Introduction
Breast cancer is one of the most common malignancies
among women, with approximately 232,340 new cases of invasive
breast cancer and 39,620 breast cancer deaths predicted to occur
among US women in 2013 (1).
Despite early diagnosis through screening programs and aggressive
therapeutic strategies, the age-standardized mortality rate still
remained at 14.1 per 100,000 individuals in 2008 (2). Therefore, precise prediction of
prognosis is important because advanced therapy is necessary for
high-risk patients. To predict prognosis, clinicians evaluate
various parameters, such as classical histological features and
hormone receptors. Recently, the expression of various molecular
markers such as HER2, TP53, Ki-67, and EGFR has contributed to
improvements in the prediction of prognosis in individual
patients.
The formation of distant metastasis is the main
cause of morbidity and mortality in patients with cancer. The
invasion or metastasis of cancer cells involves multiple steps,
including dissociation from the primary tumor, invasion into the
surrounding stroma, and intravasation into the surrounding vascular
systems (3). Each step requires a
complex network involving gene activation or repression. It has
recently been shown that epithelial-mesenchymal transition (EMT), a
mechanism important for embryonic development, plays a critical
role during malignant transformation (4,5). A
functional hallmark of the EMT program is considered the
acquisition of the ability to migrate and invade the extracellular
matrix as a single cell. Furthermore, it has also been indicated
that in molecules such as fibroblast growth receptor 2 (FGFR2),
alternative splicing of pre-messenger RNAs, in which one of two
mutually exclusive exons are included, results in differential
ligand binding specificity of the receptor during EMT (6) and contributes to the morphological
conversion accompanying EMT (7).
Accordingly, in malignant breast neoplasms, expression of the
splice variants of pre-messenger RNAs promoting EMT leads to
progression of the neoplasm, and ultimately result in poor
prognosis (8). Human ortholog of
mammalian enabled (hMena), a member of the
Ena/Vasodilator-stimulated phosphoprotein (VASP) family, is a key
molecule in cell migration. It regulates actin filament dynamics,
and protects the filaments from capping proteins at their barbed
ends and also reduces branching density (9,10).
Recently, hMena has been shown to have multiple splice variants in
tumor cells; two of the best characterized isoforms are hMena
invasive (hMenaINV), expressed exclusively in invasive
tumor cells, and hMena11a, an epithelial-specific
isoform expressed in primary breast carcinomas and downregulated in
invasive tumor cells (11–13). The exact regulation of hMena is
also unclear; however, Warzecha et al reported CD44 and
hMena transcripts undergo changes in splicing in vitro
during EMT (7).
The aim of this study was to evaluate whether the
expression of hMena isoforms hMena11a and
hMenaINV differ in non-invasive and invasive breast
cancer using breast surgical specimens and cancer cell lines. We
hypothesized that the expression of different hMena isoforms could
be a useful biomarker of malignancy and invasive or metastatic
potential.
Materials and methods
Patients and tumor tissue
Archival specimens collected between 2005 and 2012,
and 50 patients with invasive ductal carcinoma of no special type
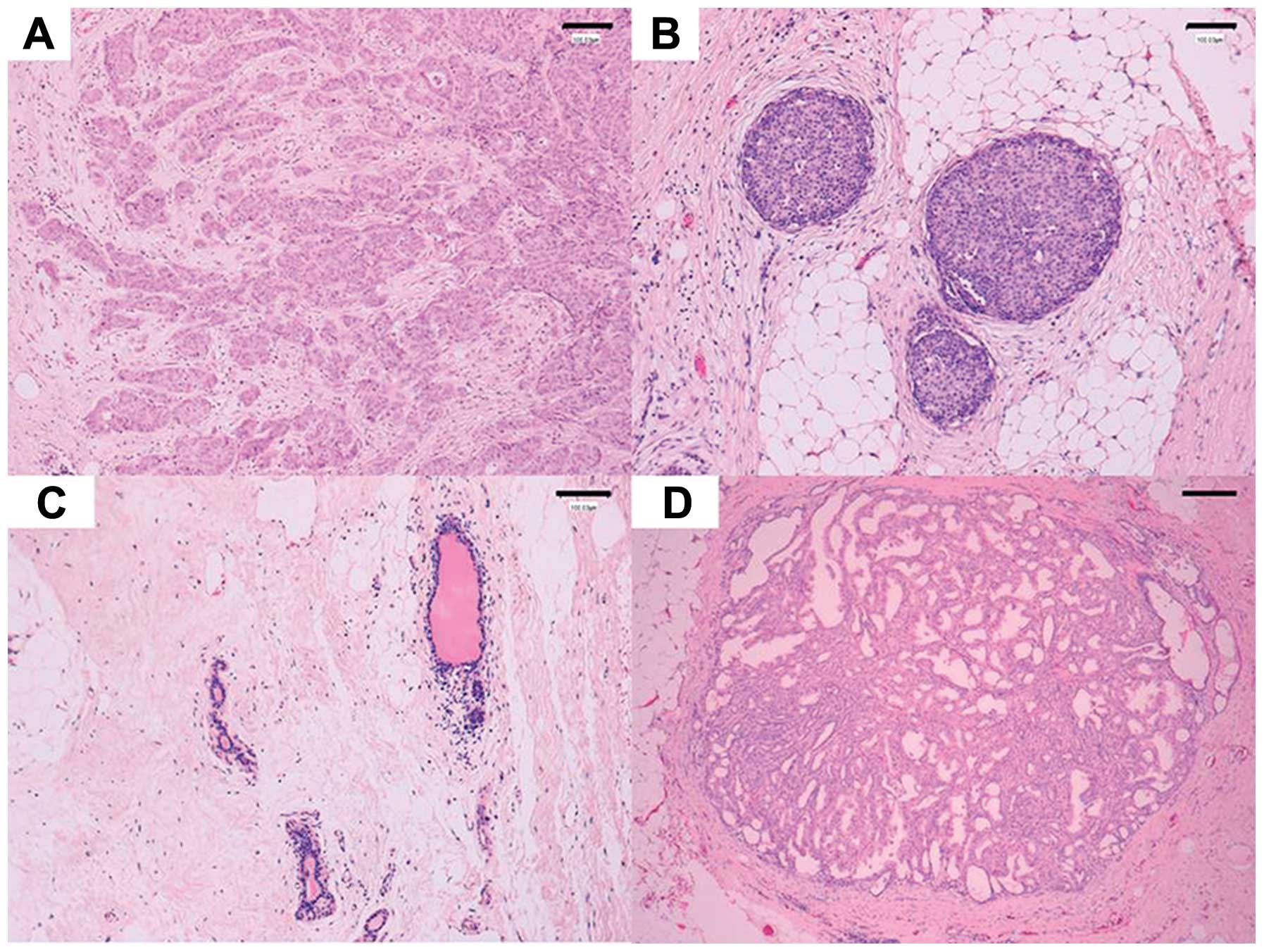
(IDC-NST) (Fig. 1A), 45 patients
with ductal carcinoma in situ (DCIS) (Fig. 1B) and 10 patients with intraductal
papilloma of the breast (Fig. 1D)
were enrolled in the study. Non-neoplastic duct tissue was obtained
from each specimen to serve as a control (Fig. 1C). All patients were surgically
treated with either breast-conserving lumpectomy or modified
radical mastectomy and axillary lymph node dissection at Asahi
General Hospital (Chiba, Japan). All samples were collected with
approval from the ethics committee of Asahi General Hospital. The
clinicopathological characteristics of each patient are shown in
Tables I and II.
