene networks and cause initiation, progression and metastasis of
human cancers (11). Therefore,
identification of aberrantly expressed miRNAs is an important
initial step in elucidating miRNA-mediated oncogenic pathways.
Based on this strategy, we analyzed the miRNA expression signature
of PCa and identified tumor-suppressive miRNAs and their associated
PCa oncogenic pathways (12–14,16).
The past studies supported the legitimacy of our strategy for miRNA
analysis and contributed to the discovery of novel
tumor-suppressive miRNAs in PCa.
In this study, we focused on the miR-29s
(miR-29a, miR-29b and miR-29c) because we observed
downregulation of these miRNAs in our PCa expression signature. Our
data confirmed that all members of the miR-29 family were
significantly downregulated in PCa tissues. Recently, we also
observed that miR-29s were downregulated in head and neck
and cervical squamous cell carcinomas (HNCSSs) (17,18).
Moreover, downregulation of miR-29-family miRNAs has been
described by other groups in several types of cancers (19); these studies are all consistent
with our results. Although the molecular mechanisms through which
miR-29s are silenced in PCa are still unknown, recent data
have suggested that transforming growth factor (TGF)-β1 inhibits
the expression of miR-29s and promotes the expression of
extracellular matrix (ECM) components (20–22).
The ECM functions as a critical source for growth, survival,
motility and angiogenic factors that significantly affect tumor
biology and progression (23–25).
Additionally, TGF-β1 signaling is known to contribute to the
epithelial-to-mesenchymal transition (EMT), an important step in
cancer progression and metastasis (26). Our data demonstrated that
restoration of miR-29s significantly inhibited cancer cell
migration and invasion. Therefore, upregulation of ECM proteins
caused by TGF-β-dependent silencing of miR-29s is an
important step for metastasis in PCa cells.
Full understanding of the targets and signaling
pathways in PCa that are regulated by the miR-29s family may
contribute to our knowledge on PCa metastasis. We categorized
miR-29s-target genes into known pathways using KEGG pathways
(17,18). From our data in this study, we
focused on the ‘focal adhesion’ pathway because restoration of
miR-29s inhibited cancer cell migration and invasion in PCa
cell lines. Furthermore, we combined the gene expression data of
upregulated genes in PCa, generating 17 candidate target genes for
miR-29s in focal adhesion pathways. The upregulation of
collagen genes in primary tumors with metastatic potential was
consistent with recent observations that epithelial-mesenchymal
interactions are critical determinants of tumor cell behavior
(27,28). High levels of type 1 collagen in
metastatic lesions and in the serum of individuals with metastatic
disease have also been reported (29,30).
In this study, we selected LAMC1, a member of
the laminin super family as a target gene that contributes to
cancer cell migration and invasion in PCa cells. Laminins are
trimeric proteins that contain α-, β-, and γ-chains and are
important ECM regulators that are and biologically active part of
the basal lamina, influencing cell differentiation, migration and
adhesion, as well as phenotype and survival (31). During tumor progression, several
members of the laminin family have been shown to be upregulated in
cancer cells and involved in cancer cell migration and invasion
(32,33). Our recent studies in HNSCC have
demonstrated that laminin-integrin signaling promotes cancer cell
migration and invasion and that these signal pathways are regulated
by tumor-suppressive miR-218 and miR-29s (17,34).
This is the first study demonstrating that
miR-29s directly regulated LAMC1 in cancer cells and
that silencing of LAMC1 inhibited cancer cell migration and
invasion. The LAMC1 chain is the most widely expressed laminin
chain, found predominantly in the basement membrane (35,36).
Upregulation of LAMC1 in meningiomas correlates with shorter times
to tumor recurrence and decreased progression-free survival
(37). Furthermore, treatment with
the specific LAMC1 peptide enhances pulmonary metastasis of B16
melanoma cells and induces the production of matrix
metalloproteinase (MMP)-9 from B16 cells (38). Some studies have suggested that
LAMC1 functions to promote metastasis and might be a novel
therapeutic target in the treatment of human cancer. Further
studies are needed to determine whether LAMC1-mediated molecular
cascades contribute to PCa metastasis. A complete understanding of
tumor-suppressive miR-29s and their regulation of
LAMC1 signaling should shed light on the mechanisms of PCa
metastasis and facilitate the development of more effective
strategies for treating PCa.
Our data showed that all members of the
miR-29-family were frequently downregulated in PCa cells.
Moreover, these miRNAs functioned as tumor suppressors and
inhibited cancer cell migration and invasion through regulation of
focal adhesion pathways, especially via LAMC1. Elucidation
of cancer pathways regulated by tumor-suppressive miR-29s
should shed light on PCa metastasis and facilitate the development
of more effective strategies for future therapeutic interventions
for this disease.
Acknowledgements
This study was supported by the
KAKENHI (C), 24592590 and (B), 25293333.
References
|
1.
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2.
|
Bott SR, Birtle AJ, Taylor CJ and Kirby
RS: Prostate cancer management: 2. An update on locally advanced
and metastatic disease. Postgrad Med J. 79:643–645. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Timsit MO, Lebret T and Méjean A:
Chemotherapy of hormonorefractory and hormonoresistant metastatic
prostate cancer. Prog Urol. 18(Suppl 7): S365–S375. 2008.(In
French).
|
|
4.
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar
|
|
11.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
12.
|
Fuse M, Kojima S, Enokida H, et al: Tumor
suppressive microRNAs (miR-222 and miR-31) regulate molecular
pathways based on microRNA expression signature in prostate cancer.
J Hum Genet. 57:691–699. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kojima S, Enokida H, Yoshino H, et al: The
tumor-suppressive microRNA-143/145 cluster inhibits cell migration
and invasion by targeting GOLM1 in prostate cancer. J Hum Genet.
