1. Introduction
Epstein-Barr virus (EBV), also known as human herpes
virus 4, is a gamma-herpes virus that consists of double-stranded
DNA of ~170 kb in length. It is one of the most common human herpes
viruses and infects >90% of the world’s population by adulthood
and establishes lifelong, latent infections. EBV was the first
virus to be associated with human malignancy, which was discovered
from a Burkitt’s lymphoma cell line in 1964 (1). Subsequent studies revealed that EBV
caused a number of different human malignancies, such as
nasopharyngeal carcinoma (NPC), Hodgkin’s lymphoma, extranodal
NK/T-cell lymphoma, nasal type and lymphoproliferative disorders of
immunocompromised hosts (2).
In 1990, Burke et al (3), first reported the association between
EBV and gastric carcinoma with characteristic
lymphoepithelioma-like histology based on polymerase chain reaction
(PCR) techniques. Subsequent development of in situ
hybridization (ISH) techniques to detect EBV-encoded small RNAs
(EBERs) facilitated the detection of EBV in cancer tissues
(4,5). In gastric carcinoma cells, EBV is not
integrated into the host genome but maintained as a type of plasmid
called an episome. The uniformity of the numbers of terminal
repeats (TRs) among EBV positive carcinoma cells reflects the
clonal origin of a tumor and suggests that EBV is a causative virus
for gastric carcinoma (6).
In spite of these findings, the importance of EBV in
gastric carcinogenesis has long been underestimated. The reason for
this is that Helicobacter Pylori (H. pylori),
discovered in 1983 by Marshall and Warren, and classified as a
definite carcinogen by World Health Organization in 1994, has been
regarded as the major factor in almost all gastric carcinomas
worldwide (7–9). Persistent infection with H.
pylori induces atrophic gastritis and intestinal metaplasia,
and subsequently leads to gastric malignancies including gastric
carcinoma and extranodal marginal zone lymphoma of
mucosa-associated lymphoid tissue (MALT lymphoma). In addition to
accumulating more case series of EBV-associated gastric carcinoma
(EBVaGC), the development of comprehensive molecular analyses has
provided evidence that EBVaGC is a distinct subset both in terms of
its clinicopathological and molecular features.
In the present review, we first describe the
clinical and histological features of EBVaGC. Furthermore, we
discuss recent findings on EBV associated gastric carcinogenesis by
focusing on the roles of latent genes, epigenetic abnormalities,
genomic alterations, and post-transcriptional regulation by
cellular and viral microRNAs (miRNAs).
2. Definitions, epidemiology and clinical
features
EBVaGC is defined by monoclonal proliferation of
carcinoma cells with latent EBV infection. The gold standard for
identifying EBV infection is ISH to detect EBER1, which is abundant
in infected cells (up to 107 molecules/cell). The
frequency of EBV infection in gastric carcinoma ranges from 2 to
20%, with a worldwide average of nearly 10% (10,11).
These differences in reported frequencies may be because of
geographical and environmental factors, although this remains
controversial. In a meta-analysis done by Murphy et al
(12), the pooled estimates of
EBVaGC frequency in American, European and Asian were 9.9, 9.2 and
8.3%, respectively, with an overall frequency of 8.7%. A recent
meta-analysis done by Camargo et al (13) revealed a similar overall frequency
(8.2%), although the frequencies they found were slightly higher in
American (12.5%) and European (13.9%) cases and lower in Asian
cases (7.5%). Based on the annual incidence of gastric carcinoma
(934,000 cases per year), nearly 70,000–80,000 people per year are
estimated to develop EBVaGC (14).
The clinical features of EBVaGC include predominance
among males and a predominant location in the proximal stomach and
remnant stomach after partial gastrectomy for gastric ulcer or
gastric carcinoma. Most published studies have shown an association
with male gender (approximately twice as many males as females),
which has been confirmed by several meta-analyses (12,15,16).
However, this male predominance decreases with age in terms of risk
estimates (15). Studies conducted
in the Americas have shown an association between EBV positive and
younger age, although this was not confirmed in a meta-analysis
(15–17).
Frequent EBV involvement in carcinomas of the
remnant stomach has been reported in several studies, with
frequencies ranging from 6 to 30% (10,12,16).
EBV-positive remnant gastric carcinoma is often found at an
anastomosis site in patients who underwent gastrojejunostomy and
Billroth II anastomosis. Chen et al (18), investigated EBV genome
polymorphisms in remnant gastric carcinoma and showed that EBV
strains were similar among carcinomas in both the remnant and
intact stomach. These findings suggest that repetitive injuries to