 | Table IClinicopathological characteristics
of IDC-NST cases (total n=50). |
Table I
Clinicopathological characteristics
of IDC-NST cases (total n=50).
| No. of cases
(%) |
|---|
| Tumor size
(max) |
| <2.5 cm | 24 (48) |
| >2.5 cm | 26 (52) |
| Lymph node
status |
| Positive | 15 (30) |
| Negative | 35 (70) |
| Molecular
subtype |
| Luminal A | 17 (34) |
| Luminal B | 28 (56) |
| Basal | 5 (10) |
| WHO histological
grade |
| Grade 1 | 17 (34) |
| Grade 2 | 18 (36) |
| Grade 3 | 15 (30) |
 | Table IIWHO histological grades of DCIS cases
(n=45). |
Table II
WHO histological grades of DCIS cases
(n=45).
| DCIS histological
grade (WHO) | No. of cases
(%) |
|---|
| Low grade | 23 (51) |
| Intermediate
grade | 15 (33) |
| High grade | 7 (15%) |
The primary tumor specimens were fixed in 10%
buffered formalin and embedded in paraffin using standard tissue
processing methods. Pathological tumor staging was determined using
the current tumor-node-metastasis system (UICC). Diagnoses and
histology were confirmed by two pathologists (Noriyuki Tanaka and
Kenichi Harigaya) who reviewed the hematoxylin-eosin
(H&E)-stained slides prepared from the paraffin blocks.
Laser capture microdissection (LCM), RNA
isolation and cDNA synthesis from FFPE sections
Ten micrometer-thick membrane sections (Leica
Microsystems) were prepared from formalin-fixed paraffin-embedded
(FFPE) tissue using standard protocols. The slides were air-dried
and stained with toluidine blue. Laser dissection was performed
using a laser capture microdissection microscope (Leica AS LMD;
Leica Microsystems, Wetzlar, Germany) with a pulsed 337 nm UV laser
according to the manufacturer’s protocols. The size of each
dissected specimen was ≥2 mm2. Total RNA was purified
from dissected tissue using an RNeasy FFPE kit (Qiagen, Venlo, The
Netherlands) according to the manufacturer’s protocols. cDNA was
synthesized from the extracted total RNA with Primescript reverse
transcript reagent (Takara) according to the manufacturer’s
protocols.
Real-time PCR analysis
Semi-quantitative real-time polymerase chain
reaction (PCR) was then performed using the ABI PRISM 7000
(Perkin-Elmer Applied Biosystems, Foster City, CA, USA) thermal
cycler with PreMix ExTaq reagent (Takara), as previously described
(14). The primers were as
follows: MENA (sense, 5′-GCGCAGAATATCAAGTGCTG-3′; antisense,
5′-TCCCAGCACAGAGTTTAGAGG-3′), MENA11a
(sense, 5′-TTTTGACAACAGGTCCTATGATTC-3′; antisense,
5′-CTTCCGTCTGGACTCCATTG-3′), MENAINV
(sense, 5′-CCATGATGCATGCCTTAGAA-3′; antisense,
5′-TCCTGGGTAGCAGTAACCTTG-3′), human GAPDH (sense,
5′-AGCCACATCGCTCAGACAC-3′; antisense, 5′-GCCCAATACGACCAAATCC-3′).
The TaqMan Probes (UPL, Roche) used for each variant were as
follows: hMena (TaqMan no. 34), hMena11a (TaqMan no.
26), hMenaINV (TaqMan no. 26), and GAPDH (TaqMan
no. 60). The PCR protocol consisted of 30 sec at 95°C followed by
60 cycles of 5 sec at 95°C and 31 sec at 60°C. The relative
expression level of hMena variants was calculated by the
comparative threshold cycle (Ct) method using
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the
internal control (15). All
experiments were performed in triplicate and the results were
expressed as mean ± SD.
Cell culture
Human breast cancer cell lines MDA-MB231, BT549,
MDA-MB468, MCF-7, T47D, ZR75-1 and human cervical cancer cell line
HeLaS3 were purchased from American Type Culture Collection
(Rockville, MD, USA). All cell lines except HeLaS3 were cultured in
Iscove’s modified Dulbecco’s medium (IMDM; Invitrogen) supplemented
with 10% fetal bovine serum. HeLaS3 cells were grown in Dulbecco’s
modified Eagle’s medium (DMEM; Niccui) supplemented with 10% calf
serum. All cell lines were maintained under 5% CO2 at
37°C.