59:78–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kojima S, Chiyomaru T, Kawakami K, et al:
Tumour suppressors miR-1 and miR-133a target the oncogenic function
of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J
Cancer. 106:405–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Chiyomaru T, Enokida H, Tatarano S, et al:
miR-145 and miR-133a function as tumour suppressors and directly
regulate FSCN1 expression in bladder cancer. Br J Cancer.
102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Fuse M, Nohata N, Kojima S, et al:
Restoration of miR-145 expression suppresses cell
proliferation, migration and invasion in prostate cancer by
targeting FSCN1. Int J Oncol. 38:1093–1101. 2011.
|
|
17.
|
Kinoshita T, Nohata N, Hanazawa T, et al:
Tumour-suppressive microRNA-29s inhibit cancer cell migration and
invasion by targeting laminin-integrin signalling in head and neck
squamous cell carcinoma. Br J Cancer. 109:2636–2645. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Yamamoto N, Kinoshita T, Nohata N, et al:
Tumor-suppressive microRNA-29a inhibits cancer cell
migration and invasion via targeting HSP47 in cervical
squamous cell carcinoma. Int J Oncol. 43:1855–1863. 2013.
|
|
19.
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Maurer B, Stanczyk J, Jüngel A, et al:
MicroRNA-29, a key regulator of collagen expression in systemic
sclerosis. Arthritis Rheum. 62:1733–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Roderburg C, Urban GW, Bettermann K, et
al: Micro-RNA profiling reveals a role for miR-29 in human and
murine liver fibrosis. Hepatology. 53:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Yang T, Liang Y, Lin Q, et al: miR-29
mediates TGFbeta1-induced extracellular matrix synthesis through
activation of PI3K-AKT pathway in human lung fibroblasts. J Cell
Biochem. 114:1336–1342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Nelson CM and Bissell MJ: Modeling dynamic
reciprocity: engineering three-dimensional culture models of breast
architecture, function, and neoplastic transformation. Semin Cancer
Biol. 15:342–352. 2005. View Article : Google Scholar
|
|
24.
|
Gassmann P, Enns A and Haier J: Role of
tumor cell adhesion and migration in organ-specific metastasis
formation. Onkologie. 27:577–582. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
White DE, Rayment JH and Muller WJ:
Addressing the role of cell adhesion in tumor cell dormancy. Cell
Cycle. 5:1756–1759. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Heldin CH, Vanlandewijck M and Moustakas
A: Regulation of EMT by TGFbeta in cancer. FEBS Lett.
586:1959–1970. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Skobe M and Fusenig NE: Tumorigenic
conversion of immortal human keratinocytes through stromal cell
activation. Proc Natl Acad Sci USA. 95:1050–1055. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Olumi AF, Grossfeld GD, Hayward SW,
Carroll PR, Tlsty TD and Cunha GR: Carcinoma-associated fibroblasts
direct tumor progression of initiated human prostatic epithelium.
Cancer Res. 59:5002–5011. 1999.PubMed/NCBI
|
|
29.
|
Brown LF, Guidi AJ, Schnitt SJ, et al:
Vascular stroma formation in carcinoma in situ, invasive carcinoma,
and metastatic carcinoma of the breast. Clin Cancer Res.
5:1041–1056. 1999.PubMed/NCBI
|
|
30.
|
Jensen BV, Johansen JS, Skovsgaard T,
Brandt J and Teisner B: Extracellular matrix building marked by the
N-terminal propeptide of procollagen type I reflect aggressiveness
of recurrent breast cancer. Int J Cancer. 98:582–589. 2002.
View Article : Google Scholar
|
|
31.
|
Aumailley M: The laminin family. Cell Adh
Migr. 7:48–55. 2013. View Article : Google Scholar
|
|
32.
|
Tzu J and Marinkovich MP: Bridging
structure with function: structural, regulatory, and developmental
role of laminins. Int J Biochem Cell Biol. 40:199–214. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Yamaguchi M, Ebihara N, Shima N, et al:
Adhesion, migration, and proliferation of cultured human corneal
endothelial cells by laminin-5. Invest Ophthalmol Vis Sci.
52:679–684. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kinoshita T, Hanazawa T, Nohata N, et al:
Tumor suppressive microRNA-218 inhibits cancer cell migration and
invasion through targeting laminin-332 in head and neck squamous
cell carcinoma. Oncotarget. 3:1386–1400. 2012.
|
|
35.
|
Nde PN, Simmons KJ, Kleshchenko YY, Pratap
S, Lima MF and Villalta F: Silencing of the laminin gamma-1 gene
blocks Trypanosoma cruzi infection. Infect Immun.
74:1643–1648. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Toti P, Villanova M, De Felice C, Megha T,
Bartolommei S and Tosi P: Expression of laminin 1 and 2 in brain
tumor vessels. An immunohistochemical study. J Submicrosc Cytol
Pathol. 30:227–230. 1998.PubMed/NCBI
|
|
37.
|
Ke HL, Ke RH, Li B, Wang XH, Wang YN and
Wang XQ: Association between laminin γ1 expression and meningioma
grade, recurrence, and progression-free survival. Acta Neurochir.
155:165–171. 2013.
|
|
38.
|
Kuratomi Y, Nomizu M, Tanaka K, et al:
Laminin gamma 1 chain peptide, C-16 (KAFDITYVRLKF), promotes
migration, MMP-9 secretion, and pulmonary metastasis of B16-F10
mouse melanoma cells. Br J Cancer. 86:1169–1173. 2002. View Article : Google Scholar : PubMed/NCBI
|