the gastric mucosa, such as bile reflux and/or changes in the
microenvironment, may be involved in the development of EBVaGC in
the remnant stomach.
Several risk factors for developing EBVaGC have been
identified (19). Eating salty or
spicy foods, frequently drinking coffee and high-temperature
drinks, and exposure to wood dust and/or iron filings are risk
factors associated with EBVaGC. Recently, smoking was also found to
be associated with EBVaGC [odds ratio (OR) of 1.5; 95% confidence
interval (CI): 1.01–2.3], after adjusting for possible confounders
(20). No significant association
has been found between EBV positive and alcohol drinking (adjusted
OR of 1.0). H. pylori infection, another strong risk factor
for gastric carcinoma, is not a risk factor for EBVaGC, which
suggests that H. pylori and EBV involve different
carcinogenic pathways (16).
The prognostic impact of EBV infection on gastric
carcinoma has long been a matter of debate. Previous studies
reported that EBVaGC exhibited a lower rate of lymph node
metastasis, especially during its early stage, and one study showed
that survival was relatively better as compared with EBV-negative
gastric carcinoma (21–24). However, several reports have shown
a greater risk of death with EBVaGC, although these results were
not statistically significant (25,26).
A recent meta-analysis provided conclusive evidence for this issue.
Camargo et al (13),
demonstrated that, in the largest series (to date) of 4,599 gastric
carcinoma cases, EBV positive was associated with a reduced
mortality rate after adjusting for the stage and other possible
confounders (Hazards ratio of 0.72; 95% CI, 0.61–0.86).
3. Histopathology
By gross appearance, EBVaGC often forms ulcerated or
saucer-like masses with marked thickening of the gastric wall. As
noted above, a tumor is frequently located in the proximal stomach
and the remnant stomach after partial gastrectomy. Multiplicity
(multiple lesions occurred synchronously or meta-chronously in the
stomach) is also a characteristic feature, as confirmed by several
studies (23,27–29).
In its early stages, EBVaGC tends to form well-demarcated, nodular
lesions in the submucosa with less fibrosis as compared with
EBV-negative gastric carcinoma, and this is beneficial for
endoscopic submucosal resection of a tumor (30).
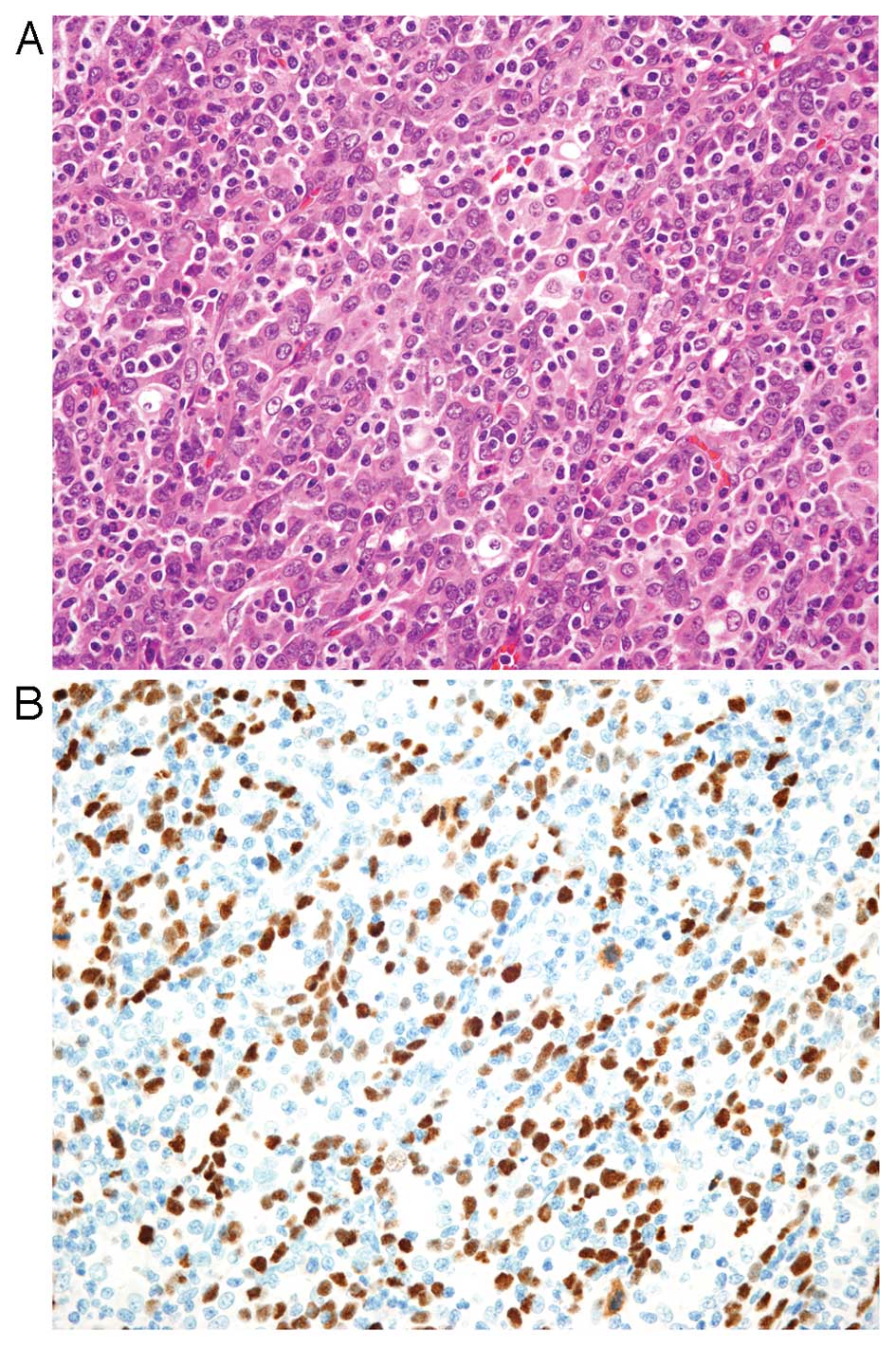
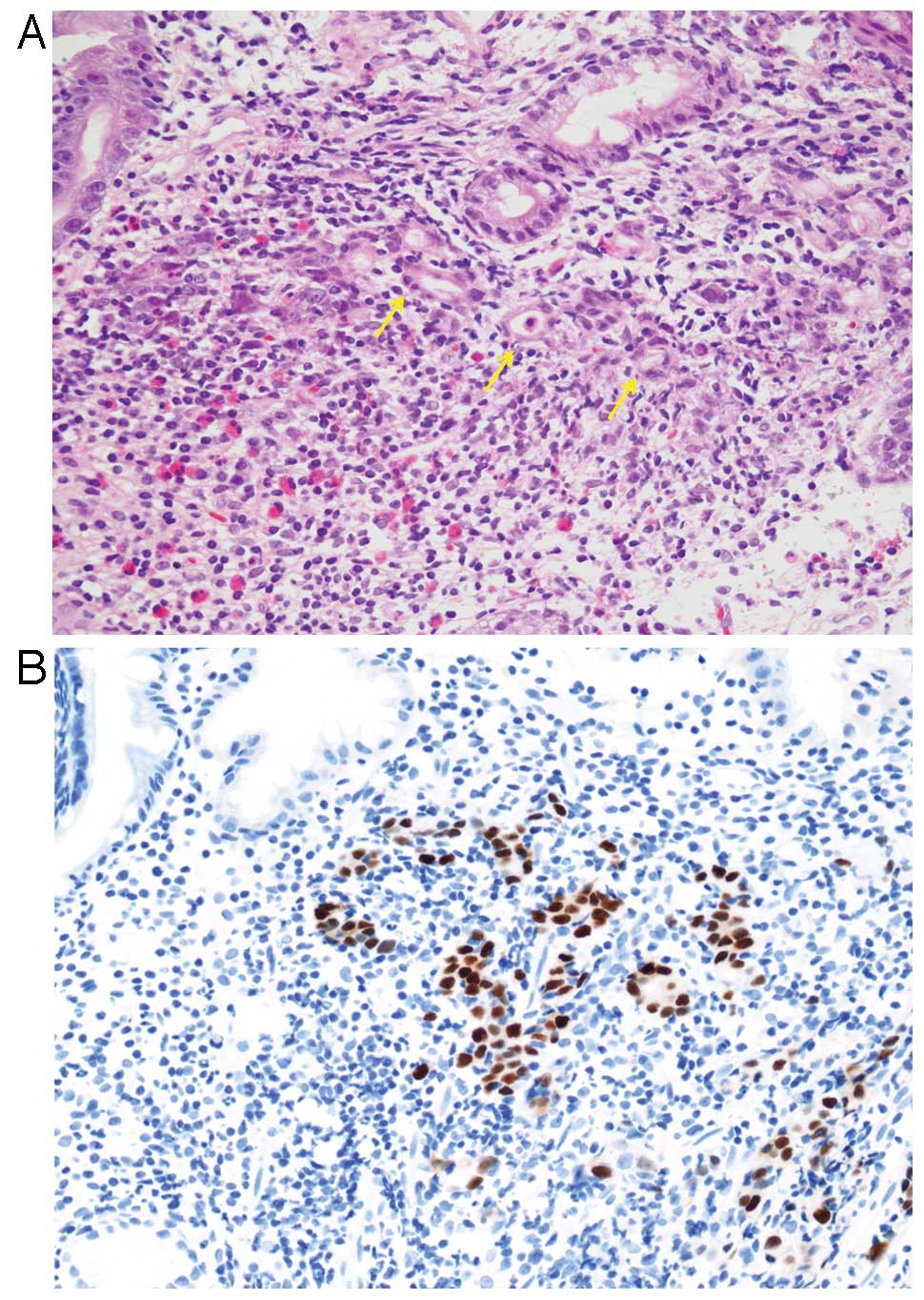
Histologically, EBVaGC is subdivided into two types;
lymphoepithelioma-like carcinoma (LELC)-type (Fig. 1) and conventional-type
adenocarcinoma (Fig. 2), although
there is a morphological continuum between these types. LELC-type
is described as a poorly differentiated carcinoma with dense
infiltration of lymphocytes, which resembles NPC. Because of the
prominent lymphocytic infiltration, it is often difficult to
identify individual carcinoma cells with routine hematoxylin and
eosin (H&E) staining. Immunohistochemistry with antibodies to
cytokeratin and EBER-ISH highlight these carcinoma cells. More than
80% of gastric carcinoma cases showing LELC-type morphology are
EBV-positive (23,31). This histological pattern is also
referred to as ‘gastric carcinoma with lymphoid stroma (GCLS)’
defined as carcinoma showing microalveolar, trabecular, or
primitive-tubular pattern with uniformly dense and diffuse lymphoid
cell infiltration, which encompasses broader morphologic variants
including LELC (32).
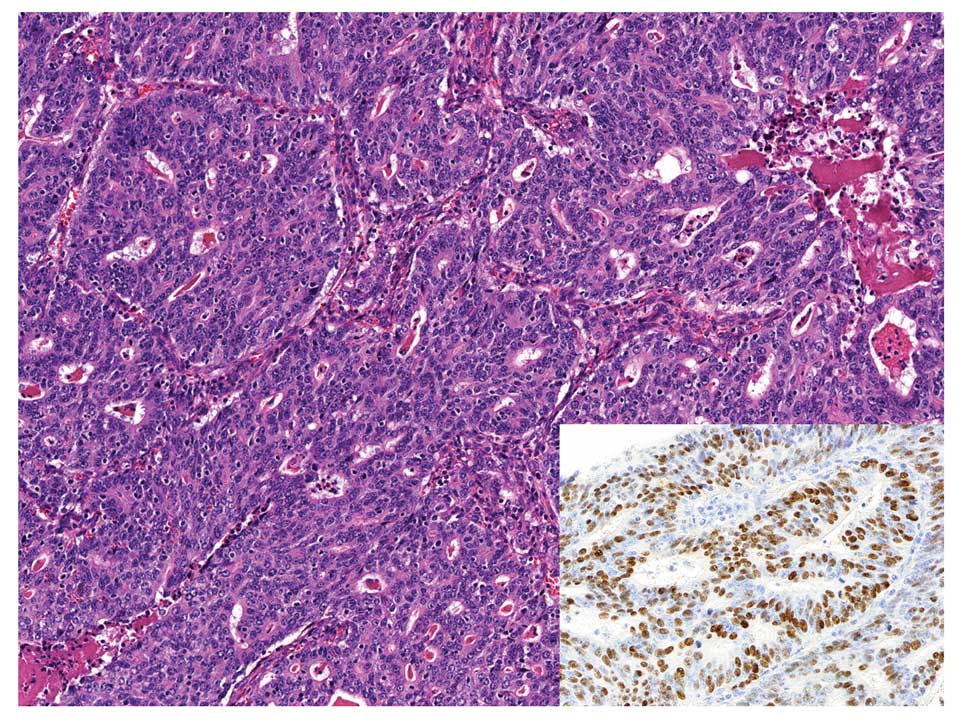
EBVaGC with conventional-type adenocarcinoma
histology shows well to moderately differentiated adenocarcinoma
with variable amount of infiltrating lymphocytes, and it is
classified as an intestinal type gastric carcinoma in Lauren’s
classification (33).
Morphologically, it is almost identical to EBV-negative gastric
carcinoma; therefore, EBER-ISH is necessary to identify the
presence of EBV in carcinoma cells. Another subtype has been
proposed called ‘carcinoma with Crohn’s disease-like lymphoid
reaction,’ which is defined as a tumor accompanied by three or more
lymphoid follicles with active germinal centers at the advancing
edge of the tumor, fewer lymphocytes than tumor cells, frequent
tubule or gland formation and minimal or no desmoplasia (34) (Fig.
3). This type represents a morphology intermediate between the
typical LELC-type and conventional-type adenocarcinoma and could be
included in GCLS.
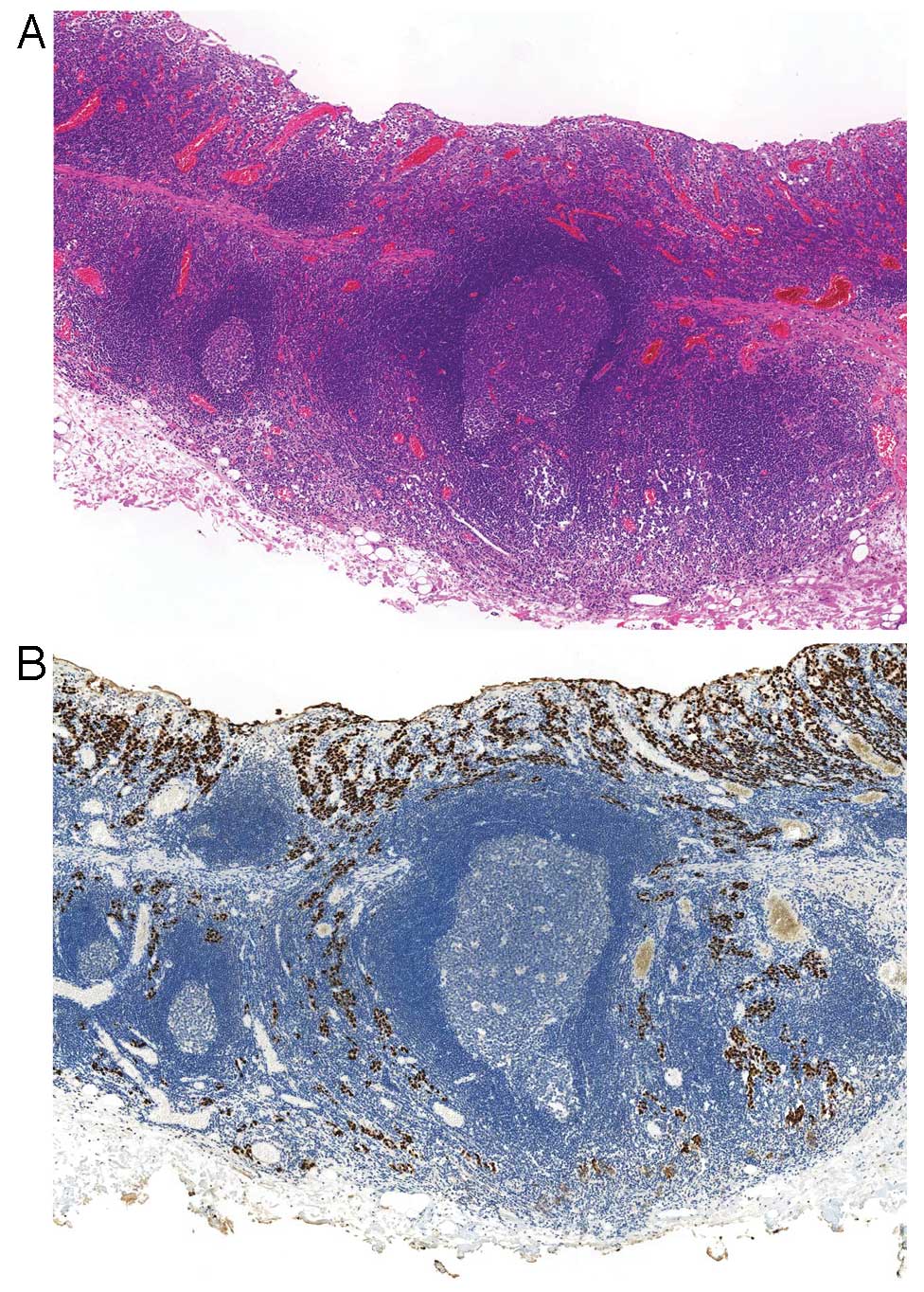
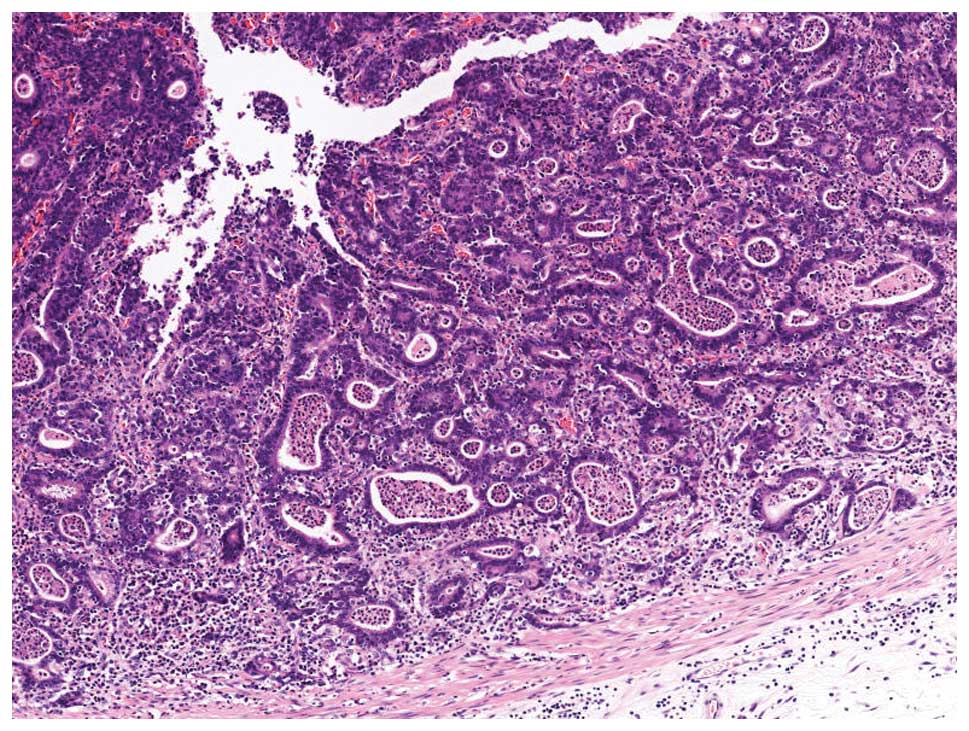
EBVaGC in its early stage shows a characteristic
histology called a ‘lace pattern’ (Fig. 4). This pattern is typically
observed in an intramucosal lesion, which shows irregularly
anastomosing tubules and cords associated with moderate to dense
lymphocytic infiltration and results in a lace-like or reticular
pattern at low magnification. When this pattern is observed in
biopsy specimens, EBER-ISH is recommended for diagnostic purposes
(Fig. 5).
