Introduction
Over the last decade, the discovery of epidermal
growth factor receptor (EGFR) gene mutations and the
development of tyrosine kinase inhibitors (TKIs) have dramatically
changed the treatment strategies for patients with advanced
non-small cell lung cancer (NSCLC) (1–5).
Therefore, EGFR mutation testing is essential for optimal
treatment selection for advanced NSCLC patients. Several methods
for detecting EGFR mutations mainly in formalin-fixed,
paraffin-embedded (FFPE) samples have already been validated and
applied in practice (6–11). However, these methods adopt
relatively complex polymerase chain reaction (PCR) technologies
with pre-designed fluorogenic probes, are packaged by
manufacturers, and are often available through outside reference
laboratories at relatively high rates. In Japan, the use of
EGFR-TKIs for chemo-naïve patients has been limited to those with
EGFR mutations since 2011. Despite this regulation, the
majority of community and university hospitals still depend on
outside laboratories for EGFR mutation testing. Accordingly,
there is a time delay between histological diagnosis and molecular
diagnosis in clinical situations. In general, obtaining PCR-based
EGFR test results from outside laboratories requires 7–14
days after tumor sampling. In cases where immediate treatment is
critical, failure to provide appropriate molecular targeted therapy
due to delayed molecular diagnostic test results may cause fatal
outcomes. Therefore, a quicker, simpler, and less expensive
point-of-care EGFR mutation testing system is needed.
In the field of infectious diseases, a more rapid
real-time PCR system for detecting pathogens has been developed
(12). Similarly, we have
developed a new, simple, high-speed real-time PCR system (referred
to as ultrarapid PCR) for the detection of the 2 most common
EGFR mutations. This assay involves a pair of
mutation-specific primers used in combination with a newly
developed PCR machine that is equipped with a novel thermo-control
mechanism that makes ultrarapid PCR cycling possible.
In-frame deletion in exon 19 (E746-A750del) and the
point mutation replacing leucine with arginine at codon 858 of exon