Antibodies
Anti-E-cadherin antibody (BD610182) was purchased
from BD Biosciences (San Jose, CA, USA). Anti-vimentin antibody
(sc-6260) was obtained from Santa Cruz Biotechnology Inc. (Santa
Cruz, CA, USA). Anti-actin antibody (A 2066) was purchased from
Sigma (St. Louis, MO, USA). Primary polyclonal antibodies against
hMena variants were raised in rabbits against amino acid sequences
AQSKV TATQD STNLR CIFC (hMenaINV) and RDSPR KNQIV FDNRS
YDSLH (hMena11a). The obtained antibodies were
affinity-purified using each immunizing peptide.
Western blotting
Whole cell lysates were prepared with ice-cold RIPA
buffer (50 mM Tris-HCl pH 7.5, 1% nonidet P-40, 150 mM NaCl, 0.1%
SDS, 0.5% deoxycholic acid) containing 1 μg/ml leupeptin, 1 μg/ml
pepstatin, 1 μg/ml aprotinin, 1 mM DTT, 1 mM NaVO4 and
0.5 mM PMSF. The supernatants were recovered as total cell lysates
following centrifugation. Aliquots of the cell lysates (50 μg of
protein) were separated by 10% SDS-PAGE and transferred to PVDF
membranes (Bio-Rad Laboratories, Hercules, CA, USA). Primary
antibodies bound to their antigens on the membranes were detected
using appropriate HRP-conjugated secondary antibodies (Amersham
Bioscience, Piscataway, NJ, USA) and a Super Signal
chemiluminescence detection system (Pierce, Rockford, IL, USA) or
the Lumi-LightPLUS western blotting substrate (Roche Diagnostics,
Basel, Switzerland) according to the manufacturer’s
instructions.
Statistical analysis
Data are summarized in bar graphs. Bars represent
the mean and whiskers the standard deviation (SD). Statistical
analysis was performed using Microsoft Excel (Microsoft Corp.,
Seattle, WA, USA). Paired-sample t-tests, χ2 test, or
Kruskal-Wallis tests were used as appropriate. Differences were
considered statistically significant when the P-value was
<0.05.
Results
hMenaINV and
hMena11a splice variant mRNA was expressed in invasive
ductal carcinoma of no special type (IDC-NST)
We sought to determine whether the mRNA expression
of hMena splice isoforms hMenaINV and
hMena11a is altered in different IDC-NST lesions
[invasive areas (IA-IDC-NST, Fig.
1A) and non-invasive intraductal lesions (NIDL-IDC-NST,
Fig. 1B)] compared with
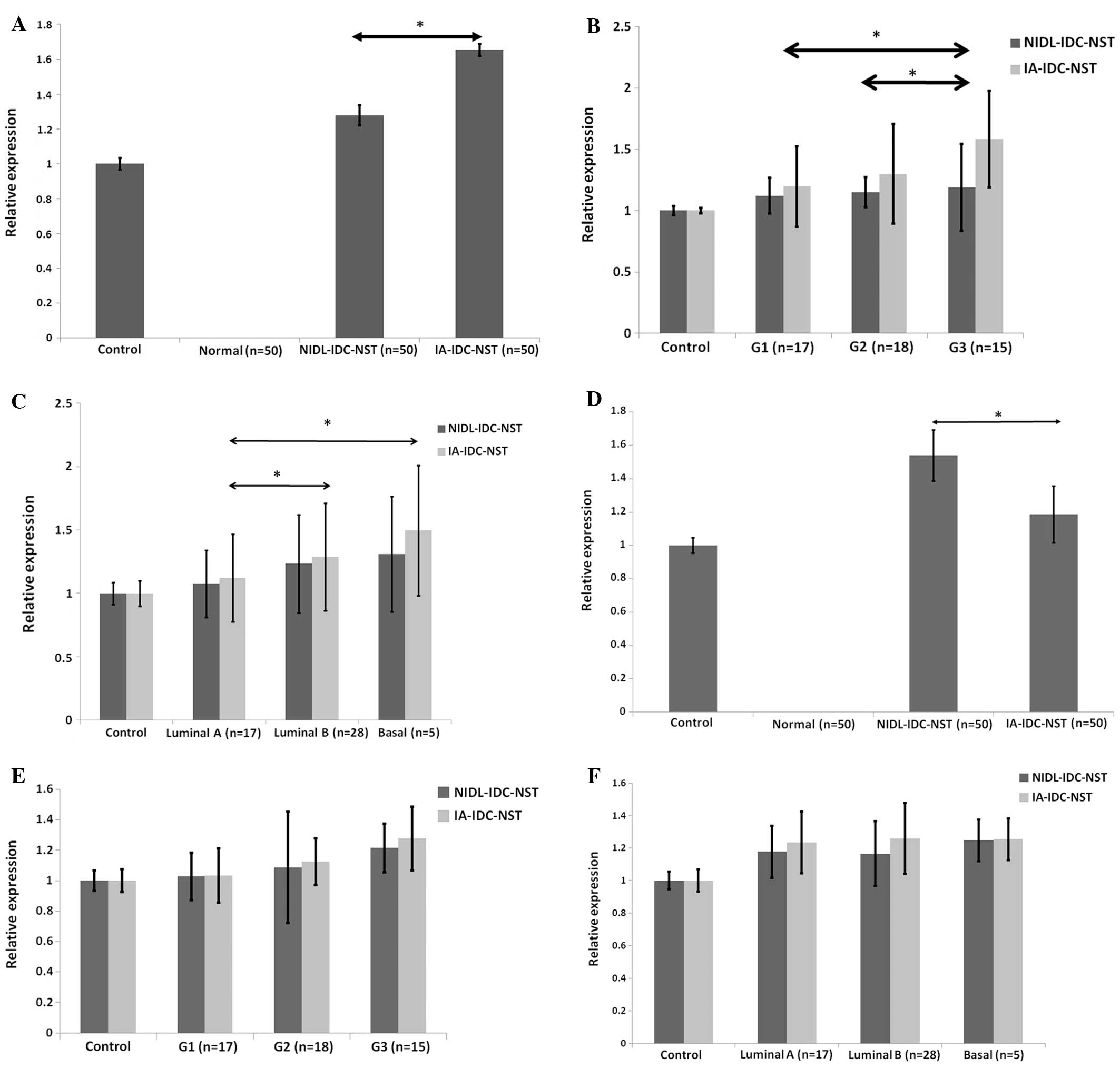
corresponding non-neoplastic duct epithelium (Fig. 1C) (50 cases). Fig. 2A demonstrated that an increase in
hMenaINV mRNA expression was found in the different
IDC-NST lesions IA-IDC-NST and NIDL-IDC-NST, while no detectable
expression was observed in non-neoplastic duct epithelia. More
intense augmentation of hMenaINV mRNA expression was
demonstrated in IA-IDC-NST than NIDL-IDC-NST (P<0.05). We
further examined whether there were any differences in
hMenaINV mRNA expression among three different subtypes
of IDC-NST according to the WHO classification (16) of histological grading (tubule
formation, nuclear pleomorphism, and mitotic accounting) or
molecular phenotype (luminal A, luminal B, and basal subtypes
according to the presence or absence of estrogen receptor,
progesterone receptor, or Her2 protein). Fig. 2B showed that higher expression of
hMenaINV mRNA was significantly detected in IA-IDC-NST
lesions of grade 3 breast carcinomas compared with either those of
grade 1 tumors (P<0.05) or those of grade 2 tumors (P<0.05).