With regard to cell differentiation, EBVaGC displays
unique features. EBVaGC immunophenotyping has shown that nearly
half of the cases have gastric-type mucin (MUC5AC and MUC6)
expression and the other half of cases are a null type; that is,
negative for gastric-type mucin or intestinal-type markers (MUC2
and CD10) (35). Another
characteristic is that >80% of EBVaGC cases express CLDN18,
while CLDN3 expression is infrequent (5%). CLDN18 and CLDN3 belong
to the claudin family that comprises tight junctions. CLDN18
expression is quite specific for the normal stomach and lung,
whereas CLDN3 is expressed in the normal small and large intestines
and in intestinal metaplasia, but is not expressed in the normal
stomach. This suggests that the targets of EBV infection and
subsequent transformation may be precursor cells that have an
intrinsic differentiation potential toward the gastric cell type,
but not the intestinal type. Notably, the differentiation toward
gastric cell type is common to both LELC-type and other morphologic
subtypes in EBVaGC, while in EBV-negative gastric carcinoma CLDN3
expression is associated with intestinal histology and CLDN18
expression with diffuse histology in Lauren’s classification.
Regardless of different morphological subtypes, all
carcinoma cells of EBVaGC are EBV-positive, which supports a causal
role for EBV in gastric carcinogenesis. Since previous studies have
not demonstrated any close associations between histological
subtypes and etiological factors, anatomical location (proximal or
distal), cellular phenotype, or genomic/epigenetic alterations, it
is currently unclear what causes the histological diversity of
EBVaGC, which is expected to be clarified in the future studies for
the better understanding of the pathogenesis of EBVaGC.
Prominent inflammatory infiltrate in the tumor is
one of the characteristic features of EBVaGC. These tumor
infiltrating cells are primarily lymphocytic, particularly
CD8-positive or CD4-positive T cells accompanied by CD68-positive
histiocytes (10,36,37).
In addition, infiltration of abundant B cells, plasma cells, or
neutrophils is often observed and these infiltrating cells rarely
masquerade as other neoplasms. We previously reported EBVaGC with
prominent Mott cell (plasma cells with multiple Russell bodies)
infiltration that mimicked plasma cell neoplasms (38). Infiltrating Mott cells and B cells
were negative for immunoglobulin light chain restrictions or heavy
chain rearrangements, which suggested that they were reactive in
nature. Another extreme variant is EBVaGC with osteoclast-like
giant cells (39). These giant
cells showed a histiocytic phenotype and were considered as a
reaction to gastric carcinoma.
The diversity of tumor infiltrating inflammatory
cells is an important feature of EBVaGC and it is also shared by
other EBV-related malignancies, such as Hodgkin’s lymphoma and
EBV-positive diffuse large B-cell lymphoma, which are often
accompanied by numerous reactive inflammatory cells. This feature
reflects the immunogenicity of EBV, and some investigators have
suspected that this immune response by the host is one reason for a
better prognosis. Further studies are needed to determine the
detailed mechanism of an immune response against EBV-positive tumor
cells in order to develop an effective treatment for this
disease.
One of the interesting findings on the background of
EBVaGC is that it is sometimes associated with gastritis cystica
profunda (GCP), which is a relatively rare, benign lesion that
is characterized by polypoid hyperplasia and cystic dilatation of
the gastric glands that extend into the stomach submucosa. Choi
et al (40), reported that
the EBV-positive rate was significantly higher in gastric carcinoma
cases with GCP (31.1%) as compared to those without GCP (5.8%). It
has been assumed that GCP may reflect chronic inflammation in the
stomach; although, it remains unclear whether the coexistence of
GCP and EBVaGC is merely coincidental, or if GCP presents as a
precancerous lesion.
4. EBV infection in gastric epithelial cells
and EBV latent genes
Previous studies have investigated how EBV infection
is established in gastric epithelial cells which lack the
expression of viral receptor CD21 (also known as CR2) through which
EBV enters B lymphocytes (41,42).
Since the efficiency of EBV infection is greatly improved by
directly co-culturing epithelial cells with EBV-producing B
lymphoblastoid cells (Akata cells) than cell-free infection, direct
cell-to-cell contact with B lymphocytes is considered to be the
major model of EBV infection in epithelial cells (43). Although quite rare, EBV infection
is found in a small fraction of non-neoplastic gastric mucosa in a
single cell or a few glands (6,44),
suggesting that EBV infection precedes the clonal growth of
EBV-infected cells and subsequently develops carcinoma. Chronic
gastritis in the background of EBVaGC might enhance the chance of
interaction between gastric epithelial cells and B lymphocytes, and
cytokines produced by inflammatory cells might support the growth
of EBV-infected gastric epithelial cells.
Most in vitro studies have utilized
cell-to-cell contact method to explore the role for EBV in gastric
carcinogenesis by secondarily infecting gastric carcinoma cell
lines with EBV. The method is quite useful in investigating how EBV
infection alters the biological nature of these cells, however,
these experiments have limits in that they use cell lines already
transformed to malignant cells by some factors other than EBV,
which should be kept in mind when applying the results to the in
vivo situation.
Once EBV infection is established in B-lymphocytes
or epithelial cells, it is maintained in a latent form. Latent EBV
infection has three distinct forms that are determined by the
expression patterns of latent genes. EBER-1 and 2, EBV-determined
nuclear antigen (EBNA)-1, BamHI A region rightward
transcripts (BARTs), and BART miRNAs (discussed later) are
expressed in all latency types. In latency type II, latent membrane
protein (LMP)-1, 2A and 2B are also expressed, and latency type III
includes all of these latent genes along with EBNA-2, 3A, 3B, 3C
and LP. These latency types differ among different EBV-associated
malignancies as shown in Table
II.
 | Table IILatency type in EBV-associated
malignancies. |
Table II
Latency type in EBV-associated
malignancies.
| Latency I | | Latency II | Latency III |
|---|
| EBERs | + | | + | + |
| EBNA-1 | + | | + | + |
| EBNA-2, 3A-C,
LP | − | | − | + |
| LMP-1 | − | | + | + |
| LMP-2A, B | − | | + | + |
| BARTs | + | | + | + |
| BART miRNAs | + | | + | + |
| Associated
malignancies | Burkitt’s
lymphoma | Gastric carcinoma
Nasopharyngeal carcinoma NK/T cell lymphoma | Hodgkin’s
lymphoma |
Immunodeficiency-associated lymphoma |
EBVaGC belongs to latency type I or II, in which
EBERs, EBNA-1, BARTs and BART miRNAs are expressed and
approximately half of EBVaGC cases express LMP-2A (45,46).
The expression pattern of these latent genes is diverse among
cases, and this is also true in NPCs and NK/T cell lymphomas. Two
studies performed comprehensive expression profiling of viral
latent and lytic transcripts in EBVaGC (47,48).
EBERs were the most abundant among all latent genes, followed by
BARTs. LMP-2A, 2B and EBNA-1 expressions were very low. Notably,
transcription of immediate early lytic genes, BZLF1 and BRLF1 was
detected in some gastric carcinoma cases without subsequent
progression of lytic cycle (47).
Similarly, BZLF1 expression was observed in gastric carcinoma cells
in vitro under long-term cultivation after EBV infection,
but very few of them progressed to late lytic phase. The authors
concluded that abortive lytic replication might somehow responsible
for EBV genome amplification (49). These findings may provide some
clues to clarify how EBV episome is maintained in EBVaGC with
low/absent EBNA-1 expression.
Although LMP-1 is a well-known oncoprotein that is
essential for EBV to efficiently transform resting primary B cells
into autonomously proliferating lymphoblastoid cell lines, its
expression is extremely low in EBVaGC and is usually undetectable
at the protein level. To investigate the oncogenic properties of
latent genes other than LMP-1, the roles of latent genes expressed
in gastric carcinoma have been investigated (Table III). These findings are discussed
separately in the following sections.
 | Table IIIRoles of EBV latent genes in
EBV-associated gastric carcinoma. |
Table III
Roles of EBV latent genes in
EBV-associated gastric carcinoma.
| Latent gene | Related molecules
(and their downstream) | Biological function
(ref.) |
|---|
| EBER-1 | IGF-1 | Autocrine growth
(51) |
| hsa-miR-200a, 200b,
(ZEB1, ZEB2, E-cadherin) |
Epithelial-to-mesenchymal transition
(52) |
| IL-6 ( STAT3, p21,
p27) | Chemoresistance
(53) |
| EBNA-1 | p53 | Tumorigenicity
(57) |
| PML (p53, p21) | Anti-apoptosis
(58) |
| LMP-2A | NF-κB
(survivin) | Anti-apoptosis
(63) |
| Cyclin E1 | Anti-apoptosis
(64) |
| Drp-1 |
Epithelial-to-mesenchymal transition
(65) |
| STAT3 (DMNT-1,
PTEN) | |
| DMNT3b | Epigenetic
silencing of tumor suppressor genes (66,67) |
| BARTs BARF1 | Bcl-2, Bax | Chemoresistance
(72) |
| Cyclin D1 | Cell proliferation
(73) |
| NF-κB/cyclin D1,
p21WAF | Cell proliferation
(74) |
EBERs
EBERs are the most abundant viral transcripts found
in latently EBV-infected cells and they have been shown to have
various effects with regard to cell proliferation,
apoptosis-resistance, production of autocrine growth factors and
interactions with host proteins to enhance cellular signaling
(50). However, only a few studies
have investigated the roles of EBERs in gastric carcinogenesis.