21 (L858R) represent >90% of oncogenic EGFR mutations.
Large clinical trials have been conducted to establish the efficacy
of EGFR-TKIs in targeting the resulting mutated EGFR proteins
(1–5). Therefore, we designed a
deletion-specific primer targeting the exon 19 E746-A750del
mutation and a point mutation-specific primer for the exon 21 L858R
mutation. PCR conditions were optimized for amplifying templates
harboring each mutation.
Endobronchial ultrasonography using a guide sheath
(EBUS-GS) combined with a virtual bronchoscopic navigation system
(VBN) is very useful approach for collecting samples from
peripheral pulmonary lesions (13–20).
However, a major disadvantage of EBUS-GS is the low sample volume
that can be obtained, leading to reduced sensitivity in molecular
testing. Therefore, we performed this validation study to determine
whether ultrarapid PCR can detect EGFR mutations with liquid
bronchial lavage fluid (BLF) samples after EBUS-GS-transbronchial
biopsies (EBUS-GS-TBBs) were taken.
Materials and methods
Patients and samples
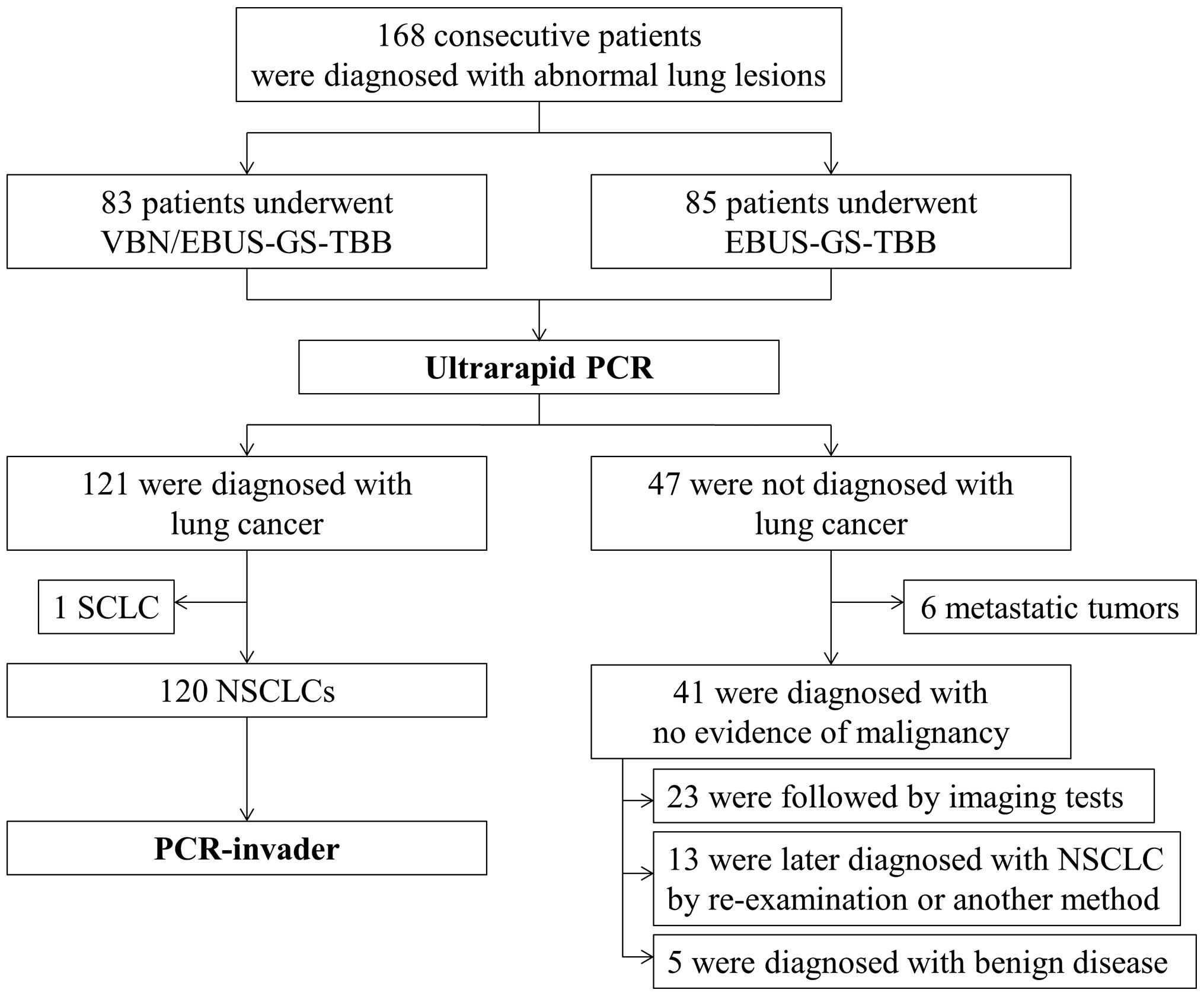
A total of consecutive 168 patients who underwent
EBUS-GS-TBB at the Tottori University Hospital (Yonago, Japan) from
November 2012 to December 2013 were enrolled prospectively
(Fig. 1). Eligible patients had
undiagnosed pulmonary lesions suspected to be lung cancer on chest
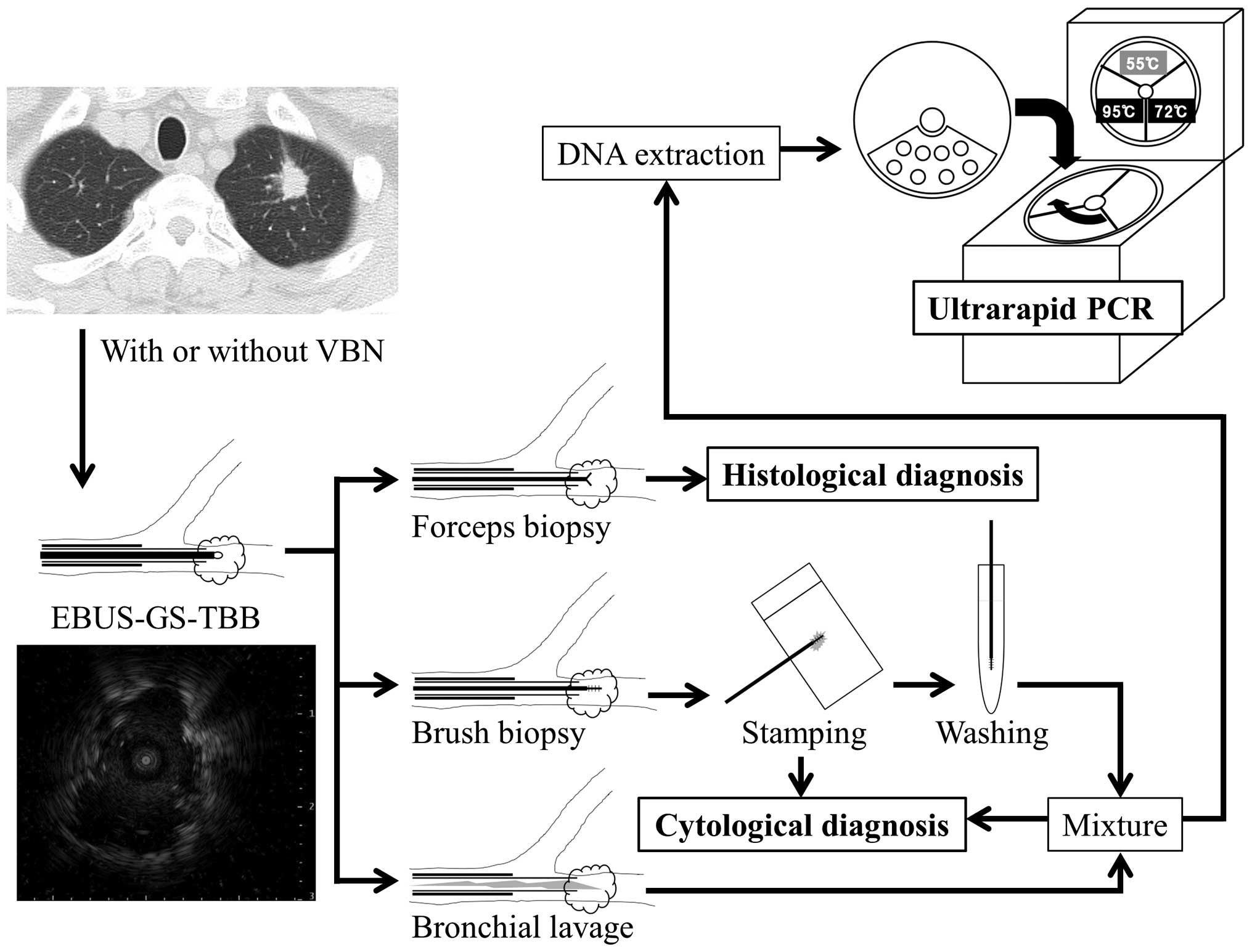
computed tomography (CT) findings. Samples were prepared by mixing
BLFs obtained during EBUS-GS-TBB procedures with saline solutions
mixed with EBUS-GS-brush biopsy samples after they were stamped on
glass slides. DNA was extracted from patient fluid samples using
the QIAamp Blood Mini kit (Qiagen, Tokyo, Japan) (Fig. 2).
Ethical approval was obtained from the Tottori
University Hospital and informed consent was obtained from all
patients involved prior to performing bronchoscopies.
VBN and EBUS-GS-TBB procedures
VBNs were performed following approval from
physicians and expert bronchoscopists, based on CT findings. CT
scan data from multi-detector chest CTs (64- or 128-row; slice
width, 0.5 mm) were acquired from all patients before EBUS-GS-TBB.