Additionally, no significant differences in hMenaINV
mRNA expression were found in NIDL-IDC-NST lesions of the three
different subclasses. Furthermore, a significant increase in
hMenaINV mRNA expression was found in IA-IDC-NST lesions
of three different molecular phenotypes; basal subtype expressed
higher levels of hMenaINV mRNA than either luminal A
subtype, or luminal B subtype (P<0.05; Fig. 2C). However, we did not find any
statistically significant difference between luminal B and basal
subtypes. In NIDL-IDC-NST lesions, there was no statistically
significant difference in hMenaINV mRNA expression among
the three different molecular phenotypes. Taken together, it would
be reasonable to conclude that higher hMenaINV mRNA
expression is found in more aggressive histological and molecular
subtypes of IA-IDC-NST lesions. Furthermore, our data suggest that
no statistically significant difference in hMenaINV
expression was found in NIDL-IDC-NST lesions, but increased
expression was found in IA-IDC-NST in accordance with tumor
progression. It is well known that determination of these subtypes
reflect patient prognosis (17),
and, therefore, our results are comparable with a previous report
that showed increased expression of hMenaINV could
confer a potent pro-metastatic phenotype when expressed in breast
cancer cells (18,19). Our results also showed that levels
of hMenaINV expression also tended to be increased in
non-invasive ductal carcinoma in clinical breast carcinoma
specimens according to histological grade, but this was not
statistically significant. These results indicate that the
measurement of hMenaINV mRNA in IA-IDC-NST but not
NIDL-IDC-NST may be useful in predicting patient prognosis from
histological samples of IDC-NST. It should be stressed that
mutually-exclusive alternative hMena mRNA splicing would not
necessarily be observed only in invasive breast carcinoma cells in
clinical specimens and that cell sheets of non-invasive intraductal
lesions in IDC-NST produced significant amounts of
hMenaINV mRNA.
Next, we examined the expression of
hMena11a mRNA at the afore-mentioned lesions of IDC-NST.
The expression of hMena11a mRNA was hardly found in
non-neoplastic breast duct epithelium but was dramatically
increased in the different lesions of IDC-NST, as in the case of
hMenaINV mRNA. Fig. 2D
shows that significant expression of hMena11a mRNA was
consistently found in NIDL-IDC-NST as well as IA-IDC-NST in
contrast to non-neoplastic duct epithelia, while the relative
expression in IA-IDC-NST was downregulated to approximately 85%
that of the level in NIDL-IDC-NST. This reduction reflects previous
reports that Mena11a is downregulated in invasive tumor
cells (11). Compared with the
elevation in hMenaINV mRNA expression in accordance to
adverse histological grade, hMena11a mRNA expression did
not change among the three subgroups classified according to WHO
histological grades or molecular phenotypes, as shown in Fig. 2E and F. Rather, grade 3 subtype of
both NIDL-IDC-NST and IA-IDC-NST tended to have increased
hMena11a mRNA expression, although this was not
statistically significant. Our results indicated that significant
hMena11a mRNA expression was found in different NIDL and
IA lesions of IDC-NST but was decreased in IA-IDC-NST, which is
supposed to undergo tumor progression. However, our results also
showed that hMena11a mRNA expression was not
downregulated in different lesions with either histological or
molecular tumor progression. Accordingly, our results do not
necessarily coincide with previous reports in vitro and
in vivo that hMena11a is downregulated in
invasive tumor cells (11,20). Collectively, our results showed
that cell sheets of non-invasive intraductal lesions in IDC-NST
produced significant amount of hMena11a mRNA as well as
hMenaINV mRNA. This would reflect whether some
populations of invasive cells are intermingled in intraductal
lesions of NIDL-IDC-NST or some populations of non-invasive
intraductal lesions could produce both hMenaINV and
hMena11a mRNA simultaneously. Nevertheless, there has
been no evidence indicating that both the INV and 11a exons are
included in the Mena mRNA at the same time or expressed at high
levels within the same cell (21).
Expression of hMena splice isoform mRNA
is increased in breast ductal carcinoma in situ
Several lines of evidence indicate that molecular
expression of hMena11a is strictly regulated during
breast carcinoma development and is predominantly found in the
non-invasive stage of breast carcinoma (12,20).
However, our results showed that both hMena splice isoforms were
dramatically increased in the different IDC-NST lesions, IA-IDC-NST
(Fig. 2A) and NIDL-IDC-NST
(Fig. 2D). Additionally, our
results also revealed that the expression of hMenaINV is
further augmented in cells at the invasive front, while
hMena11a is downregulated in cells at the invasive
front. It should be stressed that cell sheets of NIDL-IDC-NST
produced hMena11a mRNA as well as significant amounts of
hMenaINV mRNA in clinical breast carcinoma specimens.