Iwakiri et al (51),
demonstrated that EBV infection induced the expression of IGF-1 in
an EBV-negative gastric carcinoma cell line, NUGC-3 and IGF-1
functioned as an autocrine growth factor. They showed that EBERs
were responsible for these phenomena.
We recently reported that EBER-1 altered cellular
miRNA expression to suppress E-cadherin, which resulted in an
epithelial-to-mesenchymal transition (EMT) in gastric carcinoma
cell line, as will be discussed later (52). Another recent report by Banerjee
et al (53), showed that
EBERs could upregulate IL-6 expression and activated its downstream
regulator STAT3, which resulted in downregulation of the cell cycle
inhibitors p21 and p27 in a gastric carcinoma cell line, which was
associated with chemoresistance. They also showed that EBERs
induced the activation of pro-metastatic molecules, pFAK and pPAK1,
and the downregulation of anti-metastatic molecules, RhoGD1 and
KAI-1, which promoted cell migration.
EBNA-1
EBNA-1 is the only viral protein that is
consistently expressed in all types of EBV-associated malignancies,
which is essential for the replication and stable persistence of
EBV episomes. Increasing evidence has demonstrated that EBNA-1
alters the cellular environment to promote genomic instability and
may have the potential to act as an oncogene (54–56).
However, until recently, little was known regarding a role of
EBNA-1 in gastric carcinogenesis. Cheng et al (57), reported that, the gastric cell
lines SCM1 and TMC1 that were transfected with EBNA-1 had enhanced
tumorigenicity and growth rates in xenografts. Another report by
Sivachandran et al (58),
demonstrated that AGS cells that were infected with EBV had fewer
promyelocytic leukemia (PML) nuclear bodies and less PML protein
than EBV-negative cells, and that these phenomena were caused by
EBNA-1. These findings were also confirmed in biopsy samples. PML
is a tumor suppressor protein that is associated with p53
activation. They showed that by repressing PML, EBNA-1 impaired p53
acetylation, p53-dependent p21 transcription, and apoptosis, which
resulted in enhanced cell survival after DNA damage.
Considering that EBNA-1 is essential in the
maintenance of EBV genome and may also act as oncogenes, EBNA-1 is
one of the possible therapeutic targets of EBVaGC. Some previous
studies have demonstrated that suppression of EBNA-1 in lymphoma
cell lines inhibited cell proliferation (59–61).
However, as previously mentioned, expression of EBNA-1 in EBVaGC is
low, or even absent in some cases, EBNA-1-targeted therapy may not
be applicable to all EBVaGCs and other therapeutic approaches
should be sought.
LMP-2A
LMP-2A protein is expressed in about half of EBVaGC
cases and has been relatively well investigated as compared with
other latent genes with regard to gastric carcinogenesis. LMP-2A
inhibited transforming growth factor (TGF) β1-induced cellular
apoptosis in a gastric carcinoma cell line (62). We previously reported that, the
gastric cell lines that were transfected with LMP-2A had
upregulated survivin expression mediated through nuclear factor
(NF)-κB activation, which resulted in resistance against serum
deprivation-induced apoptosis (63). Similarly, Liu et al
(64), showed that transfecting
LMP-2A into a gastric carcinoma cell line improved cell growth and
reduced apoptosis, via increased cyclin E expression and the
proportion of cells in S phase. Another possible function of LMP-2A
is activating the Notch signaling pathway, which disrupts the
mitochondrial fission-fusion equilibrium by enhancing
dynamin-related protein (Drp)-1 expression, which results in
increased cell migration along with the overexpression of EMT
markers (65).
In addition to these direct modulating effects on
cell proliferation and migration, LMP-2A has a unique function
inducing epigenetic changes in the host genome. LMP-2A promotes
STAT3 phosphorylation, which activates the transcription of DNA
methyltransferase (DNMT) 1 (66).
Upregulated DNMT1 results in the epigenetic silencing of PTEN
expression through the methylation of CpG islands in its promoter
region. Zhao et al (67),
demonstrated that LMP-2A caused the upregulation of DMNT3b. As will
be discussed later, epigenetic changes, particularly
hypermethylation of the host genome, is one of the most crucial
mechanisms of EBV-induced gastric carcinogenesis, for which LMP-2A
may play a part through its activation of DNMTs.
BARTs
BARTs are multi-spliced transcripts that were
originally discovered by cDNA library analysis of NPC xenografts
and were subsequently found to be abundantly expressed in various
kinds of EBV-associated malignancies. Several of these transcripts
have open reading frames (ORFs), such as BARF0, BARF1, RPMS1 and
A73, which possibly encode for proteins. Some of these can be
artificially translated into their respective proteins in
vitro, although endogenous expression of these proteins has not
been completely confirmed in EBV-associated cancer tissues
(68–70). Several reports on BARF1 involvement
in gastric carcinogenesis have been published. The BARF1
gene is expressed in nearly 100% of EBVaGC cases (71). Expression of BARF1 is also observed
in NPC, while it is generally undetectable in EBV-positive B
lymphocytes and lymphomas, which may be crucial in EBV-associated
epithelial malignancies. Transfecting BARF1 into a gastric
carcinoma cell line induces significant alterations in host gene
expression, particularly those genes related to proliferation and
apoptosis, and BARF1-transfected cells exhibit
chemoresistance along with an increased Bcl-2-to-Bax expression
ratio (72). Wiech et al
(73), demonstrated that cyclin D1
was upregulated in BARF1-trasfected HaCaT cells and that cyclin D1
was overexpressed in EBVaGC cells, by immunohistochemistry.
Recently, Chang et al (74), demonstrated that BARF1 protein was
secreted into culture supernatants of gastric carcinoma cells that
had been transfected with BARF1, using western blot
analysis. These BARF1-expressing cells had increased cell
proliferation that was mediated via upregulated NF-κB/cyclin D1 and
reduced expression of the cell cycle inhibitor
p21WAF.
5. Epigenetic abnormalities
A number of studies have demonstrated that
epigenetic abnormalities, such as promoter hypermethylation, play
crucial roles in the carcinogenesis of EBVaGC (67,75–81).
Global and non-random CpG island methylation in the promoter
regions of many cancer-associated genes, particularly tumor
suppressor genes, is found in EBVaGC, which results in repressing
the transcription of downstream genes. Initially, assessing
methylation status was performed for individual genes using
methylation-specific PCR (MSP). EBVaGC exhibited promoter
hypermethylation in multiple genes involved in cell cycle
regulation (p14ARF, p15,
p16INK4A and p73), DNA repair
(hMLH1, MGMT and GSTP1), cell adhesion and
metastases (CDH1, TIMP1 and TIMP3), apoptosis
(DAPK and bcl-2), and signal transduction
(APC, PTEN and RASSF1A). The number of
reported hypermethylated genes in EBVaGC continues to increase
(82).
We recently performed a comprehensive analysis of
the promoter methylation status of 51 gastric carcinoma cases using
an Infinium HumanMethylation27 BeadChip (Illumina), which included
27,578 CpG sites covering 14,495 genes (83). Subsequent hierarchical clustering
analysis demonstrated that, based on methylation status, gastric
carcinoma could be subclassified into three epigenotypes,
EBV+/extensively high-methylation,
EBV−/high-methylation and
EBV−/low-methylation, which were characterized by
different sets of methylated genes: genes specific for the
EBV+ type (CXXC4, TIMP2 and
PLXND1); highly methylated in EBV+ and
EB−/high-methylation type (COL9A2, EYA1
and ZNF365); and frequently methylated in all epigenotypes
(AMPH, SORC33 and AJAP1).
Genes that were methylated in EBV-negative
carcinomas were also methylated in EBVaGC and ~270 genes were
uniquely methylated in EBVaGC. Frequent MLH1 methylation
(46%) was found in the EBV-/high-methylation type, but none of the
EBVaGC cases showed MLH1 methylation. In addition, polycomb
repressive complex (PRC)-target genes that have been reported in
embryonic stem cells were enriched in genes methylated in
EBV-negative types, regardless of methylation status. In contrast,
aberrant methylation induced by EBV was observed not only within
PRC-target genes but also within non-PRC-target genes.
EBV-infection of the low-methylation type gastric cancer cell line
MKN7, induced extensive methylation within 18 weeks to acquire an
EBV-specific methylation epigenotype.
It is worth noting that viral latent gene expression
is also suppressed by methylation. We experimentally confirmed that
viral DNA methylation preceded the methylation of host DNA by one
week (unpublished data). The methylation of viral genes might be
one host defense mechanism against foreign DNA for suppressing the
viral gene expression. However, this might result in other
outcomes. Repressing viral latent gene expression might benefit EBV
by allowing it to escape a host’s immune response. In addition,
excessive methylation may lead to repressing tumor suppressor
genes, which ultimately could give rise to carcinogenesis.
6. Somatic genomic alterations and gene
expression
Recent advances in genome-wide, high-throughput
techniques to explore genetic alterations in cancer, such as single
nucleotide polymorphism (SNP) arrays, somatic copy-number analysis,
whole-exome sequencing, mRNA and miRNA sequencing, array-based DNA
methylation profiling, and reverse-phase protein arrays provide new
means to comprehensively investigate EBVaGC at the molecular and
genetic levels. These techniques have enabled researchers to
identify relatively unknown, infrequent genetic abnormalities that
could not be found using conventional approaches (84–86)
(Table IV).
 | Table IVMutations in EBV-associated gastric
carcinoma. |
Table IV
Mutations in EBV-associated gastric
carcinoma.
| Frequently mutated
genes (ref.) |
|---|
| PIK3CA
(10.3–80%) (87–89) |
| ARID1A
(47–55%) (89,91) |
| AKT2 (38.2%)
(90) |
| TGFBR1
(25.0%) (90) |
| CCNA1
(25.0%) (90) |
| BCOR (23%)
(89) |
| MAP3K4
(20.8%) (90) |
| Other mutated genes
<10%) |
| CTNNB1(87), TP53, ITIH1,
KRAS, NRAS, PLCE1, SHOC2, GIPC1,
ITGA6, ITGB4, NRP1, PLEC, JAK2,
CSF2RB, GHR, MPL, PTPN2, SOCS3,
STAT5B, COL1A2, COL5A1, DCN, F2,
IGF1, IGF2, IGFALS, IGFBP5,
THBS1 (89) |
Lee et al (87), used a high-throughput genotyping
platform to determine the mutation status of 474 hotspots in 41
genes using 237 gastric adenocarcinomas, which included 58 EBVaGCs.
Among these, 34 cases (14.3%) harbored somatic mutations, 6 of
which concomitantly had two different mutations. Fourteen EBVaGC
cases had mutations; 6 in PIK3CA (10.3%), 1 in p53
(1.7%), 2 in APC (3.4%), 1 in STK11 (1.7%), 3 in
CTNNB1 (5.2%) and 1 in CDKN2A (1.7%). CTNNB1
mutations were significantly more frequent in EBVaGC than in
EBV-negative gastric carcinomas (one of 179 cases, 0.6%). Frequent
PIK3CA mutations were also reported in two subsequent
studies; 16.7% (of 18 EBVaGCs) in a report by Sukawa et al
(88), and 80% (of 28 EBVaGCs) in
a report by The Cancer Genome Atlas (TCGA) Research Network
(89). A recent report by Liang
et al (90), showed several
newly identified mutations in EBVaGC, including mutations in
MAP3K4 (20.8%), TGFBR1 (25.0%), CCNA1 (25.0%)
and AKT2 (38.2%). Among these, an AKT2 mutation was
associated with poor survival (90).