Individual CT data sets from VBN/EBUS-GS group were transferred to
a workstation on which VBN software (Bf-NAVI; Cybernet Systems,
Co., Ltd., Tokyo, Japan) automatically created VBN images within 15
min. VBN images could be moved multi-directionally on a monitor
beside the video-bronchoscopic monitor. All patients were
anaesthetized with midazolam and examined using a P260F video
bronchoscope (4.0 mm outer diameter; Olympus Corp., Tokyo, Japan).
The bronchoscope was introduced into the targeted bronchus with VBN
support or the guidance of 2 expert bronchoscopists based on CT
axial images. Peripheral target lesions were visualized using a 20
MHz radial-type EBUS probe (external diameter, 1.4 mm; UM-S20-17S;
Olympus) with a GS (K-201; Olympus) through a working channel.
Ultrasound images were processed in an ultrasound scanner (EU-ME-1
or EU-ME2; Olympus). Pathological samples were collected using
forceps and brushes through the GS. Biopsy samples were immediately
fixed in formalin. After biopsies were obtained, the target area
was washed with 20 ml of saline.
Mutation-specific PCR using an ultrarapid
PCR machine
EGFR exon 19 E746-A750 deletion type 1
(2235-2249del; 5′-GGAATTAAGAGAAGC-3′) and exon 21 L858R
(2573T>G) were detected using a novel high-speed real-time PCR
machine, namely a Hyper-PCR UR104MK IV (Trust Medical Co., Ltd.,
Kasai, Japan), with allele-specific primers and SpeedSTAR HS DNA
Polymerase (Takara Bio, Inc., Shiga, Japan). The UR104MK IV PCR
machine utilized a novel temperature control technology. In this
system, the PCR mixture is enclosed in a small vessel on a thin,
flexible plastic disk and sealed with adhesive film, and the disk
is rotated rapidly onto 3 separated heat elements. Rapid PCR can be
accomplished by controlling the speed of rotation and the
temperature of the 3 heat elements. The UR104MK also has the
capacity for real-time monitoring of PCR reactions with a
fluorescent probe and post-PCR melt curve analysis. The typical
time for amplification and detection when using this machine was
<10 min.
Optimized reaction mixtures contained 1.6 μl of 10X
Fast buffer I (Takara), 1.3 μl of 2.5 mM dNTPs, 0.4 μl of each
allele-specific primer (10 μM), 0.2 μl of SpeedSTAR HS DNA
Polymerase (5 U/μl; Takara), 1 μl of template DNA, 1.6 μl of
1:2,000 SYBR-Green, and 9.5 μl of ddH2O in a volume of
16 μl. Furthermore, dimethylsulfoxide was added to obtain a final
concentration of 5%. PCR thermal cycling conditions were as
follows. To amplify E746-A750del type 1, we used 1 cycle of 94°C
for 1 min, followed by 35 cycles of 98°C for 1.3 sec, 55°C for 5
sec, and 72°C for 3 sec. To amplify DNA sequences harboring the
L858R point mutation, we used 1 cycle of 94°C for 1 min, followed
by 30 cycles of 98°C for 1.3 sec, 68°C for 8 sec and 68°C for 8
sec.
Sensitivity assay
To validate the sensitivity of the PCR system,
sensitivity assays were performed using DNA mixtures extracted from
the following cell lines: PC9 (2235-2249del), H1975 (2573T>G)
and N417 (wild-type). The PC9 cell line was obtained from the RIKEN
Cell Bank (Tsukuba, Japan). The H1975 cell line was obtained from
the American Type Culture Collection (Rockville, MD, USA). The N417
cell line was provided by Dr A.F. Gazdar and Dr H. Oie (National
Cancer Institute-Navy Medical Oncology Branch, Bethesda, MD, USA).
These cell lines were mixed in different ratios. Specifically, the
PC9 and N417 cell lines were mixed in ratios of 1:0, 0.5:0.5,
0.1:0.9, 0.01:0.99 and 0:1, respectively, while the H1975 and N417
were mixed in ratios of 1:0, 0.5:0.5, 0.1:0.9, 0.01:0.99 and 0:1,
respectively. Analysis of EGFR mutations was performed as
described above.
Comparison of ultrarapid PCR with the
PCR-invader method
EGFR mutation analysis was performed with BLF
samples from all 168 patients, regardless of pathological
diagnosis, by ultrarapid PCR immediately after EBUS-GS-TBB. After
pathological diagnosis of NSCLC, the associated EGFR
mutation statuses in FFPE samples were evaluated by the PCR-invader
method (BML, Inc., Tokyo, Japan), which is used in clinical
practice at our hospital. To assess the performance of ultrarapid
PCR, we evaluated the concordance rates and calculated kappa
coefficients for both the ultrarapid PCR and PCR-invader
methods.
Statistical analysis
Average target lesion diameters and diagnostic
yields were calculated for the VBN/EBUS-GS and EBUS-GS groups,
respectively, and analyzed using the Mann-Whitney U test and the
Chi-squared test between these 2 groups. All P-values were 2-sided.
A P-value of <0.05 indicated statistical significance.
Concordance rates and Cohen’s kappa coefficients were determined
between the ultrarapid PCR and PCR-invader methods. Cohen’s kappa
coefficient was calculated as kappa = (Po-Pe)/(1-Pe), where Po is
the observed concordance rate and Pe is the expected probability of
chance agreement (21). A kappa of
zero means that there is no agreement beyond chance, and a kappa of
1.00 means that there is perfect agreement. Values ranging from
0.81 to 1.00 indicate near perfect agreements (22). All data were statistically analyzed
using IBM SPSS Statistics, ver. 22.
Results
Sensitivity
The E746-A750del mutation was detected in mixed cell
populations containing decreasing percentages (100-1%) of the
E746-A750del-positive cell line (PC9) and increasing percentages of
the N417 cell line containing 2 copies of the wild-type EGFR
gene. Similarly, the L858R mutation was detected in cell line
mixtures containing 100-1% of an L858R mutation-positive cell line
(H1975) and N417 cells (Fig.