This would reflect whether some populations of invasive breast
carcinoma cells are intermingled in non-invasive intraductal
lesions of IDC-NST or some cell populations of non-invasive
intraductal lesions could produce both MenaINV and
Mena11a, simultaneously. To explore these possibilities,
we examined whether cell populations in ductal carcinoma in
situ (DCIS) contain hMena splice isoform mRNA. We examined 45
cases of DCIS, 10 cases of intraductal papilloma, and 55 lesions of
the corresponding non-neoplastic duct epithelia around the in
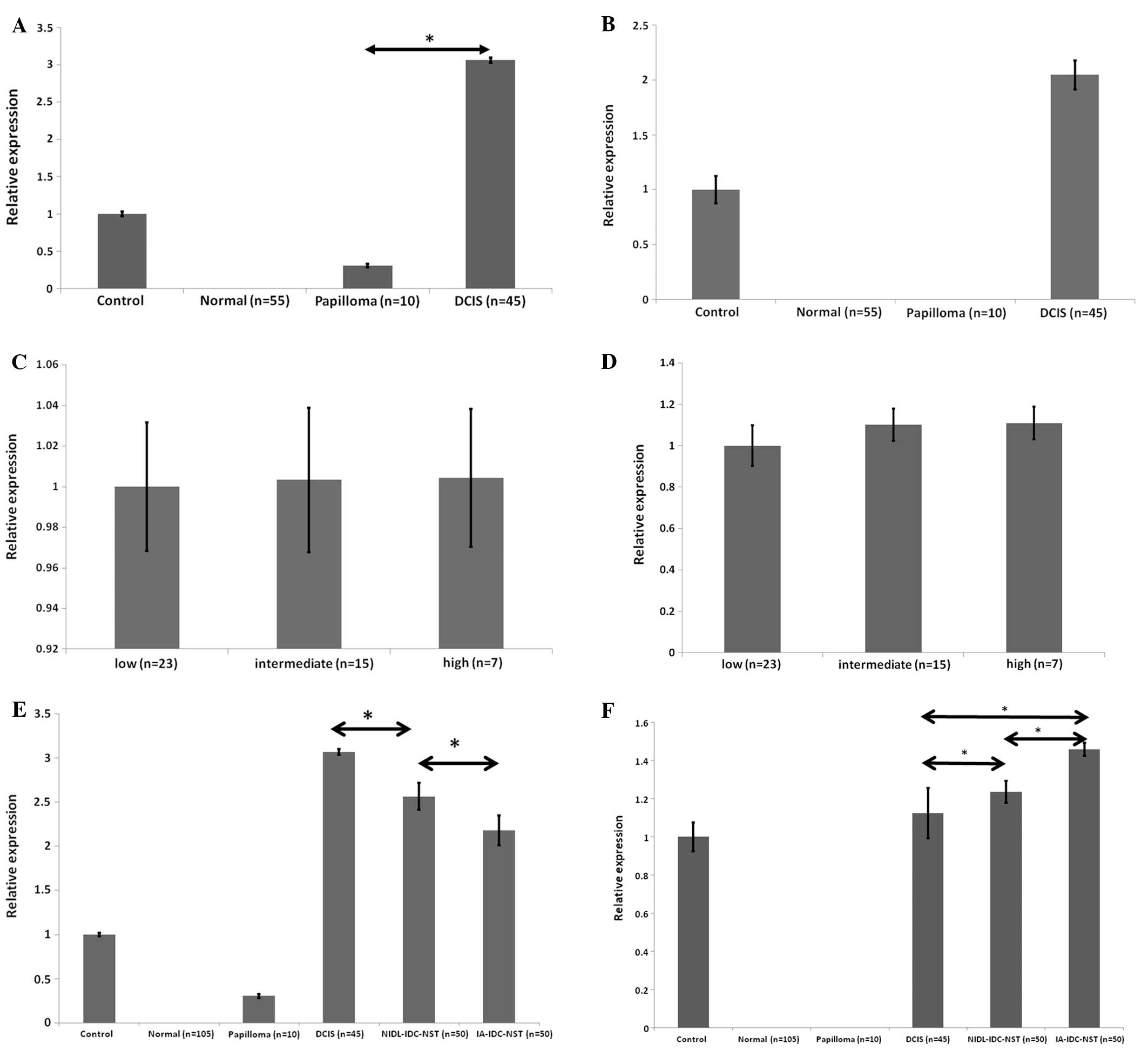
situ carcinoma and papilloma. Fig.
3A and B showed that cells of DCIS constantly expressed
significant amounts of hMena11a mRNA as well as
hMenaINV mRNA. In papilloma lesions (n=10), weak
expression of hMena11a was also found, but
hMenaINV mRNA expression was not detectable.
Accordingly, cells in DCIS produced increased amounts of both
hMenaINV and hMena11a mRNA in contrast to
non-neoplastic epithelia. We further examined the different DCIS
subtypes according to either WHO histological classification; the
expression levels of hMena11a and hMenaINV
mRNA did not differ among the three different grades of DCIS
lesions (Fig. 3C and D), although
we did not investigate the molecular phenotypes. Our results are
comparable with a recent report that showed the rate of local
recurrence for low- and intermediate-grade DCIS is not
significantly different from the rate of local recurrence for women
with high grade DCIS (15.1%) (22). Fig.
3E showed that among three different breast carcinoma lesions,
cells in DCIS always showed the highest level of
hMena11a mRNA expression, followed by those in
NIDL-IDC-NST. Cells of IA-IDC-NST consistently showed the lowest
levels of hMena11a mRNA expression among the three
different lesions. Conversely, the expression of
hMenaINV mRNA was reversed and increased in cells of
NIDL-IDC-NST, and showed the highest expression levels in cells of
IA-IDC-NST (Fig. 3F). These
results indicated that the expression of hMena11a
suppressed metastatic potential and hMenaINV expression
promoted tumor progression. Previous reports showed that
MenaINV is exclusively expressed in invasive tumor cells
in in vitro and in vivo (18,23).