Another frequently mutated gene in EBVaGC is
ARID1A. ARID1A encodes for a member of the SWI/SNF
chromatin remodeling family and is currently thought to function as
a tumor-suppressor gene. Wang et al (91), reported that 47% of EBVaGC cases (7
of 15) harbored an ARID1A mutation by exome sequencing and
73% (11/15) had reduced ARID1A protein expression by
immunohistochemical analysis. Clinically, ARID1A alterations
were associated with a better prognosis in a stage-independent
manner. Similarly, we demonstrated that loss of ARID1A expression
was frequent in EBVaGC (23/67, 34%), while ARID1A expression was
maintained in NPCs or EBV-positive lymphomas (92). Further studies are necessary to
clarify the roles of these mutations in gastric carcinogenesis by
EBV.
In a recent report by the TCGA Research Network, a
comprehensive molecular evaluation of 295 gastric carcinomas was
performed using several different modalities, including genomic
alterations, gene expression profiling and proteomic analysis. They
proposed a novel molecular classification to divide gastric
carcinomas into four types (89),
of which EBVaGC was one. The others were: ‘microsatellite
instability (MSI)’ characterized by hypermutation, gastric-CIMP
(CpG-island methylator phenotype) and MLH1 silencing; ‘genomically
stable (GS),’ which showed diffuse histology, CDH1 and
RhoA mutations, and a CLDN18-ARHGAP fusion; and
‘chromosomal instability (CIN)’ with intestinal histology, a p53
mutation, marked aneuploidy and amplification of receptor tyrosine
kinases.
Frequent PIK3CA and ARID1A mutations
in EBVaGC (80 and 55%, respectively) were also confirmed in that
study, and frequent mutations in BCOR (BCL6 interacting
co-repressor), which is a member of PRC, was also demonstrated
(23%). Similar to the results in previous reports, p53
mutations were rare in EBVaGC (82,93).
Both PIK3CA and ARID1A mutations were also frequent
in EBV-negative, MSI-high gastric carcinoma, which was confirmed in
multiple studies cited above. EBVaGC and MSI-high gastric carcinoma
also shared CIMP-high features, although epigenetic silencing of
mismatch repair genes (e.g. MLH1) was rarely observed in EBVaGC, as
previously noted. These findings were noted in several previous
reports and indicated that EBVaGC had a unique carcinogenic pathway
independent of MSI (28,79,86,94).
Another novel finding characteristic of EBVaGC was
recurrent amplification at 9p24.1 at the locus that includes
JAK2, CD274 (encoding for PD-L1) and PDCD1LG2
(encoding for PD-L2). PD-L1 and PD-L2 are ligands of PD-1 that is
expressed on T cells, B cells, monocytes and natural killer cells,
as well as tumor infiltrating lymphocytes. Upon ligation with PD-L1
and PD-L2, PD-1 suppresses downstream PI3K and Akt signaling, which
results in inhibiting T cell proliferation. Some malignancies have
been reported to have high PD-L1 expression levels, which was
associated with an aggressive behavior and poor prognosis (95). Blocking the interaction between
PD-1 and PD-L1/L2 could augment an antitumor immune response, and
clinical trials to investigate the efficacy of immunotherapy by
targeting these molecules are under way (96).
Gene expression profiling and proteomic analysis
have revealed activation in immune cell signaling and mitotic
pathways, along with inactivation of the HIF-1α transcription
factor network in EBVaGC (48,89,97,98).
In the report by the TCGA Research Network, EBVaGC exhibited high
expression of CXCL11, CXCL9, CXCL17, IDO1, CXCL10, UGT2A3,
LOC400043, CAMK2N2, DKK1 and MIA, and low expression of CLDN3,
PPP1R1B, REG4, CDH17, TFF3, SCNN1A, FUT3, MUC3A, KR7 and WFDC2 as
compared to other types of gastric cancers (89). Enhanced IL-12 mediated signaling
signatures were highly characteristic of EBVaGC. In the report by
Kim et al (98), genes
associated with cytokine activities, immune response, leukocyte
migration, hormone secretion and cholesterol transport for
lipoprotein clearance were deregulated in EBVaGC. Along with the
evidence for PD-L1/L2 overexpression, modulating immune cell
signaling may have therapeutic effects on EBVaGC (98–100).
7. miRNA abnormalities
While genetic and epigenetic alterations induce the
upregulation or downregulation of cancer-associated genes at the
transcriptional level, miRNAs are novel, post-transcriptional
regulators of gene expression. A miRNA is a small, non-coding RNA
molecule ~22 nucleotides in length and is coded in the introns or
exons of encoding genes. A miRNA precursor is processed from these
transcripts and is subsequently processed by Drosha and Dicer to a
mature form. Mature miRNA interacts with the 3′ untranslated region
(UTR) of a target mRNA and represses its translation. To date,
>2,600 human miRNAs (cellular miRNAs) have been archived in
miRBase (http://www.mirbase.org). An increasing
number of studies have shown that the dysregulation of certain
miRNAs induces carcinogenesis in various organs. Similar to
oncogenes and tumor suppressor genes, miRNAs associated with
carcinogenesis are called oncomiRs and anti-oncomiRs. Alterations
in cellular miRNAs in EBVaGC have not been intensively
investigated. We previously reported that two cellular miRNAs,
hsa-miR-200a and hsa-miR-200b, were downregulated in EBVaGC both in
tissue samples and in cell lines (52). These miRNAs targeted the
transcription repressors ZEB1 and ZEB2, which regulate E-cadherin
expression. Downregulation of these miRNAs ultimately reduced
E-cadherin expression and triggered the epithelial-to-mesenchymal
transition. EBV latent genes, BARF0, EBNA-1 and EBERs,
cooperatively suppressed hsa-miR-200a and 200b expression in cell
lines; while reduced ZEB 1, ZEB2 and E-cadherin expression was
significant only in EBERs-transfected cells.
Viral genomes also encode for miRNAs and EBV was the
first virus in which viral miRNAs were found (101–103). To date, 25 EBV miRNA precursors
and 44 mature EBV miRNAs have been registered in miRBase.
EBV-encoded miRNAs fall into two major clusters: BHRF-1 and BART.
The BHRF-1 cluster contains four mature miRNAs that are expressed
only in lytically infected cells or cells with latency type III
infections. The BART cluster is located in the non-coding region of
BARTs, and is further subdivided into subclusters 1 and 2, which
include 38 mature EBV miRNAs in total, and the miRNAs
ebv-miR-BART2-5p and ebv-miR-BART2-3p are located downstream of
these two clusters.
Several studies profiled EBV-encoded miRNA
expression in EBV-associated malignancies, including NPC and
Diffuse large B cell lymphoma (DLBCL), and showed that specific
viral miRNAs played roles in carcinogenesis (Table V) (104–121). Several reports investigated the
expression of viral miRNAs in EBVaGC tissue samples and cell lines
(97,112,122–125). EBV miRNAs were variably expressed
in EBVaGC cells, among which ebv-miR-BART1-3p, 2-5p, 3-3p, 4-5p,
5-5p, 7-3p, 9-3p, 10-3p, 17-5p and 18-5p were expressed at
relatively high levels. The ebv-miR-BART7-3p expression level was
consistently high in other EBV-associated malignancies, although
its function has not been determined. A recent study by Marquitz
et al (123), reported the
expression profiles of cellular and viral miRNAs in EBV-infected
AGS cells. By sequencing small RNA libraries created from these
cells, they showed that EBV miRNA constituted 15% of total miRNAs,
and that the remainder was derived from host cells. miRNA PCR array
analysis revealed that let-7 family members, miR-200 family
members, and several human miRNAs were downregulated by EBV
infection. In addition, EBNA-1 transfection reduced hsa-miR-143
expression in AGS cells. Another report by the same group showed
that gene expression changes induced by EBV infection of AGS cells
were highly enriched for genes involved in cell motility and
transformation pathways, and that these genes were potentially
targeted by viral miRNAs (97).
 | Table VThe roles of EBV miRNAs in
EBV-associated malignancies. |
Table V
The roles of EBV miRNAs in
EBV-associated malignancies.
| Name | Targets in EBV | Targets in host
cells | Related
malignancies (ref.) |
|---|
| miR-BHRF1-1 | | GUF1, SCRN1 | Lymphoma (126) |
| miR-BART1-5p | LMP-1 | | NPC (127) |
| | CLEC2D, LU75,
SP100, DICER1, MICB | Lymphoma (126) |
| miR-BART1-3p | | CXCL11 | Lymphoma (119) |
| miR-BART2-5p | BALF5 | | Lymphoma (128) |
| miR-BART3-3p | | DICER1, MICB | Lymphoma (126) |
| | IPO7 | Lymphoma (129) |
| miR-BART5-5p | LMP-1 | | Lymphoma (116) |
| | PUMA | NPC, GC (107) |
| miR-BART6-5p | LMP-1 | | NPC (127) |
| | DICER1 | Lymphoma, NPC
(130) |
| miR-BART6-3p | | IL6R, PTEN | Lymphoma (131) |
| miR-BART9-3p | LMP-1 | | Lymphoma (132) |
| | CDH1 | NPC (133) |
| miR-BART10-3p | BHRF1 | | Lymphoma (116) |
| miR-BART11-5p | | EBF1 | Lymphoma (134) |
| miR-BART13-3p | | CAPRIN2 | Lymphoma (116) |
| miR-BART15-3p | | NLRP3 | Lymphoma (135) |
| | BRUCE | GC (106) |
| miR-BART16 | LMP-1 | | NPC (127) |
| | TOMM22 | Lymphoma (129) |
| miR-BART17-5p | LMP-1 | | NPC (127) |
| miR-BART18-5p | | MAP3K2 | Lymphoma (121) |
| miR-BART19-3p | LMP-1 | | Lymphoma (116) |
| miR-BART20-5p | BZLF1, BRLF1 | | GC (136) |
| | TBX21 | Lymphoma (137) |
| miR-BART22 | LMP-2A | | NPC (138) |
| miR-BART
miRNAs | | BIM | GC (113) |
Viral miRNA involvement in EBVaGC remains largely
unknown. We performed comprehensive profiling of viral miRNA
expression in tissue samples of EBVaGC and identified frequently
expressed viral miRNAs. In silico analysis provided
potential targets of these miRNAs, including genes associated with
cell proliferation, apoptosis, and migration, and the direct
interaction of these target genes and specific viral miRNAs were
validated in vitro (unpublished data). Further studies to
clarify the roles of cellular and viral miRNAs and the regulatory
mechanisms of these molecules by EBV and host cells will be needed
to completely understand the carcinogenic mechanisms involved in
EBVaGC.