3).
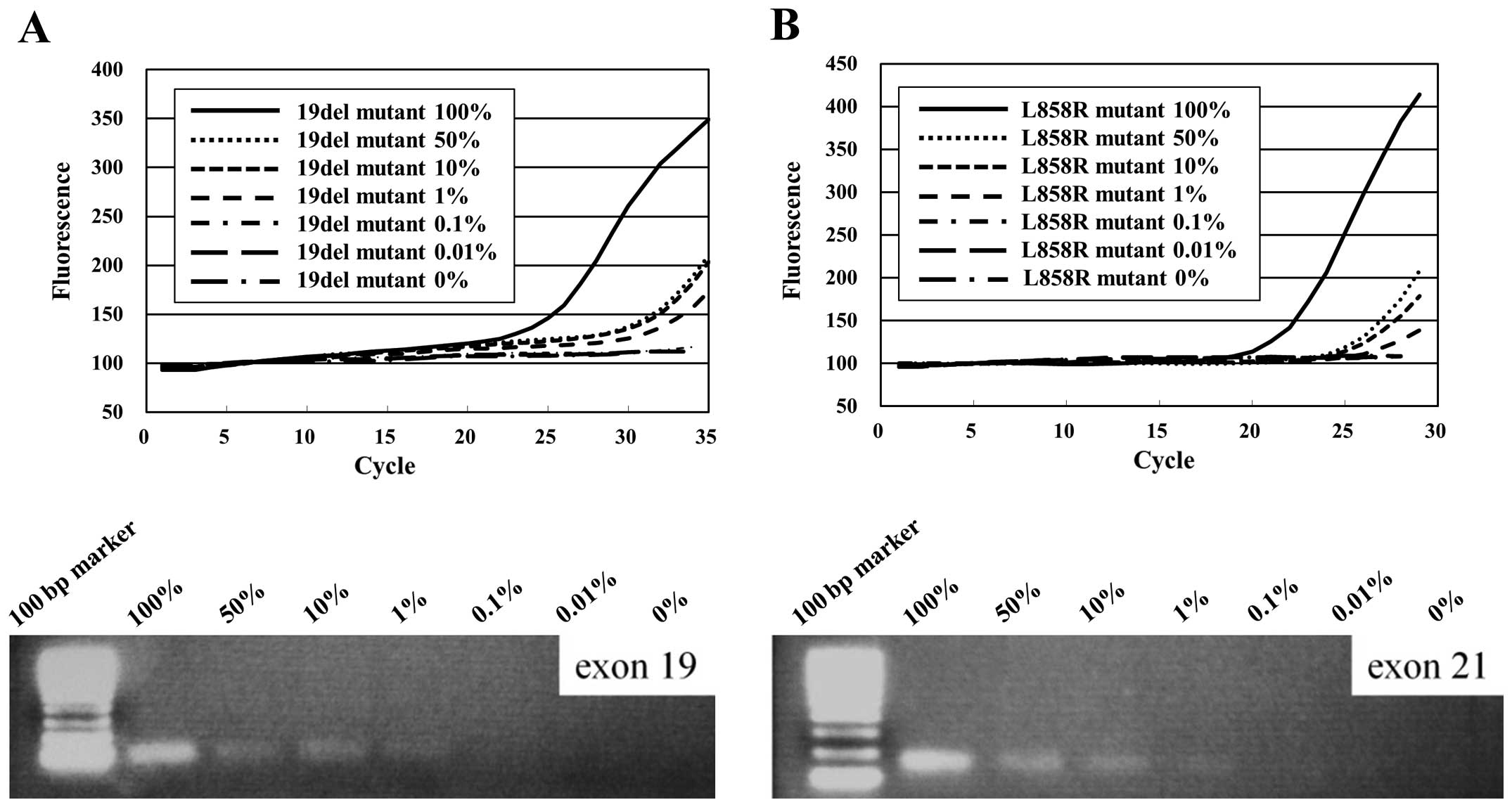 | Figure 3Sensitivity of ultrarapid PCR. (A)
Amplification of the 19del allele by ultrarapid PCR was performed
using cell samples containing 100, 50, 10, 1, 0.1, 0.01 and 0% PC14
cells, mixed with N417 cells containing 2 copies of the wild-type
EGFR gene. As few as 1% of tumor cells with the 19del
mutation could be detected. (B) Amplification of the L858R allele
by ultrarapid PCR using cell samples containing 100, 50, 10, 1,
0.1, 0.01 and 0% H1975 cells, mixed with N417 cells. As few as 1%
of tumor cells with L858R mutation could be detected. |
Characteristics of patients and patient
samples
VBN was combined with EBUS-GS for 83 out of the 168
patients enrolled in the present study. The median and average
diameters of the target lesions were 25 and 30.6 mm, respectively
(range, 8–150 mm). In the VBN/EBUS-GS group, the median and average
diameters of target lesions were 19 and 20.5 mm, respectively
(range, 8–54 mm). In the EBUS-GS group, the median and average
diameters of target lesions were 34.5 and 38.6 mm, respectively
(range, 8–150 mm; Table I). As
shown in Fig. 1, lung cancer was
diagnosed histologically in 121 patients, but not in 47 patients,
including 5 patients with benign diseases and 6 patients with
metastatic tumors. Thirteen out of the 41 patients who were not
diagnosed with NSCLC using EBUS-GS-TBB specimens were later
diagnosed with NSCLC by re-examination or using another sampling
method. Twenty-three patients were provided follow-up with imaging
examinations at fixed intervals, and did not show enlargement of
peripheral small lesions after EBUS-GS-TBB. After these 23 patients
were excluded from the analysis, the total diagnostic yield
obtained with EBUS-GS-TBB samples was 91.0% (132/145 cases). In the
EBUS-GS-TBB group, the diagnostic yield was 94.6% (70/74 cases),
while the diagnostic yield of the VBN/EBUS-GS-TBB was 87.3% (62/71
cases; Table I). Although target
lesion diameters were significantly different (P<0.001;
Mann-Whitney U test), diagnostic yields were similar in the 2
groups (P=0.18; Chi-squared test).