Nevertheless, our results indicate that cells in non-invasive
breast duct carcinoma produce Mena11a mRNA as well as
MenaINV mRNA in clinical breast carcinoma samples.
Expression of hMena splice isoforms in
cancer cell lines
To validate our results, we examined the expression
of hMena splice isoforms in several non-invasive and invasive
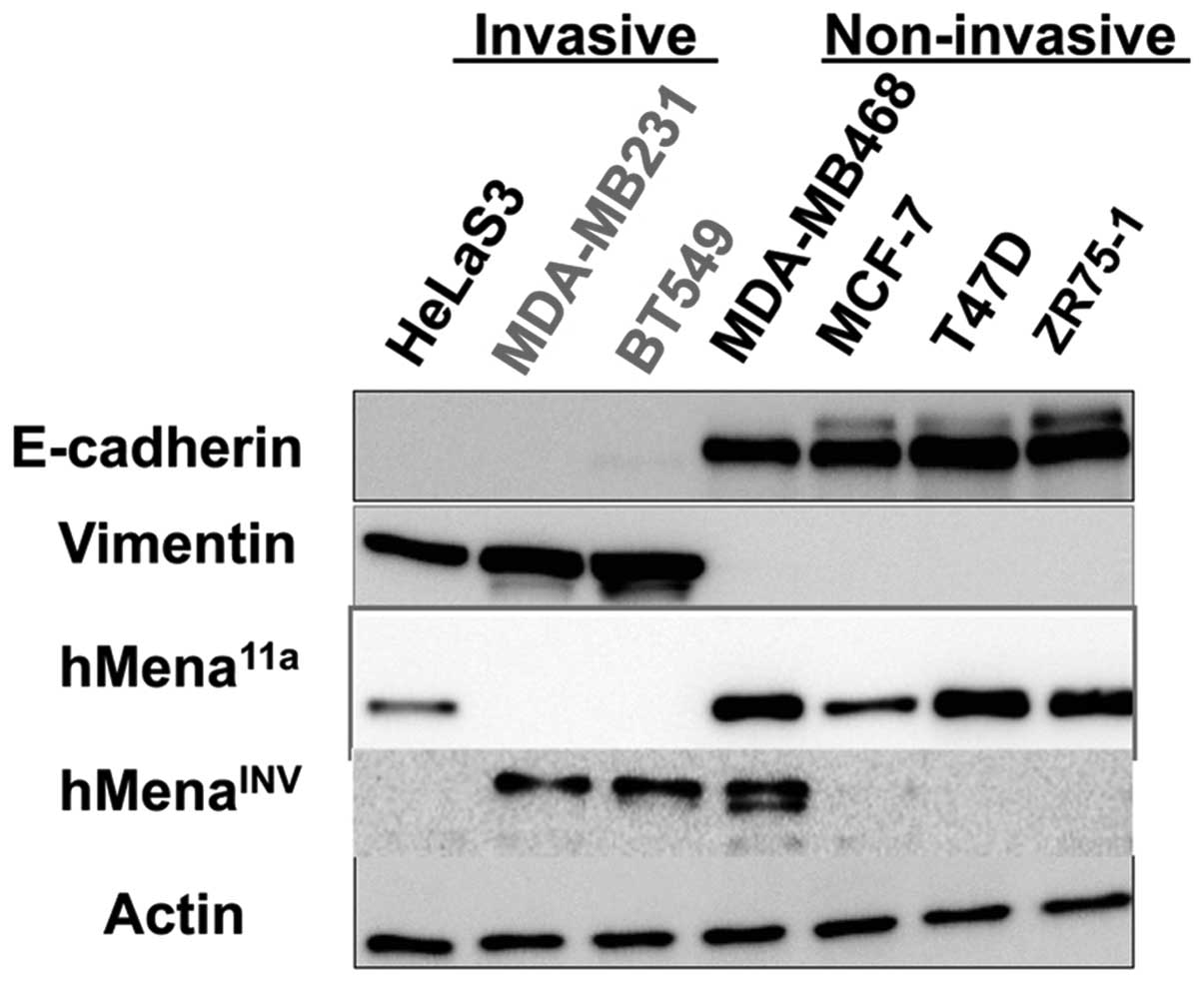
cancer cell lines by western blotting. Fig. 4 shows the expression of hMena
splice variants in several breast cancer cell lines. Seven cancer
cell lines, including four E-cadherin-positive and
vimentin-negative cell lines and three E-cadherin-negative and
vimentin-positive cell lines were used. The E-cadherin-positive
cell lines are epithelial and show non-invasive phenotypes. All
four lines expressed hMena11a protein. However, the
E-cadherin-positive and vimentin-negative non-motile breast cancer
cell line, MDA-MB468, simultaneously expressed both
hMenaINV and hMena11a. Conversely,
E-cadherin-negative and vimentin-positive HeLaS3 cells presumably
undergo EMT, and were found to express hMena11a protein
but not hMenaINV protein. These results suggest that a
proportion of the non-invasive carcinoma cells could express
hMenaINV as well as hMena11a, while invasive
carcinoma cells such as HeLaS3 express only hMena11a but
not hMenaINV. These findings are fairly consistent with
our results in clinical breast cancer samples, and suggest that a
subset of non-invasive cancer cells simultaneously express
hMena11a and hMenaINV. Accordingly, our
results indicate that the differential expression of either
hMenaINV or hMena11a in cancer cells is not
strictly regulated during tumor progression.