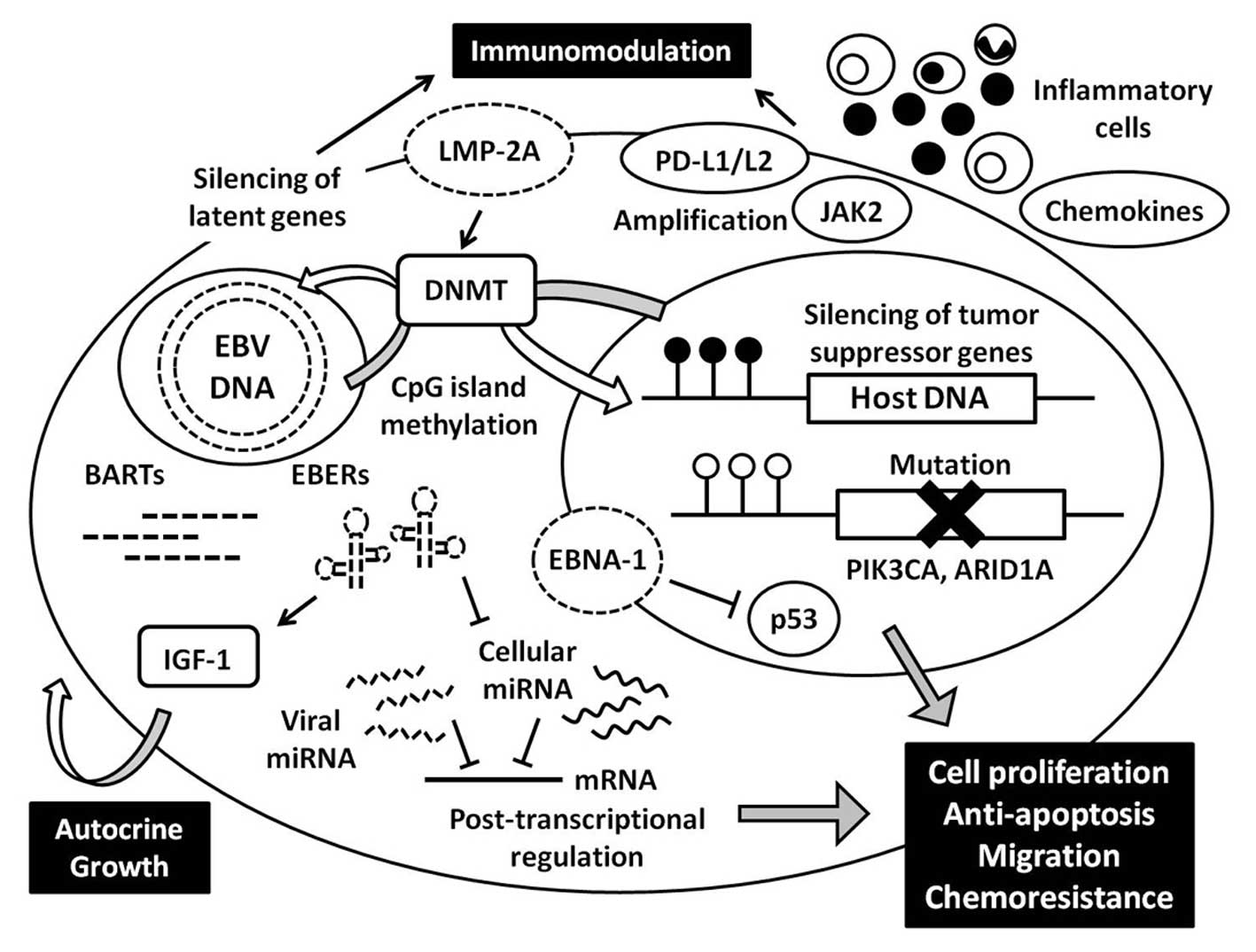
8. Conclusions
EBVaGC is a distinct subtype of gastric carcinoma
with regard to both its clinicopathological and molecular features.
Recent advances in comprehensive genome-wide analysis have provided
novel findings regarding the genetic and epigenetic abnormalities
that are unique to EBVaGC, and these various factors cooperate to
develop carcinoma (Fig. 6). Global
and non-random CpG island hypermethylation is characteristic
feature of EBVaGC, and epigenetic silencing of various genes,
especially tumor suppressor genes, play a key role in
carcinogenesis. Furthermore, increased activation of DNA
methyltransferase by LMP-2A also induces hypermethylation of EBV
genome itself, resulting in limited expression of latent genes,
which may benefit in escaping from immune response by the host.
Viral specific transcripts including latent genes and miRNAs have
oncogenic properties such as increased cell proliferation and
motility, anti-apoptotic effect, and chemoresistance, which help
tumor progression. Although EBVaGC is relatively genomically
stable, frequent mutations in the PIK3C and ARID1A
genes are found; and the roles of these mutations in carcinogenesis
are expected to be clarified in the future studies. Furthermore,
there remain questions regarding how and when these mutations
occur, what triggers hypermethylation, and how host cells regulate
the transcription of viral genes. Finally, the interaction between
carcinoma and inflammatory cells is another key to understand its
carcinogenesis, which will also benefit the development of
disease-specific therapies.
References
|
1
|
Epstein MA, Achong BG and Barr YM: Virus
particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet.
1:702–703. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young LS and Rickinson AB: Epstein-Barr
virus: 40 years on. Nat Rev Cancer. 4:757–768. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burke AP, Yen TS, Shekitka KM and Sobin
LH: Lymphoepithelial carcinoma of the stomach with Epstein-Barr
virus demonstrated by polymerase chain reaction. Mod Pathol.
3:377–380. 1990.PubMed/NCBI
|
|
4
|
Shibata D, Tokunaga M, Uemura Y, Sato E,
Tanaka S and Weiss LM: Association of Epstein-Barr virus with
undifferentiated gastric carcinomas with intense lymphoid
infiltration. Lymphoepithelioma-like carcinoma. Am J Pathol.
139:469–474. 1991.PubMed/NCBI
|
|
5
|
Shibata D and Weiss LM: Epstein-Barr
virus-associated gastric adenocarcinoma. Am J Pathol. 140:769–774.
1992.PubMed/NCBI
|
|
6
|
Fukayama M, Hayashi Y, Iwasaki Y, et al:
Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr
virus infection of the stomach. Lab Invest. 71:73–81.
1994.PubMed/NCBI
|
|
7
|
Marshall BJ and Warren JR: Unidentified
curved bacilli in the stomach of patients with gastritis and peptic
ulceration. Lancet. 1:1311–1315. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nomura A, Stemmermann GN, Chyou PH, Kato
I, Perez-Perez GI and Blaser MJ: Helicobacter pylori infection and
gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med.
325:1132–1136. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parsonnet J, Friedman GD, Vandersteen DP,
et al: Helicobacter pylori infection and the risk of gastric
carcinoma. N Engl J Med. 325:1127–1131. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fukayama M and Ushiku T: Epstein-Barr
virus-associated gastric carcinoma. Pathol Res Pract. 207:529–537.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen JN, He D, Tang F and Shao CK:
Epstein-Barr virus-associated gastric carcinoma: a newly defined
entity. J Clin Gastroenterol. 46:262–271. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murphy G, Pfeiffer R, Camargo MC and
Rabkin CS: Meta-analysis shows that prevalence of Epstein-Barr
virus-positive gastric cancer differs based on sex and anatomic
location. Gastroenterology. 137:824–833. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Camargo MC, Kim WH, Chiaravalli AM, et al:
Improved survival of gastric cancer with tumour Epstein-Barr virus
positivity: an international pooled analysis. Gut. 63:236–243.
2014.
|
|
14
|
Boyle P and Levin B: Stomach Cancer. IARC
Press; Lyon: 2008
|
|
15
|
Camargo MC, Murphy G, Koriyama C, et al:
Determinants of Epstein-Barr virus-positive gastric cancer: an
international pooled analysis. Br J Cancer. 105:38–43. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JH, Kim SH, Han SH, An JS, Lee ES and
Kim YS: Clinicopathological and molecular characteristics of
Epstein-Barr virus-associated gastric carcinoma: a meta-analysis. J
Gastroenterol Hepatol. 24:354–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Truong CD, Feng W, Li W, et al:
Characteristics of Epstein-Barr virus-associated gastric cancer: a
study of 235 cases at a comprehensive cancer center in USA. J Exp
Clin Cancer Res. 28:142009. View Article : Google Scholar
|
|
18
|
Chen JN, Jiang Y, Li HG, et al:
Epstein-Barr virus genome polymorphisms of Epstein-Barr
virus-associated gastric carcinoma in gastric remnant carcinoma in
Guangzhou, southern China, an endemic area of nasopharyngeal
carcinoma. Virus Res. 160:191–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koriyama C, Akiba S, Minakami Y and Eizuru
Y: Environmental factors related to Epstein-Barr virus-associated
gastric cancer in Japan. J Exp Clin Cancer Res. 24:547–553.
2005.
|
|
20
|
Camargo MC, Koriyama C, Matsuo K, et al:
Case-case comparison of smoking and alcohol risk associations with
Epstein-Barr virus-positive gastric cancer. Int J Cancer.
134:948–953. 2014. View Article : Google Scholar :
|
|
21
|
van Beek J, zur Hausen A, Klein Kranenbarg
E, et al: EBV-positive gastric adenocarcinomas: a distinct
clinicopathologic entity with a low frequency of lymph node
involvement. J Clin Oncol. 22:664–670. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tokunaga M and Land CE: Epstein-Barr virus
involvement in gastric cancer: biomarker for lymph node metastasis.
Cancer Epidemiol Biomarkers Prev. 7:449–450. 1998.PubMed/NCBI
|
|
23
|
Matsunou H, Konishi F, Hori H, et al:
Characteristics of Epstein-Barr virus-associated gastric carcinoma
with lymphoid stroma in Japan. Cancer. 77:1998–2004. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song HJ, Srivastava A, Lee J, et al: Host
inflammatory response predicts survival of patients with
Epstein-Barr virus-associated gastric carcinoma. Gastroenterology.
139:84–92.e82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koriyama C, Akiba S, Itoh T, et al:
Prognostic significance of Epstein-Barr virus involvement in
gastric carcinoma in Japan. Int J Mol Med. 10:635–639.
2002.PubMed/NCBI
|
|
26
|
Kijima Y, Ishigami S, Hokita S, et al: The
comparison of the prognosis between Epstein-Barr virus
(EBV)-positive gastric carcinomas and EBV-negative ones. Cancer
Lett. 200:33–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tokunaga M, Land CE, Uemura Y, Tokudome T,
Tanaka S and Sato E: Epstein-Barr virus in gastric carcinoma. Am J
Pathol. 143:1250–1254. 1993.PubMed/NCBI
|
|
28
|
Chang MS, Lee HS, Kim HS, et al:
Epstein-Barr virus and microsatellite instability in gastric
carcinogenesis. J Pathol. 199:447–452. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaizaki Y, Hosokawa O, Sakurai S and
Fukayama M: Epstein-Barr virus-associated gastric carcinoma in the
remnant stomach: de novo and metachronous gastric remnant
carcinoma. J Gastroenterol. 40:570–577. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JY, Kim KM, Min BH, Lee JH, Rhee PL
and Kim JJ: Epstein-Barr virus-associated lymphoepithelioma-like
early gastric carcinomas and endoscopic submucosal dissection: case
series. World J Gastroenterol. 20:1365–1370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakamura S, Ueki T, Yao T, Ueyama T and
Tsuneyoshi M: Epstein-Barr virus in gastric carcinoma with lymphoid
stroma. Special reference to its detection by the polymerase chain
reaction and in situ hybridization in 99 tumors, including a
morphologic analysis. Cancer. 73:2239–2249. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Watanabe H, Enjoji M and Imai T: Gastric
carcinoma with lymphoid stroma. Its morphologic characteristics and
prognostic correlations. Cancer. 38:232–243. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lauren P: The two histological main types
of gastric carcinoma: diffuse and so-called intestinal-type
carcinoma. An attempt at a histoclinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965.
|
|
34
|
Song HJ and Kim KM: Pathology of
epstein-barr virus-associated gastric carcinoma and its
relationship to prognosis. Gut Liver. 5:143–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shinozaki A, Ushiku T, Morikawa T, et al:
Epstein-Barr virus-associated gastric carcinoma: a distinct
carcinoma of gastric phenotype by claudin expression profiling. J
Histochem Cytochem. 57:775–785. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Beek J, zur Hausen A, Snel SN, et al:
Morphological evidence of an activated cytotoxic T-cell infiltrate
in EBV-positive gastric carcinoma preventing lymph node metastases.