 | Table IComparison of target lesions
diameters and diagnostic yields between VBN/EBUS-GS-TBB and
EBUS-GS-TBB. |
Table I
Comparison of target lesions
diameters and diagnostic yields between VBN/EBUS-GS-TBB and
EBUS-GS-TBB.
| VBN/EBUS-GS-TBB
(N=83) | EBUS-GS-TBB
(N=85) | P-value |
|---|
| Diameter (mm) |
| Median | 19.0 | 34.5 | |
| Average | 20.5 | 38.6 | <0.001b |
| Range | 8–54 | 8–150 | |
| Diagnostic
yielda | 87.3% (62/71
cases) | 94.6% (70/74
cases) | 0.18c |
The median age of the 121 lung cancer patients was
70 years (range, 37–97), and all of the patients were Japanese.
NSCLC specimens were classified histologically as adeno-carcinoma
in 89 patients (73.6%), squamous cell carcinoma in 22 patients
(18.2%), large-cell neuroendocrine carcinoma (LCNEC) in 4 patients
(3.3%), adenosquamous carcinoma in 2 patients (1.7%), large cell
carcinoma in 2 patients (1.7%), small cell carcinoma in 1 patient
(0.8%), and pleomorphic carcinoma in 1 patient (0.8%). The
distribution of clinical stages at the time of diagnosis was as
follows: 60 patients (49.6%) had stage I carcinoma, 13 patients
(10.7%) had stage II, 15 patients (12.4%) had stage III, and 32
patients (26.4%) had stage IV. In 1 patient, the clinical stage was
not classified (Table II).
 | Table IIPatient characteristics. |
Table II
Patient characteristics.
|
Characteristics | Diagnosed with lung
cancer by EBUS-GS-TBBa
(N=121) | Not diagnosed with
lung cancer by EBUS-GS-TBB (N=47) |
|---|
| Age (years) |
| Median | 70 | 71 |
| Range | 37–97 | 65–87 |
| Male gender, n
(%) | 75 (64.1) | 29 (56.9) |
| Smoking status, n
(%) |
| Current
smoker | 34 (28.1) | 7 (14.9) |
| Former smoker | 48 (39.7) | 22 (46.8) |
| Never smoker | 39 (32.2) | 18 (38.3) |
| Histologic type, n
(%) |
|
Adenocarcinoma | 89 (73.6) | |
| Squamous cell
carcinoma | 22 (18.2) | |
| Large cell
carcinoma | 2 (1.7) | |
| Small cell
carcinoma | 1 (0.8) | |
| Adenosquamous
carcinoma | 2 (1.7) | |
| LCNEC | 4 (3.3) | |
| Pleomorphic | 1 (0.8) | |
| Stage, n (%) |
| I | 60 (49.6) | |
| II | 13 (10.7) | |
| III | 15 (12.4) | |
| IV | 32 (26.4) | |
| Not evaluated | 1 (0.8) | |
EGFR mutation detection by ultrarapid
PCR
EGFR mutations in BLF samples were detected
by ultrarapid PCR in 26 adenocarcinoma patients among the 120 NSCLC
patients tested (21.7%), but were not detected in any of the 48
patients who were not diagnosed bronchoscopically with NSCLC.
Eleven patients (42.3%) had an EGFR 19del mutation, and 15
patients (57.7%) had an L858R EGFR point mutation (Table III).
 | Table IIIComparison of ultrarapid PCR and
PCR-invader test results found when detecting the 2 most common
EGFR mutations in samples from 120 NSCLC patients. |
Table III
Comparison of ultrarapid PCR and
PCR-invader test results found when detecting the 2 most common
EGFR mutations in samples from 120 NSCLC patients.
| PCR-invader |
|---|
|
|
|---|
| Ultrarapid PCR | Mutation (+) | Mutation (−) | Total |
|---|
| 19del |
| Mutation (+) | 11 | 0 | 11 |
| Mutation (−) | 3 | 106 | 109 |
| Total | 14 | 106 | 120 |
| L858R |
| Mutation (+) | 15 | 0 | 15 |
| Mutation (−) | 2 | 103 | 105 |
| Total | 17 | 103 | 120 |
Comparison of the ultrarapid PCR and
PCR-invader detection methods
EGFR mutations in FFPE tissues were detected
in 36 adenocarcinoma patients among 120 NSCLC patients (30.0%) by
the PCR-invader method (Table
III). Two of these patients (5.6%) had an exon 18 G719A point
mutation, 1 patient (2.8%) had a G719C point mutation and an exon
20 S768I point mutation, 1 patient (2.8%) had a G719S and a S768I
mutation, 1 patient (2.8%) had a G719C mutation and an exon 21
L858R mutation, 8 patients (22.2%) had an E746-A750del type 1
mutation, 1 patient (2.8%) had an E746-A750del type 2 mutation, 6
patients (16.7%) had low-frequency mutations in exon 19, and 16
patients (44.4%) had an L858R mutation.
As shown in Table
IV, positive concordance rates of 19del and L858R between
ultrarapid PCR and PCR-invader were both 100%, while negative
concordance rates were 97.2 and 98.1%, respectively. The kappa
coefficients for detecting the 19del and L858R mutations between
ultrarapid PCR and PCR-invader were 0.87 and 0.93, respectively.
The average turnaround time for ultrarapid PCR was only 90 min,
whereas that for the PCR-invader method by an outside laboratory
was 9 days.
 | Table IVConcordance rates and Cohen’s kappa
coefficients between the ultrarapid PCR and PCR-invader
methods. |
Table IV
Concordance rates and Cohen’s kappa
coefficients between the ultrarapid PCR and PCR-invader
methods.
| Concordance
rate | 19del (%) | L858 (%) |
|---|
| Positive | 100 | 100 |
| Negative | 97.2 | 98.1 |
| Kappa
coefficienta | 0.87 | 0.93 |
Case report
A 52-year-old non-smoking female, without previous
illness, was admitted to our hospital because of a dry cough and
dyspnea at rest. Her performance status (PS) was 3 on admission.