Whole hMena isoform expression appears
not to differ between different neoplastic or non-neoplastic breast
epithelial lesions
To examine the expression of whole hMena mRNA
expression in different non-neoplastic and neoplastic breast
epithelial lesions, we investigated 210 foci (non-neoplastic
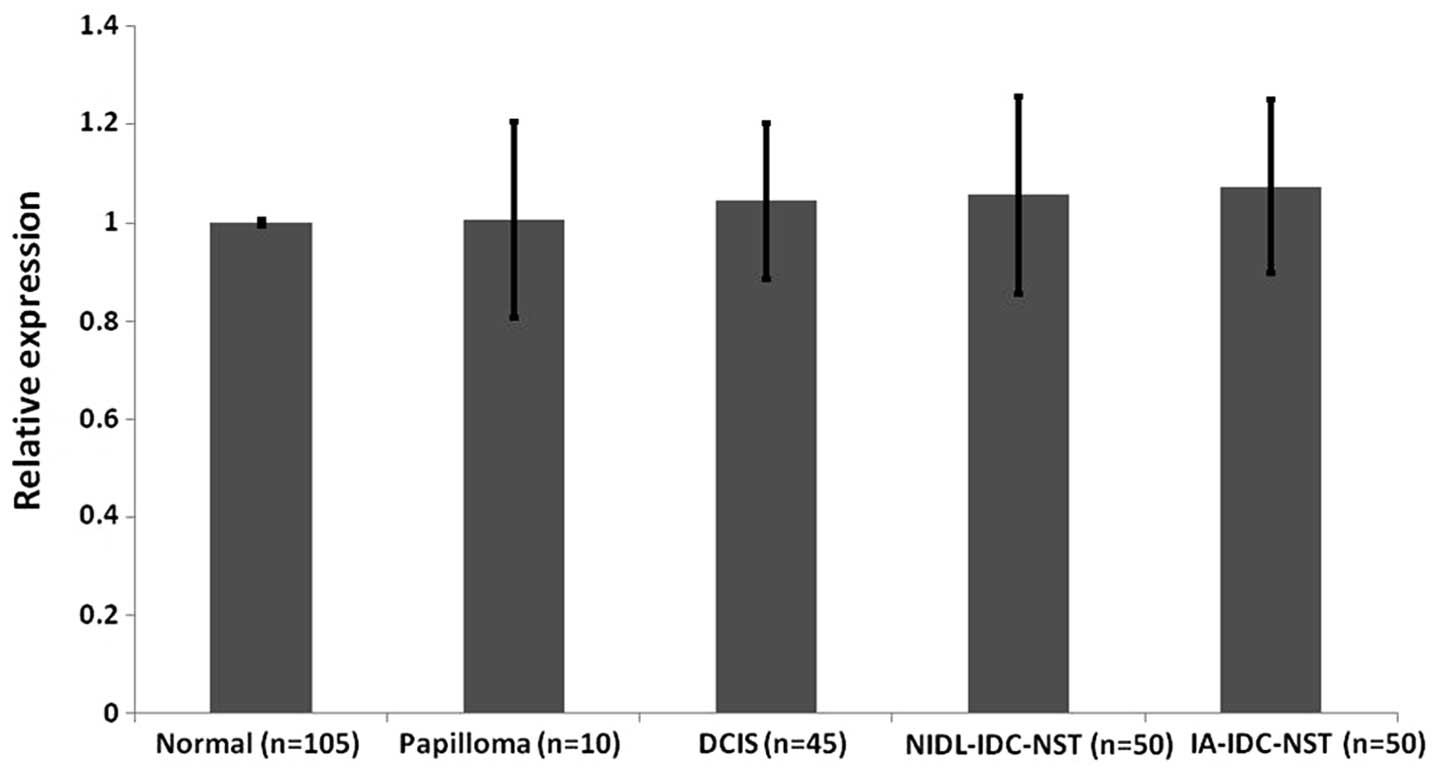
lesions: n=105; papilloma: n=10; DCIS: n=45, IDC-NST: n=50).
Fig. 5 showed that levels of whole
hMena mRNA expression were not statistically different among the
non-neoplastic and neoplastic breast epithelial foci used in this
study.
Our data resulting from semi-quantitative mRNA
expression in microdissected samples of clinical surgical specimens
are compatible with the mRNA expression of hMenaINV and
hMena11a splice variants in previous studies (18,24)
using xenograft models; however, whole hMena expression levels were
different from our results. These researchers claimed that there
was 3–4-fold augmented expression of whole hMena during tumor
progression. Although we could not determine the cause of this
discrepancy, we speculate that it stems from different model
systems; we investigated clinical human breast cancer samples while
the previous report used xenograft models.
Discussion
The Ena/VASP family of proteins is an important
regulator of actin cytoskeleton dynamics involved in cell motility.
Changes in the cellular actin network play a role in malignant
transformation and tumor progression. Previous reports have shown
that hMena variant hMena11a is predominantly
overexpressed in tumor cell lines expressing epithelial phenotypes,
while hMenaINV was shown to be overexpressed in tumor
cell lines expressing invasive phenotypes (18,22).
Furthermore, expression of ERSP1 and ERSP2 induces the inclusion of
the hMena11a exon, through which cancer cells undergo
morphological changes into an adhesive form of epithelial-like
cells. Loss of their expression induces the inclusion of the
hMenaINV exon, resulting in EMT and rendering cancer
cells motile (25). In our study,
however, we showed that simultaneous expression of
hMena11a and hMenaINV is found either in
non-invasive or invasive carcinoma lesions using FFPE breast cancer
tissue from clinical surgical specimens. In contrast, their
expression was hardly detected in normal breast tissue and benign
proliferative breast lesions. These results indicate that the
higher relative expression of hMena11a compared with
that of hMenaINV may predict malignant transformation in
breast epithelial cells, and, furthermore, a reversal in the
expression of hMena11a and hMenaINV may
dictate the state of cancer progression. Based on these results, we
suggest that differential regulation of hMena11a and
hMenaINV splice variant expression during tumor
progression is not performed in a mutually exclusive switch-on and
switch-off manner. Furthermore, using cancer cell lines, we have
shown that MDA-MB468 intermediately expresses both splice variants,
and HeLaS3 cells undergoing EMT (vimentin-positive and E-cadherin
negative) express only hMena11a without
hMenaINV. Through our studies, it appears that the
molecular mechanism is sometimes promiscuous during malignant
transformation or progression, and it seems that the invasive
potential of cancer cells is hard to predict from the expression of
a single splicing isoform such as hMenaINV.