Am J Surg Pathol. 30:59–65. 2006. View Article : Google Scholar
|
|
37
|
Kuzushima K, Nakamura S, Nakamura T, et
al: Increased frequency of antigen-specific CD8+
cytotoxic T lymphocytes infiltrating an Epstein-Barr
virus-associated gastric carcinoma. J Clin Invest. 104:163–171.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shinozaki A, Ushiku T and Fukayama M:
Prominent Mott cell proliferation in Epstein-Barr virus-associated
gastric carcinoma. Hum Pathol. 41:134–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ushiku T, Shinozaki A, Uozaki H, et al:
Gastric carcinoma with osteoclast-like giant cells.
Lymphoepithelioma-like carcinoma with Epstein-Barr virus infection
is the predominant type. Pathol Int. 60:551–558. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choi MG, Jeong JY, Kim KM, et al: Clinical
significance of gastritis cystica profunda and its association with
Epstein-Barr virus in gastric cancer. Cancer. 118:5227–5233. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsao SW, Tsang CM, Pang PS, Zhang G, Chen
H and Lo KW: The biology of EBV infection in human epithelial
cells. Semin Cancer Biol. 22:137–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rickinson AB: Co-infections, inflammation
and oncogenesis: future directions for EBV research. Semin Cancer
Biol. 26:99–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Imai S, Nishikawa J and Takada K:
Cell-to-cell contact as an efficient mode of Epstein-Barr virus
infection of diverse human epithelial cells. J Virol. 72:4371–4378.
1998.PubMed/NCBI
|
|
44
|
Hayashi K, Teramoto N, Akagi T, Sasaki Y
and Suzuki T: In situ detection of Epstein-Barr virus in the
gastric glands with intestinal metaplasia. Am J Gastroenterol.
91:14811996.PubMed/NCBI
|
|
45
|
Sugiura M, Imai S, Tokunaga M, et al:
Transcriptional analysis of Epstein-Barr virus gene expression in
EBV-positive gastric carcinoma: unique viral latency in the tumour
cells. Br J Cancer. 74:625–631. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Luo B, Wang Y, Wang XF, et al: Expression
of Epstein-Barr virus genes in EBV-associated gastric carcinomas.
World J Gastroenterol. 11:629–633. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Strong MJ, Xu G, Coco J, et al:
Differences in gastric carcinoma microenvironment stratify
according to EBV infection intensity: implications for possible
immune adjuvant therapy. PLoS Pathog. 9:e10033412013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang W, Morgan DR, Meyers MO, et al:
Epstein-barr virus infected gastric adenocarcinoma expresses latent
and lytic viral transcripts and has a distinct human gene
expression profile. Infect Agent Cancer. 7:212012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shannon-Lowe C, Adland E, Bell AI,
Delecluse HJ, Rickinson AB and Rowe M: Features distinguishing
Epstein-Barr virus infections of epithelial cells and B cells:
viral genome expression, genome maintenance, and genome
amplification. J Virol. 83:7749–7760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iwakiri D and Takada K: Role of EBERs in
the pathogenesis of EBV infection. Adv Cancer Res. 107:119–136.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Iwakiri D, Eizuru Y, Tokunaga M and Takada
K: Autocrine growth of Epstein-Barr virus-positive gastric
carcinoma cells mediated by an Epstein-Barr virus-encoded small
RNA. Cancer Res. 63:7062–7067. 2003.PubMed/NCBI
|
|
52
|
Shinozaki A, Sakatani T, Ushiku T, et al:
Downregulation of microRNA-200 in EBV-associated gastric carcinoma.
Cancer Res. 70:4719–4727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Banerjee AS, Pal AD and Banerjee S:
Epstein-Barr virus-encoded small non-coding RNAs induce cancer cell
chemoresistance and migration. Virology. 443:294–305. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gruhne B, Sompallae R, Marescotti D,
Kamranvar SA, Gastaldello S and Masucci MG: The Epstein-Barr virus
nuclear antigen-1 promotes genomic instability via induction of
reactive oxygen species. Proc Natl Acad Sci USA. 106:2313–2318.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu J, Murakami M, Verma SC, et al:
Epstein-Barr Virus nuclear antigen 1 (EBNA1) confers resistance to
apoptosis in EBV-positive B-lymphoma cells through up-regulation of
survivin. Virology. 410:64–75. 2011. View Article : Google Scholar
|
|
56
|
Saridakis V, Sheng Y, Sarkari F, et al:
Structure of the p53 binding domain of HAUSP/USP7 bound to
Epstein-Barr nuclear antigen 1 implications for EBV-mediated
immortalization. Mol Cell. 18:25–36. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheng TC, Hsieh SS, Hsu WL, Chen YF, Ho HH
and Sheu LF: Expression of Epstein-Barr nuclear antigen 1 in
gastric carcinoma cells is associated with enhanced tumorigenicity
and reduced cisplatin sensitivity. Int J Oncol. 36:151–160.
2010.
|
|
58
|
Sivachandran N, Dawson CW, Young LS, Liu
FF, Middeldorp J and Frappier L: Contributions of the Epstein-Barr
virus EBNA1 protein to gastric carcinoma. J Virol. 86:60–68. 2012.
View Article : Google Scholar :
|
|
59
|
Yin Q and Flemington EK: siRNAs against
the Epstein Barr virus latency replication factor, EBNA1, inhibit
its function and growth of EBV-dependent tumor cells. Virology.
346:385–393. 2006. View Article : Google Scholar
|
|
60
|
Hong M, Murai Y, Kutsuna T, et al:
Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA
interference inhibits proliferation of EBV-positive Burkitt’s
lymphoma cells. J Cancer Res Clin Oncol. 132:1–8. 2006. View Article : Google Scholar
|
|
61
|
Ian MX, Lan SZ, Cheng ZF, Dan H and Qiong
LH: Suppression of EBNA1 expression inhibits growth of EBV-positive
NK/T cell lymphoma cells. Cancer Biol Ther. 7:1602–1606. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fukuda M, Ikuta K, Yanagihara K, et al:
Effect of transforming growth factor-beta1 on the cell growth and
Epstein-Barr virus reactivation in EBV-infected epithelial cell
lines. Virology. 288:109–118. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hino R, Uozaki H, Inoue Y, et al: Survival
advantage of EBV-associated gastric carcinoma: survivin
up-regulation by viral latent membrane protein 2A. Cancer Res.
68:1427–1435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu X, Gao Y, Luo B and Zhao Y:
Construction and antiapoptosis activities of recombinant adenoviral
Expression vector carrying EBV latent membrane protein 2A.
Gastroenterol Res Pract. 2011:1828322011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pal AD, Basak NP, Banerjee AS and Banerjee
S: Epstein-Barr virus latent membrane protein-2A alters
mitochondrial dynamics promoting cellular migration mediated by
Notch signaling pathway. Carcinogenesis. 35:1592–1601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hino R, Uozaki H, Murakami N, et al:
Activation of DNA methyltransferase 1 by EBV latent membrane
protein 2A leads to promoter hypermethylation of PTEN gene in
gastric carcinoma. Cancer Res. 69:2766–2774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhao J, Liang Q, Cheung KF, et al:
Genome-wide identification of Epstein-Barr virus-driven promoter
methylation profiles of human genes in gastric cancer cells.
Cancer. 119:304–312. 2013. View Article : Google Scholar
|
|
68
|
Al-Mozaini M, Bodelon G, Karstegl CE, Jin
B, Al-Ahdal M and Farrell PJ: Epstein-Barr virus BART gene
expression. J Gen Virol. 90:307–316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Thornburg NJ, Kusano S and Raab-Traub N:
Identification of Epstein-Barr virus RK-BARF0-interacting proteins
and characterization of expression pattern. J Virol.
78:12848–12856. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hoebe EK, Le Large TY, Greijer AE and
Middeldorp JM: BamHI-A rightward frame 1, an Epstein-Barr
virus-encoded oncogene and immune modulator. Rev Med Virol.
23:367–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
zur Hausen A, Brink AA, Craanen ME,
Middeldorp JM, Meijer CJ and van den Brule AJ: Unique transcription
pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric
adenocarcinomas: expression of the transforming BARF1 gene. Cancer
Res. 60:2745–2748. 2000.PubMed/NCBI
|
|
72
|
Wang Q, Tsao SW, Ooka T, et al:
Anti-apoptotic role of BARF1 in gastric cancer cells. Cancer Lett.
238:90–103. 2006. View Article : Google Scholar
|
|
73
|
Wiech T, Nikolopoulos E, Lassman S, et al:
Cyclin D1 expression is induced by viral BARF1 and is overexpressed
in EBV-associated gastric cancer. Virchows Arch. 452:621–627. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chang MS, Kim DH, Roh JK, et al:
Epstein-Barr virus-encoded BARF1 promotes proliferation of gastric
carcinoma cells through regulation of NF-kappaB. J Virol.
87:10515–10523. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kaneda A, Matsusaka K, Aburatani H and
Fukayama M: Epstein-Barr virus infection as an epigenetic driver of
tumorigenesis. Cancer Res. 72:3445–3450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Matsusaka K, Funata S, Fukayama M and
Kaneda A: DNA methylation in gastric cancer, related to
Helicobacter pylori and Epstein-Barr virus. World J Gastroenterol.
20:3916–3926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yau TO, Tang CM and Yu J: Epigenetic
dysregulation in Epstein-Barr virus-associated gastric carcinoma:
disease and treatments. World J Gastroenterol. 20:6448–6456. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kusano M, Toyota M, Suzuki H, et al:
Genetic, epigenetic, and clinicopathologic features of gastric
carcinomas with the CpG island methylator phenotype and an
association with Epstein-Barr virus. Cancer. 106:1467–1479. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zong L and Seto Y: CpG island methylator
phenotype, helicobacter pylori, Epstein-Barr virus, and
microsatellite instability and prognosis in gastric cancer: a
systematic review and meta-analysis. PLoS One. 9:e860972014.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Okada T, Nakamura M, Nishikawa J, et al:
Identification of genes specifically methylated in Epstein-Barr
virus-associated gastric carcinomas. Cancer Sci. 104:1309–1314.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Saito M, Nishikawa J, Okada T, et al: Role
of DNA methylation in the development of Epstein-Barr
virus-associated gastric carcinoma. J Med Virol. 85:121–127. 2013.