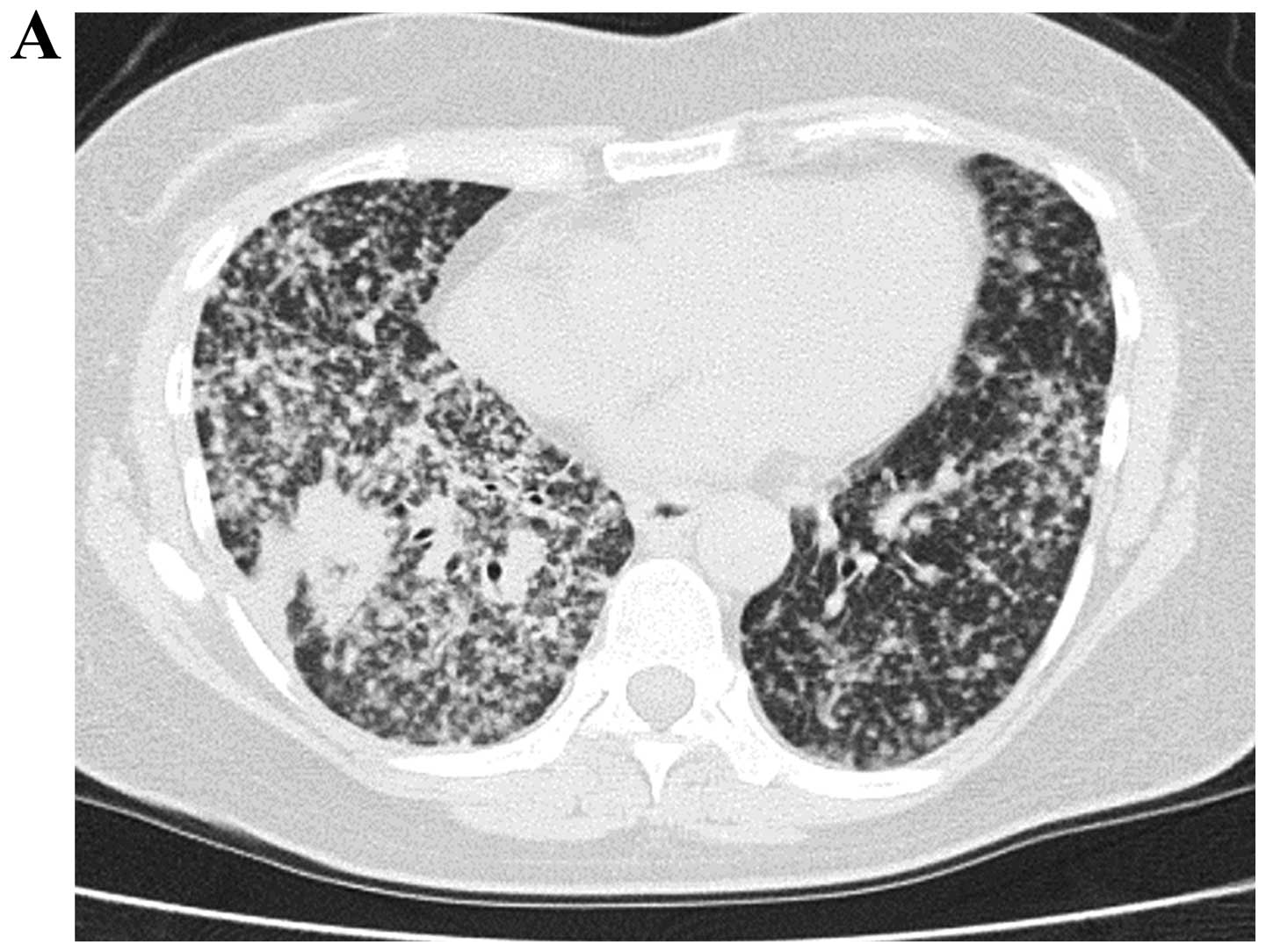
Her chest CT scan showed numerous bilateral diffuse granular lung
shadows and a 20 mm-diameter nodular shadow on the lower right lobe
(Fig. 4A). Whole body bone
scintigraphy was performed later, revealing an abnormal
accumulation in the fifth lumbar vertebra. Suspecting that she had
advanced lung cancer, we immediately performed an EBUS-GS-TBB
against the primary lesion of the lower right lobe. By 60 min after
performing the EBUS-GS-TBB procedure, we obtained a positive result
for the E746-A750del mutation by ultrarapid PCR. Because she had
respiratory failure and a poor PS on admission, she was not
eligible for cytotoxic chemotherapy. Therefore, it was deemed
appropriate to initiate EGFR-TKI therapy as soon as possible. The
following day, we started EGFR-TKI therapy (erlotinib 150 mg orally
every 24 h), after obtaining a definitive pathological diagnosis of
adenocarcinoma by an immunohistochemical method. Two weeks later,
the diffuse and numerous granular shadows of bilateral lung field
had mostly disappeared (Fig. 4B).
Moreover, her respiratory failure and poor PS score were
dramatically improved before PCR-invader results were provided.