In conclusion, we have demonstrated that
determination of hMena11a and hMenaINV
expression could be a useful biomarker in malignant transformation
and progression in breast epithelial lesions and that their
relative expression is linked to adverse prognostic factors.
Further studies may provide insights into the understanding of the
nature of cancer initiation and progression, and improve diagnosis
of non-invasive and invasive ductal carcinomas.
Acknowledgements
A portion of these data were presented at the 101st
and 102nd annual meeting of the Japanese Society of Pathology in
Tokyo and Sapporo. We are grateful to Asahi General Hospital for
surgical specimens, and T. Shida, T. Umemiya, K. Azuma and K.
Takaoka for technical help. This study was supported by
Grants-in-Aid for Scientific Research 15390122 and 22390074 from
the Japan Society for the Promotion of Science (to K.
Harigaya).
References
|
1
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar
|
|
2
|
Matsuda A, Matsuda T, Shibata A, et al:
Cancer incidence and incidence rates in Japan in 2008: a study of
25 population-based cancer registries for the Monitoring of Cancer
Incidence in Japan (MCIJ) project. Jpn J Clin Oncol. 44:388–396.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar
|
|
5
|
Rybinski B, Franco-Barraza J and Cukierman
E: The wound healing, chronic fibrosis and cancer progression
triad. Physiol Genomics. 46:223–244. 2014. View Article : Google Scholar
|
|
6
|
Savagner P, Valles AM, Jouanneau J, Yamada
KM and Thiery JP: Alternative splicing in fibroblast growth factor
receptor 2 is associated with induced epithelial-mesenchymal
transition in rat bladder carcinoma cells. Mol Biol Cell.
5:851–862. 1994. View Article : Google Scholar
|
|
7
|
Warzecha CC, Sato TK, Nabet B, Hogenesch
JB and Carstens RP: ESRP1 and ESRP2 are epithelial
cell-type-specific regulators of FGFR2 splicing. Mol Cell.
33:591–601. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shapiro IM, Cheng AW, Flytzanis NC, et al:
An EMT-driven alternative splicing program occurs in human breast
cancer and modulates cellular phenotype. PLoS Genet.
7:e10022182011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barzik M, Kotova TI, Higgs HN, et al:
Ena/VASP proteins enhance actin polymerization in the presence of
barbed end capping proteins. J Biol Chem. 280:28653–28662. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bear JE, Svitkina TM, Krause M, et al:
Antagonism between Ena/VASP proteins and actin filament capping
regulates fibroblast motility. Cell. 109:509–521. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gertler F and Condeelis J: Metastasis:
tumor cells becoming MENAcing. Trends Cell Biol. 21:81–90. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Modugno F, DeMonte L, Balsamo M, et al:
Molecular cloning of hMena (ENAH) and its splice variant
hMena+11a: epidermal growth factor increases their
expression and stimulates hMena+11a phosphorylation in
breast cancer cell lines. Cancer Res. 67:2657–2665. 2007.PubMed/NCBI
|
|
13
|
Urbanelli L, Massini C, Emiliani C and
Orlacchio A, Bernardi G and Orlacchio A: Characterization of human
Enah gene. Biochim Biophys Acta. 1759:99–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Itakura M, Terashima Y, Shingyoji M, et
al: High CC chemokine receptor 7 expression improves postoperative
prognosis of lung adenocarcinoma patients. Br J Cancer.
109:1100–1108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Monney L, Sabatos CA, Gaglia JL, et al:
Th1-specific cell surface protein Tim-3 regulates macrophage
activation and severity of an autoimmune disease. Nature.
415:536–541. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ellis I and Simpson J: Grading. WHO
Classification of Tumours of the Breast. Lakhani S, Ellis I and
Schnitt S: IARC Press; Lyon: pp. 19–23. 2012
|
|
17
|
Ellis I, Collins L and Ichihara S:
Invasive carcinoma of no special type. WHO Classification of
Tumours of the Breast. Lakhani S, Ellis I and Schnitt S: IARC
Press; Lyon: pp. 34–38. 2012
|
|
18
|
Goswami S, Philippar U, Sun D, et al:
Identification of invasion specific splice variants of the
cytoskeletal protein Mena present in mammary tumor cells during
invasion in vivo. Clin Exp Metastasis. 26:153–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Philippar U, Roussos ET, Oser M, et al: A
Mena invasion isoform potentiates EGF-induced carcinoma cell
invasion and metastasis. Dev Cell. 15:813–828. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pino MS, Balsamo M, Di Modugno F, et al:
Human Mena+11a isoform serves as a marker of epithelial
phenotype and sensitivity to epidermal growth factor receptor
inhibition in human pancreatic cancer cell lines. Clin Cancer Res.
14:4943–4950. 2008.PubMed/NCBI
|
|
21
|
Agarwal S, Gertler FB, Balsamo M, et al:
Quantitative assessment of invasive mena isoforms (Menacalc) as an
independent prognostic marker in breast cancer. Breast Cancer Res.
14:R1242012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wapnir IL, Dignam JJ, Fisher B, et al:
Long-term outcomes of invasive ipsilateral breast tumor recurrences
after lumpectomy in NSABP B-17 and B-24 randomized clinical trials
for DCIS. J Natl Cancer Inst. 103:478–488. 2011. View Article : Google Scholar
|
|
23
|
Roussos ET, Balsamo M, Alford SK, et al:
Mena invasive (MenaINV) promotes multicellular streaming
motility and transendothelial migration in a mouse model of breast
cancer. J Cell Sci. 124:2120–2131. 2011.PubMed/NCBI
|
|
24
|
Roussos ET, Goswami S, Balsamo M, et al:
Mena invasive (MenaINV) and Mena11a isoforms
play distinct roles in breast cancer cell cohesion and association
with TMEM. Clin Exp Metastasis. 28:515–527. 2011.PubMed/NCBI
|
|
25
|
Warzecha CC and Carstens RP: Complex
changes in alternative pre-mRNA splicing play a central role in the
epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol.
22:417–427. 2012. View Article : Google Scholar : PubMed/NCBI
|