View Article : Google Scholar
|
|
82
|
Chapel F, Fabiani B, Davi F, et al:
Epstein-Barr virus and gastric carcinoma in Western patients:
comparison of pathological parameters and p53 expression in
EBV-positive and negative tumours. Histopathology. 36:252–261.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Matsusaka K, Kaneda A, Nagae G, et al:
Classification of Epstein-Barr virus-positive gastric cancers by
definition of DNA methylation epigenotypes. Cancer Res.
71:7187–7197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
zur Hausen A, van Grieken NC, Meijer GA,
et al: Distinct chromosomal aberrations in Epstein-Barr
virus-carrying gastric carcinomas tested by comparative genomic
hybridization. Gastroenterology. 121:612–618. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chan WY, Liu Y, Li CY, et al: Recurrent
genomic aberrations in gastric carcinomas associated with
Helicobacter pylori and Epstein-Barr virus. Diagn Mol Pathol.
11:127–134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chong JM, Fukayama M, Hayashi Y, et al:
Microsatellite instability in the progression of gastric carcinoma.
Cancer Res. 54:4595–4597. 1994.PubMed/NCBI
|
|
87
|
Lee J, van Hummelen P, Go C, et al:
High-throughput mutation profiling identifies frequent somatic
mutations in advanced gastric adenocarcinoma. PLoS One.
7:e388922012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sukawa Y, Yamamoto H, Nosho K, et al:
Alterations in the human epidermal growth factor receptor
2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer.
World J Gastroenterol. 18:6577–6586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
The Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liang Q, Yao X, Tang S, et al: Integrative
identification of epstein-barr virus-associated mutations and
epigenetic alterations in gastric cancer. Gastroenterology.
147:1350–1362.e1354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang K, Kan J, Yuen ST, et al: Exome
sequencing identifies frequent mutation of ARID1A in molecular
subtypes of gastric cancer. Nat Genet. 43:1219–1223. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Abe H, Maeda D, Hino R, et al: ARID1A
expression loss in gastric cancer: pathway-dependent roles with and
without Epstein-Barr virus infection and microsatellite
instability. Virchows Arch. 461:367–377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Moritani S, Sugihara H, Kushima R and
Hattori T: Different roles of p53 between Epstein-Barr
virus-positive and -negative gastric carcinomas of matched
histology. Virchows Arch. 440:367–375. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Park HY, Kang SY, Kang GH, et al: EBV
infection and mismatch repair deficiency mediated by loss of hMLH1
expression contribute independently to the development of multiple
synchronous gastric carcinomas. J Surg Oncol. 106:777–782. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Dolan DE and Gupta S: PD-1 pathway
inhibitors: changing the landscape of cancer immunotherapy. Cancer
Control. 21:231–237. 2014.PubMed/NCBI
|
|
96
|
Naidoo J, Page DB and Wolchok JD: Immune
modulation for cancer therapy. Br J Cancer. 11:2214–2219. 2014.
View Article : Google Scholar
|
|
97
|
Marquitz AR, Mathur A, Shair KH and
Raab-Traub N: Infection of Epstein-Barr virus in a gastric
carcinoma cell line induces anchorage independence and global
changes in gene expression. Proc Natl Acad Sci USA. 109:9593–9598.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kim SY, Park C, Kim HJ, et al:
Deregulation of immune response genes in patients with Epstein-Barr
virus-associated gastric cancer and outcomes. Gastroenterology.
148:137–147.e139. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Chen BJ, Chapuy B, Ouyang J, et al: PD-L1
expression is characteristic of a subset of aggressive B-cell
lymphomas and virus-associated malignancies. Clin Cancer Res.
19:3462–3473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Green MR, Rodig S, Juszczynski P, et al:
Constitutive AP-1 activity and EBV infection induce PD-L1 in
Hodgkin lymphomas and posttransplant lymphoproliferative disorders:
implications for targeted therapy. Clin Cancer Res. 18:1611–1618.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kim do N and Lee SK: Biogenesis of
Epstein-Barr virus microRNAs. Mol Cell Biochem. 365:203–210. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Pfeffer S, Zavolan M, Grasser FA, et al:
Identification of virus-encoded microRNAs. Science. 304:734–736.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Barth S, Meister G and Grasser FA:
EBV-encoded miRNAs. Biochim Biophys Acta. 1809:631–640. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chan JY, Gao W, Ho WK, Wei WI and Wong TS:
Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in
undifferentiated nasopharyngeal carcinoma. Anticancer Res.
32:3201–3210. 2012.PubMed/NCBI
|
|
105
|
Chen SJ, Chen GH, Chen YH, et al:
Characterization of Epstein-Barr virus miRNAome in nasopharyngeal
carcinoma by deep sequencing. PLoS One. 5:e127452010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Choi H, Lee H, Kim SR, Gho YS and Lee SK:
Epstein-Barr virus-encoded microRNA BART15-3p promotes cell
apoptosis partially by targeting BRUCE. J Virol. 87:8135–8144.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Choy EY, Siu KL, Kok KH, et al: An
Epstein-Barr virus-encoded microRNA targets PUMA to promote host
cell survival. J Exp Med. 205:2551–2560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Cosmopoulos K, Pegtel M, Hawkins J, et al:
Comprehensive profiling of Epstein-Barr virus microRNAs in
nasopharyngeal carcinoma. J Virol. 83:2357–2367. 2009. View Article : Google Scholar :
|
|
109
|
Gourzones C, Gelin A, Bombik I, et al:
Extra-cellular release and blood diffusion of BART viral micro-RNAs
produced by EBV-infected nasopharyngeal carcinoma cells. Virol J.
7:2712010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Gourzones C, Ferrand FR, Amiel C, et al:
Consistent high concentration of the viral microRNA BART17 in
plasma samples from nasopharyngeal carcinoma patients - evidence of
non-exosomal transport. Virol J. 10:1192013. View Article : Google Scholar :
|
|
111
|
Imig J, Motsch N, Zhu JY, et al: microRNA
profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic
Acids Res. 39:1880–1893. 2011. View Article : Google Scholar :
|
|
112
|
Lung RW, Tong JH and To KF: Emerging roles
of small Epstein-Barr virus derived non-coding RNAs in epithelial
malignancy. Int J Mol Sci. 14:17378–17409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Marquitz AR, Mathur A, Nam CS and
Raab-Traub N: The Epstein-Barr Virus BART microRNAs target the
pro-apoptotic protein Bim. Virology. 412:392–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Motsch N, Alles J, Imig J, et al: MicroRNA
profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by
deep sequencing. PLoS One. 7:e421932012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nourse JP, Crooks P, Keane C, et al:
Expression profiling of Epstein-Barr virus-encoded microRNAs from
paraffin-embedded formalin-fixed primary Epstein-Barr
virus-positive B-cell lymphoma samples. J Virol Methods. 184:46–54.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Riley KJ, Rabinowitz GS, Yario TA, Luna
JM, Darnell RB and Steitz JA: EBV and human microRNAs co-target
oncogenic and apoptotic viral and human genes during latency. EMBO
J. 31:2207–2221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Vereide DT, Seto E, Chiu YF, et al:
Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene.
33:1258–1264. 2014. View Article : Google Scholar
|
|
118
|
Wong AM, Kong KL, Tsang JW, Kwong DL and
Guan XY: Profiling of Epstein-Barr virus-encoded microRNAs in
nasopharyngeal carcinoma reveals potential biomarkers and oncomirs.
Cancer. 118:698–710. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Xia T, O’Hara A, Araujo I, et al: EBV
microRNAs in primary lymphomas and targeting of CXCL-11 by
ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhu JY, Pfuhl T, Motsch N, et al:
Identification of novel Epstein-Barr virus microRNA genes from
nasopharyngeal carcinomas. J Virol. 83:3333–3341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Qiu J and Thorley-Lawson DA: EBV microRNA
BART 18-5p targets MAP3K2 to facilitate persistence in vivo by
inhibiting viral replication in B cells. Proc Natl Acad Sci USA.
111:11157–11162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Kim do N, Chae HS, Oh ST, et al:
Expression of viral microRNAs in Epstein-Barr virus-associated
gastric carcinoma. J Virol. 81:1033–1036. 2007. View Article : Google Scholar :
|
|
123
|
Marquitz AR, Mathur A, Chugh PE, Dittmer
DP and Raab-Traub N: Expression profile of microRNAs in
Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol.
88:1389–1393. 2014. View Article : Google Scholar :
|
|
124
|
Kim do N, Seo MK, Choi H, et al:
Characterization of naturally Epstein-Barr virus-infected gastric
carcinoma cell line YCCEL1. J Gen Virol. 94:497–506. 2013.
View Article : Google Scholar
|
|
125
|
Qiu J, Cosmopoulos K, Pegtel M, et al: A
novel persistence associated EBV miRNA expression profile is
disrupted in neoplasia. PLoS Pathog. 7:e10021932011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Skalsky RL, Corcoran DL, Gottwein E, et
al: The viral and cellular microRNA targetome in lymphoblastoid
cell lines. PLoS Pathog. 8:e10024842012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lo AK, To KF, Lo KW, et al: Modulation of
LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad
Sci USA. 104:16164–16169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Barth S, Pfuhl T, Mamiani A, et al:
Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the
viral DNA polymerase BALF5. Nucleic Acids Res. 36:666–675. 2008.
View Article : Google Scholar :
|
|
129
|
Dolken L, Malterer G, Erhard F, et al:
Systematic analysis of viral and cellular microRNA targets in cells
latently infected with human gamma-herpesviruses by RISC
immunoprecipitation assay. Cell Host Microbe. 7:324–334. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Iizasa H, Wulff BE, Alla NR, et al:
Editing of Epstein-Barr virus-encoded BART6 microRNAs controls
their dicer targeting and consequently affects viral latency. J
Biol Chem. 285:33358–33370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Ambrosio MR, Navari M, Di Lisio L, et al:
The Epstein Barr-encoded BART-6-3p microRNA affects regulation of
cell growth and immuno response in Burkitt lymphoma. Infect Agent
Cancer. 9:122014. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Ramakrishnan R, Donahue H, Garcia D, et
al: Epstein-Barr virus BART9 miRNA modulates LMP1 levels and
affects growth rate of nasal NK T cell lymphomas. PLoS One.
6:e272712011. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Hsu CY, Yi YH, Chang KP, Chang YS, Chen SJ
and Chen HC: The Epstein-Barr virus-encoded microRNA miR-BART9
promotes tumor metastasis by targeting E-cadherin in nasopharyngeal
carcinoma. PLoS Pathog. 10:e10039742014. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ross N, Gandhi MK and Nourse JP: The
Epstein-Barr virus microRNA BART11-5p targets the early B-cell
transcription factor EBF1. Am J Blood Res. 3:210–224.
2013.PubMed/NCBI
|
|
135
|
Haneklaus M, Gerlic M, Kurowska-Stolarska
M, et al: Cutting edge: miR-223 and EBV miR-BART15 regulate the
NLRP3 inflammasome and IL-1beta production. J Immunol.
189:3795–3799. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Jung YJ, Choi H, Kim H and Lee SK:
MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by
directly targeting BZLF1 and BRLF1. J Virol. 88:9027–9037. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Lin TC, Liu TY, Hsu SM and Lin CW:
Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation
with secondary suppression of p53 in invasive nasal NK/T-cell
lymphoma. Am J Pathol. 182:1865–1875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Lung RW, Tong JH, Sung YM, et al:
Modulation of LMP2A expression by a newly identified Epstein-Barr
virus-encoded microRNA miR-BART22. Neoplasia. 11:1174–1184.
2009.PubMed/NCBI
|