Discussion
Bronchoscopy has been used to diagnose abnormal lung
lesions for ~60 years. In recent years, the development of new
diagnostic tools, such as EBUS, GS and VBN, has substantially
improved diagnostic accuracy. Eberhardt et al (15) reported that the combination of EBUS
and VBN improved the diagnostic yield in peripheral lung lesions,
and VBN/EBUS is recommended for the diagnosis of lung peripheral
lesions in guidelines of the European Society for Medical Oncology
(23). Ishida et al
(24) reported that the diagnostic
yield of VBN combined EBUS-GS with small peripheral lesions
(diameter <30 mm) was 80%. Similarly, we found high diagnostic
yields in the present study despite the fact that most target
lesions were small, especially in the VBN/EBUS-GS group. The
appropriate decisions made regarding whether VBN should be used
reinforced the diagnostic accuracy of EBUS-GS-TBBs for small
peripheral lesions. Moreover, we usually collect at least 6 or more
tissue samples. An advantage of EBUS-GS-TBB is that it is easy to
obtain multiple biopsies through the fixed GS safely.
In this study, we validated ultrarapid PCR as a
method for detecting the 2 most common EGFR mutations in
liquid samples obtained by the EBUS-GS-TBB method. In many cases,
even though these samples contain a very small amount of tumor
cells, our method can detect the major EGFR mutations.
Previous studies have shown similar results by molecular analysis
of liquid samples collected by bronchoscopy. Yamaguchi et al
(25) concluded that the analysis
of EGFR, KRAS and TP53 mutations using curette
lavage fluids obtained by bronchoscopy was possible. Furthermore,
some reports have described the molecular analysis of lymph node
samples obtained by EBUS guided trans-bronchial needle aspiration
(26–28) or trans-esophageal ultrasound
scanning with fine needle aspiration (29,30).
Likewise, Buttitta et al (31) reported that EGFR mutation
analysis of bronchoalveolar lavage by next-generation sequencing
was possible even in cases where conventional methods failed.
Importantly, the accuracy of our method was remarkably high,
although the BLF samples contained a small amount of tumor
cells.
The greatest advantage of the ultrarapid PCR method
is its speed. To the best of our knowledge, ultrarapid PCR is the
fastest PCR system for detecting EGFR mutations at present.
Ultrarapid PCR is completed within 10 min, while other methods take
a few hours to detect mutations. This advantage can potentially
have positive effects on treatment outcomes in cases requiring
urgent treatment by early EGFR-TKI administration. Generally, the
administration of cytotoxic agents for patients with poor PS is not
recommended (32). However, some
reports indicate that the use of EGFR-TKIs in patients with poor PS
is effective and feasible because of their relatively mild
toxicities (33). It is necessary
to be careful in selecting therapeutic measures because TKIs are
associated with an increased risk for developing interstitial
pneumonitis in patients with poor PS scores (34). In addition, it will also be
important to explore therapeutic opportunities for improving
prognoses.
Most EGFR mutations are located in exon 18,
19, 20 and 21, with ~90% of these mutations occurring in exons 19
and 21 (35). In previous phase
III trials with EGFR-TKIs, patients with hotspot mutations (exon 19
deletions or exon 21 L858R) were mostly recruited. The response
rate of patients with these hotspot mutations was ~80% (2,5). In
contrast, the response rate of patients with minor mutations, such
as exon 18 point mutation G719X and exon 21 point mutation L861Q,
was only 20% (36). Moreover,
EGFR-TKIs had no proven survival benefit in patients with minor
mutations (36). Therefore, we
limited our search to these hotspot mutations in this study.
As demonstrated in our case report, ultrarapid PCR
can deliver quick results in practical clinical situations.
Patients with hotspot mutations in need of immediate care should
receive EGFR-TKI treatment as soon as possible. Failures in
providing appropriate molecular therapy due to molecular diagnosis
delays should be avoided.
Despite the promising results obtained using
ultrarapid PCR for detecting major EGFR mutations, a
limitation of this method is that it can only detect known
mutations. Detecting minor EGFR mutations in exon 18 and the
T790M point mutation associated with drug resistance (exon 20) will
require the development of additional probes. This current
limitation reduces patients’ opportunities for rapid qualification
for the third-generation EGFR-TKIs therapy, such as AZD9291
(37) by ultrarapid PCR alone.
However, this problem may be solved by the development of
additional primer sets for minor mutations in the near future.
In conclusion, it was demonstrated that ultrarapid
PCR is an extremely quick and precise method for examining clinical
liquid samples with a background of normal cells. The combination
of ultrarapid PCR and EBUS-GS-TBB methods may enable point-of-care
testing for NSCLC patient samples harboring EGFR
mutations.
References
|
1
|
Fukuoka M, Wu YL, Thongprasert S, et al:
Biomarker analyses and final overall survival results from a phase
III, randomized, open-label, first-line study of gefitinib versus
carboplatin/paclitaxel in clinically selected patients with
advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol.
29:2866–2874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
4
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou C, Wu YL, Chen G, et al: Erlotinib
versus chemotherapy as first-line treatment for patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase
3 study. Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asano H, Toyooka S, Tokumo M, et al:
Detection of EGFR gene mutation in lung cancer by mutant-enriched
polymerase chain reaction assay. Clin Cancer Res. 12:43–48. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoshi K, Takakura H, Mitani Y, et al:
Rapid detection of epidermal growth factor receptor mutations in
lung cancer by the SMart-Amplification Process. Clin Cancer Res.
13:4974–4983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hall JG, Eis PS, Law SM, et al: Sensitive
detection of DNA polymorphisms by the serial invasive signal
amplification reaction. Proc Natl Acad Sci USA. 97:8272–8277. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagai Y, Miyazawa H, Huqun, et al: Genetic
heterogeneity of the epidermal growth factor receptor in non-small
cell lung cancer cell lines revealed by a rapid and sensitive
detection system, the peptide nucleic acid-locked nucleic acid PCR
clamp. Cancer Res. 65:7276–7282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naoki K, Soejima K, Okamoto H, et al: The
PCR-invader method (structure-specific 5′ nuclease-based method), a
sensitive method for detecting EGFR gene mutations in lung cancer
specimens; comparison with direct sequencing. Int J Clin Oncol.
16:335–344. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sasaki H, Endo K, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujimoto T, Konagaya M, Enomoto M, et al:
Novel high-speed real-time PCR method (Hyper-PCR): results from its
application to adenovirus diagnosis. Jpn J infect Dis. 63:31–35.
2010.PubMed/NCBI
|
|
13
|
Asahina H, Yamazaki K, Onodera Y, Kikuchi
E, Shinagawa N, Asano F and Nishimura M: Transbronchial biopsy
using endobronchial ultrasonography with a guide sheath and virtual
bronchoscopic navigation. Chest. 128:1761–1765. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Asano F, Matsuno Y, Shinagawa N, Yamazaki
K, Suzuki T, Ishida T and Moriya H: A virtual bronchoscopic
navigation system for pulmonary peripheral lesions. Chest.
130:559–566. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eberhardt R, Anantham D, Ernst A,
Feller-Kopman D and Herth F: Multimodality bronchoscopic diagnosis
of peripheral lung lesions: a randomized controlled trial. Am J
Respir Crit Care Med. 176:36–41. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eberhardt R, Kahn N, Gompelmann D,
Schumann M, Heussel CP and Herth FJ: LungPoint - a new approach to
peripheral lesions. J Thorac Oncol. 5:1559–1563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fielding DI, Chia C, Nguyen P, Bashirzadeh
F, Hundloe J, Brown IG and Steinke K: Prospective randomised trial
of endobronchial ultrasound-guide sheath versus computed
tomography-guided percutaneous core biopsies for peripheral lung
lesions. Intern Med J. 42:894–900. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gildea TR, Mazzone PJ, Karnak D, Meziane M
and Mehta AC: Electromagnetic navigation diagnostic bronchoscopy: a
prospective study. Am J Respir Crit Care Med. 174:982–989. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seijo LM, de Torres JP, Lozano MD,
Bastarrika G, Alcaide AB, Lacunza MM and Zulueta JJ: Diagnostic
yield of electromagnetic navigation bronchoscopy is highly
dependent on the presence of a Bronchus sign on CT imaging: results
from a prospective study. Chest. 138:1316–1321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kurimoto N, Miyazawa T, Okimasa S, Maeda
A, Oiwa H, Miyazu Y and Murayama M: Endobronchial ultrasonography
using a guide sheath increases the ability to diagnose peripheral
pulmonary lesions endoscopically. Chest. 126:959–965. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kundel HL and Polansky M: Measurement of
observer agreement. Radiology. 228:303–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Landis JR and Koch GG: The measurement of
observer agreement for categorical data. Biometrics. 33:159–174.
1977. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vansteenkiste J, De Ruysscher D, Eberhardt
WE, Lim E, Senan S, Felip E and Peters S; ESMO Guidelines Working
Group. Early and locally advanced non-small-cell lung cancer
(NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 24(Suppl 6): vi89–vi98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ishida T, Asano F, Yamazaki K, et al:
Virtual bronchoscopic navigation combined with endobronchial
ultrasound to diagnose small peripheral pulmonary lesions: a
randamized trial. Thorax. 66:1072–1077. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamaguchi F, Kugawa S, Tateno H, Kokubu F
and Fukuchi K: Analysis of EGFR, KRAS and P53 mutations in lung
cancer using cells in the curette lavage fluid obtained by
bronchoscopy. Lung Cancer. 78:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jurado J, Saqi A, Maxfield R, et al: The
efficacy of EBUS-guided transbronchial needle aspiration for
molecular testing in lung adenocarcinoma. Ann Thorac Surg.
96:1196–1202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Santis G, Angell R, Nickless G, Quinn A,
Herbert A, Cane P, Spicer J, Breen R, McLean E and Tobal K:
Screening for EGFR and KRAS mutations in endobronchial ultrasound
derived trans-bronchial needle aspirates in non-small cell lung
cancer using COLD-PCR. PLoS One. 6:e251912011. View Article : Google Scholar
|
|
28
|
Tsai TH, Yang CY, Ho CC, et al: Multi-gene
analyses from waste brushing specimens for patients with peripheral
lung cancer receiving EBUS-assisted bronchoscopy. Lung Cancer.
82:420–425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewandowska MA, Jozwicki W, Jochymski C
and Kowalewski J: Application of PCR methods to evaluate EGFR, KRAS
and BRAF mutations in a small number of tumor cells in cytological
material from lung cancer patients. Oncol Rep. 30:1045–1052.
2013.PubMed/NCBI
|
|
30
|
van Eijk R, Licht J, Schrumpf M, et al:
Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle
aspirates from non-small-cell lung cancer using allele-specific
qPCR. PLoS One. 6:e177912011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Buttitta F, Felicioni L, Del Grammastro M,
et al: Effective assessment of egfr mutation status in
bronchoalveolar lavage and pleural fluids by next-generation
sequencing. Clin Cancer Res. 19:691–698. 2013. View Article : Google Scholar
|
|
32
|
Pfister DG, Johnson DH, Azzoli CG, et al:
American Society of Clinical Oncology treatment of unresectable
non-small-cell lung cancer guideline: update 2003. J Clin Oncol.
22:330–353. 2004. View Article : Google Scholar
|
|
33
|
Inoue A, Kobayashi K, Usui K, et al:
First-line gefitinib for patients with advanced non-small-cell lung
cancer harboring epidermal growth factor receptor mutations without
indication for chemotherapy. J Clin Oncol. 27:1394–1400. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kudoh S, Kato H, Nishiwaki Y, et al:
Interstitial lung disease in Japanese patients with lung cancer: a
cohort and nested case-control study. Am J Respir Crit Care Med.
177:1348–1357. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Watanabe S, Minegishi Y, Yoshizawa H, et
al: Effectiveness of gefitinib against non-small-cell lung cancer
with uncommon EGFR mutations G719X and L861Q. J Thorac Oncol.
9:189–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cross DA, Ashton SE, Ghiorghiu S, et al:
AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated
resistance to EGFR inhibitors in lung cancer. Cancer Dicov.
4:1046–1061. 2014. View Article : Google Scholar
|